Abstract
The selective expression of p53-targeted genes is central to the p53-mediated DNA damage response. It is affected by multiple factors including posttranslational modifications and cofactors of p53. Here, we proposed an integrated model of the p53 network to characterize how the cellular response is regulated by key cofactors of p53, Hzf and ASPP. We found that the sequential induction of Hzf and ASPP is crucial to a reliable cell-fate decision between survival and death. After DNA damage, activated p53 first induces Hzf, which promotes the expression of p21 to arrest the cell cycle and facilitate DNA repair. The cell recovers to normal proliferation after the damage is repaired. If the damage is beyond repair, Hzf is effectively degraded, and activated E2F1 induces ASPP, which promotes the expression of Bax to trigger apoptosis. Furthermore, interrupting the induction of Hzf or ASPP remarkably impairs the cellular function. We also proposed two schemes for the production of the unknown E3 ubiquitin ligase for Hzf degradation: it is induced by either E2F1 or p53. In both schemes, the sufficient degradation of Hzf is required for apoptosis induction. These results are in good agreement with experimental observations or are experimentally testable.
Introduction
The tumor suppressor p53 has a major role in the cellular response to various stresses including DNA damage (1). In unstressed cells, p53 is kept at a low level by its negative regulator Mdm2 (2). Upon DNA damage, p53 is stabilized and activated to transactivate a large number of genes involved in cell cycle arrest or apoptosis (3). Among the p53-targeted genes, p21 can induce cell cycle arrest, which allows time for DNA repair and promotes cell survival (4), whereas Bax is crucial to p53-dependent apoptosis (5). Although how a cell makes a decision between life and death has been a focus of intensive research, its underlying mechanism is still not completely understood.
The selective expression of target genes by p53 controls the cell fate (6). This selectivity is influenced by multiple factors including the protein level, posttranslational modifications, subcellular localization, and cofactors of p53 (7). It was proposed that low levels of p53 can transactivate genes with high-affinity promoters associated with cell cycle arrest, whereas high levels of p53 are able to activate genes with low-affinity promoters involved in apoptosis (8). The posttranslational modifications of p53 such as phosphorylation and acetylation remarkably affect its promoter selectivity (9). For example, p53 phosphorylation at Ser46 stimulates the induction of the proapoptotic gene p53AIP1 in response to genotoxic stress (10). The cofactors of p53 enhance its binding to specific promoters. Among them, ASPP1 and ASPP2, which are collectively referred to as ASPP thereafter, promote apoptosis by recruiting p53 to the promoters of proapoptotic genes such as Bax (11). By contrast, Hzf preferentially promotes the binding of p53 to the promoters of proarrest genes like p21 (12). Interestingly, upon sustained or severe DNA damage, Hzf should be sufficiently degraded to allow the activation of proapoptotic genes (12). That is, ASPP and Hzf exert opposite effects on cellular outcome. It is intriguing to explore how they interact in the DNA damage response.
A series of theoretical models has been constructed to clarify the mechanism for p53-mediated cell-fate decision after DNA damage. Previously, it was suggested that the decision between life and death is determined by the p53 level in a switchlike manner (13). In that model, activated p53 only triggers apoptosis, and thus it is impossible to distinguish cell cycle arrest from apoptosis, both mediated by p53. Recently, it was proposed that the cell fate is governed by the number of p53 pulses and apoptosis is induced only when the number of p53 pulses exceeds some threshold (14,15). Most recently, we further suggested that whereas the cell fate is determined by p53 pulses, apoptosis is evoked by high levels of p53 (16). This two-phase behavior of p53 may provide a flexible and efficient control mode. The above models focus on the influence of p53 phosphorylation (14–16), whereas little is known about the role of p53 cofactors in the decision-making process.
Motivated by the above considerations, we developed an integrated model of the p53 signaling network to explore how the dynamics of Hzf and ASPP affect the DNA damage response. The model simulations indicate that the network can make a reliable decision between survival and death depending on the extent of DNA damage. For repairable damage, p53 is primarily activated and shows a moderate level, inducing Hzf to promote the expression of p21. Consequently, the cell undergoes transient cell cycle arrest before the damage is fixed. For irreparable damage, p53 exhibits a two-phase behavior: a moderate level of p53 induces cell cycle arrest in the first phase, whereas a high level of p53 evokes apoptosis in the second phase. The sufficient degradation of Hzf is required for the activation of E2F1 and ASPP, which cooperates with p53 to induce Bax. Specifically, we proposed two schemes guaranteeing that Hzf is degraded in the presence of severe DNA damage. These results clarify how the cofactors of p53 are orchestrated to regulate the cell-fate decision.
Model and Methods
Model
We constructed an integrated model to explore the response of the p53 network to DNA damage caused by ionizing radiation (IR) (see Fig. 1). Compared with our previous models (15,16), the model network presented in this article is still composed of four modules: a DNA repair module, a sensor of DNA damage, a p53-centered feedback control module, and a cell-fate decision module. But there are four large differences: 1), activated p53 is no longer divided into p53 arrester and p53 killer; 2), the influence of p53 cofactors on gene expression is explicitly modeled; 3), the subnetwork controlling cell cycle progression is introduced; and 4), two schemes are proposed to underlie the production of an E3 ligase for Hzf degradation. The concentration of each protein is represented by a state variable in the rate equations. These ordinary differential equations and parameter values are presented in the Appendix and Table 1, respectively.
Figure 1.
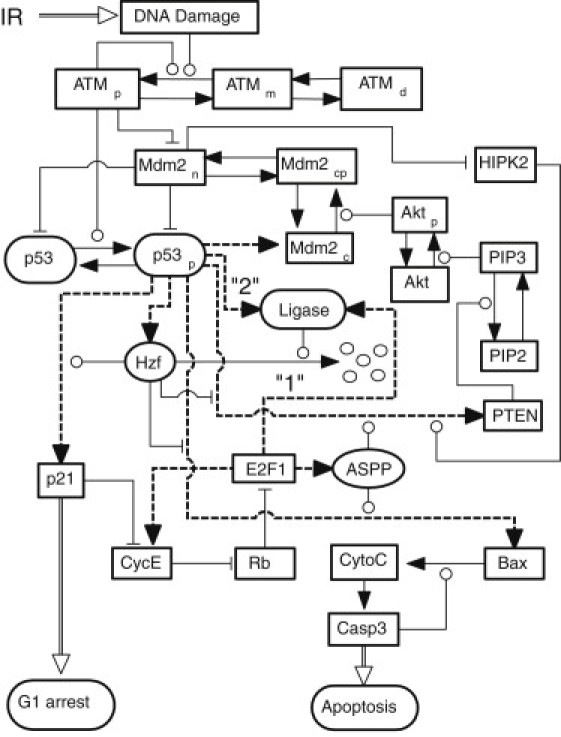
Schematic diagram of the model for p53-mediated cell-fate decision after DNA damage. There exist two p53-centered feedback loops, i.e., the p53-Mdm2 negative feedback loop and the p53-PTEN-Akt-Mdm2 positive feedback loop. Hzf and ASPP, the cofactors of p53, are induced by p53 and E2F1, respectively. Hzf is degraded by an unknown E3 ligase, which may be separately induced by E2F1 or p53 in two schemes (labeled as 1 and 2). p53 triggers cell cycle arrest and apoptosis through p21 and Bax, respectively. There exists a positive feedback loop between cytochrome c (CytoC) release and caspase-3 (Casp3) activation. (Dashed lines) Transactivation of target genes by p53 and E2F1. (Arrow-headed solid lines) State transition; (circle-headed and bar-headed lines) promotion and inhibition of state transition. (Arrow-headed double lines) Other processes.
Table 1.
Parameter values
| Parameter | Value | Parameter | Value | Parameter | Value | Parameter | Value | Parameter | Value |
|---|---|---|---|---|---|---|---|---|---|
| kdim | 10 | kundim | 1 | kacATM | 1.5 | kdeATM | 0.8 | jacATM | 1 |
| jdeATM | 2 | jnc | 4 | ATMtot | 5 | ksp53 | 0.1 | kdp53n | 0.05 |
| j1p53n | 0.1 | kdp53 | 0.7 | kdmdm2 | 0.003 | kmdm2in | 0.06 | kmdm2out | 0.09 |
| jATM | 1 | ksmdm20 | 0.002 | ksmdm2 | 0.024 | jsmdm2 | 1 | k1mdm2s | 0.3 |
| j1mdm2s | 0.1 | kmdm2s | 8 | jmdm2s | 0.3 | kacp531 | 0.2 | kdep53 | 0.1 |
| kdp53p | 0.01 | kdmdm2n1 | 0.05 | kacakt | 0.25 | kdeakt | 0.1 | jacakt | 0.1 |
| jdeakt | 0.2 | Akttot | 1 | PIPtot | 1 | kp2 | 0.1 | kp3 | 0.5 |
| jp2 | 0.2 | jp3 | 0.4 | ksHIPK2 | 0.05 | kdHIPK2 | 0.5 | jHIPK2 | 0.6 |
| ksPTEN0 | 0.01 | ksPTEN | 0.5 | jsPTEN | 1 | kdPTEN | 0.1 | kshzf0 | 0.001 |
| kshzf | 0.12 | jshzf | 0.5 | kdhzf0 | 0.02 | kdhzf | 0.45 | jligase | 2 |
| jhzf | 1.5 | ksligase0 | 0.0005 | ksligase | 0.025 | jsligase | 0.2 | kdligase | 0.005 |
| ksp210 | 0.01 | ksp21 | 0.15 | jsp21 | 0.1 | kdp21 | 0.06 | ksce0 | 0.0005 |
| ksce | 0.0275 | jsce | 0.2 | kdce | 0.005 | kasp21e | 0.5 | kdsp21e | 0.05 |
| kasre | 0.5 | kdsre | 0.05 | kprb | 0.05 | kdprb | 0.025 | jprb | 0.1 |
| jdprb | 0.1 | E2F1tot | 1 | Rbtot | 2 | ksASPP0 | 0.001 | ksASPP | 0.03 |
| jsASPP | 0.5 | kdASPP | 0.01 | jASPP | 0.5 | ksbax0 | 0.01 | ksbax | 0.3 |
| jsbax | 1 | kdbax | 0.1 | kaccytco0 | 0.001 | kaccytoc | 0.9 | kdecytoc | 0.05 |
| kaccasp30 | 0.001 | kaccasp3 | 0.9 | kdecasp3 | 0.07 | CytoCtot | 3 | jcytoc | 0.5 |
| jcasp3 | 0.5 | Casp3tot | 3 |
Note that, for the second scheme, some parameters are changed as follows: kmdm2s = 7, ksce = 0.02, jsce = 0.25, ksligase0 = 0, ksligase = 0.03, jsligase = 0.75, and kdligase = 0.006.
DNA repair and ATM activation
Upon IR, double-strand break (DSB) is the typical form of DNA damage, and repair proteins are quickly recruited to break sites, forming the DSB-protein complex (DSBC). We made the same assumptions about the generation and repair of DSBs as in our previous studies (15–17). For a population of 2000 cells exposed to the same irradiation dose of DIR, the initial numbers of DSBs obey a Poisson distribution with a mean of 35 × DIR (18). There are 20 repair proteins per cell (19). The repair process is characterized by the two-lesion kinetic model (20) and is simulated by using the Monte Carlo method (for details, see the Supplementary Information in Zhang et al. (15)).
DSBs are specifically detected by the ataxia-telangiectasia-mutated (ATM) kinase. ATM is activated by DBSCs through intermolecular phosphorylation (21); ATM converts from inactive dimer (ATMd) to inactive monomer (ATMm) and further to active, phosphorylated monomer (ATMp). In simulations, the phosphorylation and dephosphorylation processes are considered as enzyme-catalyzed reactions, which are assumed to follow the Michaelis-Menten kinetics (22). The total level of ATM is assumed constant, because it mainly undergoes posttranslational modifications after DNA damage. The dimerization rate of ATM is much larger than its undimerization rate, so that ATM dimers are predominant in unstressed cells.
p53-centered feedback loops
Here, we mainly consider two feedback loops, i.e., the p53-Mdm2 negative feedback loop and the p53-PTEN-Akt-Mdm2 positive feedback loop. Upon DNA damage, the ATM-dependent phosphorylation of p53 and Mdm2 impairs the p53-Mdm2 interaction, and thus p53 is activated (23,24). p53-induced PTEN promotes the dephosphorylation of phosphatidylinositol 3,4,5-trisphosphate (PIP3) and indirectly inhibits the phosphorylation of Akt, thereby sequestering Mdm2 in the cytoplasm (25,26). This positive feedback can promote the full activation of p53. In addition, PTEN is preferentially induced by p53 phosphorylated at Ser46 by HIPK2 (27), the degradation of which is promoted by nuclear Mdm2 (28).
We consider two forms of nuclear p53: inactive p53 and active, phosphorylated p53p. For simplicity, cytoplasmic p53 is not included in the model although it also promotes apoptosis in a transcription-independent manner (29). Three forms of Mdm2 are considered, i.e., Mdm2n (nuclear form), Mdm2c (nonphosphorylated cytoplasmic form), and Mdm2cp (phosphorylated cytoplasmic form). We assume that only Mdm2cp can enter the nucleus (25). In simulations, the activation rate of p53 and the degradation rate of Mdm2n depend on the ATMp level in the form of the Michaelis-Menten equation (Eqs. 4 and 5) (22).
Consistent with the experimental protocol (12), the presence of 10% serum is a default setting in our model, and it is sufficient for the activation of PIP3 and Akt in unstressed cells. Only phosphorylated Akt (Aktp) promotes the nuclear translocation of Mdm2 by phosphorylation. The transition from Akt to Aktp is promoted by PIP3 (30). Given no remarkable variation in the total level of Akt after DNA damage, it is assumed to be constant (31). We set kacakt > kdeakt in Eq. 11 to ensure the predominance of active Aktp in unstressed cells. The total amount of PIP2 and PIP3 is also assumed to be constant, and the dephosphorylation rate of PIP3 is proportional to the PTEN level (Eq. 13) (32). The rate constant of PTEN induction by p53 depends on the level of HIPK2 (Eq. 16) (27), whose degradation rate is assumed to be proportional to the Mdm2n level (Eq. 15) (28).
p53 can exhibit various dynamic modes; for example, p53 pulses have been observed in MCF-7 cells in response to IR (33,34). Note that the prosurvival role of Hzf was observed in several cell lines including U2OS, LNCaP, Saos2, and HCT116 (12), and no pulses in the p53 level were reported in most of these cell lines (7). Thus, here we did not mimic oscillatory behavior of p53 and ignored the p53-Wip1-ATM negative feedback loop, which is required for the initiation of p53 pulses (34).
Cell fate decision
The cofactors of p53 have important roles in the choice of cell fate between survival and death. The prosurvival cofactor Hzf, induced by p53, promotes the recruitment of p53 to proarrest genes like p21 (12). p21 inhibits the activity of CDK2 by interaction with Cyclin E (CycE), thereby preventing the phosphorylation of Rb and activation of E2F1 (4). On the other hand, the proapoptotic cofactor ASPP, induced by E2F1, contributes to apoptosis induction (35). Thus, the delicate regulation of expression of Hzf and ASPP is crucial to the cell-fate decision.
Because the E3 ubiquitin ligase for Hzf degradation (which is denoted by the term “Ligase” thereafter) is unknown, we propose two schemes ensuring that the Ligase accumulates remarkably only in the presence of severe DNA damage (12). In the first scheme, the Ligase is induced by E2F1. In the second scheme, the p53-induced Ligase accumulates slowly and becomes marked after a long time. Note that p53 and E2F1 are two important transcription factors involved in cell-cycle control and cell fate decision; very possibly, the Ligase is induced by p53 or E2F1, although other possibilities cannot be excluded.
Here, we assume that the expression of Bax is enhanced by ASPP (11). Bax promotes the release of cytochrome c (CytoC) from mitochondria (36). Released CytoC results in the activation of caspase-3 (Casp3), and activated Casp3 further promotes CytoC release by cleaving its inhibitors (37). Consequently, apoptosis ensues (38). For simplicity, we did not explicitly consider the inhibitors of apoptosis like Bcl-xL (39) and the intermediate steps between CytoC release and Casp3 activation, including the formation of apoptosome and activation of caspase-9 (40).
In simulations, p21 induction is promoted by Hzf, which is characterized by a scaling of the rate constant of production (Eq. 18). The expression of p53-mediated proapoptotic genes is inhibited by Hzf but promoted by ASPP (Eqs. 16 and 29). Thus, PTEN and Bax are induced by p53 only when Hzf drops to a low level and ASPP accumulates remarkably. Note that the transactivation of genes by p53 or E2F1 is characterized by the Hill function, and the Hill coefficient is set to four and two, respectively. The degradation of Hzf is very slow with a basal rate of kdhzf0, but it is improved with a large rate of kdhzf (i.e., kdhzf0 ≪ kdhzf in Eq. 17) in the presence of the Ligase (12). The nonphosphorylated Rb (Rb), rather than the phosphorylated Rb, inhibits E2F1 activity by association with it (Eq. 24) (41). Only free CycE, rather than p21-bound CycE, promotes the phosphorylation of Rb (Eq. 26) (4). The total levels of E2F1 and Rb are assumed to be constant, respectively (42).
Methods
The ordinary differential equations were numerically solved by using the Runge-Kutta algorithm with a time step of 0.01 min. Whereas there are two schemes for the regulation of the E3 ligase for Hzf degradation, most simulation results were based on the first scheme unless specified otherwise. The bifurcation diagrams were plotted by using the tool suite Oscill8 (http://oscill8.sourceforge.net/). The units of time and irradiation dose are minutes and Gray (Gy), respectively, and the units of other parameters ensure that the concentrations of proteins are dimensionless.
Results
Network dynamics associated with cell-fate decision
The p53 network is sufficiently complicated, and it is useful to first explore the dynamics of key proteins. We illustrate the output of each module under two typical irradiation conditions (Fig. 2). In the following, […] denotes the concentration of proteins.
Figure 2.
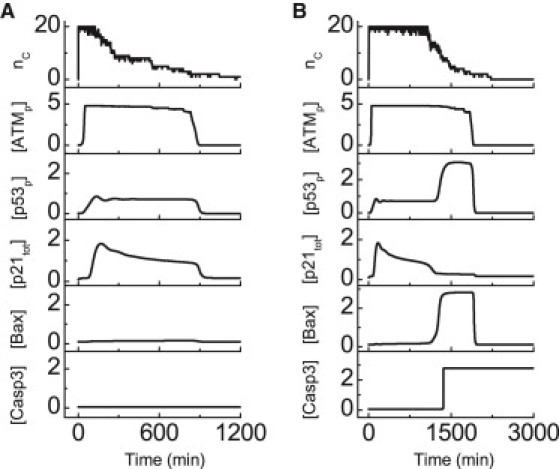
Dynamics of the p53 network after DNA damage. Shown are time courses of nc and the levels of ATMp, p53p, p21tot, Bax, and Casp3 at DIR = 2 Gy (A) or 5 Gy (B).
The number of DSBCs, nc, is used to indicate the presence of DNA damage. Upon IR, nc quickly reaches its maximum of 20 (because it is assumed that there are 20 repair proteins per cell) and remains there until the number of DSBs falls below 20 due to DNA repair. The discontinuous jumps in the traces result from the assumption that the step from DSBC to fixed DSB is irreversible. ATM is a very sensitive and reliable detector of DSBs. After DNA damage, [ATMp] rapidly rises to a high plateau and stays there until the damage is effectively repaired. The width of the plateau is positively correlated with the irradiation dose.
At DIR = 2 Gy, [p53p] is kept at a moderate level after a transient response (Fig. 2 A). Activated p53 induces p21 to arrest the cell cycle, and [p21tot] first rises and then decreases gradually. Bax and Casp3 are kept inactive. After the damage is effectively fixed, the proteins return to basal levels, and the cell recovers to normal proliferation.
At DIR = 5 Gy, sustained DNA damage leads to the two-phase dynamics of p53 (Fig. 2 B). [p53p] is still at a moderate level in the first phase and then switches to a high level in the second phase. As a result, p21 and Bax are mainly induced in the first and the second phase, respectively. Thus, Casp3 is activated, and apoptosis is triggered. Note that we did not model the events after Casp3 activation, and DNA repair would cease after Casp3 activation. Here, persistent activation of Casp3 is considered as the marker of apoptosis.
The above results indicate that the model network can make a reliable decision between cell life and death, depending on the stress level. Moderate levels of p53 induce transient cell cycle arrest to allow DNA repair, whereas high levels of p53 trigger apoptosis when the damage is beyond repair. That is, p53 is activated in a progressive manner, and the cell-fate decision is closely associated with the dynamics of p53. Essentially, this is an analog mode, in contrast to the digital mode based on p53 pulses (15,17).
Two-phase dynamics of p53
It is worthy to probe the two-phase dynamics of p53 in more detail. At DIR = 5 Gy, the proteins involved in the p53-centered feedback loops exhibit the two-phase behaviors (Fig. 3 A). In the first phase, most cytoplasmic Mdm2 molecules are phosphorylated by Aktp and translocate to the nucleus, and thus [Mdm2n] is larger than [Mdm2c]. Accordingly, p53p is at a moderate level. Moreover, PTEN is at a basal level because of low level of HIPK2, which is due to its degradation by Mdm2n. In the second phase, HIPK2 gradually accumulates and accelerates production of PTEN. Accumulation of PTEN further promotes the dephosphorylation of Aktp and sequestration of Mdm2 in the cytoplasm. Consequently, p53p and HIPK2 become more stable and get their levels elevated. Subsequently, the expression of Mdm2 and PTEN is further enhanced. By contrast, Akt activity is greatly inhibited. As a result, [p53p] is maintained at a high level. Therefore, the p53-PTEN-Akt-Mdm2 and HIPK2-PTEN-Akt-Mdm2 positive feedback loops cooperate in the switching of p53 from moderate to high levels.
Figure 3.
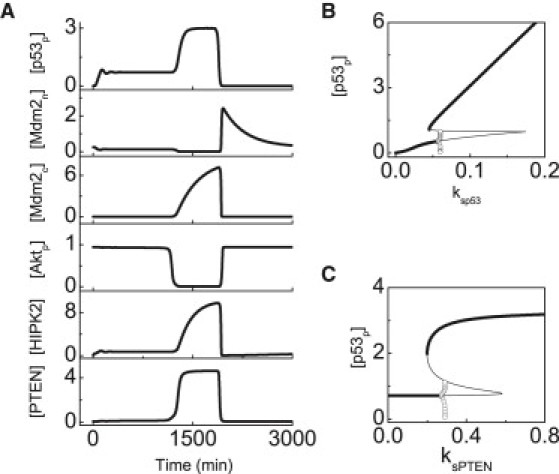
Two-phase dynamics of p53 at high damage levels. (A) Temporal evolution of the levels of the proteins involved in the p53-centered feedback loops at DIR = 5 Gy. The bifurcation diagram of [p53p] as a function of ksp53 (B) or ksPTEN (C). (Thick and thin black lines) Stable and unstable steady states, respectively. (Open circles) Minima and maxima of the limit cycles.
Now we explore the factors that remarkably influence the dynamics of p53. On one hand, we probe the dependence of the steady-state level of p53p on its production rate, ksp53, with nc set to 20. In the bifurcation diagram (Fig. 3 B), [p53p] can exhibit four distinct behaviors with increasing ksp53. For ksp53 ≤ 0.045, there is a single low-level steady state, and its level rises with ksp53. When 0.045 < ksp53 ≤ 0.057, there exist two stable steady states and one unstable steady state. For 0.057 < ksp53 ≤ 0.060, a stable limit cycle coexists with a stable steady state. When ksp53 > 0.060, there is only a stable steady state with high levels. Therefore, only when the production rate of p53 exceeds some threshold can [p53p] always be settled in a high-level state.
On the other hand, we investigate the effect of the p53-PTEN-Akt-Mdm2 feedback loop on p53 dynamics. The p53-induced production rate of PTEN, ksPTEN, is chosen as a measure of the feedback strength. In the bifurcation diagram (Fig. 3 C), [p53p] can undergo four distinct behaviors with increasing ksPTEN: a stable steady state with low levels, two stable steady states separated by one unstable steady state, the coexistence of a limit cycle with a stable steady state, and a stable steady state with high levels. In fact, increasing ksPTEN leads to the enhancement of the p53-PTEN-Akt-Mdm2 positive feedback and impairment of the p53-Mdm2 negative feedback, and thus [p53p] tends to exhibit monostability with high levels. Because PTEN is mainly induced in the second phase, its production rate specifically influences the p53 dynamics in the second phase. In sum, with our default parameter setting (ksp53 = 0.1 and ksPTEN = 0.5), [p53p] can show a two-phase behavior at high damage levels.
Role of p53 cofactors in the cell-fate decision
The cofactors of p53 modulate its selective expression of target genes and thus have a key role in determining cell fates. After DNA damage, [p53p] rises to a moderate level after a short period (Fig. 4 A). Hzf and p21 are first induced to arrest the cell cycle, whereas E2F1 activity is inhibited. At the same time, the Ligase accumulates progressively, leading to the gradual degradation of Hzf and reduction in p21 level. Meanwhile, E2F1 still accumulates slowly due to the double-negative feedback between Hzf and E2F1. The repairable DNA damage can be fixed before enough E2F1 builds up to induce ASPP, and thus there is no apoptosis. By contrast, when the DNA damage is irreparable, much E2F1 accumulates to induce ASPP, which further improves the expression of PTEN to elevate the p53p level (Fig. 4 B). Notably, the p53-dependent apoptosis requires the concurrent activation of p53 and E2F1 (17). Therefore, it is important to sequentially activate the prosurvival and proapoptotic cofactors of p53. Otherwise, the cell would become very sensitive to stress or robust to death stimuli. We will return to this point later.
Figure 4.
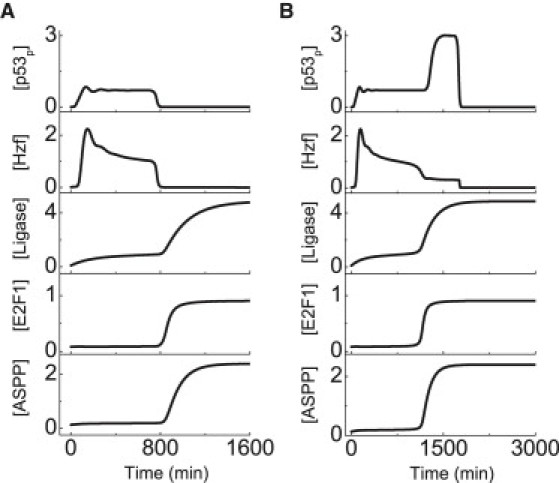
Sequential induction of the prosurvival and proapoptotic cofactors of p53. Displayed are time courses of the levels of p53p, Hzf, the Ligase, E2F1, and ASPP at DIR = 2 Gy (A) or 5 Gy (B).
In fact, the cellular outcome is closely associated with what genes are transactivated by p53. For example, the cell is seriously damaged at DIR = 6 Gy and should be eliminated timely. In this case, only Hzf and p21 are first induced by p53 in the first phase, whereas Bax is mainly induced by p53 with the help of ASPP in the second phase (Fig. 5 A). That is, p53-induced p21 leads to transient cell cycle arrest, whereas Bax activates Casp3 to initiate apoptosis. Moreover, Hzf should be sufficiently degraded before Bax induction. Therefore, if the expression of these cofactors is interrupted, the cellular function will be disrupted, as shown in the following.
Figure 5.
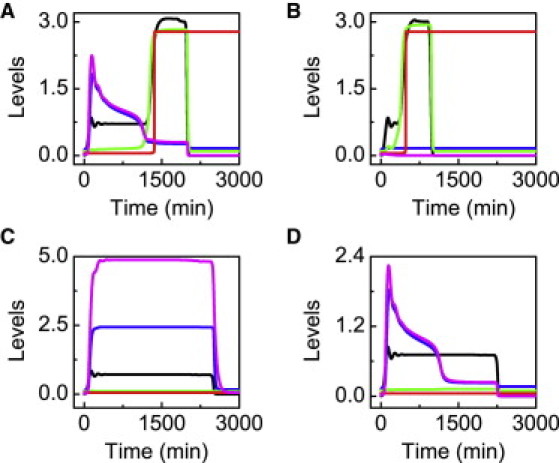
Roles of p53 cofactors in the cell-fate decision. Time courses of [p53p] (black), [Hzf] (magenta), [p21tot] (blue), [Bax] (green), and [Casp3] (red) for different cases. (A) DIR = 6 Gy; (B) DIR = 2 Gy and kshzf = 0 (i.e., with Hzf deficiency); (C) DIR = 8 Gy and kdhzf = 0 (i.e., with inhibition of the Hzf degradation by the Ligase); and (D) DIR = 8 Gy and ksASPP = 0 (i.e., with repression of E2F1-induced ASPP expression).
To mimic the case of Hzf deficiency, the rate constant for p53-induced Hzf expression, kshzf, is set to zero. Consequently, [Hzf] is kept at a very low level (Fig. 5 B). The duration of the first phase is greatly reduced because p21 is kept inactive and the activation of E2F1 becomes easier. Accordingly, the activation of Casp3 occurs much earlier, and apoptosis can be triggered even at DIR = 2 Gy. Thus, the initiation of apoptosis is significantly accelerated in Hzf-deficient cells. Indeed, Hzf-knockout mice become rather sensitive to DNA damage (12). In other words, Hzf is crucial to cell survival in the DNA damage response.
On the other hand, [Hzf] remains at high levels throughout the response when the degradation of Hzf by the Ligase is inhibited (i.e., kdhzf = 0) (Fig. 5 C). As a result, both p53p and p21 are persistently active until the damage is repaired. By contrast, Casp3 cannot be activated even at DIR = 8 Gy. These results are in good agreement with the finding that stabilization of Hzf by the proteasome inhibitor MG132 prevents p53-mediated apoptosis even in the presence of severe DNA damage (12). Therefore, the timely degradation of Hzf is essential for apoptosis induction.
In the presence of ASPP deficiency (i.e., ksASPP = 0), the proapoptotic genes cannot be induced even when Hzf is degraded sufficiently and p21 drops to low levels (Fig. 5 D). Thus, no apoptosis is triggered even at DIR = 8 Gy. Notably, ASPP has a specific role in Bax induction and apoptosis initiation but has no effect on p21-induced cell cycle arrest. These results may explain the deficiency of ASPP in some cancers and are consistent with the experimental observations (7). Taken together, both the prosurvival and proapoptotic cofactors of p53 are indispensable for a reliable cell-fate decision, avoiding unnecessary death or promoting the execution of apoptosis.
Because stochasticity exists in the generation and repair of DNA damage, cells may exhibit a large variability in outcome. To quantify such variability, we calculated the fraction of apoptotic cells, FA, among a large population. Typically, FA looks like a sigmoid function of DIR (Fig. 6 A). With the default parameter setting (kshzf = 0.12), apoptosis first appears at DIR = 1 Gy, and all cells are killed if DIR ≥ 7.5 Gy. For kshzf = 0, the curve moves leftward: apoptosis first appears in few cells at DIR = 0.4 Gy, and all cells undergo apoptosis once DIR ≥ 2.8 Gy. For kshzf = 0.14, the curve shifts rightward: apoptosis first occurs at kshzf = 6 Gy, and all cells die when kshzf ≥ 15 Gy. That is, Hzf-deficient cells become rather sensitive to death signals, whereas Hzf-proficient cells are resistant to stress. ASPP also affects the cellular outcome evidently. ASPP promotes the induction of apoptosis, and thus when increasing or decreasing the production rate of ASPP, ksASPP, the curve shifts leftward and rightward, respectively (Fig. 6 B). Therefore, controlling the levels of Hzf and ASPP within appropriate ranges is vital to a balance between cell life and death.
Figure 6.
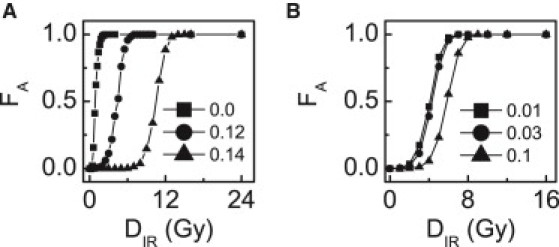
Fraction of apoptotic cells in a population of 2000 cells. The curves of FA versus DIR with different production rates of Hzf (A) or ASPP (B).
Robustness of p53 dynamics to parameter fluctuations
We have demonstrated that the degradation of Hzf is critical to p53-dependent apoptosis under the condition that the Ligase for Hzf degradation is induced by E2F1. Here, we take the second scheme where the Ligase is induced by p53 so slowly that its level becomes high enough to degrade Hzf markedly only when the DNA repair is not completed after a long time.
At DIR = 2 Gy, [p53p] reaches a moderate level during the repair process (Fig. 7 A). Because the degradation of Hzf is enhanced gradually with the accumulation of the Ligase, the p21 level drops progressively after a transient period. As the Bax level is not high enough, Casp3 cannot be activated before the DNA damage is fixed. Thus, there is no apoptosis.
Figure 7.
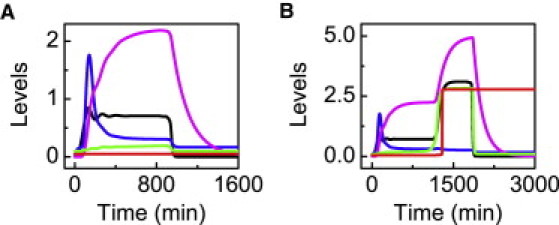
p53-mediated cell-fate decision when the Ligase for Hzf degradation is induced by p53. Shown are time courses of the levels of p53p (black), p21tot (blue), Ligase (magenta), Bax (green), and Casp3 (red) at DIR = 2 Gy (A) or 5 Gy (B).
At DIR = 5 Gy, the p53p dynamics undergo two phases (Fig. 7 B). When [p21tot] drops to rather low levels, E2F1 becomes activated, and enough ASPP accumulates to promote the induction of PTEN and Bax. Thus, [p53p] is driven to higher levels, and more Bax is induced to activate Casp3. Clearly, the level of the Ligase also exhibits a two-phase behavior, but it has a much smaller rising rate compared to the p53 dynamics. Taken together, provided Hzf is effectively degraded, the cell can make a reliable decision, which does not depend heavily on the mechanism underlying the production of the Ligase.
In the following, we analyze the robustness of p53 dynamics to parameter variations. Whereas there are 92 parameters listed in Table 1, we only select 18 parameters and increase/decrease their values by 10% with respect to their default values. The duration of the first phase, T1, and the steady-state level of p53p in the second phase, Lp53, are two crucial quantities describing p53 dynamics. With variation in the parameters, the relative changes in T1 and Lp53 are listed in Table 2.
Table 2.
Robustness of p53 dynamics to parameter variations
| Parameter | Scheme 1 |
Scheme 2 |
||||||
|---|---|---|---|---|---|---|---|---|
| −10% |
+10% |
−10% |
+10% |
|||||
| T1% | Lp53% | T1% | Lp53% | T1% | Lp53% | T1% | Lp53% | |
| kdim | 0.1 | 0.0 | −0.1 | 0.0 | −0.3 | 0.0 | 0.3 | 0.0 |
| kacATM | −0.7 | −0.2 | 0.2 | 0.1 | 1.9 | −0.2 | −0.8 | 0.1 |
| kacp530 | −2.6 | −10.0 | 2.5 | 10.0 | 5.2 | −10.0 | −3.8 | 10.0 |
| ksp53 | −7.1 | −10.7 | 7.6 | 10.7 | 17.3 | −10.6 | −8.9 | 10.6 |
| ksmdm2 | 4.3 | 0.0 | −3.3 | 0.0 | −6.0 | 0.0 | 7.3 | 0.0 |
| kmdm2s | −0.4 | 0.8 | 0.5 | −0.9 | −0.5 | 0.7 | 0.5 | −0.7 |
| kmdm2in | 0.8 | 0.7 | −0.7 | −0.7 | −1.5 | 0.6 | 1.3 | −0.6 |
| kacakt | −0.7 | 0.9 | 0.9 | −0.9 | −0.8 | 0.7 | 0.8 | −0.7 |
| kp2 | −1.0 | 0.9 | 1.2 | −1.0 | −1.0 | 0.7 | 1.2 | −0.8 |
| kshzf | −17.1 | 0.1 | 33.6 | −0.1 | −14.7 | 0.1 | 31.0 | −0.1 |
| ksligase | 55.3 | 0.0 | −17.9 | 0.0 | 30.1 | 0.0 | −12.2 | 0.0 |
| ksp21 | −13.2 | 0 | 21.2 | 0 | −9.0 | 0.0 | 12.5 | 0.0 |
| ksce | 52.3 | 0 | −18.5 | 0 | — | — | −26.2 | 0.0 |
| kprb | 20.3 | 0.0 | −11.5 | 0.0 | — | — | −23.1 | 0.0 |
| kasp21e | −4.9 | 0 | 5.1 | 0 | −4.6 | 0.0 | 4.6 | 0.0 |
| ksPTEN | 1.2 | −1.0 | −0.8 | 0.8 | 1.1 | −0.8 | −0.8 | 0.7 |
| ksHIPK2 | −0.6 | 0.0 | 0.7 | 0.0 | 0.8 | 0.0 | −0.6 | 0.0 |
| ksASPP | 0.6 | −0.1 | −0.5 | 0.1 | 0.7 | −0.1 | −0.6 | 0.1 |
For each case, the value of one of 18 parameters is increased or decreased by 10% with respect to the standard parameter set, while nc is set to 20. The bolded values in each column indicate that T1 or Lp53 is more sensitive to variation in the corresponding parameters. The long dash (—) indicates that the cell cannot commit apoptosis in this case.
For the first scheme, the p53p level is mainly sensitive to two rate constants, i.e., its production rate ksp53 and activation rate kacp530. Indeed, the change of these two parameters evokes an evident variation in Lp53. T1 is sensitive to five parameters: kshzf, ksligase, ksp21, ksce, and kprb. ksligase directly controls the production rate of the Ligase, and kshzf determines the production rate of Hzf. Thus, both the parameters are crucial to p21 regulation, thereby indirectly affecting the activation of E2F1 and the switching of p53 dynamics to the second phase. Similarly, the fluctuations in the other three parameters regulating E2F1 activation also lead to remarkable changes in T1. For the second scheme, Lp53 is also sensitive to ksp53 and kacp530. T1 is also sensitive to the above five parameters because all of them influence the activation of E2F1.
Although we only show the sensitivity of p53 dynamics to 18 parameters, its sensitivity to other parameters can be deduced from the known. For example, the cell-fate decision must be robust to the change in the undimerization rate kundim of ATM because a decrease in kundim is equivalent to an increase in kdim for the regulation of ATM dynamics. Similarly, the p53 dynamics are sensitive to the degradation rate kdp21 of the Ligase because the fluctuations in either ksp21 or kdp21 can significantly modulate the duration of cell cycle arrest. Taken together, the p53 dynamics in both the schemes are fairly robust to parameter fluctuation except for several parameters that control the p53 level and cell cycle progression.
Summary and Discussion
In this work, we developed a mathematical model to characterize the coordination of different cofactors of p53 in the DNA damage response. We found that p53 is activated in a progressive manner and undergoes a two-phase response at high damage levels. After DNA damage, activated p53 first induces Hzf and p21 to arrest the cell cycle, allowing time for DNA repair. If the damage is beyond repair, Hzf is sufficiently degraded, and activated E2F1 induces ASPP, which promotes the expression of Bax to trigger apoptosis. Meanwhile, the concentration of p53 switches from a moderate level to a high level. Therefore, the cellular outcome is closely associated with the p53 level, and the sequential expression of the prosurvival and proapoptotic cofactors of p53 is crucial to a reliable cell-fate decision.
Our results indicate that the degradation of Hzf is essential for apoptosis induction. We proposed two schemes for the production of the unknown E3 ubiquitin ligase for Hzf degradation. It was once suggested that E2F1 promotes its own activation by downregulating the transcription of p21 (43). Here, we proposed that E2F1 may inhibit the expression of p21 by inducing the Ligase for Hzf degradation. This seems plausible because E2F1 can transactivate an E3 ligase, Skp2, to degrade the inhibitors of cell cycle progression including p21 and p27 (44). We speculate that Hzf could also be degraded by Skp2 so that the expression of cell cycle inhibitors is indirectly repressed. That is, Skp2 may be a candidate for the unknown Ligase. On the other hand, the Ligase may be a target of p53, and its level becomes remarkable only after prolonged p53 activation. It is well known that p53 induces several E3 ligases including Mdm2, Pirh2, and Cop1 in the early stage of the cellular response (2,45,46). Nevertheless, they all promote cell survival, whereas the Ligase for Hzf degradation facilitates apoptosis in the later phase of the response. Indeed, Mdm2 is not the E3 ligase for Hzf degradation (12). Therefore, we prefer to expect that the Ligase is related to E2F1, which awaits experimental identification.
It is necessary to compare our model with the previous models about p53 dynamics: 1), Although a series of models has focused on oscillatory dynamics of p53 in the DNA damage response (14–17,19), we did not formulate p53 oscillations here. Typical p53 oscillations were observed in MCF-7 cells in response to IR (33,34). Here, we aimed at revealing the mechanism for cell-fate decision in several other cell lines that were used in the experiment by Das et al. (12), where no evidence supports oscillations in p53 levels. 2), Previously, active p53 was often distinguished between p53 arrester and p53 killer, which promote cell cycle arrest and apoptosis, respectively, based on its phosphorylation status. Here, we found that cooperating with its cofactors, active p53 automatically fulfills its distinct functions during different stages of the response. 3), p53 undergoes a two-phase response to severe DNA damage, switching from a low level to a high level. In our previous work, p53 also exhibit a two-phase behavior, switching from periodic oscillations to high constant levels (16). In both cases, the switching results from the predominance of the p53-PTEN-Akt-Mdm2 positive feedback. Such a two-phase response mode provides a robust mechanism for the choice of cell fate between survival and death. In sum, the p53 dynamics are diverse, depending on the cell- and stress-type. Nevertheless, some generic mechanisms may be in operation, controlling the cell fate after DNA damage.
Our work revealed how Hzf and ASPP are coordinated to affect the cell fate. Hzf is first induced to promote cell cycle arrest by facilitating the expression of p21. Only when DNA damage is severe or prolonged, Hzf is degraded and E2F1-induced ASPP promotes the expression of p53-targeted proapoptotic genes. Therefore, it is important that the prosurvival and proapoptotic cofactors of p53 are separately induced. Moreover, the concurrent activation of p53 and E2F1 is essential for p53-mediated apoptosis, consistent with Zhang et al. (17). We also simulated the effects of knockout of Hzf gene or inhibition of Hzf degradation on the cellular outcome, and our results show good agreement with the experimental observations by Das et al. (12).
Acknowledgments
This work was supported by the National Basic Research Program of China (grant No. 2007CB814806), the National Natural Science Foundation of China (grant No. 11175084), the Natural Science Foundation of Jiangsu Province (grant No. BK2009008), the Program for New Century Excellent Talents in Universities (grant No. NCET-08-0269), and the Priority Academic Program Development of Jiangsu Higher Education Institutions.
Contributor Information
Feng Liu, Email: fliu@nju.edu.cn.
Wei Wang, Email: wangwei@nju.edu.cn.
Appendix: Rate Equations
The rate equations of the model are written as
| (1) |
| (2) |
| (3) |
| (4) |
| (5) |
| (6) |
| (7) |
| (8) |
| (9) |
| (10) |
| (11) |
| (12) |
| (13) |
| (14) |
| (15) |
| (16) |
| (17) |
| (18) |
| (19) |
| (20) |
| (21) |
| (22) |
| (23) |
| (24) |
| (25) |
| (26) |
| (27) |
| (28) |
| (29) |
| (30) |
| (31) |
Note: For the second scheme, Eq. 19 should be replaced by the equation
References
- 1.Meek D.W. Tumor suppression by p53: a role for the DNA damage response? Nat. Rev. Cancer. 2009;9:714–723. doi: 10.1038/nrc2716. [DOI] [PubMed] [Google Scholar]
- 2.Wu X., Bayle J.H., Levine A.J. The p53-Mdm-2 autoregulatory feedback loop. Genes Dev. 1993;7:1126–1132. doi: 10.1101/gad.7.7a.1126. [DOI] [PubMed] [Google Scholar]
- 3.Aylon Y., Oren M. Living with p53, dying of p53. Cell. 2007;130:597–600. doi: 10.1016/j.cell.2007.08.005. [DOI] [PubMed] [Google Scholar]
- 4.Abbas T., Dutta A. p21 in cancer: intricate networks and multiple activities. Nat. Rev. Cancer. 2009;9:400–414. doi: 10.1038/nrc2657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Miyashita T., Reed J.C. Tumor suppressor p53 is a direct transcriptional activator of the human Bax gene. Cell. 1995;80:293–299. doi: 10.1016/0092-8674(95)90412-3. [DOI] [PubMed] [Google Scholar]
- 6.Vousden K.H. Outcomes of p53 activation—spoilt for choice. J. Cell Sci. 2006;119:5015–5020. doi: 10.1242/jcs.03293. [DOI] [PubMed] [Google Scholar]
- 7.Murray-Zmijewski F., Slee E.A., Lu X. A complex barcode underlies the heterogeneous response of p53 to stress. Nat. Rev. Mol. Cell Biol. 2008;9:702–712. doi: 10.1038/nrm2451. [DOI] [PubMed] [Google Scholar]
- 8.Vousden K.H., Lane D.P. p53 in health and disease. Nat. Rev. Mol. Cell Biol. 2007;8:275–283. doi: 10.1038/nrm2147. [DOI] [PubMed] [Google Scholar]
- 9.Olsson A., Manzl C., Villunger A. How important are post-translational modifications in p53 for selectivity in target-gene transcription and tumor suppression? Cell Death Differ. 2007;14:1561–1575. doi: 10.1038/sj.cdd.4402196. [DOI] [PubMed] [Google Scholar]
- 10.Oda K., Arakawa H., Taya Y. p53AIP1, a potential mediator of p53-dependent apoptosis, and its regulation by Ser-46-phosphorylated p53. Cell. 2000;102:849–862. doi: 10.1016/s0092-8674(00)00073-8. [DOI] [PubMed] [Google Scholar]
- 11.Samuels-Lev Y., O'Connor D.J., Lu X. ASPP proteins specifically stimulate the apoptotic function of p53. Mol. Cell. 2001;8:781–794. doi: 10.1016/s1097-2765(01)00367-7. [DOI] [PubMed] [Google Scholar]
- 12.Das S., Raj L., Lee S.W. Hzf determines cell survival upon genotoxic stress by modulating p53 transactivation. Cell. 2007;130:624–637. doi: 10.1016/j.cell.2007.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wee K.B., Aguda B.D. Akt versus p53 in a network of oncogenes and tumor suppressor genes regulating cell survival and death. Biophys. J. 2006;91:857–865. doi: 10.1529/biophysj.105.077693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhang T., Brazhnik P., Tyson J.J. Exploring mechanisms of the DNA-damage response: p53 pulses and their possible relevance to apoptosis. Cell Cycle. 2007;6:85–94. doi: 10.4161/cc.6.1.3705. [DOI] [PubMed] [Google Scholar]
- 15.Zhang X.P., Liu F., Wang W. Cell fate decision mediated by p53 pulses. Proc. Natl. Acad. Sci. USA. 2009;106:12245–12250. doi: 10.1073/pnas.0813088106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhang X.P., Liu F., Wang W. Two-phase dynamics of p53 in the DNA damage response. Proc. Natl. Acad. Sci. USA. 2011;108:8990–8995. doi: 10.1073/pnas.1100600108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhang X.P., Liu F., Wang W. Coordination between cell cycle progression and cell fate decision by the p53 and E2F1 pathways in response to DNA damage. J. Biol. Chem. 2010;285:31571–31580. doi: 10.1074/jbc.M110.134650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rothkamm K., Löbrich M. Evidence for a lack of DNA double-strand break repair in human cells exposed to very low x-ray doses. Proc. Natl. Acad. Sci. USA. 2003;100:5057–5062. doi: 10.1073/pnas.0830918100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ma L., Wagner J., Stolovitzky G.A. A plausible model for the digital response of p53 to DNA damage. Proc. Natl. Acad. Sci. USA. 2005;102:14266–14271. doi: 10.1073/pnas.0501352102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Stewart R.D. Two-lesion kinetic model of double-strand break rejoining and cell killing. Radiat. Res. 2001;156:365–378. doi: 10.1667/0033-7587(2001)156[0365:tlkmod]2.0.co;2. [DOI] [PubMed] [Google Scholar]
- 21.Bakkenist C.J., Kastan M.B. DNA damage activates ATM through intermolecular autophosphorylation and dimer dissociation. Nature. 2003;421:499–506. doi: 10.1038/nature01368. [DOI] [PubMed] [Google Scholar]
- 22.Kholodenko B.N. Cell-signaling dynamics in time and space. Nat. Rev. Mol. Cell Biol. 2006;7:165–176. doi: 10.1038/nrm1838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Canman C.E., Lim D.S., Siliciano J.D. Activation of the ATM kinase by ionizing radiation and phosphorylation of p53. Science. 1998;281:1677–1679. doi: 10.1126/science.281.5383.1677. [DOI] [PubMed] [Google Scholar]
- 24.Stommel J.M., Wahl G.M. Accelerated MDM2 auto-degradation induced by DNA-damage kinases is required for p53 activation. EMBO J. 2004;23:1547–1556. doi: 10.1038/sj.emboj.7600145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mayo L.D., Donner D.B. A phosphatidylinositol 3-kinase/Akt pathway promotes translocation of Mdm2 from the cytoplasm to the nucleus. Proc. Natl. Acad. Sci. USA. 2001;98:11598–11603. doi: 10.1073/pnas.181181198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mayo L.D., Dixon J.E., Donner D.B. PTEN protects p53 from Mdm2 and sensitizes cancer cells to chemotherapy. J. Biol. Chem. 2002;277:5484–5489. doi: 10.1074/jbc.M108302200. [DOI] [PubMed] [Google Scholar]
- 27.Mayo L.D., Seo Y.R., Donner D.B. Phosphorylation of human p53 at serine 46 determines promoter selection and whether apoptosis is attenuated or amplified. J. Biol. Chem. 2005;280:25953–25959. doi: 10.1074/jbc.M503026200. [DOI] [PubMed] [Google Scholar]
- 28.Rinaldo C., Prodosmo A., Soddu S. Mdm2-regulated degradation of HIPK2 prevents p53Ser46 phosphorylation and DNA damage-induced apoptosis. Mol. Cell. 2007;25:739–750. doi: 10.1016/j.molcel.2007.02.008. [DOI] [PubMed] [Google Scholar]
- 29.Green D.R., Kroemer G. Cytoplasmic functions of the tumor suppressor p53. Nature. 2009;458:1127–1130. doi: 10.1038/nature07986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Manning B.D., Cantley L.C. AKT/PKB signaling: navigating downstream. Cell. 2007;129:1261–1274. doi: 10.1016/j.cell.2007.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gottlieb T.M., Leal J.F., Oren M. Cross-talk between Akt, p53 and Mdm2: possible implications for the regulation of apoptosis. Oncogene. 2002;21:1299–1303. doi: 10.1038/sj.onc.1205181. [DOI] [PubMed] [Google Scholar]
- 32.Cantley L.C., Neel B.G. New insights into tumor suppression: PTEN suppresses tumor formation by restraining the phosphoinositide 3-kinase/AKT pathway. Proc. Natl. Acad. Sci. USA. 1999;96:4240–4245. doi: 10.1073/pnas.96.8.4240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lahav G., Rosenfeld N., Alon U. Dynamics of the p53-Mdm2 feedback loop in individual cells. Nat. Genet. 2004;36:147–150. doi: 10.1038/ng1293. [DOI] [PubMed] [Google Scholar]
- 34.Batchelor E., Mock C.S., Lahav G. Recurrent initiation: a mechanism for triggering p53 pulses in response to DNA damage. Mol. Cell. 2008;30:277–289. doi: 10.1016/j.molcel.2008.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fogal V., Kartasheva N.N., Lu X. ASPP1 and ASPP2 are new transcriptional targets of E2F. Cell Death Differ. 2005;12:369–376. doi: 10.1038/sj.cdd.4401562. [DOI] [PubMed] [Google Scholar]
- 36.Cory S., Adams J.M. The Bcl2 family: regulators of the cellular life-or-death switch. Nat. Rev. Cancer. 2002;2:647–656. doi: 10.1038/nrc883. [DOI] [PubMed] [Google Scholar]
- 37.Kirsch D.G., Doseff A., Hardwick J.M. Caspase-3-dependent cleavage of Bcl-2 promotes release of cytochrome c. J. Biol. Chem. 1999;274:21155–21161. doi: 10.1074/jbc.274.30.21155. [DOI] [PubMed] [Google Scholar]
- 38.Ow Y.P., Green D.R., Mak T.W. Cytochrome c: functions beyond respiration. Nat. Rev. Mol. Cell Biol. 2008;9:532–542. doi: 10.1038/nrm2434. [DOI] [PubMed] [Google Scholar]
- 39.Youle R.J., Strasser A. The BCL-2 protein family: opposing activities that mediate cell death. Nat. Rev. Mol. Cell Biol. 2008;9:47–59. doi: 10.1038/nrm2308. [DOI] [PubMed] [Google Scholar]
- 40.Riedl S.J., Salvesen G.S. The apoptosome: signaling platform of cell death. Nat. Rev. Mol. Cell Biol. 2007;8:405–413. doi: 10.1038/nrm2153. [DOI] [PubMed] [Google Scholar]
- 41.Yao G., Lee T.J., You L. A bistable Rb-E2F switch underlies the restriction point. Nat. Cell Biol. 2008;10:476–482. doi: 10.1038/ncb1711. [DOI] [PubMed] [Google Scholar]
- 42.Zhang T., Brazhnik P., Tyson J.J. Computational analysis of dynamical responses to the intrinsic pathway of programmed cell death. Biophys. J. 2009;97:415–434. doi: 10.1016/j.bpj.2009.04.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wu L., Timmers C., Leone G. The E2F1-3 transcription factors are essential for cellular proliferation. Nature. 2001;414:457–462. doi: 10.1038/35106593. [DOI] [PubMed] [Google Scholar]
- 44.Zhang L., Wang C. F-box protein Skp2: a novel transcriptional target of E2F. Oncogene. 2006;25:2615–2627. doi: 10.1038/sj.onc.1209286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Leng R.P., Lin Y., Benchimol S. Pirh2, a p53-induced ubiquitin-protein ligase, promotes p53 degradation. Cell. 2003;112:779–791. doi: 10.1016/s0092-8674(03)00193-4. [DOI] [PubMed] [Google Scholar]
- 46.Dornan D., Wertz I., Dixit V.M. The ubiquitin ligase COP1 is a critical negative regulator of p53. Nature. 2004;429:86–92. doi: 10.1038/nature02514. [DOI] [PubMed] [Google Scholar]


