Abstract
How rapidly does learning shape our brains? A new study using diffusion magnetic resonance imaging in both humans and rats suggests that just two hours of spatial learning is sufficient to change brain structure.
We continue to learn new skills and refine our existing abilities throughout life. To what extent does this ongoing learning shape our brain structure? We know from studies of highly skilled populations that the brains of experts are unusual – London taxi drivers have a larger posterior hippocampus for example (Maguire et al., 2000), which presumably supports their unrivalled skills in navigating the labyrinthine streets of the city. However, these experts have experienced many years of training and such cross-sectional studies can always potentially be explained by pre-existing differences in brain structure that determine our behaviour. Longitudinal studies, in which the same individuals are followed up over time, provide more direct insights into how experience shapes the brain. When novices are taught to juggle over a period of weeks to months, for example, this increases grey matter volume and changes white matter organisation in brain systems involved in visuo-motor co-ordination (Draganski et al., 2004; Scholz et al., 2009).
So experience shapes brain structure and neuroimaging provides us with a window into this structural change in humans. But how rapidly do such changes occur? Human studies of structural plasticity to date have considered periods of weeks to months of training. Yet experiments in non-human animals suggest that structural remodelling is a rapid, dynamic process that can be detected over much shorter timescales. Two photon microscopy studies in rodents, for example, reveal increases in the number of dendritic spines in motor cortex within an hour of training on a novel reaching task (Fu and Zuo, 2011).
In this issue of Neuron, Sagi and colleagues provide the first evidence that rapid structural plasticity can be detected in humans after just two hours of playing a video game (Sagi et al., 2012). The researchers used diffusion magnetic resonance imaging, which is sensitive to the self-diffusion of water molecules, to assess brain structure. Water diffusion in the brain depends on tissue architecture – if there is more space between obstacles (such as neurons, glial cells, blood vessels) then water diffuses more freely. If there is less space (as might occur if cells or blood vessels increase in size or number), then water diffuses less freely. Mean diffusivity (MD) therefore provides a probe of tissue structure. Maps of MD across the whole brain were derived from brain scans taken 2 hours apart. During the 2 hour interval, one group of participants played a car racing game that required them to repeatedly navigate around the same track; their steady improvement in performance demonstrated that they were gradually learning the layout of the track. In a control group, participants drove around a different track on each trial, so although they had a similar driving experience, they did not learn any specific spatial information. A second control group did not play the driving game during the interval period. Comparing the MD maps from the different groups revealed that the spatial learning group showed a specific decrease in MD in the hippocampus and parahippocampus, structures known to be particularly important for spatial learning and memory encoding. This decrease was behaviourally relevant – faster learners showed greater decreases in MD.
What might this decrease in MD reflect? Unfortunately, there is not a simple one to one relationship between most MRI measures and underlying tissue properties, and so interpreting any MRI change in biological terms is challenging (Zatorre et al., 2012). An impressive feature of the current study is that the authors went beyond typical speculations on possible interpretations and carried out a parallel study in rodents, in which they were able to perform both MRI and histological measures. As with the human study, just two hours of spatial learning in rats was associated with MD decrease in the hippocampus, in this case detected the following day. Subsequent histological measures allowed the authors to narrow down the possible interpretations of their MRI findings. Histology revealed that the learning group had more synaptophysin (a marker of synaptic vesicles), glial fibrillary acidic protein (GFAP) (a marker of astrocyte activation), and brain derived neurotrophic factor (BDNF) (a marker of neuronal growth that facilitates learning) in the hippocampus.
These histological results provide important clues into what cellular changes might be driving the detected MRI effect. More synaptophysin suggests an increase in synapse size or number. This agrees with Fu and Zuo's (2011) finding that post-synaptic dendritic spines change their shape over a similar time scale. However, spines are very small structures making up less than 10% of neuropil volume (Chklovskii et al., 2002). The likelihood of a small and localised increase in spine density accounting for this macroscopic MRI change seems slim. The authors suggest that the detected decrease in MD may reflect an overall shift in the ratio of extracellular to intracellular space. Extracellular space (ECS) is typically estimated at ~20% of normal adult brain tissue volume (Syková and Nicholson, 2008) and it decreases with neural activity due to swelling of cells, particularly astrocytes (MacVicar et al., 2002).
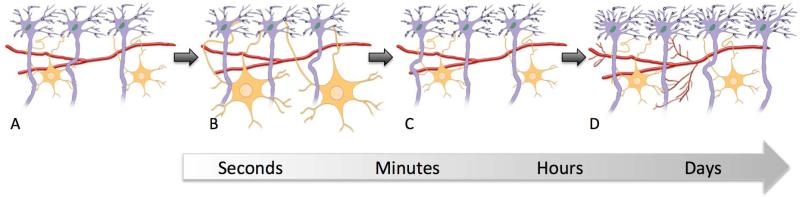
Given the higher expression of GFAP found in the rat study, rapid activity-dependent astrocyte swelling is a likely candidate to explain much of the detected MD decrease (Figure 1). Astrocytes may play an important role in learning and memory: Deleting the water channel protein aquaporin-4, that mediates astrocyte swelling, disrupts BDNF dependent long-term potentiation (LTP) (Skucas et al., 2011). Increased expression of BDNF was found in the learning group by Sagi and colleagues and the authors propose that their findings might indicate that diffusion imaging measures could be used as an indirect marker of LTP in humans. However more research is needed to test this hypothesis directly. For example, future studies could use aquaporin-4 knockout mice to block astrocyte swelling in order to test whether this mechanism is responsible for learning related decreases in MD. Importantly, however, lack of this protein does not disrupt Morris water maze learning, the task used by Sagi and colleagues, although its absence does impair other tasks of spatial memory (Skucas et al., 2011).
Figure 1.
A decrease in water diffusivity could reflect a number of different tissue changes, but not all could occur on the time scale observed in Sagi et al., (2012). Some candidate changes are illustrated here. Neurons are in purple, vasculature in red, and astrocytes in orange. Panel A illustrates the baseline state. Panel B shows the swelling of astrocytes in the presence of increased activity – a process that can occur over a course of seconds to minutes. C. Synapses (black circles) and their associated dendritic spines can be formed or modified over a period of minutes to hours. D. More elaborate structural change such as dendritic sprouting, neurogenesis and angiogenesis occurs over a period of days to weeks.
Other studies of learning-related structural change have demonstrated increases in dendritic sprouting, neurogenesis, angiogenesis, or changes in astrocyte size and number. However growth of new or even existing cells (neurons, glia or endothelial cells) takes considerable time (Figure 1), much longer than the two hours learning period in Sagi and colleagues’ study. However, processes other than growth may contribute to changing cell size or number. For example, learning may enhance the survival of recently created new neurons (Zhao et al., 2008). Hence it is possible that a decreased MD in the dentate gyrus reflects a slowing of the cell death process in the learning group while cell pruning continued at a higher rate in the control groups.
The histological results provide important evidence on what cellular changes accompany the detected MRI effects, but they cannot directly demonstrate whether any or all of these particular cellular changes drive the observed MD change. Future studies using pharmacological or genetic manipulations could test more directly the relationships between specific cellular changes and MRI effects. The rapid and perhaps transient nature of these learning related changes provides a further challenge. In vivo methodologies will be useful to fully understand how neural tissue changes in the minutes and hours after learning. Thus, molecular and optical imaging are perhaps most suited to understand how these compartments change in the living organism.
The present work, along with previous studies (Blumenfeld-Katzir et al. 2011; Lerch et al., 2010) combining imaging and histology, provides valuable insights into the types of structural changes that can be detected on different time scales with non-invasive magnetic resonance imaging (MRI). For instance 5 days of training in the water maze task increased the volume of the hippocampus as measured with MRI and produced a correlated increase in GAP-43, a marker for neuronal process remodelling (Lerch et al., 2010). In another study using 5 days of training with the same task, changes in diffusion MRI parameters were related to increases in GFAP, synaptophysin and myelin basic protein (MBP) (Blumenfeld-Katzir et al. 2011). The time-frame of these studies allows for slower remodelling mechanisms like dendritric sprouting or gliogenesis to occur (Figure 1). Such mechanisms could contribute to the structural brain changes detected using MRI in humans with long-term learning (Draganski et al., 2004; Scholz et al., 2009)
Sagi and colleagues’ results provide us with an important reminder that the brain is an extremely dynamic structure. This study used a focused period of video game playing, but presumably many of the learning experiences we undergo throughout our lives produce similar effects in task-relevant regions of our brains. The findings therefore have more general implications for human neuroimaging. Many studies that employ the standard imaging methods used here assume that human brain structure is relatively static, at least on short time scales. However, we must remember that we are merely looking at snapshots of an organ that is in a constant state of flux and these new findings demonstrate that even the relatively crude technique of MRI is sensitive to this rapid structural change.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Blumenfeld-Katzir T, Pasternak O, Dagan M, Assaf Y. PLoS One. 6:e20678. doi: 10.1371/journal.pone.0020678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chklovskii DB, Schikorski T, Stevens CF. Neuron. 2002;34:341–347. doi: 10.1016/s0896-6273(02)00679-7. [DOI] [PubMed] [Google Scholar]
- Draganski B, Gaser C, Busch V, Schuierer G, Bogdahn U, May A. Nature. 2004;427:311–312. doi: 10.1038/427311a. [DOI] [PubMed] [Google Scholar]
- Fu M, Zuo Y. Trends Neurosci. 2011;34:177–187. doi: 10.1016/j.tins.2011.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lerch JP, Yiu AP, Martinez-Canabal A, Pekar T, Bohbot VD, Frankland PW, Henkelman RM, Josselyn SA, Sled JG. Neuroimage. 2010;54:2086–2095. doi: 10.1016/j.neuroimage.2010.09.086. [DOI] [PubMed] [Google Scholar]
- MacVicar BA, Feighan D, Brown A, Ransom B. Glia. 2002;37:114–123. doi: 10.1002/glia.10023. [DOI] [PubMed] [Google Scholar]
- Maguire EA, Gadian DG, Johnsrude IS, Good CD, Ashburner J, Frackowiak RS, Frith CD. Proc Natl Acad Sci U S A. 2000;97:4398–4403. doi: 10.1073/pnas.070039597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sagi Y, Tavor I, Hofstetter S, Tzur-Moryosef S, Blumenfeld-Katzir T, Assaf Y. Neuron. 2012 doi: 10.1016/j.neuron.2012.01.025. [DOI] [PubMed] [Google Scholar]
- Scholz J, Klein MC, Behrens TE, Johansen-Berg H. Nat Neurosci. 2009;12:1370–1371. doi: 10.1038/nn.2412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skucas VA, Mathews IB, Yang J, Cheng Q, Treister A, Duffy AM, Verkman AS, Hempstead BL, Wood MA, Binder DK, Scharfman HE. J Neurosci. 2011;31:6392–6397. doi: 10.1523/JNEUROSCI.6249-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Syková E, Nicholson C. Physiological reviews. 2008;88:1277–1340. doi: 10.1152/physrev.00027.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zatorre R, Fields RD, Johansen-Berg H. Nature Neuroscience. 2012 doi: 10.1038/nn.3045. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao C, Deng W, Gage FH. Cell. 2008;132:645–660. doi: 10.1016/j.cell.2008.01.033. [DOI] [PubMed] [Google Scholar]