Protozoan parasites of the order Kinetoplastida include various species of the genera Leishmania and Trypanosoma that are responsible for substantial human morbidity and mortality in the tropics. Pathogenic Leishmania species cause a diverse group of diseases, collectively called leishmaniasis, that range in severity from spontaneously healing skin ulcers to fatal visceral disease. African and American trypanosomes cause fatal sleeping sickness and debilitating Chagas disease, respectively. More than a billion people live in areas inhabited by the insects that transmit these parasites, and millions of people are newly infected each year.
Organisms of the order Kinetoplastida have a unique organelle called the kinetoplast, an appendix of their single mitochondrion located near the basal body of the flagellum that contains a network of thousands of small interlocking circular DNAs. Kinetoplastids are among the most ancient eukaryotes, with rRNA lineages extending farther back than those of animals, plants, and even fungi (1, 2). As might be expected of such ancient organisms, the kinetoplast is only one of their many distinctive features. The top 10 list of fundamentally important biological phenomena first discovered in Leishmania and trypanosomes includes “programmed” antigenic variation of surface glycoproteins (3), glycosylphosphatidylinositol anchors of membrane proteins (4, 5), expansion/contraction of telomeric DNA repeats (6), bent DNA helices (7), eukaryotic polycistronic transcription (8), trans-splicing of precursor RNAs (9, 10), mitochondrial RNA editing (11), other unique organelles such as glycosomes (12), and distinctive metabolic pathways (13). Several of these phenomena, first unearthed because of their prominence in kinetoplastids, have subsequently been found in higher eukaryotes and have become the focus of intense research interest in those systems. In addition, the many nefarious mechanisms used by Leishmania and trypanosomes to thwart immune defenses thrown at them by their mammalian hosts have led to an enhanced appreciation of the diversity and complexity of host/parasite interactions (1).
Other surprises likely await the assiduous investigator of Leishmania and trypanosomes. The most recent surprise comes from the full DNA sequence of chromosome 1 of Leishmania major (GenBank accession no. AE001274) reported in this issue of the Proceedings by Myler et al. (14). The 34-Mb haploid genome of this diploid organism is contained in 36 chromosomes, ranging in size from the 269-kb chromosome 1 (exclusive of its telomere repeat regions) to the largest chromosome of about 2.5 Mb. Previous studies have established that 30% of the genome is composed of repeated elements, about half of which are telomeric/subtelomeric repeats and the rest of which are dispersed transposons, repeated genes, and other simple repeated sequences. None of the protein-encoding genes of Leishmania studied to date contain introns, simplifying the identification of these genes in the genomic DNA. Most, if not all, of these genes are initially transcribed into large polycistronic precursor RNAs of 60 kb or more in length that are cleaved into monocistronic mRNAs by the action of two intergenic RNA cleavage reactions, trans-splicing of a 39-nt “spliced leader” to generate the 5′ ends of all mRNAs, and 3′ cleavage/polyadenylation to create the 3′ ends (15, 16). In contrast to most other eukaryotes, no consensus 3′ poly(A) sites have been described, but efficient 5′ trans-RNA splicing typically occurs at a short consensus sequence preceded by a polypyrimidine tract (17). Little is known about the protein components of the putative trans-spliceosome, although a number of small nuclear RNAs are known to participate in the process (18). Because cis-splicing of introns in yeast/mammals and trans-splicing of the spliced leader in Leishmania/trypanosomes are mechanistically similar, however, it seems likely that similar proteins participate in these two processes.
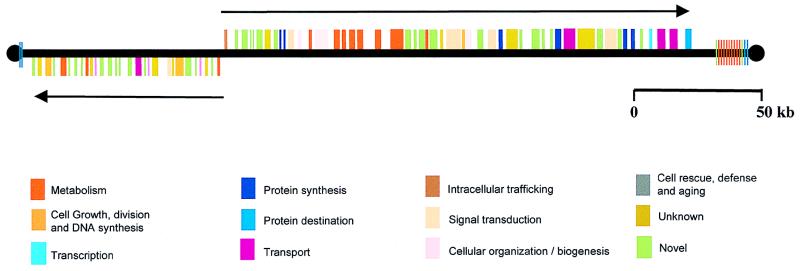
The 269-kb chromosome 1 constitutes <1% of the L. major genome and appears to contain only 79 genes, all without introns. The organization of these genes is what is surprising (Fig. 1). Fifty of the genes are lined up one after another on one strand; the other 29 are densely packed adjacent to each other on the opposite strand. All of the intergenic regions contain several tracts of 10 or more pyrimidines that are likely necessary for processing precursor transcripts into monocistronic mRNAs. The two gene clusters are separated by only 1.6 kb and are transcribed in opposite directions, toward the telomeres. At first glance, these gene clusters look more like two giant bacterial operons than eukaryotic genes. There are several lines of evidence from previous studies of other Leishmania/trypanosome genes, however, that suggest that these two gene clusters are not regulated like conventional bacterial operons. First, unlike most prokaryotic and eukaryotic organisms, where the regulation of gene expression occurs primarily at the level of transcription, in Leishmania/trypanosomes, gene regulation is largely posttranscriptional. Events and properties such as trans-RNA splicing, polyadenylation, mRNA half-lives, protein synthesis, and protein stabilities control gene expression in these organisms (19). Second, the two gene clusters of chromosome 1 do not appear to encode proteins that share a metabolic pathway or protein complex, as do the structural genes of many bacterial operons. Instead, more reminiscent of eukaryotic gene organization, the deduced protein products of these two groups of Leishmania genes have apparently unrelated functions, ranging from signal transduction and fatty acid metabolism to DNA repair and oxygen-radical defense. It is of interest that one of the genes encodes an arsenate reductase, because arsenical drugs have been used to treat leishmaniasis and are still used in advanced cases of sleeping sickness. Thirty-two of the 79 genes (41%) do not have homologues in the current nucleotide/protein databases, and some of these may be unique to Leishmania.
Figure 1.
Relative locations of the 79 genes in the smaller of the two homologues of chromosome 1 in L. major. The thick black line and black circles indicate the DNA and telomeres, respectively. The boxes depict the genes and are color-coded according to the potential functions of their protein products. The “unknown” genes have sequence similarities to genes of unknown function in other organisms. The “novel” genes have no substantive similarities with other sequences in the databases. The 50 contiguous genes on the top strand, including 5 tandem genes encoding long-chain fatty acyl CoA synthetases, are transcribed to the right (→); the 29 contiguous genes on the bottom strand are transcribed to the left (←). The colored vertical lines adjacent to the telomeres indicate telomeric and subtelomeric repeats. The larger of the two chromosome 1 homologues (not shown) has an identical gene organization, but contains an additional 20+ kb of subtelomeric repeats at the right telomere. Adapted from Myler et al. (14).
Another of the curiosities of Leishmania/trypanosomes is their apparent lack of promoters for RNA polymerase II, the enzyme that typically transcribes protein-encoding genes. Transcription in these organisms is, however, strand-specific and is performed by RNA polymerases with α-amanitin-sensitivities similar to mammalian RNA polymerases I, II, and III. Promoters for α-amanitin-resistant RNA polymerase I activity have been readily detected, as expected, in front of ribosomal RNA genes and also, quite unexpectedly, in front of developmentally regulated African trypanosome genes encoding variant surface glycoproteins and procyclins (20). Likewise, a RNA polymerase III activity with an intermediate α-amanitin sensitivity transcribes the small RNA genes classically recognized by RNA polymerase III (21). A typical α-amanitin-sensitive RNA polymerase II activity has been shown to transcribe most protein-encoding genes. However, attempts to utilize promoter-trap plasmids containing reporter genes in transfections of Leishmania/trypanosomes to identify sequences with robust polymerase II promoter activities similar to those of polymerase I promoters have not been successful. Knowledge of the sequence of the L. major chromosome 1 will facilitate the search for polymerase II promoters. The obvious place to look is in the 1.6-kb segment between the two gene clusters where bidirectional transcription presumably begins and proceeds toward the two telomeres. Alternatively, transcription may be initiated at multiple sites within the two clusters.
One approach for distinguishing between the single and multiple promoter models of transcription will be to examine the lengths of the initial transcripts from each cluster by using the technique of UV irradiation inactivation of transcription, which circumvents the limitations of promoter-trap plasmids. This method was first used on Leishmania/trypanosomes more than a decade ago (8) and is based on the fact that RNA polymerases cannot traverse pyrimidine dimers generated in DNA by UV irradiation. Thus, the farther away a gene is from its promoter, the more sensitive its transcription is to UV irradiation. An analysis of the transcription of these two clusters is amenable to this approach because unique sequences distributed throughout the clusters can be used as hybridization targets in Southern blots probed with nuclear run-on [32P]RNA. If RNA synthesis begins upstream from the two clusters, transcription of the distal genes will be more sensitive to UV irradiation than that of the proximal genes. If transcription initiation is “willy-nilly,” beginning in a strand-specific manner at random locations, then transcription of distal and proximal genes should be about equally affected by UV irradiation.
Inspection of the 1.6 kb sequence between the two clusters does not reveal any recognizably distinctive features or substantive similarities to sequences in the databases. Nevertheless, as Myler and coworkers point out (14), in addition to possibly having bidirectional promoters, this region may also have a chromosomal centromere and/or an origin of replication. Nothing is known about such elements in Leishmania/trypanosomes, but it should be experimentally straightforward to determine whether sequences in this region contribute either of these functions. Alternatively, higher order DNA structures and chromatin-associated protein complexes could define the centromeric functions and replication origin of the chromosome (22, 23).
Another interesting question to be addressed is whether the organization of genes in L. major chromosome 1 is present in other kinetoplastids. At least one recent report suggests that this is the case. Baltz and colleagues (24) recently examined the regions upstream and downstream of a group of genes encoding glucose transporters in Leishmania donovani, Trypanosoma cruzi, and several species of African trypanosomes. In all of the organisms studied, these flanking regions contain highly conserved sequences encoding a kinase, a ribosomal protein, a DnaJ homologue, and three small G proteins. Because genome sequencing projects for American and African trypanosomes are also currently in progress, a detailed comparison of their genomic organization and gene sequences with those of Leishmania should soon be possible. Because these evolutionarily related organisms have strikingly different life cycles and cause dramatically dissimilar human diseases, one interesting possibility is that they might share a similar organization of housekeeping genes but possess species-specific “pathogenicity islands” of genes, similar to those found in various bacterial pathogens (25).
The complete sequence of a chromosome in a nonkinetoplastid protozoan parasite of immense public health importance, the human malaria parasite Plasmodium falciparum, has also recently been determined (26). Similar to Leishmania, the Plasmodium genome is about 30 Mb, but a comparison of the Leishmania and Plasmodium chromosome sequences reveals that the similarity ends abruptly with genome size. The 947-kb P. falciparum chromosome 2 (GenBank accession no. AE001362) is 80% A/T and has a more typical eukaryotic gene organization than does the Leishmania chromosome. Unlike Leishmania, >40% of its 209 protein-encoding genes contain introns; these coding regions are dispersed on both strands and not arranged in polycistronic transcription units. In addition, intergenic regions of some P. falciparum genes have been shown to contain promoter sequences that can direct expression of selectable markers and transgenes after both transient and stable transfection (27), indicating that control of gene expression in these two protozoan genera may be quite different.
In summary, the genetic organization and gene sequences in L. major chromosome 1 support the view that the evolutionarily ancient Leishmania and trypanosomes are ensconced at the genetic border between prokaryotic and eukaryotic organisms. They share some features with prokaryotes (genes without introns, polycistronic transcription), other features with eukaryotes (pre-mRNA splicing), and have some features found in neither (protein-encoding genes without promoters). Myler et al. (14) indicate that the third-smallest chromosome of L. major, chromosome 3, which they have partially sequenced, also appears to have two gene clusters. In this case, however, transcription of the two clusters proceeds toward the center of the chromosome rather than in the direction of the telomeres. Thus, the Kinetoplastida surprises continue.
Footnotes
A commentary on this article begins on page 2902.
References
- 1.Beverley S M. Cell. 1996;87:787–789. doi: 10.1016/s0092-8674(00)81984-4. [DOI] [PubMed] [Google Scholar]
- 2.Fernandes A P, Nelson K, Beverley S M. Proc Natl Acad Sci USA. 1993;90:11608–11612. doi: 10.1073/pnas.90.24.11608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bridgen P J, Cross G A M, Bridgen J. Nature (London) 1976;263:613–614. doi: 10.1038/263613a0. [DOI] [PubMed] [Google Scholar]
- 4.Holder A A, Cross G A M. Mol Biochem Parasitol. 1981;2:135–150. doi: 10.1016/0166-6851(81)90095-5. [DOI] [PubMed] [Google Scholar]
- 5.Ferguson M, Cross G A M. J Biol Chem. 1984;259:3011–30115. [PubMed] [Google Scholar]
- 6.Bernards A, Michels P A, Lincke C R, Borst P. Nature (London) 1983;303:592–597. doi: 10.1038/303592a0. [DOI] [PubMed] [Google Scholar]
- 7.Marini J C, Levene S D, Crothers D M, Englund P T. Cold Spring Harbor Symp Quant Biol. 1983;47:279–283. doi: 10.1101/sqb.1983.047.01.033. [DOI] [PubMed] [Google Scholar]
- 8.Johnson P J, Kooter J M, Borst P. Cell. 1987;51:273–281. doi: 10.1016/0092-8674(87)90154-1. [DOI] [PubMed] [Google Scholar]
- 9.Boothroyd J C, Cross G A M. Gene. 1982;20:279–287. doi: 10.1016/0378-1119(82)90046-4. [DOI] [PubMed] [Google Scholar]
- 10.Walder J A, Eder P S, Engman D M, Brentano S T, Walder R Y, Knutzon D S, Dorfman D M, Donelson J E. Science. 1986;233:569–571. doi: 10.1126/science.3523758. [DOI] [PubMed] [Google Scholar]
- 11.Benne R, Van den Burg J, Brakenhoff J P J, Sloof P, Van Boom J H, Tromp M C. Cell. 1986;46:819–826. doi: 10.1016/0092-8674(86)90063-2. [DOI] [PubMed] [Google Scholar]
- 12.Opperdoes F R, Borst P. FEBS Lett. 1977;80:360–364. doi: 10.1016/0014-5793(77)80476-6. [DOI] [PubMed] [Google Scholar]
- 13.Bacchi C J, Nathan H N, Hutner S H, McCann P P, Sjoerdsma A. Science. 1980;210:332–334. doi: 10.1126/science.6775372. [DOI] [PubMed] [Google Scholar]
- 14.Myler P J, Audleman L, deVos T, Hixson G, Kiser P, Lemley C, Magness C, Rickel E, Sisk E, Sunkin S, et al. Proc Natl Acad Sci USA. 1999;96:2902–2906. doi: 10.1073/pnas.96.6.2902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ullu E, Matthews K R, Tschudi E. Mol Cell Biol. 1993;13:720–725. doi: 10.1128/mcb.13.1.720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.LeBowitz J H, Smith H Q, Rusche L, Beverley S M. Genes Dev. 1993;7:996–1007. doi: 10.1101/gad.7.6.996. [DOI] [PubMed] [Google Scholar]
- 17.Matthews K R, Tschudi C, Ullu E. Genes Dev. 1994;8:491–501. doi: 10.1101/gad.8.4.491. [DOI] [PubMed] [Google Scholar]
- 18.Tschudi C, Ullu E. Cell. 1990;61:459–466. doi: 10.1016/0092-8674(90)90527-l. [DOI] [PubMed] [Google Scholar]
- 19.Vanhamme L, Pays E. Microbiol Rev. 1995;59:223–240. doi: 10.1128/mr.59.2.223-240.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zomerdijk J C, Kieft R, Shiels P G, Borst P. Nucleic Acids Res. 1991;19:5153–5158. doi: 10.1093/nar/19.19.5153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gunzl A, Tschudi C, Nakaar V, Ullu E. J Biol Chem. 1995;270:17287–17291. doi: 10.1074/jbc.270.29.17287. [DOI] [PubMed] [Google Scholar]
- 22.Lechner J, Ortiz J. FEBS Lett. 1996;389:70–74. doi: 10.1016/0014-5793(96)00563-7. [DOI] [PubMed] [Google Scholar]
- 23.Hyman A A, Sorger P K. Annu Rev Cell Dev Biol. 1995;11:471–495. doi: 10.1146/annurev.cb.11.110195.002351. [DOI] [PubMed] [Google Scholar]
- 24.Bringaud F, Vedrenne C, Cuvillier A, Parzy D, Baltz D, Tetaud E, Pays E, Venegas J, Merlin G, Baltz T. Mol Biochem Parasitol. 1998;94:249–264. doi: 10.1016/s0166-6851(98)00080-2. [DOI] [PubMed] [Google Scholar]
- 25.Groisman E A, Ochman H. Cell. 1996;87:791–794. doi: 10.1016/s0092-8674(00)81985-6. [DOI] [PubMed] [Google Scholar]
- 26.Gardner M J, Tettelin H, Carucci D J, Cummings L M, Aravind L, Koonin E V, Shallom S, Mason T, Yu K, Fujii C, et al. Science. 1998;282:1126–1132. doi: 10.1126/science.282.5391.1126. [DOI] [PubMed] [Google Scholar]
- 27.Coppel R L, Black C G. In: Malaria, Parasite Biology, Pathogenesis, and Protection. Sherman I W, editor. Washington, DC: Am. Soc. Microbiol.; 1998. pp. 185–202. [Google Scholar]