Abstract
It has been generally accepted that the mammalian embryo starts its development with all cells identical and only when inside and outside cells form do differences between cells first emerge. However, recent findings show that cells in the mouse embryo can differ in their developmental fate and potency already by the 4-cell-stage1-4. Such differences depend on the orientation and order of the cleavage divisions that generated them2,5. Since epigenetic marks are suggested to be involved in sustaining pluripotency6,7, we considered that such developmental properties might be achieved through epigenetic mechanisms. Here, we show that modification of histone H3, through methylation of specific arginine residues, correlates with cell fate and potency. Levels of H3 methylation at specific arginines are maximal in 4-cell blastomeres that will contribute to the ICM and polar trophectoderm and undertake full development when combined together in chimeras. Arginine methylation of H3 is minimal in cells whose progeny contributes more to the mural trophectoderm and that show compromised development when combined in chimeras. This suggests that higher levels of H3 arginine methylation predispose blastomeres to contribute to the pluripotent cells of the ICM. We confirm this prediction by overexpressing the H3-specific arginine methyltransferase, CARM1, in individual blastomeres and show this directs their progeny to the ICM and results in a dramatic upregulation of Nanog and Sox2. Thus, our results identify specific histone modifications as the earliest known epigenetic marker contributing to development of ICM and show that manipulation of epigenetic information influences cell fate determination.
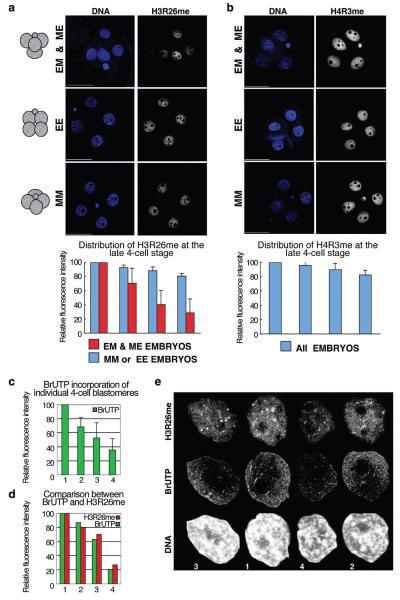
To address whether epigenetic differences exist between blastomeres at the 4-cell stage, we focused on histone methylation marks related to transcriptional activation8-10. Because the divisions of 2-cell-stage blastomeres differ in orientation in relation to the animal-vegetal axis of the egg5,11, the shape of 4-cell embryos vary: blastomeres fill the apices of a tetrahedron when they undergo one equatorial (E) and one meridional (M) division or they lie upon a similar plane when they undergo either two equatorial or two meridional divisions (Fig. 1a). Although any combination of the temporal sequence of such divisions is possible, sequential M and E divisions are most common (~80%), but they can occur in either order. We found that whereas embryos that underwent either two equatorial or two meridional divisions did not show significant variations in the levels of H3 methylation at arginine 26 (H3R26me) between 4-cell-stage blastomeres. (Fig. 1a, EE&MM embryos; blue bars), tetrahedral embryos displayed marked differences in H3R26me levels between their blastomeres (Fig.1a, EM & ME embryos; red bars). In the latter, the weakest level of H3R26me was generally less than 40% of the cell giving the strongest signal (p=0.0002). Measuring the intensity of DNA staining indicated that the variation of H3R26me levels was not related to differences in the content of DNA due to replication (not shown). We confirmed that this variation in H3R26me levels did not result from the confocal scanning of embryos of differing shapes by scanning individual cells of disaggregated embryos, where we found a similar outcome (Supplementary Figure S1). In addressing whether the methylation of other arginines also showed differences between 4-cell blastomeres, we found that levels of H3R2me and H3R17me (Supplementary Figures S2-S3a) varied between blastomeres correlating with embryo morphology in a similar way to H3R26me. This is consistent with R2 and R17 being targets for the same methyltransferase, CARM1, as R2612,13.
Figure 1.
Levels of H3R26me are different in blastomeres of 4-cell stage embryos and correlate with their spatial arrangement.
(a) 4-cell stage embryos were stained for H3R26me and grouped according to their shape in tetrahedral (EM and ME), EE (flatten,polar body on one side) or MM (flatten,polar body in the middle). Shown are projections, including all sections, of representative embryos.Fluorescence levels were quantified and normalised against the blastomere showing the highest level which was set at 100%. Decreasing values of fluorescence were normalised and averaged accordingly(n=18). Each bar represents the relative fluorescence level of each of the 4 blastomeres. Scale bar 50μm.
(b) Differences in histone arginine methylation levels in 4-cell stage blastomeres are specific:only residues that are CARM1 targets, and not PRMT1, display differential distribution.
(c) 4-cell stage blastomeres display different global transcriptional activity. BrUTP incorporation was measured in sections from nuclei of 4-cell stage embryos captured every 0.6μm (n=12). Projections were used after cropping off the nuclei using the Volocity software to quantify active (nuclear) transcription. Values were normalised as in (a).
(d-e) Global transcription levels correlate with global H3R26me levels. Quantification (d) of BrUTP incorporation (green) and H3R26me (red) of 4-nuclei of a representative embryo. Nuclei (e) are shown at the same scale,numbers at the bottom correspond to the blastomere numbers of the graph.
When we analysed CARM1 distribution in 4-cell blastomeres, we found that CARM1 levels varied with the same tendency as those of H3R26me (Supplementary Figure S4b-c). In contrast, methylation of H4R3, which is target of a different methyltransferase, PRMT114,15, appeared equivalent between blastomeres regardless of embryo morphology (Fig. 1b). Thus, the differences in the levels of histone arginine methylation in 4-cell-stage blastomeres are specific. We also observed that levels of BrUTP incorporation in late 4-cell-stage embryos were highest in the blastomeres that are enriched for H3R26me, indicative of elevated levels of global transcription in these cells (Fig. 1c-e). Since H3R26me showed the biggest difference in its distribution between blastomeres (Supplementary Figure S3b), we concentrated on analyzing this modification further.
As different developmental fate and potential can be ascribed to blastomeres depending upon the orientation and the order of division from the 2- to the 4-cell-stage in relation to the animal-vegetal axis, we wished to determine whether differences in H3R26me related to patterns of division. To this end we first grouped embryos according to their cleavage patterns to the 4-cell-stage. When the earlier of the second cleavages is M and the later E (ME embryos), the earlier dividing 2-cell blastomere contributes most of its progeny to the embryonic (ICM and adjacent polar trophectoderm) and the later one to the abembryonic part (ICM and mural trophectoderm) of the blastocyst. When the earlier second cleavage is E and the later M (EM embryos), the earlier blastomere can give rise to either the embryonic or abembryonic part of the blastocyst. By contrast, when second cleavage divisions are of similar orientation (MM or EE), the allocation of blastomere progeny is random. The ME group of embryos thus allowed the identification of individual 4-cell blastomeres that have predictable fate within the blastocyst5. Moreover, blastomeres resulting from the E division that inherit the “vegetal” cytoplasm in ME embryos tend to contribute predominantly to the mural trophectoderm and do not complete development when combined with the same type of “vegetal” cells in chimeric embryos2. In contrast, chimaeras of blastomeres that arise from early M divisions (with ‘animal–vegetal’ cytoplasm) could complete development with full success and chimeras constructed only from ‘animal’ blastomeres could also develop although with reduced success. To distinguish the progeny of 2-cell-stage blastomeres, we injected one 2-cell blastomere at random with rhodamine-dextran and then monitored the second cleavage divisions (Supplementary Figure S5a). Immunostaining then revealed similar levels of H3R26me in the progeny of both E and M divisions of EM embryos (compare M cells vs E cells; Supplementary Figure S5b-c; p=0.112). In ME embryos, however, the M sister cells displayed significantly higher levels of H3R26 methylation than both E sisters, with one of the E-derived blastomeres displaying significantly less methylation than either of the M-derived blastomeres (Supplementary Figure S5b-c; p <0.0001). In confirmation of our earlier observations (Fig.1), EE or MM embryos did not display differences in their relative H3R26me levels. Thus, in ME embryos where prediction of developmental properties is possible, higher levels of H3R26me are seen in blastomeres expected to contribute to the embryonic part of the blastocyst and lower levels in blastomeres expected to contribute to the abembryonic part.
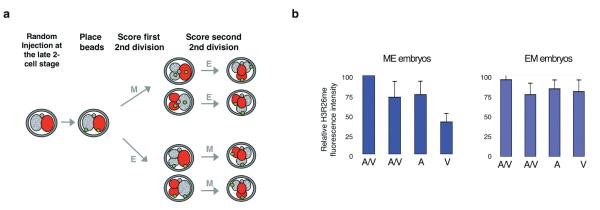
Given the reported differences in developmental potential of “animal” or “vegetal” blastomeres arising from E divisions in ME embryos2, we next examined whether these cells differed in their H3R26me levels. To this end we randomly injected a 2-cell-stage blastomere with rhodamine-dextran as before and then applied green fluorescent beads to the vegetal membranes of the two blastomeres as a second label (Fig. 2a and Ref.2). The rhodamine label allowed us to score the order and the plane of division to the 4-cell-stage and the beads served as ‘vegetal’ markers. We found that in ME embryos, the ‘vegetal’ blastomere always displayed significantly lower levels of H3R26me than the “animal” or “animal/vegetal” blastomeres (Fig. 2b; p <0.0001). In contrast, in EM embryos the ‘animal’ and ‘vegetal’ blastomeres from the E division displayed equivalent levels of H3R26me (Fig. 2b). Taken together our results indicate that more extensive H3 arginine methylation in M blastomeres of ME embryos correlates with their greater contribution to the embryonic part of the blastocyst. In contrast, blastomeres having the lowest levels of H3 arginine methylation (i.e. ‘vegetal’ blastomeres) are those predicted to contribute mainly to the abembryonic part.
Figure 2.
The ‘vegetal’ blastomere in ME embryos displays the lowest levels of H3R26me.
(a) Design to determine the identity of 4-cell-stage blastomeres according to division orientation, order, and blastomere positioning. A 2-cell stage blastomere was microinjected with rhodamine-dextran. We then placed a green fluorescent bead in the ‘vegetal’ membrane of the two blastomeres. Divisions were scored and embryos were stained for H3R26me at the late 4-cell stage.The position of the bead and the presence of rhodamine allowed identification of the blastomeres as Animal/Vegetal (A/V,derived from M divisions), Animal or Vegetal (A or V,derived from E divisions) in EM(n=10) or ME(n=9) embryos.
(b) The vegetal blastomere of ME embryos displays the lowest levels of H3R26me while the vegetal blastomere of EM embryos displays similar levels to the animal or animal/vegetal blastomeres. H3R26me levels were quantified as in Fig.1.
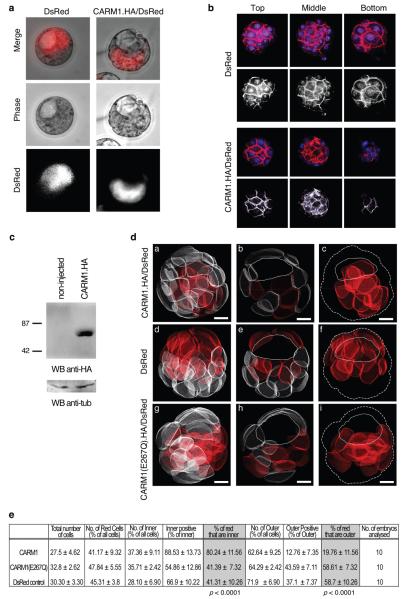
As levels of methylation at arginines 2, 17 and 26 varied similarly and as these three residues are the specific targets of CARM1/PRMT4, which is expressed maternally in mouse embryos (Supplementary Figures S4,S6), we wondered whether CARM1 might play a role in directing developmental fate and potency. To test this, we injected CARM1 mRNA into single late 2-cell-stage blastomeres aiming to elevate levels of H3 arginine methylation in the progeny of these cells from the mid 4-cell-stage. We followed the effect upon cell fate by co-injecting mRNA for DsRed as lineage tracer (Fig.3a-b). Strikingly, the labeled clone was located in the embryonic part of the blastocyst in 31 out of 35 embryos (89%) and in no embryos labeled cells were found exclusively in the abembryonic part. We randomly selected 10 of these embryos to reconstruct in 3 dimensions to locate every cell and determine to which lineage (ICM or trophectoderm) labelled cells had contributed. This showed that the ICM comprised 37% of all cells at this stage and that on average, 88.5% of ICM cells were derived from the blastomere in which CARM1 had been overexpressed (Fig. 3d, Fig.3e, Supplementary Table S1). Notably, in half of the embryos, all of the ICM comprised the exclusive progeny of the CARM1-overexpressing blastomere. By contrast, even though the majority (63%) of the blastocyst cells are outer cells, the proportion of outer cells derived from the CARM1-overexpressing blastomere was only 12% (Fig.3e, Supplementary Table S1). This was contrasted to control embryos, injected with mRNA for DsRed only, where we found that 41% and 59% of cells labeled with the lineage tracer were inner or outer cells, respectively (p<0.0001)(Fig.3e and Supplementary Table S2). Thus, we conclude that forced overexpression of CARM1 in a 2-cell blastomere leads that cell to contribute predominantly (if not exclusively) to the ICM. To establish whether the methyltransferase activity of CARM1 was required for this effect, we injected mRNA for CARM1, which contains a point mutation (E267Q) and is devoid of catalytic activity16 into single 2-cell blastomeres as above. We found that the progeny of the labelled cells could contribute to both embryonic and abembryonic regions of the blastocyst. The distribution of the progeny of the CARM1(E267Q)-injected blastomere was similar to the DsRed control (Fig.3e, Supplementary Table S3, Fig.3d). The progeny of the CARM1-overexpressing blastomere displayed increased levels of H3R26me whereas blastomeres overexpressing CARM1(E267Q) did not (Fig.4a-b). Hence, the ability of CARM1 to direct the progeny of a blastomere towards the ICM is strictly dependent on its methyltransferase activity.
Figure 3.
CARM1 overexpression in a 2-cell blastomere results in the contribution of that cell predominantly to the ICM.
A late 2-cell stage blastomere was injected with mRNA for DsRed alone (control) or in combination with mRNA for CARM1.HA. This results in CARM1 overexpression from the mid 4-cell stage since CARM1.HA/DsRed expression starts 6-8h after injection (not shown). Embryos were cultured until the blastocyst stage and observed under fluorescence microscopy. DsRed was used as a lineage tracer.
(a) Representative embryos derived from DsRed only (n=17) or DsRed/CARM1.HA-overexpression experiments (n=35).
(b) Blastocysts were stained with phallaoidin-Texas-Red and TOTO-3 (to visualize cell membranes and DNA, respectively) and analysed under confocal microscopy. Representative top, middle and bottom sections are shown. DNA is in blue; Phallaoidin (red) can be distinguished from DsRed because the latter is exclusively cytoplasmic. The red channel is shown as grayscale. The progeny of the CARM1-overexpressing blastomere is predominantly within the inner cells of the blastocyst.
(c) Overexpression of CARM1 was verified by Western blot in zygotes injected with mRNA for CARM1.HA/DsRed.
(d) Representative 3D reconstructions of blastocysts in which mRNA for CARM1.HA/DsRed (a), Ds/Red only (d), or CARM1(E267Q).HA/DsRed (g) was microinjected at the 2-cell stage. Blastocysts were stained as in (b). Confocal z-stacks were taken at 1μm intervals. IMARIS software was used to outline cell membranes to create 3D models of all cells of the embryo. Cells were then scored according to their position: cells completely surrounded by others are denoted as inner, those with an outer surface as outer. Cells were scored as either positive or negative for DsRed. Progeny of injected blastomere is shown in red. A middle slice is shown in b,e, h where the cavity is depicted with a line. In c,f ,i, only the progeny of the injected blastomere is shown, the contour of the embryo is indicated by a dashed line and the position of the cavity by a solid line. Scale bar 10μm.
Figure 4.
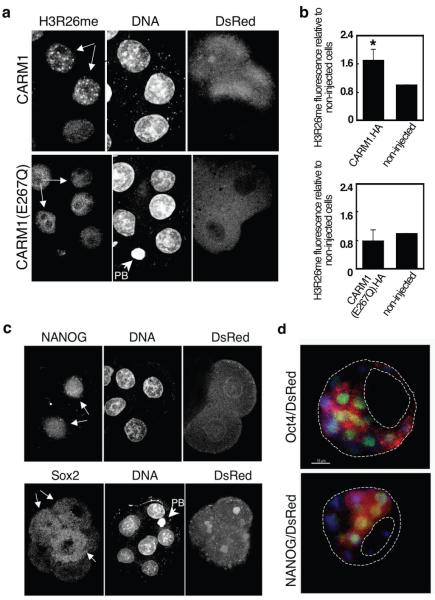
Overexpression of CARM1 results in elevated levels of arginine methylation and upregulation of Nanog and Sox2.
(a-b) A 2-cell stage blastomere was injected with mRNA for CARM1.HA/DsRed. Embryos were cultured to the 8-cell stage and stained for H3R26me. Shown are 3 nuclei of cells from a representative embryo. The progeny of the injected blastomere is indicated by arrows (note the presence of DsRed). In (b) H3R26me levels in cells overexpressing CARM1 were normalised against those of non-injected cells within the same embryo (*p=0.0006)(n=14). In the bottom, data derived from overexpression of CARM1(E267Q).HA. Here, 5 cells from the same embryo are shown(n=6). PB:polar body.
(c) Embryos were injected as in (a) and stained with a NANOG (n=5) or a Sox2 (n=11) antibody between the 6- and 8-cell stage. For NANOG, 4 nuclei of the same embryo, two of them deriving from the CARM1-overexpressing blastomere are shown(white arrows, note the presence of DsRed). NANOG is detectable only in the blastomeres deriving from the 2-cell stage blastomere injected with CARM1 mRNA. For Sox2, a representative embryo is shown. Note that Sox2 is mainly cytoplasmic at this stage18. We were unable to address CARM1 function by the converse experiment by RNAi since the protein is provided maternally and its mRNA is rapidly downregulated after fertilization. However, treatment of zygotes with specific arginine methyltransferase inhibitors25 showed that reducing levels of histone arginine methylation impaired development (Supplementary Figure S8).
(d) The progeny of CARM1-overexpressing blastomere expresses ICM markers in the blastocyst. Blastocysts were stained for Oct4/Pou5f1 (n=7) or NANOG (n=3)(green). The presence of DsRed indicates the progeny of the injected blastomere. DNA shown in blue.
We next assessed expression levels of transcription factors known to influence the development of ICM cells. Interestingly, we found that overexpression of CARM1 led to an early and dramatic upregulation of Nanog in the injected blastomeres, suggesting that the Nanog promoter is regulated by arginine methylation (Fig. 4c). In contrast, Cdx2, a trophectoderm marker, showed no induction upon CARM1 overexpression (not shown). We also detected increased levels of Sox2 in the progeny of the CARM1-injected blastomere (Fig. 4c). However, Oct4/Pou5f1 showed variable levels of expression in non-injected blastomeres as well as in CARM1 overexpressing cells (Supplementary Figure S7). Note that, in contrast to Nanog, both Oct4/Pou5f1 and Sox2 were also present in the non-injected blastomeres reflecting an earlier expression and/or their maternal inheritance17,18. The co-expression by the blastocyst stage of the ICM markers Oct4/Pou5f1 and Nanog in the progeny of the CARM1-overexpressing blastomere is consistent with the observed change in cell fate (Fig.4d). Hence, by manipulating epigenetic information through overexpression of a histone modifier it is possible to direct cells towards the ICM.
Our findings, in control experiments, that either of the 2-cell blastomeres normally contributes their progeny to inner or outer cells of the blastocyst is in accordance with our earlier findings1,2,4,5. However, it contrasts to a report that shows that as a consequence of differential Cdx2 levels between 2-cell blastomeres, one blastomere contributes exclusively to outer cells, which normally express Cdx219. Unlike these authors we do not observe expression of Cdx2 at the 2-cell-stage.
Our study provides the first demonstration that epigenetic differences develop between blastomeres by the 4-cell-stage. We cannot exclude the possibility that CARM1 mediates some of its effects through targets other than histone H3. However, the differential levels of H3 methylation between 4-cell blastomeres can account for their different cell fate and potency1,4,5. Cells with more extensive H3 arginine methylation are destined to contribute pluripotent progeny to the blastocyst. These cells show increased levels of transcription that includes expression of a select set of genes responsible for maintaining pluripotency such as Nanog and Sox218,20,21. An enrichment in modifications characteristic of euchromatin may have additional effects upon the chromatin to generate an “open” configuration that could sustain pluripotency in the embryo as has been suggested for ES cells22. Indeed, the relative importance of potential changes in chromatin structure in relation to cell plasticity demands further study.
Methods
Embryos were collected from F1 crosses (C57BL/6XCBA/H). To monitor the division from the 2-cell-stage, one blastomere was microinjected randomly with Dextran-tetramethylrhodamine (3000MW,Molecular Probes). Green fluorescent beads were placed in the membrane of blastomeres using a Piezo driller. Embryos were observed every 20 minutes to determine the plane and order of division5. Immunostaining and BrUTP labelling were performed as described23. H3 asymmetric-dimethyl-R2, H3 dimethyl-R17, H3 dimethyl-R26, H4 symmetric-dimethyl-R3 antibodies were from abcam (Supplementary Figure S9); Oct4 from R&D systems; NANOG and Sox2 from Santa Cruz. For analysis of 4-cell embryos, confocal sections were taken every 0.8μm through the whole embryo and fluorecsence signal was measured in projections using the Volocity software (Improvision). For 3D reconstructions, blastocysts were stained with phallaoidin-Texas-Red and TOTO-3. Confocal sections were captured every 1μm and processed with IMARIS (Bitplane) and 3DVirtual Embryo software24.
Supplementary Material
Acknowledgments
We would like to thank Pawel Greda for bead labeling, Caroline Lee for assistance, David Glover for comments on the manuscript, Michael Stallcup for the CARM1 expression vectors and Mark Bedford for providing the CARM1-/- MEFs and the PRMT inhibitor. METP is an EMBO long-term fellow. We are grateful to the Wellcome Trust Senior Fellowship to MZG which has supported this work.
Footnotes
Reprints and permissions information is available at npg.nature.com/reprintsandpermissions
Competing interests statement. The authors declare that they have no competing financial interests.
Supplementary Information accompanies the paper on www.nature.com/nature
References
- 1.Piotrowska K, Wianny F, Pedersen RA, Zernicka-Goetz M. Blastomeres arising from the first cleavage division have distinguishable fates in normal mouse development. Development. 2001;128:3739–48. doi: 10.1242/dev.128.19.3739. [DOI] [PubMed] [Google Scholar]
- 2.Piotrowska-Nitsche K, Perea-Gomez A, Haraguchi S, Zernicka-Goetz M. Four-cell stage mouse blastomeres have different developmental properties. Development. 2005;132:479–90. doi: 10.1242/dev.01602. [DOI] [PubMed] [Google Scholar]
- 3.Gardner RL. Specification of embryonic axes begins before cleavage in normal mouse development. Development. 2001;128:839–47. doi: 10.1242/dev.128.6.839. [DOI] [PubMed] [Google Scholar]
- 4.Fujimori T, Kurotaki Y, Miyazaki J, Nabeshima Y. Analysis of cell lineage in two- and four-cell mouse embryos. Development. 2003;130:5113–22. doi: 10.1242/dev.00725. [DOI] [PubMed] [Google Scholar]
- 5.Piotrowska-Nitsche K, Zernicka-Goetz M. Spatial arrangement of individual 4-cell stage blastomeres and the order in which they are generated correlate with blastocyst pattern in the mouse embryo. Mech Dev. 2005;122:487–500. doi: 10.1016/j.mod.2004.11.014. [DOI] [PubMed] [Google Scholar]
- 6.Li E. Chromatin modification and epigenetic reprogramming in mammalian development. Nat Rev Genet. 2002;3:662–73. doi: 10.1038/nrg887. [DOI] [PubMed] [Google Scholar]
- 7.Morgan HD, Santos F, Green K, Dean W, Reik W. Epigenetic reprogramming in mammals. Hum Mol Genet. 2005;14(Spec No 1):R47–58. doi: 10.1093/hmg/ddi114. [DOI] [PubMed] [Google Scholar]
- 8.Chen D, et al. Regulation of transcription by a protein methyltransferase. Science. 1999;284:2174–7. doi: 10.1126/science.284.5423.2174. [DOI] [PubMed] [Google Scholar]
- 9.Ma H, et al. Hormone-dependent, CARM1-directed, arginine-specific methylation of histone H3 on a steroid-regulated promoter. Curr Biol. 2001;11:1981–5. doi: 10.1016/s0960-9822(01)00600-5. [DOI] [PubMed] [Google Scholar]
- 10.Bauer UM, Daujat S, Nielsen SJ, Nightingale K, Kouzarides T. Methylation at arginine 17 of histone H3 is linked to gene activation. EMBO Rep. 2002;3:39–44. doi: 10.1093/embo-reports/kvf013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gardner RL. Experimental analysis of second cleavage in the mouse. Hum Reprod. 2002;17:3178–89. doi: 10.1093/humrep/17.12.3178. [DOI] [PubMed] [Google Scholar]
- 12.Bannister AJ, Kouzarides T. Reversing histone methylation. Nature. 2005;436:1103–6. doi: 10.1038/nature04048. [DOI] [PubMed] [Google Scholar]
- 13.Schurter BT, et al. Methylation of histone H3 by coactivator-associated arginine methyltransferase 1. Biochemistry. 2001;40:5747–56. doi: 10.1021/bi002631b. [DOI] [PubMed] [Google Scholar]
- 14.Wang H, et al. Methylation of histone H4 at arginine 3 facilitating transcriptional activation by nuclear hormone receptor. Science. 2001;293:853–7. doi: 10.1126/science.1060781. [DOI] [PubMed] [Google Scholar]
- 15.Strahl BD, et al. Methylation of histone H4 at arginine 3 occurs in vivo and is mediated by the nuclear receptor coactivator PRMT1. Curr Biol. 2001;11:996–1000. doi: 10.1016/s0960-9822(01)00294-9. [DOI] [PubMed] [Google Scholar]
- 16.Lee YH, Koh SS, Zhang X, Cheng X, Stallcup MR. Synergy among nuclear receptor coactivators: selective requirement for protein methyltransferase and acetyltransferase activities. Mol Cell Biol. 2002;22:3621–32. doi: 10.1128/MCB.22.11.3621-3632.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Scholer HR, Hatzopoulos AK, Balling R, Suzuki N, Gruss P. A family of octamer-specific proteins present during mouse embryogenesis: evidence for germline-specific expression of an Oct factor. Embo J. 1989;8:2543–50. doi: 10.1002/j.1460-2075.1989.tb08392.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Avilion AA, et al. Multipotent cell lineages in early mouse development depend on SOX2 function. Genes Dev. 2003;17:126–40. doi: 10.1101/gad.224503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Deb K, Sivaguru M, Yong HY, Roberts RM. Cdx2 gene expression and trophectoderm lineage specification in mouse embryos. Science. 2006;311:992–6. doi: 10.1126/science.1120925. [DOI] [PubMed] [Google Scholar]
- 20.Mitsui K, et al. The homeoprotein Nanog is required for maintenance of pluripotency in mouse epiblast and ES cells. Cell. 2003;113:631–42. doi: 10.1016/s0092-8674(03)00393-3. [DOI] [PubMed] [Google Scholar]
- 21.Chambers I, et al. Functional expression cloning of Nanog, a pluripotency sustaining factor in embryonic stem cells. Cell. 2003;113:643–55. doi: 10.1016/s0092-8674(03)00392-1. [DOI] [PubMed] [Google Scholar]
- 22.Meshorer E, et al. Hyperdynamic plasticity of chromatin proteins in pluripotent embryonic stem cells. Dev Cell. 2006;10:105–16. doi: 10.1016/j.devcel.2005.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Torres-Padilla ME, Zernicka-Goetz M. Role of TIF1alpha as a modulator of embryonic transcription in the mouse zygote. J Cell Biol. 2006;174:329–38. doi: 10.1083/jcb.200603146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tassy O, Daian F, Hudson C, Bertrand V, Lemaire P. A quantitative approach to the study of cell shapes and interactions during early chordate embryogenesis. Curr Biol. 2006;16:345–58. doi: 10.1016/j.cub.2005.12.044. [DOI] [PubMed] [Google Scholar]
- 25.Cheng D, et al. Small molecule regulators of protein arginine methyltransferases. J Biol Chem. 2004;279:23892–9. doi: 10.1074/jbc.M401853200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.