Abstract
Arabidopsis seedling development is controlled by many regulatory genes involved in multiple signaling pathways. The functional relationships of these genes working in multiple signaling cascades have started to be unraveled. Arabidopsis HY1/HO1 is a rate-limiting enzyme involved in biosynthesis of phytochrome chromophore. HY5 (a bZIP protein) promotes photomorphogenesis, however ZBF1/MYC2 (a bHLH protein) works as a negative regulator of photomorphogenic growth and light regulated gene expression. Further, MYC2 and HY1 have been shown to play important roles in jasmonic acid (JA) signaling pathways. Here, we show the genetic interactions of HY1 with two key transcription factor genes of light signaling, HY5 and MYC2, in Arabidopsis seedling development. Our studies reveal that although HY1 acts in an additive manner with HY5, it is epistatic to MYC2 in light-mediated seedling growth and gene expression. This study further demonstrates that HY1 additively or synergistically functions with HY5, however it works upstream to MYC2 in JA signaling pathways. Taken together, this study demonstrates the functional interrelations of HY1, MYC2 and HY5 in light and JA signaling pathways.
Background
Light is one of the most important environmental factors for plant growth and development throughout its life cycle [1,2]. Plants have evolved with multiple photoreceptor-systems to monitor the surrounding light quality, quantity, and direction. In Arabidopsis, these photoreceptors include the blue/UV-A light absorbing cryptochromes (CRY1 to CRY3) and phototropins (PHOT1 and PHOT2); the red/far-red light absorbing phytochromes (phy: phyA to phyE) [3-7]. Arabidopsis phytochromes form homo and hetero dimers with each other [8-10]. Formation of such heteromeric photoreceptors increases the potential complexity of R/FR light sensing and signaling mechanism in plants. Similarly, light induced activation of cryptochromes leads to possible autophosphorylation and dimerization [11]. Moreover, phytochromes and cryptochromes work together either by interaction with each other in a light-dependent and interdependent manner [12,13].
Arabidopsis seedlings exhibit two distinct developmental patterns, photomorphogenesis or skotomorphogenesis depending on the presence or absence of light, respectively [13-15]. Skotomorphogenesis is the strategy followed under dark conditions where Arabidopsis seedlings exhibit elongated hypocotyl, closed cotyledons with apical hooks; whereas in presence of light, photomorphogenesis is initiated, characterized by short hypocotyl with fully developed cotyledons. This developmental change from skotomorphogenesis to photomorphogenesis is carried out by different classes of photoreceptors, and characterised by a change in the expression of about one-third of genes in the Arabidopsis genome [16-18].
Genetic screen of Arabidopsis seedlings for developmental defects under light conditions have led to the identification of several transcription factors that either act as a positive or negative regulator downstream to specific photoreceptor or a set of photoreceptors [19-27]. Recently, a DNA-ligand binding screen has led to the identification of three Z-box binding factors, ZBF1/MYC2, ZBF2/GBF1 and ZBF3/CAM7 [28-33]. MYC2 is a bHLH transcription factor that acts downstream to cry1 and cry2 photoreceptors, and negatively regulates blue light-mediated photomorphogenic growth and blue and far red-light regulated gene expression [29]. MYC2 also functions as a transcriptional regulator for ABA and JA signaling pathways [29,34-37].
HY5 is one of the first known and most extensively studied bZIP transcription factor involved in promoting photomorphogenesis. Arabidopsis seedlings mutant for HY5 exhibit elongated hypocotyl under various wavelengths of light, suggesting that functionally HY5 is downstream to multiple photoreceptors [19,38-40]. Further, hy5 mutant seedlings exhibit defects in root growth and reduction in chlorophyll and anthocyanin accumulation [19,41]. In addition, studies have shown the involvement of HY5 in both auxin and cytokinin signaling pathways [42-44], suggesting that HY5 might be a common intermediate in light and hormone signaling pathways. The chromatin immunoprecipitation (CHIP) assays have revealed that HY5 preferentially binds to more than 3000 chromosomal sites that were distributed in all the five chromosomes [45].
Arabidopsis HY1 encodes heme oxygenase (HO) that catalyses the committed step in the conversion of heme to biliveridin IXα (BV), which is further converted to photochromobilin through sequential steps and exported to cytoplasm where it binds to the newly synthesized apo-phys by an autocatalytic process to form functional holo-phytochrome. The HOs are encoded by a small gene family that includes HY1, HO2, HO3 and HO4. Among all the four members of the HO family HY1 is highly expressed in almost all the tissues and plays a major role in synthesis of holo-phytochrome [46]. Seedlings mutant for HY1 exhibits elongated hypocotyl in red and far red light, and display defects in root development. Further, the light inducible genes such as CAB, RBCS and CHS are under-expressed in hy1 mutant background [47-49]. Recently, it has been reported that seedlings mutant for HY1 show elevated levels of JA and expression of JA-inducible defense genes [50].
In this study, in order to identify genes that might be working parallel to HY5, a genetic screen was set up using hy5-ks50 mutant lines through EMS mutagenesis. Gene cloning and genetic complementation analysis revealed that one of these mutants (enhancer of HY5: ehy5) contains a mutation in the HY1gene. We have investigated the interrelations of HY1 with two transcription factors, HY5 and MYC2, with respect to light-controlled Arabidopsis seedling development and JA responsiveness.
Results
Mutations in EHY5 modulate HY5-controlled hypocotyl elongation
HY5 is a key transcription factor in light signaling pathways that promote photomorphogenesis under a broad spectrum of light [32,40]. Although the hy5 mutant seedlings display elongated hypocotyl in light, the seedlings are not completely etiolated similar to dark grown seedlings. Therefore, there might be additional factors present that are involved in the promotion of photomorphogenesis under various wavelengths of light [39]. Recent studies have shown that CAM7/ZBF3 works in various wavelengths of light to promote photomorphogenic growth and light regulated gene expression. Further, HY5 and CAM7 work synergistically or additively in the promotion of photomorphogenesis [32].
In order to find additional factors that promote photomorphogenesis in concert with HY5, an extragenic enhancer screen was set up using hy5 mutant lines (hy5-ks50 mutant; 19) through EMS mutagenesis. Several double mutant lines that showed enhanced hypocotyl growth as compared to that of hy5 mutants were identified. One such mutant line, hy5 ehy5 (ehy5: enhancer of HY5) double mutant, was selected for further study. The segregated ehy5 line (obtained in F2 population from a back cross with wild type (Ws) was repeatedly backcrossed with wild type (Ws) to purify the mutation from any other back ground mutations.
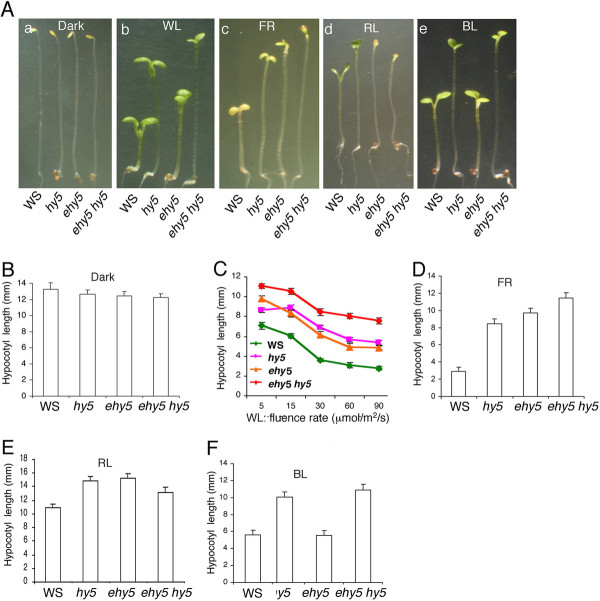
The examination of seedling morphology revealed that neither ehy5 alone nor ehy5 hy5 double mutants exhibited any altered morphology in the dark (Figure 1A (a) and 1B). The characteristic long hypocotyl phenotype of hy5 in WL (white light) irradiation was further enhanced in ehy5 hy5 double mutants, exhibiting a super tall phenotype under various fluences of WL (Figure 1A (b) and 1C). To determine whether this reduced sensitivity of ehy5 hy5 phenotype is specific to a particular wavelength of light, the growth of 6-day-old ehy5 hy5 double mutant seedlings was tested in various wavelengths of light. As shown in Figure 1A (c) and 1D, ehy5 hy5 double mutants displayed further reduced sensitivity to far-red light (FR) as compared to ehy5 and hy5 single mutants, suggesting that EHY5 and HY5 additively control the hypocotyl growth in FR. On the other hand, hypocotyl length of ehy5 hy5 double mutants was found to be closer to either of the single mutants in red light (RL), suggesting that EHY5 and HY5 are likely to work in the same branched pathways in controlling the hypocotyl length in RL (Figure 1A (d) and 1E). The ehy5 mutants exhibited similar hypocotyl length to that of wild type in blue light (BL), and the hypocotyl length of ehy5 hy5 double mutants was similar to that of hy5 single mutants, suggesting that additional mutation in EHY5 does not affect the hy5 phenotype in BL (Figure 1A (e) and 1F).
Figure 1.
The ehy5 mutants display elongated hypocotyl. A, Phenotype of segregated wild-type (WS), ehy5, hy5, and ehy5 hy5 double mutants in dark and different light conditions. Six-day old constant dark, WL (90 μmol m-2 s-1), FR (90 μmol m-2 s-1), RL (90 μmol m-2 s-1) and BL (45 μmol m-2 s-1) grown seedlings (a to e). B-C, Quantification of hypocotyl length of 6-day-old seedlings grown in constant dark and various fluences of WL, respectively. D-F, Quantification of hypocotyl length of 6-day-old constant FR (90 μmol m-2 s-1), RL (90 μmol m-2 s-1) and BL (45 μmol m-2 s-1) grown seedlings, respectively. The error bar indicates standard deviation (SD). The experiment was repeated more than twice and similar results were obtained each time. A representative result is presented. For measuring hypocotyl length, ~30 seedlings were used in each genotype.
Map based cloning reveals that EHY5 encodes HY1
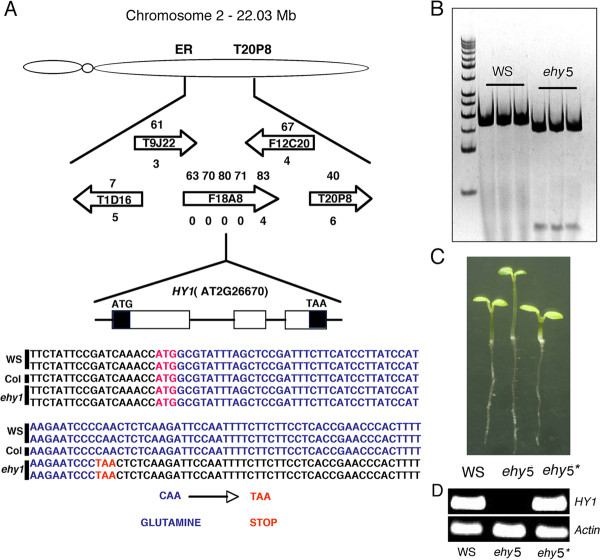
To determine the genetic basis of EHY5 mutation, we followed map-based cloning strategy. The ehy5 mutants (WS) were genetically crossed to wild type (Col), and the resulting F1 progeny showed wild type phenotype. F1 plants were self-pollinated and since the ehy5 long hypocotyl phenotype is easy to score at the seedling stage, the EHY5 locus has served as a useful landmark for classical mapping. For fine mapping, the segregating F2 populations with the ehy5 phenotype were used for mapping with Simple Sequence Length Polymorphism (SSLP) and Cleaved and Amplified Polymorphic Sequence (CAPS) markers that we developed during this study and also that are available in the database at the Arabidopsis Information Resource (TAIR). Initially, the target locus was mapped between the markers ER and T20P8 on Chromosome 2 (Figure 2A). Further fine mapping with seven genetic markers delimited the target gene to a 20-Kb region on the F18A8 BAC clone. To further identify the exact position of the EMS mutation, we have sequenced the genomic DNA fragment of the 20-Kb region from the ehy5 background and compared with that of wild type (WS) genomic DNA sequence, which revealed that a single C to T nucleotide substitution in the first exon of the HY1 (AT2G26670) DNA leads to the conversion of Glutamine (CAA) to stop codon (TAA), resulting in the premature termination of the protein translation (Figure 2A). This EMS induced substitution in HY1 first exon introduces a DdeI recognition site adjacent to the mutation region. We developed a dCAPS marker to confirm the mutation in ehy5 (Figure 2B).
Figure 2.
Positional cloning and molecular identification of EHY5. A, Map-based cloning. The genetic locus of ehy5 mutation was first mapped between markers ER and T20P8 on chromosome 2. Fine mapping using genetic markers designed from BAC clones. The direction of the BAC clone is indicated by the arrow. The numbers above and below the arrow indicate the marker number and the corresponding recombinants for the respective marker. Sequence of the genomic DNA fragment from wild-type (WS) and ehy5 mutant plants and comparison with wild-type (Col) genomic DNA sequence indicate C to T mutation. B, DNA polymorphism between ehy5 and wild-type (WS) plants. The C to T mutation in ehy5 genomic DNA adds a DdeI recognition site. The DNA fragments flanking the DdeI site were amplified from the wild-type and ehy5 plants, digested with DdeI, and separated on native PAGE. C, Genetic complementation of ehy5. Phenotypes of 6-day-old wild-type (WS), ehy5 and ehy5* (HY1 complemented) are shown. D, RT- PCR results show the expression of HY1 in wild-type (WS), ehy5 and ehy5*. Actin bands show the loading control. The RT-PCR experiment was repeated thrice and a representative result is shown.
As a final step to establish that the EHY5 locus encodes HY1 transcript, we tested whether a wild type genomic fragment containing the entire HY1 gene could complement ehy5. Fragment containing entire HY1 coding region with its native promoter was introduced into ehy5mutant background. As shown in Figure 2C, ehy5 seedlings transformed with full length HY1 genomic DNA fragment exhibited wild-type phenotype. The positive transformants were confirmed by RT-PCR (Figure 2D). These results indicate that the ehy5 mutant is an allele of hy1 mutant, and henceforth we refer to ehy5 as hy1.
HY1 and HY5 additively regulate the expression of light regulated genes and accumulation of chlorophyll and anthocyanin during early seedling development
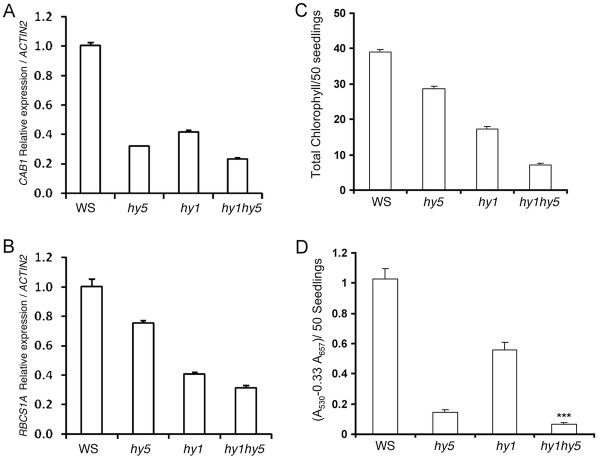
The loss-of-function mutants of HY5 display partial photomorphogenic growth at various wavelengths of light with reduced expression of light-regulated genes such as CAB1 and RBCS-1A. Similarly, hy1 also shows reduced accumulation of CAB and RBCS transcripts [51]. To examine how HY5 and HY1 genetically interact to regulate the expression of light inducible genes, we monitored the expression of CAB1 and RBCS-1Aby real time PCR. As shown in Figure 3A-B, the expression of CAB1 and RBCS-1A was reduced in both hy1 and hy5 single mutants as compare to wild-type, and the accumulation of transcript was further reduced in hy1 hy5 double mutants compared to either of the single mutants. These results indicate that HY1 and HY5 act in an additive manner to regulate the expression of CAB1 and RBCS-1A genes.
Figure 3.
HY1 and HY5 additively regulate the light-induced gene expression. A - B, Relative expression of CAB1and RBCS-1A in 6-day-old seedlings grown in WL (90 μmol m-2 s-1). C, Accumulation of chlorophyll in 6-day-old wild-type and mutant seedlings grown in WL (90 μmol m-2 s-1). D, Accumulation of anthocyanin in 6-day-old wild-type and mutant seedlings grown in WL (90 μmol m-2 s-1). The error bars indicate SD. *** - indicates significant difference from hy5 (p > 0.001 student's t-test, n = 30, number of seedlings used for hypocotyl measurement). Real-time PCR was repeated more than thrice and in each biological experiment three technical replicates were used. Similar results were obtained in all the experiments. A representative figure is shown here. For chlorophyll and anthocyanin estimation, 50 seedlings was used in each genotype and the experiment was repeated thrice and in each biological experiment, four technical replicated were used. Similar results were obtained in all the experiments. A representative figure is presented.
Earlier studies have shown that hy5 and hy1 mutant seedlings display reduction in the accumulation of chlorophyll and anthocyanin. To determine the genetic interaction of HY1 and HY5 for chlorophyll and anthocyanin accumulation, chlorophyll and anthocyanin contents were estimated from six-day-old WL grown seedlings. As shown in Figure 3C and 3D, the hy1 hy5 double mutants showed less accumulation of chlorophyll and anthocyanin as compared to that of hy1 and hy5 single mutants, suggesting that HY1 and HY5 act in an additive manner to control the accumulation of chlorophyll and anthocyanin in WL.
HY1 and HY5 work in an additive or synergistic manner to control JA responsiveness
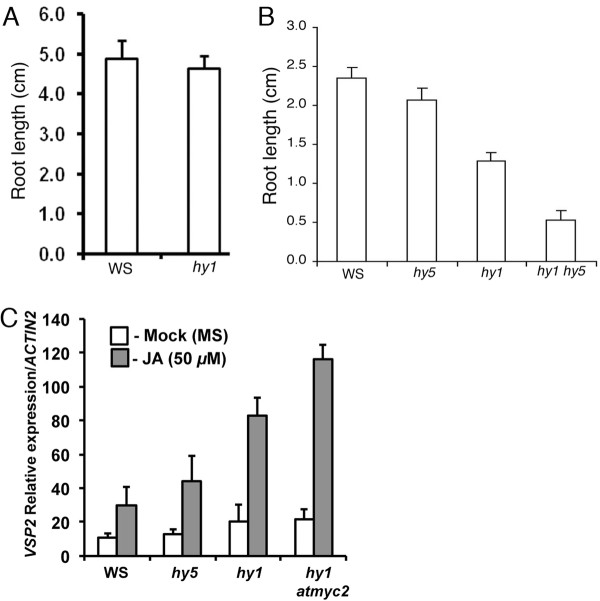
In the presence of jasmonic acid (JA), hy1 mutants have stunted root growth and expression of JA-inducible defence genes [50]. We asked whether mutation in HY5 can modulate the JA sensitiveness of hy1 mutants. To examine that, we grew the seedlings in the presence or absence of JA and examined the root growth. Although very little difference, if any, was observed between the wild type and hy1 mutants in the absence of JA (Figure 4A), 15 μM JA caused root growth retardation in hy1 mutant seedlings as compared to the wild type (Figure 4B). The effect was more severe in hy1 hy5 double mutants (Figure 4B). These results suggest a synergistic function of HY1 and HY5 in JA-mediated root growth. To determine whether the expression of JA regulated genes is affected in hy1 hy5 double mutants, the transcript accumulation of JA-responsive marker gene VSP2 was determined [35,50]. The real time PCR analyses had shown that JA treatment induced the expression of VSP2 both in hy1 and hy5 mutants, and the level of expression was further increased in hy1 hy5 background (Figure 4C). These results indicate that HY1 and HY5 function in an additive manner to regulate the expression of VSP2 in response to JA.
Figure 4.
JA responsiveness of hy1 hy5 double mutants. A, Quantification of root length of 16-day-old wild-type and hy1 mutant plants grown in constant WL (90 μmol m-2 s-1) without hormone (JA). B, Quantification of root length of A. C, Relative induction of VSP2 expression by JA in wild-type and mutant plants. Six-day-old wild-type and mutant seedlings were treated with MS (Mock) or with JA (50 μM) for 5-hours and total RNA was isolated from 100 mg of tissue and used for quantitative real-time PCR analysis. ACTIN2 was used as internal control. Approximately 25 to 30 seedlings were used for the root growth measurement. The error bars indicate standard error (SE) of three biological replicates.
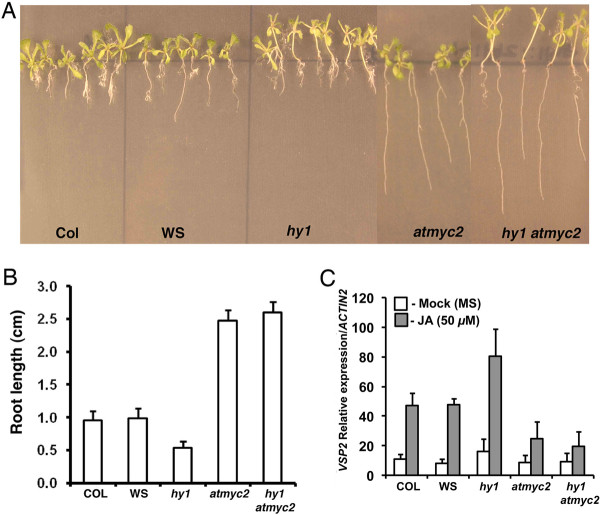
Additional mutation in MYC2 abolishes the hyper-sensitive responses of hy1 to JA
MYC2, a bHLH transcription factor, acts as a negative regulator of blue light mediated photomorphogenic growth and cross talks with JA and ABA signaling pathways [29,33,36,37,52,53]. MYC2 positively regulates the expression of JA-responsive genes such as VSP2 by directly binding to the G-box motif present in the promoter of VSP2 [35,36]. Furthermore, atmyc2 mutant seedlings are insensitive to JA-induced inhibition of root growth. To investigate the interaction between HY1 and MYC2 with respect to JA-responses, we constructed hy1 atmyc2 double mutants through genetic crosses. The root growth of hy1 atmyc2 double mutant plants was monitored in the absence or presence of 15 μM JA. No significant difference in root length was observed among the mutants and wild type in the absence of JA. JA caused severe root growth retardation in wild-type and hy1 mutants, however the effect was drastically reduced in atmyc2 and hy1 atmyc2 mutant plants (Figure 5A and 5B). These results indicate that MYC2 works downstream to HY1 in JA-mediated inhibition of root growth. We then examined the expression of one of the JA-inducible marker genes VSP2 by real time PCR in various mutant backgrounds. As shown in Figure 5C, whereas there was very little expression of VSP2 in the absence of JA, the expression of VSP2 was increased in the presence of JA in wild-type and hy1 mutant plants. Further, the hy1 mutants showed significantly higher level of accumulation of VSP2 transcript as compared to wild-type background. However, the expression of VSP2 was less in atmyc2 plants, as expected from its less sensitiveness to JA, and was found to be similar to hy1 atmyc2 double mutants. These results suggest that MYC2 works downstream to HY1 in JA-induced expression of VSP2 gene.
Figure 5.
JA responsiveness of hy1 atmyc2 double mutants. A, Root growth of 16-day-old wild-type and various mutant plants grown in constant WL (90 μmol m-2 s-1) in presence of 15 μM JA. B, Quantification of root length of A. C, Relative induction of VSP2 expression by JA in wild-type and mutant plants. For experimental detail, see legend to Figure 4C.
Overlapping functions of HY1 and MYC2 in Arabidopsis seedling development
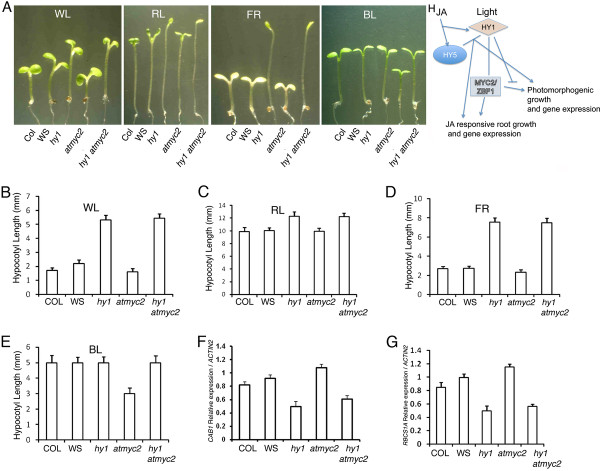
The atmyc2 mutants display hypersensitive response to BL, and are epistatic to cry1 and cry2 [29]. In order to determine how these two light signaling components, HY1 and MYC2, genetically interact to control early seedling development, we measured the hypocotyl length of atmyc2 hy1 double mutants in various light conditions. Similar to hy1 or atmyc2 single mutants, hy1 atmyc2 double mutants did not show any altered growth in the dark. However, under WL conditions, hy1 atmyc2 double mutants displayed hypocotyl length similar to hy1 single mutants (Figure 6A and 6B). Furthermore, as shown in Figure 6A and 6C to 6E, hy1 atmyc2 double mutants displayed hypocotyl length similar to hy1 single mutants in RL, FR and BL conditions. These results indicate that although additional mutation in MYC2 does not affect the phenotype of hy1 mutants in RL and FR, it is able to suppress the atmyc2 phenotype in BL. MYC2 acts as a negative regulator of light induced gene expression such as CAB1 and RBCS-1A. We examined how the additional mutation in MYC2 affects the expression of light-inducible genes in hy1 mutant background. The real time PCR analysis revealed that the expression of CAB1 and RCBS-1A was similar to that of hy1 single mutant in hy1 atmyc2 background (Figure 6F and 6G). These results suggest that HY1 is epistatic to MYC2 in controlling the light induced gene expression.
Figure 6.
Light-mediated seedling development of hy1 atmyc2. A, Phenotype of wild-type and various mutant seedlings in different light conditions. Six-day-old constant WL (90 μmol m-2 s-1), RL (90 μmol m-2 s-1), FR (90 μmol m-2 s-1) and BL (45 μmol m-2 s-1) grown seedlings. B-E, Quantification of hypocotyl length of 6-day-old constant WL (90 μmol m-2 s-1), RL (90 μmol m-2 s-1), FR (90 μmol m-2 s-1) and BL (45 μmol m-2 s-1) grown seedlings, respectively. F-G, The relative expression of CAB1 and RBCS-1A in 6-day-old seedlings grown in WL (90 μmol m-2 s-1). The error bars indicate SD. Approximately 25 to 30 seedlings were used for hypocotyl length measurement. For gene expression studies, total RNA was isolated from 100 mg of tissue was used for cDNA preparation. The real-time PCR experiments were repeated more than twice and three technical replicates were used for each genotype. Similar results were obtained in all the experiments. A representative graph is shown. H, Working model shows the role of HY1, HY5 and MYC2 in photomorphogenesis and JA responsiveness. HY1 and HY5 act additively in response to JA and light signaling pathways. MYC2 acts downstream to HY1 in JA responsiveness, and HY1 acts negatively to MYC2-mediated BL specific photomorphogenic growth.
Discussion
Although many components of light signaling pathways are known, the interconnections of these components in Arabidopsis seedling development is unclear. Moreover, very little information is available on cross talks of various components of light signaling with other signaling cascades and vice versa. In this study, we have demonstrated the genetic interactions of HY1 with two other light-signaling components, HY5 and MYC2, which belong to two important families of transcription factors, bZIP and bHLH, respectively, in Arabidopsis seedling development. Furthermore, this study reveals that HY1, HY5 and MYC2 are functionally connected in JA signaling pathways.
An attempt to identify new genes that might enhance hy5 phenotype, similar to CAM7/ZBF3 led to the identification of EHY5 [32]. Map based cloning and genetic complementation of ehy5 mutants reveal that EHY5 codes for HY1 (HO1), a rate-limiting enzyme that catalyzes the conversion of heme to biliverdin IXα (BV) in the chromophore biosynthesis pathway [54]. Phenotypic analyses under various light conditions have revealed that HY1 and HY5 function in an additive manner resulting in a super tall phenotype in WL. Similar additive function of HY5 and HY1 was also observed in the regulation of hypocotyl growth in FR. Genetic interaction studies between HY1 and HY5 reveal that they are likely to work in the same branched pathways of light signaling. On the other hand, mutations in HY1 does not affect the hy5 phenotype in BL. However, the additional mutation in HY1 is able to suppress the atmyc2 phenotype in BL. These results strongly suggest the wavelength specific interdependent functions of HY1, HY5 and MYC2 in the regulation of hypocotyl growth in Arabidopsis seedling development.
The expression of light regulated genes is down-regulated in hy1 mutant background. HY5 directly binds to the G-box present in the promoters of light regulated genes and promote their expression [39]. MYC2/ZBF1 also interacts with the Z-/G-box LRE present in the light-inducible promoters such as CAB1 and RBCS1A, however down-regulates their expression [29,33,45]. Analysis of light-regulated gene expression in hy1 hy5 double mutants reveal that HY1 and HY5 function in an additive manner and elevate the expression of light regulated genes. These two proteins also function in an additive manner to regulate the accumulation of chlorophyll and anthocyanin. On the other hand, the expression of CAB1 and RBCS-1A in hy1 atmyc2 double mutant seedlings was similar to that of hy1 single mutants, and thus suggesting that HY1 works downstream to MYC2 in the regulation of CAB1 and RBCS-1A expression. It has been shown earlier that although atmyc2 works downstream to cry1 and cry2 photoreceptors, phyA is epistatic to atmyc2 in BL [29].
Plant growth and development is a complex phenomenon, which is likely to be regulated through interactions between light and phytohormone signaling pathways. Recent studies have shown that signals from light and multiple hormonal signaling pathways cross talk through common downstream regulatory proteins such as MYC2 and HY5 [29,34-36,42-44,52,55]. For example, seedlings mutant for HY5 show altered balance of auxin and cytokinin signaling and also has decreased expression of two negative regulators of auxin signaling pathways such as AXR2/IAA7 and SLR/IAA14. The functional overlap of light and JA signaling in defence, wound and shade response has been reported [56,57]. MYC2 regulates JA responses via differential regulation of an intermediate spectrum of transcription factors with activating or repressing roles. Furthermore, a JA activated MKK3-MPK6 pathway negatively regulates the expression of MYC2 [53]. It has been shown that phytochorme deficient hy1 mutant seedlings overproduce JA and also display constant expression of JA inducible defense related genes such VSP1. The possible reason may be that there is reduction in the total photoactive phytochrome pool in the hy1 mutant background and thereby resulting an altered light sensitivity. This may lead to photo-oxidative stress resulting in upregulation of JA synthesis in hy1 mutants [50]. The cross talks among multiple signaling pathways occur at the level of intermediate components of the signaling pathways rather than at the receptor level. For example, cross talks between light and JA signaling is mediated by the transcription factor (intermediate component) MYC2/ZBF1. MYC2/ZBF1 works in cryptochrome mediated blue light signaling pathways [29], however cry1/cry2 mutants do not have altered JA responses.
It is worth mentioning here that it has earlier been reported that hy1 mutants display shorter roots than wild type plants [50]. However, this study does not find such difference in the absence of JA. The apparent discrepancy may be attributed to the developmental stages the observations were made. Whereas Zhai et al., 2007 found the difference at the early seedling stage, this study demonstrates the results of 16-day-old young adult plants, where the altered hypocotyl length was fairly maintained. In this study, our results demonstrate that HY5 and HY1 act additively or synergistically to regulate the JA-induced root-growth-inhibition and expression of JA-responsive genes. Although hy5 mutants do not show altered root growth in JA, the JA inducible gene VSP2 was upregulated in hy5 mutants in the presence of JA. These results indicate a negative regulatory role of HY5 in JA-mediated regulation of VSP2. On the other hand, MYC2 which acts as a negative regulator of light signaling, acts as a positive regulator of JA-mediated VSP2 expression Figure 5; [35]. Thus, both these transcription factors work in an opposite manner in light and JA signaling pathways.
Conclusions
This study demonstrates an overlapping function of HY1 with two important transcription factors of light signaling, HY5 and MYC2, in light and JA signaling pathways. The findings in this work will help to better understand the light signaling in Arabidopsis, and the cross talk of light and JA signaling pathways.
Methods
Plant materials, growth conditions and generation of double mutants
Arabidopsis (Arabidopsis thaliana) seeds were surface sterilized and sown on Murashige and Skoog plates, then kept at 4°C in darkness for 3 to 5 days, and transferred to specific light conditions at 22°C. The intensities of WL and various colour lights (in the light-emitting diode chamber, Q-Beam 3200-A; (Quantum Devices)) used were described in Yadav et al. (2002). For the generation of double mutants such as hy1 atmyc2, homozygous hy1 (WS) mutant plants were genetically crossed with atmyc2-1 (Col-0) homozygous mutant lines. In the F2 generation, seedlings were grown in WL (90 μmol m-2 s-1) for the identification of hy1 homozygous lines, and long hypocotyl hy1 mutants were selected and transferred to soil. To determine the genotype of AtMYC2 locus, about 40 seedlings from each line were tested by genomic PCR. F3 progenies that were homozygous for atmyc2 mutant plants were further examined by RT-PCR and considered as hy1 atmyc2 double mutants. For measurement of hypocotyl length, ~30 seedlings were used in each genotype. The hypocotyl length measurement was repeated more than twice with similar results.
Mutant screen and map-based cloning
Ethyl methanesulfonate (EMS)-mutagenized, hy5KS50 M2 seeds of Arabidopsis thaliana ecotype Wassilewskija (Ws) were grown on MS media under WL conditions and hypocotyl length was compared with that of wild type and hy5KS50 mutant lines. Seedlings that showed enhanced and elongated hypocotyl length (as compared to hy5KS50) under all light conditions tested were selected and used for further studies. To identify the genetic basis of the EMS mutation in ehy5, we isolated the new ehy5 mutant, from the hy5KS50 ehy5 double mutant background. The double mutant plants were back-crossed successively to wild-type (WS) and the segregated ehy5 mutant seedlings in the F2 generation was selected and used for further back-crosses with wild type (WS) for four generations (to purify the background mutations in the EMS treated hy5-ks50 mutant plants) before physiological and genetic analysis. The ehy5 mutant was out-crossed with Wild-type (Col) ecotype, and the mapping population was selected from F2 generation. A total of 823 individual F2 plants showing the ehy5 phenotype were selected for genetic mapping. Genomic DNA was prepared using the protocol described by Edwards et al. (1991). Cleaved amplified polymorphic sequence (CAPS) and simple sequence length polymorphism (SSLP) markers between Col and Ws were used for mapping EHY5. For genetic complementation analysis, a genomic fragment containing the entire HY1/At2g26670 coding sequence along with its promoter was amplified from the wild-type (Ws) by PCR and the PCR product was digested with HindIII and SmaI and inserted into same sites of modified pBI121 binary vector. The construct obtained was then introduced into ehy5 (hy1) mutant plants using A. tumefaciens-mediated transformation. Transformants were selected based on their resistance to kanamycin.
Root growth measurement
Seeds were on MS media in vertical square plates and stratified at 4°C in dark conditions for 4 days to induce uniform germination. The plates were placed vertically in racks, and the seedlings were grown under constant white light conditions (90 μmol m-2 s-1) for 16 days. The root length of wild type, single and double mutants was measured. Approximately 25 to 30 seedlings were used for the root length measurement. The experiments were repeated for three times with similar results.
Root growth response to methyl jasmonate
Seeds of wild type and mutant plants were plated on MS with 15 μM of methyl jasmonate (Sigma) in square plates, after four days of stratification in cold (4°C), plates were placed vertically in racks, and the seedlings were grown under constant white light conditions (90μmol m-2 s-1) for 16 days. The root length of wild type, single and double mutants was measured. For determining the VSP2 expression, six-day-old white-light grown seedlings were mock (only MS solution) or JA treated (50 μM JA in MS solution) for 5 hour. After the time period, seedlings were washed with sterile milliQ, excess water was removed with the tissue paper and the tissue was harvested and snap freeze in liquid nitrogen and total RNA was extracted from 100 mg of tissue, using the RNeasy plant mini kit (Quaigen), and cDNA were synthesized from total RNA using titan one-tube RT-PCR system (Roche Applied Science) following the manufacturer's instructions. Real-time PCR analysis of gene expression was carried out by using LightCycler-FastStart DNA Master-PLUS SYBR Green (Roche Applied Science) and was performed using StepOne Real-Time PCR system (ABI). CT values of VSP2 were normalized, relative to that of ACTIN2 (Internal control).
The following primers were used for the experiment
VSP2-FP: 5' GGCCTTGCATCTTTACCAAAAC 3'
VSP2-RP: 5' GTAGTAGAGTGGATTTGGGAGC 3'
ACTIN2-FP: 5' AAAGGCTTAAAAAGCTGGGG 3'
ACTIN2-RP: 5' GGGACTAAAACGCAAAACGA 3'
Real-time PCR analysis
Total RNA was extracted from 100 mg of tissue, using the RNeasy plant mini kit (Quaigen), according to manufacturer's protocol. RT-AMV reverse transcriptase (Roche Applied Science) was used for both semi-quantitative RT-PCR and cDNA synthesis. Real-time PCR analysis of gene expression was carried out by using LightCycler-FastStart DNA Master-PLUS SYBR Green (Roche Applied Science) and was performed using Step-one Real-Time PCR system (ABI). CT values of CAB1 and RBCS1A were normalised, relative to that of ACTIN2 (Internal control). Real-time PCR was repeated more than thrice and in each biological experiment three technical replicates were used.
The following primers were used for the experiment
HY1-FP: 5' GTGTATCCCTCTTCTCTATTCC 3'
HY1-RP: 5' TCTGAATCCTAGGTCGAGG 3'
CAB1- FP: 5' GTTAACAACAACGCATGGC 3'
CAB1-RP: 5' CCTCTCACACTCACGAAGCA 3'
RBCS1A-FP: 5' TCGGATTCTCAACTGTCTGATG 3'
RBCS1A-RP: 5' ATTTGTAGCCGCATTGTCCT 3'
ACTIN2-FP: 5' TGATGCACTTGTGTGTGACAA 3'
ACTIN2-RP: 5' GGGACTAAAACGCAAAACGA 3'
Chlorophyll and anthocyanin measurements
Chlorophyll and anthocyanin contents were measured following essentially the same protocols as described in [41]. For chlorophyll and anthocyanin estimation, 50 seedlings were used in each genotype and the experiment was repeated thrice; and in each biological experiment, four technical replicated were used.
Authors' contributions
VBRP was involved in map-based cloning of EHY1/HY1, generation of double mutants, phenotypic characterization, JA responsiveness and gene expression study. VSK carried out the hy5 enhancer screen and identified and partly characterize the ehy5 mutant. AN helped in the map-based cloning and participated in the design of the manuscript. SC conceived of the study, participated in its design and coordination, and helped to draft the manuscript. All authors read and approved the final manuscript.
Contributor Information
Babu Rajendra V Prasad, Email: prasad.pratima@gmail.com.
Selva V Kumar, Email: sclerotinia@gmail.com.
Ashis Nandi, Email: ashis_nandi@yahoo.com.
Sudip Chattopadhyay, Email: sudipchatto@yahoo.com.
Acknowledgements
This work is financially supported by Department of Biotechnology, Government of India to S.C and A.N.; V.B.R.P is a recipient of UGC fellowship, Government of India.
References
- Smith H. Phytochromes and light signal perception by plants-an emerging synthesis. Nature. 2002;407:585–591. doi: 10.1038/35036500. [DOI] [PubMed] [Google Scholar]
- Sullivan JA, Deng XW. From seed to seed: The role of photoreceptors in Arabidopsi development. Dev Biol. 2003;260:289–297. doi: 10.1016/S0012-1606(03)00212-4. [DOI] [PubMed] [Google Scholar]
- Cashmore AR, Jarillo JA, Wu YJ, Liu D. Cryptochromes: Blue light receptors for plants and animals. Science. 1999;284:760–765. doi: 10.1126/science.284.5415.760. [DOI] [PubMed] [Google Scholar]
- Briggs WR, Christie JM. Phototropins1 and 2: versatile plant blue light receptors. Trends Plant Sci. 2002;7:204–210. doi: 10.1016/S1360-1385(02)02245-8. [DOI] [PubMed] [Google Scholar]
- Quail PH. Phytochrome photosensory signalling networks. Nat Rev Mol Cell Biol. 2002;3:85–93. doi: 10.1038/nrm728. [DOI] [PubMed] [Google Scholar]
- Lin C, Shalitin D. Cryptochrome structure and signal transduction. Annu Rev Plant Biol. 2003;2003(54):469–496. doi: 10.1146/annurev.arplant.54.110901.160901. [DOI] [PubMed] [Google Scholar]
- Li QH, Yang HQ. Cryptochrome signaling in plants. Photochem Photobiol. 2007;83:94–101. doi: 10.1562/2006-02-28-IR-826. [DOI] [PubMed] [Google Scholar]
- Wagner D, Koloszvari M, Quail PH. Two small spatially distinct regions of phytochrome B are required for efficient signaling rates. Plant Cell. 1996;8:859–719. doi: 10.1105/tpc.8.5.859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JI, Bhoo SH, Han YJ, Zarate X, Furuya M, Song PS. The PAS2 domain is required for dimerization of phytochromeA. J Photochem Photobiol. 2006;178:115–121. doi: 10.1016/j.jphotochem.2005.10.028. [DOI] [Google Scholar]
- Ted C, Ahmed S, Matt M, Peng L, Michael F, Robert AS. Obligate Heterodimerization of Arabidopsi Phytochromes C and E and interaction with the PIF3 Basic Helix-Loop - Helix Transcription Factor. Plant Cell. 2009;21:786–799. doi: 10.1105/tpc.108.065227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sang Y, Li QH, Rubio V, Zhang YC, Mao J, Deng XW, Yang HQ. N-terminal domain-mediated homodimerization is required for photoreceptor activity of Arabidopsi CRYPTOCHROME 1. Plant Cell. 2005;17:1569–1584. doi: 10.1105/tpc.104.029645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo H, Yang H, Mockler TC, Lin C. Regulation of flowering time by Arabidopsi photoreceptors. Science. 1998;279:1360–1363. doi: 10.1126/science.279.5355.1360. [DOI] [PubMed] [Google Scholar]
- Mas P, Devlin PF, Panda S, Kay SA. Functional interaction of phytochrome B and cryptochrome 2. Nature. 2000;408:207–211. doi: 10.1038/35041583. [DOI] [PubMed] [Google Scholar]
- Nagy F, Schafer E. Phytochromes control photomorphogenesis by differentially regulated, interacting signaling pathways in higher plants. Annu Rev Plant Biol. 2002;53:329–355. doi: 10.1146/annurev.arplant.53.100301.135302. [DOI] [PubMed] [Google Scholar]
- Chen M, Chory J, Frankhauser C. Light signal transduction in higher Plants. Ann Rev Genet. 2004;38:87–117. doi: 10.1146/annurev.genet.38.072902.092259. [DOI] [PubMed] [Google Scholar]
- Ma L, Li J, Qu L, Hager J, Chen Z, Zhao H, Deng XW. Light control of Arabidopsi development entails coordinated regulation of genome expression and cellular pathways. Plant Cell. 2001;13:2589–2607. doi: 10.1105/tpc.010229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tepperman JM, Zhu T, Chang HS, Wang X, Quail PH. Multiple transcription factors genes are early targets of phytochrome A signalling. Proc Natl Sci USA. 2001;98:9437–9442. doi: 10.1073/pnas.161300998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiao Y, Yang H, Ma L, Sun N, Yu H, Liu T, Gao Y, Gu H, Chen Z, Wada M. A Genome-Wide Analysis of Blue-Light Regulation of Arabidopsis Transcription Factor Gene Expression during Seedling Development. Plant Physiol. 2003;133:1480–1493. doi: 10.1104/pp.103.029439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oyama T, Shimura Y, Okada K. The Arabidopsi HY5 gene encodes a bZIP protein that regulates stimulus-induced development of root and hypocotyl. Genes Dev. 1997;1997(11):2983–2995. doi: 10.1101/gad.11.22.2983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hudson M, Ringli C, Boylan MT, Quail PH. The FAR1 locus encodes a novel nuclear protein specific to phytochrome A signaling. Genes Dev. 1999;13:2017–2027. doi: 10.1101/gad.13.15.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ballesteros ML, Bolle C, Lois LM, Moore JM, Vielle-Calzada JP, Grossniklaus U, Chua NH. LAF1, a MYB transcription activator for phytochrome A signaling. Genes Dev. 2001;15:2613–2625. doi: 10.1101/gad.915001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Deng XW. Arabidopsi FHY3 defines a key phytochrome A signaling component directly interacting with its homologous partner FAR1. EMBO J. 2002;21:1339–1349. doi: 10.1093/emboj/21.6.1339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim YM, Woo JC, Song PS, Soh MS. HFR1, a phytochrome A signaling component, acts in a separate pathway from HY5, downstream of COP1 in Arabidopsi. Plant J. 2002;30:711–719. doi: 10.1046/j.1365-313X.2002.01326.x. [DOI] [PubMed] [Google Scholar]
- Yang KY, Kim YM, Lee S, Song PS, Soh MS. Overexpression of a mutant basic helix-loop-helix protein HFR1, HFR1-N105, activates a branch pathway of light signaling in Arabidopsis. Plant Physiol. 2003;2003(133):1630–1642. doi: 10.1104/pp.103.029751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parks HD, Ok PL, Sik JK, Shik DC, Hyun HS, Gil HN. The Arabidopsi COG1 gene encodes a Dof domain transcription factor and negatively regulates phytochrome signaling. Plant J. 2003;34:161–171. doi: 10.1046/j.1365-313X.2003.01710.x. [DOI] [PubMed] [Google Scholar]
- Ward JM, Cufr AC, Denzel AM, Neff MM. The Dof Transcription factor OBP3 modulates phytochrome and cryptochrome signaling in Arabidopsi. Plant Cell. 2005;17:475–485. doi: 10.1105/tpc.104.027722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jang IC, Yang SW, Yang JY, Chua NH. Independent and interdependent functions of LAF1 and HFR1 in phytochrome A signaling. Genes Dev. 2007;21:2100–2111. doi: 10.1101/gad.1568207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yadav V, Kundu S, Chattopadhyay D, Negi P, Wei N, Deng XW, Chattopadhyay S. Light regulated modulation of Z-box containing promoters by photoreceptors and downstream regulatory components, COP1 and HY5, in Arabidopsi. Plant J. 2002;31:741–753. doi: 10.1046/j.1365-313X.2002.01395.x. [DOI] [PubMed] [Google Scholar]
- Yadav V, Mallappa C, Gangappa NS, Bhatia S, Chattopadhyay S. A basic Helix-loop-helix transcription factor in Arabidopsi, MYC2, acts as a repressor of blue light-mediated photomorphogenic growth. Plant Cell. 2005;17:1953–1966. doi: 10.1105/tpc.105.032060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mallappa C, Yadav V, Negi P, Chattopadhyay S. A basic leucine zipper transcription factor, G-box binding Factor 1, regulates blue light-mediated photomorphogenic growth in Arabidopsi. J Biol Chem. 2006;281:22190–22199. doi: 10.1074/jbc.M601172200. [DOI] [PubMed] [Google Scholar]
- Mallappa C, Singh A, Gangappa SN, Chattopadhyay S. GBF1, a Transcription Factor of Blue Light Signaling in Arabidopsis, is Degraded in the Dark by a Proteasome-mediated Pathway Independent of COP1 and SPA1. J Biol Chem. 2008;283:35772–35782. doi: 10.1074/jbc.M803437200. [DOI] [PubMed] [Google Scholar]
- Kushwaha R, Singh A, Chattopadhyay S. Calmodulin7 plays an important role as transcriptional regulator in Arabidopsi seedling development. Plant Cell. 2008;20:1747–1759. doi: 10.1105/tpc.107.057612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gangappa SN, Prasad VBR, Chattopadhyay S. Functional Interconnection of MYC2 and SPA1 in the Photomorphogenic Seedling Development of Arabidopsi. Plant Physiol. 2010;154:1210–1219. doi: 10.1104/pp.110.163717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abe H, Urao T, Ito T, Seki M, Shinozaki K, Yamaguchi-Shinozaki K. Arabidopsi AtMYC2 (bHLH) and AtMYB2 (MYB) function as transcriptional activators in Abscisic acid signaling. Plant Cell. 2003;15:63–78. doi: 10.1105/tpc.006130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boter M, Ruiz-Rivero O, Abdeen A, Prat S. Conserved MYC transcription factors play a key role in jasmonate signaling both in tomato and Arabidopsi. Genes Dev. 2004;8:1577–1591. doi: 10.1101/gad.297704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorenzo O, Chico JM, Sanchez-Serrano JJ, Solano R. JASMONATE-INSENSITIVE1 encodes a MYC transcription factor essential to discriminate between different jasmonate-regulated defense responses in Arabidopsi. Plant Cell. 2004;2004(16):1938–1950. doi: 10.1105/tpc.022319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dombrecht B, Xue GP, Sprague SJ, Krikegaard JA, Ross JJ, Reid JB, Fit GP, Sewelam N, Schenk PM, Manners JM, Kazan K. MYC2 differentially modulates diverse jasmonate-dependent functions in Arabidopsi. Plant Cell. 2007;19:2225–2245. doi: 10.1105/tpc.106.048017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koornneef M, Rolff E, Spruit CJP. Genetic control of light-inhibited hypocotyl elongation in Arabidopsis thalian. Z Pflanzenphysiol. 1980;100:147–160. [Google Scholar]
- Chattopadhyay S, Ang LH, Puente P, Deng XW, Wei N. Arabidopsi bZIP protein HY5 directly interacts with light-responsive promoters in mediating light control of gene expression. Plant Cell. 1998;10:673–683. doi: 10.1105/tpc.10.5.673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ang LH, Chattopadhyay S, Wei N, Oyama T, Okada K, Batschauer A, Deng XW. Molecular interaction between COP1 and HY5 defines a regulatory switch for light control of Arabidopsi development. Mol Cell. 1998;1:213–222. doi: 10.1016/S1097-2765(00)80022-2. [DOI] [PubMed] [Google Scholar]
- Holm M, Ma LG, Qu LJ, Deng XW. Two interacting bZIP proteins are direct targets of COP1-mediated control of light-dependent gene expression in Arabidopsi. Genes Dev. 2002;16:1247–1259. doi: 10.1101/gad.969702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cluis CP, Mouchel CF, Hardtke CS. The Arabidopsi transcription factor HY5 integrates light and hormone signaling pathways. Plant J. 2004;38:332–347. doi: 10.1111/j.1365-313X.2004.02052.x. [DOI] [PubMed] [Google Scholar]
- Sibout R, Sukumar P, Hettiarachchi C, Holm M, Muday GK, Hardtke CS. Opposite root growth phenotypes of hy versus hy5 hy mutants correlate with increased constitutive auxin signaling. PLoS Genet. 2006;2:e202. doi: 10.1371/journal.pgen.0020202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandenbussche F, Habricot Y, Condiff AS, Maldiney R, Straeten DD, Ahmad M. HY5 is a point of convergence between cryptochrome and cytokinin signaling pathways in Arabidopsis thalian. Plant J. 2007;49:428–441. doi: 10.1111/j.1365-313X.2006.02973.x. [DOI] [PubMed] [Google Scholar]
- Lee J, He K, Stolc V, Lee H, Figueroa P, Gao Y, Tongprasit W, Zhao H, Lee I, Deng XW. Analysis of transcription factor HY5 genomic binding sites revealed its hierarchical role in light regulation of development. Plant Cell. 2007;19:731–749. doi: 10.1105/tpc.106.047688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emborg TJ, Walker JM, Noh B, Vierstra RD. Multiple heme oxygenase family members contribute to the biosynthesis of the phytochrome chromophore in Arabidopsi. Plant Physiol. 2006;140:856–868. doi: 10.1104/pp.105.074211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terry MJ. Phytochrome chromophore-deficient mutants. Plant Cell Environ. 1997;20:740–745. doi: 10.1046/j.1365-3040.1997.d01-102.x. [DOI] [Google Scholar]
- Muramoto T, Kohchi T, Yokota A, Hwang I, Goodman HM. The Arabidopsi photomorphogenetic mutant hy is deficient in phytochrome chromophore biosynthesis as a result of a mutation in a plastid heme oxygenase. Plant Cell. 1999;11:335–348. doi: 10.1105/tpc.11.3.335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis SJ, Kurepa J, Vierstra RD. The Arabidopsis thalian HY1 locus, required for phytochrome chromophore biosynthesis, encodes a protein related to heme oxygenases. Proc Natl Acad Sci USA. 1999;96:6541–6546. doi: 10.1073/pnas.96.11.6541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhai Q, Li CB, Zheng W, Wu X, Zhao J, Zhou G, Jiang H, Sun J, Lou Y, Li C. Phytochrome chromophore deficiency leads to overproduction of jasmonic acid and elevated expression of jasmonate-responsive genes in Arabidopsi. Plant Cell Phy. 2007;48:1061–1071. doi: 10.1093/pcp/pcm076. [DOI] [PubMed] [Google Scholar]
- Lopez-Juez E, Jarvis RP, Takeuchi A, Page MA, Chory J. New Arabidopsis cu mutants suggest a close connection between plastid- and phytochrome regulation of nuclear gene expression. Plant Physiol. 1998;1998(118):803–815. doi: 10.1104/pp.118.3.803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chini A, Fonseca S, Fernandez GO, Garcia-Casado G, Lopez-Vidriero I, Lozano FM, Ponce MR, Micol JL, Solano R. The JAZ family of repressors is the missing link in jasmonate signalling. Nature. 2007;448:666–671. doi: 10.1038/nature06006. [DOI] [PubMed] [Google Scholar]
- Takahashi F, Yoshida R, Ichimura K, Mizoguchi T, Seo S, Yonezawa M, Maruyama K, Yamaguchi-Shinisali Y, Shinozaki K. The mitogen-activated Protein kinase cascade MKK3-MPK6 is an important part of the jasmonate signal transduction pathway in Arabidopsi. Plant Cell. 2007;19:805–818. doi: 10.1105/tpc.106.046581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osterlund MT, Wei N, Deng XW. The roles of photoreceptor systems and the COP1-targeted destabilization of HY5 in light control of Arabidopsis seedling development. Plant Plysiology. 2000;124:1520–1524. doi: 10.1104/pp.124.4.1520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson JP, Badruzsaufari E, Schenk PM, Manners JM, Desmond OJ, Ehlert C, Maclean DJ, Ebert PR, Kazan K. Antagonistic interaction between Abscisic acid and jasmonate-ethylene signaling pathways modulates defense gene expression and disease resistance in Arabidopsis. Plant Cell. 2004;16:3460–3479. doi: 10.1105/tpc.104.025833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kazan K, Manners JM. The interplay of light and jasmonate signaling during defence and development. J Expt Bot. 2011;62:4087–4100. doi: 10.1093/jxb/err142. [DOI] [PubMed] [Google Scholar]
- Robson F, Okamoto H, Patrick E, Harris S-R, Wasternack C, Brearley C, Turner JG. Jasmonate and phytochrome A signaling in Arabidopsis wound and shade responses are integrated through JAZ1 stability. Plant Cell. 2010;22:1143–1160. doi: 10.1105/tpc.109.067728. [DOI] [PMC free article] [PubMed] [Google Scholar]