Abstract
The Cancer Genome Atlas (TCGA) project has generated gene expression data that divides glioblastoma (GBM) into four transcriptional classes: proneural, neural, classical, and mesenchymal. Because transcriptional class is only partially explained by underlying genomic alterations, we hypothesize that the tumor microenvironment may also have an impact. In this study, we focused on necrosis and angiogenesis because their presence is both prognostically and biologically significant. These features were quantified in digitized histological images of TCGA GBM frozen section slides that were immediately adjacent to samples used for molecular analysis. Correlating these features with transcriptional data, we found that the mesenchymal transcriptional class was significantly enriched with GBM samples that contained a high degree of necrosis. Furthermore, among 2422 genes that correlated with the degree of necrosis in GBMs, transcription factors known to drive the mesenchymal expression class were most closely related, including C/EBP-β, C/EBP-δ, STAT3, FOSL2, bHLHE40, and RUNX1. Non-mesenchymal GBMs in the TCGA data set were found to become more transcriptionally similar to the mesenchymal class with increasing levels of necrosis. In addition, high expression levels of the master mesenchymal factors C/EBP-β, C/EBP-δ, and STAT3 were associated with a poor prognosis. Strong, specific expression of C/EBP-β and C/EBP-δ by hypoxic, perinecrotic cells in GBM likely account for their tight association with necrosis and may be related to their poor prognosis.
See related Commentary on page 1768
Glioblastoma (GBM) (World Health Organization, grade IV) is the most common and highest grade astrocytoma.1,2 Currently incurable, it has a mean survival that only slightly exceeds 1 year following standard surgical and adjuvant therapies.3 Analyses of large scale gene expression and genomic datasets have indicated that this disease represents multiple molecular subclasses, raising the possibility that future therapies could be directed at underlying class-specific mechanisms. Phillips et al4 and Verhaak et al5 have each shown that unsupervised clustering of GBM gene expression profiles results in three or four distinct transcriptional classes.
Epigenetic changes and genetic alterations, including mutations, amplifications, and deletions of established tumor suppressors and oncogenes, account for at least some transcriptional class identity of GBM. For example, among The Cancer Genome Atlas (TCGA) tumors, which contain proneural, neural, classical, and mesenchymal transcriptional classes, IDH1 mutations and the CpG island methylator phenotype (G-CIMP+) are seen almost exclusively in the proneural transcriptional class, whereas nearly all tumors with NF1 mutations or deletions are within the mesenchymal class.5–7 However, some of the best characterized genomic alterations in GBM, including TP53 and PTEN mutation, EGFR and PDGFR amplification and CDKN2A deletion are noted in multiple transcriptional classes, indicating that cell-intrinsic genetic defects only partially explain the class-specific gene expression patterns.
A recent analysis of GBM expression classes used a novel algorithm to reconstruct transcriptional interactions and uncovered a small set of transcription factors that regulate the transition to the mesenchymal class, including C/EBP-β, C/EBP-δ, STAT3, FOSL2, bHLHE40, and RUNX1. Among these, C/EBP-β, C/EBP-δ, and STAT3 were found to be master transcriptional regulators, controlling the expression of other key regulators, and accounting for the majority of downstream signaling events and the mesenchymal gene signature.8 Underlying genetic alterations or pathophysiological triggers of these master transcriptional regulators were not uncovered. It remains possible that elements of the tumor microenvironment, including tumor hypoxia, necrosis, angiogenesis, or inflammatory cell infiltrates, could strongly impact both transcriptional regulators and gene expression class. Microenvironmental contributions to expression class, as well as the tissue sampling considerations that are intimately related, will need to be carefully considered as molecular profiles are used to direct therapies.
To address these issues, we performed an integrated morphological and molecular analysis of microenvironmental factors as they relate to GBM transcriptional class. We analyzed gene expression and genetic correlates of angiogenesis and necrosis in GBM using molecular data and the digitized images from corresponding frozen sections used for quality assurance by the TCGA. We found that the mesenchymal class of GBM was enriched with samples displaying a high degree of necrosis, and that the expression of transcriptional regulators of the mesenchymal transition, C/EBP-β, C/EBP-δ, STAT3, FOSL2, bHLHE40, and RUNX1, were tightly correlated with the extent of necrosis. Nonmesenchymal GBMs became more transcriptionally similar to the mesenchymal class with increasing levels of necrosis. Using human GBM tissue sections, we demonstrated that C/EBP-β and CEBP-δ were specifically expressed by hypoxic, peri-necrotic pseudopalisading cells, accounting for the association of these factors with necrosis. Our finding that the high expression of C/EBP-β, C/EBP-δ, and STAT3 portends a poor prognosis suggests that these key signaling nodes may hold potential for targeted therapies.
Materials and Methods
Digitized Images used for Morphological Analysis
A complete description of samples, and related imaging, molecular, and pathology data are provided in Supplemental Tables 1–6 (available at http://ajp.amjpathol.org). Whole-slide digitized images of GBM frozen sections were obtained from the TCGA portal (http://tcga-data.nci.nih.gov/tcga/tcgaHome2.jsp; last accessed June 6, 2011). Frozen section slides were scanned and digitized at 20× resolution on an Aperio scanner at the TCGA Biospecimen Core Resource located at the International Genomics Consortium (Intgen, Phoenix, AZ). The frozen section slides were those reviewed by Intgen contracted neuropathologists for quality assurance to ensure that the tissue between the top section and bottom section were adequate for molecular studies. Importantly, the tissue used for molecular analysis by TCGA was immediately adjacent to the tissue used to create slides for this investigation. We used these digitized images as our primary source of data and annotated necrosis on 177 slides from 99 samples corresponding to 91 patients. Angiogenesis was annotated on 168 slides from 95 samples corresponding to 88 patients. As a secondary source of data on necrosis in GBM samples, we used semi-quantitative annotations of 293 GBMs that were recorded by TCGA neuropathologists after reviewing frozen section slides for quality assurance. For these images, the percent necrosis was recorded as a visual estimate of total tissue area involved. Angiogenesis was recorded only as “present” or “absent” and was not useful in our analysis. There were 70 samples with overlap between the two sources of data.
Quantification of Percent Necrosis and Angiogenesis in GBM Samples
Regions of necrosis and angiogenesis were manually outlined using ImageScope software (Aperio, Vista CA) by two pathologists working together and reaching a consensus on the features to include (CA, DJB).9 The boundaries of tissue sections were outlined to calculate total tissue area. Duplicate adjacent sections within the same slide were not analyzed. Regions identified as either necrosis or angiogenesis were exhaustively outlined within the tissue boundaries (Figure 1). Angiogenic regions were identified as those vascular regions departing from normal, and exhibiting characteristics of cellular hypertrophy, cellular hyperplasia, or microvascular proliferation.1,10,11 In addition to the endothelial compartment, the perivascular cells, including pericytes, fibroblasts, and any inflammatory infiltrates, were included within marked angiogenic regions. Luminal areas were automatically subtracted from angiogenic regions using computer-based color segmentation. The extent of necrosis and the extent of angiogenesis were calculated as a percentage of whole tissue by taking the ratios of necrosis or angiogenesis surface areas to the total tissue section area. Areas were summed over multiple slides before ratio calculation to determine per-sample and per-patient percentages.
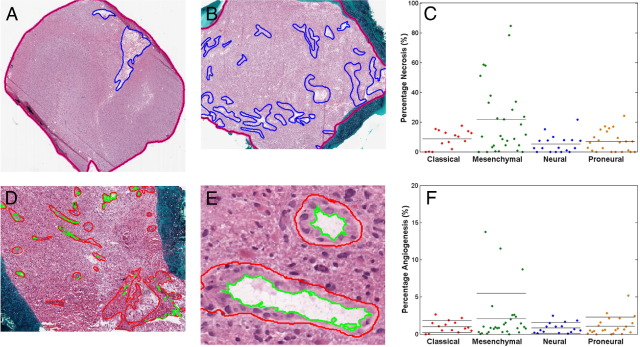
Figure 1.
Representative examples of The Cancer Genome Atlas (TCGA) glioblastoma (GBM) frozen sections marked up for (A, B) necrosis and (D, E) angiogenesis and the distribution of cases within TCGA gene expression classes based on their percent necrosis (C) and percent angiogenic vessels (F). The blue line surrounds regions of necrosis in (A, B) and the pink line surrounds the entire tissue and represents the total tissue area. The red line surrounds angiogenic vessels in (D, E) and the green line represents the lumen, which is subtracted from the vessel area. Each dot in (C, F) represents a GBM sample. Solid horizontal lines within each category represent the mean value. The mean value of necrosis was larger in the mesenchymal class than in the other three classes (one-way analysis of variance, P = 8.7e-4). There was no statistically significant difference in mean angiogenic areas among expression classes.
Correlation of Necrosis and Angiogenesis with GBM Transcriptional Class, Copy Number Alterations, Gene Expression, and Methylation Status
Transcriptional class labels for TCGA patients were obtained from the TCGA Advanced Working Group. This labeling extends the original set of samples labeled by Verhaak et al5 using Affymetrix HT_HG-U133A data to classify previously unlabeled TCGA samples given centroids derived from the original labeled set. Patient-reduced percentages for necrosis and angiogenesis were tested for association with transcriptional class with one-way analysis of variance tests. Copy number data were obtained from the GBM Pathway analysis at the Memorial Sloan Kettering Cancer Genomics Portal (http://www.cbioportal.org/public-portal; last accessed June 6, 2011). Level 3 sequence data obtained from the TCGA portal was filtered to remove silent mutations. Necrosis was compared between amplified/normal, deleted/normal, and mutant/wild-type samples using two-way t-tests. Level 2 robust multichip average normalized gene expression data from the Affymetrix U133A platform was averaged over samples with multiple arrays to create patient-reduced expression profiles. Probes with an unlogged expression range of <20 and a fold change of <1.5 were removed. Percentage necrosis and angiogenesis were correlated with expression profiles using the Cox proportional hazards within the significance analysis of microarray procedure.12 Gene-centric profiles of C/EBP-β, C/EBP-δ, and STAT3 were obtained by averaging over the corresponding probes. Methylation phenotype G-CIMP status was calculated, as previously described.7,13
Pathway Analysis
Ingenuity Pathway Analysis (IPA) (Ingenuity Systems, Redwood City, CA) was applied to gene lists generated by significance analysis of microarray analysis of necrosis and angiogenesis, as previously described.13 Both direct and indirect relationships were included. Data sources were restricted to human species and cell lines. Network significance was assigned by hypergeometric scoring.
Correlation of Macrophages with GBM Transcriptional Class and Gene Expression
We obtained TCGA neuropathologists ratings of pathological categories for 112 GBM samples with either gene expression or transcriptional class assignments (http://tcga-data.nci.nih.gov/tcga/tcgaHome2.jsp; last accessed June 6, 2011). Association between transcriptional class and the presence of macrophages (0, 1+, or 2+) was performed using Fisher's test. Expression differences for C/EBP-β, C/EBP-δ, and STAT3 were examined using two-way t-tests with unequal variance.
Mesenchymal Signature Analysis
The mesenchymal signature was calculated using the classification to nearest centroid procedure.14 A set of 250 probes that distinguish mesenchymal samples were selected by ranking t-statistics computed by comparing sample-reduced expression profiles of mesenchymal and nonmesenchymal samples. The mesenchymal signature was calculated as the average expression of these 250 probes of all mesenchymal sample profiles. Transcriptional distance of each nonmesenchymal sample to the mesenchymal signature was calculated as the Euclidean distance normalized by the SD of each probe. Correlation between transcriptional distance and percent necrosis were calculated using Spearman rank correlation.
Immunohistochemistry
Ten archived surgically resected GBM specimens were retrieved from Emory University Hospital Department of Pathology. These GBMs had not been previously treated with radiation or chemotherapy and blocks contained regions of high-grade glioma with foci of necrosis, infiltrating glioma, and adjacent non-neoplastic brain. GBM tissues were fixed in 10% buffered formalin, routinely processed, and paraffin-embedded. Immunohistochemical (IHC) studies were performed on 6-μm sections as described.15,16 Sections were deparaffinized and subjected to heat-induced epitope retrieval by steaming for 15 minutes. Slides were then incubated with antibodies directed toward Stat3 (rabbit polyclonal, 1:100; Cell Signaling, Beverly, MA) and phospho-Stat3 (rabbit polyclonal, 1:100; Abcam, Cambridge, MA), C/EBP-β (monoclonal, 1:100; Abcam), C/EBP-δ (polyclonal, 1:100, Novus Biologicals, Littleton, CO), CD68 (monoclonal, 1:100; Biocare Medical, Concord, CA), and CD163 (polyclonal, 1:100; Novus Biologicals). Antibodies were detected using the avidin-biotin-peroxidase complex method using 3,3′-diaminobenzidine as the chromogen. Normal sera served as the negative control. Sections were counterstained with hematoxylin.
Glioblastoma Cell Lines and Culture Conditions
Human GBM cell line U87MG cell culture conditions have been previously described.15,16 Cells used in experiments were grown to 80% confluence in 100-mm culture dishes, placed in serum free media in conditions of 21% O2 (normoxia) or 1% O2 (hypoxia)17,18. For experiments in 1% O2, culture dishes were placed in incubators that are dedicated to hypoxia (94% N2, 5% CO2, and 1% O2 at 37°C). Exposure to hypoxia lasted 24 hours.
Western Blot Analysis
Immunoblots were performed on proteins from cell lysates of the indicated cell lines. The NE-PER Nuclear and Cytoplasmic Extraction Reagents (Pierce Biotechnology, Rockford, IL) was used for separation of nuclear and cytoplasmic protein fractions. Protein concentrations were determined by a Bradford assay (Bio-Rad Laboratories, Hercules, CA). Equal amounts of protein (30 μg) were resolved on a 10% SDS-PAGE and transferred to nitrocellulose membranes. Blots were incubated in blocking solution (PBS containing 0.02% Tween-20 and 5% nonfat milk) and incubated overnight at 4°C with antibodies specific for Stat3 (monoclonal, 1:2000, Cell Signaling) and phospho-Stat3 (monclonal, 1:1000; Cell Signaling), C/EBP-β (polycolonal, 1:1000, Cell Signaling), C/EBP-δ (polyclonal, 1:1000, Novus Biologicals), and HIF-1α (monoclonal, 1:1000, BD Transduction Lab, Research Triangle Park, NC). Blots were washed and incubated with horseradish peroxidase conjugated to goat anti-mouse or goat anti-rabbit antibodies (1:2000, Bio-Rad, Hercules, CA) for 1 hour at room temperature and developed by enhanced chemiluminescence reagents (Pierce Biotechnology). Histone H1 (monoclonal, 1:4000; Santa Cruz Biotechnology, Santa Cruz, CA) was used as loading control for the nuclear compartment.
Survival Analysis
Association between C/EBP-β, C/EBP-δ, and STAT3 expression and survival were examined using the log rank test to compare the upper and lower quartiles of gene-centric profiles. Survival was taken as “days to death” for uncensored patients and “days to last follow-up” for right-censored patients.
Reporting
All qualitative analysis were repeated three times. Quantitative data are expressed as mean ± SEM. Significance was defined as P < 0.05.
Results
Extent of Necrosis and Angiogenesis Vary within Frozen TCGA GBM Samples
We investigated potential correlation of microenvironmental features in GBM with gene expression and genomic patterns, and we primarily focused on the two dominant pathological findings of GBM: necrosis and angiogenesis.11,19,20 Two sources of data were used as measures of the degree of necrosis and angiogenesis in frozen section slides of TCGA samples analyzed for quality assurance before molecular analysis. As a primary source, we downloaded digitized images from all 177 available frozen section slides, corresponding to 99 samples and 91 patients, and marked up images for degree of necrosis and angiogenesis using a computer-human interface (Figure 1). The percentage necrosis varied from 0 to 84.7% with a mean of 13.6 ± 1.9%. The degree of angiogenesis varied from 0 to 13.7% with a mean of 1.4 ± 0.2%. There was a weak positive relationship between the percent necrosis and angiogenesis within each sample (Spearman's rho = 0.29).
As a secondary source, we used annotations of 293 GBMs, which were recorded by TCGA neuropathologists who reviewed frozen section slides for quality assurance. Percent necrosis was recorded as a visual estimate of total tissue area involved. Angiogenesis was recorded only as “present” or “absent,” and was not useful in our analysis. Percent necrosis in these cases varied from 0 to 87.5% with a mean of 14.5 ± 1.6%. Among 70 samples with overlap between the two sources of data, there was strong agreement on percent necrosis (Spearman's rho = 0.71).
Mesenchymal GBMs Are Enriched with Samples with High Necrosis
To determine whether there was a correlation between transcriptional class and extent of necrosis or angiogenesis, we examined each class for the distributions of these features. In the set of 85 GBMs manually marked for necrosis with associated molecular data, we found that the mesenchymal class was significantly enriched with samples that had a high degree of necrosis (31% of cases with >25% necrosis) compared to the other three tumor subtypes (0 cases with >25% necrosis) (Figure 1C). The mesenchymal class also had a higher mean percent necrosis (21.5 ± 4.5%) than the other three classes combined (6.9 ± 0.9%; one-way analysis of variance; P = 8.7 × 10−4). Extent of angiogenesis was not significantly associated with GBM subtype, yet all extreme outliers (angiogenesis >8%) were mesenchymal (Figure 1F). We validated these findings using the set of GBMs, which were visually estimated for necrosis by TCGA neuropathologists (291 samples; not shown). In this set, the mesenchymal class was also significantly enriched in samples with high necrosis compared to other classes (one-way analysis of variance; P = 6.9 × 10−5).
Because genetic alterations may play a role in the gene expression class and could be related to the extent of necrosis and angiogenesis, we determined if the level of necrosis was related to TP53, PTEN, EGFR, NF1, or IDH1 mutations, EGFR or PDGFRA amplification, CDKN2A deletion or G-CIMP+ status. In analysis of 189 samples with DNA methylation data, we found that low necrosis was significantly associated with G-CIMP+ status (two-way t-test; P = 0.0064), but not with the proneural class as a whole. Analysis of a more limited set of 114 cases with mutation data did not reveal a significant association among IDH1 mutants and wild-type cases. No other significant correlations were noted between angiogenesis and necrosis, and frequent genetic alterations in GBM (TP53, PTEN, EGFR, or NF1 mutations, EGFR or PDGFRA amplification, CDKN2A deletion).
Next we compared extremely necrotic mesenchymal samples to GBMs from other transcriptional classes to determine whether they had a distinctive spectrum of genomic alterations.6 Because those mesechnymal GBMs with >40% necrosis represented statistical outliers (Figure 1C), we compared this subset to all nonmesenchymal GBMs. These outlying necrotic mesenchymal GBMs did not harbor a particular set of genetic alterations when compared to samples in the other three transcriptional classes (Table 1). Thus, there was not a defining set of genetic alterations associated with the highly necrotic mesenchymal cases.
Table 1.
Mutation and Copy Number Alterations in Highly Necrotic GBMs Compared to Low Necrosis, Nonmesenchymal GBMs
| Genes mutated | High necrosis, MS; n (%) | Low necrosis, non-MS; n (%) |
|---|---|---|
| TP53 | 1/4 (25) | 15/70 (21.4) |
| PTEN | 0/4 (0) | 14/70 (20.0) |
| NF1 | 1/4 (25) | 3/70 (4.3) |
| EGFR | 1/4 (25) | 14/70 (20) |
| ERBB2 | 0/4 (0) | 5/70 (7.1) |
| RB1 | 0/4 (0) | 4/70 (5.7) |
| PIK3R1 | 0/4 (0) | 10/70 (14.3) |
| PIK3CA | 1/4 (25) | 3/70 (4.3) |
| IDH1 | 0/4 (0) | 7/70 (10.0) |
| Copy number alterations | n (%) | n (%) |
| EFGR amplification | 3/4 (75) | 50/66 (75.8) |
| PDGFRA amplification | 1/4 (25) | 12/66 (18.2) |
| CDKN2A deletion | 2/4 (50) | 2/66 (63.6) |
Frequency of mutations and copy number variation from the TP53, RB, and receptor tyrosine kinase pathways in MS samples with more than 40% necrosis and samples from all other transcriptional classes.
GBMs, glioblastomas; MS, mesenchymal; RB, retinoblastoma.
Extent of Necrosis Is Correlated with the Expression of Master Regulators of the Mesenchymal Transition and Specific Canonical Signaling Networks
Next we used significance analysis of microarrays Cox proportional hazards modeling of gene expression from the HT-HGU133A platform to identify mRNAs significantly associated with the extent of necrosis and/or angiogenesis using a false discovery rate cutoff of <5%. Significance analysis of microarray analysis of necrosis extent identified 2422 significantly correlated genes. Gene lists are provided in Supplemental Tables S7 and S8 (available at http://ajp.amjpathol.org). Among the positively correlated genes were factors identified as master regulators of the mesenchymal tumor class, including C/EBP-β (ranked 4), C/EBP-δ (ranked 60), STAT3 (ranked 213), FOSL2 (ranked 10), bHLHE40 (ranked 221), and RUNX1 (ranked 366) (Table 2).8 The master regulator ZNF238 (ranked 363), identified as a repressor, was listed among the negatively correlated genes (see Supplemental Table S7 at http://ajp.amjpathol.org).
Table 2.
Master Transcriptional Regulators of the Mesenchymal Transition are Strongly Associated with Extent of Necrosis in TCGA GBMs
| Gene symbol | q value (corrected P value) | Rank among 2422 genes |
|---|---|---|
| C/EBP-β | <0.0017 | 4 |
| FOSL2 | <0.0017 | 10 |
| C/EBP-δ | <0.0017 | 60 |
| STAT3 | <0.0017 | 213 |
| bHLHE40 | <0.0017 | 221 |
| ZNF238 | 0.0091 | 363 |
| RUNX1 | 0.0017 | 366 |
GBMs, glioblastomas; TCGA, The Cancer Genome Atlas.
We further analyzed gene expression and the extent of necrosis specifically within the mesenchymal expression class (28 cases), splitting these into those with <15% necrosis (16 cases) and those with >15% (12 cases). Analysis of the 2422 genes correlated with necrosis identified 49 probes significantly upregulated in the high necrosis group, including C/EBP-β (ranked 3) and matrix metalloproteinase-19.
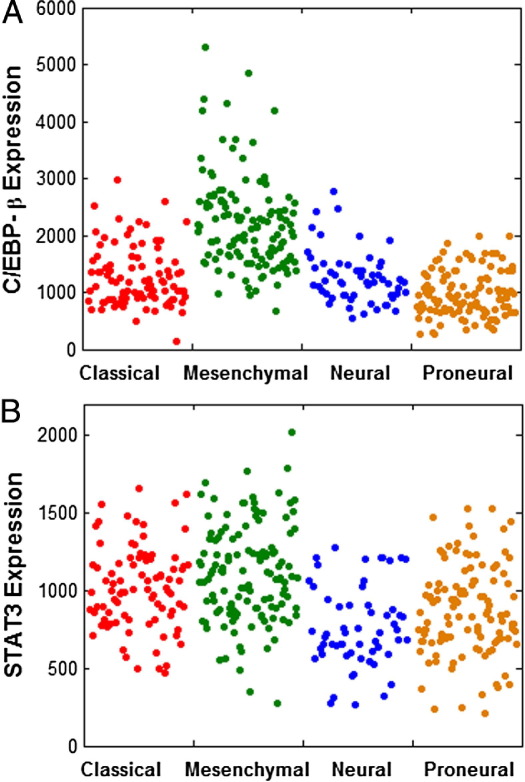
Because the master regulators of the mesenchymal transcriptional class were defined using data from Phillips et al4, we determined whether these transcription factors were also enriched within the mesenchymal class defined by TCGA data.5 We found that C/EBP-β, C/EBP-δ, STAT3, FOSL2, RUNX1, and bHLHE40 were all significantly upregulated when comparing mesenchymal tumors to other classes (Figure 2). In order of enrichment, C/EBP-β was most highly enriched, followed by FOSL2, RUNX1, bHLHE40, C/EBP-δ, and STAT3 (Table 3). The repressor ZNF238 was also significantly down-regulated in the mesenchymal class. However, in comparison of transcription factor expression among individual expression classes, we noted that C/EBP-δ, STAT3, and ZNF238 showed similar expression in classical and mesenchymal tumors (Table 3).
Figure 2.
Distribution of CEBP-β and STAT3 gene expression levels within The Cancer Genome Atlas gene expression classes. Each dot represents one glioblastoma (GBM) sample. Compared to the neural, proneural, and classical gene expression classes combined, the mesenchymal class showed enrichment for tumors with high CEBP-β (A) and STAT3 (B). All six activators and the repressor (ZNF238) are significantly differentially expressed when comparing mesenchymal to non-mesenchymal GBMs (two-way t-test, unequal variances; see Table 3).
Table 3.
Transcriptional Regulators of the MS Transition are Differentially Expressed in MS and Non-MS GBM
| P value | C/EBP-β | C/EBP-δ | STAT3 | FOSL2 | bHLHE40 | RUNX1 | ZNF238 |
|---|---|---|---|---|---|---|---|
| MS vs non-MS | 2.5 × 10−29 | 4.12 × 10−16 | 1.33 × 10−9 | 5.27 × 10−22 | 6.67 × 10−16 | 1.89 × 10−18 | 5.85 × 10−15 |
| MS vs CL | 8.27 × 10−21 | 0.0115 | 0.0238 | 8.49 × 10−13 | 3.89 × 10−6 | 9.80 × 10−9 | 0.0825 |
The differential expression of C/EBP-δ, STAT3, and ZNF238 was less significant when comparing mesenchymal (MS) and classical (CL) tumors.
GBM, glioblastoma.
Genes that were significantly associated with the extent of necrosis were submitted to IPA to identify pathways enriched in gene sets associated with necrosis (Figure 3). IPA analysis of genes identified significantly enriched canonical pathways associated with acute response, migration, invasion, thrombosis, cell survival, and hypoxia including Rac (P = × 10−6), Rho (P = 1.1 × 10−6), PI3K/AKT (P = 4.1 × 10−6), complement system (P = 1.3 × 10−4), NFκB (P = 1.8 × 10−4), IL-6 (P = 4.3 × 10−5), ERK/MAPK (P = 4.9 × 10−4), JAK/STAT (P = 0.0083), and hypoxia signaling (P = 0.015). Significance analysis of microarray for angiogenesis identified only 18 significantly correlated genes, none of which were mesenchymal master regulators (see Supplemental Table S8 at http://ajp.amjpathol.org). The intersection of the necrosis and angiogenesis correlated gene sets contained 8 significantly correlated genes, including the positively correlated gene FN1. We attribute the lack of significant findings on angiogenesis-related genes to the limitations of the methods used for measurement. Given that the surface area of angiogenesis represents only a small percentage of the total tissue (<4% for the large majority of samples), meaningful correlations between angiogenesis and gene expression were not observed, most likely due to low signal-to-noise ratios in the microarray experiments and imaging quantifications.
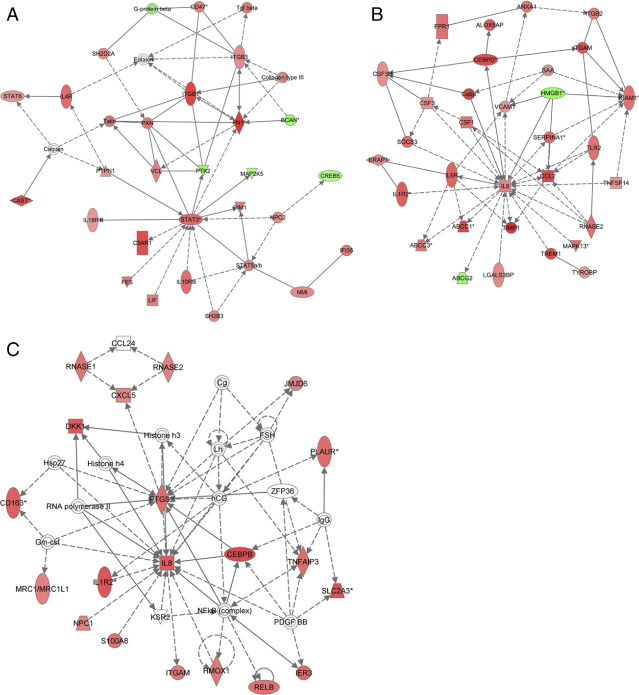
Figure 3.
Ingenuity pathway analysis (IPA) identified critical expression networks containing genes that are significantly correlated with the extent of necrosis in The Cancer Genome Atlas (TCGA) glioblastoma (GBM) samples from all subclasses (A and B). The STAT3 network was among the most closely associated with necrosis (A). The C/EBP gene network was also highly correlated with the extent of necrosis and demonstrates the central importance of IL-6. (B). Gene networks that were differentially expressed between mesenchymal GBMs with high (>15%) and low extent of necrosis (≤15%) included those that were centered around CEBP-β and were strongly associated with upregulated IL-8 signaling (C).
We also performed a pathway analysis on genes differentially expressed in high and low necrosis mesenchymal GBMs. Within this set, IPA identified significantly enriched canonical pathways associated with inflammation, cell death, and hypoxia, including IL-6 (P = 1.4 × 10−4), IL-8 (P = 0.0013), and hypoxia-inducible factor-1α (P = 0.0029) (Figure 3).21,22
High Co-Expression of C/EBP-β, C/EBP-δ and STAT3 Is Associated with Poor Prognosis
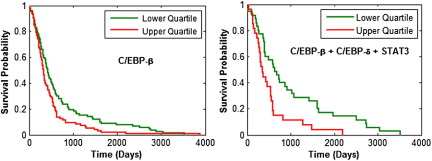
To determine whether expression of master regulators of the mesenchymal transition was correlated with clinical outcome, we used TCGA data to compare the survival of patients with high expression (upper quartile) to those with low expression (lower quartile) of C/EBP-β, C/EBP-δ, and STAT3 (Figure 4). We examined the impact of individual gene expression, as well as their co-expression. GBMs with high expression of C/EBP-β were associated with a significantly shorter survival (50% survival, 11 months) than those with low expression (50% survival, 14 months) (log rank test, P = 0.0071). High expression of C/EBP-δ showed a trend toward shorter survival (log rank test, P = 0.0564), whereas high expression of STAT3 was significantly associated with a shorter survival (P = 5.73 × 10−4) (not shown). The co-expression of high C/EBP-β, C/EBP-δ, and STAT3 was associated with a substantially shorter survival (50% survival, 12 months) compared to those that showed all low expression (50% survival, 21 months) (log rank test, P = 0.0021).
Figure 4.
Glioblastomas (GBMs) with high expression of C/EBP-β (upper quartile) were associated with a significantly shorter survival (50% survival, 11 months) than those with low expression (50% survival, 14 months) (log rank test P = 0.0071), and high co-expression of C/EBP-β, C/EBP-δ, and STAT3 was associated with a shorter survival (50% survival, 12 months) than those that had low expression of all three (50% survival, 21 months) (log rank test P = 0.0021).
Extent of Necrosis Is Correlated with Proximity to Mesenchymal Transcriptional Signature
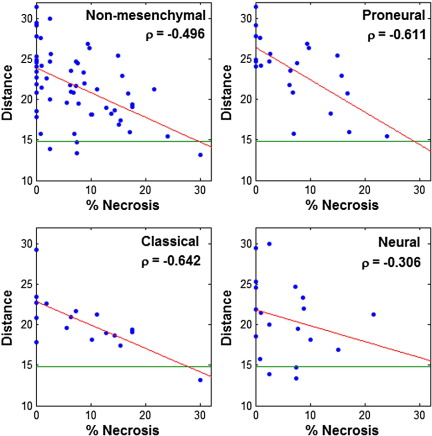
Molecular classes identified by Verhaak et al5 are defined purely by transcription and are characterized by the formation of clusters in transcriptional space. To probe the relationship between necrosis on transcriptional class, we compared the extent of necrosis with distance to a characteristic mesenchymal gene expression signature in transcriptional space.14 We first identified a set of 250 genes that distinguish the mesenchymal class using a t-statistic to rank and select probes with the greatest differential expression between mesenchymal and nonmesenchymal samples. Expression of this gene set was averaged over all mesenchymal samples to calculate a representative mesenchymal signature. Then we calculated the distance in terms of gene expression for each nonmesenchymal sample to the mesenchymal signature as a measure of similarity to the mesenchymal class. Among nonmesenchymal GBMs, we found that the distance to the mesenchymal signature was inversely related to the extent of necrosis (Figure 5). Using the mark-up data for necrosis, we found an overall correlation of −0.50 among all nonmesenchymal samples, −0.61 in proneural samples, −0.64 in classical samples, and −0.31 in neural samples. This trend was verified in the visually estimated necrosis dataset, which uncovered correlations of −0.27 among nonmesenchymal samples, −0.24 in proneural samples, −0.31 in classical samples, and −0.32 in neural samples. Using the mean distance of mesenchymal samples to the mesenchymal signature as a baseline (14.8 standard units), we found that many necrosis-rich nonmesenchymal samples are remarkably close to the mesenchymal class. Thus, the gene expression signatures of nonmesenchymal GBMs became more similar to the mesenchymal signature with increasing levels of necrosis.
Figure 5.
Influence of necrosis on transcriptional distance to mesenchymal class signature. Euclidean distance to the mesenchymal class signature was computed for all nonmesenchymal samples, and the Spearman correlations between transcriptional distance and extent of necrosis were computed. The median distance between mesenchymal samples and the mesenchymal signature (indicated in green) is 14.8. There is an inverse relation between the extent of necrosis and the distance to the mesenchymal gene signature.
We also found that 94% of the 250 characteristic mesenchymal probes were present in the necrosis-correlated gene set (average rank, 544), suggesting that the with the most distinguishing mesenchymal class genes are also significantly correlated with necrosis. Overall, these data suggest that the extent of necrosis strongly impacts the expression of genes that define the mesenchymal class.
Macrophage Infiltrates Are Not Associated with Mesenchymal Class or Master Regulators
We explored potential microenvironmental explanations for the association between necrosis and master transcriptional regulators (C/EBP-β, C/EBP-δ, and STAT3).8 We considered whether macrophage infiltrates might be responsible, because C/EBP-β, C/EBP-δ, and STAT3 are expressed by inflammatory cells; IPA identified a pro-inflammatory signature associated with necrosis (NFκB, IL-6); and macrophage infiltrates might increase with the extent of tissue damage.23–25 Using the set of 112 TCGA GBMs reviewed by a panel of TCGA neuropathologists and annotated for macrophage infiltrate as 0 (90 cases), 1+ (21 cases), or 2+ (1 case) for molecular correlates, we noted an enrichment of GBMs with macrophage infiltrates (1+ or 2+) within the proneural transcriptional class (Fisher's exact P = 0.026), but not in the mesenchymal class. When we compared the gene expression of C/EBP-β, C/EBP-δ, STAT3, FOSL2, bHLHE40, and RUNX1 in those cases with macrophage infiltrates (1+ or 2+) to those without, we did not find any statistically significant differences.
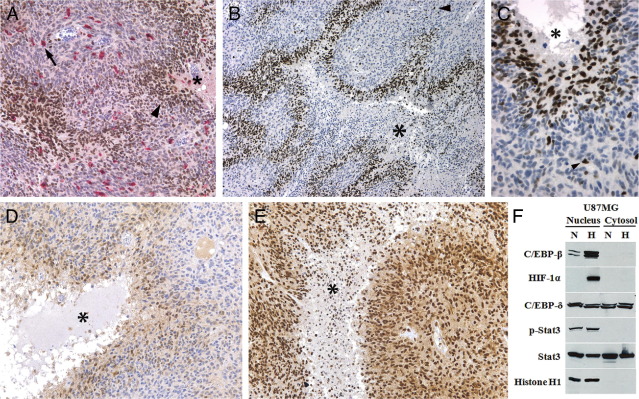
To explore the pattern and degree of macrophage infiltrate in GBMs, especially related to necrosis and to the expression of C/EBP-β, C/EBP-δ, and STAT3, we performed IHC on 10 cases selected to contain infiltrating glioma, high grade glioma with necrosis and angiogenesis, and adjacent non-neoplastic brain. Double immunostaining was used to estimate the percentage of CD68+ or CD163+ macrophages within the tumor sample and to determine the strength of transcription factor expression in this cell population (Figure 6). The percentage of macrophages within GBM samples varied from 5% to 24% (mean, 14.5% ± 2.2%), whereas the large majority of cells were neoplastic (>70% in all cases). Macrophages were seen clustered around blood vessels, within brain tumor stroma individually, and in small collections within necrotic foci (Figure 6). Pseudopalisading cells directly surrounding necrosis were not enriched with macrophages, consistent with our prior studies.26 The extent of necrosis within the tissue sections of these GBMs varied from 5% to 59% (mean, 24.0% ± 5.2%). We found that the expression of STAT3 and pSTAT3 was consistently strong in macrophages (2+), whereas the expression of both C/EBP-δ and C/EBP-β was moderate (1+). There was a weak positive correlation between extent of necrosis and percent macrophages within these 10 GBM samples (Spearman's rho = 0.33). Thus, C/EBP-β, C/EBP-δ, and STAT3 were expressed by macrophages, but these represented only a minor percentage of total cells within GBM samples and exhibiting only a modest relationship with the extent of necrosis.
Figure 6.
Double immunohistochemistry for macrophages (CD163, red) and C/EBP-β (brown) in human glioblastoma (GBM) samples demonstrates that macrophages represent a minority of the cells in GBM and are scattered throughout the tumor stroma (arrow) but do not accumulate around necrosis (A, asterisk). C/EBP-β (A–C) and C/EBP-δ (D) expression was increased in perinecrotic pseudopalisading cells directly surrounding necrosis (asterisk) of GBM, whereas expression of pSTAT3 (E) and STAT 3 was uniform in tumor cells and did not show microregional variation. Expression of CEBP-β was restricted to the nucleus of tumor cells within 4 to 6 cell layers from necrosis (A, arrowhead). Only rare cells away from necrosis showed expression (B, C, arrowhead). Hypoxia causes increased expression of C/EBP-β in GBM cells (F). U87MG cells grown for 24 hours under hypoxia (H) (1% O2) show increased C/EBP-β protein in the nuclear compartment by Western blot, with low protein expression under normoxia (N) (21% O2). Protein expression of C/EBP-δ and STAT3 was seen in both the nuclear and cytoplasmic compartments under normoxia and hypoxia. pSTAT3 was expressed exclusively in the nucleus and did not show any hypoxic upregulation. Increased nuclear HIF-1α expression under hypoxia serves as a positive control for hypoxic conditions, with histone H1 serving as a loading control for the nuclear compartment.
C/EBP-β and C/EBP-δ Are Strongly Upregulated in Hypoxic, Perinecrotic “Pseudopalisading” Cells in GBM
In our IHC study of C/EBP-β, C/EBP-δ, STAT, and pSTAT3, we noticed a consistent pattern of protein expression of C/EBP-β and C/EBP-δ that strongly correlated with the presence and degree of necrosis (Figure 6). C/EBP-β and C/EBP-δ were spatially restricted to the pseudopalisading cells that surround necrosis in GBMs.19 These cells surrounding the necrosis are hypoxic and express angiogenic factors such as vascular endothelial growth factor and IL-8, proteases, and coagulation factors.16,27 C/EBP-β was specifically expressed in the 2 to 5 cell layers directly surrounding necrosis, and it was exclusively nuclear in location. C/EBP-δ was not as strongly upregulated in perinecrotic cells, yet extended a greater distance from necrosis, and displayed nuclear and cytoplasmic staining. In regions of neoplasm between foci of necrosis, protein expression was noted only infrequently (<5% of cells) and weakly (0 to 1+) for both C/EBP-β and C/EBP-δ. There was no significant protein expression in neurons or glia within the adjacent non-neoplastic brain tissue. STAT3 and pSTAT3 did not demonstrate a perinecrotic expression pattern. Rather, these proteins were moderately to strongly expressed by the majority of neoplastic cells (2+ to 3+) and did not show substantial microregional variation. STAT3 was expressed in both the cytoplasm and nucleus, whereas pSTAT3 was predominantly nuclear.
C/EBP-β Is a Hypoxia-Inducible Transcription Factors in Vitro
Given the strong expression of C/EBP-β and C/EBP-δ in hypoxic pseudopalisading cells, we investigated whether these factors were hypoxia-inducible.28 We exposed the human GBM cell line U87MG to either 20% O2 (normoxia) or 1% O2 (hypoxia) for 24 hours and measured the expression of C/EBP-β, C/EBP-δ, STAT3, pSTAT3, and HIF-1α by Western blot. We noted upregulation of C/EBP-β under hypoxic conditions by U87MG (Figure 6). Similar to IHC staining, expression was restricted to the nuclear compartment. C/EBP-δ, STAT3, and pSTAT3 were not induced by hypoxia in this cell line.
Discussion
This study represents the first correlative investigation of TCGA molecular data with tumor microenvironmental features. One of its key strengths is the use of the frozen section slides from TCGA samples, which are immediately adjacent to the tissue used to generate corresponding molecular data. Frozen sections do not allow detailed visualization of cytologic fine structures, although, in the present case they greatly improved the ability to detect molecular correlates of microenvironmental features by minimizing sampling error. Using this approach, we found that the extent of necrosis within TCGA GBM tissue samples was associated with a specific transcriptional class defined by Verhaak et al.5 The mesenchymal transcriptional class was highly enriched with tumor samples that contained greater levels of necrosis than those in the proneural, neural, and classical classes. In contrast, a transcriptional class of GBM associated with low levels of necrosis was not identified.
Genes that showed among the strongest correlations between their expression and the extent of necrosis included transcription factors that were recently identified as regulators of the mesenchymal transition, C/EBP-β, C/EBP-δ, STAT3, FOSL2, bHLH-B2, and RUNX1.8 Using a network analysis algorithm and gene expression data from Phillips et al4, these factors were found to account for 74% of the expressed genes within the mesenchymal gene signature.4,8 Consistent with their critical role in determining the transcriptional signature of the mesenchymal class, our findings using TCGA data demonstrated that C/EBP-β, C/EBP-δ, STAT3, FOSL2, bHLH-B2, and RUNX1 were highly overexpressed in the mesenchymal GBMs as compared to the other three classes.
Explanations for the clustering of GBM samples into specific expression classes are currently incomplete, yet there are likely multiple contributing factors. It is clear that epigenetic changes and genetic alterations, including mutations and copy number alterations, have an impact on the transcriptional signatures. For example, GBMs with IDH1 mutations and/or the G-CIMP+ methylator phenotype are nearly all proneural, whereas those with NF1 deletions or mutations are enriched in the mesenchymal class.5,7 Those tumors with genetic alterations in TP53, EGFR, PDGFRA, PTEN, and CDK2A, however, are found in multiple transcriptional classes. We did not find an association between these common genetic alterations and the extent of necrosis in GBM that might explain the relation of high necrosis and mesenchymal class. The mesenchymal GBMs with the highest levels of necrosis had a similar spectrum of alterations in these genes as those with low necrosis in all transcriptional classes. These findings indicate that similar genetic alterations can be associated with dissimilar transcriptional signatures and suggest that some other biological parameters associated with necrosis (hypoxia, low pH, immune cell infiltrates, and so forth) may impact transcriptional class independently.
Among the transcription factors identified by Carro et al8, as key mediators of the mesenchymal transition, it was demonstrated by in silico and in vitro approaches that C/EBP-β, C/EBP-δ, and STAT3 were master transcriptional regulators and together controlled the others in a hierarchical manner. These findings were compelling, yet they did not demonstrate genetic or pathophysiological triggers or explore microregional distributions that may be important to tumor progression.
Because C/EBP-β, C/EBP-δ, and STAT3 are known for their roles in inflammation, we considered whether their correlation with necrosis in GBM was due to expression by macrophages, which are infrequent compared to neoplastic cells, yet increase in numbers with necrosis.23–25 In our analysis of TCGA GBMs annotated by a panel of neuropathologists, we found no association between the degree of macrophage infiltrate and the mesenchymal transcriptional class. Neither was there a relationship noted between degree of macrophage infiltrate and the expression of C/EBP-β, C/EBP-δ, and STAT3. In our IHC analysis, macrophages showed a moderate level of expression of these transcription factors, yet this cell type accounted for a minor percentage of total cells in GBM specimens, with the vast majority of cells being neoplastic. Thus, the expression of C/EBP-β, C/EBP-δ, and STAT3 by macrophages may contribute to the mesenchymal signature, but most likely in a minor fashion. In addition to macrophages, there may be other types of non-neoplastic cells that could contribute to the mesenchymal signature and are deserving of further investigation.
We also explored whether the necrosis that emerges in GBM could account for the upregulation of mesenchymal transcription factors. Necrosis and its associated severe hypoxia are critical pathophysiological features that arise in the microenvironment as astrocytomas progress to GBM and are strongly associated with aggressive biological behavior.19,22,29 The dense collection of pseudopalisading cells that surround necrotic foci in GBM are hypoxic, express HIF-1α, and secrete pro-angiogenic factors including vascular endothelial growth factor and IL-8, which lead to an angiogenic response that drives rapid tumor growth.19,26,30 We noted that the expression of C/EBP-β was strong and highly specific within these pseudopalisading cells in GBM samples. C/EBP-δ was also seen in peri-necrotic cells, but its upregulation was not as dramatic or specific. Neoplastic cells that were not surrounding necrosis showed only infrequent expression of C/EBP-β and C/EBP-δ. Given this pattern, it would be expected that C/EBP-β and C/EBP-δ gene and protein expression would correlate with the degree of necrosis in GBMs, as noted in our analysis. STAT3 expression did not show a spatial association with necrosis in GBM tissue sections and its correlation with necrosis by gene expression analysis could not be explained morphologically.
Given the correlation between gene expression signatures and degree of necrosis, it could be argued that the transcriptional program gives rise to tumors that are more prone to develop necrosis, or that the development of necrosis causes a change in transcriptional signature.31 We find it more likely that necrosis impacts the transcriptional class. Two of the master transcriptional regulators, C/EBP-β and C/EBP-δ, which sit atop the hierarchy of critical signaling networks in the mesenchymal transition, were found to be specifically expressed by perinecrotic cells in human GBM samples, and C/EBP-β was upregulated by hypoxia in vitro, suggesting that the emergence of necrosis leads to upregulation of these factors. Among the two C/EBP isoforms, C/EBP-β was the most tightly associated with the mesenchymal transcription class and with the extent of necrosis within the mesenchymal GBM samples. C/EBP-β also showed the most specific expression in pseudopalisading cells in GBM samples and was hypoxia-inducible in vitro. Surprisingly, C/EBP-δ was not upregulated by hypoxia in cell culture, which may indicate that its induction in perinecrotic cells is due to other physiological factors, such as high pH or regional cytokines. Combined, these data suggest that C/EBP-β may be most critical for the tight association between necrosis and the mesenchymal gene signature.
Our results imply that GBM transcriptional signatures are strongly impacted by the development of necrosis and could evolve within a duration of time as tumors progress. Nonmesenchymal GBMs were noted to be more transcriptionally similar to the mesenchymal class with increasing levels of necrosis, suggesting that there may be transitions between transcriptional classes that depend on necrosis and potentially other microenvironmental influences. It remains possible that differing transcriptional classes may be found within the same GBM and depend on the degree of necrosis within the tissue sampled. These implications deserve further study and will need to be carefully considered as therapies are directed toward specific molecular profiles in the future.
Beyond their significance to transcriptional class identity, it is clear that these master transcriptional regulators are strong drivers of tumor biology. Within the TCGA dataset, and including all transcriptional classes, GBMs with high expression of C/EBP-β, C/EBP-δ, and STAT3 had much shorter survivals than those with low expression. These data suggest that these master regulators or their immediate downstream signaling nodes may be appropriate therapeutic targets.
Acknowledgments
The authors thank Kathryn Brunn and Xin Jiang for their technical assistance with the experiments and the Research Pathology Laboratory of Winship Cancer Institute, including Dianne Alexis and Jennifer Shelton, for their assistance with histology and immunohistochemistry.
Footnotes
This work was supported by the United States Public Health Service, NIH grants CA149107 (D.J.B.), LM011119 (J.H.S.), CA86335 and CA116804 (E.G.V.M.), CA138292 (Winship Cancer Center Support Grant), the National Cancer Institute's Cancer Bioinformatics Grid (CaBIG), In Silico Research Center of Excellence ContractHHSN261200800001E and NO1-CO-12400 (T.K., E.G.V.M., S.H.S., C.S.M., D.J.B.), the Georgia Research Alliance (J.H.S.), and the Georgia Cancer Coalition (J.H.S., D.J.B.).
CME Disclosure: None of the authors disclosed any relevant financial relationships.
Supplemental material for this article can be found on http://ajp.amjpathol.org or at doi: 10.1016/j.ajpath.2012.01.040.
Supplementary data
References
- 1.Louis D.N., Ohgaki H., Wiestler O.D., Cavenee W.K. 4th ed. Intl. Agency for Research; Lyon: 2007. WHO classification of tumours of the central nervous system. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.CBTRUS . Central Brain Tumor Registry of the United States; Hinsdale, IL: 2010. CBTRUS statistical report: primary brain and central nervous system tumors in the United States in 2004–2006. [Google Scholar]
- 3.Stupp R., Mason W.P., van den Bent M.J., Weller M., Fisher B., Taphoorn M.J., Belanger K., Brandes A.A., Marosi C., Bogdahn U., Curschmann J., Janzer R.C., Ludwin S.K., Gorlia T., Allgeier A., Lacombe D., Cairncross J.G., Eisenhauer E., Mirimanoff R.O. Radiotherapy plus concomitant and adjuvant temozolomide for glioblastoma. N Engl J Med. 2005;352:987–996. doi: 10.1056/NEJMoa043330. [DOI] [PubMed] [Google Scholar]
- 4.Phillips H.S., Kharbanda S., Chen R., Forrest W.F., Soriano R.H., Wu T.D., Misra A., Nigro J.M., Colman H., Soroceanu L., Williams P.M., Modrusan Z., Feuerstein B.G., Aldape K. Molecular subclasses of high-grade glioma predict prognosis, delineate a pattern of disease progression, and resemble stages in neurogenesis. Cancer Cell. 2006;9:157–173. doi: 10.1016/j.ccr.2006.02.019. [DOI] [PubMed] [Google Scholar]
- 5.Verhaak R.G., Hoadley K.A., Purdom E., Wang V., Qi Y., Wilkerson M.D., Miller C.R., Ding L., Golub T., Mesirov J.P., Alexe G., Lawrence M., O'Kelly M., Tamayo P., Weir B.A., Gabriel S., Winckler W., Gupta S., Jakkula L., Feiler H.S., Hodgson J.G., James C.D., Sarkaria J.N., Brennan C., Kahn A., Spellman P.T., Wilson R.K., Speed T.P., Gray J.W., Meyerson M., Getz G., Perou C.M., Hayes D.N. Integrated genomic analysis identifies clinically relevant subtypes of glioblastoma characterized by abnormalities in PDGFRA. IDH1, EGFR, and NF1. Cancer Cell. 2010;17:98–110. doi: 10.1016/j.ccr.2009.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cancer Genome Atlas Network Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature. 2008;455:1061–1068. doi: 10.1038/nature07385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Noushmehr H., Weisenberger D.J., Diefes K., Phillips H.S., Pujara K., Berman B.P., Pan F., Pelloski C.E., Sulman E.P., Bhat K.P., Verhaak R.G., Hoadley K.A., Hayes D.N., Perou C.M., Schmidt H.K., Ding L., Wilson R.K., Van Den Berg D., Shen H., Bengtsson H., Neuvial P., Cope L.M., Buckley J., Herman J.G., Baylin S.B., Laird P.W., Aldape K. Identification of a CpG island methylator phenotype that defines a distinct subgroup of glioma. Cancer Cell. 2010;17:510–522. doi: 10.1016/j.ccr.2010.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Carro M.S., Lim W.K., Alvarez M.J., Bollo R.J., Zhao X., Snyder E.Y., Sulman E.P., Anne S.L., Doetsch F., Colman H., Lasorella A., Aldape K., Califano A., Iavarone A. The transcriptional network for mesenchymal transformation of brain tumours. Nature. 2010;463:318–325. doi: 10.1038/nature08712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cooper L.A., Kong J., Gutman D.A., Wang F., Cholleti S.R., Pan T.C., Widener P.M., Sharma A., Mikkelsen T., Flanders A.E., Rubin D.L., Van Meir E.G., Kurc T.M., Moreno C.S., Brat D.J., Saltz J.H. An integrative approach for in silico glioma research. IEEE Trans Biomed Eng. 2010;57:2617–2621. doi: 10.1109/TBME.2010.2060338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Perry A., Brat D.J. Elsevier; Philadelphia: 2010. Practical surgical pathology: a diagnostic approach. [Google Scholar]
- 11.Brat D.J., Prayson R.A., Ryken T.C., Olson J.J. Diagnosis of malignant glioma: role of neuropathology. J Neurooncol. 2008;89:287–311. doi: 10.1007/s11060-008-9618-1. [DOI] [PubMed] [Google Scholar]
- 12.Tusher V.G., Tibshirani R., Chu G. Significance analysis of microarrays applied to the ionizing radiation response. Proc Natl Acad Sci USA. 2011;98:5116–5121. doi: 10.1073/pnas.091062498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cooper L.A., Gutman D.A., Long Q., Johnson B.A., Cholleti S.R., Kurc T., Saltz J.H., Brat D.J., Moreno C.S. The proneural molecular signature is enriched in oligodendrogliomas and predicts improved survival among diffuse gliomas. PLoS ONE. 2010;5:e12548. doi: 10.1371/journal.pone.0012548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dabney A.R. Classification of microarrays to nearest centroids. Bioinformatics. 2005;21:4148–4154. doi: 10.1093/bioinformatics/bti681. [DOI] [PubMed] [Google Scholar]
- 15.Rong Y., Belozerov V.E., Tucker-Burden C., Chen G., Durden D.L., Olson J.J., Van Meir E.G., Mackman N., Brat D.J. Epidermal growth factor receptor and PTEN modulate tissue factor expression in glioblastoma through JunD/activator protein-1 transcriptional activity. Cancer Res. 2009;69:2540–2549. doi: 10.1158/0008-5472.CAN-08-1547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rong Y., Post D.E., Pieper R.O., Durden D.L., Van Meir E.G., Brat D.J. PTEN and hypoxia regulate tissue factor expression and plasma coagulation by glioblastoma. Cancer Res. 2005;65:1406–1413. doi: 10.1158/0008-5472.CAN-04-3376. [DOI] [PubMed] [Google Scholar]
- 17.Rong Y., Hu F., Huang R., Mackman N., Horowitz J.M., Jensen R.L., Durden D.L., Van Meir E.G., Brat D.J. Early growth response gene-1 regulates hypoxia-induced expression of tissue factor in glioblastoma multiforme through hypoxia-inducible factor-1-independent mechanisms. Cancer Res. 2006;66:7067–7074. doi: 10.1158/0008-5472.CAN-06-0346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Post D.E., Sandberg E.M., Kyle M.M., Devi N.S., Brat D.J., Xu Z., Tighiouart M., Van Meir E.G. Targeted cancer gene therapy using a hypoxia inducible factor dependent oncolytic adenovirus armed with interleukin-4. Cancer Res. 2007;67:6872–6881. doi: 10.1158/0008-5472.CAN-06-3244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rong Y., Durden D.L., Van Meir E.G., Brat D.J. “Pseudopalisading” necrosis in glioblastoma: a familiar morphologic feature that links vascular pathology, hypoxia, and angiogenesis. J Neuropathol Exp Neurol. 2006;65:529–539. doi: 10.1097/00005072-200606000-00001. [DOI] [PubMed] [Google Scholar]
- 20.Kaur B., Tan C., Brat D.J., Post D.E., Van Meir E.G. Genetic and hypoxic regulation of angiogenesis in gliomas. J Neurooncol. 2004;70:229–243. doi: 10.1007/s11060-004-2752-5. [DOI] [PubMed] [Google Scholar]
- 21.Van Meir E., Sawamura Y., Diserens A.C., Hamou M.F., de Tribolet N. Human glioblastoma cells release interleukin 6 in vivo and in vitro. Cancer Res. 1990;50:6683–6688. [PubMed] [Google Scholar]
- 22.Kaur B., Khwaja F.W., Severson E.A., Matheny S.L., Brat D.J., Van Meir E.G. Hypoxia and the hypoxia-inducible-factor pathway in glioma growth and angiogenesis. Neuro Oncol. 2005;7:134–153. doi: 10.1215/S1152851704001115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bradley M.N., Zhou L., Smale S.T. C/EBPbeta regulation in lipopolysaccharide-stimulated macrophages. Mol Cell Biol. 2003;23:4841–4858. doi: 10.1128/MCB.23.14.4841-4858.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Roy S.K., Hu J., Meng Q., Xia Y., Shapiro P.S., Reddy S.P., Platanias L.C., Lindner D.J., Johnson P.F., Pritchard C., Pages G., Pouyssegur J., Kalvakolanu D.V. MEKK1 plays a critical role in activating the transcription factor C/EBP-beta-dependent gene expression in response to IFN-gamma. Proc Natl Acad Sci USA. 2002;99:7945–7950. doi: 10.1073/pnas.122075799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yu H., Pardoll D., Jove R. STATs in cancer inflammation and immunity: a leading role for STAT3. Nat Rev Cancer. 2009;9:798–809. doi: 10.1038/nrc2734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Brat D.J., Castellano-Sanchez A.A., Hunter S.B., Pecot M., Cohen C., Hammond E.H., Devi S.N., Kaur B., Van Meir E.G. Pseudopalisades in glioblastoma are hypoxic, express extracellular matrix proteases, and are formed by an actively migrating cell population. Cancer Res. 2004;64:920–927. doi: 10.1158/0008-5472.can-03-2073. [DOI] [PubMed] [Google Scholar]
- 27.Desbaillets I., Diserens A.C., de Tribolet N., Hamou M.F., Van Meir E.G. Regulation of interleukin-8 expression by reduced oxygen pressure in human glioblastoma. Oncogene. 1999;18:1447–1456. doi: 10.1038/sj.onc.1202424. [DOI] [PubMed] [Google Scholar]
- 28.Brat D.J., Castellano-Sanchez A., Kaur B., Van Meir E.G. Genetic and biologic progression in astrocytomas and their relation to angiogenic dysregulation. Adv Anat Pathol. 2002;9:24–36. doi: 10.1097/00125480-200201000-00004. [DOI] [PubMed] [Google Scholar]
- 29.Spence A.M., Muzi M., Swanson K.R., O'Sullivan F., Rockhill J.K., Rajendran J.G., Adamsen T.C., Link J.M., Swanson P.E., Yagle K.J., Rostomily R.C., Silbergeld D.L., Krohn K.A. Regional hypoxia in glioblastoma multiforme quantified with [18F]fluoromisonidazole positron emission tomography before radiotherapy: correlation with time to progression and survival. Clin Cancer Res. 2008;14:2623–2630. doi: 10.1158/1078-0432.CCR-07-4995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Brat D.J., Bellail A.C., Van Meir E.G. The role of interleukin-8 and its receptors in gliomagenesis and tumoral angiogenesis. Neuro Oncol. 2005;7:122–133. doi: 10.1215/S1152851704001061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Raza S.M., Fuller G.N., Rhee C.H., Huang S., Hess K., Zhang W., Sawaya R. Identification of necrosis associated genes in glioblastoma by cDNA micro-array analysis. Clin Cancer Res. 2004;10:212–221. doi: 10.1158/1078-0432.ccr-0155-3. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.