Abstract
Significance: Derived from the inner cell mass of the preimplantation embryo, embryonic stem cells are prototype pluripotent stem (PS) cells that have the ability of self-renewal and differentiation into almost all cell types. Exploration of the mechanisms governing this pluripotency is important for understanding reprogramming mechanisms and stem cell behavior of PS cells and can lead to enhancing reprogramming efficiency and other applications. Recent Advances: Induced pluripotent stem cells are recently discovered PS cells that can be derived from somatic cells by overexpression of pluripotency-related transcription factors. Recent studies have shown that transcription factors and their epigenetic regulation play important roles in the generating, maintaining, and differentiating these PS cells. Recent advances in sequencing technologies allow detailed analysis of target epigenomes and microRNAs (miRs), and have revealed unique epigenetic marks and miRs for PS cells. Critical Issues: Epigenetic modifications of genes include histone modifications, DNA methylation, and chromatin remodeling. Working closely with epigenetic modifiers, miRs play an important role in inducing and maintaining pluripotency. Future Directions: The dynamic changes in epigenetic marks during reprogramming and their role in cell fate changes are being uncovered. This review focuses on these new advances in the epigenetics of PS cells. Antioxid. Redox Signal. 17, 205–223.
Introduction
Embryonic stem (ES) cells, which develop from embryonic inner cell mass during the stage of preimplantation, can proliferate indefinitely maintaining their phenotype and differentiate into any cell types of the three germ layers (41, 172), a characteristic known as pluripotency. These self-renewing capabilities and pluripotency of ES cells are mediated by several transcription factors: OCT4, SOX2, and NANOG (17, 99), which are highly expressed in undifferentiated ES cells (175, 180). Pluripotency-related transcription factors co-occupy promoters of KLF4 and other genes that are involved in self-renewal (17, 77, 99). These factors also bind to the promoters of regulator genes of developmental lineage commitment and cellular differentiation (17, 99) to inhibit the expression in ES cells (“silencing”). In 2006, Takahashi and Yamanaka discovered that overexpression of four transcription factors, Oct3/4, Sox2, Klf4, and c-Myc, can reprogram fibroblasts into ES-like cells, referred to as induced pluripotent stem (iPS) cells. iPS cells closely resemble ES cells in genomic, cell, and molecular biologic characteristics. Remarkably, the same four factors identified in the murine system were able to confer pluripotency in human cells, indicating that the fundamental transcriptional network governing pluripotency is conserved across species. In contrast to ES cells, direct reprogramming provides a convenient and ethical means of generating pluripotent stem (PS) cells. Within just a few years of their discovery, iPS cells have been shown to hold incredible potential for research and therapeutic applications in regenerative medicine. Intriguingly, iPS cells were found to have very similar genetic and epigenetic features to ES cells. Gene regulation is affected not only by the DNA sequence that carries heritable information, but also by epigenetic modifications that alter DNA and the chromatin proteins. Epigenetic modifications include histone modifications, DNA methylation, and nucleosome rearrangement. Because gene regulation is influenced by epigenetic modifications, the stemness of PS cells, ES cells, and iPS cells can be characterized by a unique epigenetic signature. Therefore, exploring the epigenetic landscape of PS cells will be useful for understanding their biologic characteristics and differentiation and therapeutic potentials. This review focuses on the role of epigenetic regulation in PS cells.
Key Factors of Epigenetic Regulation
As the substrate of transcription, chromatin is subjected to various forms of epigenetic regulation, including histone modifications, DNA methylation, and chromatin remodeling.
Histone modifications
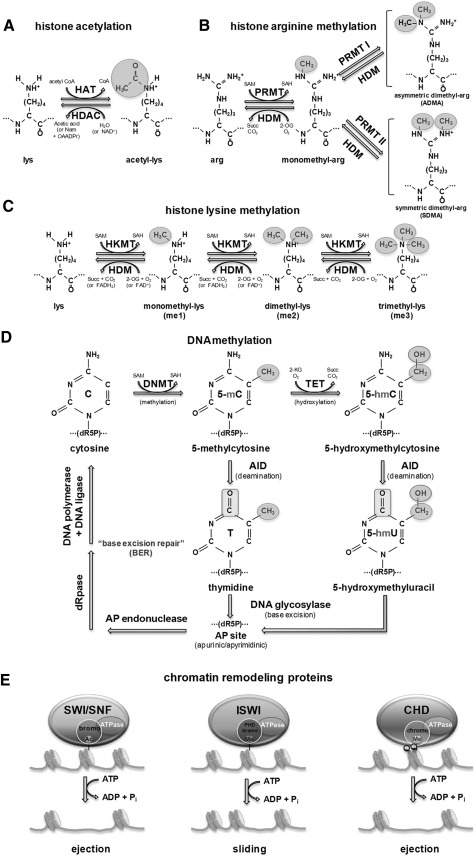
Histones, DNA-wrapping proteins, are subjected to posttranslational modifications including acetylation and methylation by enzymes primarily on their N-terminal tails for regulating chromatin structure. The core of histones H2A, H2B, H3, and H4, which comprise high-order DNA structure units called nucleosomes, can be modified by these posttranslational additions of chemical groups. Acetylation of histones occurs at multiple lysine sites by the balanced actions of histone acetyltransferase (HAT) and histone deacetylase (HDAC), and the resultant acetylated lysine residues can interfere with the positive charge of histones, resulting in weakening the interaction with negatively charged DNAs that can directly influence chromatin structure and fluidity (Fig. 1A). Histone methylation is the modification of histone proteins by the addition of mono- (me1), symmetrical di- (me2s), and asymetrical di- (me2a) methyl groups at arginine residues (Fig. 1B), and mono- (me1), di- (me2), and tri- (me3) methyl groups at lysine residues (Fig. 1C). Unlike acetylation, methylation is highly site-specific, and is maintained by histone methyltransferase (HMT) and histone demethylase (HDM) (Fig. 1B, C). Histone methylation is generally associated with repression of gene expression. For example, trimethylation of lysine 9 and lysine 27 of histone 3 (H3K9 and H3K27) is correlated with inactive regions of chromatin. However, methylation of lysine 4 of histone 3 (H3K4) or arginine on H3 and H4 result in transcriptional activation.
FIG. 1.
Key factors of epigenetic regulation. (A) Histone acetylation. Histones are acetylated at amino groups on lysine residues in N-terminal tail by HAT, with acetyl-CoA serving as the acetyl donor. Deacetylation of histones is mediated by classical HDAC class I, II, and IV using hydrolysis of acetate from lysine residues, or sirtuins, HDAC class III, which remove acetyl groups with NAD+ releasing nicotinamide and the unique metabolite O-acetyl-ADP-ribose. lys, lysine; HAT, histone acetyltransferase; HDAC, histone deacetylase; acetyl-CoA, acetyl coenzyme A; NAD+, nicotinamide adenine dinucleotide; Nam, nicotinamide; OAADPr, 2′-O-acetyl-ADP-ribose. (B) Histone arginine methylation. Arginine residues on histones can be monomethylated by PRMTs using SAM as methyl donor, and can be catalyzed through serial addition of methyl groups from SAM to ADMA by type I PRMT or SDMA by type II PRMT. HDM JMJD6 catalyzes the hydroxylation of methyl-arginine residues by consumption of both 2-OG and O2, releasing succinate and CO2. arg, arginine; PRMT, protein arginine methyltransferase; HDM, histone demethylase; SAM, S-adenosyl methionine; SAH, S-adenosyl homocysteine; 2-OG, 2-oxoglutarate; Succ, succinate; ADMA, asymmetric dimethyl-arg; SDMA, symmetric dimethyl-arg. (C) Histone lysine methylation. Lysine can be mono-, di-, or tri-methylated by serial iteration of HKMT using SAM as a methyl donor. Monomethyl and dimethyl-lysine can be hydroxylated by HDMs in a 2-OG-dependent manner (JmjC proteins) or FAD-dependent manner (LSD1). Trimethyl-lysine can be demethylated by JMJD6, which has arginine demethylase activity HKMT, in a 2-OG-dependent manner. HKMT, histone lysine methyltransferase; FAD+, flavin adenine dinucleotide; jmjC, jumonji-domain containing; LSD1, lysine-specific demethylase 1. (D) DNA methylation. A methyl group from SAM is attached to the 5th carbon from the nitrogen atom in the base of cytosine by DNMT resulting in 5-mC. 5-mC can be further hydroxylated by TET family enzymes to form 5-hmC. Demethylation of 5-mC and 5-hmC base can be initiated by deamination through the activity of AID followed by the BER pathway, which is the stepwise enzymatic mechanism for repairing damaged DNA using DNA glycosylase, AP endonuclease, dRpase, DNA polymerase, and DNA ligase. BER, base excision repair; DNMT, DNA methyltransferase; TET, ten–eleven translocation; dR5P, deoxyribose-5-phosphate; 5-mC, 5-methylcytosine; 5-hmC, 5-hydroxymethylcytosine; AID, activation-induced cytidine deaminase; AP, apurinic/apyrimidinic; dRpase, deoxyribophosphodiesterase. (E) Chromatin remodeling proteins. The binding of the chromatin remodeling complex, SWI/SNF, ISWI, and CHD, to chromatin is mediated by the unique domain (bromo, PHD bromo, and chromo) of each protein, which can interact with specific modified histones (acetylated or methylated) and methylated DNA in an ATP-independent manner. The steps of chromatin remodeling, including disruption of histone-DNA contacts, ejecting the nucleosome (ejection), and repositioning (sliding), are dependent on the hydrolysis of ATP. SWI/SNF and CHD family proteins can trigger ejection of the nucleosome, while ISWI can slide a nucleosome to another position. SWI/SNF, switch/sucrose nonfermentable; ISWI, imitation switch; CHD, chromodomain helicase DNA-binding.
By regulating chromatin structure, epigenetic modifications play an essential role in controlling access to genes and regulatory elements in the genome (20). The differences in epigenetic status between a somatic cell and a PS cell are evident, and dedifferentiation requires global epigenetic reprogramming. For instance, PS cells contain bivalent domains that are characteristic chromatin signatures (10, 11). These are regions enriched for both repressive histone H3 lysine 27 trimethylation (H3K27me3) and active histone H3 lysine 4 trimethylation (H3K4me3) (116). It was assumed initially that bivalent domains might be ES-cell specific because they were first identified using chromatin-immunoprecipitation (ChIP) followed by hybridization to microarrays (ChIP-Chip) that featured key developmental regulators. All of these resolved either to a univalent (H3K4me3 only or H3K27me3 only) state or lost both marks in differentiated cells (10). Using ChIP followed by high-throughput sequencing (ChIP-seq) technology, Mikkelsen and colleagues showed that bivalent domains are more generally indicative of genes that remain in a poised or balanced state characterizing their plasticity. Pluripotent cells were found to contain large numbers of bivalent domains (∼2500) compared with multipotent neural progenitor cells (NPCs) (∼200) that still retain multilineage potential but are more restricted than ES cells (109).
DNA methylation
A second component of the epigenetic marks is DNA methylation, which is a stable and heritable mark that is involved in gene silencing, including genomic imprinting and X-chromosome inactivation. DNA methylation patterns are dynamic during early embryonic development and are essential for normal development (141). Overall DNA methylation levels remain stable during ES-cell differentiation, although they are not static at any individual gene level (109). The 5′-promoter regions of many transcriptional units contain clusters of the dinucleotide CpG, which are methylated at transcriptionally silent genes and demethylated upon activation. In differentiated cells, the Oct4, Nanog, and Sox2 promoter regions are highly methylated and in an inactivated state, whereas in ES cells these promoters are unmethylated and in an activated state. During reprogramming, almost complete demethylation of these promoters has been observed (102, 115, 121, 184). Therefore, the loss of DNA methylation at the promoters of pluripotency-related genes appears essential for achieving reprogramming to pluripotency state. Interestingly, loss of DNA methylation at this class of genes seems to be a rather late event in the reprogramming process because cells that have already acquired self-renewing properties still showed high levels of DNA methylation (115).
DNA methyltransferase (Dnmt)1, Dnmt3a, and Dnmt3b are three enzymes necessary for DNA methylation (14, 53) (Fig. 1D). Dnmt1 maintains DNA methylation at hemi-methylated DNA after DNA replication during cell divisions, whereas Dnmt3a and Dnmt3b are responsible for establishing de novo DNA methylation (28). Lack or mutation of Dnmt1 causes a two-third loss of DNA methylation or embryonic lethality (96). Embryos deficient in Dnmt1 are smaller and seem to have delays in developmental processes (96). Embryos with mutant Dnmt3b seem to proceed normally in their developmental stages initially but show multiple defects in their later stages (126). In mouse embryonic fibroblasts (MEFs), conditional deletion of Dnmt3b leads to partial loss of methylation (38). These findings show that Dnmt1 and Dnmt3b play a significant role in maintenance of proliferating cells and their epigenomic landscape. In contrast, lack of Dnmt3a does not seem to cause obvious developmental defects. A study showed that homozygous Dnmt3a knockout mice developed to term but died within ∼1 month after birth (126). Dnmt3a seems to function as a co-regulator of Dnmt1 and Dnmt3b, and interacts with the N-terminus of histone H3 (67, 127). It has been suggested that it functions as a co-regulator of both Dnmt3a and Dnmt3b and has recently been shown to interact with the N-terminal tail of histone H3 when it lacks methylation at lysine 4 (67, 127).
Chromatin remodeling
Chromatin remodeling enzymes add or remove histone covalent modifications and utilize the energy of ATP hydrolysis to disrupt chromatin structure. This makes DNA/chromatin available to proteins that need to access DNA or histones directly during cellular processes. There are three well-characterized families of ATP-dependent nucleosome remodeling enzymes, SWI/SNF, ISWI, and chromodomain helicase DNA-binding (CHD). These are multi-subunit complexes containing a conserved ATPase domain as the catalytic subunit, with additional components to form large multiprotein-complexes, which promote specific histone posttranslational modifications and incorporation of histone variants (75, 145) (Fig. 1E). The initial creation of the DNA loop or bulge is the result of the translocase activity that has been described to SWI/SNF and ISWI (143, 186, 200). The ATPase of SWI/SNF binds to a specific location on the nucleosome, from which it utilizes its 3′ to 5′ translocase activity to draw DNA into one entry/exit and pump it to the other in a directional wave (144). Binding of the complex to the nucleosome creates significant rearrangement of the DNA with respect to the histone octamer even in the absence of ATP hydrolysis, which may facilitate the creation of the DNA bulge required for ATP-dependent translocation (101). In contrast to SWI/SNF, the smaller ISWI chromatin remodeling complexes make limited contacts with the nucleosome and the extranucleosomal DNA (33, 48, 71, 149). These complexes bind their substrate as a dimeric motor to facilitate the bidirectional and processive translocation of DNA over the nucleosome (15, 47, 133). This is consistent with the role of these remodelers in nucleosome spacing, and their ability to sample DNA linker lengths to position nucleosomes equidistant from either end (48, 165, 195, 199). In this case, ATP hydrolysis by one of the two ATPases results in loosened DNA-histone contacts that may act similarly to the ATP-independent conformational change upon SWI/SNF binding to promote DNA translocation following ATP hydrolysis by the second ATPase (49). ATPases of the CHD family are characterized by N-terminal tandem chromodomains in addition to the conserved DEAD/H-related ATPase domain (132, 173). CHD is targeted to sites of active transcription through PHD-mediated recognition of H3K4me3 (45, 156), and associates with other preinitiation factors to facilitate transcriptional elongation and splicing (157). Despite the similarity between their core enzymatic subunits, their common affinity for nucleosomes, and their common ability to disrupt nucleosomal templates in vitro, there is little functional similarity between members in vivo and as a result, very little predictive power for members of unknown function.
Epigenetic Landscape in Pluripotent Stem Cells
There are unique epigenetic marks in PS cells, ES cells, and iPS cells. It is well known that certain epigenetic marks are critical for maintaining pluripotency, which include histone modifications, DNA methylation of pluripotency genes, and regulation of pluripotency- or differentiation-specific microRNA (miRs) expression. However, other studies still claim that a majority of epigenetic marks are unnecessary for ES cell survival. Although it is possible to maintain PS cells without all of their epigenetic marks, this leads to a decrease in their differentiation potential. Thus, the degree of in vitro epigenetic influence in pluripotency of ES cells needs to be assessed by further studies.
Chromatin structure in PS cells
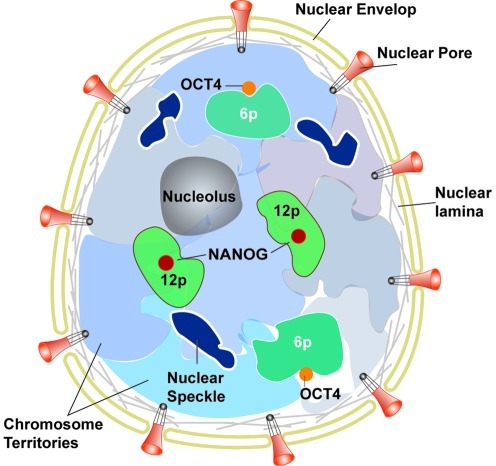
PS cells, including ES and iPS cells, have a dynamic and specialized higher order chromatin structure that is highly organized for the regulation of gene expression programs during development and differentiation, and is established by histone modification, DNA methylation, and the activity of ATP-dependent nucleosome remodeling complexes (107). PS cells have distinctive chromosomal regions that contain the genes that maintain pluripotency. A region of clustered pluripotency genes, including NANOG at chromosome 12p, has more central nuclear localization in ES cells than in differentiated cells (187). Although the functional significance of positioning in the nuclear center is unknown, the presence of the nuclear spots within the most gene-dense human chromosomes suggests that it may confer some transcriptional advantage (19). Chromosome 12p contains other pluripotency-related genes, STELLA and GDF3, which are also expressed in ES cells and downregulated upon differentiation (31). Another pluripotency gene, OCT4, at chromosome 6p forms a loop to relocalize to a position outside its chromosome territory (187). The data showing increased DNA methylation and histone deacetylation of the OCT4 enhancer/promoter in trophoblast cells compared with ES cells are consistent with a more long-range remodeling of chromatin architecture around OCT4, which might also contribute to its transcriptional regulation (58). Preferential association of inactive genes with the nuclear periphery has been reported in differentiated cells; however, neither NANOG nor OCT4 is associated with the nuclear periphery of clusters of inactive genes (198) (Fig. 2). In addition, genes and chromosome activities are directly related to their spatial positioning within the nucleus that is prone to alterations during early differentiation (78, 104). Furthermore, studies have indicated that nuclear structures such as nucleolus, heterochromatin (129), nuclear speckles (domains enriched in splicing factors) (32), and nuclear lamina experience morphological changes during the early stage of differentiation.
FIG. 2.
Chromatin structures in pluripotent stem cells. The nucleus of the cell is composed of a nuclear envelope that contains pores, a meshwork of intermediate filaments (nuclear lamina), nucleolus, subnuclear bodies including nuclear speckle, which is enriched in pre-mRNA splicing factors, and distinct territories of each chromosome that occupy a defined volume of the nucleus. Chromosome 12p (green) including NANOG, which is located well within the 12p chromosome territory, has a more central nuclear localization in ES cells, while chromosome 6p (dark green) with OCT4 (orange), which forms a loop to relocalize to a position outside the 6p chromosome territory, is located at an intermediate position between the nuclear periphery and center. (To see this illustration in color the reader is referred to the web version of this article at www.liebertonline.com/ars).
Histone modification in PS cells
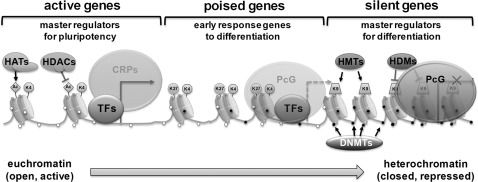
N-terminal tail residues of histones can undergo posttranslational modifications such as acetylation, methylation, and phosphorylation (81). Given that ES and somatic cells contain almost identical genomic DNA, epigenetic regulation is one of the major influences on their pluripotency and differentiation potential. To maintain pluripotency in ES cells, differentiation-triggering genes should be inactive. Polycomb group proteins (PcGs) play important roles in silencing these developmental regulators. They form multiple polycomb repressive complexes (PRCs), the components of which are conserved from Drosophila to humans (148). The PcG proteins function in two distinct PRCs, PRC1 and PRC2. Genome-wide binding site analyses of PRC1 and PRC2 in mouse ES cells and PRC2 in human ES cells (18, 93) showed that the genes regulated by the PcG proteins are co-occupied by nucleosomes with trimethylated H3K27. These genes are transcriptionally repressed in ES cells and are preferentially activated when differentiation is induced. The pluripotency factors Oct4, Sox2, and Nanog co-occupy a significant fraction of the PcG protein-regulated genes when acting as transcription factors (18, 93). Most of the transcriptionally silent developmental regulators targeted by Oct4, Sox2, and Nanog are also occupied by the PcGs (11, 18, 93). PRC2 catalyzes H3K27 methylation, an enzymatic activity required for PRC2-mediated epigenetic gene silencing. H3K27 methylation is thought to provide a binding surface for PRC1, which facilitates oligomerization, condensation of chromatin structure, and inhibition of chromatin remodeling activity in order to maintain silencing. PRC1 also contains a histone ubiquitin ligase, Ring1b, whose activity appears to contribute to silencing of genes in ES cells (162). How the PcG is recruited to genes encoding developmental regulators in ES cells is unknown. Some of the most conserved vertebrate sequences are associated with genes encoding developmental regulators, and some of these may be sites for DNA-binding proteins that recruit PcG proteins (Fig. 3).
FIG. 3.
Epigenetic status of pluripotent stem cells. Pluripotent stem cells have appropriate epigenetic marks for different genes. Master regulators for pluripotency-related genes are highly activated in PS cells by an open chromatin structure consisting of an unmethylated promoter region (small white circles on DNA strands), acetylated histones (Ac) that are mediated by the regulation of HATs and HDACs, and H3K4 methylation (K4). The open chromatin structure for active promoter regions is also remodeled by ATP-dependent CRPs, and permissible for binding of TFs to recruit RNA polymerase for transcription of genes. The bivalent domains at the promoters of early response genes to differentiation, consisting of both repressive H3K27 methylation (K27) and active K4 marks, keep them poised for activation in pluripotent stem (PS) cells with the binding of PcG proteins long-term silencing of differentiation-related genes through DNA methylation are performed by DNMTs. DNA methylation (small black circles on DNA strands) is often accompanied by the repressive H3K9 methylation (K9), on promoters, which is mediated by HMTs and leads to chromatin compaction. DNA methylated-silenced promoters show a loss of H3K4 methylation and histone de-acetylation. CRPs, chromatin remodeling proteins; TFs, transcription factors; Ac, acetylation; K4, H3K4 methylation; K27, H3K27 methylation; PcG, polycomb group protein; HMTs, histone methyltransferases; K9, H3K9 methylation.
Recent studies demonstrated that the silent developmental genes that are occupied by Oct4, Sox2, Nanog, and PcG proteins experience an unusual form of transcriptional regulation (55). These genes undergo transcription initiation but not productive transcript elongation in ES cells. The transcription initiation apparatus is recruited to developmental gene promoters, but RNA polymerase is incapable of fully transcribing these genes, presumably because of repression mediated by the PcG. This explains why the silent genes encoding developmental regulators are generally organized in bivalent domains that are occupied by nucleosomes with histone H3K4me3 and H3K27me3 (4, 11, 55).
The presence of inactive RNA polymerase at the promoters of genes encoding developmental regulators may explain why these genes are especially poised for transcription activation during differentiation (18, 93). PcG complexes and associated proteins may serve to pause RNA polymerase machinery at key regulators of development in PS cells and lineage-committed cells where they are not expressed. When the cells are activated, PcGs and nucleosomes with H3K27 methylation are lost (18, 93, 116), allowing the transcription apparatus to fully function and transcribe these genes. The mechanisms that lead to selective activation of genes encoding specific developmental regulators are not yet understood (87). Beyond the specific regulation of development-related genes, ES cells maintain chromatin in a highly dynamic and transcriptionally permissive state. Fewer heterochromatin foci are detected in ES cell nuclei compared with differentiated cells. Fluorescence recovery after photobleaching and biochemical analyses reveal that ES cells, compared with differentiated cells, have an increased fraction of loosely bound or soluble architectural chromatin proteins, including core and linker histones and heterochromatin protein HP1. A hyperdynamic chromatin structure is functionally important for pluripotency (113). The status of histone modifications also indicates that the chromatin in ES cells is more transcriptionally permissive than in differentiated cells. Consistent with the global dynamics of chromatin, ES cell differentiation is associated with a decrease in global levels of active histone marks, such as acetylated histone H3 and H4, and an increase in repressive histone marks, histone H3 lysine 9 methylation (91, 113). Taken together, these unique epigenetic characteristics of ES cells facilitate rapid but regulated transcription, allowing differentiation down different cell fate pathways as needed by the organism.
Several studies of murine iPS cells have identified a small number of representative loci that have consistent chromatin and DNA methylation patterns (102, 170, 184). A ChIP-Chip study investigating H3K4me3 and H3K27me3 in the promoter regions of 16500 genes showed that iPS cells were highly similar to ES cells in epigenetic state (102). The H3K4me3 pattern was similar across all samples, indicating that reprogramming was largely associated with changes in H3K27me3 rather than H3K4me3 (102). Another ChIP-Seq study exploring genome-wide chromatin maps in several iPS lines derived by different methods (110, 184) demonstrated that global levels of repressive H3K27me3 and the characteristic bivalent chromatin structure are retained in the various iPS cell lines. The restoration of repressive chromatin marks appears crucial to stably silence lineage-specific genes that are active in somatic cells and inactive in pluripotent cells, suggesting that failure to establish the repressive marks results in incompletely reprogrammed cells. Activating H3K4me3 patterns are also crucial for complete reprogramming and have been observed to be restored genome wide, in particular around the promoters of pluripotency-associated genes such as Oct4 and Nanog, in the fully reprogrammed iPS lines (115).
Histone demethylation plays a critical role in ES cell epigenesis and pluripotency. The mechanism of histone demethylation was discovered only recently (81, 150, 151). Removal of a specific H3K27 tri-methylation can be achieved by two enzyme families: Jumonji (JMJD) and UTX (1, 35, 92). Jmjd1a and Jmjd2c genes, which are responsible for H3K9me2 and H3K9me3 demethylation, are up-regulated by Oct4. Jmjd2a induces Nanog up-regulation and Jmjd1a demethylates H3K9me2 at the Tc11, Zfp57, and Tcfcp211 promoters (100) (Fig. 1C).
Histone acetylation activates transcription. The HAT p300 regulates the state of pluripotency by activating transcription. p300 is recruited to sites of multiple pluripotency factors such as Nanog, Oct4, and Sox2 (27). These multiple binding sites are involved in activation of the H3K4me3 mark. In ES cells, Tip60-p400 is the HAT and chromatin remodeling complex that positively or negatively regulates gene expression and DNA repair (146, 160). The complex is also a transcriptional activator that incorporates the histone H2AZ and catalyzes histone acetylation at target promoters (160). ES cells with Tip60-p400 deficiency show abnormal morphology and cannot self-renew or differentiate. There are significant similarities between genes affected by Tip60-p400 depletion and Nanog depletion. Furthermore, Tip60-p400 binding, Nanog binding, and the H3K4me3 mark have strong correlations with one another. Tip60-p400 localizes to both highly expressed genes and poised genes marked by bivalent modifications (43). Tip60-p400 binding is necessary in regulating histone H4 acetylation. In ES cells, Tip60-p400 can act as a repressor by catalyzing histone acetylation.
Histone deacetylation is mediated by nucleosome remodeling deacetylase (NuRD) complexes, which are responsible for HDAC activities as well as ATP-dependent chromatin remodeling. The NuRD complex consists of SNF2-like ATPase subunits, Mi2α or -β, deacetylase subunits HDAC1/2, and associated subunits including methyl-CpG-binding proteins MBD 1/2/3 and metastasis associated proteins MTA1/2/3 (37). Mi2β, in association with HDACs and MBD subunits, plays an important role as a transcriptional repressor but can also be an activator and is required for hematopoietic stem cell self-renewal and differentiation (179, 189, 196). Pluripotency requires MBD3, as ES cells without MBD3 cannot commit to specific developmental lineages (72). However, MBD3 is not required for expression of pluripotency factors but only for repression of gene transcription. Unrepressed promoters become hyperacetylated at H3K9 and H4, whereas there is no change in the acetylation status of other inactive promoters, suggesting that the NuRD complex is not globally required for ES cell gene silencing. Furthermore, MBD3-depleted ES cells can undergo initial stages of differentiation by forming embryoid bodies and trophectoderm but fail to differentiate further except when induced with retinoic acid. MBD3-depleted ES cells also fail to develop in utero as chimeric embryos when aggregated with wild-type morulae. Thus, MBD3 as part of the NuRD complex facilitates ES cell differentiation but is not absolutely required for ES cells to respond to all inductive signals.
DNA methylation in PS cells
DNA methylation suppresses gene expression; thus, in ES cell status, most of the pluripotency-related transcription factors are hypomethylated and lineage-committed genes are heavily methylated at their promoter regions (Fig. 3). H3K4me3 and DNA methylation are known to be linked. DNA methylation maps have been created for mouse and human pluripotent cells (89, 97, 109). High-CpG promoters of ES cells have H3K4me3 histone modifications and entirely lack DNA methylation (109,116). This may be related to the ability of Dnmt3 to bind to unmodified H3K4 (127). In ES cells, CpGs in low-CpG promoters, which are generally associated with tissue-specific genes (183), are mostly methylated, with the exception of a small subset (<10%) that are enriched for H3K4me3 or H3K4me2 (109). All pluripotency genes in ES cells have H3K4 methylation and DNA hypomethylation (65, 115, 116). However, the loci that are devoid of H3K4 and H3K27 methylation show extensive DNA methylation instead (46). Genome-wide bisulfite sequencing in human ES cells (89, 97) has confirmed the findings of these previous, more limited mouse (46, 109, 118) and human studies (183).
The Dnmt family maintains and directs catalysis of DNA methylation. The methyl-CpG-binding domain (MBD) family recognizes DNA sequences with high CpG content and mediates the effects of DNA methylation (60). During the division of the zygote, paternal and maternal methylation patterns disintegrate by down-regulating the expression of the Dnmt1 gene (136). During embryogenesis and formation of germ layers, there is a re-establishment of a new DNA methylation pattern. During this period, the epigenomic status of ES cells inside ICM is restored to redefine pluripotency. Studies of Dnmt1-deficient (Dnmt1−/−) and Dnmt3a/Dnmt3b-deficient (Dnmt[3a−/−,3b−/−]) ES cells have demonstrated that restoring DNA methylation is essential for development (94, 126). These Dnmt mutant ES cells exhibited an early-lethal phenotype, defects in differentiation during embryoid body formation of Dnmt1−/− (128), and teratoma formation of late-passage Dnmt[3a−/−,3b−/−] (29). Down-regulation of Oct4 and Nanog expression during the differentiation of NT2 cells, a neuronally committed human teratocarcinoma cell line, is related to methylation of 5′-flanking regions of both these genes (36).
While the role of non–CpG methylation is unclear in somatic cells, recent evidence suggests that the role of non–CpG methylation is important in maintenance of ES cell pluripotency. In adult somatic tissues, DNA methylation typically occurs at the site of the CpG dinucleotide. However, non–CpG methylation, including CpA and CpT methylation, is prevalent in mouse ES cells (108, 109) and human ES cells (89, 97). Non–CpG methylation level is mediated by Dnmt3a and Dnmt3b (39). Overexpression of Dnmt3a in Drosophila, which has no Dnmts or DNA methylation (53), induces CpG as well as non–CpG methylation. These data support the notion that non–CpG methylation results from de novo methylation activity of Dnmt3a, which is highly expressed in ES cells and poorly expressed in somatic tissues (135). In both mouse and human, non–CpG methylation is low in differentiated cells (97, 135) and reappears in iPS cell generation (97).
Recent studies have shown the importance of DNA hydroxymethylation in regulating gene expression. In fact, DNA hydroxymethylation is highly prevalent in ES cells compared with somatic cells. The ten–eleven translocation (TET) family direct catalysis of 5-methyl cytosine to 5-hydroxymethylcytosine (5hmC) (168) (Fig. 1D). 5hmC is detectable in undifferentiated mouse ES cells as well as in Purkinje neurons and granule cells (82). During the differentiation of ES cells, the amount of Tet1 and 5hmC decreases (168), and Tet1 knockdown by shRNA damages ES cells' ability to maintain and self-renew with diminishing Nanog expression, suggesting a crucial role of Tet1 in maintaining pluripotency (66). However, a more recent study showed that Tet1-deficient (Tet1−/−) mouse ES cells still maintain pluripotency with expression of pluripotency markers Oct4, Sox2, and Nanog, and can drive normal embryo development in tetraploid complementation assays, although these mutant mice have a partial reduction of 5hmC and a slightly small body size at birth (34). Another study using shRNAs to knock down Tet1 and Tet2 also showed that depletion of these genes did not affect Nanog expression, self-renewal, or pluripotency (80).
Chromatin remodeling enzymes
ES cells are rich in euchromatin and diffused heterochromatin. Chromatin structural remodeling leads to highly compact heterochromatin during differentiation. The change from lower to a higher chromatin structure state is helped by chromatin remodeling protein complexes. Either the remodeling enzymes have the ability to alter the N-terminal characteristics of histones by adding or removing acetyl, methyl, ubiquitin, and sumo groups, or the remodeling proteins are composed of ATP-dependent enzymes, which use energy to transiently separate the association between DNA and histones. These modifications, in turn, may induce a conformational change in nucleosome and chromatin condensation state. This kind of ATP-mediated remodeling provides an accessible DNA, which is more prone to gene activation or repression by the transcriptional apparatus (154) (Fig. 3). An example is the SWI/SNF protein complex in mammals, which is composed of several different subunits such as Snf2. Different chromatin remodeling enzymes that are ATP-dependent are highly expressed in mouse ES cells and are down-regulated after leukemia inhibitory factor removal (84). Mice that lack remodeling proteins SNF2H (163), BRG1 (21), SNF5 (79), or SSRP (24) die before implantation. The SWI/SNF mechanism of catalysis and regulation of ES cell pluripotency is not very well understood and requires further investigation. It has been found that the SWI/SNF chromatin remodeling component, BAF250, is vital for ES cell pluripotency (50). BAF250b dysfunction lowers expression of pluripotency factors and increases that of lineage markers such as Gata2. The SWI/SNF complex also regulates nucleosome sliding and eviction (56, 95).
miRs in PS cells
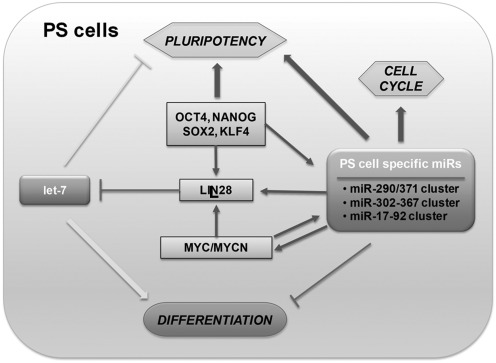
The function of miRs in stem cells has been established in many systems, where disruption of Dicer or DGCR8, which are miR-processing enzymes, leads to defects in embryo development (12, 57, 73, 120, 123, 182, 188). Dicer-null mouse is embryonic lethal, indicating its critical role in early embryonic development (12). However, Dicer-null mouse ES cells are viable but show defective cell cycle progression. Dicer-null ES cells are defective in differentiation and fail to express differentiation markers such as HNF4a, BMP4, and GATA1 (73). Dicer-deficient cells further show decreased levels of DNA methylation and Dnmts (9, 158) and increases in telomerase recombination and elongation (9). This defect in DNA methylation leads to lack of differentiation by incomplete and reversible silencing of Oct4 (9, 158). DGCR8 knock-out mouse ES cells show phenotypes similar to Dicer-deficient mouse ES cells, showing reduced cell proliferation, abnormal cell cycle control, and defects in differentiation (182). DGCR8-null ES cells arrest in G1 phase and do not differentiate into teratoma, a feature characteristic of normal ES cells (16, 76, 112). Blelloch and colleagues have discovered specific functions for families of miRs controlling cell cycle and self-renewal in DGCR8-null mouse ES cells (111, 181) (Fig. 4).
FIG. 4.
microRNAs in pluripotent stem cells. microRNAs (miRs) regulate both self-renewal and differentiation pathways of PS cells by regulating various factors that mediate these processes. PS cell-specific miRs inhibit the transcriptional repressors that target pluripotency-related genes, including OCT4, NANOG, SOX2, KLF4, and MYC, or cell cycle-related genes to promote self-renewal, and directly inhibit the expression of differentiation-related genes. Another mechanism of promoting self-renewal is the regulation of let-7 miR processing by LIN28, which directly binds to a consensus sequence in the pre-let-7 miR loop region. Positive and negative regulations are shown by arrows and T lines, respectively.
ES cells have been reported to express a small subset of unique miRs (23, 62, 86, 119, 166). The human miR-371 cluster is located on chromosome 19 and is analogous to the mouse miR-290 cluster, and the miR-302 cluster located on chromosome 4 is associated with both murine and human ES cells (62, 164, 166). Two additional clusters, miR-17 on chromosome 13 and the miR-106a cluster on chromosome X, have also been shown to be upregulated in ES cells (90).
miR-290 and miR-371 clusters
The miR-290 cluster is comprised of six miRs (miR-290 to miR-295), all of which are expressed at high levels in undifferentiated mouse ES cells but decreasing upon differentiation (62). Exogenous miR-290 family members can partially rescue self-renewal of Dicer-null cells (9, 158). The human homolog of the miR-290 cluster includes miR-371, -372, -373, and -373*. These miRs were identified as embryonic carcinoma and ES cell-specific miRs (69, 86, 166). The cluster is transcribed as a single polycistronic transcript and regulated by a common promoter (166). Mouse mutants with a homozygous deletion of the miR-290–295 cluster are embryonic lethal, demonstrating its importance during embryo development (2). The ES core transcriptional regulatory circuit, Oct4, Sox2, and Nanog, has recently been shown to be physically associated with promoters for the miR-290 cluster (105).
miR-302-367 cluster
Another ES cell-specific miRs cluster is the miR-302–367 cluster, shown to be highly and specifically expressed in undifferentiated ES cell (69, 86, 119). This cluster, encoded by human chromosome 4, consists of nine different miRs: miR-302a, -302a*, -302b, -302b*, -302c, -302c*, -302d, -367, and -367* (88). The ES cell-specific transcription factors, Oct4, Nanog, Sox2, and Rex1, were shown to be upstream regulators of the miR-302–367 promoter (8, 25). In addition, miR-302–367 has been identified to posttranscriptionally regulate cyclin D1 and Cdk4, affecting cell cycle progression (25). In addition to their role in self-renewal and proliferation, miR-302–367 may indirectly induce the TGFβ/Nodal/Activin pathway via inhibition of intermediate negative regulators to maintain cells in the undifferentiated state (7).
miR-17–92 cluster
The miR-17–92 cluster is highly expressed in ES cells, and a single polycistronic transcript generates six miRs: miR-17, -18a, -19a, -20a, -19b-1, and -92a-1 (54, 62, 119). This cluster is a known OncomiR that promotes cell proliferation in several forms of cancer (59, 85, 124, 137, 176) and is activated by c-Myc, a repressor of other miRs such as miR-15 and let-7 (26). miR-20a is thought to play a role in cell proliferation and inhibition of apoptosis at the G1 to S transition via a feedback loop with E2F factors (124, 167, 174, 191). The role of c-Myc in combination with Oct4, Sox2, and Klf4 in inducing somatic cells to iPS cells suggests that these miRs play a key role in stem cell renewal and pluripotency. Indeed, Melton and colleagues demonstrated that a let-7 inhibitor was able to replace exogenous c-Myc in the production of iPS cell cultures (111).
let-7 family
ES cells are characterized by the presence of low levels of let-7, which is highly expressed in differentiating cell types (54, 86). Let-7g levels have been shown to be regulated by LIN28, a gene highly expressed in pluripotent cells (177). The LIN28 gene is in turn targeted by let-7 via target sites in its 3′-UTR (122, 130, 142, 177). Both LIN28 and let-7g genes are associated with Oct4/Sox2/Nanog/Tcf3 binding in ES cells, resulting in a regulatory loop where the core pluripotent transcription factors activate the LIN28 gene associated with maintenance of self-renewal and pluripotency while repressing let-7g to prevent differentiation (105). Exogenous addition of let-7 miRs can suppress self-renewal in DGCR8 null mouse ES cells but not in wild-type ES cells, and this suppression can be inhibited by introduction of ES cell–cell cycle-regulating miRs such as miR-294 (111).
Epigenetic Changes During Differentiation
The investigation of lineage specification and global epigenetic remodeling for specific cell types in vivo is a difficult challenge despite advances in experimental technologies. Therefore, PS cells have emerged as a powerful system to investigate epigenetic changes during differentiation. As human PS cells are the only available model system to study the mechanism of differentiation and cell fate decision in human developmental biology, they have been widely used for such purposes. Most of genome-wide DNA methylation and histone-modification studies were conducted comparing epigenetic changes during differentiation of PS cells into their differentiated lineages. During this cell fate change, PS cells lose their pluripotency by silencing pluripotency-related genes and gain differentiated cell phenotype by activating silenced or poised lineage-specific genes as the promoters and enhancers of these genes become accessible by epigenetic changes. The promoter-specific regulation of chromatin structure by specific enzymes leads to proper differentiation into specific cell types. However, the mechanism of recruiting enzymes that regulate chromatin remodeling and histone modification needs to be further elucidated.
Neural differentiation
During the differentiation of PS cells into the neural lineage, the promoters of many genes related to the neural lineage, including Nestin, Otx2, Nkx2-2, Astn, Olig1, and the miRNA mir-9-3, lose a repression mark, H3K27me3, and retain a monovalent activation mark, H3K4me3, resulting in triggering expression of these genes (116). This removal of H3K27me3 is mediated by upregulation of H3K27-specific HDM, Kdm6b (also known as Jmjd3), with downregulation of specific HMT, enhancer of zeste homolog 2 (Ezh2) at the promoters of neural genes (22). During the transition from human ES cells to NPCs, pluripotency-related genes such as NANOG, TERT, and LIN28, and non-neuronal lineage genes, including GATA4 and NODAL, gain another repressive mark, H3K9me2, concomitantly with the loss of H3K4me3, leading to their long-term repression (52). The transcriptional repressor RE1-silencing transcription factor (REST, also known as NRSF) targets a group of neuronal genes through its interaction with the 21-base pair RE1 element. It represses neuronal differentiation by recruiting histone modifiers and chromatin-binding proteins (5). A subset of REST target genes escape the repression and are selectively induced during the transition from ES cells to NPCs. Degradation of REST by β-transducin repeat-containing protein 1 (β-TRCP1) has been shown to be essential for ES cell commitment to a neural fate (185). Also, DNAs at the loci of neural-related distal regulatory elements is demethylated at CpG site upon commitment to the neural lineage, whereas non-neural gene loci gain CpG methylation in NPCs (118). The targeting mechanism of histone modifiers and DNMTs to specific loci is not well understood. However, it has been shown that PcG proteins preferentially target highly conserved promoters with long unmethylated CpG islands in ES cells (138, 147).
Endothelial differentiation
The role of histone acetylation in endothelial differentiation was identified by the studies using small molecule HDAC inhibitors such as trichostatin A (TSA). These inhibitors block the endothelial differentiation of adult progenitor cells by downregulation of HOXA9 expression (140). Other studies also showed similar negative effects of TSA or valproic acid (VPA), another HDAC inhibitor, on the expression of endothelial nitric oxide synthase (eNOS) at a high concentration (∼0.3 μg/ml) of inhibitors and on angiogenesis of human endothelial cells (114, 139). In contrast, HDAC inhibitors at a low concentration (0.1 μg/ml) in combination with DNMT inhibitor, 5′-aza-2′-deoxycystidine (5-aza-dC), induced differentiation into endothelial cells from bone-marrow-derived multipotent adult progenitor cells (103). This inhibitor of DNMT, 5-aza-dC, can also induce endothelial cell differentiation from mouse ES cells (6). Interestingly, nitric oxide (NO), which is generated by eNOS, results in the activation of class IIa HDACs and subsequently induces mesoderm differentiation of mouse ES cells (159). The elevated level of NO in endothelial cells by shear stress and direct exposure of mouse ES cells to NO upregulated vascular gene expression, including Acta2 (also known as smooth muscle actin), Taglin (smooth muscle protein 22-alpha), Pecam1 (CD31), Kdr (Flk-1), Mef2c, and Acta1 (alpha-sarcomeric actin) (64, 159).
Cardiac differentiation
Inhibition of DNA methylation by small molecule chemical, 5-aza-dC, also promotes cardiac differentiation of ES cells in a time-dependent manner (193). In addition, inhibition of HDAC by TSA promotes cardiac differentiation by increased expression of acetylated Gata4, Nkx2-5, and Mef2c (74). In Nxk2.5-GFP mouse ES cells, TSA exposure between days 7 and 8 of differentiation doubled the percentage of GFP+ cells, and boosted expression of Nkx2-5, Myh7 (β-MHC), and Naap (atrial natriuretic factor [ANF]). TSA-treated mouse EBs, which showed enhanced acetylation of histone 3 and 4, have increased GATA4 acetylation and DNA binding to the ANF promoter and enhancedcardiomyocyte differentiation (74). Another cardiac differentiation-related histone methylatransferase is Dot1L, which catalyzes H3K79 methylation despite lacking a SET domain (44). Dot1L knockout mice are embryonically lethal due to dilated heart and angiogenesis defects in yolk sac. ES cells from Dot1L knockout blastocysts show global loss of H3K79 methylation as well as other repressive histone marks, H3K9me2 and H4K20me3, and display aneuploidy, telomere elongation, and proliferation defects (68). In addition, overexpression of cardiac-enriched subunit of the SWI/SNF-like chromatin remodeling BAF complex, Baf60c (also known as Smarcd3), when combined with ectopic expression of Gata4 and Tbx5, was able to induce cardiac differentiation of mouse embryos (171).
Reprogramming of Cell Fate and the Role of the Epigenome
In 2006, Shinya Yamanaka discovered that adult somatic cells can be reprogrammed to ES-like cells called “iPS cells” using viral transduction of pluripotency-related transcription factor genes (169, 170). The mechanism by which ectopic transcription factors override the existing epigenetic state and change it into a specific alternative state without passing through normal development or complete resetting of all marks is still largely unknown; however this triumph suggests the flexibility of the epigenome and its influence on cell fate determination. The key factors involved in the epigenetic remodeling have not been identified, and many questions regarding the dynamics of this process still remain.
Epigenetic changes during reprogramming
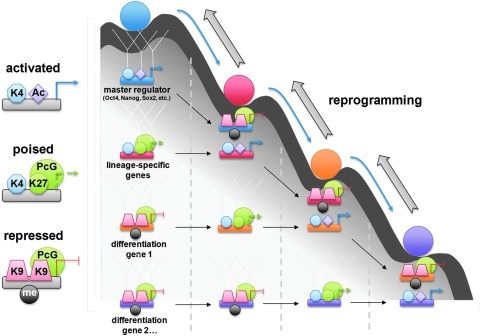
The term “epigenetics” was first coined by Waddington in 1942 to describe the role of genetic interactions in phenotype. Later, he presented the epigenetic landscape model in 1957 by using the metaphor of a ball traveling down a canal that starts from the stage of the fertilized totipotent embryo and ends with various lineage-specific cells. In this concept, cells move downward through different one-way branched valleys inside a canal and select their ultimate irreversible cellular fates (178). The four transcription factors essential in reprogramming reverse the cells' path in this canal by unlocking specific epigenetic gateways, which normally induces stabilization into a differentiated form (194) (Fig. 5).
FIG. 5.
Epigenetic changes in pluripotent stem cells and reprogramming. Epigenetic landscape of PS cells showing cell populations with different developmental potentials is illustrated as marbles rolling down a landscape into one of levels at each cell fates. At each developmental stage, the combinations of changing of epigenetic marks for different sets of genes determine the epigenetic landscape of cells. Thick arrows show examples of reprogramming processes. (To see this illustration in color the reader is referred to the web version of this article at www.liebertonline.com/ars).
Yamanaka proposed a stochastic model for iPS cell generation. A group of cells are prevented from moving back up the slope toward the state of pluripotency by some epigenetic barrier; once they overcome such barriers, they will achieve the ability to self-renew. The second group goes through partial reprogramming. This will cause them to lose their pluripotency and roll toward a specific lineage. The third group with improper expression of ectopic factors trans-differentiates. The fourth group undergoes apoptosis or death instead of traveling on the slope. Due to large differences in epigenetic status between ES cells and their differentiated progeny, Oct4 and Sox2 cannot find their targets in somatic cells. It has been proposed that c-Myc and Klf4 alter the structure of chromatin, enabling these two core factors access to their targets, thereby increasing the expression of downstream genes (170). c-Myc induces up-regulation of Gcn5 (histone acetyl transferase gene), which is a key player in histone structure, and therefore might improve the accessibility of target genes to Oct4. Klf4 is also acetylated by p300 (acetyl transferase protein) and has the ability to control gene transcription through regulation of histone acetylation (42).
Histone modification during reprogramming
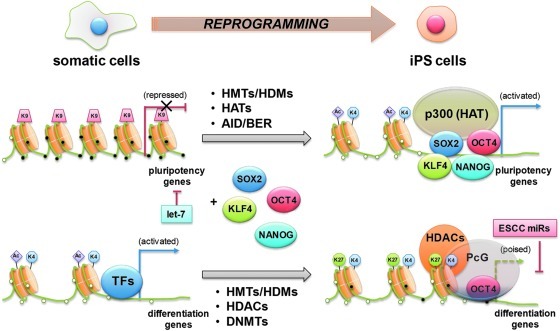
Studies using cDNA microarray and ChIP approaches revealed that Oct4 regulates the expression of over 350 genes in ES cells, including several epigenetic modifiers (106). Two HDMs, Jmjd1a and Jmjd2c, have been identified to be part of the groups of the genes regulated by Oct4 (100), and Jmdj2c is recruited to the Nanog promoter. In this way, ES cell transcription factors regulate the expression of chromatin remodeling genes and, in turn, help unveil chromatin conformation in promoter regions of target genes allowing self-regulation of the epigenetic network. During the reprogramming of fibroblasts into pluripotent cells, genes in an open chromatin state, which is marked by H3K4me3 or H3K4me3 and H3K27me3 (bivalent), are efficiently reactivated, whereas genes that are silenced by H3K27me3 alone remain mostly repressed. In both near promoters and intergenic regions, most of the high CpG promoters are enriched with monovalent H3K4me3 (activation mark) or bivalent H3K4me3 and H3K27me3 (repression mark) in ES cells (Fig. 6). However, MEFs show only H3K27 (115). One group of genes shows the reactivation and associated gain of H3K4me3 at key pluripotency factors. These can be further subdivided into two classes: the first includes genes such as Lin28, Sox2, and Fgf4, which are repressed by H3K27 and lack detectable H3K4me3; the second includes the genes Oct4, Nanog, and Dppa, which are repressed by DNA methylation and also lack detectable H3K4me3. The other major group describes genes, including MyoD, which are repressed by H3K27me3 and highly enriched for developmental transcription factors. To create a truly pluripotent cell line, these loci must remain repressed but need to acquire H3K4me3 to reestablish their bivalency and thus their developmental competence for all germ layers and cell types (115).
FIG. 6.
Reprogramming of cell fate by epigenetic changes. The reprogramming factors OCT4, SOX2, and KLF4 bind promoter regions of pluripotency-related genes, which is repressed by let-7 miRs in differentiated somatic cells, together with other transcription factors, including NANOG, and recruit co-activators, such as the HAT, p300, during reprogramming. This binding pattern is found in transcriptionally active genes in ES cells as open euchromatin structure with histone Ac, K4, and unmethylated DNA (small white circles). In reprogrammed iPS cells, genes that are bound by OCT4 and inhibited by ESCC miRs are often repressed potentially through the recruitment of PcG proteins, HDACs, or DNMTs (resulting DNA methylation as shown with small black circles), but become activated upon differentiation. ESCC, embryonic stem cell-specific cell cycle regulating; iPS, induced pluripotent stem. (To see this illustration in color the reader is referred to the web version of this article at www.liebertonline.com/ars).
DNA methylation during reprogramming
The efficiency of iPS cell reprogramming is extremely low (0.01–0.1%) (61). Treating differentiated cells with DNA methylation inhibitor 5-Aza-CR or siRNA or shRNA against Dnmt1 to revert repressive epigenetic marks of pluripotency genes can significantly increase the efficiency of iPS generation (115). These data suggest that epigenetic modifications are acquired to overcome the epigenetic repression of one or more key loci during the artificial process of reprogramming. DNA methylation is a critical epigenetic barrier to cellular reprogramming, and the removal of DNA methylation (i.e., demethylation) at the promoter of pluripotency genes is the key mechanism of reprogramming (155) (Fig. 6). The molecular mechanisms of DNA demethylation have been proposed as passive or active processes. Passive DNA demethylation occurs by DNA replication in the absence of methylation of newly synthesized DNA strands when maintenance methylation is impaired (197). Active DNA demethylation depends on demethylating enzymes, which is independent of replication (197). Activation-induced cytidine deaminase (AID, also known as AICDA) is the only enzyme that has been proposed as a DNA demethylase in mammalian reprogramming (13, 131). It is suggested that AID-mediated DNA demethylation occurs due to deamination of methylated cytidine residues in single-stranded DNA, followed by DNA repair (134). A recent study using a cell fusion reprogramming strategy showed that a rapid increase in Oct4 and Nanog expression was accompanied by DNA demethylation at their promoters (13). Moreover, AID knockdown by siRNA decreases the expression of pluripotency genes such as Oct4 and Nanog by reducing DNA demethylation (13). These data support the importance of active DNA demethylation by AID in reprogramming.
Role of miRs in reprogramming
The importance of miRs for ES cells is further indicated by the discovery that specific miRs mimics and inhibitors induce the reprogramming of somatic cells into iPS cells (3, 70, 117). Several miRs, including ES cell-specific cell cycle-regulating (ESCC) miRs (miR-291a-3p, miR-291b-3p, miR-294, miR-295, and miR-302), Myc-induced miRs (miR-200, miR-141, miR-429, and miR-17-92 clusters), miR-92b, and the miR-520 cluster, have been shown to positively regulate the self-renewal and pluripotency of ES cells. Among these, ESCC miRs (miR-291-3p, miR-294, and miR-295) increase the efficiency of reprogramming of fibroblasts into iPS cells induced by Oct4, Sox2, and Klf4 (70). Additionally, the inhibition of tissue-specific miRs, let-7, has been shown to promote reprogramming (111). miR-125, which inhibits the expression of Lin28, is also expected to positively influence reprogramming (190, 192) (Fig. 4). More importantly, ectopic expression of only miRs (miR-302/367 and miR-200c/302/369) without transcription factor over-expression was shown to have sufficient capability for rapid and efficient reprogramming of mouse and human somatic cells into iPS cells (3, 117).
The equivalency of ES cells and iPS cells is controversial, and this discrepancy is possibly mediated by the difference in the epigenetic status between these two types of PS cells (30, 125). During the reprogramming, miRs also have roles in epigenetic modifications that favor reprogramming. For example, the Dlk1–Dio3 gene cluster, which includes ∼50 conserved miRs, is often silenced in iPS cells (98, 161), and the treatment by HDAC inhibitors of iPS cells led to activation of the Dlk1–Dio3 gene miR cluster to enhance the developmental potential to be equivalent to that of ES cells (161). These findings suggest that the appropriate epigenetic status of cells needs to be achieved during the reprogramming, and further reinforce the critical roles of miRs for establishing and maintaining pluripotency. Additionally, inhibition or knock-down of Dnmt enhanced iPS cell generation by inducing the demethylation of promoters of pluripotency-related genes (115). Dnmt inhibitors were also used to generate iPS cells with only two factors (Oct4 and Klf4) or three factors (Sox2, Oct4, and Klf4) (63, 152). The targets of miR-29b, which induces global DNA hypomethylation and re-expression of Ink4b tumor suppressor gene, are Dnmt1, 3a, and 3b (51). Therefore, miR-29b expression and Dnmt inhibitors is expected to enhance reprogramming efficiency by promoting demethylation of promoter regions of pluripotency-related genes. These results indicate a critical role for miRs in reprogramming somatic cells into iPS cells.
Epigenome reprogramming by small molecules
Strong evidence that epigenetic modifications are closely related to the reprogramming of somatic cells is augmentation of reprogramming efficiency by the use of small molecules in generating iPS cells. In somatic cells, reprogramming factors have highly methylated endogenous loci. However, in ES cells and iPS cells, these factors are hypomethylated (65). Their promoters need to be demethylated in order to be reactivated and thereby derive iPS cells. Direct reprogramming steps require downstream epigenetic modifiers because the direct reprogramming factors lack demethylation activity. Differences in epigenetic modifiers such as CpG methylation between ES cells and lineage-specific ES cells emphasize importance of epigenetic landscape in iPS cell generation. Doi et al. (40) found that characterization of CpG methylation can help in distinguishing the identity of cell types such as fibroblasts, ES cells, and iPS cells. Because chromatin remodeling is important for reprogramming, small molecules are being used to overcome these epigenetic blocks and enhance iPS cell generation. Consequently, DNA and HMTs, chemical inhibitors of HDACs, and genetic factors were used in different combinations (63, 153). Ding and colleagues (83) found that a small-molecule inhibitor of G9a HMT, BIX-01294, could enhance the induction of reprogramming in neural stem cells while replacing Oct4. Since G9a is a down-regulator of Oct4 during early development, they suggested that BIX-01294 enhances iPS cell formation by inhibiting G9a and subsequently releasing Oct4 from negative regulation. iPS cells were generated from MEFs with only two factors, Oct4 and Klf4, in the presence of BIX-01294.
In a four-factor system, VPA increases the efficiency and kinetics of reprogramming by a hundred fold. VPA can chemically induce iPS cell generation with only Oct4 and Sox2 from human fibroblasts (63) and it could replace either Klf4 or c-Myc in reprogramming steps. These results indicate that DNA methylation, histone methylation, and histone deacetylation contribute to epigenetic hurdles which limit effective iPS cell generation.
Conclusion
Recent advances in unbiased next-generation sequencing methods for whole epigenome sequencing allowed mapping of epigenetic marks at single-nucleotide resolution and led to novel insights into the role of epigenetic changes in transcriptional regulation and cell fate determination. In particular, epigenome sequencing of ES and iPS cells shed light on the mechanisms governing maintenance and induction of pluripotency. In addition, other advances have been made in regard to the identification of PS cell-related miRs, their role in reprogramming and maintenance of PS cells, and their interaction with other epigenetic modulators.
However, much more work needs to be done to fully understand the role of epigenetic modifications in PS cells. Particularly, the dynamic changes of chromatin state in responses to the ectopic expression of transcription factors during the generation of iPS cells should be comprehensively investigated to develop more efficient methods of reprogramming. New approaches for defining the epigenetic landscape are needed for a better understanding of the chromatin structure and its role in establishment of cell identity. A comprehensive epigenomic map will also be helpful to determine the quality and utility of directly reprogrammed or in vitro differentiated cells. Epigenome reference maps and data by the enormous increase in sequencing capabilities will significantly facilitate our understanding of the epigenetic landscape of PS cells and their dynamic changes during reprogramming and differentiation.
Abbreviations Used
- Ac
acetylation
- acetyl-CoA
acetyl coenzyme A
- ADMA
asymmetric dimethyl-arg
- AID
activation-induced cytidine deaminase
- AP
apurinic/apyrimidinic
- arg
arginine
- BER
base excision repair
- CHD
chromodomain helicase DNA-binding
- ChIP
chromatin-immunoprecipitation
- CRPs
chromatin remodeling proteins
- DNMT
DNA methyltransferase
- dR5P
deoxyribose-5-phosphate
- dRpase
deoxyribophosphodiesterase
- eNOS
endothelial nitric oxide synthase
- ES
embryonic stem
- ESCC
embryonic stem cell-specific cell cycle regulating
- FAD+
flavin adenine dinucleotide
- 5-hmC
5-hydroxymethylcytosine
- 5-mC
5-methylcytosine
- HAT
histone acetyltransferase
- HDAC
histone deacetylase
- HDM
histone demethylase
- HKMT
histone lysine methyltransferase
- HMTs
histone methyltransferases
- iPS
induced pluripotent stem
- ISWI
imitation switch
- jmjC
jumonji-domain containing
- K4
H3K4 methylation
- K9
H3K9 methylation
- K27
H3K27 methylation
- LIF
leukemia inhibitory factor
- LSD1
lysine-specific demethylase 1
- lys
lysine
- MEFs
mouse embryonic fibroblasts
- miRs
microRNAs
- NAD+
nicotinamide adenine dinucleotide
- Nam
nicotinamide
- NGS
next-generation sequencing
- NPCs
neural progenitor cells
- NuRD
nucleosome remodeling deacetylase
- OAADPr
2′-O-acetyl-ADP-ribose
- PcG
polycomb group protein
- PRCs
polycomb repressive complexes
- PRMT
protein arginine methyltransferase
- PS
pluripotent stem
- SAH
S-adenosyl homocysteine
- SAM
S-adenosyl methionine
- SDMA
symmetric dimethyl-arg
- Succ
succinate
- SWI/SNF
switch/sucrose nonfermentable
- TET
ten–eleven translocation
- TFs
transcription factors
- TSA
trichostatin A
- 2-OG
2-oxoglutarate
- VPA
valproic acid
Acknowledgments
This work was supported in part by NIH grants DP3DK094346, RC1GM092035, and NIH contract, HHSN268201000043C (Program of Excellence in Nanotechnology Award); NSF-EBICS (Emergent Behaviors of Integrated Cellular Systems) grant; and Stem Cell Research Center of the 21st Century Frontier Research Program grant SC4300, funded by the Ministry of Science and Technology, Republic of Korea. We would like to thank Andrea Wecker and Julie J. Kim for editing of the article and Juhyun Kim for assisting in preparation of figures.
References
- 1.Agger K. Cloos PA. Christensen J. Pasini D. Rose S. Rappsilber J. Issaeva I. Canaani E. Salcini AE. Helin K. UTX and JMJD3 are histone H3K27 demethylases involved in HOX gene regulation and development. Nature. 2007;449:731–734. doi: 10.1038/nature06145. [DOI] [PubMed] [Google Scholar]
- 2.Ambros V. Chen X. The regulation of genes and genomes by small RNAs. Development. 2007;134:1635–1641. doi: 10.1242/dev.002006. [DOI] [PubMed] [Google Scholar]
- 3.Anokye-Danso F. Trivedi CM. Juhr D. Gupta M. Cui Z. Tian Y. Zhang Y. Yang W. Gruber PJ. Epstein JA. Morrisey EE. Highly efficient miRNA-mediated reprogramming of mouse and human somatic cells to pluripotency. Cell Stem Cell. 2011;8:376–388. doi: 10.1016/j.stem.2011.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Azuara V. Perry P. Sauer S. Spivakov M. Jorgensen HF. John RM. Gouti M. Casanova M. Warnes G. Merkenschlager M. Fisher AG. Chromatin signatures of pluripotent cell lines. Nat Cell Biol. 2006;8:532–538. doi: 10.1038/ncb1403. [DOI] [PubMed] [Google Scholar]
- 5.Ballas N. Grunseich C. Lu DD. Speh JC. Mandel G. REST and its corepressors mediate plasticity of neuronal gene chromatin throughout neurogenesis. Cell. 2005;121:645–657. doi: 10.1016/j.cell.2005.03.013. [DOI] [PubMed] [Google Scholar]
- 6.Banerjee S. Bacanamwo M. DNA methyltransferase inhibition induces mouse embryonic stem cell differentiation into endothelial cells. Exp Cell Res. 2010;316:172–180. doi: 10.1016/j.yexcr.2009.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Barroso-del Jesus A. Lucena-Aguilar G. Menendez P. The miR-302-367 cluster as a potential stemness regulator in ESCs. Cell Cycle. 2009;8:394–398. doi: 10.4161/cc.8.3.7554. [DOI] [PubMed] [Google Scholar]
- 8.Barroso-del Jesus A. Romero-Lopez C. Lucena-Aguilar G. Melen GJ. Sanchez L. Ligero G. Berzal-Herranz A. Menendez P. Embryonic stem cell-specific miR302-367 cluster: human gene structure and functional characterization of its core promoter. Mol Cell Biol. 2008;28:6609–6619. doi: 10.1128/MCB.00398-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Benetti R. Gonzalo S. Jaco I. Munoz P. Gonzalez S. Schoeftner S. Murchison E. Andl T. Chen T. Klatt P. Li E. Serrano M. Millar S. Hannon G. Blasco MA. A mammalian microRNA cluster controls DNA methylation and telomere recombination via Rbl2-dependent regulation of DNA methyltransferases. Nat Struct Mol Biol. 2008;15:268–279. doi: 10.1038/nsmb.1399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bernstein BE. Meissner A. Lander ES. The mammalian epigenome. Cell. 2007;128:669–681. doi: 10.1016/j.cell.2007.01.033. [DOI] [PubMed] [Google Scholar]
- 11.Bernstein BE. Mikkelsen TS. Xie X. Kamal M. Huebert DJ. Cuff J. Fry B. Meissner A. Wernig M. Plath K. Jaenisch R. Wagschal A. Feil R. Schreiber SL. Lander ES. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell. 2006;125:315–326. doi: 10.1016/j.cell.2006.02.041. [DOI] [PubMed] [Google Scholar]
- 12.Bernstein E. Kim SY. Carmell MA. Murchison EP. Alcorn H. Li MZ. Mills AA. Elledge SJ. Anderson KV. Hannon GJ. Dicer is essential for mouse development. Nat Genet. 2003;35:215–217. doi: 10.1038/ng1253. [DOI] [PubMed] [Google Scholar]
- 13.Bhutani N. Brady JJ. Damian M. Sacco A. Corbel SY. Blau HM. Reprogramming towards pluripotency requires AID-dependent DNA demethylation. Nature. 2010;463:1042–1047. doi: 10.1038/nature08752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bird A. DNA methylation patterns and epigenetic memory. Genes Dev. 2002;16:6–21. doi: 10.1101/gad.947102. [DOI] [PubMed] [Google Scholar]
- 15.Blosser TR. Yang JG. Stone MD. Narlikar GJ. Zhuang X. Dynamics of nucleosome remodelling by individual ACF complexes. Nature. 2009;462:1022–1027. doi: 10.1038/nature08627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bodnar MS. Meneses JJ. Rodriguez RT. Firpo MT. Propagation and maintenance of undifferentiated human embryonic stem cells. Stem Cells Dev. 2004;13:243–253. doi: 10.1089/154732804323099172. [DOI] [PubMed] [Google Scholar]
- 17.Boyer LA. Lee TI. Cole MF. Johnstone SE. Levine SS. Zucker JP. Guenther MG. Kumar RM. Murray HL. Jenner RG. Gifford DK. Melton DA. Jaenisch R. Young RA. Core transcriptional regulatory circuitry in human embryonic stem cells. Cell. 2005;122:947–956. doi: 10.1016/j.cell.2005.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Boyer LA. Plath K. Zeitlinger J. Brambrink T. Medeiros LA. Lee TI. Levine SS. Wernig M. Tajonar A. Ray MK. Bell GW. Otte AP. Vidal M. Gifford DK. Young RA. Jaenisch R. Polycomb complexes repress developmental regulators in murine embryonic stem cells. Nature. 2006;441:349–353. doi: 10.1038/nature04733. [DOI] [PubMed] [Google Scholar]
- 19.Boyle S. Gilchrist S. Bridger JM. Mahy NL. Ellis JA. Bickmore WA. The spatial organization of human chromosomes within the nuclei of normal and emerin-mutant cells. Hum Mol Genet. 2001;10:211–219. doi: 10.1093/hmg/10.3.211. [DOI] [PubMed] [Google Scholar]
- 20.Brambrink T. Foreman R. Welstead GG. Lengner CJ. Wernig M. Suh H. Jaenisch R. Sequential expression of pluripotency markers during direct reprogramming of mouse somatic cells. Cell Stem Cell. 2008;2:151–159. doi: 10.1016/j.stem.2008.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bultman S. Gebuhr T. Yee D. La Mantia C. Nicholson J. Gilliam A. Randazzo F. Metzger D. Chambon P. Crabtree G. Magnuson T. A Brg1 null mutation in the mouse reveals functional differences among mammalian SWI/SNF complexes. Mol Cell. 2000;6:1287–1295. doi: 10.1016/s1097-2765(00)00127-1. [DOI] [PubMed] [Google Scholar]
- 22.Burgold T. Spreafico F. De Santa F. Totaro MG. Prosperini E. Natoli G. Testa G. The histone H3 lysine 27-specific demethylase Jmjd3 is required for neural commitment. PLoS One. 2008;3:e3034. doi: 10.1371/journal.pone.0003034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Calabrese JM. Seila AC. Yeo GW. Sharp PA. RNA sequence analysis defines Dicer's role in mouse embryonic stem cells. Proc Natl Acad Sci U S A. 2007;104:18097–18102. doi: 10.1073/pnas.0709193104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cao S. Bendall H. Hicks GG. Nashabi A. Sakano H. Shinkai Y. Gariglio M. Oltz EM. Ruley HE. The high-mobility-group box protein SSRP1/T160 is essential for cell viability in day 3.5 mouse embryos. Mol Cell Biol. 2003;23:5301–5307. doi: 10.1128/MCB.23.15.5301-5307.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Card DA. Hebbar PB. Li L. Trotter KW. Komatsu Y. Mishina Y. Archer TK. Oct4/Sox2-regulated miR-302 targets cyclin D1 in human embryonic stem cells. Mol Cell Biol. 2008;28:6426–6438. doi: 10.1128/MCB.00359-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chang TC. Yu D. Lee YS. Wentzel EA. Arking DE. West KM. Dang CV. Thomas-Tikhonenko A. Mendell JT. Widespread microRNA repression by Myc contributes to tumorigenesis. Nat Genet. 2008;40:43–50. doi: 10.1038/ng.2007.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chen L. Daley GQ. Molecular basis of pluripotency. Hum Mol Genet. 2008;17:R23–R27. doi: 10.1093/hmg/ddn050. [DOI] [PubMed] [Google Scholar]
- 28.Chen T. Li E. Structure and function of eukaryotic DNA methyltransferases. Curr Top Dev Biol. 2004;60:55–89. doi: 10.1016/S0070-2153(04)60003-2. [DOI] [PubMed] [Google Scholar]
- 29.Chen T. Ueda Y. Dodge JE. Wang Z. Li E. Establishment and maintenance of genomic methylation patterns in mouse embryonic stem cells by Dnmt3a and Dnmt3b. Mol Cell Biol. 2003;23:5594–5605. doi: 10.1128/MCB.23.16.5594-5605.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chin MH. Mason MJ. Xie W. Volinia S. Singer M. Peterson C. Ambartsumyan G. Aimiuwu O. Richter L. Zhang J. Khvorostov I. Ott V. Grunstein M. Lavon N. Benvenisty N. Croce CM. Clark AT. Baxter T. Pyle AD. Teitell MA. Pelegrini M. Plath K. Lowry WE. Induced pluripotent stem cells and embryonic stem cells are distinguished by gene expression signatures. Cell Stem Cell. 2009;5:111–123. doi: 10.1016/j.stem.2009.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Clark AT. Rodriguez RT. Bodnar MS. Abeyta MJ. Cedars MI. Turek PJ. Firpo MT. Reijo Pera RA. Human STELLAR, NANOG, and GDF3 genes are expressed in pluripotent cells and map to chromosome 12p13, a hotspot for teratocarcinoma. Stem Cells. 2004;22:169–179. doi: 10.1634/stemcells.22-2-169. [DOI] [PubMed] [Google Scholar]
- 32.Constantinescu D. Gray HL. Sammak PJ. Schatten GP. Csoka AB. Lamin A/C expression is a marker of mouse and human embryonic stem cell differentiation. Stem Cells. 2006;24:177–185. doi: 10.1634/stemcells.2004-0159. [DOI] [PubMed] [Google Scholar]
- 33.Dang W. Kagalwala MN. Bartholomew B. The Dpb4 subunit of ISW2 is anchored to extranucleosomal DNA. J Biol Chem. 2007;282:19418–19425. doi: 10.1074/jbc.M700640200. [DOI] [PubMed] [Google Scholar]
- 34.Dawlaty MM. Ganz K. Powell BE. Hu YC. Markoulaki S. Cheng AW. Gao Q. Kim J. Choi SW. Page DC. Jaenisch R. Tet1 is dispensable for maintaining pluripotency and its loss is compatible with embryonic and postnatal development. Cell Stem Cell. 2011;9:166–175. doi: 10.1016/j.stem.2011.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.De Santa F. Totaro MG. Prosperini E. Notarbartolo S. Testa G. Natoli G. The histone H3 lysine-27 demethylase Jmjd3 links inflammation to inhibition of polycomb-mediated gene silencing. Cell. 2007;130:1083–1094. doi: 10.1016/j.cell.2007.08.019. [DOI] [PubMed] [Google Scholar]
- 36.Deb-Rinker P. Ly D. Jezierski A. Sikorska M. Walker PR. Sequential DNA methylation of the Nanog and Oct-4 upstream regions in human NT2 cells during neuronal differentiation. J Biol Chem. 2005;280:6257–6260. doi: 10.1074/jbc.C400479200. [DOI] [PubMed] [Google Scholar]
- 37.Denslow SA. Wade PA. The human Mi-2/NuRD complex and gene regulation. Oncogene. 2007;26:5433–5438. doi: 10.1038/sj.onc.1210611. [DOI] [PubMed] [Google Scholar]
- 38.Dodge JE. Okano M. Dick F. Tsujimoto N. Chen T. Wang S. Ueda Y. Dyson N. Li E. Inactivation of Dnmt3b in mouse embryonic fibroblasts results in DNA hypomethylation, chromosomal instability, and spontaneous immortalization. J Biol Chem. 2005;280:17986–17991. doi: 10.1074/jbc.M413246200. [DOI] [PubMed] [Google Scholar]
- 39.Dodge JE. Ramsahoye BH. Wo ZG. Okano M. Li E. De novo methylation of MMLV provirus in embryonic stem cells: CpG versus non-CpG methylation. Gene. 2002;289:41–48. doi: 10.1016/s0378-1119(02)00469-9. [DOI] [PubMed] [Google Scholar]
- 40.Doi A. Park IH. Wen B. Murakami P. Aryee MJ. Irizarry R. Herb B. Ladd-Acosta C. Rho J. Loewer S. Miller J. Schlaeger T. Daley GQ. Feinberg AP. Differential methylation of tissue- and cancer-specific CpG island shores distinguishes human induced pluripotent stem cells, embryonic stem cells and fibroblasts. Nat Genet. 2009;41:1350–1353. doi: 10.1038/ng.471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Evans MJ. Kaufman MH. Establishment in culture of pluripotential cells from mouse embryos. Nature. 1981;292:154–156. doi: 10.1038/292154a0. [DOI] [PubMed] [Google Scholar]
- 42.Evans PM. Zhang W. Chen X. Yang J. Bhakat KK. Liu C. Kruppel-like factor 4 is acetylated by p300 and regulates gene transcription via modulation of histone acetylation. J Biol Chem. 2007;282:33994–34002. doi: 10.1074/jbc.M701847200. [DOI] [PubMed] [Google Scholar]
- 43.Fazzio TG. Huff JT. Panning B. An RNAi screen of chromatin proteins identifies Tip60-p400 as a regulator of embryonic stem cell identity. Cell. 2008;134:162–174. doi: 10.1016/j.cell.2008.05.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Feng Q. Wang H. Ng HH. Erdjument-Bromage H. Tempst P. Struhl K. Zhang Y. Methylation of H3-lysine 79 is mediated by a new family of HMTases without a SET domain. Curr Biol. 2002;12:1052–1058. doi: 10.1016/s0960-9822(02)00901-6. [DOI] [PubMed] [Google Scholar]
- 45.Flanagan JF. Mi LZ. Chruszcz M. Cymborowski M. Clines KL. Kim Y. Minor W. Rastinejad F. Khorasanizadeh S. Double chromodomains cooperate to recognize the methylated histone H3 tail. Nature. 2005;438:1181–1185. doi: 10.1038/nature04290. [DOI] [PubMed] [Google Scholar]
- 46.Fouse SD. Shen Y. Pellegrini M. Cole S. Meissner A. Van Neste L. Jaenisch R. Fan G. Promoter CpG methylation contributes to ES cell gene regulation in parallel with Oct4/Nanog, PcG complex, and histone H3 K4/K27 trimethylation. Cell Stem Cell. 2008;2:160–169. doi: 10.1016/j.stem.2007.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Fyodorov DV. Kadonaga JT. Dynamics of ATP-dependent chromatin assembly by ACF. Nature. 2002;418:897–900. doi: 10.1038/nature00929. [DOI] [PubMed] [Google Scholar]
- 48.Gangaraju VK. Bartholomew B. Dependency of ISW1a chromatin remodeling on extranucleosomal DNA. Mol Cell Biol. 2007;27:3217–3225. doi: 10.1128/MCB.01731-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gangaraju VK. Prasad P. Srour A. Kagalwala MN. Bartholomew B. Conformational changes associated with template commitment in ATP-dependent chromatin remodeling by ISW2. Mol Cell. 2009;35:58–69. doi: 10.1016/j.molcel.2009.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gao X. Tate P. Hu P. Tjian R. Skarnes WC. Wang Z. ES cell pluripotency and germ-layer formation require the SWI/SNF chromatin remodeling component BAF250a. Proc Natl Acad Sci U S A. 2008;105:6656–6661. doi: 10.1073/pnas.0801802105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Garzon R. Liu S. Fabbri M. Liu Z. Heaphy CE. Callegari E. Schwind S. Pang J. Yu J. Muthusamy N. Havelange V. Volinia S. Blum W. Rush LJ. Perrotti D. Andreeff M. Bloomfield CD. Byrd JC. Chan K. Wu LC. Croce CM. Marcucci G. MicroRNA-29b induces global DNA hypomethylation and tumor suppressor gene reexpression in acute myeloid leukemia by targeting directly DNMT3A and 3B and indirectly DNMT1. Blood. 2009;113:6411–6418. doi: 10.1182/blood-2008-07-170589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Golebiewska A. Atkinson SP. Lako M. Armstrong L. Epigenetic landscaping during hESC differentiation to neural cells. Stem Cells. 2009;27:1298–1308. doi: 10.1002/stem.59. [DOI] [PubMed] [Google Scholar]
- 53.Goll MG. Bestor TH. Eukaryotic cytosine methyltransferases. Annu Rev Biochem. 2005;74:481–514. doi: 10.1146/annurev.biochem.74.010904.153721. [DOI] [PubMed] [Google Scholar]
- 54.Gu P. Reid JG. Gao X. Shaw CA. Creighton C. Tran PL. Zhou X. Drabek RB. Steffen DL. Hoang DM. Weiss MK. Naghavi AO. El-daye J. Khan MF. Legge GB. Wheeler DA. Gibbs RA. Miller JN. Cooney AJ. Gunaratne PH. Novel microRNA candidates and miRNA-mRNA pairs in embryonic stem (ES) cells. PLoS One. 2008;3:e2548. doi: 10.1371/journal.pone.0002548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Guenther MG. Levine SS. Boyer LA. Jaenisch R. Young RA. A chromatin landmark and transcription initiation at most promoters in human cells. Cell. 2007;130:77–88. doi: 10.1016/j.cell.2007.05.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Gutierrez JL. Chandy M. Carrozza MJ. Workman JL. Activation domains drive nucleosome eviction by SWI/SNF. EMBO J. 2007;26:730–740. doi: 10.1038/sj.emboj.7601524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hatfield SD. Shcherbata HR. Fischer KA. Nakahara K. Carthew RW. Ruohola-Baker H. Stem cell division is regulated by the microRNA pathway. Nature. 2005;435:974–978. doi: 10.1038/nature03816. [DOI] [PubMed] [Google Scholar]
- 58.Hattori N. Nishino K. Ko YG. Ohgane J. Tanaka S. Shiota K. Epigenetic control of mouse Oct-4 gene expression in embryonic stem cells and trophoblast stem cells. J Biol Chem. 2004;279:17063–17069. doi: 10.1074/jbc.M309002200. [DOI] [PubMed] [Google Scholar]
- 59.Hayashita Y. Osada H. Tatematsu Y. Yamada H. Yanagisawa K. Tomida S. Yatabe Y. Kawahara K. Sekido Y. Takahashi T. A polycistronic microRNA cluster, miR-17-92, is overexpressed in human lung cancers and enhances cell proliferation. Cancer Res. 2005;65:9628–9632. doi: 10.1158/0008-5472.CAN-05-2352. [DOI] [PubMed] [Google Scholar]
- 60.Hendrich B. Tweedie S. The methyl-CpG binding domain and the evolving role of DNA methylation in animals. Trends Genet. 2003;19:269–277. doi: 10.1016/S0168-9525(03)00080-5. [DOI] [PubMed] [Google Scholar]
- 61.Hochedlinger K. Plath K. Epigenetic reprogramming and induced pluripotency. Development. 2009;136:509–523. doi: 10.1242/dev.020867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Houbaviy HB. Murray MF. Sharp PA. Embryonic stem cell-specific MicroRNAs. Dev Cell. 2003;5:351–358. doi: 10.1016/s1534-5807(03)00227-2. [DOI] [PubMed] [Google Scholar]
- 63.Huangfu D. Maehr R. Guo W. Eijkelenboom A. Snitow M. Chen AE. Melton DA. Induction of pluripotent stem cells by defined factors is greatly improved by small-molecule compounds. Nat Biotechnol. 2008;26:795–797. doi: 10.1038/nbt1418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Illi B. Scopece A. Nanni S. Farsetti A. Morgante L. Biglioli P. Capogrossi MC. Gaetano C. Epigenetic histone modification and cardiovascular lineage programming in mouse embryonic stem cells exposed to laminar shear stress. Circ Res. 2005;96:501–508. doi: 10.1161/01.RES.0000159181.06379.63. [DOI] [PubMed] [Google Scholar]
- 65.Imamura M. Miura K. Iwabuchi K. Ichisaka T. Nakagawa M. Lee J. Kanatsu-Shinohara M. Shinohara T. Yamanaka S. Transcriptional repression and DNA hypermethylation of a small set of ES cell marker genes in male germline stem cells. BMC Dev Biol. 2006;6:34. doi: 10.1186/1471-213X-6-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Ito S. D'Alessio AC. Taranova OV. Hong K. Sowers LC. Zhang Y. Role of Tet proteins in 5mC to 5hmC conversion, ES-cell self-renewal and inner cell mass specification. Nature. 2010;466:1129–1133. doi: 10.1038/nature09303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Jia D. Jurkowska RZ. Zhang X. Jeltsch A. Cheng X. Structure of Dnmt3a bound to Dnmt3L suggests a model for de novo DNA methylation. Nature. 2007;449:248–251. doi: 10.1038/nature06146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Jones B. Su H. Bhat A. Lei H. Bajko J. Hevi S. Baltus GA. Kadam S. Zhai H. Valdez R. Gonzalo S. Zhang Y. Li E. Chen T. The histone H3K79 methyltransferase Dot1L is essential for mammalian development and heterochromatin structure. PLoS Genet. 2008;4:e1000190. doi: 10.1371/journal.pgen.1000190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Josephson R. Ording CJ. Liu Y. Shin S. Lakshmipathy U. Toumadje A. Love B. Chesnut JD. Andrews PW. Rao MS. Auerbach JM. Qualification of embryonal carcinoma 2102Ep as a reference for human embryonic stem cell research. Stem Cells. 2007;25:437–446. doi: 10.1634/stemcells.2006-0236. [DOI] [PubMed] [Google Scholar]
- 70.Judson RL. Babiarz JE. Venere M. Blelloch R. Embryonic stem cell-specific microRNAs promote induced pluripotency. Nat Biotechnol. 2009;27:459–461. doi: 10.1038/nbt.1535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Kagalwala MN. Glaus BJ. Dang W. Zofall M. Bartholomew B. Topography of the ISW2-nucleosome complex: insights into nucleosome spacing and chromatin remodeling. EMBO J. 2004;23:2092–2104. doi: 10.1038/sj.emboj.7600220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Kaji K. Caballero IM. MacLeod R. Nichols J. Wilson VA. Hendrich B. The NuRD component Mbd3 is required for pluripotency of embryonic stem cells. Nat Cell Biol. 2006;8:285–292. doi: 10.1038/ncb1372. [DOI] [PubMed] [Google Scholar]
- 73.Kanellopoulou C. Muljo SA. Kung AL. Ganesan S. Drapkin R. Jenuwein T. Livingston DM. Rajewsky K. Dicer-deficient mouse embryonic stem cells are defective in differentiation and centromeric silencing. Genes Dev. 2005;19:489–501. doi: 10.1101/gad.1248505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Kawamura T. Ono K. Morimoto T. Wada H. Hirai M. Hidaka K. Morisaki T. Heike T. Nakahata T. Kita T. Hasegawa K. Acetylation of GATA-4 is involved in the differentiation of embryonic stem cells into cardiac myocytes. J Biol Chem. 2005;280:19682–19688. doi: 10.1074/jbc.M412428200. [DOI] [PubMed] [Google Scholar]
- 75.Keenen B. de la Serna IL. Chromatin remodeling in embryonic stem cells: regulating the balance between pluripotency and differentiation. J Cell Physiol. 2009;219:1–7. doi: 10.1002/jcp.21654. [DOI] [PubMed] [Google Scholar]
- 76.Keller G. Embryonic stem cell differentiation: emergence of a new era in biology and medicine. Genes Dev. 2005;19:1129–1155. doi: 10.1101/gad.1303605. [DOI] [PubMed] [Google Scholar]
- 77.Kim J. Chu J. Shen X. Wang J. Orkin SH. An extended transcriptional network for pluripotency of embryonic stem cells. Cell. 2008;132:1049–1061. doi: 10.1016/j.cell.2008.02.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Kim SH. McQueen PG. Lichtman MK. Shevach EM. Parada LA. Misteli T. Spatial genome organization during T-cell differentiation. Cytogenet Genome Res. 2004;105:292–301. doi: 10.1159/000078201. [DOI] [PubMed] [Google Scholar]
- 79.Klochendler-Yeivin A. Fiette L. Barra J. Muchardt C. Babinet C. Yaniv M. The murine SNF5/INI1 chromatin remodeling factor is essential for embryonic development and tumor suppression. EMBO Rep. 2000;1:500–506. doi: 10.1093/embo-reports/kvd129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Koh KP. Yabuuchi A. Rao S. Huang Y. Cunniff K. Nardone J. Laiho A. Tahiliani M. Sommer CA. Mostoslavsky G. Lahesmaa R. Orkin SH. Rodig SJ. Daley GQ. Rao A. Tet1 and Tet2 regulate 5-hydroxymethylcytosine production and cell lineage specification in mouse embryonic stem cells. Cell Stem Cell. 2011;8:200–213. doi: 10.1016/j.stem.2011.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Kouzarides T. Chromatin modifications and their function. Cell. 2007;128:693–705. doi: 10.1016/j.cell.2007.02.005. [DOI] [PubMed] [Google Scholar]
- 82.Kriaucionis S. Heintz N. The nuclear DNA base 5-hydroxymethylcytosine is present in Purkinje neurons and the brain. Science. 2009;324:929–930. doi: 10.1126/science.1169786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Kubicek S. O'Sullivan RJ. August EM. Hickey ER. Zhang Q. Teodoro ML. Rea S. Mechtler K. Kowalski JA. Homon CA. Kelly TA. Jenuwein T. Reversal of H3K9me2 by a small-molecule inhibitor for the G9a histone methyltransferase. Mol Cell. 2007;25:473–481. doi: 10.1016/j.molcel.2007.01.017. [DOI] [PubMed] [Google Scholar]
- 84.Kurisaki A. Hamazaki TS. Okabayashi K. Iida T. Nishine T. Chonan R. Kido H. Tsunasawa S. Nishimura O. Asashima M. Sugino H. Chromatin-related proteins in pluripotent mouse embryonic stem cells are downregulated after removal of leukemia inhibitory factor. Biochem Biophys Res Commun. 2005;335:667–675. doi: 10.1016/j.bbrc.2005.07.128. [DOI] [PubMed] [Google Scholar]
- 85.Kutay H. Bai S. Datta J. Motiwala T. Pogribny I. Frankel W. Jacob ST. Ghoshal K. Downregulation of miR-122 in the rodent and human hepatocellular carcinomas. J Cell Biochem. 2006;99:671–678. doi: 10.1002/jcb.20982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Lakshmipathy U. Love B. Goff LA. Jornsten R. Graichen R. Hart RP. Chesnut JD. MicroRNA expression pattern of undifferentiated and differentiated human embryonic stem cells. Stem Cells Dev. 2007;16:1003–1016. doi: 10.1089/scd.2007.0026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Lan F. Bayliss PE. Rinn JL. Whetstine JR. Wang JK. Chen S. Iwase S. Alpatov R. Issaeva I. Canaani E. Roberts TM. Chang HY. Shi Y. A histone H3 lysine 27 demethylase regulates animal posterior development. Nature. 2007;449:689–694. doi: 10.1038/nature06192. [DOI] [PubMed] [Google Scholar]
- 88.Landgraf P. Rusu M. Sheridan R. Sewer A. Iovino N. Aravin A. Pfeffer S. Rice A. Kamphorst AO. Landthaler M. Lin C. Socci ND. Hermida L. Fulci V. Chiaretti S. Foa R. Schliwka J. Fuchs U. Novosel A. Muller RU. Schermer B. Bissels U. Inman J. Phan Q. Chien M. Weir DB. Choksi R. De Vita G. Frezzetti D. Trompeter HI. Hornung V. Teng G. Hartmann G. Palkovits M. Di Lauro R. Wernet P. Macino G. Rogler CE. Nagle JW. Ju J. Papavasiliou FN. Benzing T. Lichter P. Tam W. Brownstein MJ. Bosio A. Borkhardt A. Russo JJ. Sander C. Zavolan M. Tuschl T. A mammalian microRNA expression atlas based on small RNA library sequencing. Cell. 2007;129:1401–1414. doi: 10.1016/j.cell.2007.04.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Laurent L. Wong E. Li G. Huynh T. Tsirigos A. Ong CT. Low HM. Kin Sung KW. Rigoutsos I. Loring J. Wei CL. Dynamic changes in the human methylome during differentiation. Genome Res. 2010;20:320–331. doi: 10.1101/gr.101907.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Laurent LC. Chen J. Ulitsky I. Mueller FJ. Lu C. Shamir R. Fan JB. Loring JF. Comprehensive microRNA profiling reveals a unique human embryonic stem cell signature dominated by a single seed sequence. Stem Cells. 2008;26:1506–1516. doi: 10.1634/stemcells.2007-1081. [DOI] [PubMed] [Google Scholar]
- 91.Lee JH. Hart SR. Skalnik DG. Histone deacetylase activity is required for embryonic stem cell differentiation. Genesis. 2004;38:32–38. doi: 10.1002/gene.10250. [DOI] [PubMed] [Google Scholar]
- 92.Lee MG. Villa R. Trojer P. Norman J. Yan KP. Reinberg D. Di Croce L. Shiekhattar R. Demethylation of H3K27 regulates polycomb recruitment and H2A ubiquitination. Science. 2007;318:447–450. doi: 10.1126/science.1149042. [DOI] [PubMed] [Google Scholar]
- 93.Lee TI. Jenner RG. Boyer LA. Guenther MG. Levine SS. Kumar RM. Chevalier B. Johnstone SE. Cole MF. Isono K. Koseki H. Fuchikami T. Abe K. Murray HL. Zucker JP. Yuan B. Bell GW. Herbolsheimer E. Hannett NM. Sun K. Odom DT. Otte AP. Volkert TL. Bartel DP. Melton DA. Gifford DK. Jaenisch R. Young RA. Control of developmental regulators by Polycomb in human embryonic stem cells. Cell. 2006;125:301–313. doi: 10.1016/j.cell.2006.02.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Lei H. Oh SP. Okano M. Juttermann R. Goss KA. Jaenisch R. Li E. De novo DNA cytosine methyltransferase activities in mouse embryonic stem cells. Development. 1996;122:3195–3205. doi: 10.1242/dev.122.10.3195. [DOI] [PubMed] [Google Scholar]
- 95.Li B. Carey M. Workman JL. The role of chromatin during transcription. Cell. 2007;128:707–719. doi: 10.1016/j.cell.2007.01.015. [DOI] [PubMed] [Google Scholar]
- 96.Li E. Bestor TH. Jaenisch R. Targeted mutation of the DNA methyltransferase gene results in embryonic lethality. Cell. 1992;69:915–926. doi: 10.1016/0092-8674(92)90611-f. [DOI] [PubMed] [Google Scholar]
- 97.Lister R. Pelizzola M. Dowen RH. Hawkins RD. Hon G. Tonti-Filippini J. Nery JR. Lee L. Ye Z. Ngo QM. Edsall L. Antosiewicz-Bourget J. Stewart R. Ruotti V. Millar AH. Thomson JA. Ren B. Ecker JR. Human DNA methylomes at base resolution show widespread epigenomic differences. Nature. 2009;462:315–322. doi: 10.1038/nature08514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Liu L. Luo GZ. Yang W. Zhao X. Zheng Q. Lv Z. Li W. Wu HJ. Wang L. Wang XJ. Zhou Q. Activation of the imprinted Dlk1-Dio3 region correlates with pluripotency levels of mouse stem cells. J Biol Chem. 2010;285:19483–19490. doi: 10.1074/jbc.M110.131995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Loh YH. Wu Q. Chew JL. Vega VB. Zhang W. Chen X. Bourque G. George J. Leong B. Liu J. Wong KY. Sung KW. Lee CW. Zhao XD. Chiu KP. Lipovich L. Kuznetsov VA. Robson P. Stanton LW. Wei CL. Ruan Y. Lim B. Ng HH. The Oct4 and Nanog transcription network regulates pluripotency in mouse embryonic stem cells. Nat Genet. 2006;38:431–440. doi: 10.1038/ng1760. [DOI] [PubMed] [Google Scholar]
- 100.Loh YH. Zhang W. Chen X. George J. Ng HH. Jmjd1a and Jmjd2c histone H3 Lys 9 demethylases regulate self-renewal in embryonic stem cells. Genes Dev. 2007;21:2545–2557. doi: 10.1101/gad.1588207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Lorch Y. Maier-Davis B. Kornberg RD. Mechanism of chromatin remodeling. Proc Natl Acad Sci U S A. 2010;107:3458–3462. doi: 10.1073/pnas.1000398107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Maherali N. Sridharan R. Xie W. Utikal J. Eminli S. Arnold K. Stadtfeld M. Yachechko R. Tchieu J. Jaenisch R. Plath K. Hochedlinger K. Directly reprogrammed fibroblasts show global epigenetic remodeling and widespread tissue contribution. Cell Stem Cell. 2007;1:55–70. doi: 10.1016/j.stem.2007.05.014. [DOI] [PubMed] [Google Scholar]
- 103.Mahpatra S. Firpo MT. Bacanamwo M. Inhibition of DNA methyltransferases and histone deacetylases induces bone marrow-derived multipotent adult progenitor cells to differentiate into endothelial cells. Ethn Dis. 2010;20:S1-60–S1-64. [PMC free article] [PubMed] [Google Scholar]
- 104.Marshall WF. Gene expression and nuclear architecture during development and differentiation. Mech Dev. 2003;120:1217–1230. doi: 10.1016/j.mod.2003.05.001. [DOI] [PubMed] [Google Scholar]
- 105.Marson A. Levine SS. Cole MF. Frampton GM. Brambrink T. Johnstone S. Guenther MG. Johnston WK. Wernig M. Newman J. Calabrese JM. Dennis LM. Volkert TL. Gupta S. Love J. Hannett N. Sharp PA. Bartel DP. Jaenisch R. Young RA. Connecting microRNA genes to the core transcriptional regulatory circuitry of embryonic stem cells. Cell. 2008;134:521–533. doi: 10.1016/j.cell.2008.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Matoba R. Niwa H. Masui S. Ohtsuka S. Carter MG. Sharov AA. Ko MS. Dissecting Oct3/4-regulated gene networks in embryonic stem cells by expression profiling. PLoS One. 2006;1:e26. doi: 10.1371/journal.pone.0000026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Meissner A. Epigenetic modifications in pluripotent and differentiated cells. Nat Biotechnol. 2010;28:1079–1088. doi: 10.1038/nbt.1684. [DOI] [PubMed] [Google Scholar]
- 108.Meissner A. Gnirke A. Bell GW. Ramsahoye B. Lander ES. Jaenisch R. Reduced representation bisulfite sequencing for comparative high-resolution DNA methylation analysis. Nucleic Acids Res. 2005;33:5868–5877. doi: 10.1093/nar/gki901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Meissner A. Mikkelsen TS. Gu H. Wernig M. Hanna J. Sivachenko A. Zhang X. Bernstein BE. Nusbaum C. Jaffe DB. Gnirke A. Jaenisch R. Lander ES. Genome-scale DNA methylation maps of pluripotent and differentiated cells. Nature. 2008;454:766–770. doi: 10.1038/nature07107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Meissner A. Wernig M. Jaenisch R. Direct reprogramming of genetically unmodified fibroblasts into pluripotent stem cells. Nat Biotechnol. 2007;25:1177–1181. doi: 10.1038/nbt1335. [DOI] [PubMed] [Google Scholar]
- 111.Melton C. Judson RL. Blelloch R. Opposing microRNA families regulate self-renewal in mouse embryonic stem cells. Nature. 2010;463:621–626. doi: 10.1038/nature08725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Menendez P. Bueno C. Wang L. Human embryonic stem cells: a journey beyond cell replacement therapies. Cytotherapy. 2006;8:530–541. doi: 10.1080/14653240601026654. [DOI] [PubMed] [Google Scholar]
- 113.Meshorer E. Misteli T. Chromatin in pluripotent embryonic stem cells and differentiation. Nat Rev Mol Cell Biol. 2006;7:540–546. doi: 10.1038/nrm1938. [DOI] [PubMed] [Google Scholar]
- 114.Michaelis M. Michaelis UR. Fleming I. Suhan T. Cinatl J. Blaheta RA. Hoffmann K. Kotchetkov R. Busse R. Nau H. Cinatl J., Jr. Valproic acid inhibits angiogenesis in vitro and in vivo. Mol Pharmacol. 2004;65:520–527. doi: 10.1124/mol.65.3.520. [DOI] [PubMed] [Google Scholar]
- 115.Mikkelsen TS. Hanna J. Zhang X. Ku M. Wernig M. Schorderet P. Bernstein BE. Jaenisch R. Lander ES. Meissner A. Dissecting direct reprogramming through integrative genomic analysis. Nature. 2008;454:49–55. doi: 10.1038/nature07056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Mikkelsen TS. Ku M. Jaffe DB. Issac B. Lieberman E. Giannoukos G. Alvarez P. Brockman W. Kim TK. Koche RP. Lee W. Mendenhall E. O'Donovan A. Presser A. Russ C. Xie X. Meissner A. Wernig M. Jaenisch R. Nusbaum C. Lander ES. Bernstein BE. Genome-wide maps of chromatin state in pluripotent and lineage-committed cells. Nature. 2007;448:553–560. doi: 10.1038/nature06008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Miyoshi N. Ishii H. Nagano H. Haraguchi N. Dewi DL. Kano Y. Nishikawa S. Tanemura M. Mimori K. Tanaka F. Saito T. Nishimura J. Takemasa I. Mizushima T. Ikeda M. Yamamoto H. Sekimoto M. Doki Y. Mori M. Reprogramming of mouse and human cells to pluripotency using mature microRNAs. Cell Stem Cell. 2011;8:633–638. doi: 10.1016/j.stem.2011.05.001. [DOI] [PubMed] [Google Scholar]
- 118.Mohn F. Weber M. Rebhan M. Roloff TC. Richter J. Stadler MB. Bibel M. Schubeler D. Lineage-specific polycomb targets and de novo DNA methylation define restriction and potential of neuronal progenitors. Mol Cell. 2008;30:755–766. doi: 10.1016/j.molcel.2008.05.007. [DOI] [PubMed] [Google Scholar]
- 119.Morin RD. O'Connor MD. Griffith M. Kuchenbauer F. Delaney A. Prabhu AL. Zhao Y. McDonald H. Zeng T. Hirst M. Eaves CJ. Marra MA. Application of massively parallel sequencing to microRNA profiling and discovery in human embryonic stem cells. Genome Res. 2008;18:610–621. doi: 10.1101/gr.7179508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Murchison EP. Partridge JF. Tam OH. Cheloufi S. Hannon GJ. Characterization of Dicer-deficient murine embryonic stem cells. Proc Natl Acad Sci U S A. 2005;102:12135–12140. doi: 10.1073/pnas.0505479102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Nakagawa M. Koyanagi M. Tanabe K. Takahashi K. Ichisaka T. Aoi T. Okita K. Mochiduki Y. Takizawa N. Yamanaka S. Generation of induced pluripotent stem cells without Myc from mouse and human fibroblasts. Nat Biotechnol. 2008;26:101–106. doi: 10.1038/nbt1374. [DOI] [PubMed] [Google Scholar]
- 122.Newman MA. Thomson JM. Hammond SM. Lin-28 interaction with the Let-7 precursor loop mediates regulated microRNA processing. RNA. 2008;14:1539–1549. doi: 10.1261/rna.1155108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Nimmo RA. Slack FJ. An elegant miRror: microRNAs in stem cells, developmental timing and cancer. Chromosoma. 2009;118:405–418. doi: 10.1007/s00412-009-0210-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.O'Donnell KA. Wentzel EA. Zeller KI. Dang CV. Mendell JT. c-Myc-regulated microRNAs modulate E2F1 expression. Nature. 2005;435:839–843. doi: 10.1038/nature03677. [DOI] [PubMed] [Google Scholar]
- 125.Ohi Y. Qin H. Hong C. Blouin L. Polo JM. Guo T. Qi Z. Downey SL. Manos PD. Rossi DJ. Yu J. Hebrok M. Hochedlinger K. Costello JF. Song JS. Ramalho-Santos M. Incomplete DNA methylation underlies a transcriptional memory of somatic cells in human iPS cells. Nat Cell Biol. 2011;13:541–549. doi: 10.1038/ncb2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Okano M. Bell DW. Haber DA. Li E. DNA methyltransferases Dnmt3a and Dnmt3b are essential for de novo methylation and mammalian development. Cell. 1999;99:247–257. doi: 10.1016/s0092-8674(00)81656-6. [DOI] [PubMed] [Google Scholar]
- 127.Ooi SK. Qiu C. Bernstein E. Li K. Jia D. Yang Z. Erdjument-Bromage H. Tempst P. Lin SP. Allis CD. Cheng X. Bestor TH. DNMT3L connects unmethylated lysine 4 of histone H3 to de novo methylation of DNA. Nature. 2007;448:714–717. doi: 10.1038/nature05987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Panning B. Jaenisch R. DNA hypomethylation can activate Xist expression and silence X-linked genes. Genes Dev. 1996;10:1991–2002. doi: 10.1101/gad.10.16.1991. [DOI] [PubMed] [Google Scholar]
- 129.Park SH. Kook MC. Kim EY. Park S. Lim JH. Ultrastructure of human embryonic stem cells and spontaneous and retinoic acid-induced differentiating cells. Ultrastruct Pathol. 2004;28:229–238. doi: 10.1080/01913120490515595. [DOI] [PubMed] [Google Scholar]
- 130.Piskounova E. Viswanathan SR. Janas M. LaPierre RJ. Daley GQ. Sliz P. Gregory RI. Determinants of microRNA processing inhibition by the developmentally regulated RNA-binding protein Lin28. J Biol Chem. 2008;283:21310–21314. doi: 10.1074/jbc.C800108200. [DOI] [PubMed] [Google Scholar]
- 131.Popp C. Dean W. Feng S. Cokus SJ. Andrews S. Pellegrini M. Jacobsen SE. Reik W. Genome-wide erasure of DNA methylation in mouse primordial germ cells is affected by AID deficiency. Nature. 2010;463:1101–1105. doi: 10.1038/nature08829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Pray-Grant MG. Daniel JA. Schieltz D. Yates JR., 3rd Grant PA. Chd1 chromodomain links histone H3 methylation with SAGA- and SLIK-dependent acetylation. Nature. 2005;433:434–438. doi: 10.1038/nature03242. [DOI] [PubMed] [Google Scholar]
- 133.Racki LR. Yang JG. Naber N. Partensky PD. Acevedo A. Purcell TJ. Cooke R. Cheng Y. Narlikar GJ. The chromatin remodeller ACF acts as a dimeric motor to space nucleosomes. Nature. 2009;462:1016–1021. doi: 10.1038/nature08621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Rai K. Huggins IJ. James SR. Karpf AR. Jones DA. Cairns BR. DNA demethylation in zebrafish involves the coupling of a deaminase, a glycosylase, and gadd45. Cell. 2008;135:1201–1212. doi: 10.1016/j.cell.2008.11.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Ramsahoye BH. Biniszkiewicz D. Lyko F. Clark V. Bird AP. Jaenisch R. Non-CpG methylation is prevalent in embryonic stem cells and may be mediated by DNA methyltransferase 3a. Proc Natl Acad Sci U S A. 2000;97:5237–5242. doi: 10.1073/pnas.97.10.5237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Reik W. Dean W. Walter J. Epigenetic reprogramming in mammalian development. Science. 2001;293:1089–1093. doi: 10.1126/science.1063443. [DOI] [PubMed] [Google Scholar]
- 137.Rinaldi A. Poretti G. Kwee I. Zucca E. Catapano CV. Tibiletti MG. Bertoni F. Concomitant MYC and microRNA cluster miR-17-92 (C13orf25) amplification in human mantle cell lymphoma. Leuk Lymphoma. 2007;48:410–412. doi: 10.1080/10428190601059738. [DOI] [PubMed] [Google Scholar]
- 138.Ringrose L. Paro R. Polycomb/Trithorax response elements and epigenetic memory of cell identity. Development. 2007;134:223–232. doi: 10.1242/dev.02723. [DOI] [PubMed] [Google Scholar]
- 139.Rossig L. Li H. Fisslthaler B. Urbich C. Fleming I. Forstermann U. Zeiher AM. Dimmeler S. Inhibitors of histone deacetylation downregulate the expression of endothelial nitric oxide synthase and compromise endothelial cell function in vasorelaxation and angiogenesis. Circ Res. 2002;91:837–844. doi: 10.1161/01.res.0000037983.07158.b1. [DOI] [PubMed] [Google Scholar]
- 140.Rossig L. Urbich C. Bruhl T. Dernbach E. Heeschen C. Chavakis E. Sasaki K. Aicher D. Diehl F. Seeger F. Potente M. Aicher A. Zanetta L. Dejana E. Zeiher AM. Dimmeler S. Histone deacetylase activity is essential for the expression of HoxA9 and for endothelial commitment of progenitor cells. J Exp Med. 2005;201:1825–1835. doi: 10.1084/jem.20042097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Rougier N. Bourc'his D. Gomes DM. Niveleau A. Plachot M. Paldi A. Viegas-Pequignot E. Chromosome methylation patterns during mammalian preimplantation development. Genes Dev. 1998;12:2108–2113. doi: 10.1101/gad.12.14.2108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Rybak A. Fuchs H. Smirnova L. Brandt C. Pohl EE. Nitsch R. Wulczyn FG. A feedback loop comprising lin-28 and let-7 controls pre-let-7 maturation during neural stem-cell commitment. Nat Cell Biol. 2008;10:987–993. doi: 10.1038/ncb1759. [DOI] [PubMed] [Google Scholar]
- 143.Saha A. Wittmeyer J. Cairns BR. Chromatin remodeling by RSC involves ATP-dependent DNA translocation. Genes Dev. 2002;16:2120–2134. doi: 10.1101/gad.995002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Saha A. Wittmeyer J. Cairns BR. Chromatin remodeling through directional DNA translocation from an internal nucleosomal site. Nat Struct Mol Biol. 2005;12:747–755. doi: 10.1038/nsmb973. [DOI] [PubMed] [Google Scholar]
- 145.Saladi SV. de la Serna IL. ATP dependent chromatin remodeling enzymes in embryonic stem cells. Stem Cell Rev. 2010;6:62–73. doi: 10.1007/s12015-010-9120-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Sapountzi V. Logan IR. Robson CN. Cellular functions of TIP60. Int J Biochem Cell Biol. 2006;38:1496–1509. doi: 10.1016/j.biocel.2006.03.003. [DOI] [PubMed] [Google Scholar]
- 147.Schuettengruber B. Cavalli G. Recruitment of polycomb group complexes and their role in the dynamic regulation of cell fate choice. Development. 2009;136:3531–3542. doi: 10.1242/dev.033902. [DOI] [PubMed] [Google Scholar]
- 148.Schuettengruber B. Chourrout D. Vervoort M. Leblanc B. Cavalli G. Genome regulation by polycomb and trithorax proteins. Cell. 2007;128:735–745. doi: 10.1016/j.cell.2007.02.009. [DOI] [PubMed] [Google Scholar]
- 149.Schwanbeck R. Xiao H. Wu C. Spatial contacts and nucleosome step movements induced by the NURF chromatin remodeling complex. J Biol Chem. 2004;279:39933–39941. doi: 10.1074/jbc.M406060200. [DOI] [PubMed] [Google Scholar]
- 150.Seward DJ. Cubberley G. Kim S. Schonewald M. Zhang L. Tripet B. Bentley DL. Demethylation of trimethylated histone H3 Lys4 in vivo by JARID1 JmjC proteins. Nat Struct Mol Biol. 2007;14:240–242. doi: 10.1038/nsmb1200. [DOI] [PubMed] [Google Scholar]
- 151.Shi Y. Histone lysine demethylases: emerging roles in development, physiology and disease. Nat Rev Genet. 2007;8:829–833. doi: 10.1038/nrg2218. [DOI] [PubMed] [Google Scholar]
- 152.Shi Y. Desponts C. Do JT. Hahm HS. Scholer HR. Ding S. Induction of pluripotent stem cells from mouse embryonic fibroblasts by Oct4 and Klf4 with small-molecule compounds. Cell Stem Cell. 2008;3:568–574. doi: 10.1016/j.stem.2008.10.004. [DOI] [PubMed] [Google Scholar]
- 153.Shi Y. Do JT. Desponts C. Hahm HS. Scholer HR. Ding S. A combined chemical and genetic approach for the generation of induced pluripotent stem cells. Cell Stem Cell. 2008;2:525–528. doi: 10.1016/j.stem.2008.05.011. [DOI] [PubMed] [Google Scholar]
- 154.Sif S. ATP-dependent nucleosome remodeling complexes: enzymes tailored to deal with chromatin. J Cell Biochem. 2004;91:1087–1098. doi: 10.1002/jcb.20005. [DOI] [PubMed] [Google Scholar]
- 155.Simonsson S. Gurdon J. DNA demethylation is necessary for the epigenetic reprogramming of somatic cell nuclei. Nat Cell Biol. 2004;6:984–990. doi: 10.1038/ncb1176. [DOI] [PubMed] [Google Scholar]
- 156.Sims RJ., 3rd Chen CF. Santos-Rosa H. Kouzarides T. Patel SS. Reinberg D. Human but not yeast CHD1 binds directly and selectively to histone H3 methylated at lysine 4 via its tandem chromodomains. J Biol Chem. 2005;280:41789–41792. doi: 10.1074/jbc.C500395200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Sims RJ., 3rd Millhouse S. Chen CF. Lewis BA. Erdjument-Bromage H. Tempst P. Manley JL. Reinberg D. Recognition of trimethylated histone H3 lysine 4 facilitates the recruitment of transcription postinitiation factors and pre-mRNA splicing. Mol Cell. 2007;28:665–676. doi: 10.1016/j.molcel.2007.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Sinkkonen L. Hugenschmidt T. Berninger P. Gaidatzis D. Mohn F. Artus-Revel CG. Zavolan M. Svoboda P. Filipowicz W. MicroRNAs control de novo DNA methylation through regulation of transcriptional repressors in mouse embryonic stem cells. Nat Struct Mol Biol. 2008;15:259–267. doi: 10.1038/nsmb.1391. [DOI] [PubMed] [Google Scholar]
- 159.Spallotta F. Rosati J. Straino S. Nanni S. Grasselli A. Ambrosino V. Rotili D. Valente S. Farsetti A. Mai A. Capogrossi MC. Gaetano C. Illi B. Nitric oxide determines mesodermic differentiation of mouse embryonic stem cells by activating class IIa histone deacetylases: potential therapeutic implications in a mouse model of hindlimb ischemia. Stem Cells. 2010;28:431–442. doi: 10.1002/stem.300. [DOI] [PubMed] [Google Scholar]
- 160.Squatrito M. Gorrini C. Amati B. Tip60 in DNA damage response and growth control: many tricks in one HAT. Trends Cell Biol. 2006;16:433–442. doi: 10.1016/j.tcb.2006.07.007. [DOI] [PubMed] [Google Scholar]
- 161.Stadtfeld M. Apostolou E. Akutsu H. Fukuda A. Follett P. Natesan S. Kono T. Shioda T. Hochedlinger K. Aberrant silencing of imprinted genes on chromosome 12qF1 in mouse induced pluripotent stem cells. Nature. 2010;465:175–181. doi: 10.1038/nature09017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Stock JK. Giadrossi S. Casanova M. Brookes E. Vidal M. Koseki H. Brockdorff N. Fisher AG. Pombo A. Ring1-mediated ubiquitination of H2A restrains poised RNA polymerase II at bivalent genes in mouse ES cells. Nat Cell Biol. 2007;9:1428–1435. doi: 10.1038/ncb1663. [DOI] [PubMed] [Google Scholar]
- 163.Stopka T. Skoultchi AI. The ISWI ATPase Snf2h is required for early mouse development. Proc Natl Acad Sci U S A. 2003;100:14097–14102. doi: 10.1073/pnas.2336105100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Strauss WM. Chen C. Lee CT. Ridzon D. Nonrestrictive developmental regulation of microRNA gene expression. Mamm Genome. 2006;17:833–840. doi: 10.1007/s00335-006-0025-7. [DOI] [PubMed] [Google Scholar]
- 165.Strohner R. Wachsmuth M. Dachauer K. Mazurkiewicz J. Hochstatter J. Rippe K. Langst G. A ‘loop recapture’ mechanism for ACF-dependent nucleosome remodeling. Nat Struct Mol Biol. 2005;12:683–690. doi: 10.1038/nsmb966. [DOI] [PubMed] [Google Scholar]
- 166.Suh MR. Lee Y. Kim JY. Kim SK. Moon SH. Lee JY. Cha KY. Chung HM. Yoon HS. Moon SY. Kim VN. Kim KS. Human embryonic stem cells express a unique set of microRNAs. Dev Biol. 2004;270:488–498. doi: 10.1016/j.ydbio.2004.02.019. [DOI] [PubMed] [Google Scholar]
- 167.Sylvestre Y. De Guire V. Querido E. Mukhopadhyay UK. Bourdeau V. Major F. Ferbeyre G. Chartrand P. An E2F/miR-20a autoregulatory feedback loop. J Biol Chem. 2007;282:2135–2143. doi: 10.1074/jbc.M608939200. [DOI] [PubMed] [Google Scholar]
- 168.Tahiliani M. Koh KP. Shen Y. Pastor WA. Bandukwala H. Brudno Y. Agarwal S. Iyer LM. Liu DR. Aravind L. Rao A. Conversion of 5-methylcytosine to 5-hydroxymethylcytosine in mammalian DNA by MLL partner TET1. Science. 2009;324:930–935. doi: 10.1126/science.1170116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Takahashi K. Tanabe K. Ohnuki M. Narita M. Ichisaka T. Tomoda K. Yamanaka S. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell. 2007;131:861–872. doi: 10.1016/j.cell.2007.11.019. [DOI] [PubMed] [Google Scholar]
- 170.Takahashi K. Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126:663–676. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- 171.Takeuchi JK. Bruneau BG. Directed transdifferentiation of mouse mesoderm to heart tissue by defined factors. Nature. 2009;459:708–711. doi: 10.1038/nature08039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Thomson JA. Itskovitz-Eldor J. Shapiro SS. Waknitz MA. Swiergiel JJ. Marshall VS. Jones JM. Embryonic stem cell lines derived from human blastocysts. Science. 1998;282:1145–1147. doi: 10.1126/science.282.5391.1145. [DOI] [PubMed] [Google Scholar]
- 173.Tran PT. Liskay RM. Functional studies on the candidate ATPase domains of Saccharomyces cerevisiae MutLalpha. Mol Cell Biol. 2000;20:6390–6398. doi: 10.1128/mcb.20.17.6390-6398.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Trimarchi JM. Lees JA. Sibling rivalry in the E2F family. Nat Rev Mol Cell Biol. 2002;3:11–20. doi: 10.1038/nrm714. [DOI] [PubMed] [Google Scholar]
- 175.van den Berg DL. Snoek T. Mullin NP. Yates A. Bezstarosti K. Demmers J. Chambers I. Poot RA. An Oct4-centered protein interaction network in embryonic stem cells. Cell Stem Cell. 2010;6:369–381. doi: 10.1016/j.stem.2010.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Venturini L. Battmer K. Castoldi M. Schultheis B. Hochhaus A. Muckenthaler MU. Ganser A. Eder M. Scherr M. Expression of the miR-17-92 polycistron in chronic myeloid leukemia (CML) CD34+ cells. Blood. 2007;109:4399–4405. doi: 10.1182/blood-2006-09-045104. [DOI] [PubMed] [Google Scholar]
- 177.Viswanathan SR. Daley GQ. Gregory RI. Selective blockade of microRNA processing by Lin28. Science. 2008;320:97–100. doi: 10.1126/science.1154040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Waddington CH, editor. The Strategy of the Genes; a Discussion of Some Aspects of Theoretical Biology. London, UK: Allen & Unwin; 1957. p. 262. [Google Scholar]
- 179.Wade PA. Gegonne A. Jones PL. Ballestar E. Aubry F. Wolffe AP. Mi-2 complex couples DNA methylation to chromatin remodelling and histone deacetylation. Nat Genet. 1999;23:62–66. doi: 10.1038/12664. [DOI] [PubMed] [Google Scholar]
- 180.Wang J. Rao S. Chu J. Shen X. Levasseur DN. Theunissen TW. Orkin SH. A protein interaction network for pluripotency of embryonic stem cells. Nature. 2006;444:364–368. doi: 10.1038/nature05284. [DOI] [PubMed] [Google Scholar]
- 181.Wang Y. Blelloch R. Cell cycle regulation by MicroRNAs in embryonic stem cells. Cancer Res. 2009;69:4093–4096. doi: 10.1158/0008-5472.CAN-09-0309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Wang Y. Medvid R. Melton C. Jaenisch R. Blelloch R. DGCR8 is essential for microRNA biogenesis and silencing of embryonic stem cell self-renewal. Nat Genet. 2007;39:380–385. doi: 10.1038/ng1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Weber M. Hellmann I. Stadler MB. Ramos L. Paabo S. Rebhan M. Schubeler D. Distribution, silencing potential and evolutionary impact of promoter DNA methylation in the human genome. Nat Genet. 2007;39:457–466. doi: 10.1038/ng1990. [DOI] [PubMed] [Google Scholar]
- 184.Wernig M. Meissner A. Foreman R. Brambrink T. Ku M. Hochedlinger K. Bernstein BE. Jaenisch R. In vitro reprogramming of fibroblasts into a pluripotent ES-cell-like state. Nature. 2007;448:318–324. doi: 10.1038/nature05944. [DOI] [PubMed] [Google Scholar]
- 185.Westbrook TF. Hu G. Ang XL. Mulligan P. Pavlova NN. Liang A. Leng Y. Maehr R. Shi Y. Harper JW. Elledge SJ. SCFbeta-TRCP controls oncogenic transformation and neural differentiation through REST degradation. Nature. 2008;452:370–374. doi: 10.1038/nature06780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Whitehouse I. Stockdale C. Flaus A. Szczelkun MD. Owen-Hughes T. Evidence for DNA translocation by the ISWI chromatin-remodeling enzyme. Mol Cell Biol. 2003;23:1935–1945. doi: 10.1128/MCB.23.6.1935-1945.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Wiblin AE. Cui W. Clark AJ. Bickmore WA. Distinctive nuclear organisation of centromeres and regions involved in pluripotency in human embryonic stem cells. J Cell Sci. 2005;118:3861–3868. doi: 10.1242/jcs.02500. [DOI] [PubMed] [Google Scholar]
- 188.Wienholds E. Koudijs MJ. van Eeden FJ. Cuppen E. Plasterk RH. The microRNA-producing enzyme Dicer1 is essential for zebrafish development. Nat Genet. 2003;35:217–218. doi: 10.1038/ng1251. [DOI] [PubMed] [Google Scholar]
- 189.Williams CJ. Naito T. Arco PG. Seavitt JR. Cashman SM. De Souza B. Qi X. Keables P. Von Andrian UH. Georgopoulos K. The chromatin remodeler Mi-2beta is required for CD4 expression and T cell development. Immunity. 2004;20:719–733. doi: 10.1016/j.immuni.2004.05.005. [DOI] [PubMed] [Google Scholar]
- 190.Wilson KD. Venkatasubrahmanyam S. Jia F. Sun N. Butte AJ. Wu JC. MicroRNA profiling of human-induced pluripotent stem cells. Stem Cells Dev. 2009;18:749–758. doi: 10.1089/scd.2008.0247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Woods K. Thomson JM. Hammond SM. Direct regulation of an oncogenic micro-RNA cluster by E2F transcription factors. J Biol Chem. 2007;282:2130–2134. doi: 10.1074/jbc.C600252200. [DOI] [PubMed] [Google Scholar]
- 192.Wu L. Belasco JG. Micro-RNA regulation of the mammalian lin-28 gene during neuronal differentiation of embryonal carcinoma cells. Mol Cell Biol. 2005;25:9198–9208. doi: 10.1128/MCB.25.21.9198-9208.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Xu C. Police S. Rao N. Carpenter MK. Characterization and enrichment of cardiomyocytes derived from human embryonic stem cells. Circ Res. 2002;91:501–508. doi: 10.1161/01.res.0000035254.80718.91. [DOI] [PubMed] [Google Scholar]
- 194.Yamanaka S. Elite and stochastic models for induced pluripotent stem cell generation. Nature. 2009;460:49–52. doi: 10.1038/nature08180. [DOI] [PubMed] [Google Scholar]
- 195.Yang JG. Madrid TS. Sevastopoulos E. Narlikar GJ. The chromatin-remodeling enzyme ACF is an ATP-dependent DNA length sensor that regulates nucleosome spacing. Nat Struct Mol Biol. 2006;13:1078–1083. doi: 10.1038/nsmb1170. [DOI] [PubMed] [Google Scholar]
- 196.Yoshida T. Hazan I. Zhang J. Ng SY. Naito T. Snippert HJ. Heller EJ. Qi X. Lawton LN. Williams CJ. Georgopoulos K. The role of the chromatin remodeler Mi-2beta in hematopoietic stem cell self-renewal and multilineage differentiation. Genes Dev. 2008;22:1174–1189. doi: 10.1101/gad.1642808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Zhu JK. Active DNA demethylation mediated by DNA glycosylases. Annu Rev Genet. 2009;43:143–166. doi: 10.1146/annurev-genet-102108-134205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Zink D. Amaral MD. Englmann A. Lang S. Clarke LA. Rudolph C. Alt F. Luther K. Braz C. Sadoni N. Rosenecker J. Schindelhauer D. Transcription-dependent spatial arrangements of CFTR and adjacent genes in human cell nuclei. J Biol Chem. 2004;166:815–825. doi: 10.1083/jcb.200404107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Zofall M. Persinger J. Bartholomew B. Functional role of extranucleosomal DNA and the entry site of the nucleosome in chromatin remodeling by ISW2. Mol Cell Biol. 2004;24:10047–10057. doi: 10.1128/MCB.24.22.10047-10057.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Zofall M. Persinger J. Kassabov SR. Bartholomew B. Chromatin remodeling by ISW2 and SWI/SNF requires DNA translocation inside the nucleosome. Nat Struct Mol Biol. 2006;13:339–346. doi: 10.1038/nsmb1071. [DOI] [PubMed] [Google Scholar]