Abstract
Epigenetic changes have been implicated in the pathogenesis of asthma. We sought to determine if IL13, a key cytokine in airway inflammation and remodeling, induced epigenetic DNA methylation and miRNAs expression changes in the airways in conjunction with its transcriptional gene regulation. Inducible expression of an IL13 transgene in the airways resulted in significant changes in DNA methylation in 177 genes, most of which were associated with the IL13 transcriptional signature in the airways. A large number of genes whose expression was induced by IL13 were found to have decreased methylation, including those involved in tissue remodeling (Olr1), leukocyte influx (Cxcl3, Cxcl5, CSFr2b), and the Th2 response (C3ar1, Chi3l4). Reciprocally, some genes whose expression was suppressed were found to have increased methylation (e.g. Itga8). In addition, miRNAs were identified with targets for lung development and Wnt signaling, amongst others. These results indicate that IL13 confers an epigenetic methylation and miRNA signature that accompanies its transcriptional program in the airways, which may play a critical role in airway inflammation and remodeling.
Keywords: Epigenetics, miRNA, DNA methylation, allergic airway disease
Introduction
Asthma is a common chronic inflammatory airway condition with a strong genetic and inheritability component, as siblings and first-degree relatives of those with the disease are often affected [1,2]. However, the remodeling seen in asthma likely reflects a combination of genetic predisposition and environmental exposures. Studies have shown the importance of environmental exposures on the development of asthma. For example, studies have revealed that children exposed to prenatal polycyclic aromatic hydrocarbons and postnatal tobacco smokers were more likely to have difficulty breathing and probable asthma [3,4]. Studies on pregnant mice demonstrated that exposure to aerosolized leachate of residual oil fly ash would lead to the development of several asthma-like symptoms in their offspring, suggesting that inherited epigenetic changes may lead to asthma [5]. This link between environmental pollutants and airway inflammation could result from epigenetic modifications, as cells alter their epigenetic profiles following exposure to environmental toxins [6].
Epigenetic changes such as DNA methylation of CpG dinucleotides within the transcription start sites are common mechanisms for gene expression regulation [7,8]. Many studies have reported that asthma risk and airway inflammation can be influenced by epigenetic regulation [4,9]. It has been shown that methylation of a highly conserved CpG in the Ifng (interferon gamma) promoter is associated with polarizing naïve T cells into the pro-allergic T helper type 2 (Th2) cells [10]. These Th2 cells play a major role in the development of asthma due to their production of pro-inflammatory cytokines [11]. Methylation of CpG islands within the promoter region of ADAM33, a gene important in the development of asthma in human, was shown to regulate the transcription of the gene [12,13]. Apart from DNA methylation, microRNAs (miRNAs) can also regulate gene expression in an epigenetic manner by targeting and degrading specific messenger RNAs [14]. It has been shown that miRNA expression is dysregulated in asthma, and this could be one of the major mechanisms for the initiation and development of the disease [15,16]. Other epigenetic mechanisms in inflammatory cells, such as histone modifications and chromatin remodeling, have also been implicated to play a role in the development of airway inflammation [17,18].
Interleukin 13 (IL13) is a pleiotropic 12 kDa cytokine that is produced and secreted in large quantities by Th2 cells [19-21]. A large number of studies have demonstrated that IL13 is overproduced in asthma and have implicated IL13 in the pathogenesis of Th2 inflammation and airway remodeling [22,23]. Lung-specific constitutive overexpression of IL13 produces airway epithelial cell hypertrophy, macrophage-rich inflammation, subepithelial airway fibrosis, mucus metaplasia, airways hyperresponsiveness, and other physiological changes observed in asthma [24]. IL13 instillation or expression in the airways also induces a recognizable set of genes that mediate various aspects of tissue inflammation and remodeling [25-28]. Importantly, even after the cessation of IL13 treatment, a subgroup of induced genes remain expressed, suggesting the possibility that they maintain autonomous expression through epigenetic modifications [26].
For our studies, we used a well-characterized transgenic mouse model of allergic airway inflammation induced by IL13 [29]. In this model, IL13 is conditionally overexpressed in the mouse lung when treated with doxycycline. Upon IL13 induction, these mice showed inflammatory cell infiltration, pronounced emphysema, increased pulmonary compliance, lung volume enlargement, mucus metaplasia, and increased expression of matrix metalloproteinases and cathepsins in the lung [29]. We used this mouse model to examine genes that were epigenetically regulated by DNA methylation and miRNAs in allergic airway inflammation. To accomplish this, we performed gene expression arrays, methylated DNA immunoprecipitation (MeDIP) arrays, and miRNA expression arrays with RNA or DNA isolated from whole lungs. We found gene expression patterns that correlated with DNA methylation status in genes associated with allergic airway inflammatory processes. We also identified miRNAs that were differentially expressed with targets for inflammatory genes. These results suggested that IL13 rapidly induced epigenetic responses that could contribute to the regulation of genes involved in allergic airway inflammation.
Materials and methods
Transgenic mice
Mice were housed and bred under the regulation of the Division of Laboratory Animal Medicine at the University of California, Los Angeles. The CC10-rtTA-IL13 transgenic (TG) mouse is a well-characterized model of allergic airway disease [29]. Briefly, the Clara cell 10-kDa (CC10) gene promoter was used to conditionally express IL13 in the mouse lung in a doxycycline-inducible fashion. For these experiments, mice were exposed to doxycycline for a time period of either 1 week or 4 weeks. The control group consisted of wild type (WT) mice that were exposed to doxycycline. Baseline leakiness of the IL13 transgene expression was found with these mice so that even in the absence of doxycycline they have slightly elevated IL13 and mild allergic airway inflammation [29]. Therefore, doxycycline untreated transgenic mice were not used as controls in our studies.
After exposure to the appropriate time period of doxycycline, mice were euthanized and the lungs were removed for subsequent studies described below.
Gene expression analysis
TG and WT mice were treated with doxycycline for one week. Mice were euthanized and the left upper lobes from all mice were removed for RNA extraction using the TRIzol method. The RNA quality was checked using an Agilent 2100 Bioanalyzer, prior to amplification of RNA and subsequent microarray hybridization on the Affymetrix 430 2.0 array, following standard Affymetrix protocols. Raw gene expression data (.cel files) were generated by standard Affymetrix protocols and deposited in the Gene Expression Omnibus of the National Center for Biotechnology Information (accession number GSE37085). The import and normalization of the raw data were completed by Bioconductor in the R software environment. Two types of statistical tests (t-test and rank product) were performed for each probe set in order to detect differentially expressed genes between the TG and WT lung tissue. For both methods, only genes with an adjusted P-value < 0.01 and fold change ≥ 2 based on the false discovery rate were considered as differentially expressed genes and were annotated.
MeDIP method and data analysis
TG and WT mice were treated with doxycycline for one week. Mice were then euthanized and lungs were surgically removed. The DNA was extracted from the left lower lobes of the lungs. Methylated DNA immunoprecipitation (MeDIP) was performed per the Nimblegen MeDIP array protocol. Briefly, an antibody that recognizes 5-methylcytosine was used to target the methylated fraction of the genome resulting in enrichment of CpG islands [30]. The MeDIP fractions were hybridized to Nimblegen 2.1M Deluxe Promoter Arrays that cover ~10 kb of all annotated promoters. The MeDIP data was visualized using the Affymetrix Integrated Genome Browser (www.affymetrix.com/partners_programs/programs/developer/tools/download_igb.affx)
Correlation of MeDIP methylation data and gene expression data
To identify regions of significant enrichment in MeDIP, we used a z-score analysis to reflect methylation status for a selected region. For Z-score calculations, the log ratios for each Nimblegen array were normalized by subtracting their mean log ratio values. Outlier probes with log ratio values greater than two were set to two. To measure changes in methylation between samples we computed the average log ratios of all probes within a 2 kb window centered on each probe; only windows with 5 or more probes were considered for analysis. These were then normalized to generate a Z-score for the window using the following equation: z = MeanWindow *SquareRoot (#probes in window)/Standard Deviation of Array.
The probes within these windows were then averaged, and a z-score was computed by subtracting the average value of probes on the entire array and dividing by the standard deviation of probes in the array. The resulting value was then multiplied by the square root of the number of probes within the window to obtain the final normalized z-score. We computed the z-scores for both samples, and reported the difference in z-scores between them. Each of these regions was associated with the nearest gene, whose transcriptional start site was within 2 kb. We then generated a list of regions that had significant differences in z-scores (indicating a change in methylation) along with a significant change in gene expression.
Statistics
All experiments were performed with at least three different primary cultures or mice in independent experiments except for the MeDIP experiment, which was performed once as a discovery experiment for validation in more tissue. Significance was evaluated by Student’s t-test.
MicroRNA expression analysis and correlation with gene expression
The CC10-rtTA-IL13 TG mice were treated with doxycycline for 1 week. The control group consisted of WT mice treated with doxycycline. After exposure, lungs were removed and RNA was extracted from left upper lobes in all mice. RNA extracts were amplified followed by microarray hybridization on Affymetrix GeneChip miRNA array (released in 2009) to profile the miRNA expressions in WT and TG mice. The raw expression data were first processed for background estimation and correction, normalization, and summarization using miRNAQCTools, an Affymetrix software specifically developed for the GeneChip miRNA arrays (http://www.affymetrix.com/estore/browse/level_seven_software_products_only.jsp?productId=131558&categoryId =cat50004&productName=miRNA-QC-Tool#1_1). The processed expression data were then log2-transformed and analyzed with the significant analysis of microarray (SAM) software [31]. The miRNA genes with q-value less than 0.05 and fold change above 2 were chosen as significantly regulated and used for further analysis. Matlab was used to analyze mRNA data. Raw data from six microarrays were first normalized followed by a filter to eliminate probes with a maximum intensity less than 100. The gene fold changes between TG and WT groups were calculated. Targetscan database was used to predict the targets of miRNAs, and only targets with the context+ score less than - 0.35 were considered. The correlations of the miRNAs on gene expression were estimated by computing the z-score, which is the averaged expression fold change of targeted genes between TG and WT subtracted by the average expression fold change of all genes, multiplied by the square root of the number of targets, and divided by the standard deviation of the expression fold change of all genes. DAVID Bioinformatics Resources were used to match microarray probe to genes and to obtain gene functional annotations. Published data set (GSE18010) by Tachdjian et al. [27], which used identical mice treated with the same conditions, was used to compare and verify our results.
Quantitative real-time PCR
For the validation of gene expression data by quantitative real-time PCR, TaqMan primers and probes (for Cxcl3) and SYBR Green primers and probes (for Pdgfra, Prkca) were obtained from Applied Biosystems. RNA was isolated from left upper lobes of the lungs from TG and WT mice treated with doxycycline for one week. Standard protocols recommended by Applied Biosystems were followed for the experiments. The expression levels of the candidate genes were compared to that of Gapdh, which was used as an endogenous control. The data are presented in ΔCT, which is obtained by subtracting the CT values of Gapdh from the CT values of the candidate genes. A smaller ΔCT value indicates higher expression level. The PCR reactions were performed on the Applied Biosystems Step One Plus Real-Time PCR System.
Methylation-specific PCR
For the validation of MeDIP data, we compared the promoter region of Cxcl3 in TG and WT mice by methylation-specific PCR (MS-PCR). Mice were treated the same way as those used in the MeDIP experiment, and genomic DNA was extracted from the left lower lobes of the lungs. Bisulfite conversion was performed on the extracted genomic DNA using the Zymo Research EZ DNA Methylation-Direct Kit. Bisulfite-converted DNA was used as template for MS-PCR. The primers were designed using the MethPrimer website (www.urogene.org/methprimer/index1.html). We searched for potential primer sites within the 2 kb window where the z-score was calculated for Cxcl3 in MeDIP data analysis. Two sets of primers were designed, one primed for fully methylated DNA (M primers), while the other primed for fully unmethylated DNA (U primers). The primer sequences are, methylated forward primer: 5’-AAT AGA AAT AAA TGT AC GGG GAT TC-3’; methylated reverse primer: 5’-AAT AGA AAT AAA TGT ACG GGG ATT C-3’; unmethylated forward primer: 5’-TAG AAA TAA ATG TAT GGG GAT TTG A-3’; unmethylated reverse primer: 5’- TAGAA ATA AAT GTA TGG GGA TTT GA-3’. The thermal cycler setting was as follow: 2 minutes at 95°C, 40 cycles of [30 seconds at 95°C, 30 seconds at 53°C, 1 minute at 72°C], 5 minutes at 72°C, hold at 4° C. Amplified products were visualized on a 2% agarose gel stained with Safe View Classic. Expected sizes for the amplified regions are 185 bp for the M primers and 182 bp for the U primers.
Results
Identification of methylation patterns that correspond with gene expression levels in IL13-induced allergic airway inflammation
Gene expression profiling of CC10-raTA-IL13 TG mouse lung revealed a similar gene signature of allergic airway inflammation that has been described previously [27]. IL13 induction with doxycycline for 7 days significantly modified DNA methylation patterns in 177 unique genes, most of which were associated with the IL13 transcriptional signature in the airways. A list of selected genes with differential expression level and DNA methylation status is shown in Table 1. The z-scores reflect methylation status: a negative number indicates a hypomethylated state; positive number indicates a methylated state. The z-scores were computed within a 2 kb window that contained 5 or more probes, centered on the probe location listed in the table.
Table 1.
Gene expression and promoter methylation profiling in TG and WT mice treated with doxycycline for 1 week
| Gene | Transcript fold change (log2) | WT z-score | TG z-score | z-score difference | Probe location |
|---|---|---|---|---|---|
| Fcer2a | 4.53 | 4.77 | 3.61 | -1.16 | 3695142 |
| Cxcl5 | 4.42 | 2.20 | 0.60 | -1.60 | 91191474 |
| Cxcl3 | 4.39 | 2.61 | 1.36 | -1.25 | 91217994 |
| Slc39a2 | 4.38 | 4.69 | 3.29 | -1.40 | 52515016 |
| Olr1 | 4.25 | 0.45 | -1.51 | -1.96 | 129457700 |
| Csf2rb2 | 4.06 | 4.61 | 2.47 | -2.14 | 78136104 |
| Msr1 | 3.97 | 1.01 | -1.37 | -2.39 | 40729897 |
| C3ar1 | 3.75 | -0.72 | -2.08 | -1.36 | 122807674 |
| Pik3r5 | 3.48 | 2.62 | 0.98 | -1.65 | 68311677 |
| Lilrb3 | 3.29 | 0.03 | -1.06 | -1.09 | 3672988 |
| Fcgr3 | 3.18 | 1.16 | -1.11 | -2.27 | 172991171 |
| Abcd2 | 3.12 | -0.01 | -1.79 | -1.78 | 91023786 |
| Igf2bp3 | 3.07 | 0.21 | -1.53 | -1.74 | 49165953 |
| Chi3l4 | 2.72 | -1.73 | -3.13 | -1.41 | 106023657 |
A large number of genes whose expression was induced by IL13 were found to have decreased methylation, including those involved in tissue remodeling (Olr1), leukocyte influx (Cxcl3, Cxcl5, CSFr2b), and the Th2 response (C3ar1, Chi3l4). Reciprocally, some genes whose expression is suppressed were found to have increased methylation. Among them are Itga8, Rbpms, Bex2, and Palld (z-score difference: 1.99, 0.93, 4.37, and 0.68, respectively).
Validation of gene expression and DNA promoter methylation for Cxcl3
We chose Cxcl3 as a candidate for the validation of gene expression from the microarray data and promoter methylation identified by MeDIP. To validate gene expression, we performed quantitative real-time PCR for Cxcl3 using RNA isolated from lungs of 3 TG mice and 3 WT mice treated with doxycycline for a week. We found increased expression of the gene in the lungs of TG mice compared to that of WT mice (p = 0.0034) (Figure 1A). The real-time PCR data thus confirmed the gene expression microarray data.
Figure 1.
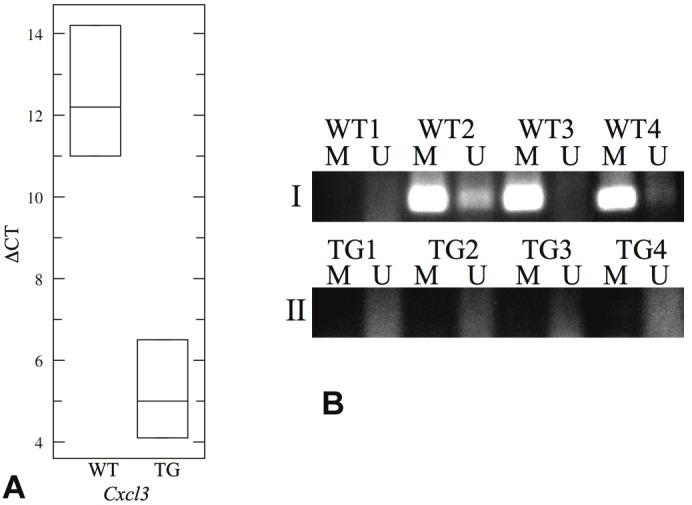
Data validation of Cxcl3 gene expression and DNA methylation status in WT and TG mice. (A) Gene expression data of Cxcl3 from the Affymetrix 430 2.0 array was validated by quantitative real-time PCR. The box-plot presents the data in ΔCT, which is the difference between the CT values of Cxcl3 and Gapdh. A higher ΔCT value indicates lower gene expression. Cxcl3 expression was found to be higher in the TG lungs compared to the WT lungs (p = 0.0034), which validated our gene expression array data. (B) Methylation-specific PCR was performed to validate MeDIP array data for Cxcl3. Panel I shows PCR-amplified fragments from WT mice while Panel II shows amplified fragments from TG mice. M columns represent amplified products by the M primers while U columns represent amplified products by the U primers. Three out of 4 WT mice harbored methylated CpGs in the investigated Cxcl3 promoter region, while all 4 of the TG mice did not. These data correlate with the MeDIP array data that promoter CpG methylation is lost in the transition from WT lungs to IL13-induced TG lungs, which explains the increased Cxcl3 expression in the TG lungs.
To validate the methylation status of Cxcl3 promoter that was determined by MeDIP, we performed bisulfite conversion on the genomic DNA extracted from 4 TG mice and 4 WT mice, followed by methylation-specific PCR on the promoter region used for z-score calculation in the MeDIP array data. There were more methylated CpG sites than unmethylated CpG sites in 3 out of 4 WT lungs, whereas all TG lungs had more unmethylated CpG sites in the same promoter region (Figure 1B). These findings verified the results from MeDIP, which indicated that CpG methylation of the Cxcl3 promoter was higher in the WT than in the TG lungs shown in Table 1. These results suggested that IL13 induces methylation modifications in the genome within a week of exposure to overexpression of IL13 and that this is reflected by a change in the transcriptional signature.
Identification of differentially expressed miRNAs and their target genes in IL13-induced allergic airway inflammation
Expression of seven miRNAs from the Affymetrix GeneChip miRNA array were significantly induced in the TG lungs compared to WT (Table 2). Two of the miRNAs examined in the array, mmu-miR-685 and mmu-miR-699, however, were discovered to be fragments of RNase P RNA and RNase MRP RNA, respectively. These two miRNAs were since withdrawn from the miRBase and were not included in our subsequent analysis. Using the Targetscan database, the potential targets of the miRNAs were predicted. Correlating back to the Affymetrix 430 2.0 array for gene expression analysis, we computed the z-score for each miRNA, which determined the degree of differential expression of the predicted target genes for that particular miRNA in terms of standard deviations away from the mean expression level of all genes on the array. With the exception of mmu-miR-127, which did not return any target that passed the cutoff of context+ score < -0.35, all 4 of the remaining miRNAs had negative z-scores, indicating the predicted target genes had lower expression levels compared to the overall genome-wide expression levels (Table 2). Of the target genes downregulated by the induced miRNAs, some were involved in translation regulation (Tnrc6b, El24), transcription regulation (Foxn3, Plagl2), signal transduction (Prkca, Lphn3), and proteolysis (Ube2e3, Dpp8). A list of selected target genes downregulated in TG lungs with q-values less than 0.05 is shown in Table 3.
Table 2.
miRNAs with significant differential expression (fold change > 2 and q-value < 0.05) and corresponding z-scores for their targeted genes.
| miRNA | Fold change (log2) | z-score |
|---|---|---|
| mmu-miR-685 | 2.0244 | --- |
| mmu-miR-379 | 1.188 | -4.1521 |
| mmu-miR-699 | 1.0953 | --- |
| mmu-miR-362 | 1.0657 | -0.9579 |
| mmu-miR-146b | 1.5057 | -0.078687 |
| mmu-miR-127 | 2.062 | --- |
| mmu-miR-34a | 1.5613 | -0.64695 |
Table 3.
List of selected genes targeted by miRNAs significantly overexpressed in TG mice.
| Target gene | Fold change (log2) | Corresponding miRNA | Description of function |
|---|---|---|---|
| Lphn3 | -1.29984 | mmu-miR-379 | Cell surface receptor linked signal transduction, neuropeptide signaling |
| Prkca | -0.9328 | mmu-miR-362 | Wnt signaling pathway, MAPK signaling pathway |
| Pdgfra | -0.9101 | mmu-miR-34a | Lung development, respiratory tube development |
| Tnrc6B | -0.65567 | mmu-miR-379 | Translation regulation |
| Znrf3 | -0.59586 | mmu-miR-146 | Posttranslational modification, chaperones |
| Zfp644 | -0.50939 | mmu-miR-362 | Zinc ion binding, transition metal ion binding |
| Klhl2 | -0.43732 | mmu-miR-362 | Cytoskeleton actin filament bundle |
| Ube2e3 | -0.42316 | mmu-miR-379 | Proteolysis, catabolic processes |
| Eif4g2 | -0.12771 | mmu-miR-379 | Translation regulation |
Validation of miRNA target genes expression
We performed quantitative real-time PCR to validate the downregulation of 2 target genes shown in Table 3. Pdgfra and Prkca, both targets of miRNAs induced in TG mice, were chosen as candidates for the validation study. Four TG mice and four WT mice were sacrificed after being treated with doxycycline for 7 days, and RNA was isolated from the upper left lobes of the lungs. From the real-time PCR data, both Pdgfra and Prkca showed lower expression levels in the lungs of TG mice compared to WT mice, validating the expression patterns indicated in the Affymetrix 430 2.0 array data (Figure 2).
Figure 2.
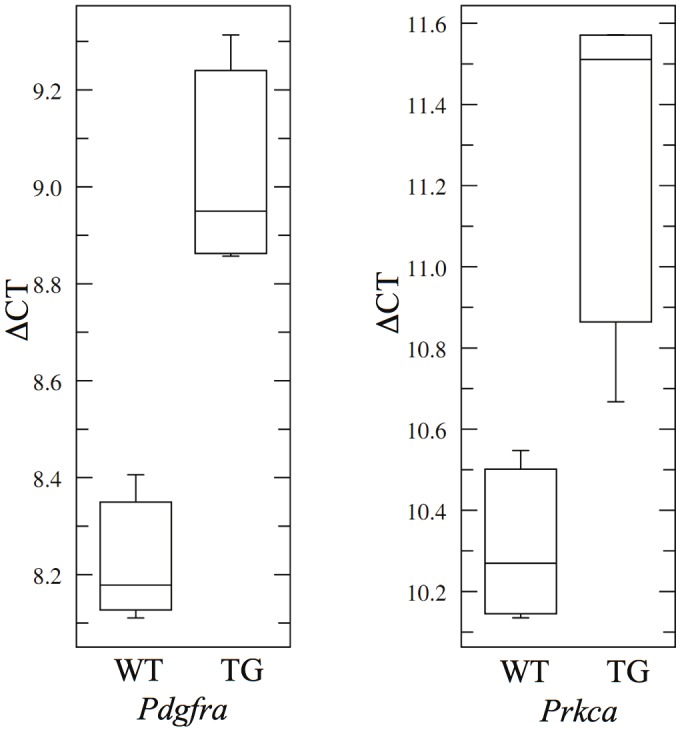
Quantitative real-time PCR validating the expression differences between TG and WT lungs for selected genes targeted by miRNAs that were upregulated in TG mice. Genes chosen for validation study were Pdgfra (targeted by mmu-miR-34a) and Prkca (targeted by mmu-miR-362). Both of these genes were found to be downregulated in the TG lungs (p = 0.0006 and 0.0054, respectively), confirming the data from the gene expression array. The ΔCT values were calculated by subtracting the CT values of the endogenous control Gapdh from the CT values of the target genes.
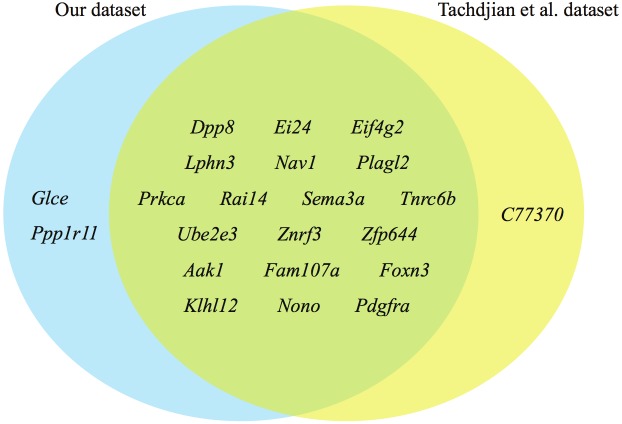
To validate the expression patterns of more target genes, we examined the array dataset published by Tachdjian et al. (GSE18010) [27]. We compared the downregulated genes targeted by the miRNAs listed in Table 2 with the list generated by our dataset (Figure 3). We saw a significant overlap of downregulated target genes between the two datasets, with only one uniquely downregulated target gene in Tachdjian dataset, while 2 uniquely downregulated target genes in our dataset. This analysis showed that in IL13-induced allergic airway inflammation, these genes were consistently downregulated in the lungs, most probably due to epigenetic regulation of miRNAs.
Figure 3.
Comparison of downregulated genes that were potential targets of overexpressed miRNAs in the TG mice. The blue region shows 2 genes unique to our dataset; the yellow region shows a single gene unique to Tachdjian et al’s dataset; the green region shows common genes downregulated in both our datasets that are potential targets of overexpressed miRNAs in the TG mice.
Discussion
In this study, we demonstrate that IL13 expression in the airways induces epigenetic changes, involving coordinate changes in the DNA methylation profile of target gene promoters and the induction of a miRNA transcriptome, that may play a critical role in the establishment of allergic airway inflammation and remodeling. Although there have been some studies focusing on DNA methylation of specific genes in asthma, to our knowledge this is the first genome-wide analysis of DNA methylation status in experimental asthma, with the results contextualized by linking them to the pro-asthmatic transcriptional changes induced by IL13 [13,32].
From our MeDIP array data and the gene expression array data, we found genes with increased expression in the TG lungs compared to the WT lungs that corresponded to the loss of DNA promoter methylation. A case in point is the macrophage scavenger receptor 1 (Msr1), which is important in COPD and is upregulated in smokers [33,34]. MSR1 is recently shown to be physically interacting with TRAF6 in human [35], which is a signal transducer for interleukin-1 (IL1) [36]. IL1 is a pro-inflammatory cytokine pivotal in the development of Th2 allergic responses [37]. Here we showed that Msr1 expression was upregulated during allergic airway inflammation, coordinate with the demethylation of its promoter. We also discovered some genes that were hypermethylated and that correlated with decreased gene expression in the TG lungs. One example is the Itga8 gene, which is highly expressed in rat alveolar interstitial fibroblasts under normal conditions [38,39]. Its expression was repressed in the bacterial endotoxin mouse model of bronchopulmonary dysplasia [40]. ITGA8 was also shown to regulate lung morphogenesis by controlling the migration and adhesion of mesenchymal cells [40]. Furthermore, CpG methylation of the Itga8 promoter has been described in some cases of human ovarian cancer [41]. Our data suggests that Itga8 expression is repressed in asthma via DNA promoter methylation, leading to dysregulated lung morphogenesis. Overall, our results implicate epigenetic methylation changes in the coordination and consolidation of IL13-regulated transcriptional circuits involving both induced and repressed genes.
Another aspect of epigenetic regulation, the expression of miRNAs, was also examined in this study. There are a few published studies on miRNAs in asthma using different methodologies and producing different results. In one study in which a real-time PCR approach was employed to investigate the expression of 227 miRNAs in airway biopsies procured from normal subjects and patients with mild asthma, no differential miRNA expression was found [42]. Lu et al. used the same doxycycline inducible IL13 transgenic mice but examined miRNA expression after 28 days of doxycycline on a different microarray platform. This study found 21 miRNAs that were differentially expressed in the doxycycline treated versus untreated mice [43]. In our study, we compared miRNA expression levels in doxycycline-treated inducible IL13 transgenic mice as compared to wild type syngeneic mice due to leakiness of the rtTA-CMV cassette driving IL13 expression. After correlating miRNA expression with mRNA expression, we found that only one miRNA, mir146b, overlapped with the miRNAs identified by Lu et al. We examined miRNAs whose expression was induced in the TG lungs and identified their predicted target genes that were downregulated in the disease state. From these data, we compiled a list of miRNA-regulated genes that play a role in the development of allergic airway inflammation.
One example of a gene regulated by an IL13-induced miRNA is Prkca, encoding protein kinase C alpha. This protein kinase is involved in various biological pathways within the cell, including apoptosis, cell cycle regulation, proliferation, cell motility, and cell morphology [44,45]. Single-nucleotide polymorphisms (SNPs) within the PRKCA gene in humans is significantly associated with asthma [46]. Another miRNA-targeted gene is Pdgfra, encoding platelet-derived growth factor receptor alpha, a cell surface catalytic tyrosine kinase receptor that has intracellular activity. The platelet-derived growth factor (PDGF) signaling pathway is important in lung development, especially lung growth and the formation of alveoli [47,48]. SNPs in the promoter region of human PDGFRA have been linked to the severity of childhood asthma [49]. We showed here that Prkca and Pdgfra expression were decreased in IL13-induced allergic airway inflammation via miRNA regulation. Thus, our results further implicate miRNA induction as yet another epigenetic mechanism by which IL13 controls gene expression circuitries.
In summary, we have found that IL13 expression in the airways is associated with coordinate changes in the methylation status of promoters of a large number of gene components of the IL13 transcriptome, consistent with an epigenetic regulatory function of gene methylation in allergic airway inflammation. We similarly found miRNAs with predicted targets for genes that are important in maintaining expression of transcriptional circuitries associated with chronic airway disease. Other studies have also implicated additional mechanisms, such as histone modifications, in promoting gene expression changes in asthma [18,50]. These findings point to the involvement of multiple mechanisms of epigenetic regulation in the establishment of allergic airway inflammation and in the evolution of its chronic phenotypes and manifestations. Overall, our studies and those from other groups, suggest that interventions that manipulate the epigenome of the asthmatic lung may provide highly effective therapies in the future.
Acknowledgments
We thank Dr. Jack Elias for the provision of the CC10-rtTA-IL13 mice. This work was supported by NIH grant R01 HL094561-01 (to BG) and 2R01AI065617-11A1 and U19 AI070453 (to TAC).
References
- 1.Cantani A, Micera M. A study on 300 asthmatic children, 300 controls and their parents confirms the genetic transmission of allergy and asthma. Eur Rev Med Pharmacol Sci. 2011;15:1051–1056. [PubMed] [Google Scholar]
- 2.Kurzius-Spencer M, Guerra S, Sherrill DL, Halonen M, Elston RC, Martinez FD. Familial aggregation of allergen-specific sensitization and asthma. Pediatr Allergy Immunol. 2012;23:21–27. doi: 10.1111/j.1399-3038.2011.01220.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Miller RL, Garfinkel R, Horton M, Camann D, Perera FP, Whyatt RM, Kinney PL. Polycyclic aromatic hydrocarbons, environmental tobacco smoke, and respiratory symptoms in an inner-city birth cohort. Chest. 2004;126:1071–1078. doi: 10.1378/chest.126.4.1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Miller RL, Ho SM. Environmental epigenetics and asthma: current concepts and call for studies. Am J Respir Crit Care Med. 2008;177:567–573. doi: 10.1164/rccm.200710-1511PP. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hamada K, Suzaki Y, Leme A, Ito T, Miyamoto K, Kobzik L, Kimura H. Exposure of pregnant mice to an air pollutant aerosol increases asthma susceptibility in offspring. J Toxicol Environ Health A. 2007;70:688–695. doi: 10.1080/15287390600974692. [DOI] [PubMed] [Google Scholar]
- 6.Bollati V, Baccarelli A. Environmental epigenetics. Heredity (Edinb) 2010;105:105–112. doi: 10.1038/hdy.2010.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Weber M, Hellmann I, Stadler MB, Ramos L, Paabo S, Rebhan M, Schubeler D. Distribution, silencing potential and evolutionary impact of promoter DNA methylation in the human genome. Nat Genet. 2007;39:457–466. doi: 10.1038/ng1990. [DOI] [PubMed] [Google Scholar]
- 8.Reik W, Dean W. DNA methylation and mammalian epigenetics. Electrophoresis. 2001;22:2838–2843. doi: 10.1002/1522-2683(200108)22:14<2838::AID-ELPS2838>3.0.CO;2-M. [DOI] [PubMed] [Google Scholar]
- 9.Kabesch M, Michel S, Tost J. Epigenetic mechanisms and the relationship to childhood asthma. Eur Respir J. 2010;36:950–961. doi: 10.1183/09031936.00019310. [DOI] [PubMed] [Google Scholar]
- 10.Jones B, Chen J. Inhibition of IFN-gamma transcription by site-specific methylation during T helper cell development. EMBO J. 2006;25:2443–2452. doi: 10.1038/sj.emboj.7601148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Barnes PJ. Th2 cytokines and asthma: an introduction. Respir Res. 2001;2:64–65. doi: 10.1186/rr39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shapiro SD, Owen CA. ADAM-33 surfaces as an asthma gene. N Engl J Med. 2002;347:936–938. doi: 10.1056/NEJMcibr022144. [DOI] [PubMed] [Google Scholar]
- 13.Yang Y, Haitchi HM, Cakebread J, Sammut D, Harvey A, Powell RM, Holloway JW, Howarth P, Holgate ST, Davies DE. Epigenetic mechanisms silence a disintegrin and metalloprotease 33 expression in bronchial epithelial cells. J Allergy Clin Immunol. 2008;121:1393–1399. doi: 10.1016/j.jaci.2008.02.031. 1399 e1391-1314. [DOI] [PubMed] [Google Scholar]
- 14.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 15.Garbacki N, Di Valentin E, Huynh-Thu VA, Geurts P, Irrthum A, Crahay C, Arnould T, Deroanne C, Piette J, Cataldo D, Colige A. MicroRNAs profiling in murine models of acute and chronic asthma: a relationship with mRNAs targets. PLoS One. 2011;6:e16509. doi: 10.1371/journal.pone.0016509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Polikepahad S, Knight JM, Naghavi AO, Oplt T, Creighton CJ, Shaw C, Benham AL, Kim J, Soibam B, Harris RA, Coarfa C, Zariff A, Milosavljevic A, Batts LM, Kheradmand F, Gunaratne PH, Corry DB. Proinflammatory role for let-7 microRNAS in experimental asthma. J Biol Chem. 2010;285:30139–30149. doi: 10.1074/jbc.M110.145698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Agarwal S, Rao A. Modulation of chromatin structure regulates cytokine gene expression during T cell differentiation. Immunity. 1998;9:765–775. doi: 10.1016/s1074-7613(00)80642-1. [DOI] [PubMed] [Google Scholar]
- 18.Adcock IM, Tsaprouni L, Bhavsar P, Ito K. Epigenetic regulation of airway inflammation. Curr Opin Immunol. 2007;19:694–700. doi: 10.1016/j.coi.2007.07.016. [DOI] [PubMed] [Google Scholar]
- 19.McKenzie AN, Culpepper JA, de Waal Malefyt R, Briere F, Punnonen J, Aversa G, Sato A, Dang W, Cocks BG, Menon S, de Vries JE, Banchereau J, Zurawski G. Interleukin 13, a T-cell-derived cytokine that regulates human monocyte and B-cell function. Proc Natl Acad Sci USA. 1993;90:3735–3739. doi: 10.1073/pnas.90.8.3735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Minty A, Chalon P, Derocq JM, Dumont X, Guillemot JC, Kaghad M, Labit C, Leplatois P, Liauzun P, Miloux B, Casellas P, Loison G, Lupker J, Shire D, Ferrara P, Caput D. Interleukin-13 is a new human lymphokine regulating inflammatory and immune responses. Nature. 1993;362:248–250. doi: 10.1038/362248a0. [DOI] [PubMed] [Google Scholar]
- 21.Wynn TA. IL-13 effector functions. Annu Rev Immunol. 2003;21:425–456. doi: 10.1146/annurev.immunol.21.120601.141142. [DOI] [PubMed] [Google Scholar]
- 22.Kotsimbos TC, Ernst P, Hamid QA. Interleukin-13 and interleukin-4 are coexpressed in atopic asthma. Proc Assoc Am Physicians. 1996;108:368–373. [PubMed] [Google Scholar]
- 23.Wills-Karp M, Luyimbazi J, Xu X, Schofield B, Neben TY, Karp CL, Donaldson DD. Interleukin-13: central mediator of allergic asthma. Science. 1998;282:2258–2261. doi: 10.1126/science.282.5397.2258. [DOI] [PubMed] [Google Scholar]
- 24.Zhu Z, Homer RJ, Wang Z, Chen Q, Geba GP, Wang J, Zhang Y, Elias JA. Pulmonary expression of interleukin-13 causes inflammation, mucus hypersecretion, subepithelial fibrosis, physiologic abnormalities, and eotaxin production. J Clin Invest. 1999;103:779–788. doi: 10.1172/JCI5909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Follettie MT, Ellis DK, Donaldson DD, Hill AA, Diesl V, DeClercq C, Sypek JP, Dorner AJ, Wills-Karp M. Gene expression analysis in a murine model of allergic asthma reveals overlapping disease and therapy dependent pathways in the lung. Pharmacogenomics J. 2006;6:141–152. doi: 10.1038/sj.tpj.6500357. [DOI] [PubMed] [Google Scholar]
- 26.Fulkerson PC, Fischetti CA, Hassman LM, Nikolaidis NM, Rothenberg ME. Persistent effects induced by IL-13 in the lung. Am J Respir Cell Mol Biol. 2006;35:337–346. doi: 10.1165/rcmb.2005-0474OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tachdjian R, Mathias C, Al Khatib S, Bryce PJ, Kim HS, Blaeser F, O'Connor BD, Rzymkiewicz D, Chen A, Holtzman MJ, Hershey GK, Garn H, Harb H, Renz H, Oettgen HC, Chatila TA. Pathogenicity of a disease-associated human IL-4 receptor allele in experimental asthma. J Exp Med. 2009;206:2191–2204. doi: 10.1084/jem.20091480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zimmermann N, Doepker MP, Witte DP, Stringer KF, Fulkerson PC, Pope SM, Brandt EB, Mishra A, King NE, Nikolaidis NM, Wills-Karp M, Finkelman FD, Rothenberg ME. Expression and regulation of small proline-rich protein 2 in allergic inflammation. Am J Respir Cell Mol Biol. 2005;32:428–435. doi: 10.1165/rcmb.2004-0269OC. [DOI] [PubMed] [Google Scholar]
- 29.Zheng T, Zhu Z, Wang Z, Homer RJ, Ma B, Riese RJ Jr, Chapman HA Jr, Shapiro SD, Elias JA. Inducible targeting of IL-13 to the adult lung causes matrix metalloproteinase- and cathepsin-dependent emphysema. J Clin Invest. 2000;106:1081–1093. doi: 10.1172/JCI10458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li N, Ye M, Li Y, Yan Z, Butcher LM, Sun J, Han X, Chen Q, Zhang X, Wang J. Whole genome DNA methylation analysis based on high throughput sequencing technology. Methods. 2010;52:203–212. doi: 10.1016/j.ymeth.2010.04.009. [DOI] [PubMed] [Google Scholar]
- 31.Tusher VG, Tibshirani R, Chu G. Significance analysis of microarrays applied to the ionizing radiation response. Proc Natl Acad Sci USA. 2001;98:5116–5121. doi: 10.1073/pnas.091062498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Koppelman GH, Nawijn MC. Recent advances in the epigenetics and genomics of asthma. Curr Opin Allergy Clin Immunol. 2011;11:414–419. doi: 10.1097/ACI.0b013e32834a9573. [DOI] [PubMed] [Google Scholar]
- 33.Heguy A, O'Connor TP, Luettich K, Worgall S, Cieciuch A, Harvey BG, Hackett NR, Crystal RG. Gene expression profiling of human alveolar macrophages of phenotypically normal smokers and nonsmokers reveals a previously unrecognized subset of genes modulated by cigarette smoking. J Mol Med (Berl) 2006;84:318–328. doi: 10.1007/s00109-005-0008-2. [DOI] [PubMed] [Google Scholar]
- 34.Ohar JA, Hamilton RF Jr, Zheng S, Sadeghnejad A, Sterling DA, Xu J, Meyers DA, Bleecker ER, Holian A. COPD is associated with a macrophage scavenger receptor-1 gene sequence variation. Chest. 2010;137:1098–1107. doi: 10.1378/chest.09-1655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yu X, Yi H, Guo C, Zuo D, Wang Y, Kim HL, Subjeck JR, Wang XY. Pattern recognition scavenger receptor CD204 attenuates Toll-like receptor 4-induced NF-kappaB activation by directly inhibiting ubiquitination of tumor necrosis factor (TNF) receptor-associated factor 6. J Biol Chem. 2011;286:18795–18806. doi: 10.1074/jbc.M111.224345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cao Z, Xiong J, Takeuchi M, Kurama T, Goeddel DV. TRAF6 is a signal transducer for interleukin-1. Nature. 1996;383:443–446. doi: 10.1038/383443a0. [DOI] [PubMed] [Google Scholar]
- 37.Schmitz N, Kurrer M, Kopf M. The IL-1 receptor 1 is critical for Th2 cell type airway immune responses in a mild but not in a more severe asthma model. Eur J Immunol. 2003;33:991–1000. doi: 10.1002/eji.200323801. [DOI] [PubMed] [Google Scholar]
- 38.Levine D, Rockey DC, Milner TA, Breuss JM, Fallon JT, Schnapp LM. Expression of the integrin alpha8beta1 during pulmonary and hepatic fibrosis. Am J Pathol. 2000;156:1927–1935. doi: 10.1016/s0002-9440(10)65066-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Schnapp LM, Breuss JM, Ramos DM, Sheppard D, Pytela R. Sequence and tissue distribution of the human integrin alpha 8 subunit: a beta 1-associated alpha subunit expressed in smooth muscle cells. J Cell Sci. 1995;108:537–544. doi: 10.1242/jcs.108.2.537. [DOI] [PubMed] [Google Scholar]
- 40.Benjamin JT, Gaston DC, Halloran BA, Schnapp LM, Zent R, Prince LS. The role of integrin alpha8beta1 in fetal lung morphogenesis and injury. Dev Biol. 2009;335:407–417. doi: 10.1016/j.ydbio.2009.09.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cai LY, Abe M, Izumi S, Imura M, Yasugi T, Ushijima T. Identification of PRTFDC1 silencing and aberrant promoter methylation of GPR150, ITGA8 and HOXD11 in ovarian cancers. Life Sci. 2007;80:1458–1465. doi: 10.1016/j.lfs.2007.01.015. [DOI] [PubMed] [Google Scholar]
- 42.Williams AE, Larner-Svensson H, Perry MM, Campbell GA, Herrick SE, Adcock IM, Erjefalt JS, Chung KF, Lindsay MA. MicroRNA expression profiling in mild asthmatic human airways and effect of corticosteroid therapy. PLoS One. 2009;4:e5889. doi: 10.1371/journal.pone.0005889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lu TX, Munitz A, Rothenberg ME. MicroRNA-21 is up-regulated in allergic airway inflammation and regulates IL-12p35 expression. J Immunol. 2009;182:4994–5002. doi: 10.4049/jimmunol.0803560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nishizuka Y. Protein kinase C and lipid signaling for sustained cellular responses. FASEB J. 1995;9:484–496. [PubMed] [Google Scholar]
- 45.Michie AM, Nakagawa R. The link between PKCalpha regulation and cellular transformation. Immunol Lett. 2005;96:155–162. doi: 10.1016/j.imlet.2004.08.013. [DOI] [PubMed] [Google Scholar]
- 46.Murphy A, Tantisira KG, Soto-Quiros ME, Avila L, Klanderman BJ, Lake S, Weiss ST, Celedon JC. PRKCA: a positional candidate gene for body mass index and asthma. Am J Hum Genet. 2009;85:87–96. doi: 10.1016/j.ajhg.2009.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bostrom H, Gritli-Linde A, Betsholtz C. PDGF -A/PDGF alpha-receptor signaling is required for lung growth and the formation of alveoli but not for early lung branching morphogenesis. Dev Dyn. 2002;223:155–162. doi: 10.1002/dvdy.1225. [DOI] [PubMed] [Google Scholar]
- 48.Souza P, Kuliszewski M, Wang J, Tseu I, Tanswell AK, Post M. PDGF-AA and its receptor influence early lung branching via an epithelial-mesenchymal interaction. Development. 1995;121:2559–2567. doi: 10.1242/dev.121.8.2559. [DOI] [PubMed] [Google Scholar]
- 49.Wu LS, Tan CY, Wang LM, Lin CG, Wang JY. Variant in promoter region of platelet-derived growth factor receptor-alpha (PDGFRalpha) gene is associated with the severity and allergic status of childhood asthma. Int Arch Allergy Immunol. 2006;141:37–46. doi: 10.1159/000094180. [DOI] [PubMed] [Google Scholar]
- 50.Durham AL, Wiegman C, Adcock IM. Epigenetics of asthma. Biochim Biophys Acta. 2011;1810:1103–1109. doi: 10.1016/j.bbagen.2011.03.006. [DOI] [PubMed] [Google Scholar]