Abstract
Antimicrobial use in food animals may contribute to antimicrobial resistance in bacteria of animals and humans. Commensal bacteria of animal intestine may serve as a reservoir of resistance-genes. To understand the dynamics of plasmid-mediated resistance to cephalosporin ceftiofur in enteric commensals of cattle, we developed a deterministic mathematical model of the dynamics of ceftiofur-sensitive and resistant commensal enteric Escherichia coli (E. coli) in the absence of and during parenteral therapy with ceftiofur. The most common treatment scenarios including those using a sustained-release drug formulation were simulated; the model outputs were in agreement with the available experimental data. The model indicated that a low but stable fraction of resistant enteric E. coli could persist in the absence of immediate ceftiofur pressure, being sustained by horizontal and vertical transfers of plasmids carrying resistance-genes, and ingestion of resistant E. coli. During parenteral therapy with ceftiofur, resistant enteric E. coli expanded in absolute number and relative frequency. This expansion was most influenced by parameters of antimicrobial action of ceftiofur against E. coli. After treatment (>5 weeks from start of therapy) the fraction of ceftiofur-resistant cells among enteric E. coli, similar to that in the absence of treatment, was most influenced by the parameters of ecology of enteric E. coli, such as the frequency of transfer of plasmids carrying resistance-genes, the rate of replacement of enteric E. coli by ingested E. coli, and the frequency of ceftiofur resistance in the latter.
Introduction
The emergence and spread of antimicrobial resistance (AMR) is progressively demarcating the epochal success of antimicrobial therapies of bacterial infections. Some classes of antimicrobials are used in both human and veterinary medicines; among antibiotics these are β-lactams, including cephalosporins, as well as aminoglycosides, macrolides, tetracyclines, sulphonamides, and in some countries fluoroquinolones [1]. Humans may be exposed to AMR-bacteria from food animals via occupational exposure or contaminated food products. In the 1990s in the USA, a domestically-acquired infection of a boy with ceftriaxone-resistant Salmonella was traced to cattle carrying ceftiofur-resistant Salmonella after the boy’s father had treated the diarrheic calves [2]. Human food-borne infections with AMR-bacteria are clinically challenging [1], [3]. Furthermore, ingested strains can become a part of the human enteric microflora [4], and transmit AMR-genetic determinants to other human bacteria [5]. For cephalosporins, the principal mechanism via which resistance disseminates is horizontal transfer of AMR-genes encoded on conjugative plasmids [6], [7], [8]. The AMR-strains occasionally demonstrate a higher transmissibility via the food chain, e.g., an AMR-strain of Escherichia coli (E. coli) on pig carcasses has survived processing and chilling better than the parental antimicrobial-sensitive strain [9]. Cattle meat products can be contaminated by animals’ feces, and so the enteric microflora [10]. Therefore, minimizing the frequency of AMR in cattle enteric bacteria en masse can aid in decreasing human exposure to AMR-strains.
Within animal hosts, enteric commensals may also transmit AMR-genetic determinants to pathogens, e.g., E. coli can transmit plasmidic AMR-genes to Salmonella [11], [12]. However, the in-vivo frequency of such transfer is unknown, and may be limited by the number of plasmids shared [12], differences in plasmid developments between bacterial species [11], or restrictions on plasmid establishment in the heterologous recipients [13]. The frequency of plasmid transfer from E. coli to Salmonella is much lower compared to promiscuous plasmid sharing between E. coli cells [12]. However, occasionally AMR-strains themselves exhibit a higher virulence for [14], or a greater ability to colonize animal hosts [15], [16]. This necessitates the use of even newer drugs to combat animal infections [1].
A complete cessation of antimicrobial therapies in food animals is impractical [1], and, in the absence of alternatives, unethical [17]. The real challenge is to implement therapies that minimize emergence and spread of AMR [17], [18]. Also, farm animals present a model system where the potential of candidate policies for reduction of antimicrobial usage can be evaluated at the population level, with further relevance to policies in humans [19].
The containment of resistance to 3rd generation cephalosporins is categorized by the World Health Organization as critically important. Ceftiofur is the only drug in this class licensed to treat food animals in the USA. Ceftiofur’s chemical structure is close to that of ceftriaxone, which is used to treat bacterial meningitis and salmonellosis in humans. Ceftiofur is administered parenterally to individual cattle to treat interdigital necrobacillosis, pneumonia or metritis, and to groups of beef calves for metaphylaxis of bovine respiratory disease (BRD). The drug can also be applied intramammary to treat mastitis or as a dry-off therapy.
Resistance to ceftiofur in enteric bacteria of cattle in the USA is mediated predominantly by plasmid-encoded gene blaCMY-2 [12], [20], which codes for a cephamycinase [21], [22], [23]. The gene has been reported in Salmonella and E. coli isolates from feces of food animals and meat products in retail [24], and in Salmonella isolates responsible for human illness [25], [26]. The resistant E. coli have been isolated from feces of beef and dairy cattle, sewage and ground beef [24], [27]. Between bacteria, both inter-generational and horizontal transfers of plasmidic blaCMY-2 occur. In the enteric environment, the horizontal plasmid transfer is the main mechanism of AMR-gene spread within and between bacterial species, both Gram-positive and Gram-negative [13]. E. coli can constitute up to 86% of the fecal Gram-negative bacteria in dairy cattle [28], and act as a donor of plasmidic AMR-genes [13]. Recent field studies demonstrate that a fraction of enteric E. coli carry plasmidic blaCMY-2 even in cattle not known to be treated with ceftiofur [27], [29], [30]. Enteric E. coli are primarily commensal and are genetically diverse [10], [31]; among them, E. coli carrying blaCMY-2 are not strongly clonal at either serotype or PFGE levels [6], [12]. The “background” resistant fraction can have mixed origins. Ecological origins may include adaptation of bacteria to co-exist with fungi that are natural producers of β-lactams, and subsequent transfer of chromosomal AmpC locus from Citrobacter freundii to other Enterobacteriaceae as a plasmidic gene [21], [32]. Also, exposure to resistant E. coli can occur on the farm when post-weaned calves are colonized with ruminant-specific microflora (Tom Besser, personal communication). Similarly, ceftiofur-resistant E. coli in broilers is associated with its presence at the hatchery and on the farm [33].
During parenteral treatment with ceftiofur, a decline in the numbers of enteric E. coli is reported in healthy 3-4 mos old calves [27], healthy adult cattle [34], and lactating dairy cattle treated for metritis or interdigital necrobacillosis [35] (Table S1). Studies employing genetic methods to examine the effects on entire enteric bacterial populations have arrived at similar conclusions [36]. This strongly suggests that parenteral treatment of cattle with ceftiofur results in exposure of their enteric bacteria to antimicrobially-active drug metabolites, with the dose and duration sufficient for prominent effects on the enteric bacteria.
The objectives of this modeling study were to analyze, first, whether the reported fractions of blaCMY-2-carrying commensal enteric E. coli in cattle could be maintained in the absence of immediate ceftiofur pressure; and, second, how the dynamics of the resistant and sensitive enteric E. coli changed during parenteral ceftiofur treatment depending on the treatment protocol.
Materials and Methods
Dynamics of Ceftiofur-sensitive and Resistant Commensal Enteric E. coli in the Absence of Immediate Ceftiofur Pressure
Ecology of commensal enteric E. coli
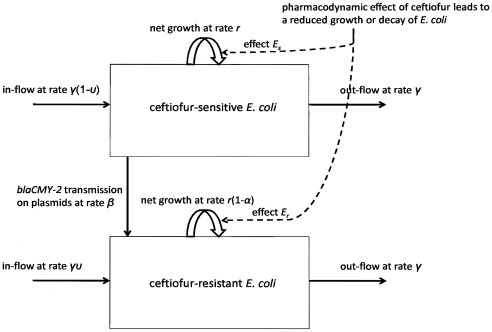
A flow-chart of the model is given in Figure 1. Due to unfavorable conditions for E. coli growth in the upper parts of the cattle gastrointestinal tract, only E. coli in the large intestine was considered (referred to as “commensal enteric E. coli”). These may exist in a planktonic “free-living” mode of growth, or by being incorporated into intestinal biofilms [37]. The biofilm-trapped latter likely constitute a small fraction of the total, hence enteric E. coli were considered to be free-living. Population growth of E. coli, as a facultative anaerobe, slows in anaerobic conditions [38]; the maximum net growth rate (in exponential growth phase) in numbers of enteric E. coli, r, was parameterized accordingly. E. coli growth in the enteric environment is likely further restricted by intra-specific competition, inter-specific competition with other microflora, and feces substrate composition [39]. A logistic model of bacterial growth was used to reflect the intra-specific competition. The upper limit for total E. coli per g of feces, Nmax, was parameterized from the reported numbers of viable E. coli in cattle feces (Table 1), and so bore the expected effects of the inter-specific competition.
Figure 1. Flow-chart of the model of the dynamics of ceftiofur-sensitive and resistant commensal E. coli in the cattle large intestine.
Bacterial growth is density-dependent with fractional net growth rate r; fitness cost for cells with blaCMY-2-carrying plasmids manifests as a reduction α in r. Resistant cells transfer blaCMY-2 to the progeny during cell division. Horizontally, blaCMY-2 is transferred to the sensitive cells at rate β; the transmission is frequency-dependent with the total of β*Nr*Ns/N. There is fractional in-flow and out-flow of E. coli at rate γ; fraction υ of in-flowing E. coli are ceftiofur-resistant. Antimicrobial action of ceftiofur metabolites, depending on their concentration, results in either reduced growth or decay in number of E. coli.
Table 1. Parameter definitions and values.
| Parameter | Definition, units | Value | Reference | |
| Bacteria | ||||
| r | Specific growth rate, h−1 | 0.17 | estimated from [39] | |
| γ | Fractional in-flow/out flow, h−1 | 0.01 | estimated from [27] | |
| Nmax | Max E. coli, log CFU/g of feces | |||
| 6-mos beef (220 kg) | 5.5 | [31] | ||
| 6-mos dairy (180 kg) | 6.5 | [27] | ||
| adult dairy (600 kg) | 4.3 | [90] | ||
| post-partum/lactating dairy (600 kg) | 4.3 | – | ||
| AMR | ||||
| pAMR | Fraction of ceftiofur-resistant enteric E. coli at start of treatment | |||
| 6-mos beef (220 kg) | 0.018 | [80] | ||
| 6-mos dairy (180 kg) | 0.050 | [78] | ||
| adult dairy (600 kg) | 0.007 | [79] | ||
| post-partum/lactating dairy (600 kg) | 0.018 | – | ||
| β | Plasmid transmission term, h-1 | 0.004 | [27], [42] | |
| a | Resistance fitness cost as fraction of r | 0.05 | [42] | |
| υ | Resistant E. coli fraction in in-flow(pAMR*0.6 based on [31]) | |||
| 6-mos beef (220 kg) | 0.0110 | |||
| 6-mos dairy (180 kg) | 0.0310 | |||
| adult dairy (600 kg) | 0.0042 | |||
| post-partum/lactating dairy (600 kg) | 0.0110 | |||
| Biliary ceftiofur metabolites | ||||
| p | Bile-excreted fraction of injected dose | 0.37 | [47], [51] | |
| Tδ | Passage time to large intestine, h | 6 | – | |
| V | Volume of large intestine, L | |||
| 6-mos beef or dairy | 5 | – | ||
| adult cattle | 20 | – | ||
| λ | Biodegradation decay constant, h−1 | 0.2 | estimated from [48], [60] | |
| H | Hill coefficient in Emax model | 1.5 | estimated using [68] | |
| MICs | MIC for sensitive E. coli, µg/mL | 1 | – | |
| MICr | MIC for resistant E. coli, µg/mL | 8 | – | |
E. coli is capable of replicating outside animal hosts; commensal E. coli circulate between cattle hosts and their environment [40]. In beef cattle reared at either pasture or feedlot, ∼60% of fecal E. coli are genetically related to those in animals’ oral cavities [31]. From in vivo experiments in post-weaned calves [27], an estimated ∼20–30% of fecal coliforms are E. coli strains fed to the animals on the day of measure or the preceding day. The in-flow of ingested bacteria and the out-flow of bacteria with feces likely ensures a regular partial replacement of E. coli “free-living” in the large intestine. To reflect this, the rates of hourly fractional in-flow and outflow of enteric E. coli were taken to be equal, both γ. A fraction υ of the ingested bacteria was assumed to carry plasmids with blaCMY-2. In-flowing bacteria would mix homogeneously with those already in the intestine.
Plasmid transfer and fitness cost of plasmid-mediated resistance
Various conjugative plasmids of E. coli can carry blaCMY-2 [12]. There is no evidence of enhanced plasmid transfer in enteric E. coli during parenteral ceftiofur therapy [27]. The maximum number of cells to which a donor E. coli can transfer a plasmid per unit time is inherently restricted by biology of conjugation; the transconjugant (recipient cell) undergoes a 40-80 minutes maturation before becoming a proficient donor [41]. Therefore, the transfer was modeled as a contagious process [65], [66] with frequency-dependent transmission. β was the transmission term for blaCMY-2-carrying plasmids from resistant donor to sensitive cells, Nr - number of resistant cells, Ns - number of sensitive cells, and N - total number of E. coli cells. Then, “force of transfer” per a sensitive cell per unit time was β*Nr/N, and the total transfer was β*Nr*Ns/N.
The growth rate of a bacterial strain is considered to represent its evolutionary fitness [38]. Having blaCMY-2-carrying plasmids is associated with either a fitness cost, i.e., reduced growth [42], or a fitness gain, i.e., enhanced growth [43]; or no change in growth [44]. The fitness cost appears more often, and was modeled as a fractional reduction, α, in net growth rate, r [45].
The fate of AMR-bacterial strains in the absence of antimicrobial pressure is unclear. In some laboratory experiments, a gradual loss of AMR-gene-carrying plasmids during cell divisions after thousands of bacterial generations (several months) is reported [42]; others, however, report maintenance of the plasmid profile, in particular by E. coli [44]. The AMR-strains can acquire compensatory mutations to restore fitness without losing resistance, e.g., a better growth performance of E. coli with chromosomal-encoded resistance to streptomycin [46], or plasmid-encoded resistance to tetracycline [43]. Notably, these processes occur over extended time horizons. The period of parenteral treatment of cattle with ceftiofur is at most 7 days, followed by at most a 13-day pre-slaughter withdrawal period. Hence the possibility of loss of plasmidic blaCMY-2 by enteric E. coli was not considered in this analysis.
Model for dynamics without treatment
The ordinary differential Equations [1] and [2] described the changes in Ns and Nr, respectively, over time in the absence of immediate ceftiofur pressure:
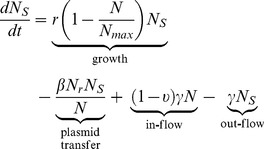 |
(1) |
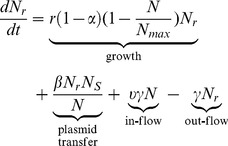 |
(2) |
Dynamics of Ceftiofur-sensitive and Resistant Commensal Enteric E. coli During Parenteral Ceftiofur Treatment
Pharmacokinetics and biodegradation of ceftiofur metabolites
Ceftiofur in cattle is metabolized shortly post injection (p.i.) [47]. Major ceftiofur metabolites retain the β-lactam ring [48]; their antimicrobial activity against E. coli is close to that of ceftiofur [49], [50]. The total of ceftiofur and its active metabolites is termed the concentration of ceftiofur equivalents (CE) [51]. The pharmacokinetics of ceftiofur in cattle following an intramuscular (IM) or a subcutaneous (SC) injection in cattle are similar in terms of CE-pattern in the plasma [52]. The pharmacokinetics of formulations containing ceftiofur sodium and ceftiofur hydrochloride salts are similar in terms of CE-pattern in the plasma of pigs [53]; this is considered to hold for cattle [48].
In humans, ceftriaxone (its structure is close to that of ceftiofur) is excreted via both renal and hepatic pathways [54]. There is no evidence of a correlation between ceftriaxone concentrations in bile (bile metabolite is structurally similar to ceftriaxone) and in plasma [54], or of ceftriaxone intestinal absorption and enterohepatic circulation [55]. The rate of ceftriaxone biliary excretion in humans positively correlates with the rate of bile acid secretion [56]; experimental data in rats suggest a common mechanism for hepatic transport of ceftriaxone and bile acids [57]. There is inter-individual variability in achieved ceftriaxone concentrations in bile [54], [56], [58], and in feces [54] of humans.
In cattle, ceftiofur administered parenterally is also excreted via both urine (∼65%) and, through bile, feces (∼35%) [47]. There are no published data on the pattern or inter-individual variability of the biliary excretion. We assumed that in cattle, as in humans, there was no enterohepatic ceftiofur circulation, and CE-concentration in the intestine was independent of that in systematic distribution. Of (radio-labeled) ceftiofur dose injected IM, 29% is detected in cattle feces in 8 hours, and 37% in 12 hours p.i. [51]. The exact structure of intestinal metabolites is unknown; although it is likely that they enter the large intestine having an intact β-lactam ring. However, most of the (radio-labeled) amounts in feces lack antimicrobial activity [48], [59]. This is attributed to enteric bacteria biodegrading metabolites within and outside the intestinal environment [60], because of variable timing of metabolite degradation in normal vs. sterilized cattle feces [60]. In normal cattle feces fortified with 100 µg/g of CE, under aerobic conditions it takes ∼8 hours for these to entirely degrade to antimicrobially inactive compounds [48], [60]. The dynamics of decline in CE-concentration appears to be an exponential decay [48], [60]. The exact enteric species producing β-lactamases involved are unknown [48]; different species may be involved, as plasmid-mediated β-lactamases are widely produced by Gram-negative bacteria [61].
Let D denote ceftiofur dose in one injection. Fraction p of D was excreted in bile, and the volume of the animal’s large intestine was V.
First consider a therapy with repeated injections of a non-sustained-release ceftiofur formulation (Table S1, scenarios R1-R3). D was, and p and V were taken to be equal for every injection nj, which occurred at time Tj since start of treatment, t = 0. As the pattern of ceftiofur biliary excretion is unknown, two possibilities were explored. Under pattern 1, amount D*p was excreted at 1 hour p.i. After a passage time Tδ, biliary metabolites entered the large intestine. At entry, for a given nj, CE-concentration per g of feces (assuming weight-to-volume ratio of feces of 1) was D*p/V, then decayed exponentially due to the biodegradation at rate λ. Total CE-concentration per g of feces in large intestine, C (CE µg/g), at time t was:
| (3) |
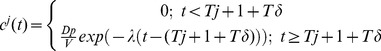 |
(4) |
where  ; j = injection number: 1, …, n, n = 5
; j = injection number: 1, …, n, n = 5
Under pattern 2, m = 6 equal fractions of D*p were excreted hourly at hour 1 to 6 p.i. (similarly to uniform patterns of ceftriaxone biliary excretion in rabbits [62], and of bile flow in cattle [63]). k was excretion fraction number. The choice of hours 1 to 6 p.i. was based on the working hypothesis that Tδ = 6 hours, thus the entire amount D*p would reach the large intestine by 12 hours p.i (as in experimental observations [51]). Initial CE µg/g feces was D*p/6*V. The concentration C (CE µg/g), at time t was as in Equation [3]; cj for a given nj was:
| (5) |
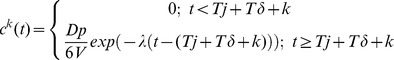 |
(6) |
where  , j = injection number: 1, …, n, n = 5 and k = 1,…m, m = 6.
, j = injection number: 1, …, n, n = 5 and k = 1,…m, m = 6.
Now consider a SC injection of a sustained-release ceftiofur formulation (Table S1, scenarios SB1-SB3 and SD1). According to the data published by the drug manufacturer, plasma CE-concentration peaks at hours 1 to 2 p.i., then declines but remains above the therapeutic threshold (0.2 µg/mL) for ∼10 days [64], [65]. Due to the quality of these data, data for 60 time points over 10 days p.i. were extracted to detail the plasma pattern. We assumed that the plasma pattern paralleled the pattern of drug release, and that entire drug dose, D, was released within 10 days p.i. At each time point i, amount di of ceftiofur (so that  ; what fraction of D was di was determined by the drug release pattern) was released at the site of injection. Fraction p of di was excreted in bile 1 hour later. The passage of metabolites to the large intestine, and decay in their concentration due to the biodegradation were modeled similarly to the above.
; what fraction of D was di was determined by the drug release pattern) was released at the site of injection. Fraction p of di was excreted in bile 1 hour later. The passage of metabolites to the large intestine, and decay in their concentration due to the biodegradation were modeled similarly to the above.
Pharmacodynamic effect
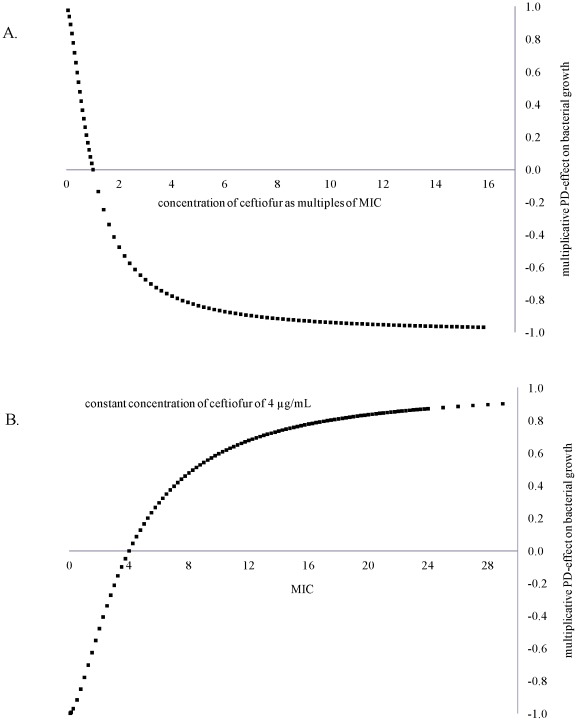
Antimicrobial action of β-lactams results in the death of growing, preparing to divide bacteria (both the dividing cell and its “daughter” cell are killed); unaffected growing cells replicate (survive and produce “daughter” cells) [66]. What fraction of growing cells is killed vs. is replicating at a given time, and so what is net growth or decline of the bacterial population, depends on the concentration of β-lactams [67], [68]. The changes in net growth of ceftiofur-sensitive and resistant enteric E. coli depending on CE-concentration were modeled using a fractional inhibitory Emax pharmacodynamic (PD) model, where Emax term specifies the maximum possible PD-effect [69], [70]. The 50% PD-effect was with stationary concentration of CE, at which half of the growing cells were replicating, and half were killed (no net change in number of bacteria) [71]. In the case of β-lactams, for a given drug and microbe, the stationary concentration is close to a commonly measured minimum inhibitory concentration (MIC) [71]. Therefore, in this PD-model at a CE-concentration <MIC, net population growth was positive (as more growing cells were able to replicate than were killed). At a CE-concentration >MIC, the population declined. The maximum decline was when all growing cells were killed; the population declined at the rate of attempted growth. This was specified by setting Emax = 2 (giving -1 as the multiplier for growth rate at a sufficiently high CE-concentration). If CE-concentration rose further, the PD-effect saturated, as no more cells could be killed than those growing to divide. The PD-model behavior is illustrated in Figure 2. The total kill depended on how long CE-concentration was at or above that producing maximum effect. The model therefore depicted time-dependent PD of cephalosporins [68], [72], [73], with a point of maximum effect (at a drug concentration of low multiples of MIC) after which further concentration rise does not enhance the rate of killing [68], [72], [74]. The model also accounted for that for antimicrobial resistance via enzymatic deactivation that can be surmounted by a higher drug dose, the change in antimicrobial activity against resistant bacteria should be reflected as an increase in the drug concentration producing the 50% PD-effect [75].
Figure 2. Pharmacodynamic model.
A. Multiplicative pharmacodynamic effect on E. coli net growth with a constant minimum inhibitory concentration (MIC = 1 µg/mL) and changing ceftiofur concentration expressed as multiples of MIC; Hill coefficient = 1.5. B: Multiplicative pharmacodynamic effect on E. coli net growth with changing MIC and a constant ceftiofur concentration; Hill coefficient = 1.5.
Denoting MICs for ceftiofur-sensitive and MICr for resistant E. coli, Es in Equation [7] and Er in Equation [8] described fractional changes in net growth of ceftiofur-sensitive and resistant E. coli, respectively, at CE-concentration C:
| (7) |
| (8) |
where Emax = 2, and H is Hill coefficient.
Postantibiotic effect, the period post exposure to antibiotic after which surviving bacteria begin to multiply normally, in Gram-negative bacilli after the majority of β-lactams is from none to brief [68], [76], [77], and so was not considered.
Model for dynamics during treatment
The ordinary differential Equations [9] and [10] described the changes in Ns and Nr, respectively, over time of parenteral ceftiofur treatment:
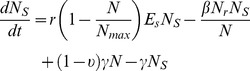 |
(9) |
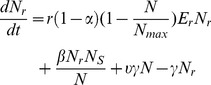 |
(10) |
Post treatment, when ceftiofur metabolites had been eliminated from the large intestine (C = 0, Es = 1, Er = 1), the dynamics reverted to Equations [1] and [2].
Parameterization
Table 1 details the parameters and their values. The maximum number of E. coli per g of feces in large intestine, Nmax, was based on reported numbers of viable fecal E. coli (colony-forming units, CFU, of E. coli or fecal coliforms were considered as a measurement of viable E. coli). Starting values of Ns and Nr were calculated using N (set at 90% of Nmax), and pAMR - fraction of ceftiofur-resistant E. coli at start of treatment. pAMR estimates were adopted from tests of E. coli sensitivity at start of or without connection to ceftiofur treatments. The available estimates varied depending on cattle age and purpose: 6% of ceftriaxone-resistant E. coli for breakpoint ≥16 µg/mL in 2-6 mos post-weaned dairy calves (estimated from [78]); 7.4% of ceftiofur-resistant E. coli for breakpoint ≥16 µg/mL in dairy cattle [28]; 0.7% of ceftazidime-resistant coliforms for breakpoint ≥8 µg/mL across samples from 39 dairy herds [79]; and 1.8% of ceftazidime-resistant E. coli for breakpoint ≥8 µg/mL in feedlot steers [80].
E. coli doubling time in the large intestine was assumed to be 4 hours [39]; hence hourly net growth rate in the exponential phase of population growth (in bacteriological terms, the specific growth rate), r, was 0.17. The fitness cost of resistance (fractional decrease in r) was parameterized from in vitro competition assays between E. coli strains carrying plasmids with blaCMY-2 and those that do not; a crude average of experimental data α = 0.05 was used [42].
Rates of horizontal transfer of individual plasmids with blaCMY-2 in vitro vary from 10−8 to 10−3 [42], [81]. In vivo in post-weaned calves fed a donor and a recipient E. coli strains, the overall rate of generation of blaCMY-2-transconjugates in fecal E. coli is 8−5 to 2−3 [27].
The rates of hourly fractional in-flow and out-flow of E. coli “free-living” in the large intestine, γ = 0.01 (to the daily total of 0.24), were estimated from an in vivo study in post-weaned dairy calves [27]. The fraction of ceftiofur-resistant cells in in-flow was set at 0.6*pAMR, based on 60% genetic similarity of E. coli in oral cavities and in feces of beef cattle reared at either pasture or feedlot [31].
Of parenteral ceftiofur dose, D, fraction p = 0.37 was excreted in bile within 6 hours p.i. (under excretion pattern 1 or 2) [51]; metabolites reached the large intestine in Tδ = 6 hours post excretion. Volume of the large intestine was 20L in an adult cattle, and 5L in a 6-mos calf. The rate of exponential decay in CE-concentration in the large intestine was twice lower compared to feces under aerobic conditions [48], [60].
As there are no published data from a time-kill experiment for E. coli and ceftiofur, the PD-model was applied to reproduce the data from in vitro time-kill experiments for E. coli and β-lactam ticarcillin [68], and performed well; H of 1.5 performed optimally for both concentrations below and above MIC. Under aerobic conditions, ceftiofur and its major metabolites are highly active against veterinary isolates of E. coli, with MIC50 = 0.25 µg/mL and MIC90 = 0.50 µg/mL [50]. Decrease in activity under anaerobic conditions appears to be limited (“one 2-fold dilution” in vitro), but the data are scarce. For the PD-model, MICs = 1 µg/mL and MICr = 8 µg/mL were used.
Sensitivity of methods based on bacteriological culture to detect a strain of E. coli in bovine feces is generally restricted to when >100 CFU/g is present [82]. We processed model outputs, Ns and Nr, to separate scenarios when ceftiofur-resistant E. coli likely would not be detected by culturing the feces.
Model Solving, and Uncertainty and Global Parameter Sensitivity Analysis
Solutions of the ordinary differential equations were approximated numerically using the fourth-order Runge–Kutta method implemented in Vensim® PLE Plus software (Ventana Systems, Inc.; Harvard, MA, USA). In the deterministic analysis, first, for each treatment scenario (Table S1), the model without treatment (Equations [1] and [2]) was solved varying parameter values to reproduce the reported pAMR (Table 1). Then concentration C, and PD-effects Es and Er were calculated. These were introduced into the model (Equations [9] and [10]) and the equations were solved. The models were solved starting with total E. coli, N, at 90% of Nmax.
The analysis of uncertainty and global parameter sensitivity of the model outputs was conducted for the treatment scenarios R2 and SB2 (Table S1). Given the dynamics of ceftiofur metabolites in the large intestine during therapy (section Pharmacokinetics and biodegradation of ceftiofur metabolites above), the sensitivity analysis was targeted at how the model outputs correlated with changes in the parameters of ecology of enteric E. coli (r, α, β, γ and υ), and of pharmacodynamics of the metabolites against E. coli (MICs, MICr and H). A uniform distribution (U) was assumed for all, because of the lack of knowledge of distributions of individual parameters. The minimum and maximum values were specified based on the literature review (all rates h−1): r∼U(0.05, 0.5), α∼U(−0.2, 0.2), β∼U(10−5, 0.01), γ∼U(10−3, 0.02), υ∼U(10−5, 0.10), MICs∼U(0.2, 1.9), MICr∼U(2, 16), and H∼U(0.5, 4). For each of the two treatment scenarios, 500 Monte Carlo simulations were performed with Latin Hypercube sampling of each parameter space at each time point over 90 days from start of therapy for the model with treatment, and for 180 days for the model without treatment. This was implemented in Vensim® PLE Plus software. The uncertainty in model outputs was explored graphically. The sensitivity of model outputs to changes in values of each parameter was evaluated with the Spearman correlation coefficient (ρ) [83]. Whether ρ was significantly different from zero was tested with a Student’s t-test, with the test statistics, denoting the number of simulations as w, calculated as  and assumed to follow a t-distribution with (w-2) degrees of freedom. If the test’s p-value was ≤0.05, the correlation was considered as significant.
and assumed to follow a t-distribution with (w-2) degrees of freedom. If the test’s p-value was ≤0.05, the correlation was considered as significant.
Results
Deterministic Analysis
Maintenance of ceftiofur resistance in commensal enteric E. coli in the absence of immediate ceftiofur pressure
In the model without treatment (Equations [1]–[2]), for every scenario (Table S1) the reported pAMR could be reproduced with r = 0.17, α = 0.05, β = 4−3, γ = 0.01, and υ = 0.6*pAMR. (These parameter values were used for the deterministic analysis of treatment scenarios: Table 1). Nmax corresponded to the scenario, and had no influence on the dynamics observed. pAMR was most sensitive to β and υ (or, if υ was kept constant, to γ). β>0.01 (somewhat unrealistic, see the discussion below) allowed reproducing the reported pAMR if there was no ceftiofur resistance among ingested E. coli, υ = 0. In all the scenarios, the resulting Nr was over 100; hence, culture-based methods would likely detect the presence of resistance.
Effects of parenteral ceftiofur therapy on commensal enteric E. coli: repeated ceftiofur administration
Models with both hypothesized biliary excretion patterns produced outputs resembling experimental data (Table S1); thus, biliary excretion of ceftiofur likely occurs within the first several hours p.i. Further results and discussion refer to the model with excretion pattern 2 (uniform excretion hourly at hour 1 to 6 p.i.).
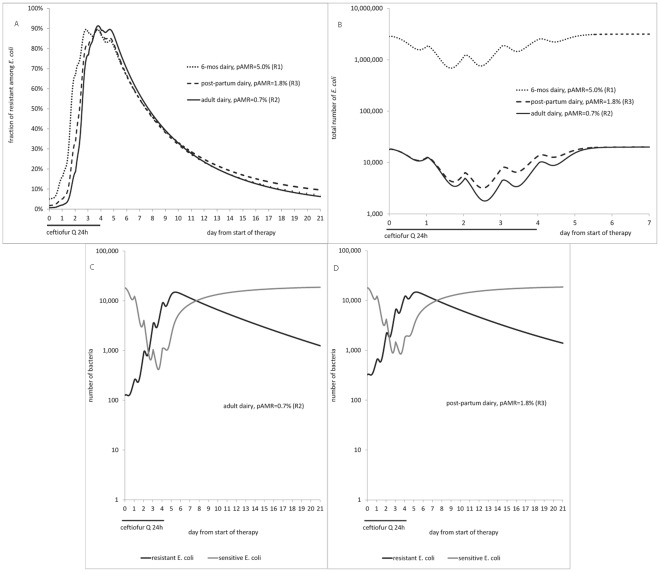
For all the treatment scenarios, the model (Equations [9]–[10]) outputs showed a decrease in counts of total and of ceftiofur-sensitive enteric E. coli, and a rise in the fraction of resistant among E. coli during therapy, similar to experimental observations (Fig. 3 cf. Table S1, scenarios R1-R3). For a 5-day therapy in an adult dairy, the model showed a decrease in total count from day 1, to a minimum on day 3, remaining decreased until day 4, and then returning to pre-treatment level on day 7 (Fig. 3B). The timing was close to the results for an on-farm 5-day ceftiofur therapy reporting fecal E. coli decreasing from day 1, to a minimum on day 6 (the samples were not collected every day), returning to pre-treatment level by day 9 (Table S1, R2). The maximum drop differed between the model, ∼1.05 log CFU/g, and the on-farm study, estimated ∼4.0 log CFU/g. However, the count change in the model corresponded to a 91% reduction in number of E. coli. Also, there may have been inter-individual variability in the count and its dynamics in the cattle treated on-farm, but only average numbers were reported. For a 5-day therapy in a 6-mos dairy, the model output of a 0.66 log CFU/g drop in total E. coli resembled well the field data of 0.5-1 log CFU/g drop in culturable fecal bacteria in individual 2-6 mos calves [78] (Fig. 3B cf. Table S1, R1). In both these scenarios, the total E. coli count dropped starting from day 1 of therapy.
Figure 3. Effect of therapy with repeated ceftiofur administration on enteric E. coli in the deterministic case considered (r = 0.17, α = 0.05, β = 4−3, γ = 0.01; all h−1).
A: Fraction of ceftiofur-resistant among E. coli. B: Total number of E. coli. C: Dynamics of ceftiofur-sensitive and resistant E. coli in an adult dairy. D: Dynamics of ceftiofur-sensitive and resistant E. coli in a post-partum dairy. pAMR = frequency of resistance at start of therapy.
As total E. coli was composed of both ceftiofur-sensitive and resistant cells, propagation of the latter offset the decline in total count, but this took time. E.g, in an adult dairy the total E. coli count was lowest on day 3 of therapy, dropping by 1.05 log CFU/g (Fig. 3B); but the count of ceftiofur-sensitive cells was lowest on day 4, dropping by 1.68 log CFU/g (Fig. 3C). These corresponded to a 91% and 98% reduction in number of bacteria, respectively. The fraction of resistant among E. coli peaked on day 4 (Fig. 3A). At this point, resistant cells filled most of the “carrying capacity”, sensitive cells grew less and were less exposed to antimicrobial action (hence no further reduction in sensitive counts). Similarly, in a 6-mos calf the number of sensitive E. coli dropped to its minimum and the fraction of resistant E. coli rose to its maximum (Fig. 3A) the next day after the maximum drop in total E. coli (Fig. 3B).
In cattle of a given age treated under a given protocol, the lower was the initial frequency of resistance, pAMR, the larger was the decline in total count of E. coli (Fig. 3B, R2 vs. R3) and of sensitive E. coli (Fig. 3C vs. 3D) during therapy. However, the maximum fraction of resistant among E. coli during therapy did not seem to depend on pAMR in the range explored, peaking to 90-91% in either an adult (pAMR = 0.7%), post-partum (pAMR = 1.8%), or 6-mos (pAMR = 5.0%) dairy (Fig. 3A). Yet it took longer for the fraction to completely return to pre-treatment level if this already had been elevated, e.g., ∼16 weeks in a 6-mos (pAMR = 5.0%) cf. ∼10 weeks in an adult (pAMR = 0.7%) dairy.
Scenarios R1-R3 were repeated with ceftiofur dosage 1.1 mg CE/kg (with biliary excretion pattern 1). In an adult dairy, including post-partum, there was a slightly lesser peak in number and relative fraction of resistant E. coli, and a day later in therapy compared to using 2.2 mg CE/kg. There was however a difference in a 6-mos dairy; the drop in total E. coli was 26% with 1.1 mg vs. 64% with 2.2 mg dosage, and the fraction of resistant cells rose to 27% vs. 80%, returning to pre-treatment level in 104 vs. 110 days, respectively.
Effects of parenteral ceftiofur therapy on commensal enteric E. coli: sustained-release ceftiofur formulation
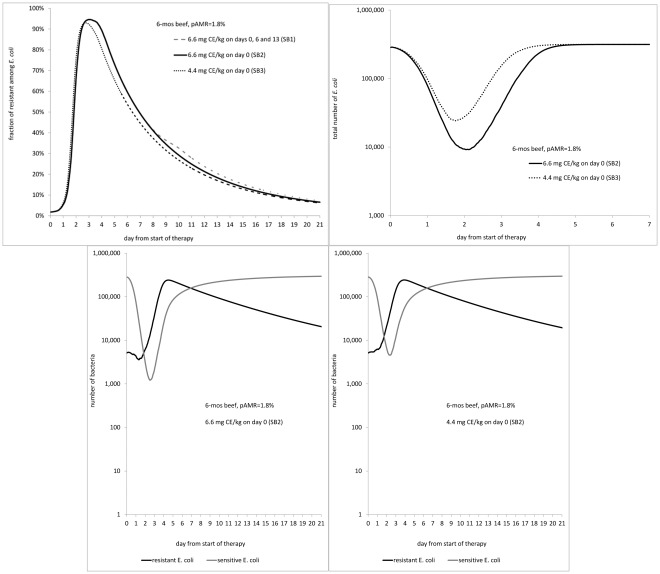
For a 6-mos beef administered a sustained-release ceftiofur formulation, lowering the dosage to 4.4 from 6.6 mg CE/kg (Table S1, SB3 vs. SB2), resulted in a smaller drop in total E. coli, 1.11 vs.1.54 log CFU/g, occurring a day earlier, day 2 vs. day 3, of therapy (Fig. 4B). The count of sensitive E. coli dropped by 1.83 log CFU/g with 4.4 mg vs. by 2.4 log CFU/g with 6.6 mg (98.5% vs. 99.6% reduction in number of sensitive bacteria), in either case being the lowest on day 3 (Fig. 4D vs. 4C). Quantitatively, the decrease in total E. coli matched field data; the timing was not contrasted because in the field study available for comparison with the scenarios SB1-SB3, the fecal samples were only obtained on days 0, 2, 6, 9, 13, 16, 20, and 28 of therapy [84]. The fraction of resistant E. coli, however, still rose to its highest of 93% on day 3 with 4.4 mg, cf. 95% on day 4 with 6.6 mg (Fig. 4A). The fraction reported by the field study varied from 40% to 90% [84]. In both scenarios SB2 and SB3, it took 111-112 days for the fraction of resistant E. coli to completely return to pre-treatment level.
Figure 4. Effect of therapy using a sustained-release ceftiofur formulation on enteric E. coli in the deterministic case considered (r = 0.17, α = 0.05, β = 4−3, γ = 0.01; all h−1).
A: Fraction of ceftiofur-resistant among E. coli. B: Total number of E. coli. Dynamics of ceftiofur-sensitive and resistant E. coli in a 6-mos beef treated with C: 6.6 mg CE/kg dosage, and D: 4.4 mg CE/kg dosage. pAMR = frequency of resistance at start of therapy.
With 6.6 mg CE/kg dosage, whether this was administered once on day 0, or 3 times on days 0, 6 and 13 (Table S1, SB1 vs. SB2) had a very limited effect on the dynamics observed (Fig. 4A). This is because the fraction of resistant E. coli was still high, and the number of sensitive E. coli depressed, following the 1st dose at the time the 2nd dose was given; similarly for doses 2 and 3. This agreed with field observations [84].
For an adult dairy (SD1), the model outputs were similar to a single administration of sustained-release ceftiofur formulation in 4.4 mg CE/kg dosage to a 6-mos beef (SB3).
Sensitivity of model outputs to variability in parameter values
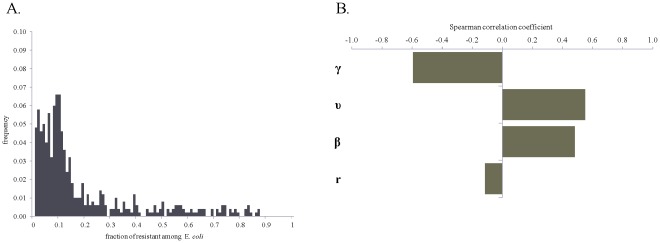
In the absence of immediate ceftiofur pressure, the maintenance of a fraction of ceftiofur-resistant cells among commensal enteric E. coli depended on the rate of in-flow and out-flow of E. coli, the prevalence of ceftiofur resistance in the in-flow, and the rate of transfer of blaCMY-2-carrying plasmids between E. coli within the intestine, and, to a lesser extent, on the rate of bacterial growth (Fig. 5B). Certain, although infrequent, combinations of the parameters produced a largely elevated frequency of resistance (Fig. 5A).
Figure 5. Fraction of ceftiofur-resistant among enteric E. coli in long-term absence of ceftiofur pressure.
A: Uncertainty: frequency histogram for 500 model simulations with randomly varying parameters of bacterial ecology. B: Sensitivity: significant linear correlations (p-value ≤0.05) between ranked-transformed values of the parameters of bacterial ecology and the fraction of resistance.
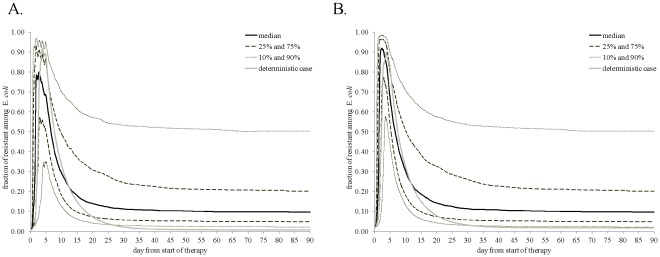
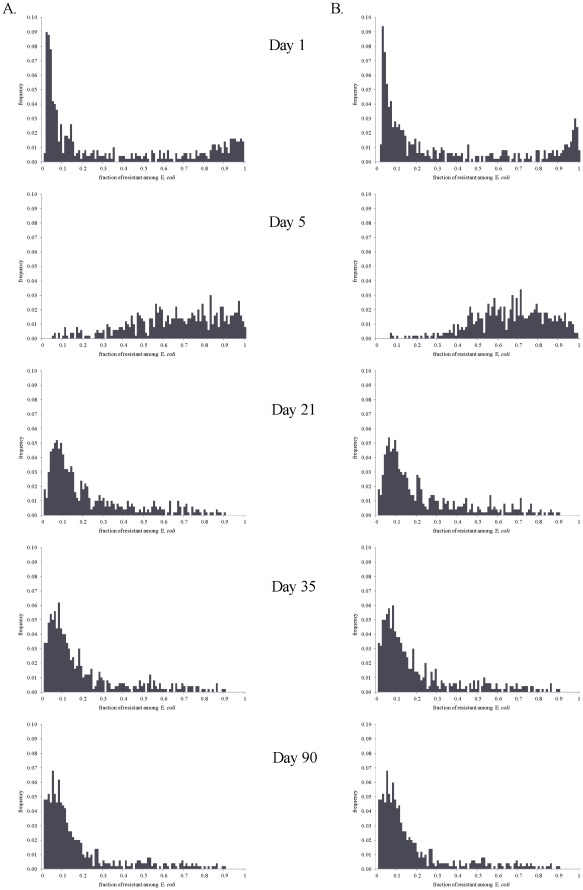
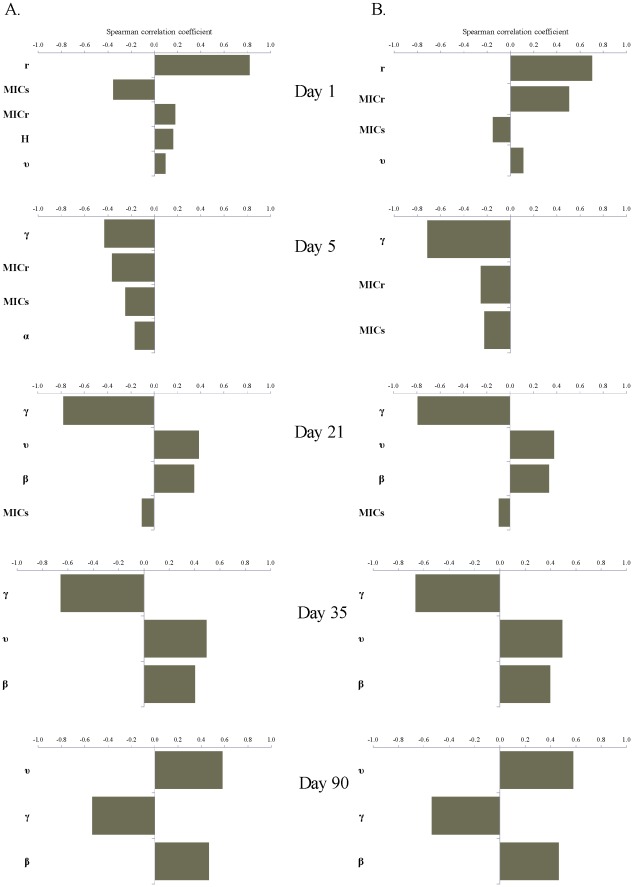
There were similar tendencies in the dynamics of ceftiofur-resistant enteric E. coli under the scenarios of a 5-day treatment of an adult dairy with a non-sustained release ceftiofur formulation in 2.2 mg CE/kg dosage (R2), and a single injection of a sustained-release ceftiofur formulation to a 6-mos beef in 6.6 mg CE/kg dosage (SB2) (Fig. 6). However, the median (over 500 model simulations) peak fraction of resistant among enteric E. coli was in the upper 70% range in the former vs. over 90% in the latter scenario (Fig. 6A vs.6B). There was a substantial uncertainty in the fraction of resistant E. coli, as during so after treatment, with the explored parameter ranges (Figs. 6 and 7). The largest uncertainty was observed on days 1 and 5 from start of therapy (Fig. 7). At those time points the variability in both the ecology of enteric E. coli, and in antimicrobial action of enteric ceftiofur metabolites against E. coli contributed to the outcome. On day 1 (Fig. 8), a higher fraction of resistance strongly correlated with a higher rate of bacterial growth, and a lower MICs, which would correspond to a larger kill of growing sensitive E. coli, hence a larger niche for expansion of the resistant cells, in turn ensured by the higher growth potential. A higher fraction of resistance also correlated with a higher MICr, i.e., a larger expansion of resistant E. coli if these were less sensitive at the start of therapy. The importance of individual parameters changed by day 5 when the fraction of resistance, after reaching its maximum, was still correlated with the pharmacodynamical parameters, but also became influenced again by the rate of fractional replacement of enteric E. coli (Fig. 8). By day 35, the fraction of resistance tended to settle at lower than 20%, further clustering toward lower values by day 90 (Fig. 7). This outcome depended on the same parameters that were most important for resistance maintenance in the absence of treatment: the rate of horizontal transfer of blaCMY-2, the rate of in-flow and out-flow of enteric E. coli, and the prevalence of ceftiofur-resistance in the in-flow (Fig. 8, Days 35 and 90 vs. Fig. 5B).
Figure 6. Uncertainty in the fraction of ceftiofur-resistant among enteric E. coli during 90 days from start of therapy.
Statistics are for 500 model simulations at each time point randomly varying parameters of bacterial ecology and pharmacodynamics. A: A 5-day repeated ceftiofur administration to an adult dairy (frequency of resistance at start of therapy 0.7%). B: An injection of a sustained-release ceftiofur formulation to a 6-mos beef (frequency of resistance at start of therapy 1.8%).
Figure 7. Frequency histograms for the fraction of ceftiofur-resistant among enteric E. coli on days 1, 5, 21, 35 and 90 from start of therapy.
Histograms are of 500 model simulations at each time point randomly varying parameters of bacterial ecology and pharmacodynamics. A: A 5-day repeated ceftiofur administration to an adult dairy (frequency of resistance at start of therapy 0.7%). B: An injection of a sustained-release ceftiofur formulation to a 6-mos beef (frequency of resistance at start of therapy 1.8%).
Figure 8. Significant linear correlations (p-value ≤0.05) between ranked-transformed values of the parameters of bacterial ecology and pharmacodynamics and the frequency of ceftiofur-resistant among enteric E. coli on days 1, 5, 21, 35 and 90 from start of therapy.
A: A 5-day repeated ceftiofur administration to an adult dairy (frequency of resistance at start of therapy 0.7%). B: An injection of a sustained-release ceftiofur formulation to a 6-mos beef (frequency of resistance at start of therapy 1.8%).
Discussion
This modeling study suggested that ceftiofur-resistant commensal enteric E. coli in cattle could persist between treatments. A low but stable fraction of resistance can be maintained even if the number of resistant E. coli grows slower than that of the sensitive ones, when the rate of blaCMY-2 transfer in enteric E. coli is sufficiently high or a sufficient fraction of ingested E. coli is ceftiofur-resistant. The latter could occur if the conditions on the farm allow for a close circulation of commensal E. coli between cattle and their environment.
The values reported by field studies of the fraction of ceftiofur-resistant cells among fecal E. coli in the absence of immediate ceftiofur pressure were reproduced in the deterministic analysis with a transfer rate for blaCMY-2-carrying plasmids of 4−3. For individual plasmids, in vitro transfer rates are up to 2−3 [42]. In vivo in post-weaned calves fed a donor and a recipient strain, a rate of blaCMY-2-transconjugant generation in fecal E. coli of 2−3 has been reported [27]. Several plasmids may be present in enteric E. coli, and the blaCMY-2-transfer rate in vivo may be the cumulative of those for individual plasmids. Generally, conjugation frequency may depend on the physiological status of donor cells, phase of growth of donor population [85], and physical conditions [13], [86], [87]. As the enteric environment is E. coli’s ecological niche, the cells are likely in normal physiological condition; this, coupling with nutrient availability for population growth, absence of light and favorable temperatures, may allow for the bacterial conjugation, and so plasmid transfer rate, to be at the high end of the biological maximum [87], [88].
Ceftiofur-resistant E. coli in this study were defined as cells having a plasmid with blaCMY-2. A possibility of variability in the degree of resistance conferred by presence of more than one copy of the gene, or by presence of another mechanism of resistance, and how this may be reflected in MIC-values, was not considered; neither are such data available in experimental literature. This complicated interpretation of the correlations of MICr with the fraction of resistant among E. coli during therapy (Fig. 8).
The increase in absolute number of E. coli cells carrying blaCMY-2 per g of feces during parenteral ceftiofur therapy could lead to a higher frequency of horizontal transfer of this plasmidic gene to the other enteric bacteria, including potential zoonotic pathogens. However, this would require not only the donor but also the recipient populations to be present in sufficient numbers [89]. The numbers of ceftiofur-sensitive cells among the other bacteria may be diminishing during therapy, similar to the numbers of ceftiofur-sensitive E. coli. Hence, the net effect on the frequency of blaCMY-2-transmission from E. coli to the other bacteria would depend on the degree of sensitivity of the latter to antimicrobial action of enteric ceftiofur metabolites. Importantly, the only origin and spread of AMR considered here for ceftiofur and E. coli were the a-priori presence of plasmidic blaCMY-2, and its vertical and horizontal transfers, respectively. Other mechanisms, e.g. resistance mediated by chromosomal genes or that due to plasmid-mediated extended-spectrum beta-lactamases, may need to be considered to understand AMR dynamics across the species. Therefore, based on the current model, we could not infer what effect ceftiofur therapy might have on inter-species spread of blaCMY-2. Furthermore, evaluating the potential for spread of AMR-determinants requires accounting for not only the within-host dynamics addressed here, but also how the resistant bacteria spread among the hosts, and between the hosts and their environment.
This study highlighted that results of ceftiofur treatment trials would be more informative for modeling if the data reported would include the dynamical change in fraction of the resistant fecal bacteria, and description of variability among individual cattle. Frequent sampling during treatment would help with detailing the length of time available for expansion of resistant bacteria; continuing sampling post treatment would help with understanding the mechanisms involved in resistance maintenance. On the epidemiological side, important knowledge gaps are details of E. coli cycling between cattle and their environment, including the degree of replacement of enteric E. coli, the prevalence of ceftiofur resistance in E. coli ingested by cattle, and the rates of horizontal blaCMY-2-transfer in vivo. Absence of publicly accessible data on the concentration and antimicrobial activity of ceftiofur metabolites in cattle intestine during parenteral therapy hinders more detailed research into the selective pressure experienced by enteric bacteria. On the pharmacological side, time-kill experiments (as opposed to experiments establishing MIC-values) mimicking intestinal conditions are needed to describe the pharmacodynamics of ceftiofur metabolites against enteric bacteria.
To conclude, first, the results showed that reported low fractions of ceftiofur-resistant commensal enteric E. coli in cattle could be maintained without immediate ceftiofur pressure. Second, during parenteral ceftiofur therapy there likely are antibiotically-active drug metabolites in the large intestine, circumventing a slash in the number of ceftiofur-sensitive enteric E. coli. These conclusions are strongly supported by the concordance of the model outputs with experimental data. Hence, there is a window during therapy when ceftiofur-resistant E. coli could expand in absolute number and relative frequency; the degree of expansion depends on the parameters of antimicrobial action of the metabolites against E. coli, as well as on the rates of enteric E. coli growth and replacement. However, whether the post-treatment fraction of resistance would remain elevated in the long-term depends on a present combination of the parameters of bacterial ecology, the same parameters that are important for maintenance of resistance in the absence of ceftiofur pressure. Namely, these are the rate of horizontal transfer of plasmids with blaCMY-2 between enteric E. coli, which may be determined by which plasmids are present, and the frequency of resistance in E. coli ingested by cattle, which may be determined by the extent of E. coli circulation between cattle and their environment.
Supporting Information
Modeled scenarios of treatment of cattle with ceftiofur, and experimental data considered for comparison.
(DOCM)
Acknowledgments
We are grateful to Craig Altier of Cornell University and Thomas Besser of Washington State University for generous sharing of their expertise in biology of ceftiofur resistance; and to Morgan Scott of Kansas University for informative discussions. We thank scholars of the Leadership Program for Veterinary Scholars at Cornell University: summer 2010 scholar Clinton Doering for starting-up literature research on the topic, and summer 2011 scholar Sarah Wood for interactive discussions.
Footnotes
Competing Interests: The authors have declared that no competing interests exist.
Funding: This work was funded by the National Institute of Food and Agriculture of the United States Department of Agriculture (grant # 2010-51110-21083). The funders had no role in study design, analysis, decision to publish, or preparation of the manuscript.
References
- 1.McDermott PF, Zhao S, Wagner DD, Simjee S, Walker RD, et al. The food safety perspective of antibiotic resistance. Animal Biotechnology. 2002;13:71–84. doi: 10.1081/ABIO-120005771. [DOI] [PubMed] [Google Scholar]
- 2.Fey PD, Safranek TJ, Rupp ME, Dunne EF, Ribot E, et al. Ceftriaxone-resistant salmonella infection acquired by a child from cattle. N Engl J Med. 2000;342:1242–1249. doi: 10.1056/NEJM200004273421703. [DOI] [PubMed] [Google Scholar]
- 3.Cohen ML, Tauxe RV. Drug-resistant Salmonella in the United States: an epidemiologic perspective. Science. 1986;234:964–969. doi: 10.1126/science.3535069. [DOI] [PubMed] [Google Scholar]
- 4.Linton AH, Howe K, Bennett PM, Richmond MH, Whiteside EJ. The colonization of the human gut by antibiotic resistant Escherichia coli from chickens. J Appl Bacteriol. 1977;43:465–469. doi: 10.1111/j.1365-2672.1977.tb00773.x. [DOI] [PubMed] [Google Scholar]
- 5.Salyers AA, Gupta A, Wang Y. Human intestinal bacteria as reservoirs for antibiotic resistance genes. Trends Microbiol. 2004;12:412–416. doi: 10.1016/j.tim.2004.07.004. [DOI] [PubMed] [Google Scholar]
- 6.Winokur PL, Vonstein DL, Hoffman LJ, Uhlenhopp EK, Doern GV. Evidence for transfer of CMY-2 AmpC beta-lactamase plasmids between Escherichia coli and Salmonella isolates from food animals and humans. Antimicrob Agents Chemother. 2001;45:2716–2722. doi: 10.1128/AAC.45.10.2716-2722.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Boerlin P, Reid-Smith RJ. Antimicrobial resistance: its emergence and transmission. Anim Health Res Rev. 2008;9:115–126. doi: 10.1017/S146625230800159X. [DOI] [PubMed] [Google Scholar]
- 8.McCuddin Z, Carlson SA, Rasmussen MA, Franklin SK. Klebsiella to Salmonella gene transfer within rumen protozoa: Implications for antibiotic resistance and rumen defaunation. Veterinary Microbiology. 2006;114:275–284. doi: 10.1016/j.vetmic.2005.12.004. [DOI] [PubMed] [Google Scholar]
- 9.Delsol AA, Halfhide DE, Bagnall MC, Randall LP, Enne VI, et al. Persistence of a wild type Escherichia coli and its multiple antibiotic-resistant (MAR) derivatives in the abattoir and on chilled pig carcasses. International Journal of Food Microbiology. 2010;140:249–253. doi: 10.1016/j.ijfoodmicro.2010.03.023. [DOI] [PubMed] [Google Scholar]
- 10.Aslam M, Nattress F, Greer G, Yost C, Gill C, et al. Origin of contamination and genetic diversity of Escherichia coli in beef cattle. Appl Environ Microbiol. 2003;69:2794–2799. doi: 10.1128/AEM.69.5.2794-2799.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Call DR, Kang MS, Daniels J, Besser TE. Assessing genetic diversity in plasmids from Escherichia coli and Salmonella enterica using a mixed-plasmid microarray. J Appl Microbiol. 2006;100:15–28. doi: 10.1111/j.1365-2672.2005.02775.x. [DOI] [PubMed] [Google Scholar]
- 12.Daniels JB, Call DR, Besser TE. Molecular epidemiology of blaCMY-2 plasmids carried by Salmonella enterica and Escherichia coli isolates from cattle in the Pacific Northwest. Appl Environ Microbiol. 2007;73:8005–8011. doi: 10.1128/AEM.01325-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Licht TR, Wilcks A. Conjugative gene transfer in the gastrointestinal environment. Adv Appl Microbiol. 2006;58:77–95. [PubMed] [Google Scholar]
- 14.Ravel J, Fricke WF, McDermott PF, Mammel MK, Zhao SH, et al. Antimicrobial Resistance-Conferring Plasmids with Similarity to Virulence Plasmids from Avian Pathogenic Escherichia coli Strains in Salmonella enterica Serovar Kentucky Isolates from Poultry. Applied and Environmental Microbiology. 2009;75:5963–5971. doi: 10.1128/AEM.00786-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Khachatryan AR, Hancock DD, Besser TE, Call DR. Role of calf-adapted Escherichia coli in maintenance of antimicrobial drug resistance in dairy calves. Appl Environ Microbiol. 2004;70:752–757. doi: 10.1128/AEM.70.2.752-757.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Khachatryan AR, Hancock DD, Besser TE, Call DR. Antimicrobial drug resistance genes do not convey a secondary fitness advantage to calf-adapted Escherichia coli. Appl Environ Microbiol. 2006;72:443–448. doi: 10.1128/AEM.72.1.443-448.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Apley MD, Brown SA, Fedorka-Cray PJ, Ferenc S, House JK, et al. Role of veterinary therapeutics in bacterial resistance development: animal and public health perspectives. Journal of the American Veterinary Medical Association. 1998;212:1209–1213. [PubMed] [Google Scholar]
- 18.Morley PS, Apley MD, Besser TE, Burney DP, Fedorka-Cray PJ, et al. Antimicrobial drug use in veterinary medicine. Journal of Veterinary Internal Medicine. 2005;19:617–629. doi: 10.1892/0891-6640(2005)19[617:aduivm]2.0.co;2. [DOI] [PubMed] [Google Scholar]
- 19.Lanzas C, Ayscue P, Ivanek R, Grohn YT. Model or meal? Farm animal populations as models for infectious diseases of humans. Nature Reviews Microbiology. 2010;8:139–148. doi: 10.1038/nrmicro2268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bauernfeind A, Stemplinger I, Jungwirth R, Ernst S, Casellas JM. Sequences of beta-lactamase genes encoding CTX-M-1 (MEN-1) and CTX-M-2 and relationship of their amino acid sequences with those of other beta-lactamases. Antimicrob Agents Chemother. 1996;40:509–513. doi: 10.1128/aac.40.2.509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Barlow M, Hall BG. Origin and evolution of the AmpC beta-lactamases of Citrobacter freundii. Antimicrob Agents Chemother. 2002;46:1190–1198. doi: 10.1128/AAC.46.5.1190-1198.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bush K, Jacoby GA, Medeiros AA. A functional classification scheme for beta-lactamases and its correlation with molecular structure. Antimicrob Agents Chemother. 1995;39:1211–1233. doi: 10.1128/aac.39.6.1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bauernfeind A, Stemplinger I, Jungwirth R, Giamarellou H. Characterization of the plasmidic beta-lactamase CMY-2, which is responsible for cephamycin resistance. Antimicrob Agents Chemother. 1996;40:221–224. doi: 10.1128/aac.40.1.221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Heider LC, Hoet AE, Wittum TE, Khaitsa ML, Love BC, et al. Genetic and phenotypic characterization of the bla(CMY) gene from Escherichia coli and Salmonella enterica isolated from food-producing animals, humans, the environment, and retail meat. Foodborne Pathogens and Disease. 2009;6:1235–1240. doi: 10.1089/fpd.2009.0294. [DOI] [PubMed] [Google Scholar]
- 25.Dunne EF, Fey PD, Kludt P Reporter R, Mostashari F, et al. Emergence of domestically acquired ceftriaxone-resistant Salmonella infections associated with AmpC beta-lactamase. JAMA. 2000;284:3151–3156. doi: 10.1001/jama.284.24.3151. [DOI] [PubMed] [Google Scholar]
- 26.Dutil L, Irwin R, Finley R, Ng LK, Avery B, et al. Ceftiofur resistance in Salmonella enterica serovar Heidelberg from chicken meat and humans, Canada. Emerging Infectious Diseases. 2010;16:48–54. doi: 10.3201/eid1601.090729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Daniels JB, Call DR, Hancock D, Sischo WM, Baker K, et al. Role of ceftiofur in selection and dissemination of blaCMY-2-mediated cephalosporin resistance in Salmonella enterica and commensal Escherichia coli isolates from cattle. Appl Environ Microbiol. 2009;75:3648–3655. doi: 10.1128/AEM.02435-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sawant AA, Hegde NV, Straley BA, Donaldson SC, Love BC, et al. Antimicrobial-resistant enteric bacteria from dairy cattle. Appl Environ Microbiol. 2007;73:156–163. doi: 10.1128/AEM.01551-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tragesser LA, Wittum TE, Funk JA, Winokur PL, Rajala-Schultz PJ. Association between ceftiofur use and isolation of Escherichia coli with reduced susceptibility to ceftriaxone from fecal samples of dairy cows. American Journal of Veterinary Research. 2006;67:1696–1700. doi: 10.2460/ajvr.67.10.1696. [DOI] [PubMed] [Google Scholar]
- 30.Morley PS, Dargatz DA, Hyatt DR, Dewell GA, Patterson JG, et al. Effects of restricted antimicrobial exposure on antimicrobial resistance in fecal Escherichia coli from feedlot cattle. Foodborne Pathogens and Disease. 2011;8:87–98. doi: 10.1089/fpd.2010.0632. [DOI] [PubMed] [Google Scholar]
- 31.Aslam M, Greer GG, Nattress FM, Gill CO, McMullen LM. Genetic diversity of Escherichia coli recovered from the oral cavity of beef cattle and their relatedness to faecal E. coli. Lett Appl Microbiol. 2004;39:523–527. doi: 10.1111/j.1472-765X.2004.01620.x. [DOI] [PubMed] [Google Scholar]
- 32.Philippon A, Arlet G, Jacoby GA. Plasmid-determined AmpC-type beta-lactamases. Antimicrob Agents Chemother. 2002;46:1–11. doi: 10.1128/AAC.46.1.1-11.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Persoons D, Haesebrouck F, Smet A, Herman L, Heyndrickx M, et al. Risk factors for ceftiofur resistance in Escherichia coli from Belgian broilers. Epidemiol Infect. 2011;139:765–771. doi: 10.1017/S0950268810001524. [DOI] [PubMed] [Google Scholar]
- 34.Singer RS, Patterson SK, Wallace RL. Effects of therapeutic ceftiofur administration to dairy cattle on Escherichia coli dynamics in the intestinal tract. Appl Environ Microbiol. 2008;74:6956–6962. doi: 10.1128/AEM.01241-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mann S, Siler JD, Jordan D, Warnick LD. Foodborne Pathogens and Disease; 2011. Antimicrobial susceptibility of fecal Escherichia coli isolates in dairy cows following systemic treatment with ceftiofur or penicillin. [DOI] [PubMed] [Google Scholar]
- 36.Alali WQ, Scott HM, Norby B, Gebreyes W, Loneragan GH. Quantification of the bla(CMY-2) in feces from beef feedlot cattle administered three different doses of ceftiofur in a longitudinal controlled field trial. Foodborne Pathogens and Disease. 2009;6:917–924. doi: 10.1089/fpd.2009.0271. [DOI] [PubMed] [Google Scholar]
- 37.Ketyi I. Effectiveness of antibiotics on the autochthonous Escherichia coli of mice in the intestinal biofilm versus its planktonic phase. Acta Microbiol Immunol Hung. 1994;41:189–195. [PubMed] [Google Scholar]
- 38.Durso LM, Smith D, Hutkins RW. Measurements of fitness and competition in commensal Escherichia coli and E. coli O157:H7 strains. Appl Environ Microbiol. 2004;70:6466–6472. doi: 10.1128/AEM.70.11.6466-6472.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Freter R, Brickner H, Botney M, Cleven D, Aranki A. Mechanisms That Control Bacterial-Populations in Continuous-Flow Culture Models of Mouse Large Intestinal Flora. Infection and Immunity. 1983;39:676–685. doi: 10.1128/iai.39.2.676-685.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ayscue P, Lanzas C, Ivanek R, Grohn YT. Modeling On-Farm Escherichia coli O157:H7 Population Dynamics. Foodborne Pathogens and Disease. 2009;6:461–470. doi: 10.1089/fpd.2008.0235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Andrup L, Andersen K. A comparison of the kinetics of plasmid transfer in the conjugation systems encoded by the F plasmid from Escherichia coli and plasmid pCF10 from Enterococcus faecalis. Microbiology-Uk. 1999;145:2001–2009. doi: 10.1099/13500872-145-8-2001. [DOI] [PubMed] [Google Scholar]
- 42.Subbiah M, Top EM, Shah DH, Call DR. Selection Pressure Required for Long-Term Persistence of blaCMY-2-Positive IncA/C Plasmids. Appl Environ Microbiol. 2011;77:4486–4493. doi: 10.1128/AEM.02788-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lenski RE, Simpson SC, Nguyen TT. Genetic analysis of a plasmid-encoded, host genotype-specific enhancement of bacterial fitness. J Bacteriol. 1994;176:3140–3147. doi: 10.1128/jb.176.11.3140-3147.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Poole TL, Brichta-Harhay DM, Callaway TR, Beier RC, Bischoff KM, et al. Persistence of resistance plasmids carried by beta-hemolytic Escherichia coli when maintained in a continuous-flow fermentation system without antimicrobial selection pressure. Foodborne Pathogens and Disease. 2011;8:535–540. doi: 10.1089/fpd.2010.0732. [DOI] [PubMed] [Google Scholar]
- 45.Bergstrom CT, Lipsitch M, Levin BR. Natural selection, infectious transfer and the existence conditions for bacterial plasmids. Genetics. 2000;155:1505–1519. doi: 10.1093/genetics/155.4.1505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Schrag SJ, Perrot V, Levin BR. Adaptation to the fitness costs of antibiotic resistance in Escherichia coli. Proc Biol Sci. 1997;264:1287–1291. doi: 10.1098/rspb.1997.0178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Jaglan PS, Kubicek MF, Arnold TS, Cox BL, Robins RH, et al. Metabolism of ceftiofur - nature of urinary and plasma metabolites in rats and cattle. Journal of Agricultural and Food Chemistry. 1989;37:1112–1118. [Google Scholar]
- 48.Hornish RE, Kotarski SF. Cephalosporins in veterinary medicine - ceftiofur use in food animals. Curr Top Med Chem. 2002;2:717–731. doi: 10.2174/1568026023393679. [DOI] [PubMed] [Google Scholar]
- 49.Ritter L, Kirby G, Cerniglia C. Toxicological evaluation of certain veterinary drug residues in food. (857) Ceftiofur WHO Food Additives Series. 1996;36 [Google Scholar]
- 50.Salmon SA, Watts JL, Yancey RJ. In vitro activity of ceftiofur and its primary metabolite, desfuroylceftiofur, against organisms of veterinary importance. Journal of Veterinary Diagnostic Investigation. 1996;8:332–336. doi: 10.1177/104063879600800309. [DOI] [PubMed] [Google Scholar]
- 51.Beconi-Barker MG, Roof RD, Vidmar TJ, Hornish RE, Smith EB, et al. Ceftiofur sodium: Absorption, distribution, metabolism, and excretion in target animals and its determination by high-performance liquid chromatography. Veterinary Drug Residues. 1996;636:70–84. [Google Scholar]
- 52.Brown SA, Chester ST, Speedy AK, Hubbard VL, Callahan JK, et al. Comparison of plasma pharmacokinetics and bioequivalence of ceftiofur sodium in cattle after a single intramuscular or subcutaneous injection. Journal of Veterinary Pharmacology and Therapeutics. 2000;23:273–280. doi: 10.1046/j.1365-2885.2000.00271.x. [DOI] [PubMed] [Google Scholar]
- 53.Brown SA, Hanson BJ, Mignot A, Millerioux L, Hamlow PJ, et al. Comparison of plasma pharmacokinetics and bioavailability of ceftiofur sodium and ceftiofur hydrochloride in pigs after a single intramuscular injection. J Vet Pharmacol Ther. 1999;22:35–40. doi: 10.1046/j.1365-2885.1999.00182.x. [DOI] [PubMed] [Google Scholar]
- 54.Arvidsson A, Alvan G, Angelin B, Borga O, Nord CE. Ceftriaxone - Renal and Biliary-Excretion and Effect on the Colon Microflora. Journal of Antimicrobial Chemotherapy. 1982;10:207–215. doi: 10.1093/jac/10.3.207. [DOI] [PubMed] [Google Scholar]
- 55.Bakken JS, Cavalieri SJ, Gangeness D. Influence of plasma exchange pheresis on plasma elimination of ceftriaxone. Antimicrob Agents Chemother. 1990;34:1276–1277. doi: 10.1128/aac.34.6.1276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Arvidsson A, Leijd B, Nord CE, Angelin B. Interindividual Variability in Biliary-Excretion of Ceftriaxone - Effects on Biliary Lipid-Metabolism and on Intestinal Microflora. European Journal of Clinical Investigation. 1988;18:261–266. doi: 10.1111/j.1365-2362.1988.tb01256.x. [DOI] [PubMed] [Google Scholar]
- 57.Xia Y, Lambert KJ, Schteingart CD, Gu JJ, Hofmann AF. Concentrative biliary secretion of ceftriaxone. Inhibition of lipid secretion and precipitation of calcium ceftriaxone in bile. Gastroenterology. 1990;99:454–465. doi: 10.1016/0016-5085(90)91029-6. [DOI] [PubMed] [Google Scholar]
- 58.Hoffmann-La Roche Limited. Prepared June 16, 1987. In: 9 RevisedFebruary., editor. 2010. Product monograph: Rocephin (sterile ceftriaxone sodium). 46 pp; 2010. [Google Scholar]
- 59.United States Food and Drug Administration. 46 pp; 1990. Environmental Assessment of Excenel Sterile Suspension (Ceftiofur Hydrochloride). [Google Scholar]
- 60.Gilbertson TJ, Hornish RE, Jaglan PS, Koshy KT, Nappier JL, et al. Environmental Fate of Ceftiofur Sodium, a Cephalosporin Antibiotic - Role of Animal Excreta in Its Decomposition. Journal of Agricultural and Food Chemistry. 1990;38:890–894. [Google Scholar]
- 61.Livermore DM. Mechanisms of resistance to beta-lactam antibiotics. Scand J Infect. 1991;Dis(Suppl 78):7–16. [PubMed] [Google Scholar]
- 62.Merle-Melet M, Seta N, Farinotti R, Carbon C. Reduction in biliary excretion of ceftriaxone by diclofenac in rabbits. Antimicrob Agents Chemother. 1989;33:1506–1510. doi: 10.1128/aac.33.9.1506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Symonds HW, Mather DL, Hall ED. Surgical procedure for modifying the duodenum in cattle to measure bile flow and the diurnal variation in biliary manganese, iron, copper and zinc excretion. Res Vet Sci. 1982;32:6–11. [PubMed] [Google Scholar]
- 64. Pfizer Animal Health website. Graph of plasma concentrations of ceftiofur equivalents following a single injection of a sustained-release ceftiofur formulation in lactating dairy: Available: http://www.excede.com/Excede.aspx?country=US&drug=XT&species=DA&sec=310. Accessed 1 Oct 2011.
- 65. Pfizer Animal Health website. Graph of plasma concentrations of ceftiofur equivalents following a single injection of a sustained-release ceftiofur formulation in beef: Available: http://www.excede.com/Excede.aspx?country=US&drug=XT&species=BF&sec=300. Accessed 1 Oct 2011.
- 66.Tipper DJ. Mode of action of beta-lactam antibiotics. Pharmacol Ther. 1985;27:1–35. doi: 10.1016/0163-7258(85)90062-2. [DOI] [PubMed] [Google Scholar]
- 67.Mattie H, van Dokkum AM, Brus-Weijer L, Krul AM, van Strijen E. Antibacterial activity of four cephalosporins in an experimental infection in relation to in vitro effect and pharmacokinetics. J Infect Dis. 1990;162:717–722. doi: 10.1093/infdis/162.3.717. [DOI] [PubMed] [Google Scholar]
- 68.Craig WA, Ebert SC. Killing and regrowth of bacteria in vitro: a review. Scand J Infect. 1990;Dis(Suppl 74):63–70. [PubMed] [Google Scholar]
- 69.Mouton JW, Dudley MN, Cars O, Derendorf H, Drusano GL. Standardization of pharmacokinetic/pharmacodynamic (PK/PD) terminology for anti-infective drugs: an update. J Antimicrob Chemother. 2005;55:601–607. doi: 10.1093/jac/dki079. [DOI] [PubMed] [Google Scholar]
- 70.Goutelle S, Maurin M, Rougier F, Barbaut X, Bourguignon L, et al. The Hill equation: a review of its capabilities in pharmacological modelling. Fundam Clin Pharmacol. 2008;22:633–648. doi: 10.1111/j.1472-8206.2008.00633.x. [DOI] [PubMed] [Google Scholar]
- 71.Mouton JW, Vinks AA. Pharmacokinetic/pharmacodynamic modelling of antibacterials in vitro and in vivo using bacterial growth and kill kinetics: the minimum inhibitory concentration versus stationary concentration. Clin Pharmacokinet. 2005;44:201–210. doi: 10.2165/00003088-200544020-00005. [DOI] [PubMed] [Google Scholar]
- 72.Drusano GL. Antimicrobial pharmacodynamics: critical interactions of ‘bug and drug’. Nat Rev Microbiol. 2004;2:289–300. doi: 10.1038/nrmicro862. [DOI] [PubMed] [Google Scholar]
- 73.Drusano GL. Scandinavian Journal of Infectious Diseases; 1991. Human Pharmacodynamics of Beta-Lactams, Aminoglycosides and Their Combination. pp. 235–248. [PubMed] [Google Scholar]
- 74.Hanberger H. Pharmacodynamic effects of antibiotics. Studies on bacterial morphology, initial killing, postantibiotic effect and effective regrowth time. Scand J Infect. 1992;Dis(Suppl 81):1–52. doi: 10.3109/inf.1992.24.suppl-81.01. [DOI] [PubMed] [Google Scholar]
- 75.Czock D, Keller F. Mechanism-based pharmacokinetic–pharmacodynamic modeling of antimicrobial drug effects. J Pharmacokinet Biopharm. 2007;34:727–751. doi: 10.1007/s10928-007-9069-x. [DOI] [PubMed] [Google Scholar]
- 76.Ambrose PG, Bhavnani SM, Rubino CM, Louie A, Gumbo T, et al. Pharmacokinetics-pharmacodynamics of antimicrobial therapy: it’s not just for mice anymore. Clinical Infectious Diseases. 2007;44:79–86. doi: 10.1086/510079. [DOI] [PubMed] [Google Scholar]
- 77.Vogelman B, Gudmundsson S, Turnidge J, Leggett J, Craig WA. Invivo Postantibiotic Effect in a Thigh Infection in Neutropenic Mice. Journal of Infectious Diseases. 1988;157:287–298. doi: 10.1093/infdis/157.2.287. [DOI] [PubMed] [Google Scholar]
- 78.Jiang X, Yang H, Dettman B, Doyle MP. Analysis of fecal microbial flora for antibiotic resistance in ceftiofur-treated calves. Foodborne Pathogens and Disease. 2006;3:355–365. doi: 10.1089/fpd.2006.3.355. [DOI] [PubMed] [Google Scholar]
- 79.Daniels JB, Call DR, Hancock D, Sischo WM, Baker K, et al. Role of Ceftiofur in Selection and Dissemination of bla(CMY-2)-Mediated Cephalosporin Resistance in Salmonella enterica and Commensal Escherichia coli Isolates from Cattle. Applied and Environmental Microbiology. 2009;75:3648–3655. doi: 10.1128/AEM.02435-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Alexander TW, Yanke LJ, Topp E, Olson ME, Read RR, et al. Effect of subtherapeutic administration of antibiotics on the prevalence of antibiotic-resistant Escherichia coli bacteria in feedlot cattle. Applied and Environmental Microbiology. 2008;74:4405–4416. doi: 10.1128/AEM.00489-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Fricke WF, Welch TJ, McDermott PF, Mammel MK, LeClerc JE, et al. Comparative genomics of the IncA/C multidrug resistance plasmid family. J Bacteriol. 2009;191:4750–4757. doi: 10.1128/JB.00189-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.LeJeune JT, Hancock DD, Besser TE. Sensitivity of Escherichia coli O157 detection in bovine feces assessed by broth enrichment followed by immunomagnetic separation and direct plating methodologies. Journal of Clinical Microbiology. 2006;44:872–875. doi: 10.1128/JCM.44.3.872-875.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Marino S, Hogue IB, Ray CJ, Kirschner DE. A methodology for performing global uncertainty and sensitivity analysis in systems biology. Journal of Theoretical Biology. 2008;254:178–196. doi: 10.1016/j.jtbi.2008.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Lowrance TC, Loneragan GH, Kunze DJ, Platt TM, Ives SE, et al. Changes in antimicrobial susceptibility in a population of Escherichia coli isolated from feedlot cattle administered ceftiofur crystalline-free acid. American Journal of Veterinary Research. 2007;68:501–507. doi: 10.2460/ajvr.68.5.501. [DOI] [PubMed] [Google Scholar]
- 85.Muela A, Pocino M, Arana I, Justo JI, Iriberri J, et al. Effect of growth phase and parental cell survival in river water on plasmid transfer between Escherichia coli strains. Appl Environ Microbiol. 1994;60:4273–4278. doi: 10.1128/aem.60.12.4273-4278.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Fernandez-Astorga A, Muela A, Cisterna R, Iriberri J, Barcina I. Biotic and abiotic factors affecting plasmid transfer in Escherichia coli strains. Appl Environ Microbiol. 1992;58:392–398. doi: 10.1128/aem.58.1.392-398.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Arana II, Justo JI, Muela A, Pocino M, Iriberri J, et al. Influence of a Survival Process in a Freshwater System upon Plasmid Transfer Between Escherichia coli Strains. Microb Ecol. 1997;33:41–49. doi: 10.1007/s002489900006. [DOI] [PubMed] [Google Scholar]
- 88.Arana I, Pocino M, Muela A, Fernandez-Astorga A, Barcina I. Detection and enumeration of viable but non-culturable transconjugants of Escherichia coli during the survival of recipient cells in river water. J Appl Microbiol. 1997;83:340–346. doi: 10.1046/j.1365-2672.1997.00244.x. [DOI] [PubMed] [Google Scholar]
- 89.Stecher B, Denzler R, Maier L, Bernet F, Sanders MJ, et al. Gut inflammation can boost horizontal gene transfer between pathogenic and commensal Enterobacteriaceae. Proc Natl Acad Sci U S A. 2012;109:1269–1274. doi: 10.1073/pnas.1113246109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Sorum H, Sunde M. Resistance to antibiotics in the normal flora of animals. Vet Res. 2001;32:227–241. doi: 10.1051/vetres:2001121. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Modeled scenarios of treatment of cattle with ceftiofur, and experimental data considered for comparison.
(DOCM)