Abstract
Molecular identification of eighteen Termitomyces species collected from two states, Ondo and Ekiti in Nigeria was carried out using the internal transcribed spacer (ITS) region. The amplicons obtained from rDNA of Termitomyces species were compared with existing sequences in the NCBI GenBank. The results of the ITS sequence analysis discriminated between all the Termitomyces species (obtained from Ondo and Ekiti States) and Termitomyces sp. sequences obtained from NCBI GenBank. The degree of similarity of T1 to T18 to gene of Termitomyces sp. obtained from NCBI ranges between 82 and 99 percent. Termitomyces species from Garbon with ascension number AF321374 was the closest relative of T1 to T18 except T12 that has T. eurhizus and T. striatus as the closet relative. Phylogenetic tree generated with ITS sequences obtained from NCBI GenBank data revealed that T1 to T18 are more related to Termitomyces species indigenous to African countries such as Senegal, Congo, and Gabon.
1. Introduction
Termitomyces species belongs to a group called “termitophilic Agaricales” This group was created for these fungi by Heim [1]. There is symbiosis association that exists between the termite and the fungus, Termitomyces, since neither of the two partners can exist without the other. Hence, artificial cultivation had been difficult. Termitomyces species is a well known edible mushroom in Nigeria.
These mushrooms make their appearance after heavy rains [2] and grow in contact with termite nests in forest soil. They usually appear between the months of April through October. Termitomyces species is an important source of enzymes of industrial importance such as xylanase, amylase, and cellulase [3], antioxidant compounds such as polyphenol and vitamin C [4]; protein (31.4–36.4%) [5] and immunostimulatory agent [6]. There is evidence that the extract can activate splenocytes [6].
For a long time, most researchers in Nigeria examine mushrooms with the naked eye based on phenotypic characters. It has been impossible to distinguish between genetically related species by this method. Morphologically, mushrooms belonging to different genera may look similar. The present study mainly focuses on ascertaining the phylogenetic relationship between Termitomyces species found in Ondo and Ekiti States Nigeria by sequencing of their ITS zone. Moreover, comparing the gene sequence of Termitomyces species from Ondo and Ekiti States Nigeria with sequences obtained from the NCBI GeneBank.
2. Materials and Methods
2.1. Fungal Material
Fungal material Fresh fruiting body of Termitomyces species were collected from Ekiti and Ondo States (Figure 1), Nigeria (Table 1). The fruitbodies were kept dry by wrapping in tissue paper and keeping in a polythene paper containing silica gel. The polythene bags containing the samples were well labeled for easy identification.
Figure 1.
Map of Nigeria (a) and map of the states Ondo and Ekiti States (b) where Termitomyces samples were collected.
Table 1.
Information on Termitomyces species collected from Ondo and Ekiti States.
| Termitomyces sp. | Location where collection was made | State | Year of collection | Name of collector |
|---|---|---|---|---|
| T1 | Ado Ekiti | Ekiti | October, 2006 | Oyetayo, V. O. |
| T2 | Ado Ekiti | Ekiti | October, 2006 | Oyetayo, V. O. |
| T3 | FUT, Akure | Ondo | September, 2006 | Oyetayo, V. O. |
| T4 | FUT, Akure | Ondo | September, 2006 | Oyetayo, V. O. |
| T5 | Ado Ekiti | Ekti | September, 2006 | Oyetayo, V. O. |
| T6 | Akure | Ondo | September, 2006 | Oyetayo, V. O. |
| T7 | Aule | Ondo | October, 2007 | Fakoya, S. |
| T8 | Aule | Ondo | October, 2007 | Fakoya, S. |
| T9 | Igbatoro | Ondo | July, 2009 | Oyetayo, V. O. |
| T10 | Igbatoro | Ondo | July, 2009 | Oyetayo, V. O. |
| T11 | UNAD Road | Ekiti | October, 2009 | Oyetayo, V. O. |
| T12 | Orita Obele, Akure | Ondo | July, 2009 | Oyetayo, V. O. |
| T13 | Obanla, FUTA | Ondo | September, 2008 | Oyetayo, V. O. |
| T14 | Orita Obele, Akure | Ondo | September, 2009 | Fakoya, S. |
| T15 | Ilara Mokin | Ondo | August, 2009 | Fakoya, S. |
| T16 | Ogbese | Ondo | August, 2009 | Fakoya, S. |
| T17 | Owena | Ondo | August, 2009 | Fakoya, S. |
| T18 | Obanla, FUTA | Ondo | September, 2009 | Fakoya, S. |
2.2. Extraction of DNA
Standard DNA isolation methods employing CTAB lysis buffer [7] was used. Briefly, dried Termitomyces fruitbodies were ground in mortal. The grinded materials were transferred into well-labeled tube. Prewarmed extraction buffer (CTAB) was added, and the tubes were incubated at 65°C for 30 to 60 minutes. Equal volume of chloroform and alcohol (24 : 1) was added and mixed by inverting tubes for 15 minutes. The tubes were centrifuged for 10 minutes at 10,000 g (13000 rpm). The process was repeated, but the time of mixing was 3 minutes and time of centrifugation was 5 minutes at the same speed as above. Upper aqueous layers were removed into clean tubes, and 40 μL NaAc was added followed by 260 μL of cold isopropanol. This was gently mixed by inverting tubes. The tubes were incubated at −20°C overnight. On the second day, the mixture was centrifuged at 10,000 g (13000 rpm) for 10 minutes. The supernatant was discarded and pellets rinsed with 70% alcohol and mixed for sometimes. This procedure was repeated three times. After discarding the supernatant, the sample was dried in a dryer for 20 minutes at room temperature. Pellets were resuspended in 30 μL TE. DNA concentration and quality was checked on an ethidium-stained agarose gel (0.7%) using 0.2 μL of each sample.
2.3. PCR Amplification of the ITS Region
The entire region of ITS4 and ITS5 was amplified by PCR. The reaction mix was made up to a total volume of 25 μL, composed of 23 μL of Taq polymerase “Ready to Go” (Pharmacia) with 0.2 μL of each primer (100 pM) and 2 μL of DNA solution. The tubes were placed in a thermal cycler (GenAmp PCR System 2400; Perkin-Elmer) for amplification under the following conditions: 30 cycles of (1) denaturation at 95°C for 30 s, (2) annealing at 50°C for 1 min, (3) extension at 72°C for 1 min. The amplification products were purified using a PCR Purification Kit and electrophoresed on agarose gel. The amplified products were purified using a PCR Purification Kit and electrophoresed on ethidium-stained agarose gel (0.7%) to check the purity. DNA sequencing was performed using the primers (ITS 4 and ITS 5) in an Applied Biosystem DNA Analyser.
2.4. Sequencing of DNA and Alignment of Sequence
Alignments were performed with the Clustal W package [8]. The aligned sequences were corrected manually, focusing on gap positions. DNA sequence data were analyzed to provide pairwise percentage sequence divergence. The data obtained from the sequence alignment were used to plot a tree diagram (Tree View, Win 32).
3. Results and Discussion
The results of the ITS sequence analysis discriminated between all the 18 Termitomyces species obtained from Ondo and Ekiti States, Nigeria (T1 to T18) and Termitomyces sp. sequences obtained from NCBI GenBank. The ITS region of the rDNA is the most used genomic region for molecular characterization of fungi [9, 10] (Gardes and Bruns, 1993). The degree of similarity of T1 to T18 to gene of Termitomyces sp. Obtained from NCBI ranges between 82 and 99 percent (Table 2). Termitomyces species from Garbon with ascension number AF321374 was the closest relative of T1 to T18 (Table 3).
Table 2.
Genomic identification based on the ITS gene sequences of Termitomyces species collected from Ondo and Ekiti States, Nigeria.
| Termitomyces | Phenotypic identity | Closest relative in NCBI GenBank | Ascension number of closest relative | % Identity with sequence from NCBI GenBank |
|---|---|---|---|---|
| T1 | T. clypeatus | T. striatus | AB073519 | 89 |
| T2 | T. clypeatus | T. striatus | AF321367 | 91 |
| T3 | T. robustus | T. eurhizus | AF321366 | 91 |
| T4 | T. robustus | T. striatus | AF321367 | 93 |
| T5 | T. rubustus | T. striatus | AB073519 | 89 |
| T6 | T. robustus | T. striatus | AF321374 | 93 |
| T7 | T. clypeatus | T. striatus | AF321367 | 93 |
| T8 | T. robustus | T. striatus | AB073519 | 93 |
| T9 | T. clypeatus | T. striatus | AF321367 | 91 |
| T10 | T. clypeatus | T. striatus | AF321367 | 93 |
| T11 | T. clypeatus | T. striatus | AF321367 | 93 |
| T12 | T. clypeatus | T. eurhizus | AB073529 | 88 |
| T13 | T. clypeatus | T. striatus | AF321374 | 98 |
| T14 | Termitomyces sp. | T. striatus | AF321374 | 99 |
| T15* | T. microcarpus | T. microcarpus | AB073529 | 82 |
| T16 | Termitomyces sp. | T. striatus | AF321374 | 99 |
| T17 | Termitomyces sp. | T. striatus | AF321374 | 99 |
| T18 | Termitomyces sp. | T. striatus | AF321374 | 99 |
*Phenotypic identification confirmed with genomic data.
Table 3.
Information on gene sequence of Termitomyces species from NCBI GenBank with close identity with T1 to T18.
| Ascension number | Name | Location |
|---|---|---|
| AB073519 | Termitomyces sp. group3 | Thailand: Saraburi |
| AF321367 | Termitomyces striatus | Republic of Congo |
| AF321366 | Termitomyces eurhizus | Republic of Congo |
| AF321374** | Termitomyces sp. AGI | Gabon |
| AB073529 | Termitomyces sp. group 8 | Thailand: Khao Kitchagoot |
| AF321364 | Termitomyces sp. OSI | Senegal |
| AF321365 | Termitomyces sp. ASI | Senegal |
**Gene sequence of Termitomyces sp. from NCBI GenBank with the closest identity with most Termitomyces sp. from Ondo and Ekiti states, Nigeria.
Phylogenetic tree generated with ITS sequences obtained from NCBI GenBank data base revealed that T1 to T18 are more related to Termitomyces species indigenous to African countries such as Senegal, Congo, and Gabon (Figure 2). Five clades were observed in the final phylogenetic tree; Clade 1 was made up of Termitomyces species (Con 1 and Con 2) from Congo DR. Clade 2 was made up of Termitomyces species (Gab 1) from Gabon and Termitomyces species (T4, T7, T8, T9, T10, T11, T12, T13, T14, T16, T17, and T18) from Nigeria. This implies that the Termitomyces species from Nigeria and Gab 1 may be from the same ancestral stock. Clade 3 was made up of Termitomyces species from Gabon (Gab2) and Senegal (Sen 1 and 2). Clade 4 was made up of only Termitomyces species T12 while clade 5 was made up of Termitomyces species (T1, T2, T3, and T5) from Nigeria. This suggests that they may be new species.
Figure 2.
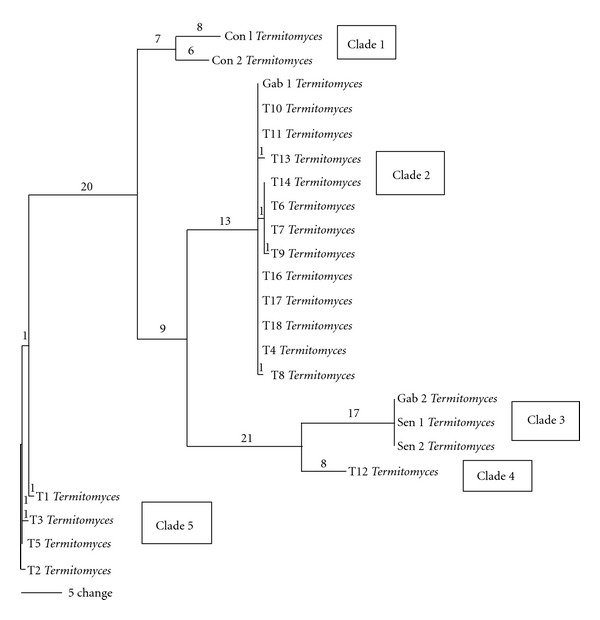
Phylogenetic tree showing positions of Termitomyces species collected from Akure and Ado Ekiti (T1 to T18) relative to existing sequences obtained from NCBI Genbank ITS sequence data.
The closest relatives of T1 to T18 which were phenotypically identified as T. clypeatus and T. robustus were T. striatus and T. eurhizus except T15 which was T. microcarpus as revealed by BLAST search (Table 2). Earlier report by Frøslev et al. [11] showed that Sinotermitomyces carnosus, S. griseus and S. rugosiceps are synonyms of T. mammiformis. Moreover, Frøslev et al. [11] also found that S. cavus and S. taiwanensis are, respectively, conspecific with T. heimii and T. clypeatus. Another study by Oyetayo [12] revealed that phylogenetic tree generated from the ITS sequence obtained from Termitomyces species earlier identified phenotypically as T. clypeatus was found to be 100% homologous to T. eurhizus found in NCBI GenBank. This shows that T. clypeatus from Nigeria may be conspecific of T. eurhizus.
This study showed that not all the gene sequence of Termitomyces species indigenous to Nigeria are 100% homologous with existing gene sequences in NCBI GenBank. Termitomyces species from some countries in Africa such as Congo, Gabon, and Senegal are more closely related to Termitomyces species indigenous to Nigeria. This may suggest common origin. An earlier phylogenetic study of some African Termitomyces revealed that they are from monophyletic origin [13]. Clades 4 and 5 shows that Termitomyces species (T1, T2, T3, T5, and T12) are totally different from others species whose gene sequences are already in NCBI GenBank.
Acknowledgments
The author wishes to acknowledge the financial support of CAS-TWAS. V. O. Oyetayo is a recipient of the CAS-TWAS Postdoctoral fellowship to China. Yao is also gratefully acknowledged for hosting V. O. Oyetayo in his laboratory (Key Laboratory of Systematic Mycology and Lichenology, Institute of Microbiology, Chinese Academy of Sciences, Beijing, China).
References
- 1.Heim R. Etudes descriptives et expérimentales sur les agarics termitophiles d'Afrique tropicale. Mémoire de l'Académie des Sciences. 1941;64:25–29. [Google Scholar]
- 2.Alasoadura SO. Studies in the higher fungi of Nigeria. 1-The Genus Termitomyces Heim. Journal of West African Science Association. 1967;12(2):136–146. [Google Scholar]
- 3.Khowala S, Sengupta S. Secretion of β-glucosidase by Termitomyces clypeatus: regulation by carbon catabolite products. Enzyme and Microbial Technology. 1992;14(2):144–149. [Google Scholar]
- 4.Mau JL, Chang CN, Huang SJ, Chen CC. Antioxidant properties of methanolic extracts from Grifola frondosa, Morchella esculenta and Termitomyces albuminosus mycelia. Food Chemistry. 2004;87(1):111–118. [Google Scholar]
- 5.Ogundana SK, Fagade OE. Nutritive value of some Nigerian edible mushrooms. Food Chemistry. 1982;8(4):263–268. [Google Scholar]
- 6.Mondal S, Chakraborty I, Rout D, Islam SS. Isolation and structural elucidation of a water-soluble polysaccharide (PS-I) of a wild edible mushroom, Termitomyces striatus . Carbohydrate Research. 2006;341(7):878–886. doi: 10.1016/j.carres.2006.02.004. [DOI] [PubMed] [Google Scholar]
- 7.Zolan ME, Pukkila PJ. Inheritance of DNA methylation in Coprinus cinereus. Molecular and Cellular Biology. 1986;6(1):195–200. doi: 10.1128/mcb.6.1.195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG. The CLUSTAL X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Research. 1997;25(24):4876–4882. doi: 10.1093/nar/25.24.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gardes M, Bruns TD. Rapid characterization of ectomycorrhizae using RFLP pattern of their PCR amplified-ITS. Mycological Society Newsletter. 1991;41:44–45. [Google Scholar]
- 10.Anderson JB, Stasovski E. Molecular phylogeny of Northern Hemisphere species of Armillaria . Mycologia. 1992;84:505–516. [Google Scholar]
- 11.Frøslev TG, Aanen DK, Læssøe T, Rosendahl S. Phylogenetic relationships of Termitomyces and related taxa. Mycological Research. 2003;107(11):1277–1286. doi: 10.1017/s0953756203008670. [DOI] [PubMed] [Google Scholar]
- 12.Oyetayo VO. Molecular characterisation of Termitomyces species collected from Ado Ekiti and Akure, Nigeria. Nigerian Journal of Microbiology. 2009;23(1):1933–1938. [Google Scholar]
- 13.Rouland-Lefevre C, Diouf MN, Brauman A, Neyra M. Phylogenetic relationships in Termitomyces (family agaricaceae) based on the nucleotide sequence of ITS: a first approach to elucidate the evolutionary history of the symbiosis between fungus-growing termites and their fungi. Molecular Phylogenetics and Evolution. 2002;22(3):423–429. doi: 10.1006/mpev.2001.1071. [DOI] [PubMed] [Google Scholar]


