Summary
Cone photoreceptors have faster light responses than rods and a higher demand for 11-cis retinal (11cRAL), the chromophore of visual pigments. RPE65 is the isomerohydrolase in the retinal pigment epithelium (RPE) which converts all-trans retinyl ester (atRE) to 11-cis retinol, a key step in the visual cycle to regenerate 11cRAL. Accumulating evidence suggests that cone-dominant species express an alternative isomerase, likely in retinal Müller cells, in order to meet the high demand for the chromophore by cones. Herein we describe the identification and characterization of a novel isomerohydrolase, RPE65c, from the cone-dominant zebrafish retina. RPE65c shares 78% amino acid sequence identity with RPE-specific zebrafish RPE65a (orthologue of human RPE65) and retains all of the known key residues for the enzymatic activity of RPE65. Similar to the other RPE-specific RPE65, RPE65c was present in both the membrane and cytosolic fractions, used atRE as its substrate and required iron for its enzymatic activity. However, immunohistochemistry detected RPE65c in the inner retina including Müller cells, but not in the RPE. Furthermore, double-immunostaining of dissociated retinal cells using antibodies for RPE65c and glutamine synthetase (a Müller cell marker), showed that RPE65c co-localized with the Müller cell marker. These results suggest that RPE65c is the alternative isomerohydrolase in the intra-retinal visual cycle, providing 11cRAL to cone photoreceptors in cone-dominant species. Identification of an alternative visual cycle will contribute to the understanding of the functional differences of rod and cone photoreceptors.
Keywords: retinoids, cone-dominant retina, isomerohydrolase, Müller cell, visual cycle
Introduction
Both rod and cone visual pigments in vertebrates require 11-cis retinal (11cRAL) as the chromophore. Isomerization of 11cRAL to all-trans retinal (atRAL) by a photon induces a conformation change of the visual pigments, triggers the phototransduction cascade and initiates vision [1, 2]. The retinoid visual cycle means the recycling of 11cRAL through a process involving multiple enzymes and retinoid-binding proteins between photoreceptors and retinal pigment epithelium (RPE); it is essential for maintaining normal vision [3, 4]. The key step in the retinoid visual cycle is the conversion of all-trans retinyl ester (atRE) to 11-cis retinol (11cROL). This conversion is catalyzed by a membrane-associated enzyme predominantly expressed in the RPE [5–7]. An RPE-specific 65 kDa protein (RPE65) was identified to have isomerohydrolase activity [8–10] that is both iron-dependent and requires retinyl ester as its substrate [11, 12]. The RPE65 knockout mouse (RPE65−/−) showed no detectable 11-cis retinoids and over accumulation of atRE in the RPE [13]. Furthermore, RPE65 gene mutations are associated with inherited retinal degenerations such as Retinitis Pigmentosa (RP) and Leber’s Congenital Amaurosis (LCA) [14–16]. We have shown that purified RPE65 has isomerohydrolase activity after it is reconstituted into liposomes, confirming that RPE65 is the isomerohydrolase in the RPE [17]. Finally, RPE65 was crystallized and its 3-D structure revealed [18], which confirmed the key enzymatic residues previously identified by site-directed mutagenesis and in vitro enzymatic activity assay [11, 19–21].
Cone photoreceptors have faster responses to light than rod photoreceptors and thus, demand more chromophore supplies [22, 23]. It has been suggested that the cone-dominant retina has an alternative visual cycle independent of the RPE [24–27]. Several studies suggested that this RPE-independent retinoid visual cycle may be present in the Müller glia cells of the cone-dominant chicken retina to provide additional 11cRAL for cones [24–27]. The Müller cell is the principal glial cell type in the vertebrate retina and is a specialized radial cell which spans the entire thickness of the inner retina. The Müller cell constitutes an anatomical link between the retinal neurons and supports their activities by exchanging molecules between the other retinal layers [28]. In addition, it has been shown that several retinoid-binding proteins and enzymes involved in vitamin A metabolism are present in Müller cells [29–32]. Thus, it has been proposed that Müller cells could be a possible alternative source of 11-cis retinoids, and may play an important role in 11cRAL recycling.
Recently, Kefalov’s group demonstrated that cone photoreceptors recovered light sensitivity following photobleaching when the cone photoreceptors are connected with other retinal cells, but not with the RPE; rod photoreceptors did not recover under the same conditions [33, 34]. In addition, Müller cell-specific gliotoxin (L-α-AAA) inhibited the functional recovery of cone photoreceptors [33, 34], providing further evidence that a cone-specific visual cycle is dependent on Müller cells. However, an alternative isomerase which converts all-trans retinoids to 11-cis retinoids in the retina has not been identified from any species, and RPE65 remained the only known isomerohydrolase that can generate 11cROL.
Zebrafish is a commonly used model in vision research [35–37]. The retina of the zebrafish is cone-dominant, with a composition of 79% cones and 21% rods based on immunohistochemical analysis at 7 days post-fertilization (dpf) [38]. It was recently shown that morpholino-mediated knockdown of zebrafish RPE65a (an orthologue of human RPE65) did not completely attenuate 11cRAL regeneration in the zebrafish eye [39]. This study provided evidence that there is another isomerohydrolase in the zebrafish retina and that it is RPE-independent. Therefore, the present study used zebrafish as a cone-dominant model and identified the alternative isomerohydrolase.
Results
Cloning and amino acid sequence analyses of a novel isomerohydrolase in the zebrafish eye
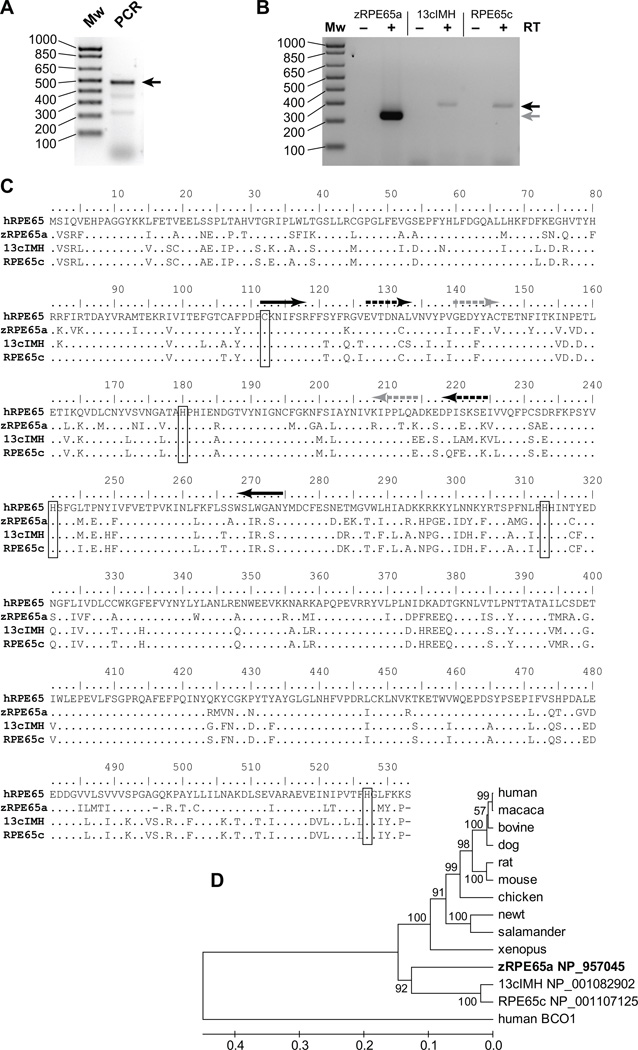
We performed PCR using zebrafish retina cDNA and a set of degenerate primers at the well-conserved regions of the RPE65 sequence (see Table 1 and Fig. 1C, black arrows). PCR amplified a fragment of the expected size (Fig. 1A). At the level of deduced amino acid sequences, one of the clones was identical to a “novel protein similar to vertebrate RPE65 (Genbank accession number; NP_001107125)”. The cloned fragment showed 79.9 % and 78.5 % amino acid sequence identities to previously reported zebrafish RPE65a (Genbank accession number; NP_957045, [39]) and human RPE65, respectively, and thus is named RPE65c. The RPE65c fragment showed 94.0% amino acid sequence identity to another recently identified orthologous form of RPE65, 13-cis specific isomerohydrolase (13cIMH: original name is retinal pigment epithelium-specific protein b (rpe65b; accession number in GenBank NP_001082902)), which is expressed in the zebrafish brain and converts atRE exclusively to 13-cis retinol (13cROL) [40]. Furthermore, we determined the expression of zebrafish RPE65a, 13cIMH and RPE65c separately in the eye by RT-PCR using gene-specific primers based on the sequences in GenBank (Fig. 1B). The specificity of the primers was confirmed by PCR using each cDNA clone as the template (see Fig. S1). The sequences of the PCR products were confirmed by direct DNA sequencing. An amino acid sequence alignment of RPE65c with human RPE65, zebrafish RPE65a and 13cIMH (Fig. 1C), showed that RPE65c shares 75.6%, 78.0% and 96.7% overall amino acid sequence identities to human RPE65, zebrafish RPE65a and 13cIMH, respectively. The known key residues in RPE65, including four His residues for iron binding and a palmitylated Cys residue for membrane association [21], were conserved in RPE65c, suggesting that RPE65c is likely an enzyme in the zebrafish eye.
TABLE 1.
Primer sets in this manuscript.
| Primer names | Accession number |
Sequences |
|---|---|---|
| Deg RPE65-Fwd | N/A | 5’-TGCARRAAYATHTTYTCCAG-3’ |
| Deg RPE65-Rev | 5’-AYRAAYTCRWRBCCYTTCCA-3’ | |
| RPE65a-Fwd | NM_200751 | 5’-GCGGCCGCCACCATGGTCAGCCGTTTTGAACAC-3’ |
| RPE65a-Rev | 5’-GATATCTTATGGTTTGTACATCCCATGGAAAG-3’ | |
| RPE65c-Fwd | NM_001113653 | 5’-GCGGCCGCCACCATGGTCAGCCGTCTTGAACAC-3’ |
| RPE65c-Rev | 5’-AAGCTTCTAAGGTTTGTAGATGCCGTGGAG-3’ | |
| RPE65a GSP-Fwd | NM_200751 | 5’-TGGGGAGGACTTTTATGCTGT-3’ |
| RPE65a GSP-Rev | 5’-CTTTTGTGTAGGTGGGATTCG-3’ | |
| 13cIMH GSP-Fwd | NM_001089433 | 5’-CTGAGGTTACAGACAACTGTTC-3’ |
| 13cIMH GSP-Rev | 5’-CCTTTGACATCGCAAGTGGATCA-3’ | |
| RPE65c GSP-Fwd | NM_001113653 | 5’-TTGAGGTGACAGACAATTGCCT-3’ |
| RPE65c GSP-Rev | 5’-TCTTTGACTTCTCAAACTGATCG-3’ | |
| RPE65a-His-Fwd | NM_200751 | 5’-GCGGCCGCCACCATGCATCATCACCATCACCATGTCAGCCGTTTTGAACAC-3’ |
| RPE65c-His-Fwd | NM_001113653 | 5’-GCGGCCGCCACCATGCATCATCACCATCACCATGTCAGCCGTCTTGAACAC-3’ |
Figure 1. Cloning of zebrafish RPE65c and sequence comparisons with RPE65 isoforms.
(A) PCR products using degenerate primers and zebrafish eyecup cDNA were confirmed by 2.0% agarose gel electrophoresis. The PCR product with the expected size is indicated by an arrow. (B) To verify expression of RPE65a and its homologs in the zebrafish eyecup, RT-PCR analysis was performed using a set of gene-specific primers for zebrafish RPE65a, 13cIMH and RPE65c and zebrafish eyecup cDNA either in the absence (indicated by “−“) or the presence (“+”) of reverse transcriptase (RT) to exclude possible genomic DNA contamination. The PCR products were confirmed by 2.0% agarose gel electrophoresis. Black and gray arrows indicate the PCR products with expected sizes for RPE65c, zebrafish RPE65a and 13cIMH, respectively. (C) Alignment of amino acid sequences of human RPE65 (hRPE65), zebrafish RPE65a (zRPE65a), 13cIMH and RPE65c. The human RPE65 sequence was used as the template; amino acid residues identical to human RPE65 are represented by dots. The known key residues (four His residues forming an iron binding domain and a palmitylated Cys residue for membrane association) are boxed. The black arrows indicate the positions of degenerate primers. The black and grey arrows with broken lines show the positions of gene-specific primers to amplify specific PCR products. (D) A phylogenetic tree constructed by the UPGMA method in MEGA application version 4.02 [52]. Human BCO1 was used as an out-group. The numbers on the branches are the mean clustering probabilities from 1000 bootstrap resamplings.
Phylogenetic tree analysis suggested that the ancestral form of zebrafish RPE65c and 13cIMH were generated by gene duplication prior to the divergence of the ancestral amphibian, and the divergence of RPE65c and 13cIMH may have occurred more recently (Fig. 1D). In addition, based on GenBank information, zebrafish RPE65c and 13cIMH are encoded by distinct neighbor genes on chromosome 8, whereas the zebrafish RPE65a gene is on chromosome 18. Taken together, this further supports the proposal that zebrafish RPE65c, RPE65a and 13cIMH are three homologous proteins encoded by distinct genes.
Zebrafish RPE65c exhibits isomerhydrolase activity
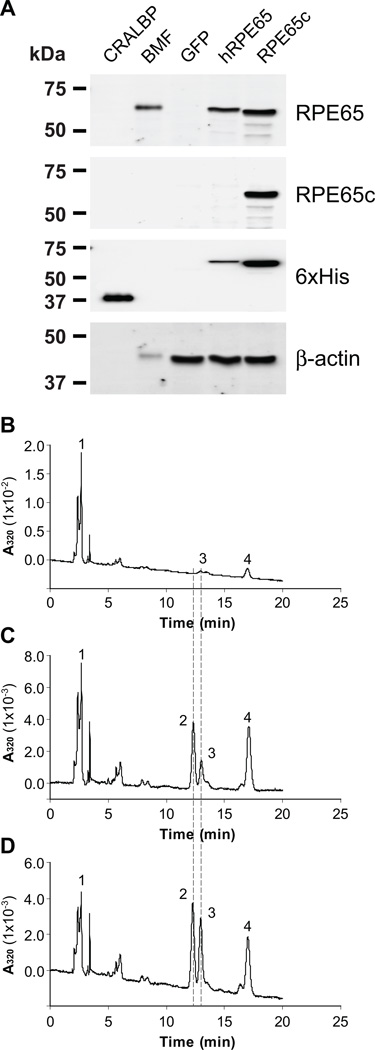
In order to study its enzymatic activity, we cloned full-length zebrafish RPE65c into a shuttle vector and generated a recombinant adenovirus expressing RPE65c (Ad-RPE65c) as previously described [19]. The 293A cells were separately infected at MOI (multiplicity of infection) 100 by adenoviruses expressing GFP (green fluorescence protein, negative control), human RPE65 (positive control) or RPE65c and cultured for 24 hrs. Protein expression was confirmed by Western blot analysis (Fig. 2A). In order to evaluate the isomerohydrolase activity of zebrafish RPE65c, equal amounts of total cellular proteins (125 µg) from the infected cells were incubated with atRE incorporated into liposomes as described previously [17]. Under the same assay conditions, the cell lysate expressing GFP did not generate any detectable 11cROL, although a very minor 13cROL peak (peak 3) was detected (possibly via thermal isomerization), as shown in the high-performance liquid chromatography (HPLC) profiles (Fig. 2B). In contrast, human RPE65 generated a dominant 11cROL peak and a minor 13cROL peak (Fig. 2C, peak 2 and 3). Similar to human RPE65, zebrafish RPE65c catalyzed the generation of both 11cROL and 13cROL (Fig. 2D), suggesting that zebrafish RPE65c is a second isomerohydrolase identified in the eye. It is noteworthy that 13cROL production by zebrafish RPE65c was more prominent than that by human RPE65 under the same assay conditions (Fig. 2).
Figure 2. Isomerohydrolase activity of zebrafish RPE65c.
The adenovirus expressing GFP (negative control), human RPE65 and zebrafish RPE65c were separately infected in 293A cells at MOI 100. (A) Protein expression was confirmed by Western blot analyses. CRALBP (0.5 µg of 6×His-tagged recombinant CRALBP as the positive control for His-tagged protein blot), BMF (2.5 µg of bovine RPE microsomal fraction as the positive control for RPE65 blot), GFP, hRPE65 and RPE65c; 25 µg of total cellular protein expressing GFP, human RPE65 or RPE65c. (B–D) Equal amounts of total cellular proteins from the cells (125 µg) expressing GFP (B), human RPE65 (C) and RPE65c (D) were incubated with liposomes containing atRE (250 µM lipids, 3.3 µM atRE) for 1 hr at 37°C, and the generated retinoids were analyzed by HPLC. The peaks were identified as follows: 1, retinyl esters; 2, 11cROL; 3, 13cROL; 4, atROL.
Substrate specificity of zebrafish RPE65c
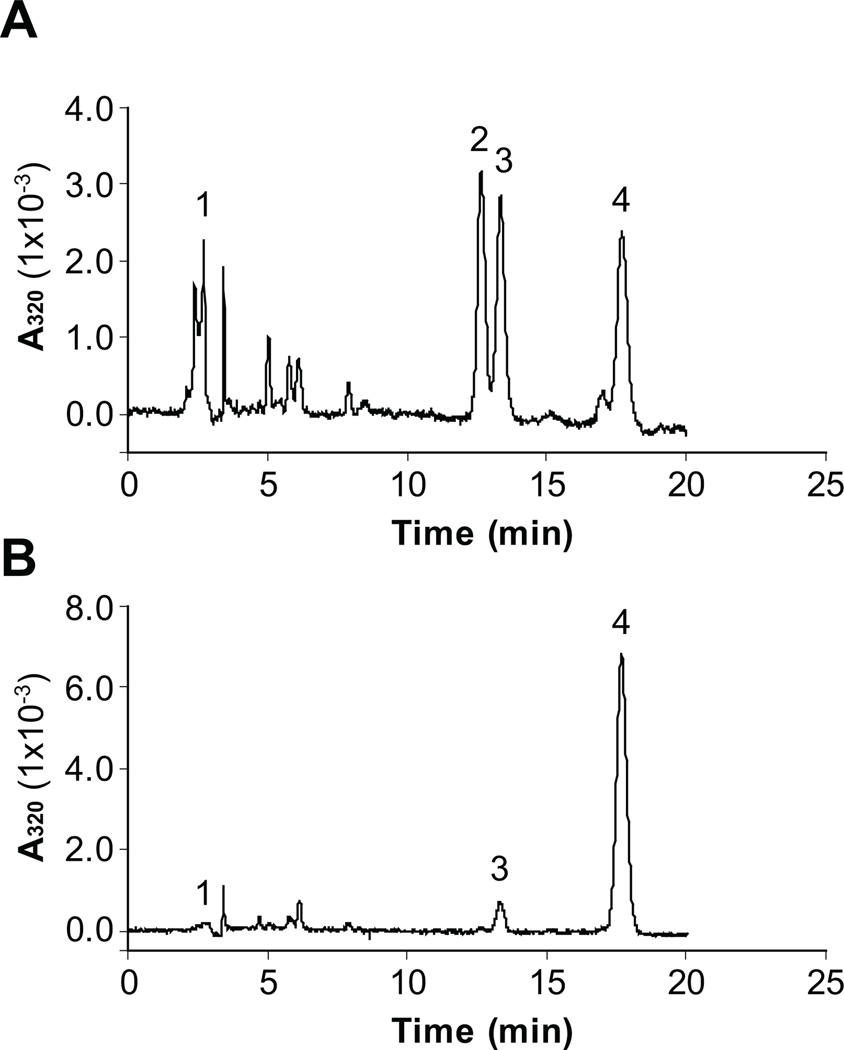
It was proposed that the potential alternative isomerase in retinal Müller cells may use all-trans retinol (atROL) as the substrate for the conversion of 11cROL [24–27]; the substrate of RPE65 in the RPE is atRE [12]. To verify the substrate specificity of zebrafish RPE65c, total cell lysates expressing RPE65c were incubated with either atRE or atROL incorporated into liposomes. HPLC analysis of the generated retinoids showed that RPE65c converted atRE to both 11cROL and 13cROL (Fig. 3A). However, RPE65c did not generate any detectable 11cROL from atROL (Fig. 3B), suggesting that RPE65c requires atRE as its intrinsic substrate.
Figure 3. AtRE is the substrate of zebrafish RPE65c.
Equal amounts of total cellular proteins from the cells (125 µg) expressing RPE65c were incubated with liposomes containing atRE (A) or atROL (B). The generated retinoids were extracted and analyzed by HPLC. The peaks were identified as follows: 1, retinyl esters; 2, 11cROL; 3, 13cROL; 4, atROL.
The isomerohydrolase activity of zebrafish RPE65c is dependent on iron
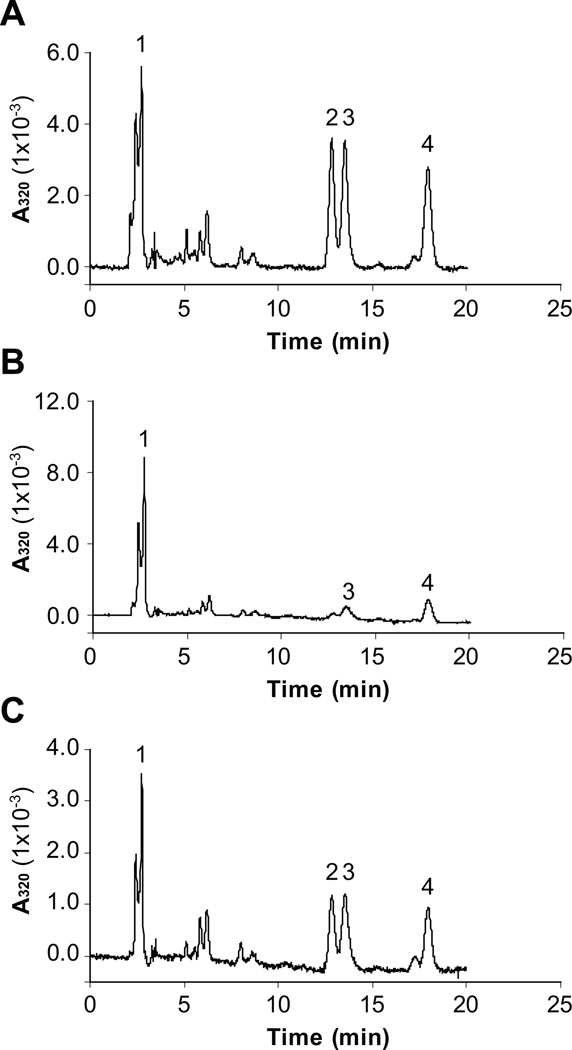
Since zebrafish RPE65c contains the conserved His residues which form the iron-binding site in RPE65 [9, 11, 18, 19], we determined if RPE65c is an iron-dependent enzyme. The cell lysate expressing RPE65c was incubated with liposomes containing atRE, and the generated retinoids from the reaction were analyzed by HPLC. In the absence of a metal chelator, RPE65c catalyzed the production of 11cROL and 13cROL from atRE (Fig. 4A). However, in the presence of 1 mM of the metal chelator, bipyridine, the enzymatic activity of RPE65c was almost completely abolished (Fig. 4B). Supplement of 6 mM FeSO4 to the reaction together with bipyridine partially restored the impaired enzymatic activity (Fig. 4C), suggesting that the enzymatic activity of RPE65c is iron-dependent.
Figure 4. Zebrafish RPE65c is an iron-dependent enzyme.
The 293A cell lysate expressing RPE65c was incubated with liposomes containing atRE (A), liposomes containing atRE in the presence of 1 mM bipyridine (B) and liposomes containing atRE, in the presence of 1 mM bipyridine and 6 mM FeSO4 (C). The generated retinoids were analyzed by HPLC. The peaks were identified as follows: 1, retinyl esters; 2, 11cROL; 3, 13cROL; 4, atROL.
Characterization of the kinetic parameters of zebrafish RPE65c’s enzymatic activity
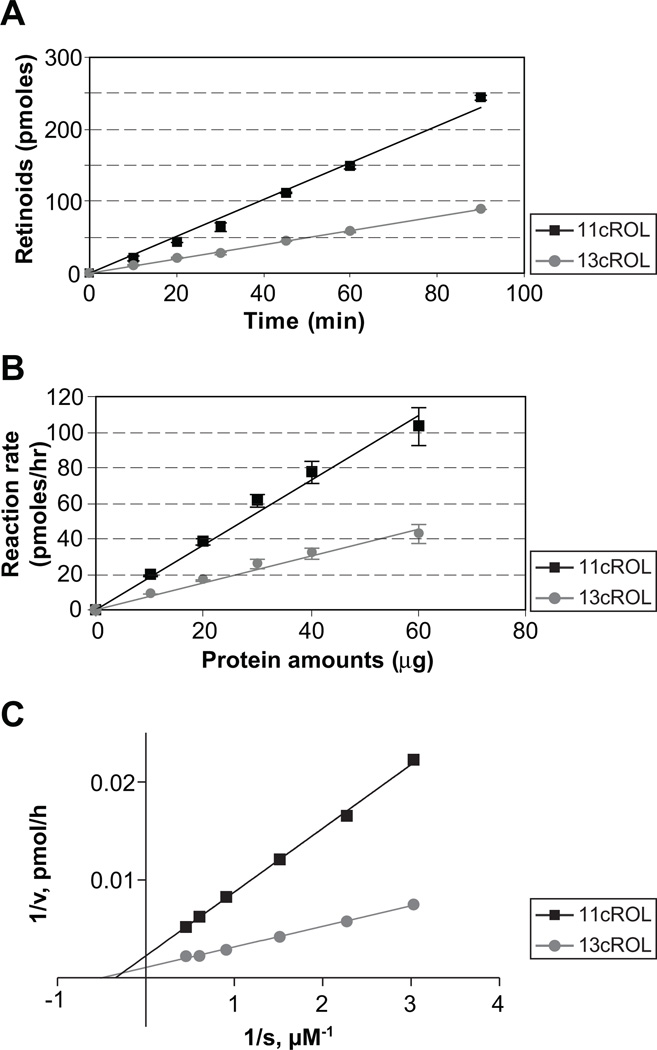
To determine the steady-state kinetics of the enzymatic activity of zebrafish RPE65c, the assay conditions were optimized to ensure that all of the measurements were taken within the linear range. First, we plotted the time course of 11cROL and 13cROL generation after incubation of the atRE-liposomes with 125 µg of total cell lysate expressing RPE65c. The time courses of 11cROL and 13cROL production appeared linear in its initial phase (Fig. 5A). Therefore, all of the further experiments in this study were conducted within this range. Secondly, to establish the dependence of 11cROL and 13cROL production on the level of RPE65c protein, 293A cells were infected with Ad-RPE65c at MOI 100 and then cultured for 24 hrs. Increasing amounts of total cellular proteins expressing RPE65c (10, 20, 30, 40, 60 µg) were incubated with the same amount of liposomes containing atRE. The production of 11cROL and 13cROL were found to be dependent on the RPE65c protein levels (Fig. 5B). Finally, to analyze the substrate dependence of RPE65c activity, we measured the initial reaction velocity using different concentrations of atRE-liposomes (Fig. 5C). Lineweaver-Burk analysis of the data yielded the kinetic parameters for the reaction catalyzed by RPE65c: for 11cROL, the Michaelis-Menten constant (Km) = 1.91 µM and the Vmax = 1.82 nmol/mg of total protein/hr; for 13cROL, the Km = 2.95 µM and the Vmax = 0.91 nmol/mg of total protein/hr.
Figure 5. Enzymatic parameters of zebrafish RPE65c.
Cells were infected with adenovirus expressing RPE65c at MOI 100 and cultured for 24 hr. Equal amounts of total cellular proteins (125 µg) were incubated with liposome containing atRE for the indicated time intervals. (A) Time courses of 11cROL and 13cROL production were separately plotted. Total cellular protein expressing RPE65c was incubated with liposomes containing atRE (250 µM lipids, 3.3 µM atRE) for 1 hr at 37 °C, and the generated retinoids were analyzed by HPLC. (B) Dependence of production of 11cROL and 13cROL on RPE65c protein levels. The increasing amounts of total cellular proteins expressing RPE65c (10, 20, 30, 40, 60 µg) were incubated with the same amount of liposomes containing atRE for 1 hr. The produced 11cROL and 13cROL were separately quantified from the area of the 11cROL and 13cROL peaks, respectively (mean ± SD, n=3), and plotted against protein concentration of the cell lysate expressing RPE65c. (C) Lineweaver-Burk plot of 11cROL and 13cROL generation by RPE65c. Liposomes with increasing concentrations (S) of atRE were incubated with equal amounts of cell lysate (125 µg) expressing RPE65c by adenovirus at MOI 100 for 1 hr. Initial rates (V) of 11cROL and 13cROL generation were calculated based on 11cROL and 13cROL production recorded by HPLC.
Localization of zebrafish RPE65c in the retina and its subcellular fractionation
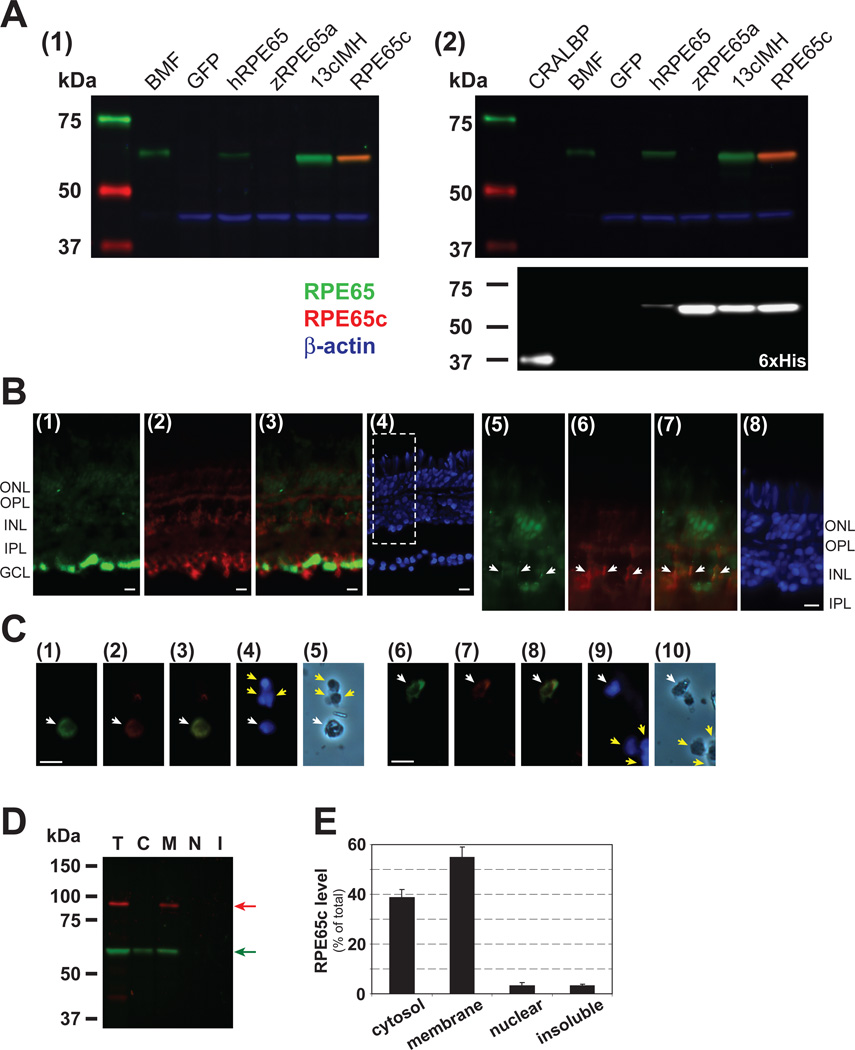
To analyze the cellular localization of zebrafish RPE65c in the retina, we generated an antibody using a specific zebrafish RPE65c peptide, and the specificity of the antibody was confirmed using recombinant RPE65c, human RPE65, zebrafish RPE65a and 13cIMH. As shown by Western blot analysis, the antibody specifically recognized RPE65c but not human RPE65, zebrafish RPE65a or 13cIMH (Fig. 6, A1, A2). Using this antibody, we examined the localization of RPE65c in the zebrafish retina by immunohistochemistry. Intense RPE65c signal was detected in the inner retina near the ganglion cell layer, in a region where the Müller end feet are located, and weak signal was observed between the outer nuclear and ganglion cell layers (Fig. 6, B1). Double immunostaining using the anti-RPE65c antibody and an antibody for a Müller cell marker, glutamine synthetase (GS), showed the RPE65c signal was co-localized with the GS signal in the region between the ganglion cell layer and the outer nuclear layer, in which the Müller cell processes are located (Fig. 6, B5–B7). This suggests that RPE65c may be expressed in Müller cells. Under the same conditions, no RPE65c signal was detected in the RPE (Fig. S2). To provide conclusive evidence supporting RPE65c expression in Müller cells, we performed double immunostaining of dissociated retinal cells with antibodies for RPE65c and GS. The signals of RPE65c and GS were found co-localized in a number of dissociated retinal cells (Fig. 6C), confirming that RPE65c is expressed in Müller cells.
Figure 6. Cellular localization of zebrafish RPE65c in the eye and subcellular fractionation of RPE65c in cultured cells.
(A) Specificity of the antibody for RPE65c was evaluated by Western blot analysis. (A1) Bovine microsomal fraction (BMF, 2.5 µg) and equal amount of total cellular protein (25 µg) of the 293A cells expressing GFP, human RPE65 (hRPE65), zebrafish RPE65 (zRPE65a), 13cIMH and RPE65c were blotted with mouse anti-RPE65 monoclonal (green signals), rabbit anti-RPE65c (red) and goat anti-β-actin (blue) antibodies. (A2) Purified his-tagged cellular retinal-aldehyde binding protein (CRALBP, 0.5 µg), BMF (2.5 µg), and 293A cell lysates (25 µg) infected by adenovirus expressing GFP, hRPE65-His, zRPE65a-His, 13cIMH-His and RPE65c-His were blotted with the same set of antibodies (left panel). Then, the membrane was stripped and re-blotted with a mouse anti-6×His-tag monoclonal antibody. (B) Immunohisotochemistry of RPE65c in the zebrafish retina. The zebrafish retinal section was double-stained with antibodies for RPE65c (green) and GS (Müller cell marker, red). (B1–4) The images show immunostaining of RPE65c (B1), GS (B2), merged RPE65c and GS staining (B3), and DAPI staining (B4), respectively. (B5–8) High magnification images of the boxed area in panel (B4) show RPE65c staining (B5), GS staining (B6), merged RPE65c and GS staining (B7), and DAPI staining (B8), respectively. White arrows indicate overlapped signals in the area of Müller cell processes. (C) Immunostaining of dissociated retinal cells using antibodies for RPE65c and GS. (C1–10) The representative images of RPE65c staining (C1, C6), GS staining (C2, C7), merged RPE65c and GS staining (C3, C8), DAPI staining (C4, C9), and phase contrast images (C5, C10), respectively. Representative cells with RPE65c and GS staining were indicated by white arrows and negative cells were indicated by yellow arrows. ONL, outer nuclear layer; OPL, outer plexiform layer; INL, inner nuclear layer; IPL, inner plexiform layer; GCL, ganglion cell layer. Scale bar = 10 µm. (D) Subcellular localization of RPE65c in cultured cells. Forty-eight hours post-transfection of the RPE65c expression plasmid, the cells were harvested and separated into 4 subcellular fractions by a FractionPrep™ kit. Equal amounts of fractionated proteins (25 µg of total protein, 5 µg each fraction) were applied for Western blot analyses using antibodies specific for RPE65c (green arrow) and Calnexin (ER membrane marker, red arrow). T; total cell lysates, C; cytosol, M; membrane, N; nuclear fractions and I; detergent-insoluble fraction including cytoskeleton and inclusion body. (E) The level of RPE65c in each fraction was quantified by densitometry and expressed as % of total RPE65c (means ± SD, n=3) from 3 independent experiments.
Moreover, we examined the subcellular distribution of RPE65c expressed in 293A cells using a subcellular fractionation kit (FractionPrepTM: Biovision, Mountain View, CA). Western blot analysis of different subcellular fractions showed that RPE65c was present in both the membrane and cytosolic fractions (Fig. 6D, E), similar to that of recombinant human RPE65 [20, 21].
Discussion
It has been known for a long time that cone photoreceptors have faster recovery of light sensitivity from desensitization than rod photoreceptors due to the higher regeneration rates of visual pigments [41, 42]. This faster recovery demands faster recycling of 11cRAL, the chromophore of visual pigments [22, 23]. It has been speculated that there may be an alternative visual cycle in cone-dominant retinas, to meet the high demand for 11cRAL by cones [24–27]. Several lines of evidence have suggested that there is an alternative isomerase in the retina of cone-dominant species, likely in retinal Müller cells [33, 34]. Here, we reported cloning and characterization of a novel isomerohydrolase expressed in the Müller cells of cone-dominant zebrafish. This is the first enzyme, except for RPE65 in the RPE, which can generate 11-cis retinoid from its all-trans isoform in the eye. This enzyme may serve as a key component of the alternative visual cycle and contribute to the fast resensitization of cone photoreceptors [2].
Schonthaler and co-workers reported that although the generation of 11cROL is reduced by morpholino-mediated knockdown of zebrafish RPE65a in the RPE (orthologous to human RPE65), a significant amount of 11cROL is still generated. Based on this observation, the authors proposed that there is an RPE65-independent regeneration of 11-cis retinoids in zebrafish eyes [39]. Our results suggest that zebrafish RPE65c identified in this study can contribute to this RPE65-independent isomerohydrolase activity. Although it shares significant sequence homology with zebrafish RPE65a, RPE65c is encoded by a gene located on a different chromosome than the RPE65a gene. This observation suggests that RPE65c is not from a polymorphism or alternative splicing product of the RPE65a gene. Furthermore, we recently reported that another isoform of RPE65a, 13cIMH (RPE65b), is expressed in the zebrafish brain and exclusively generates 13cROL, not 11cROL in our in vitro assay system [40]. Even though the amino acid identities of 13cIMH and RPE65c are extremely high, they are encoded by two distinct genes located on chromosome 8. Furthermore, the products of these enzymes and their tissue localizations were clearly different.
RPE65c has multiple structural and functional features similar to RPE65. RPE65c has the conserved key residues of RPE65 including four His residues known for iron binding and a palmitylated Cys residue responsible for membrane association [11, 19–21]. Furthermore, RPE65c is present in both the membrane and cytosolic fractions, is an iron-dependent enzyme and requires atRE as its substrate, simlar to RPE65. On the other hand, RPE65c localization is different from RPE65 in that it is expressed in retinal Müller cells as opposed to the RPE.
We previously showed that RPE65 predominantly generates 11cROL in the presence of lecithin retinol acyltransferase (LRAT) and cellular retinaldehyde-binding protein (CRALBP) under our in vitro assay conditions (at 37 °C for 1 hr) [8, 19, 21, 43]. In this case, CRALBP may stabilize the RPE65-generated 11cROL so that the other isoforms of retinol, including 13cROL, can be re-esterified by LRAT to become 13-cis retinyl ester. Once it is in the ester form, it cannot be separated from atRE and 11-cis retinyl ester (11cRE) (Fig. S3A). It was reported that 11cROL is not as favorable a substrate of LRAT as atROL and 13cROL [44]. As a result, 11cROL is detected as the major product in the presence of LRAT. This may explain the phenomenon of why RPE65 generates predominantly 11cROL in the presence of LRAT in our previous assays [8, 11, 19, 20] and under actual physiological conditions in the RPE (Fig. S3A). In the absence of LRAT, as used in this study, RPE65 generates a slightly higher level of 13cROL, as there is no ester synthetase to esterify the 13cROL generated in the reaction. Therefore, 13cROL is accumulated in the absence of LRAT in the reaction (Fig. S3B).
Under our current in vitro assay conditions, RPE65c generated both 11cROL and 13cROL in the absence of LRAT. Thus, we separately calculated the Km of RPE65c, determining that they were 1.91 µM for 11cROL and 2.95 µM for 13cROL production. The Km of RPE65c for 11cROL generation was 3.7-fold and 1.9-fold lower than that of recombinant bovine RPE65 [10] and purified recombinant chicken RPE65 [17], respectively. Likewise, the catalytic efficiencies (Vmax/Km) for 11cROL and 13cROL were 0.953 and 0.308, respectively. This suggests that RPE65c is still an efficient enzyme to catalyze 11cROL production, even if RPE65c produces more 13cROL than human RPE65.
Since RPE65c requires atRE as its substrate, one question is how atRE is generated in the inner retina. It remains unclear where RPE65c obtains its substrate in Müller cells. Although LRAT, the enzyme known to generate atRE for RPE65, is expressed in the RPE and was not found in the retina [26], another ester synthetase’s (Acyl CoA-dependent retinol acyl transferase; ARAT) activity was observed in primary retinal Müller cell culture [26, 45]. It is likely that atRE, the substrate for RPE65c, is generated by ARAT in Müller cells.
Previous evidence has suggested that CRALBP is expressed in both the RPE and Müller cells of mammals [31, 32]. In addition, it was reported that CRALBP-b, an isoform of CRALBP, is specifically expressed in zebrafish Müller cells [46, 47]. Our previous studies have shown that RDH10, which catalyzes oxidation of 11cROL to 11cRAL [48], is also expressed in Müller cells [30]. These studies suggest that, in addition to RPE65c, Müller cells produce other components of the visual cycle essential for the regeneration of the chromophore. Our immunohistochemistry analysis showed RPE65c in the inner retina, including Müller cells, even though RPE65c did not completely colocalize with GS, which is a commonly used Müller cell marker. It should be noted that GS is a cytosolic protein, while RPE65c is a protein associated with the membrane, likely with the endoplasmic reticulum membrane similar to RPE65 [21]. The different subcellular localizations of GS and RPE65c may account for the fact that RPE65c and GS signals are not completely overlapped. To further confirm that RPE65c is expressed in Müller cells, we performed immunostaining of dissociated retinal cells. The result clearly showed that RPE65c is expressed in GS positive Müller cells. Based on this result, we conclude that RPE65c is indeed expressed in the retinal Müller cells of zebrafish. In addition to Müller cells, RPE65c may also be expressed in ganglion cells, since immunohistochemistry showed intense immunosignals in the ganglion cell layer. It has been shown that a small portion of ganglion cells express an opsin-like photosensitive molecule, melanopsin, which uses 11cRAL as its chromophore [49], same as visual pigments. It is likely that RPE65c expressed in ganglion cells might contribute to providing chromophore to melanopsin to maintain their light sensitivities. The expression and function of RPE65c in other cell types in the retina remains to be investigated.
Earlier studies showed that primary chicken Müller cells contain an enzyme to catalyze atROL into 11cROL and 11cRE [24–27]. However, the enzyme catalyzing the conversion of atROL into 11cROL in Müller cells has not yet been identified. RPE65c used atRE as the substrate and generated 11cROL (Fig. 3). A recent study suggested that the potential isomerase in chicken Müller cells may not be RPE65, since 11cRE generation in the retina homogenate was not inhibited by the metal chelator, bipyridine [50]. It is not clear whether the isomerase in chicken Müller cells is an orthologue of zebrafish RPE65c or not, since the chicken enzyme has not been cloned yet. The difference in iron dependency between the potential isomerase in chicken Müller cells and zebrafish RPE65c suggests that they may not be orthologous enzymes.
In summary, the present study has identified the alternative isomerohydrolase in the retinal Müller cells of a cone-dominant species, which may play a key role in the intra-retinal visual cycle. Further studies are warranted to establish the function of this isomerohydrolase in the cone visual cycle.
Materials and Methods
Cloning and construction of zebrafish RPE65c expression vectors
The cornea and lens were removed from enucleated zebrafish eyes, and total RNA was extracted from the eyecups using Trizol reagent (Invitrogen, Carlsbad, CA), and further purified by an RNeasy kit (Qiagen, Valencia, CA). The cDNA was synthesized using the TaqMan reverse transcriptase system (Applied Biosystems Inc., Foster City, CA) with an oligo-dT primer and random hexamer. PCR was performed with PCR master mix (Roche, Indianapolis, IN) at 94°C for 5 min followed by 35 cycles of 94°C for 30 sec, 48°C for 30 sec, and 72°C for 30 sec using a set of degenerate primers (Deg RPE65-Fwd and Rev, see Table 1). The PCR products were confirmed by 2.0% agarose gel electrophoresis, and cloned into pGEM T-Easy vector (Promega, Madison, WI). The inserts were sequenced using an ABI-3770 automated DNA sequencer (Applied Biosystems Inc., Foster City, CA). The sequenced clones were verified by “BLAST search” in GenBank, which identified a novel sequence (NM_001113653).
To clone the full-length RPE65c, RT-PCR was performed with Pfu-Turbo (Stratagene, La Jolla, CA) and gene-specific primers (see Table 1, RPE65c-Fwd; containing a NotI site and the Kozak sequence [51] and RPE65c-Rev; containing a HindIII site) at 94°C for 5 min followed by 35 cycles of 94°C for 1 min, 58°C for 1 min, and 72°C for 2 min. Following agarose gel electrophoresis, the PCR product was purified and cloned into the pGEM-T Easy vector (Promega, Madison, WI). The insert was sequenced from both directions to exclude any mutations. The confirmed RPE65c cDNA was cloned into a pcDNA3.1(−) expression vector. Following sequence confirmation, the expression constructs were purified by a QIAfilter Maxi Prep kit (Qiagen, Valencia, CA). Furthermore, we constructed adenovirus vectors expressing RPE65c with 6×His-tag at the 5’ end, as described previously [17, 19, 40].
RT-PCR
The zebrafish eyes were dissected, and total RNA was extracted from eyecups using Trizol reagent (Invitrogen, Carlsbad, CA), and further purified by RNeasy kit (Qiagen, Valencia, CA). The cDNA was synthesized using the TaqMan reverse transcriptase system (Applied Biosystems Inc., Foster City, CA) with an oligo-dT primer and random hexamer. In order to eliminate potential genomic DNA contamination, the RNA from the eyecups were treated with DNase I and cDNA synthesis was carried out with (+) or without (−) reverse transcriptase (RT). Based on the sequences in GenBank, we designed gene-specific primers (see Fig. 1C broken line arrows s and Table 1), and RT-PCR was performed with Taq DNA polymerase (Roche, Indianapolis, IN) at 94°C for 5 min followed by 35 cycles of 94°C for 30 sec, 58°C for 30 sec, and 72°C for 30 sec. The sizes of the PCR products were confirmed by 2.0 % of agarose gel electrophoresis and further confirmed by direct sequencing of PCR products.
Amino acid sequence comparisons and phylogenetic tree analyses of RPE65
Amino acid sequences of human RPE65 (NP_000320), zebrafish RPE65a (NP_957045), 13cIMH (NP_001082902) and RPE65c (NP_001107125) were aligned using the “Clustal-W” program in BioEdit (Ibis Therapeutics, Carlsbad, CA). A phylogenetic tree was constructed using the UPGMA method with 1000 times bootstrap re-sampling in MEGA application version 4.02 [52]. The known RPE65 sequences of human, macaque monkey (XP_001095946), bovine (NP_776878), dog (NP_001003176), rat (NP_446014), mouse (NP_084263), chicken (NP_990215), Japanese fireberry newt (BAC41351), tiger salamander (AAD12758), African clawed frog (AAI25978), zebrafish RPE65a, 13cIMH and RPE65c were used for phylogenetic analysis. Human BCO1 (NP_059125) was used as the out-group of the tree.
Western blot analysis
Briefly, protein concentration was measured by Bradford assay [53]. Equal amounts of protein (25 µg) from each sample were resolved by SDS-PAGE and blotted with a 1:1000 dilution of a rabbit polyclonal antibody to RPE65 [54] and a 1:5000 dilution of mouse monoclonal antibody for β-actin (Abcam, Cambridge, MA) as a loading control. Following three washes with TBST, the membrane was then incubated for 1.5 hr with 1:25000 dilution of anti-mouse IgG conjugated with DyLight 549 and anti-rabbit IgG conjugated with DyLight 649 (Pierce, Rockford, IL), and the bands were detected using the FluorChem Q imaging system (AlphaInnotech, San Leandro, CA). The bands (intensity × area) were semi-quantified by densitometry using AlphaView Q software (AlphaInnotech, San Leandro, CA), and averaged from at least 3 independent experiments.
The antibody for zebrafish RPE65c was raised against the synthesized peptide SDQFEKSKILVQF (residues from 217 to 229) from a specific region in zebrafish RPE65c. As was necessary, different combinations of antibodies were used to detect the 3 proteins at once. The primary antibodies were as follows: a rabbit anti-RPE65 polyclonal antibody (1:1000 dilution [54]), a rabbit anti-RPE65c polyclonal antibody (1:500, generated in this study), a mouse anti-6×His-tag monoclonal antibody (1:3000, Millipore, Billerica, MA), a mouse anti-RPE65 monoclonal antibody (1:5000, Millipore, Billerica, MA) and a goat anti-β-actin polyclonal antibody (1:1000, Santa Cruz, Santa Cruz, CA). The secondary antibodies were: donkey anti-goat IgG conjugated with DyLight 488, donkey anti-mouse IgG conjugated with DyLight 549, donkey anti-rabbit IgG conjugated with DyLight 649 (1:25000 dilution, Jackson ImmunoResearch, West Grove, PA) and horse anti-mouse IgG conjugated with Horseradish Peroxidase (Vector Labs, Burlingame, CA). When further protein detections on the same membrane were necessary, the membrane was stripped using a stripping buffer (Pierce, Rockford, IL) and re-blotted with the desired antibodies.
In vitro isomerohydrolase activity assay
The 293A cells were separately infected with adenoviruses expressing human RPE65, RPE65c or GFP as a negative control at MOI 100. The isomerohydrolase activity assay was carried out as described previously [17]. The blank HPLC profile (background) was obtained using proteins without substrate under the same reaction conditions and extraction procedures. The peak of each retinoid isomer was identified based on its characteristic retention time and the absorption spectrum of each retinoid standard. The isomerohydrolase activity was calculated from the area of the 11cROL and 13cROL peaks and expressed as mean ± SD from 3 independent measurements.
Retinal cell dissociation
The zebrafish eyes were enucleated, and the corneas and lens were removed. Ten retinas were carefully isolated from the eyecups and rinsed 3 times with PBS supplemented with 10 mM glucose. The retinas were treated with PBS containing papain (15 U/mL) and 10 mM glucose for 40 min at 25 °C. Then, the retinas were dissociated by gentle pipetting, passed through 70 µm mesh and centrifuged 1000×g for 5 min. Finally, the dissociated retinal cells were suspended with PBS, directly applied to positively charged glass slides (VWR, Radnor, PA) and air dried at 25 °C. The dissociated retinal cells on the slides were fixed with 4% paraformaldehyde in PBS for 20 min at 25 °C. Immunostaining was performed as explained in “Immunohistochemistry”.
Immunohistochemistry
The zebrafish eyes were fixed in 100 mM phosphate buffer containing 4% paraformaldehyde. The fixed tissues were used for frozen sections. Following blocking with 1% bovine serum albumin, the slides were double-stained with a rabbit anti-RPE65c polyclonal antibody (1:200 dilution) and a mouse anti-GS monoclonal antibody (1:1000 dilution, Millipore, Billerica, MA). After 3 washes, the slides were incubated with Cy3-labeled anti-rabbit IgG and Cy5-labeled anti-mouse IgG (1:200 dilution, Jackson ImmunoResearch Laboratories, West Grove, PA). Following 3 washes, the slides were treated with mounting medium containing DAPI (Vectorlab, San Diego, CA). The fluorescent signals were observed using a Zeiss Axio Observer Z1 (Carl Zeiss, Thromwood, NY).
Subcellular fractionation of RPE65c in cultured cells
The 293A cells expressing RPE65c were harvested and washed twice with ice-cold PBS. Subcellular fractionation analysis was performed as described previously [21]. Anti-RPE65 and anti-calnexin (ER membrane marker, 1:2500 dilution, Abcam, Cambridge, MA) antibodies were used to identify the subcellular localization of RPE65c and to verify the ER membrane preparation. Distribution of RPE65c in each fraction was analyzed by densitometry and expressed as the mean ± SD from 3 independent experiments.
Supplementary Material
Acknowledgements
We thank Dr. Tomoko Obara (University of Oklahoma Health Sciences Center) for providing the zebrafish, Dr. John Crabb for providing the CRALBP expression vector and Drs. Krysten Farjo and Anne R. Murray for critical review of the manuscript. This study was supported by NIH grants EY018659, EY012231, EY019309, a grant (P20RR024215) from the National Center For Research Resources, and a grant from OCAST.
Abbreviation
- 11cRAL
11-cis retinal
- atRAL
all-trans retinal
- RPE
retinal pigment epithelium
- atRE
all-trans retinyl ester
- 11cROL
11-cis retinol
- RPE65
retinal pigment epithelium specific 65-kDa protein
- RP
retinitis pigmentosa
- LCA
Leber’s congenital amaurosis
- RPE65a
zebrafish RPE65 (orthologue of human RPE65) in the RPE
- 13cIMH
13-cis isomerohydrolase (original name is RPE65b mainly expressing in the brain)
- RPE65c
an novel isoform of RPE65 expressed in the retina
- Ad-RPE65c
adenovirus expressing RPE65c
- MOI
multiplicity of infection
- GFP
green fluorescence protein
- 13cROL
13-cis retinol
- HPLC
high-performance liquid chromatography
- atROL
all-trans retinol
- GS
glutamine synthetase
- LRAT
lecithin retinol acyltransferase
- CRALBP
cellular retinaldehyde-binding protein
- 11cRE
11-cis retinyl ester
- ARAT
Acyl CoA-dependent retinol acyl transferase
- RT
reverse transcriptase
- BCO1
β-carotene 15,15’-monooxygenase
- DAPI
4',6-diamidino-2-phenylindole.
References
- 1.Bridges CDB. The rhodopsin-porphyropsin system. In: Dartnall HJA, editor. Handbook of Sensory Physiology. Berlin: Springer; 1972. pp. 417–480. [Google Scholar]
- 2.Ebrey T, Koutalos Y. Vertebrate photoreceptors. Prog Retin Eye Res. 2001;20:49–94. doi: 10.1016/s1350-9462(00)00014-8. [DOI] [PubMed] [Google Scholar]
- 3.McBee JK, Palczewski K, Baehr W, Pepperberg DR. Confronting complexity: the interlink of phototransduction and retinoid metabolism in the vertebrate retina. Prog Retin Eye Res. 2001;20:469–529. doi: 10.1016/s1350-9462(01)00002-7. [DOI] [PubMed] [Google Scholar]
- 4.Rando RR. The biochemistry of the visual cycle. Chem Rev. 2001;101:1881–1896. doi: 10.1021/cr960141c. [DOI] [PubMed] [Google Scholar]
- 5.Bernstein PS, Law WC, Rando RR. Biochemical characterization of the retinoid isomerase system of the eye. J Biol Chem. 1987;262:16848–16857. [PubMed] [Google Scholar]
- 6.Bernstein PS, Law WC, Rando RR. Isomerization of all-trans-retinoids to 11-cis-retinoids in vitro. Proc Natl Acad Sci USA. 1987;84:1849–1853. doi: 10.1073/pnas.84.7.1849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rando RR. Membrane phospholipids as an energy source in the operation of the visual cycle. Biochemistry. 1991;30:595–602. doi: 10.1021/bi00217a001. [DOI] [PubMed] [Google Scholar]
- 8.Moiseyev G, Chen Y, Takahashi Y, Wu BX, Ma JX. RPE65 Is the Isomerohydrolase in the Retinoid Visual Cycle. Proc Natl Acad Sci USA. 2005;102:12413–12418. doi: 10.1073/pnas.0503460102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Redmond TM, Poliakov E, Yu S, Tsai JY, Lu Z, Gentleman S. Mutation of key residues of RPE65 abolishes its enzymatic role as isomerohydrolase in the visual cycle. Proc Natl Acad Sci USA. 2005;102:13658–13663. doi: 10.1073/pnas.0504167102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jin M, Li S, Moghrabi WN, Sun H, Travis GH. Rpe65 is the retinoid isomerase in bovine retinal pigment epithelium. Cell. 2005;122:449–459. doi: 10.1016/j.cell.2005.06.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Moiseyev G, Takahashi Y, Chen Y, Gentleman S, Redmond TM, Crouch RK, Ma JX. RPE65 is an iron(II)-dependent isomerohydrolase in the retinoid visual cycle. J Biol Chem. 2006;281:2835–2840. doi: 10.1074/jbc.M508903200. [DOI] [PubMed] [Google Scholar]
- 12.Moiseyev G, Crouch RK, Goletz P, Oatis J, Jr, Redmond TM, Ma JX. Retinyl esters are the substrate for isomerohydrolase. Biochemistry. 2003;42:2229–2238. doi: 10.1021/bi026911y. [DOI] [PubMed] [Google Scholar]
- 13.Redmond TM, Yu S, Lee E, Bok D, Hamasaki D, Chen N, Goletz P, Ma JX, Crouch RK, Pfeifer K. Rpe65 is necessary for production of 11-cis-vitamin A in the retinal visual cycle. Nat Genet. 1998;20:344–351. doi: 10.1038/3813. [DOI] [PubMed] [Google Scholar]
- 14.Morimura H, Fishman GA, Grover SA, Fulton AB, Berson EL, Dryja TP. Mutations in the RPE65 gene in patients with autosomal recessive retinitis pigmentosa or leber congenital amaurosis. Proc Natl Acad Sci USA. 1998;95:3088–3093. doi: 10.1073/pnas.95.6.3088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Thompson DA, Gyurus P, Fleischer LL, Bingham EL, McHenry CL, Apfelstedt-Sylla E, Zrenner E, Lorenz B, Richards JE, Jacobson SG, et al. Genetics and phenotypes of RPE65 mutations in inherited retinal degeneration. Invest Ophthalmol Vis Sci. 2000;41:4293–4299. [PubMed] [Google Scholar]
- 16.Thompson DA, Gal A. Vitamin A metabolism in the retinal pigment epithelium: genes, mutations, and diseases. Prog Retin Eye Res. 2003;22:683–703. doi: 10.1016/s1350-9462(03)00051-x. [DOI] [PubMed] [Google Scholar]
- 17.Nikolaeva O, Takahashi Y, Moiseyev G, Ma JX. Purified RPE65 shows isomerohydrolase activity after reassociation with a phospholipid membrane. FEBS J. 2009;276:3020–3030. doi: 10.1111/j.1742-4658.2009.07021.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kiser PD, Golczak M, Lodowski DT, Chance MR, Palczewski K. Crystal structure of native RPE65, the retinoid isomerase of the visual cycle. Proc Natl Acad Sci USA. 2009;106:17325–17330. doi: 10.1073/pnas.0906600106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Takahashi Y, Moiseyev G, Chen Y, Ma JX. Identification of conserved histidines and glutamic acid as key residues for isomerohydrolase activity of RPE65, an enzyme of the visual cycle in the retinal pigment epithelium. FEBS Lett. 2005;579:5414–5418. doi: 10.1016/j.febslet.2005.09.002. [DOI] [PubMed] [Google Scholar]
- 20.Takahashi Y, Moiseyev G, Chen Y, Ma JX. The roles of three palmitoylation sites of RPE65 in its membrane association and isomerohydrolase activity. Invest Ophthalmol Vis Sci. 2006;47:5191–5196. doi: 10.1167/iovs.06-0614. [DOI] [PubMed] [Google Scholar]
- 21.Takahashi Y, Moiseyev G, Ablonczy Z, Chen Y, Crouch RK, Ma JX. Identification of a novel palmitylation site essential for membrane association and isomerohydrolase activity of RPE65. J Biol Chem. 2009;284:3211–3218. doi: 10.1074/jbc.M807248200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Baylor DA. Photoreceptor signals and vision. Proctor lecture. Invest Ophthalmol Vis Sci. 1987;28:34–49. [PubMed] [Google Scholar]
- 23.Kawamura S, Tachibanaki S. Rod and cone photoreceptors: molecular basis of the difference in their physiology. Comp Biochem Physiol A Mol Integr Physiol. 2008;150:369–377. doi: 10.1016/j.cbpa.2008.04.600. [DOI] [PubMed] [Google Scholar]
- 24.Das SR, Bhardwaj N, Kjeldbye H, Gouras P. Muller cells of chicken retina synthesize 11-cis-retinol. Biochem J. 1992;285:907–913. doi: 10.1042/bj2850907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Muniz A, Villazana-Espinoza ET, Hatch AL, Trevino SG, Allen DM, Tsin AT. A novel cone visual cycle in the cone-dominated retina. Exp Eye Res. 2007;85:175–184. doi: 10.1016/j.exer.2007.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mata NL, Ruiz A, Radu RA, Bui TV, Travis GH. Chicken retinas contain a retinoid isomerase activity that catalyzes the direct conversion of all-trans-retinol to 11-cis-retinol. Biochemistry. 2005;44:11715–11721. doi: 10.1021/bi050942m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mata NL, Radu RA, Clemmons RC, Travis GH. Isomerization and oxidation of vitamin a in cone-dominant retinas: a novel pathway for visual-pigment regeneration in daylight. Neuron. 2002;36:69–80. doi: 10.1016/s0896-6273(02)00912-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bringmann A, Pannicke T, Grosche J, Francke M, Wiedemann P, Skatchkov SN, Osborne NN, Reichenbach A. Muller cells in the healthy and diseased retina. Prog Retin Eye Res. 2006;25:397–424. doi: 10.1016/j.preteyeres.2006.05.003. [DOI] [PubMed] [Google Scholar]
- 29.Pandey S, Blanks JC, Spee C, Jiang M, Fong HK. Cytoplasmic retinal localization of an evolutionary homolog of the visual pigments. Exp Eye Res. 1994;58:605–613. doi: 10.1006/exer.1994.1055. [DOI] [PubMed] [Google Scholar]
- 30.Wu BX, Moiseyev G, Chen Y, Rohrer B, Crouch RK, Ma JX. Identification of RDH10, an All-trans Retinol Dehydrogenase, in Retinal Muller Cells. Invest Ophthalmol Vis Sci. 2004;45:3857–3862. doi: 10.1167/iovs.03-1302. [DOI] [PubMed] [Google Scholar]
- 31.Bunt-Milam AH, Saari JC. Immunocytochemical localization of two retinoid-binding proteins in vertebrate retina. J Cell Biol. 1983;97:703–712. doi: 10.1083/jcb.97.3.703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bok D, Ong DE, Chytil F. Immunocytochemical localization of cellular retinol binding protein in the rat retina. Invest Ophthalmol Vis Sci. 1984;25:877–883. [PubMed] [Google Scholar]
- 33.Wang JS, Estevez ME, Cornwall MC, Kefalov VJ. Intra-retinal visual cycle required for rapid and complete cone dark adaptation. Nat Neurosci. 2009;12:295–302. doi: 10.1038/nn.2258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang JS, Kefalov VJ. An alternative pathway mediates the mouse and human cone visual cycle. Curr Biol. 2009;19:1665–1669. doi: 10.1016/j.cub.2009.07.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Neuhauss SC. Behavioral genetic approaches to visual system development and function in zebrafish. J Neurobiol. 2003;54:148–160. doi: 10.1002/neu.10165. [DOI] [PubMed] [Google Scholar]
- 36.Tsujikawa M, Malicki J. Genetics of photoreceptor development and function in zebrafish. Int J Dev Biol. 2004;48:925–934. doi: 10.1387/ijdb.041890mt. [DOI] [PubMed] [Google Scholar]
- 37.Fleisch VC, Neuhauss SC. Parallel visual cycles in the zebrafish retina. Prog Retin Eye Res. 2010;29:476–486. doi: 10.1016/j.preteyeres.2010.05.001. [DOI] [PubMed] [Google Scholar]
- 38.Doerre G, Malicki J. Genetic analysis of photoreceptor cell development in the zebrafish retina. Mech Dev. 2002;110:125–138. doi: 10.1016/s0925-4773(01)00571-8. [DOI] [PubMed] [Google Scholar]
- 39.Schonthaler HB, Lampert JM, Isken A, Rinner O, Mader A, Gesemann M, Oberhauser V, Golczak M, Biehlmaier O, Palczewski K, et al. Evidence for RPE65-independent vision in the cone-dominated zebrafish retina. Eur J Neurosci. 2007;26:1940–1949. doi: 10.1111/j.1460-9568.2007.05801.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Takahashi Y, Moiseyev G, Chen Y, Farjo KM, Nikolaeva O, Ma JX. An Enzymatic Mechanism for Generating the Precursor of Endogenous 13-cis Retinoic Acid in the Brain. FEBS J. 2011;278:973–987. doi: 10.1111/j.1742-4658.2011.08019.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yoshizawa T, Imamoto Y. Structure and photobleaching process of chicken iodopsin. Biophys Chem. 1995;56:57–62. doi: 10.1016/0301-4622(95)00015-p. [DOI] [PubMed] [Google Scholar]
- 42.Shichida Y, Imai H, Imamoto Y, Fukada Y, Yoshizawa T. Is chicken green-sensitive cone visual pigment a rhodopsin-like pigment? A comparative study of the molecular properties between chicken green and rhodopsin. Biochemistry. 1994;33:9040–9044. doi: 10.1021/bi00197a002. [DOI] [PubMed] [Google Scholar]
- 43.Moiseyev G, Takahashi Y, Chen Y, Kim S, Ma JX. RPE65 from Cone-dominant Chicken Is a More Efficient Isomerohydrolase Compared with That from Rod-dominant Species. J Biol Chem. 2008;283:8110–8117. doi: 10.1074/jbc.M703654200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bok D, Ruiz A, Yaron O, Jahng WJ, Ray A, Xue L, Rando RR. Purification and characterization of a transmembrane domain-deleted form of lecithin retinol acyltransferase. Biochemistry. 2003;42:6090–6098. doi: 10.1021/bi0342416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Muniz A, Villazana-Espinoza ET, Thackeray B, Tsin AT. 11-cis-Acyl-CoA:retinol O-acyltransferase activity in the primary culture of chicken Muller cells. Biochemistry. 2006;45:12265–12273. doi: 10.1021/bi060928p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Fleisch VC, Schonthaler HB, von Lintig J, Neuhauss SC. Subfunctionalization of a retinoid-binding protein provides evidence for two parallel visual cycles in the cone-dominant zebrafish retina. J Neurosci. 2008;28:8208–8216. doi: 10.1523/JNEUROSCI.2367-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Collery R, McLoughlin S, Vendrell V, Finnegan J, Crabb JW, Saari JC, Kennedy BN. Duplication and divergence of zebrafish CRALBP genes uncovers novel role for RPE- and Muller-CRALBP in cone vision. Invest Ophthalmol Vis Sci. 2008;49:3812–3820. doi: 10.1167/iovs.08-1957. [DOI] [PubMed] [Google Scholar]
- 48.Farjo KM, Moiseyev G, Takahashi Y, Crouch RK, Ma JX. The 11-cis-retinol dehydrogenase activity of RDH10 and its interaction with visual cycle proteins. Invest Ophthalmol Vis Sci. 2009;50:5089–5097. doi: 10.1167/iovs.09-3797. [DOI] [PubMed] [Google Scholar]
- 49.Walker MT, Brown RL, Cronin TW, Robinson PR. Photochemistry of retinal chromophore in mouse melanopsin. Proc Natl Acad Sci USA. 2008;105:8861–8865. doi: 10.1073/pnas.0711397105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Muniz A, Betts BS, Trevino AR, Buddavarapu K, Roman R, Ma JX, Tsin AT. Evidence for two retinoid cycles in the cone-dominated chicken eye. Biochemistry. 2009;48:6854–6863. doi: 10.1021/bi9002937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kozak M. An analysis of 5'-noncoding sequences from 699 vertebrate messenger RNAs. Nucleic Acids Res. 1987;15:8125–8148. doi: 10.1093/nar/15.20.8125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kumar S, Nei M, Dudley J, Tamura K. MEGA: a biologist-centric software for evolutionary analysis of DNA and protein sequences. Brief Bioinform. 2008;9:299–306. doi: 10.1093/bib/bbn017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 54.Ma JX, Zhang J, Othersen KL, Moiseyev G, Ablonczy Z, Redmond TM, Chen Y, Crouch RK. Expression, purification, and MALDI analysis of RPE65. Invest Ophthalmol Vis Sci. 2001;42:1429–1435. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.