Abstract
Cancer cells adapt their metabolic processes to drive macromolecular biosynthesis for rapid cell growth and proliferation (1,2). RNAi-based loss of function screening has proven powerful for the identification of novel and interesting cancer targets, and recent studies have used this technology in vivo to identify novel tumor suppressor genes (3). Here, we developed a method for identifying novel cancer targets via negative selection RNAi screening in solid tumours. Using this method, we screened a set of metabolic genes associated with aggressive breast cancer and stemness to identify those required for in vivo tumourigenesis. Among the genes identified, phosphoglycerate dehydrogenase (PHGDH) is in a genomic region of recurrent copy number gain in breast cancer and PHGDH protein levels are elevated in 70% of ER-negative breast cancers. PHGDH catalyzes the first step in the serine biosynthesis pathway, and breast cancer cells with high PHGDH expression have elevations in serine synthesis flux. Suppression of PHGDH in cell lines with elevated PHGDH expression, but not those without, causes a strong decrease in cell proliferation and a reduction in serine synthesis. We find that PHGDH suppression does not affect intracellular serine levels, but causes a drop in the levels of alpha-ketoglutarate, another output of the pathway and a TCA cycle intermediate. In cells with high PHGDH expression, the serine synthesis pathway contributes approximately 50% of the total anaplerotic flux of glutamine into the TCA cycle. These results reveal that certain breast cancers are dependent upon increased serine pathway flux caused by PHGDH over-expression and demonstrate the utility of in vivo negative selection RNAi screens for finding potential anticancer targets.
As a starting point for identifying metabolic genes required for tumourigenesis, we cross-referenced maps of metabolic pathways with the KEGG database to compile a comprehensive list of 2,752 genes encoding all known human metabolic enzymes and transporters (Supplementary Table 1). Public oncogenomic data were analyzed to score genes based on three properties: (i) higher expression in tumours versus normal tissues, (ii) high expression in aggressive breast cancer, or (iii) association with the stem cell state (Fig. 1a). Genes scoring in two of these three categories as well as those at the top of each category were selected to define a high priority set of 133 metabolic enzyme and transporter genes (Supplementary Table 2). We assembled lentiviral shRNA vectors targeting these genes (median 5 shRNAs per gene) and used them to generate two libraries of shRNA-expressing lentiviruses, one containing 235 distinct shRNAs (targeting transporters and control genes) and the other 516 distinct shRNAs (targeting metabolic enzymes and control genes) (4).
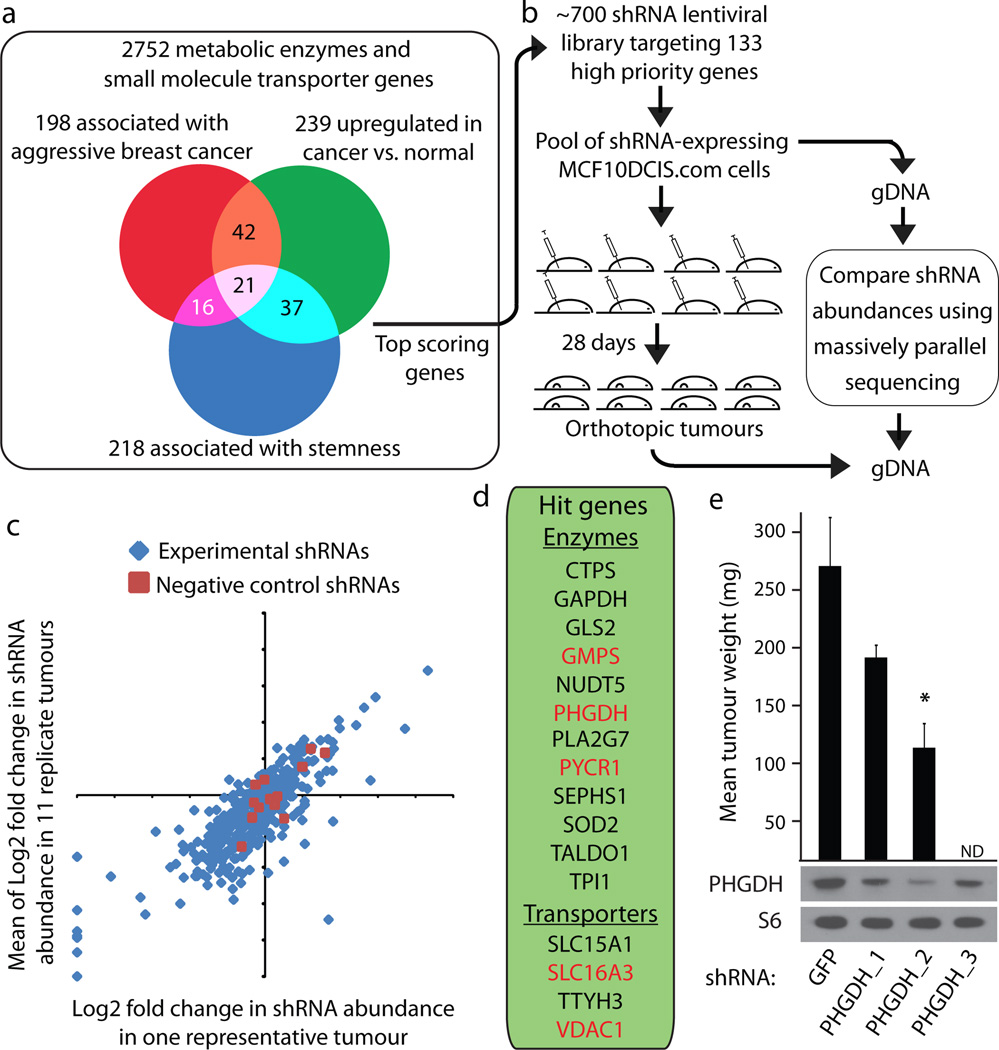
Figure 1. Outline of in vivo pooled screening strategy identifying PHGDH as essential for tumourigenesis.
a, Venn Diagram outlining meta-analysis. b, Outline of experimental design. c, Log2 fold change in shRNAs abundance of experimental (blue) or neutral shRNAs (red) for a single tumour (X-axis) compared to an average of eleven tumours (Y-axis). d, Genes scoring in vivo. e, Average weight of tumours from MCF10DCIS.com cells expressing shRNAs targeting PHGDH (PHGDH_1, PHGDH_2 and PHGDH_3) or control (GFP) and protein expression of PHGDH or RPS6 (S6). Error bars are SEM (n=4). Asterisks indicate probability value (p) < 0.05. ND = Not Done.
To identify genes that may be essential for tumourigenesis, the libraries were screened for shRNAs that become depleted during breast tumour formation in mice. Human MCF10DCIS.COM cells (5) were chosen for the screens because, of several breast cancer lines examined, these were capable of forming tumours upon injection of the fewest number of cells. 1.5 million MCF10DCIS.COM cells were infected with each library so that each cell carried one viral integrant, and ~500–1000 cells per shRNA (100,000–1 million cells total) were injected into mouse mammary fat pads at two sites per animal (Supplementary Discussion). Twenty-eight days later orthotopic tumours were harvested and massively parallel DNA sequencing was used to determine the abundance of each shRNA in genomic DNA from tumours and initially injected cells (Fig. 1b). shRNA abundances correlated well between replicate tumours (Fig. 1c) and 5 or 12 tumours per library were analyzed to identify shRNAs that became significantly depleted during tumour formation. Sixteen genes were designated hits in the screen, with at least 75% of the shRNAs targeting these genes scoring (Fig. 1d and Supplementary Table 3).
Several genes previously shown to have important roles in cancer emerged as hits, including the mitochondrial ATP transporter VDAC1; the lactic acid transporter SLC16A3; and the nucleotide synthesis genes GMPS and CTPS. The hit list also includes genes involved in the control of oxidative stress (SOD2, GLS2, SEPHS1), the pentose phosphate pathway (TALDO1) glycolysis (GAPDH, TPI1), and in the proline (PYCR1) and serine (PHGDH) biosynthetic pathways. An analogous pooled screen carried out in MCF10DCIS.com cells grown in culture rather than in tumor xenografts revealed that of 20 genes that scored in the in vitro screen, 10 also scored in the in vivo screen (Supplementary Fig. 2a, Supplementary Table 3 and Supplementary Discussion). Interestingly, AK2, which encodes an adenylate kinase that generates ADP from ATP and AMP, was required for in vitro but not in vivo growth (Supplementary Fig. 2b).
For five hit genes (PHGDH, GMPS, SLC16A3, PYCR1, and VDAC1), two scoring shRNAs were tested for their effects on tumour formation. Each of these shRNAs suppressed expression of their targets in MCF10DCIS.com cells and reduced tumor forming capacity. (Fig. 1e, Supplementary Fig. 2c). For reasons discussed below, PHGDH was of particular interest, and the three shRNAs that scored in the in vivo screen also decreased PHGDH protein expression and two shRNAs of differing knockdown efficacies inhibited tumour growth consistent with their capacity to suppress PHGDH expression (Fig. 1e). Moreover, tumours derived from cells that in culture had confirmed reductions in PHGDH levels had, in immunohistochemical (Supplementary Fig. 3a) and immunoblotting assays (Supplementary Fig. 3b), PHGDH staining or levels similar to control tumours, suggesting that tumourigenesis selected for cells that lost shRNA-mediated PHGDH suppression.
To prioritize genes for follow up studies we consulted a recently available analysis of copy number alteration across cancer genomes (6). Indeed, PHGDH exists in a region of chromosome 1p commonly amplified in breast cancer and melanoma (Fig. 2a), as well as in several other cancer types (not shown). In total, 18% of patient derived breast cancer cell lines and 6% of primary tumours have amplifications in PHGDH. In the datasets examined, none of the other hit genes are in genomic regions of focal and recurrent copy number gain.
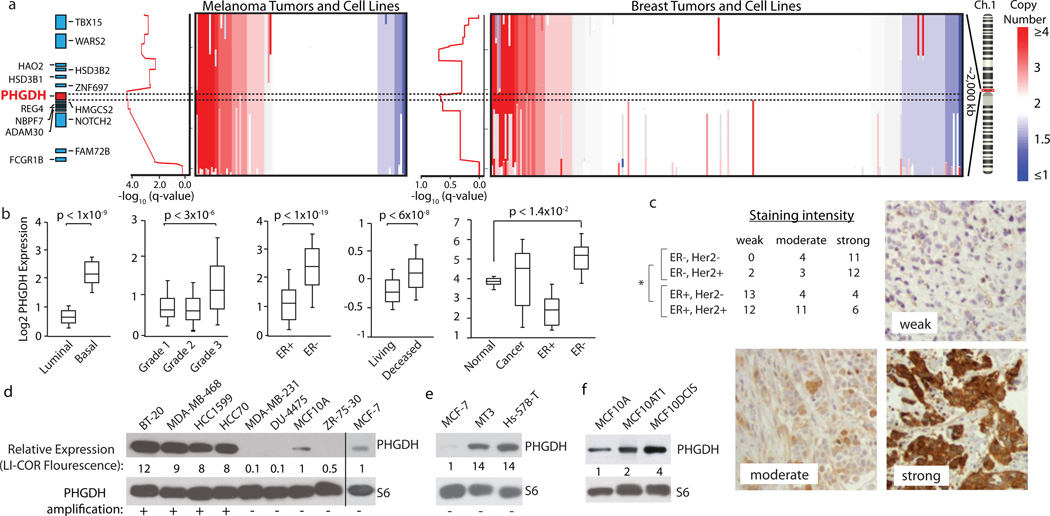
Figure 2. Genomic amplifications of PHGDH in cancer and association of PHGDH expression with aggressive breast cancer markers.
a, PHGDH vicinity copy number (CN) data for melanoma (left, n=111) and breast cancer (BC, right, n=243) samples. Coloured bar indicates degree of CN loss (blue) or gain (red). Samples sorted by CN at PHGDH locus (dotted lines). Graphs at left of CN data shows amplification significance (−log10(q-value), ~0.60 is the significance threshold for amplification). b, Representative PHGDH gene expression data for indicated BC groups. Whiskers indicate 91st and 9th percentile. c, Table reports numbers of human BC samples with “weak”, “moderate”, or “strong” PHGDH staining from BC subgroups indicated. Representative staining intensities shown in images. Asterisk indicates p<0.0001 comparing ER-positive versus ER-negative classes (Fisher’s exact test). d–f, PHGDH protein levels are shown for (d) PHGDH amplified versus non-amplified (annotated with “+” or “−“), (e) PHGDH non-amplified, over-expressing, and (f) MCF10A derived cell lines. Values below PHGDH immunoblots are normalized immunoflourescent quantification (LI-COR) of PHGDH levels relative to actin control and MCF10A and MCF7.
Our meta analysis for genes associated with aggressive breast cancer is corroborated by a previous study which found elevated PHGDH mRNA levels in breast cancers that are ER-negative, of the basal type, and associated with poor 5-year survival (7). We confirmed these associations in distinct gene expression datasets (Fig. 2b) and additionally found that PHGDH is elevated in ER-negative breast cancer relative to normal breast tissue (Fig. 2b). Of all the genes identified as hits in our screen, PHGDH has the most significantly elevated expression in ER-negative breast cancer (Supplementary Fig. 4). Moreover, by analyzing 82 human breast tumour samples with an immunohistochemical assay for PHGDH, we found that PHGDH protein levels correlate significantly with ER-negative status (Fig. 2c). In total, compared to ER-positive breast tumours, ~68% and ~70% of ER-negative breast tumours have elevations of PHGDH at the mRNA and protein levels, respectively (Fig. 2b, 2c and Supplementary Methods). ER-negative breast cancer comprises approximately 20–25% of all breast cancer cases, but as many as 50% of all breast cancer deaths within 5 years of diagnosis (8), underscoring the importance of identifying additional drug targets for this class of breast cancer.
Across a set of breast cancer lines, four lines with PHGDH amplifications had 8–12 fold higher PHGDH protein expression compared to non-transformed MCF10A and ER-positive MCF7 cell lines, which do not have PHGDH amplifications (Fig. 2d). Mechanisms other than gene copy number increases must also exist for boosting PHGDH expression because PHGDH protein levels were also elevated in two ER-negative cell lines (MT3, Hs578T) lacking the PHGDH amplification (Fig. 2e). This is consistent with the finding that PHGDH expression is upregulated at the mRNA and protein level in a higher fraction of ER-negative breast cancers than the fraction exhibiting amplification at the DNA level. Interestingly, PHGDH is also expressed 4-fold more in the MCF10DCIS.COM cells used in the in vivo screen than in two parental lines (MCF-10A; MCF10AT) that exhibit no or lower tumourigenicity (9) (Fig. 2f).
PHGDH encodes 3-phosphoglycerate dehydrogenase, the first enzyme branching from glycolysis in the three-step serine biosynthetic pathway (10) (Fig. 3a). PHGDH uses NAD as a cofactor to oxidize the glycolytic intermediate 3-phosphoglycerate into phospho-hydroxypyruvate (11, 12), which subsequent enzymes in the pathway convert into serine via transamination (PSAT1) and phosphate ester hydrolysis (PSPH) reactions (10) (Fig. 3a). Serine is essential for synthesis of proteins and other biomolecules needed for cell proliferation, including nucleotides, phosphatidyl-serine, and sphingosine (Supplementary Fig. 1). Classic studies show elevated serine biosynthetic activity, as determined by enzyme assays, in rat tumour lysates (10, 13), and suggest that PSPH is the rate-limiting enzyme of this pathway in the liver (14). Interestingly, we find that numerous genes that are expected to promote serine biosynthesis or are involved in the subsequent metabolism of serine for biosynthesis are elevated in ER-negative breast cancer (Supplementary Fig. 5), demonstrating that PHGDH elevation occurs in the context of upregulation of a broader pathway.
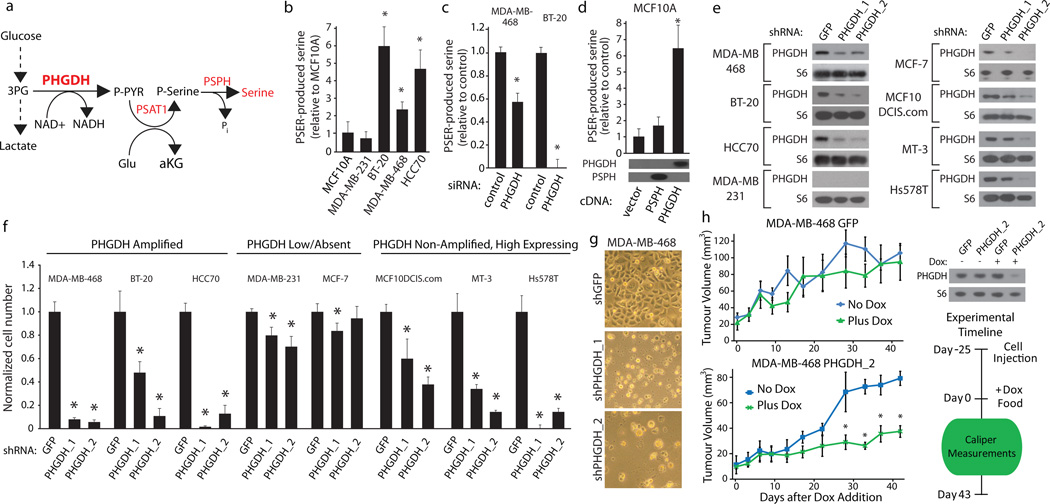
Figure 3. Cell lines with elevated PHGDH expression have increased serine biosynthetic pathway activity and are sensitive to PHGDH suppression.
a, Serine biosynthesis pathway (SBP). b–d, Serine production by SBP in (b) indicated breast cell lines, (c) after PHGDH suppression by siRNA, and (d) MCF-10A cells expressing PHGDH or PSPH cDNAs with associated immunoblots. e–f, Immunoblots of indicated proteins (e) for indicated cell lines expressing control shRNA (GFP) or shRNAs against PHGDH (PHGDH_1 and PHGDH_2). Relative proliferation (f) of cells transduced with shRNA constructs after seven days. g, Images showing cellular morphology of MDA-MB-468 at day seven of (f). h, Tumour growth of MDA-MB-468 cells expressing doxycycline inducible control shRNA (GFP) or shRNA against PHGDH (shPHGDH_2) in mice fed doxycycline (Dox, 2mg/kg, green lines, n=5) or normal (blue lines, n=4) diet after initial tumour palpation (Day 0). Immunoblots of PHGDH or RPS6 (S6) shown for cells in vitro. Asterisks indicate p < 0.05 relative to control. Error bars for metabolite measurements (n=4) and tumour size indicate SEM and for cell number indicate SD (n=3).
To understand the metabolic consequences of increased PHGDH expression we used metabolite profiling and serine synthesis pathway flux analysis to examine breast cancer cells with and without PHGDH amplifications. We found that cells with PHGDH amplifications (BT-20, MDA-MB-468, HCC70), had elevated flux through the serine synthesis pathway compared to those without PHGDH amplifications (MDA-MB-231, MCF7 and MCFC10A) (Fig. 3b and Supplementary Fig 6a). Cells with elevated PHGDH and high pathway flux were capable of robust proliferation in medium lacking serine, while in cells with low levels of PHGDH, the deprivation of serine caused a significant blunting or even cessation of proliferation (Supplementary Fig. 6b).
PHGDH is required for the increased serine pathway flux of cells with elevated PHGDH because RNAi-mediated PHGDH suppression significantly reduced flux in MDA-MB-468 and BT-20 cells (Fig. 3c). Conversely, in MCF-10A human mammary cells engineered to overexpress PHGDH, serine pathway flux increased to levels similar to those in MDA-MB-468, BT-20, and HCC70 cells (Fig. 3d). Furthermore, MCF-10A cells overexpressing PHGDH had increased proliferation in the absence of serine, indicating that PHGDH overexpression is sufficient to drive flux through the pathway (Supplementary Fig. 6c). Interestingly, overexpression of PSPH, considered the rate-limiting serine biosynthetic enzyme in the liver, did not increase pathway flux in MCF-10A cells (Fig. 3d). The observations that PSPH is rate limiting in the liver while PHGDH is rate limiting in MCF10A cells can be reconciled by the observation that serine levels in the liver (2 mM) are well above the concentration at which PSPH is feedback inhibited by serine (500 µM), but low in cell lines in culture (~100 µM), a concentration at which PSPH should be active 14. These data demonstrate that PHGDH is a key enzyme controlling flux through the serine biosynthetic pathway in cancer cells.
We next asked if cells with an increase in PHGDH expression require it for cell proliferation and survival. In cell lines with elevated PHGDH expression (BT-20, MDA-MB-468, HCC70, Hs578T and MT3), but not without (MDA-MB-231, MCF-7), RNAi-mediated suppression of PHGDH caused a dramatic decrease in cell number (Fig. 3e–f, Supplementary Fig. 6d) and cell death (Fig. 3g and Supplementary Fig. 6e) in the absence of apoptotic markers (Supplementary Fig. 6f). This sensitivity to PHGDH suppression was observed both in cells with PHGDH amplifications (BT-20, MDA-MB-468, HCC70) and in those with high PHGDH expression but lacking the amplifications (MT3, Hs578T). Consistent with flux through the serine synthesis pathway being important in cells with high PHGDH expression, suppression of the other two enzymes in the pathway (PSAT1 and PSPH) inhibited the proliferation of MDA-MB-468 and BT-20, but not MCF7, cells (Supplementary Fig. 6g). Moreover, inhibition of PSPH inhibited tumour formation by MCF10DCIS.com cells (Supplementary Fig. 6h). Therefore, elevated PHGDH expression defines a set of breast cancer cell lines with increased serine pathway flux that are dependent upon PHGDH, PSAT1, and PSPH for proliferation. This finding suggests that many ER-negative breast cancers that express PHGDH at high levels (~70% of all ER-negative disease in our dataset, Fig. 2c) may be sensitive to inhibitors of the serine synthesis pathway.
To investigate whether PHGDH suppression can affect the growth of established tumours, we generated an inducible shRNA (15) that, upon doxycycline treatment, reduced PHGDH protein levels in MDA-MB-468 cells (Fig. 3h). MDA-MB-468 cells transduced with this shRNA were allowed to form murine mammary fat pad tumors for 25 days before introduction of doxycycline in a subset of mice (Fig. 3h). Compared to control mice, those given doxycycline exhibited substantially reduced tumour growth, while tumours made from cells transduced with a control inducible shRNA grew equally well in the presence or absence of doxycycline (Fig. 3h). These results indicate that PHGDH suppression can adversely affect growth in existing tumours (Supplementary Discussion).
Serine is a central metabolite for biosynthetic reactions, and we find that overexpression of PHGDH contributes significantly to biosynthetic flux to serine. However, PHGDH suppression inhibited proliferation even in cells growing in media containing normal levels of extracellular serine (Fig. 3f), and supplementation with additional serine or a cell-permeable methyl-serine-ester did not blunt the effects of the PHGDH knockdown (Fig. 4a, 4b). Intracellular and extracellular serine are in equilibrium (Supplementary Fig. 7a), and import of extracellular serine was not defective in the cell lines studied (Supplementary Fig. 7b). These findings suggest that serine production may not be the only important role of PHGDH in cell lines with high PHGDH expression. We considered three hypotheses to explain our observations: (1) serine produced via the PHGDH pathway is utilized in a different manner than exogenous serine, (2) suppression of PHGDH adversely affects glycolysis, or (3) the PHGDH, PSAT1 and PSPH reactions produce metabolites besides serine that are also critical for cell proliferation. The first hypothesis was deemed unlikely because serine synthesized intracellularly is in equilibrium with extracellular serine (Supplementary Fig. 7a). The second hypothesis was also unlikely because PHGDH suppression did not affect glucose uptake or lactate production (Supplementary Fig. 7c).
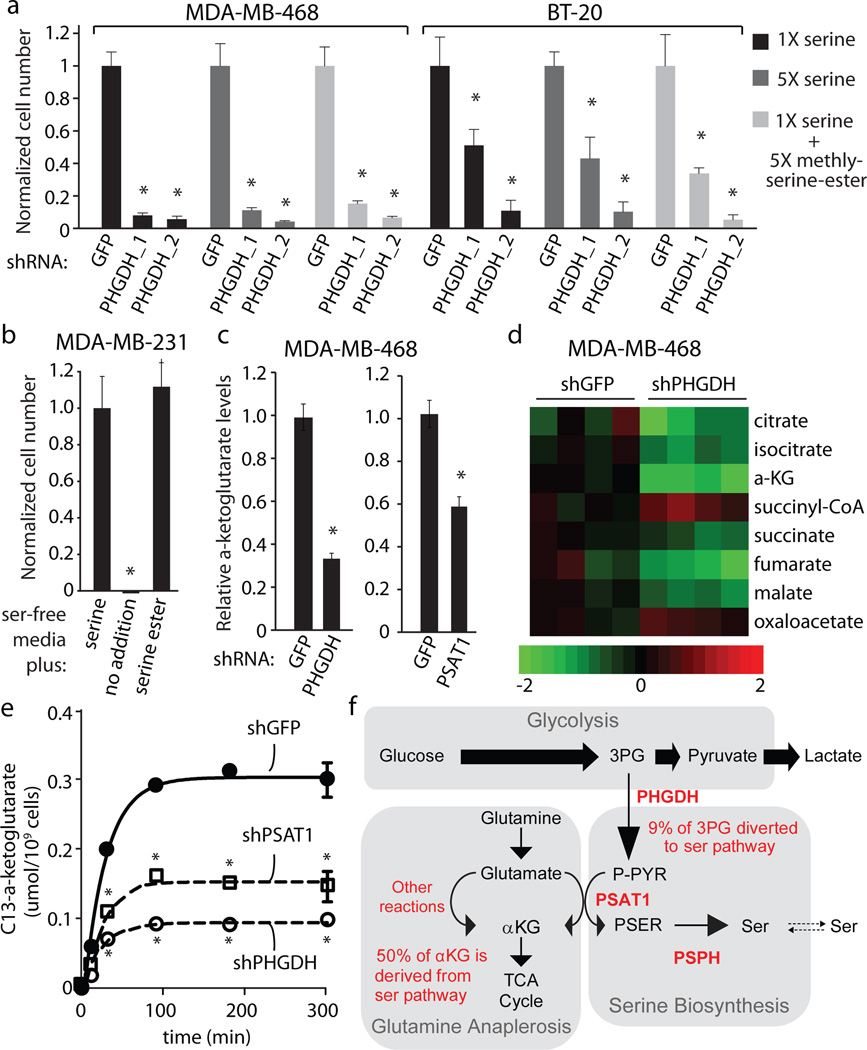
Figure 4. Suppression of PHGDH results a deficiency in anaplerosis of glutamine to alpha-ketoglutarate.
a, Relative proliferation of cell lines indicated expressing control shRNA (GFP) or shRNAs against PHGDH (PHGDH_1 and PHGDH_2) after seven days of growth under conditions indicated. b, Relative proliferation of MDA-MB-231 cells under conditions indicated. c, Intracellular alpha-ketoglutarate (aKG) four days after treatment with shRNA against PHGDH or PSAT1 cell number normalized relative to control shRNA (GFP). d, Citric acid cycle intermediate levels four days after treatment with shRNA against PHGDH or GFP (n=4). Color bar shows Log2 scale. e, aKG isotopic labeling at indicated time points after treatment with isotopically labeled glutamine four days after treatment with shRNA against PHGDH, PSAT1 or GFP. f, Model of relative metabolite fluxes for indicated pathways. Asterisks indicate p < 0.05 relative to control. Error bars indicate SEM (n=4).
To pursue the third hypothesis, we considered which additional metabolites the serine synthesis pathway might produce in significant levels in cells with high PHGDH expression. The serine pathway produces equimolar amounts of serine and alpha-ketoglutarate (aKG, Supplementary Fig. 1). Proliferating cells utilize intermediates of the citric acid (TCA) cycle, such as aKG, as biosynthetic precursors, and upregulate anaplerotic reactions that drive glutamine derived carbon into the TCA cycle, counterbalancing biosynthetic efflux (16, Supplementary Discussion). We hypothesized that in cells with high PHGDH expression, the PSAT1 reaction might contribute a significant fraction of glutamate to aKG flux. If true, the serine biosynthesis pathway would play an important role in TCA anaplerosis of glutamine-derived carbon. Consistent with this possibility, suppression of PHGDH in MDA-MB-468 cells caused a large reduction in the levels of aKG (Fig. 4c, Supplementary Fig. 7d). In fact, of the major metabolites measured, aKG was the one with the most significant and largest change upon PHGDH suppression, whereas serine levels were not significantly changed (Supplementary Fig. 8). PHGDH suppression also caused a significant reduction in other TCA components (Fig. 4d, Supplementary Fig. 8). Like suppression of PHGDH, suppression of PSAT1 also caused a significant reduction in serine pathway flux and aKG levels (Fig. 4c, Supplementary Fig. 7d–e). Furthermore, labeling studies using U-C13-glutamine revealed that the absolute flux from glutamine to aKG and other TCA intermediates was significantly reduced in cells with RNAi-mediated suppression of PHGDH or PSAT1 (Fig. 4e, Supplementary Fig. 9a–b). These data indicate that in cell lines with high PHGDH expression, the serine synthesis pathway is responsible for approximately 50% of the net conversion of glutamate to aKG and that suppression of PHGDH results in a significant loss of TCA intermediate flux and steady state levels of TCA intermediates (Fig. 4f, Supplementary Fig. 9a–b). Furthermore, labeling studies using U-13C-glucose in cell lines with PHGDH amplification (MDA-MB-468) and without (MDA-MB-231) revealed that in cells with high PHGDH expression, flux through the serine biosynthesis pathway shunts 8–9% of the glycolytic flux towards serine production, compared to 1–2% in the cell line with low PHGDH expression (Fig. 4f and Supplementary Fig. 9a). Therefore, increased flux through the serine biosynthesis pathway has a major impact on aKG production, but a smaller effect on glycolysis or serine availability in these cells (Supplementary Discussion). In contrast, another prominent aKG-producing transaminase, alanine aminotransferase, does not contribute significantly to aKG production in PHGDH-amplified cells (Supplementary Fig. 10).
We find that PHGDH expression is a critical part of a cellular program promoting serine pathway flux (Supplementary Fig. 5) and is responsible for a considerable portion of anaplerosis of glutamate into the TCA cycle as aKG (Supplementary Fig. 1). As ~70% of ER-negative breast cancers exhibit elevated PHGDH (Fig. 2c), our work suggests that targeting the serine synthesis pathway may be therapeutically valuable in breast cancers with elevated PHGDH expression or PHGDH amplifications (Supplementary Discussion). Lastly, we anticipate that the screening approach described here may be applicable to other cancer types or gene sets, enabling the identification of novel cancer targets directly in an in vivo context.
Methods Summary
To undertake negative selection RNAi screening in solid tumours, pools of MCF10DCIS.com cells expressing an shRNA library were injected into the 4th mammary fat pad of immunocompromised mice and allowed to form tumours. Abundances of shRNAs in the tumours was determined using massively parallel sequencing and compared to shRNA abundance in the injected cells. Genes targeted by shRNAs that were significantly depleted during tumour growth were considered hits and prioritized by analyzing gene copy number data from human tumours and cancer cell lines. Lentiviral shRNAs were used to suppress PHGDH expression in breast cancer cell lines with and without PHGDH genomic amplification. Serine synthesis pathway activity and anaplerosis were measured via flux analyses utilizing isotopically labeled molecules.
Supplementary Material
Acknowledgements
We thank members of the Sabatini Lab, Manjae Kwon, Biao Luo, and Ferenc Reinhardt for assistance. This research is supported by fellowships from Susan G. Komen for the Cure to R.L.P. and the Life Science Research Foundation to Y.D.S. and grants from the Keck Foundation, the David H. Koch Institute for Integrative Cancer Research at MIT, The Alexander and Margaret Stewart Trust Fund, and NIH Grant CA103866 to D.M.S. D.M.S. is an investigator of the Howard Hughes Medical Institute.
Footnotes
Supplementary Information is linked to the online version of the paper at www.nature.com/nature.
Author Contributions R.L.P. and D.M.S. conceived the project and designed the experiments. R.L.P performed the screening, knockdown, cell proliferation and tumour formation experiments. K.B., Y.D.S., F.G.B., M.E.P., P.P.S., K.O., Z-Y.T., N.Y.K. and W.W.C assisted with experiments. E.F.B. collected breast cancer patient samples and scored PHGDH IHC. D.K. performed and M.M-K. assisted with interpretation of PHGDH IHC. K.M., S.S., H-K.W., A.K.J. and E.D. performed metabolite profiling and flux experiments. N.S., J.B. and L.A.G. assisted with amplification data. G.S.C and D.E.R. assisted with screening technology. B.Y. and A.M.C. provided bioinformatic support. R.L.P wrote and D.M.S. edited the manuscript.
Author Information Reprints and permissions information is available at www.nature.com/reprints. The authors declare competing financial interests: details accompany the full-text HTML version of the paper at www.nature.com/nature.
References
- 1.Jones RG, Thompson CB. Tumor suppressors and cell metabolism: a recipe for cancer growth. Genes Dev. 2009;23(5):537–548. doi: 10.1101/gad.1756509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hsu PP, Sabatini DM. Cancer cell metabolism: Warburg and beyond. Cell. 2008;134(5):703–707. doi: 10.1016/j.cell.2008.08.021. [DOI] [PubMed] [Google Scholar]
- 3.Bric A, et al. Functional identification of tumor-suppressor genes through an in vivo RNA interference screen in a mouse lymphoma model. Cancer Cell. 2009;16(4):324–335. doi: 10.1016/j.ccr.2009.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Moffat J, et al. A lentiviral RNAi library for human and mouse genes applied to an arrayed viral high-content screen. Cell. 2006;124(6):1283–1298. doi: 10.1016/j.cell.2006.01.040. [DOI] [PubMed] [Google Scholar]
- 5.Miller FR, Santner SJ, Tait L, Dawson PJ. MCF10DCIS.com xenograft model of human comedo ductal carcinoma in situ. J Natl Cancer Inst. 2000;92(14):1185–1186. doi: 10.1093/jnci/92.14.1185a. [DOI] [PubMed] [Google Scholar]
- 6.Beroukhim R, et al. The landscape of somatic copy-number alteration across human cancers. Nature. 463(7283):899–905. doi: 10.1038/nature08822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pollari S, et al. Enhanced serine production by bone metastatic breast cancer cells stimulates osteoclastogenesis. Breast Cancer Res Treat. doi: 10.1007/s10549-010-0848-5. [DOI] [PubMed] [Google Scholar]
- 8.van de Vijver MJ, et al. A gene-expression signature as a predictor of survival in breast cancer. N Engl J Med. 2002;347(25):1999–2009. doi: 10.1056/NEJMoa021967. [DOI] [PubMed] [Google Scholar]
- 9.Miller FR, et al. Xenograft model of progressive human proliferative breast disease. J Natl Cancer Inst. 1993;85(21):1725–1732. doi: 10.1093/jnci/85.21.1725. [DOI] [PubMed] [Google Scholar]
- 10.Snell K. Enzymes of serine metabolism in normal, developing and neoplastic rat tissues. Adv Enzyme Regul. 1984;22:325–400. doi: 10.1016/0065-2571(84)90021-9. [DOI] [PubMed] [Google Scholar]
- 11.Achouri Y, Rider MH, Schaftingen EV, Robbi M. Cloning, sequencing and expression of rat liver 3-phosphoglycerate dehydrogenase. Biochem J. 1997;323(Pt 2):365–370. doi: 10.1042/bj3230365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Walsh DA, Sallach HJ. Purification and properties of chicken liver D-3-phosphoglycerate dehydrogenase. Biochemistry. 1965;4(6):1076–1085. doi: 10.1021/bi00882a015. [DOI] [PubMed] [Google Scholar]
- 13.Knox WE, Herzfeld A, Hudson J. Phosphoserine phosphatase distribution in normal and neoplastic rat tissues. Arch Biochem Biophys. 1969;132(2):397–403. doi: 10.1016/0003-9861(69)90381-6. [DOI] [PubMed] [Google Scholar]
- 14.Lund K, Merrill DK, Guynn RW. The reactions of the phosphorylated pathway of L-serine biosynthesis: thermodynamic relationships in rabbit liver in vivo. Arch Biochem Biophys. 1985;237(1):186–196. doi: 10.1016/0003-9861(85)90268-1. [DOI] [PubMed] [Google Scholar]
- 15.Wiederschain D, et al. Single-vector inducible lentiviral RNAi system for oncology target validation. Cell Cycle. 2009;8(3):498–504. doi: 10.4161/cc.8.3.7701. [DOI] [PubMed] [Google Scholar]
- 16.DeBerardinis RJ, et al. Beyond aerobic glycolysis: transformed cells can engage in glutamine metabolism that exceeds the requirement for protein and nucleotide synthesis. Proc Natl Acad Sci U S A. 2007;104(49):19345–19350. doi: 10.1073/pnas.0709747104. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.