Abstract
Activation of the epidermal growth factor receptor (EGFR) in glioblastoma (GBM) occurs through mutations or deletions in the extracellular (EC) domain. Unlike lung cancers with EGFR kinase domain (KD) mutations, GBMs respond poorly to the EGFR inhibitor erlotinib. Using RNAi, we show that GBM cells carrying EGFR EC mutations display EGFR addiction. In contrast to KD mutants found in lung cancer, glioma-specific EGFR EC mutants are poorly inhibited by EGFR inhibitors that target the active kinase conformation (e.g., erlotinib). Inhibitors which bind to the inactive EGFR conformation, on the other hand, potently inhibit EGFR EC mutants and induce cell death in EGFR mutant GBM cells. Our results provide first evidence for single kinase addiction in GBM, and suggest that the disappointing clinical activity of first-generation EGFR inhibitors in GBM versus lung cancer may be attributed to the different conformational requirements of mutant EGFR in these two cancer types.
INTRODUCTION
Glioblastoma (GBM) is the most common malignant brain tumor in adults. Most GBM patients succumb to their disease within two years and there is a dire need for the development of novel therapeutics (1). Inhibitors of deregulated signaling pathways are active agents in a variety of human cancers (2, 3) and represent a compelling area of drug development for GBM because many of these tumors harbor genetic alterations in growth factor signaling pathways (4, 5).
The epidermal growth factor receptor (EGFR) is a member of the EGFR family of receptor tyrosine kinases which also includes HER2 (ErbB2), HER3 (ErbB3), and HER4 (ErbB4) (6). EGFR has generated particular interest as a drug target in GBM because of the high frequency of EGFR alterations in this disease (7) and because ATP-site competitive EGFR kinase inhibitors are active agents in patients with EGFR-mutant lung cancer (8). EGFR kinase inhibitors which received regulatory approval for the treatment of lung cancer (erlotinib, gefitinib), however, have shown disappointing results in patients with GBM (9). Reasons for this lack of response in GBM remain poorly understood and include redundancy in signaling pathways (10) and intratumoral heterogeneity (11).
One key difference between EGFR in GBM and lung cancer is the distribution of mutations within the EGFR coding sequence. EGFR mutations in lung cancer reside in the intracellular kinase domain (KD) (12). EGFR mutations in GBM cluster in the extracellular (EC) domain and include in-frame deletions (such as the common “variant III”) (7) and missense mutations (13)(Fig. 1A). Both EGFR ectodomain and kinase domain mutations encode oncoproteins with the ability to transform NIH-3T3 cells in the absence of ligand (13–15). In this study, we examined the role of EGFR for the survival of GBM cells harboring EGFR ectodomain mutations. We demonstrate that EGFR signals are essential for the survival of these cells and that EGFR EC mutants differ markedly from EGFR KD mutants in their sensitivity to ATP-site competitive EGFR kinase inhibitors.
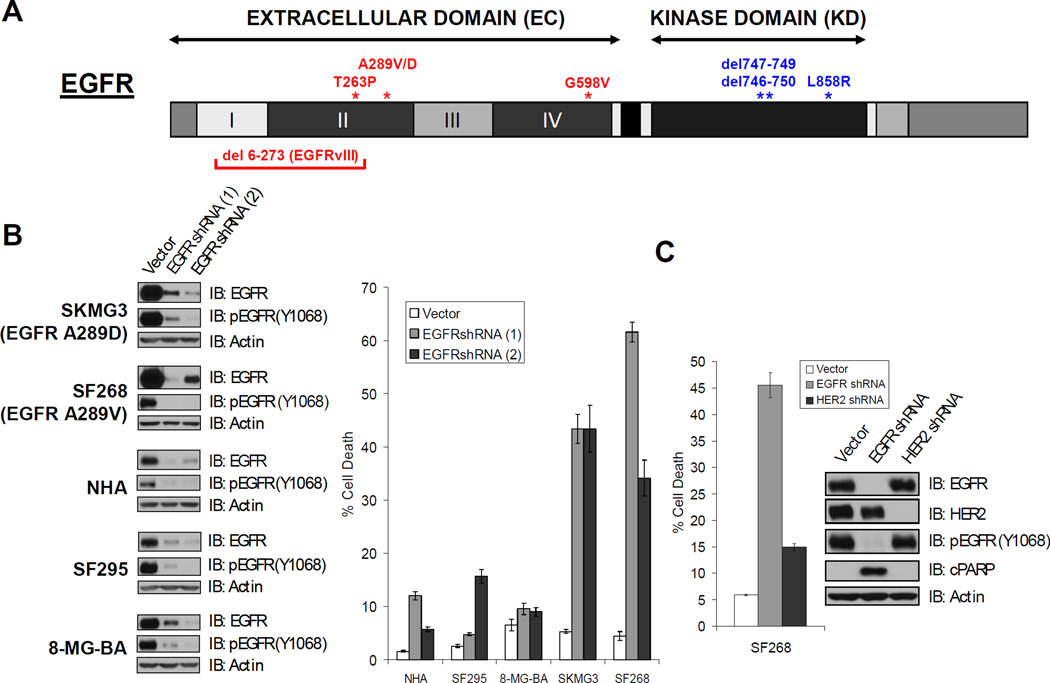
FIGURE 1. EGFR-knockdown induces cell death in GBM cells with EGFR EC mutations.
A. EGFR domain structure. Mutations indicated in red have been documented in glioblastoma (GBM) but not in lung cancer, while those indicated in blue are seen in non-small cell lung cancer (NSCLC) and not in GBM. Roman numerals indicate subdomains within the EC domain. B. EGFR-mutant GBM lines are sensitive to EGFR knockdown. The indicated cell lines were acutely transduced with control or two different EGFR-targeted shRNAs. The extent of EGFR knockdown was assessed by immunoblot (left panel). The effects of the hairpins on cell death was assessed by trypan blue exclusion 5 days post-infection (right panel). (NHA, normal human astrocytes). C. HER2 knockdown only induces minimal cell death in EGFR mutant SF268 GBM cells. Cells were acutely transduced with control, EGFR-targeted, or HER2-targeted shRNAs. The extent of EGFR and HER2 knockdown was evaluated by immunoblot (inset). Cell death was assessed as in B. Confer Suppl. Figure 2 for results of HER2 knockdown in EGFR mutant SKMG3 GBM cells.
RESULTS
1. EGFR mutant GBM cells are EGFR addicted
Missense mutations in the EGFR extracellular (EC) domain are found in 10–15 % of GBMs (4, 5, 13). To determine whether EGFR signals are essential for the survival of GBM cells endogenously expressing such mutations, we first sequenced the coding region of EGFR in a panel of GBM cell lines. We found two lines with EGFR EC mutations. Both mutations resulted in amino acid substitutions at alanine 289, the most common site of extracellular EGFR missense mutations in human GBMs (Fig. 1A). Alanine was substituted by valine (A289V) in SF268 cells and by aspartic acid (A289D) in SKMG3 cells (Suppl. Figure 1). We tested whether depletion of the EGFR protein was sufficient to induce cell death in these lines. Acute infection of SKMG3 and SF268 cells with retroviral shRNA constructs targeting two distinct areas of the EGFR mRNA resulted in loss of EGFR protein expression within 72 hours of infection and robust cell death induction after 5 days. EGFR knockdown in human astrocytes (NHAs)(16) and two GBM cell lines without EGFR mutation (SF295, 8-MG-BA) did not induce cell death (Fig. 1B). Of note, SKMG3 cells do not express the tumor suppressor protein Phosphatase and Tensin homolog (PTEN), confirming our earlier findings that PTEN inactivation is not sufficient to relieve EGFR mutant cancer cells from their dependence on EGFR for survival (17).
We conducted similar experiments with shRNA constructs targeting the EGF receptor family member HER2 because HER2 can heterodimerize with EGFR and transmit oncogenic signals in certain cellular contexts (18). HER2 knockdown did not induce a significant amount of cell death as measured by the trypan-blue dye exclusion assay and immunoblotting for the cleaved Caspase3 substrate Poly (ADP-ribose) polymerase (PARP) (Fig. 1C and Suppl. Fig. 2). HER2 depletion also did not affect EGFR phosphorylation at tyrosine 1068, suggesting that basal EGFR phosphorylation in SF268 and SKMG3 cells is not the result of trans-phosphorylation by the HER2 kinase.
Several prosurvival functions of EGFR have been attributed to kinase independent properties of the receptor protein (19). To assess whether EGFR kinase activity is required for the survival of SKMG3 (A289D-EGFR) and SF268 (A289V-EGFR) cells, we treated them with the “second-generation” EGFR kinase inhibitor HKI-272 (20). This drug irreversibly inhibits EGFR (and other ErbB family members) because it forms covalent interactions with cysteines in the ATP cleft of the kinase domain (21). HKI-272 induced cell death in SF268 and SKMG3 cells, but not in EGFR wildtype GBM (SF295, 8-MG-BA), lung cancer cells (H460), or human astrocytes (NHAs)(Fig. 2A).
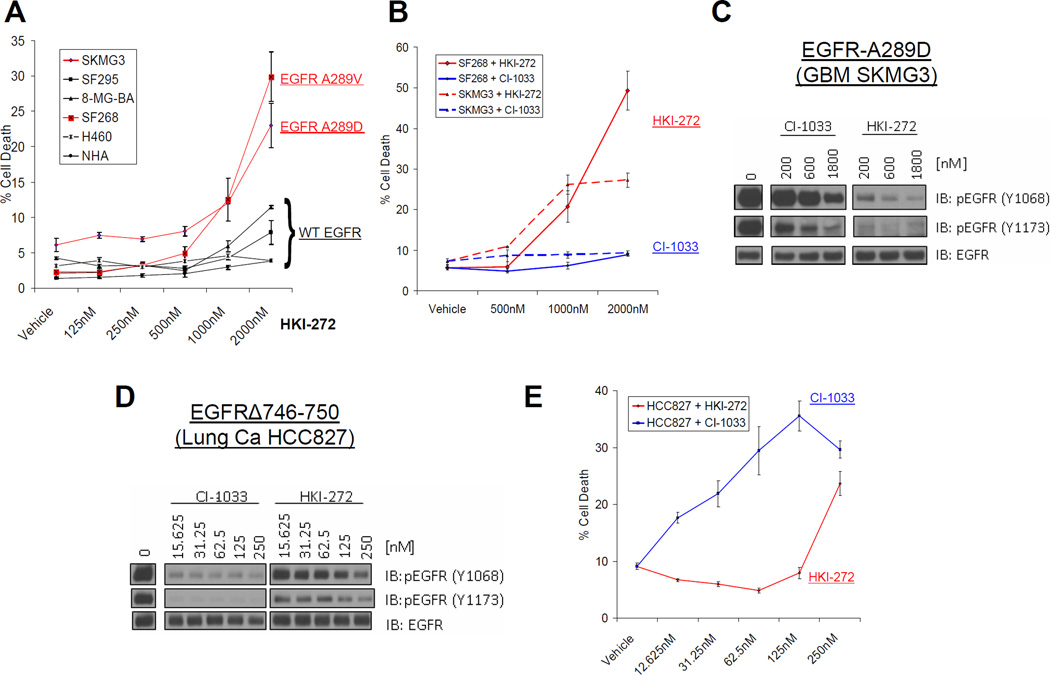
FIGURE 2. Differential sensitivity of EGFR mutant glioma and lung cancer cell lines to the irreversible EGFR inhibitors HKI-272 and CI-1033.
A. HKI-272 induces cell death in GBM cells with EGFR EC mutation (SKMG3, SF268), but not EGFR wildtype cancer cell lines or astrocytes (NHA). Cell death was assessed by trypan blue exclusion following five days of inhibitor treatment. Cells lines in black express wild type EGFR, while those in red contain EGFR EC mutations. B. CI-1033, unlike HKI-272, does not induce cell death in GBM cells with EGFR EC mutation. SF268 (solid) or SKMG3 (dashed) cells were treated with the indicated doses of HKI-272 (red) or CI-1033 (blue) for 5 days. Cell death was evaluated at day 5 by trypan blue exclusion. C. HKI-272 is more potent than CI-1033 in blocking EGFR phosphorylation in SKMG3 cells with EGFR EC mutation. SKMG3 cells were treated with the indicated doses of CI-1033 or HKI-272 and whole lysates were analyzed by immunoblot with the indicated antibodies. D. CI-1033 is more potent than HKI-272 in blocking EGFR phosphorylation in HCC827 lung cancer cells harboring the (Δ746-750) EGFR kinase domain mutant. HCC827 were treated with the indicated doses of HKI-272 or CI-1033. Lysates of these cells were made and analyzed by immunoblot as indicated. E. CI-1033 is more potent than HKI-272 in inducing cell death in HCC827 lung cancer cells. Cell death was also assessed five days after treatment as before.
To extend our observations with HKI-272 to a second EGFR kinase inhibitor, we repeated our experiments with CI-1033. Like HKI-272, CI-1033 is an irreversible, ATP-site competitive inhibitor of ErbB receptors and inhibits phosphorylation of wildtype EGFR in intact cells with similar potency (IC50: 7.4 nM)(22) as HKI-272 (IC50: 3 nM)(20). To our surprise, CI-1033 failed to induce cell death in either SF268 or SKMG3 cells (Fig. 2B). Immunoblots of whole cell lysates from SKMG3 cells treated with either inhibitor showed that CI-1033 inhibited EGFR phosphorylation less effectively than HKI-272 (Fig. 2C).
We wondered whether the differential effect of HKI-272 and CI-1033 on EGFR was unique to GBM cells with EGFR EC mutations. We therefore also compared the activity of both compounds in HCC827 lung cancer cells which harbor a deletion in the EGFR kinase domain (EGFRΔ746-750). In contrast to our findings in GBM cells, CI-1033 more potently inhibited EGFR phosphorylation (Fig. 2D) and more potently induced cell death (Fig. 2E) than HKI-272. Both inhibitors induced cell death at submicromolar concentrations in HCC827 cells, consistent with the reported hypersensitivity of the EGFRΔ746-750 mutant to ATP-site competitive EGFR kinase inhibitors in vitro and in lung cancer patients (23–26). In summary, these results indicate that EGFR mutant GBM cell lines require EGFR kinase activity for survival and point toward differences in EGFR kinase inhibitor responsiveness between EGFR ectodomain mutants and EGFR kinase domain mutants.
2. Enhanced sensitivity of EGFR ectodomain mutants to lapatinib
Crystal structures of the EGFR catalytic domain in complex with ATP-site competitive EGFR kinase inhibitors have identified different receptor conformations (27, 28). In complex with the FDA-approved drug lapatinib/GW572016 (tykerb), the EGFR kinase domain is in an inactive conformation (also called “type II” conformation) (29). In complex with erlotinib/OSI-74 (tarceva), the EGFR kinase domain adopts an active (or “type I”) conformation (30)(Fig. 3A). Since HKI-272 binds the inactive conformation of the EGFR kinase domain (“type II”)(31) and CI-1033 likely binds the active conformation (based on analogy to compounds with similar chemical structure), we hypothesized that conformation-specific binding to EGFR might explain the differential response of GBM cell lines with EGFR EC mutants to these two compounds. If correct, lapatinib (“type II inhibitor”) should also show superior activity against EGFR EC mutants than erlotinib (“type I inhibitor”).
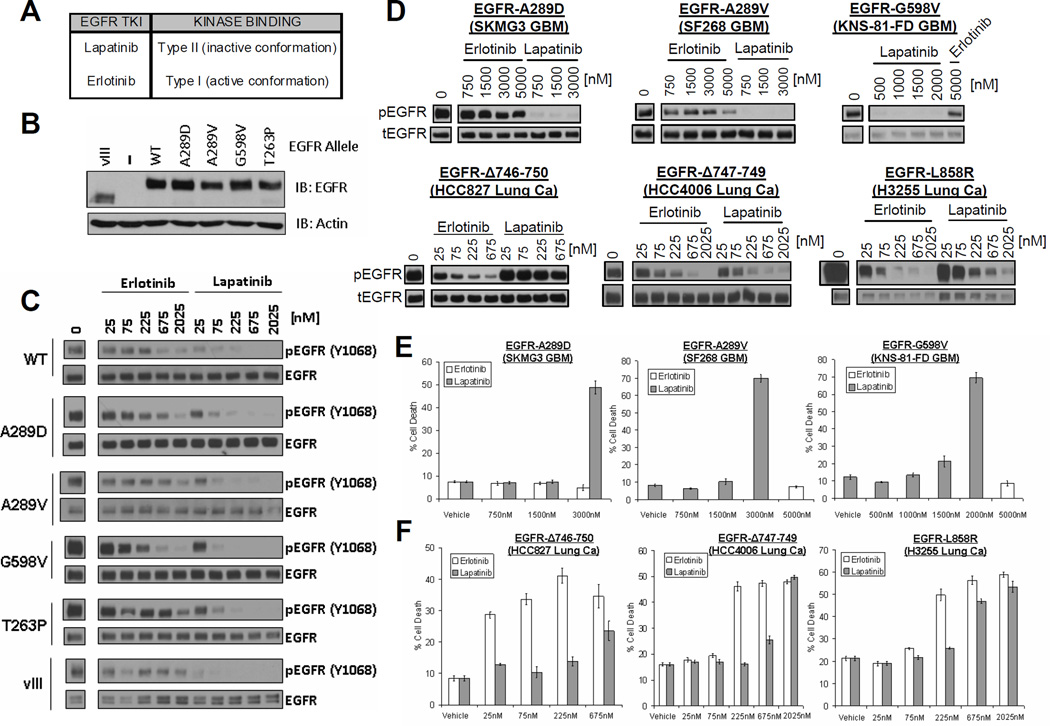
FIGURE 3. Differential sensitivity of glioma- versus lung cancer-specific EGFR mutants to the reversible EGFR inhibitors lapatinib and erlotinib.
A. Classification of reversible ATP-competitive EGFR kinase inhibitors based on crystal structures. B. Ectopic expression of glioma-specific EGFR EC mutants in EGFR-deficient (EGFR-NEG) NR6 fibroblasts. Shown are immunoblots of whole cell lystates. C. Enhanced sensitivity of EGFR EC mutants to lapatinib in NR6 cells. NR6 cells expressing the indicated alleles of EGFR were treated with increasing doses of erlotinib or lapatinib as shown. Cell lysates were made and analyzed by immunoblot with the indicated antibodies. D. Differential sensitivity of endogenously expressed glioma- versus lung cancer-specific EGFR mutants to lapatinib versus erlotinib. The indicated GBM (upper panels) and lung cancer (Lung Ca, lower panels) cell lines were treated with various doses of lapatinib or erlotinib. Cells were harvested and analyzed by western blot using antibodies for tyrosine-phosphorylated EGFR (pEGFR, top) or total EGFR (tEGFR, bottom). E and F. Differential cell death response of GBM (E) and lung cancer (F) cell lines to lapatinib versus erlotinib. Cell death was assessed by trypan blue exclusion assay after 5 days of incubation with the indicated drug.
To examine this question, we first expressed several EGFR ectodomain mutants in NR6 fibroblasts which do not detectably express EGFR or other ErbB family members and are widely used for the biochemical characterization of EGFR family members (24, 32, 33). After deriving stable sublines for each EGFR allele (Fig. 3B), we examined changes in EGFR phosphorylation in response to equimolar concentrations of erlotinib or lapatinib. While both inhibitors lowered EGFR phosphorylation in a dose-dependent fashion, lapatinib showed significantly greater potency against all examined EGFR ectodomain mutants (A289D, A289D, G598V, T263P, vIII) and, less dramatically, also against wildtype EGFR (Fig. 3C). We obtained similar results in human astrocytes which do express endogenous wildtype EGFR and which we further engineered to overexpress either wildtype EGFR or the two most common EGFR ectodomain mutants in GBM (A289V and EGFRvIII)(Suppl. Figure 3).
We next extended our comparison between lapatinib and erlotinib to GBM cell lines endogenously expressing EGFR ectodomain mutants. These included SKMG3 (A289D EGFR) and SF268 (A289V EGFR) cells as well as a third line (KNS-81-FD) recently reported to harbor the G598V EGFR ectodomain mutant (COSMIC database). To benchmark our results against previous work on EGFR kinase domain mutants, our experiments also included the lung cancer cell lines HCC827 (EGFRΔ746-750), HCC4006 (EGFRΔ747-749), and H3255 (EGFR L858R). Similar to our results in NR6 cells and astrocytes, lapatinib was more potent than erlotinib at inhibiting basal phosphorylation of all examined EGFR ectodomain mutants. Erlotinib, on the other hand, was more potent than lapatinib at inhibiting EGFR in lung cancer cell lines with the EGFR kinase domain mutants EGFRΔ746-750 and EGFR L858R (Fig. 3D), consistent with previous studies (34). Akt and Erk, two well-documented effector kinases of the examined EGFR kinase domain mutants, were also more potently inhibited by erlotinib compared to lapatinib in these lines (Suppl. Fig. 4A–B). Interestingly, inhibition of EGFR in SKMG3 GBM cells did not result in Akt or Erk inhibition, suggesting that the A289D mutant utilizes other downstream effector pathways (Suppl. Fig. 4C).
We also examined the effects of lapatinib and erlotinib on cell death. Lapatinib, but not erlotinib, induced cell death in all examined GBM cell lines with EGFR ectodomain mutants (Fig. 3E). In EGFR mutant lung cancer cell lines, erlotinib induced cell death at lower concentrations than lapatinib (Fig. 3F).
3. Type II EGFR inhibitors effectively displace ATP from EGFR EC mutants
Our results with four different EGFR kinase inhibitors suggested that the catalytic domain of EGFR ectodomain mutants might favor an inactive-like conformation that is more accessible to lapatinib or HKI-272 than to erlotinib or CI-1033. To further test this model, we developed an assay that measures the ability of EGFR kinase inhibitors to compete in whole cell lysates with ATP for binding to the ATP-cleft of the EGFR kinase domain (Fig. 4A).
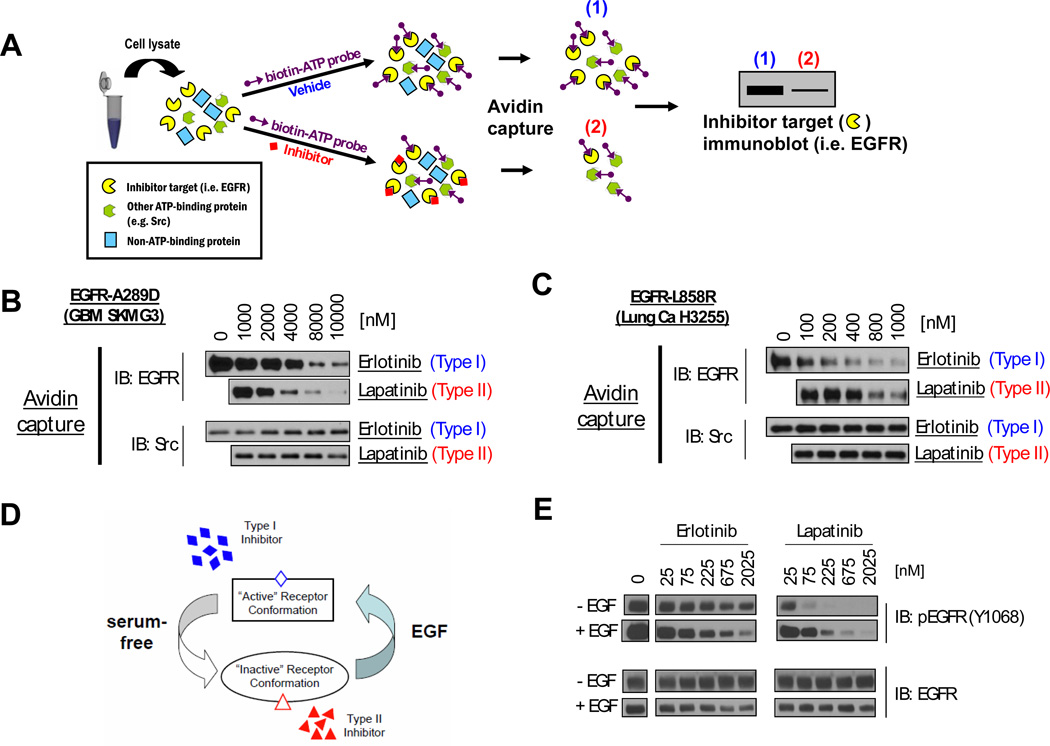
FIGURE 4. Type II EGFR kinase inhibitors more effectively displace ATP from the EGFR kinase domain of EGFR ectodomain mutants than type I inhibitors.
A. Schematic of ATP competition-binding assessment. Lysates of cells expressing the ATP-binding protein under study are simultaneously treated with a targeted agent (e.g. EGFR TKI) or vehicle, and a biotinylating ATP probe. All ATP-binding proteins will be biotinylated unless the ATP-binding pocket is occupied (e.g. with EGFR TKI). Lysates are subjected to avidin pulldown. The ability of the test compound to compete with ATP for binding to the target protein is assessed by immunoblot of the pulldown using antibodies against the target protein (e.g. EGFR). B. and C. Results of ATP competition assay in lysates from (B) EGFR EC mutant GBM cells and (C) EGFR KD mutant lung cancer cells. (B) Lapatinib more effectively competes with ATP for binding to the EGFR-TK in SKMG3 (EGFR A289D) cell lysates than erlotinib. (C.) Erlotinib more effectively competes with ATP for binding to the EGFR-TK in H3255 cell lysates (EGFR L858R KD mutation) than lapatinib. Cell lysates were carried through the assay described in A. The ATP probe was competed with the indicated doses of erlotinib or lapatinib. Following the avidin pulldown, samples were analyzed by immunoblot with antibodies for EGFR. Immunoblots were also probed with antibodies for Src as a control. D. A model of ligand-induced changes in EGFR conformation. In the absence of ligand (serum-free), the conformational equilibrium of EGFR-EC mutants is shifted towards to “inactive” conformation which is preferentially bound by type II inhibitors. In ligand-occupied receptor (EGF), the conformational equilibrium shifts towards the “active” conformation, which is the preferred substrate of type I inhibitors. E. EGF “desensitizes” the A289D EGFR EC mutant from lapatinib and “sensitizes” it to erlotinib. SKMG3 GBM cells were serum-starved, stimulated with EGF or vehicle, and subsequently treated with the indicated doses of erlotinib and lapatinib (while still under EGF treatment). Cell were lysed 30 minutes of drug treatment and analyzed by immunoblot with the indicated antibodies.
Coincubation of whole cell lysates from A289D-EGFR mutant SKMG3 cells with biotinylated ATP and erlotinib demonstrated decreased ATP-binding with increasing erlotinib concentrations. Coincubation of a replicate sample of the same whole cell lysate with increasing concentrations of lapatinib blocked ATP binding at lower concentrations of lapatinib than erlotinib. As a specificity control, we determined ATP binding to the kinase domain of SRC and found no displacement of ATP-binding by either lapatinib or erlotinib (Fig. 4B). We also repeated these experiments with whole cell lysates from H3255 lung cancer cells (EGFR-L858R KD mutant), and found that erlotinib blocked ATP binding to the EGFR kinase domain more effectively than lapatinib (Fig. 4C).
Since differences in off-rates between the reversible EGFR kinase inhibitors lapatinib and erlotinib might affect results of the ATP-competition assay, we performed additional experiments with the irreversible EGFR kinase inhibitors CI-1033 and HKI-272. In whole cell lysates from A289D-EGFR SKMG3 cells, HKI-272 more effectively blocked ATP binding to the EGFR kinase domain than CI-1033 (Suppl. Fig. 6), consistent with our model. Lastly, we explored whether a forced change in receptor conformation, induced by ligand binding, might alter the ability of EGFR inhibitors to gain access to the kinase domain (Fig. 4D) and block EGFR phosphorylation. We were able to examine this question in SKMG3 cells harboring the EGFR-A289D mutant, because we had previously shown that this mutant, unlike EGFRvIII, does not abrogate the ability of EGFR to respond to EGF (13). When we treated EGFR A289D-mutant SKMG3 cells with lapatinib or erlotinib in the presence of EGF, we indeed found that EGF “desensitized” EGFR to lapatinib and sensitized EGFR to erlotinib: higher lapatinib (right-shift) and lower erlotinib (left-shift) concentrations were required to achieve a similar degree of EGFR inhibition than in the absence of EGF (Fig. 4E). We obtained similar results in receptor-negative NR6 cells reconstituted with EGFR-A289D (Suppl. Fig 5).
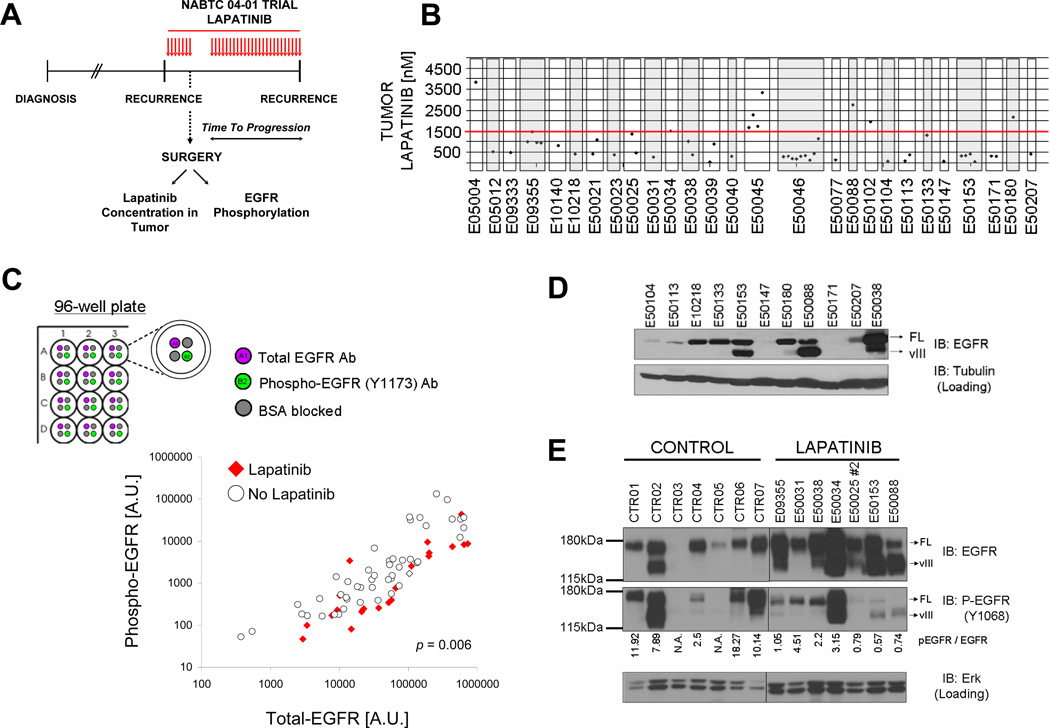
4. Lapatinib fails to achieve sufficient intratumoral concentrations in GBM patients
Clinical trials with type I EGFR kinase inhibitors (erlotinib, gefitinib) in GBM demonstrated poor inhibition of the EGFR signaling axis in tumor tissue (35, 36). To determine the ability of lapatinib to penetrate into GBM tumor tissue and inhibit EGFR phosphorylation, we conducted a multicenter clinical trial in which patients received 750 mg of lapatinib orally for 7 days prior to a surgical procedure that was required for tumor recurrence (Fig. 5A)(Suppl. Information). 44 patients with recurrent GBM enrolled into the study and underwent surgery (Suppl. Tables 1 and 2). Lapatinib was generally well tolerated (Suppl. Table 3 and Suppl. Information).
FIGURE 5. Lapatinib fails to achieve sufficient intratumoral concentrations in GBM patients.
A. Design of the multicenter NABTC 04-01 biomarker trial for patients with recurrent GBM. GBM patients requiring tumor resection for recurrent disease received preoperative lapatinib (750mg p.o. BID). Lapatinib concentrations in tumor tissue and EGFR phosphorylation were assessed in surgical specimens. See Suppl. Information for details. B. Intratumoral lapatinib concentrations in GBM are below lapatinib concentrations required to induce cell death in EGFR mutant GBM cells (red line, 1.5µM). C. Incomplete inhibition of EGFR phosphorylation in GBM tumor tissue by lapatinib. Shown are levels of total EGFR (x-axis) and phosphorylated EGFR (y-axis) in tumors from GBM patients who received preoperative lapatinib (red diamonds) versus tumors from GBM patients who did not receive any EGFR kinase inhibitor prior to surgery (empty circles). Levels of tEGFR phosphorylation and total levels were measured concurrently with a multi-array immunoassay using electrochemiluminescence detection (see top panel). D. EGFR expression in GBMs from patients enrolled in the NABTC 04-01 study. Shown are immunoblots of tumor lysates probed with EGFR or a loading control. Full length (FL) and truncated EGFRvIII (vIII) mutant forms of EGFR are indicated by arrows. E. Incomplete EGFR inhibition in GBM tumor tissue by lapatinib. Shown are immunoblots of EGFR overexpressing GBMs on the NABTC 04-01 study (labeled “LAPATINIB”) versus tumors from GBM patients who did not receive an EGFR kinase inhibitor prior to surgery (“CONTROL”). Total levels of EGFR and EGFR tyrosine phosphorylation were analyzed by immunoblot. Total Erk levels were also examined as a loading control.
Lapatinib concentrations in the plasma sample collected during surgery varied considerably between patients (0.188 to 2.319 µg/mL)(Suppl. Table 4) with mean plasma concentrations (mean ± SD: 1.203 ± 0.518 µg/mL) similar to plasma levels reported in the literature for this dosing schedule (37). Tumor concentrations of lapatinib varied considerably between patients (70-3826 nM). The median concentrations for the entire cohort (497 nM) was above the IC50 for inhibition of EGFR phosphorylation (~ 200nM) but below drug concentrations reported to induce cell death in cancer cell lines (see below) (38) (Fig. 5B)(Suppl. Table 4 and 5).
We assessed EGFR phosphorylation on tyrosine 1173 in all patient samples for which residual frozen tumor was available and compared it to EGFR phosphorylation in 49 tumor samples from GBM patients who had not received any EGFR kinase inhibitor prior to surgery (referred to as “no lapatinib” controls)(Suppl. Table 6). Since EGFR levels in GBM range over two to three orders of magnitude (39), we chose an electrochemiluminescent detection method with a broad linear range of detection (MSD mesoscale). This platform offered the additional advantage that it allowed us to determine total and phospho-EGFR signal for each sample in a single well and run all clinical trial and control samples together in a 96-well format. Compared to control samples (Fig. 5C, empty circles), the group of lapatinib-treated tumors (Fig. 5C, red diamonds) showed less EGFR phosphorylation per total EGFR signal (p=0.006). However, all lapatinib-treated tumors showed residual EGFR phosphorylation above levels seen in lapatinib-naïve tumors not overexpressing EGFR.
For all tumors with sufficient residual sample, we also performed immunoblot analysis (n=27). EGFR immunoblot analysis showed EGFR overexpression in 12/27 (44.4 %) tumors; a 140 KDa band, consistent with the EGFRvIII deletion, was detected in 7/27 (25.9 %) of tumors, all within the group of tumors overexpressing EGFR (examples are shown in Fig. 5D). Only one of these tumors (1/27)(E0038) harbored a missense mutation in the EGFR ectodomain (T263P)(Suppl. Table 7). A comparison of EGFR phosphorylation between lapatinib treated tumors with EGFR overexpression and control tumors showed that lapatinib-treated GBMs showed lower levels of EGFR phosphorylation than controls with similar levels of EGFR overexpression (Fig. 5E). All lapatinib treated tumors showed residual EGFR phosphorylation above levels seen in GBM controls lacking EGFR overexpression, consistent with our ELISA results.
Since all patients underwent surgical tumor resection, we could not evaluate the radiographic tumor responses to lapatinib.
5. Level of EGFR inhibition determines cell death response in EGFR mutant GBM cells
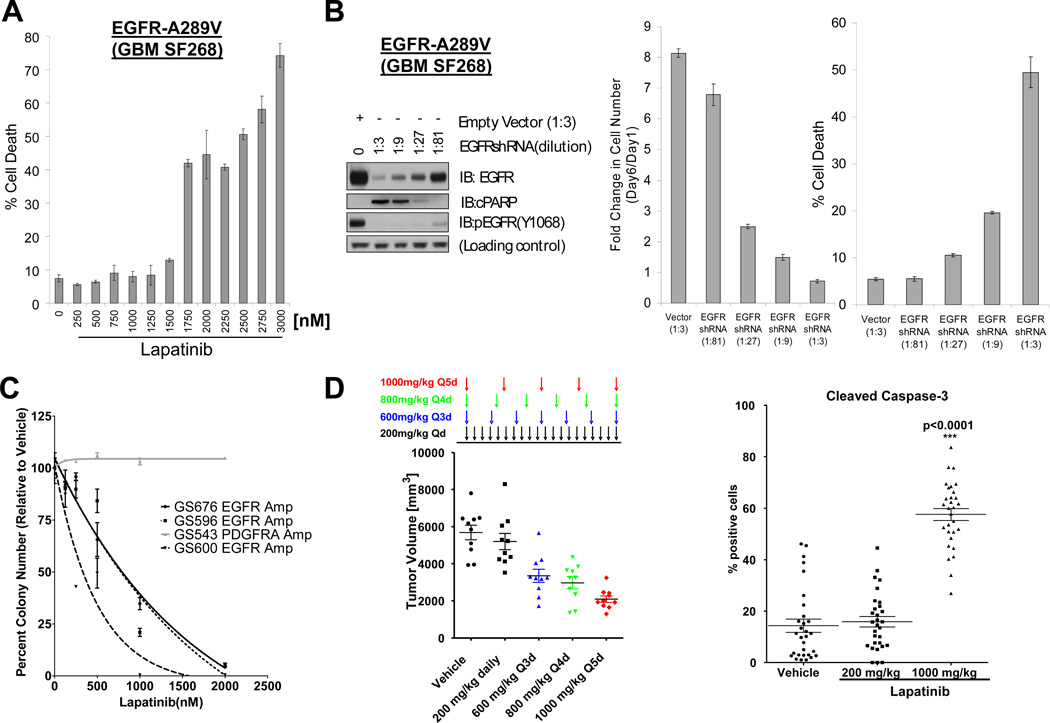
Studies in cancer cell lines have shown that cell death induction by lapatinib requires drug concentrations of 2–3 µM, drug concentrations above the IC50s for inhibition of EGFR phosphorylation and inhibition of cell proliferation (38). Detailed dose-response experiments in EGFR mutant SF268 (Fig. 6A), SKMG3 (Suppl. Fig. 7A) and KNS-81-FD (Suppl. Fig.7B) GBM cells similarly showed dose-dependent cell death induction only above lapatinib concentrations of 1500–1750 nM (Suppl. Fig.7).
FIGURE 6. Level of EGFR inhibition determines cell death response in EGFR mutant GBM cells.
A. Lapatinib induces cell death in SF268 GBM cells (A289V EGFR) at concentrations above 1.5µM. Cell death was assessed 5 days after treatment by trypan blue exclusion. Similar results are shown in Suppl. Figure 7A for SKMG3 cells (A289D-EGFR) and in KNS-81-FD cells (G598V-EGFR)(Suppl. Figure 7B). B. Cell death induction in SF268 GBM cells (A289V EGFR) requires near complete EGFR inactivation. SF268 cells were acutely transduced with a dilution series of a lentiviral EGFR-targeted shRNA (dilution factor is indicated in parenthesis) or with a control shRNA (empty vector). A fraction of the cells from each infection was used for immunoblot analysis with the indicated antibodies (left panel). The remaining cells were re-seeded and allowed to grow for 5 days post-infection. The fold number of viable cells (relative to day 1) and fraction of dead cells are shown in the middle and right panel, respectively. Similar results were obtained in SKMG3 GBM cells (Suppl. Figure 8). C. Lapatinib inhibits anchorage-independent growth of EGFR amplified (GS676, GS596, GS600), but not PDGFRA amplified, GBM tumor sphere cultures. Anchorage independent growth of four freshly derived GBM tumor sphere cultures was assessed in soft agar in the presence of the indicated concentrations of lapatinib. See also Suppl. Figure 9. D. Pulsatile lapatinib dosing (1000 mpk every 5 days) is superior to daily lapatinib dosing (200 mpk qd) in inducing growth inhibition and cell death induction in EGFR amplified GS676 GBM tumor sphere lines. GS676 cells were injected subcutaneously into SCID mice and treatment initiated at the indicated lapatinib dosing schedules after tumors were established. Tumor volumes (left panel) were evaluated at the end of treatment by caliper measurements. The right panel shows quantification of cleaved Caspase3 staining in tumor sections from the indicated cohorts, i.e. vehicle, 200mg/kg lapatinib, and 1000mg/kg lapatinib. IHC images are shown in Suppl. Figure 10.
While lapatinib ranks amongst the most selective ATP-site competitive kinase inhibitors (40), we sought to confirm that this cell death “threshold” reflected a requirement for near complete EGFR inhibition rather than potential off-target effects of lapatinib. We performed titration experiments with a retroviral EGFR shRNA construct in GBM cells with EGFR EC mutations. At a virus dilution of 1:27, SF268 (EGFR-A289V) GBM cells showed clear reductions in EGFR protein levels and EGFR phosphorylation and greater than 50 % growth inhibition, but no evidence for cell death (PARP cleavage, trypan blue exclusion). When EGFR protein levels were almost undetectable by immunoblotting (1:3 dilution of the EGFRshRNA virus), on the other hand, we observed robust cell death induction and PARP cleavage (Fig. 6B). We observed similar results in A289D-EGFR mutant SKMG3 cells (Suppl. Fig. 8). These results demonstrate that even low levels of EGFR activity, which cannot accurately be quantified by immunoblotting using phosphospecific EGFR antibodies, are sufficient to sustain the survival of EGFR mutant glioma cells.
To further explore the biological significance of potent EGFR blockade in-vivo, we extended our experiments to GBM tumor sphere cultures freshly derived from GBM patients. Unlike SF268 and SKMG3 cells, these cells form aggressive tumors in immunodeficient mice.
In preliminary experiments, we compared the effects of erlotinib and lapatinib on in vitro cell viability in two EGFR-amplified GBM tumor sphere lines (GS676 and GS600), and again, found that only lapatinib was able to effectively induce cell death (Suppl. Fig. 9A–B). We also assessed the effects of lapatinib on anchorage-independent growth in a slightly larger panel of glioma sphere lines. In all three lines with EGFR gene amplification (GS676, GS596, GS600), lapatinib reduced colony formation in a dose-dependent fashion with complete abrogation of colony growth above 2 µM lapatinib (Fig. 6C)(Suppl. Fig.9C). Lapatinib had no effect on colony formation of a PDGFRA-amplified (GS543) glioma sphere line (Fig. 6C).
We then compared the effectiveness of different lapatinib dosing schedules on the growth of subcutaneous GS676 GBM xenografts. After tumors were established, mice were assigned to either treatment with vehicle or four different oral lapatinib dosing schedules: 200 mg/kg daily, 600 mg every third day, 800 mg every fourth day, or 1000 mg every fifth day. We designed this dosing schedule based on previous reports that transient potent blockade of oncogenic kinases is able to irreversibly commit cancer cells to cell death (41, 42). We observed maximal growth inhibition (Fig. 6D) and caspase activation (Fig. 6E)(Suppl. Fig. 10) in the cohort receiving 1000 mg/kg every fifth day.
DISCUSSION
The EGFR kinase inhibitor erlotinib has received regulatory approval for the treatment of EGFR mutant lung cancer (8, 43), but results with this agent in GBM have been disappointing. Our study provides a potential explanation for the differential activity of erlotinib against these two cancer types. In contrast to the most common EGFR kinase mutants in lung cancer, the most common oncogenic EGFR alterations in glioblastoma are relatively insensitive to erlotinib. Instead, these mutants are preferentially inhibited by EGFR inhibitors that can only be accommodated by the inactive conformation of the EGFR catalytic pocket due to their bulky aniline substituents (lapatinib, HKI-272)(29, 44). While many novel EGFR kinase inhibitors distinguish themselves from first-generation EGFR kinase inhibitors by their irreversible mode of EGFR binding or activity against selected kinases in addition to EGFR (8, 45), our results argue for focused clinical development of type II EGFR kinase inhibitors for EGFR mutant GBM.
The molecular mechanisms for the inhibitor selectivity of EGFR extracellular versus EGFR kinase domain mutants require further study. Studies of (near) full length EGFR receptors are beginning to uncover details of the relationship between the extracellular and kinase domains of receptor tyrosine kinases (46) It seems unlikely that the conformation of extracellular EGFR mutants is identical to the inactive-like conformation (i.e. catalytically incompetent) described in structural studies of the isolated kinase domain (29), especially when considering that these mutants possess ligand-independent constitutive activity and transforming ability (13). Instead, we propose that the unliganded extracellular-domain mutant receptors exist in a dimeric state that retains enough flexibility within the kinase domain to accommodate lapatinib and other type II EGFR kinase inhibitors. This flexibility appears to be compromised in EGFR kinase domain mutants (34).
While our study uncovered a relative vulnerability of “glioma-relevant” EGFR genotypes (wildype, EGFRvIII, A289V/D, G598V, T263P) to lapatinib, oral lapatinib therapy at a dose of 750 mg twice daily failed to prolong progression-free survival in patients with recurrent GBM in our study and another recent phase I/I trial (47). Neither of the two GBM patients whose tumors showed intratumoral drug concentrations above 1500 nM (E50088 and E50034) and also overexpressed EGFR could be evaluated for therapeutic response (Suppl. Table 8). This results highlights the need to enrich clinical trials with targeted agents in GBM for patients whose tumors harbor the drug-relevant oncogenic lesion, a strategy that is already pursued in the development of kinase inhibitors for several other human cancer types (48).
The experience with BRAF-mutant melanoma illustrates the importance of effective kinase inhibition for therapeutic response (49). Such potent EGFR inhibition is readily achievable in lung cancer because of the direct effects of kinase domain mutations on inhibitor and ATP affinity (44). Further clinical trials are required to explore whether a similar degree of EGFR kinase inhibition can be achieved in EGFR mutant GBM through alternative lapatinib dosing schedules (e.g., pulsatile dosing) (50), type II EGFR inhibitors with improved CNS penetration, or perhaps combination therapies converging on the mutant EGFR protein and its effectors.
MATERIALS AND METHODS
Cell lines and reagents
SF295 and SF268 cells were obtained from the NCI. H460, HCC827, and HCC4006 cells were purchased from ATCC. KNS-81-FD cells were purchased from JCRB. 8-MG-BA and H3255 cells were kindly provided by Dr. Rameen Beroukhim (Broad Institute). SKMG3 cells were provided by Conforma Therapeutics. Normal human astrocytes (NHA) were kindly provided by Dr. Russell Pieper (UCSF). NR6 cells were kindly provided by Dr. Harvey Herschman (UCLA). DNA fingerprinting was used for authentication of all glioma cell lines; no further validation was performed. All antibodies with the exception of anti-Actin and Ki-67 were purchased from Cell Signaling Technologies. Anti-Actin antibody was purchased from Sigma. Ki-67 antibody was purchased from Dako. Erlotinib and lapatinib were purchased from LC Laboratories. CI-1033 and HKI-272 were purchased from Selleck Chemicals.
Electrochemiluminescent detection of EGFR and pEGFR in tumor samples
Phospho(Tyr1173)/Total EGFR Assay was purchased from Meso Scale Discovery and assay was carried out as described in the product insert (Cat# K15104D) using a SECTOR Imager 2400 instrument.
Plasmids
Wild type EGFR was shuttled from pLXSN-EGFR (kindly provided by Dr. David Riese, Purdue University) into pLNCX2 as a XhoI restriction fragment. pLHCX-EGFRvIII was kindly provided by Dr. Paul Mischel (UCLA). pLNCX2-EGFR was used as template to generate A289D, A289V, G598V, and T263P point mutants using Quickchange (Agilent). Lentiviral shRNA constructs targeting EGFR and ErbB2 were purchased from Sigma (EGFRshRNA(1), TRCN0000010329; EGFRshRNA(2), TRCN0000121068; ErbB2, TRCN0000195369).
Retroviral infections
For transduction of wild type and mutant EGFR into NR6 fibroblasts, pan-tropic retrovirus was generated using the Pantropic Retroviral Expression System from Clontech. Briefly, EGFR cDNAs were co-transfected with pVSGV into the GP2-293 packaging cell line. Viral particles were collected 36 and 60 hours post-transfection and target cells were infected for 18 hours with each virus collection. Stable expressors were derived through antibiotic selection. Knockdown of EGFR and ErbB2 was done using lentiviral shRNAs. Viral particles were produced by cotransfection of shRNA constructs (empty pLKO vector was used as control) with two packaging plasmids (pMD2G and pPAX2) into 293T cells. Viral particles were collected at 36 and 60 hours after transfection. Each virus was diluted 1:3 with collection media (Iscove’s media + 10% fetal bovine serum) and infections were carried out with diluted virus for 3 hours. Where noted, virus stock was further diluted as indicated.
Assessment of cell death induction
Cells were seeded on 6cm dishes and allowed to attach overnight. Cells were then treated with the indicated drugs at the indicated doses for 5 days. Each treatment group was seeded in triplicate. Following treatment, both attached and unattached cells were harvested and counted on a ViCell Cell Viability analyzer. The instrument uses trypan blue to assess cell death. Cell death was expressed as the fraction of trypan-blue-positive cells over the total number of cells.
Soft agar colony formation assay
Cells were seeded at 5000 (GS543), 25000 (GS596), or 50000 (GS600 and GS676) cells/plate based on pre-determined colony formation efficiencies of untreated cells such that each cell line would give rise to similar numbers of colonies under vehicle control conditions. Cells were plated in Neurocult media (Stem Cell Technologies) containing 0.65% nobel agar and growth factor supplements and each treatment group was done in duplicate. Colonies were stained with crystal violet (0.005 %) three weeks after plating, imaged in a Gel Count (Oxford Optronix), and images processed using the Charm algorithm (Oxford Optronix) to obtain colony number and colony size distributions.
ATP competition assay
The ability of EGFR TKIs to compete with ATP for binding to EGFR was measured using the Pierce Kinase Enrichment Kit with ATP Probe and was carried out according to the manufacturer’s protocol with the following modifications. Briefly, cells are harvested and lysed. Lysates are then passed through a desalting column to remove ATP. Following this buffer exchange, lysates are incubated with a pre-made mixture of the appropriate inhibitor at the desired concentration and desthiobiotin-ATP probe to a final concentration of 5µM. This mixture is then incubated for 5 minutes at room temperature. The reaction is terminated by addition of 4M urea. Avidin agarose beads are then added to the reaction mixtures and allowed to pulldown biotinylated proteins for 1 hour at room temperature. Beads are washed 3 times and eluted with 3X Laemmli sample buffer. Pulldowns are then analyzed by immunoblot.
Immunohistochemistry and computer-assisted image analysis
Paraffin-embedded sections of tumor xenografts were obtained at 5µm/slide. Antigen retrieval, immunohistochemical detection and counter staining were performed using the Ventana Discovery Ultra autostainer (Ventana) using primary antibodies against cleaved caspase-3 at a 1:1000 dilution. To determine apoptotic index we used total number of nuclei with positive cleaved-Caspase-3 labeling x100/ total number of nuclei on H&E staining. Histological fields were captured with a camera (SPOT Imaging Solutions, Sterling Heights, Michigan). Digitized images were segmented using segmentation techniques such as density and size thresholding to distinguish negative from positive objects using image analysis software (ImageJ, NIH, USA). The segmentation process resulted in the generation of binary images from which the number of stained objects and total numbers of nuclei were determined. Three separate regions were analyzed from in each tumor sample.
Tumor xenografts
Mice are restrained using IACUC approved restraint techniques to expose the flank. The hair is removed with an electric razor and the injection site is disinfected with 70% ethanol. Then 106 cells, in 100 uL of a 50:50 mixture of growth media and in Matrigel (BD, catalog #356237), is injected under the skin. Mice are monitored to ensure that tumor growth does not exceed 1.5 cm in diameter.
SIGNIFICANCE.
About 40 % of human GBMs harbor oncogenic EGFR alterations but attempts to therapeutically target EGFR with first-generation EGFR kinase inhibitors have failed. Here we demonstrate selective sensitivity of glioma-specific EGFR mutants to ATP-site competitive EGFR kinase inhibitors which target the inactive conformation of the catalytic domain.
Supplementary Material
ACKNOWLEDGEMENTS
We thank members of the Mellinghoff laboratory and Dr. Charles Sawyers for helpful discussions in the course of this work. We thank Dr. William Weiss (UCSF) and Dr. Kevan Shokat (UCSF) for sharing unpublished results.
GRANT SUPPORT
This work was supported through U54CA143798 (IKM) and U01 CA141502 from the National Cancer Institute. Further funding support was provided by the Leon Levy foundation, the Sontag Foundation, the Doris Duke Charitable Foundation, and an Advanced Clinical Research Award from the American Society of Clinical Oncology (IKM). CG was supported through an American Brain Tumor Association Basic Research Fellowship Award, MGC was supported through NIH-5K08NS062907, BO was supported through grants from the American Italian Cancer Foundation and a MSKCC Brain Tumor Center grant, and YN was the recipient of an American Brain Tumor Association Medical Student Summer Fellowship. Investigators of the NABTC-04-01 Clinical Trial were supported through the following funding sources: 5-U01CA62399-09 (A.B.L., and L.M.D.); NABTC # CA62399 and Member # CA62422, GCRC Grant # M01-RR00079 (S.M.C., K.R.L., and M.D.P.); CA62426 (J.G.K.); CA62412, GCRC Grant # CA16672 (W.K.A.Y. and M.R.G.); U01CA62407-08 (P.Y.W.); U01CA62421-08, GCRC Grant # M01 RR03186 (M.M. and H.I.R.); U01CA62405, GCRC Grant # M01-RR00056 (F.L.);U01 CA62399, GCRC Grant # M01-RR0865 (T.F.C.).
REFERENCES
- 1.Wen PY, Kesari S. Malignant gliomas in adults. N Engl J Med. 2008 Jul 31;359(5):492–507. doi: 10.1056/NEJMra0708126. [DOI] [PubMed] [Google Scholar]
- 2.Sawyers CL. Shifting paradigms: the seeds of oncogene addiction. Nat Med. 2009 Oct;15(10):1158–1161. doi: 10.1038/nm1009-1158. [DOI] [PubMed] [Google Scholar]
- 3.Sellers WR. A blueprint for advancing genetics-based cancer therapy. Cell. 2011 Sep 30;147(1):26–31. doi: 10.1016/j.cell.2011.09.016. [DOI] [PubMed] [Google Scholar]
- 4.McLendon R, Friedman A, Bigner D, Van Meir EG, Brat DJ, Mastrogianakis M, et al. Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature. 2008 Oct 23;455(7216):1061–1068. doi: 10.1038/nature07385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Parsons DW, Jones S, Zhang X, Lin JC, Leary RJ, Angenendt P, et al. An integrated genomic analysis of human glioblastoma multiforme. Science. 2008 Sep 26;321(5897):1807–1812. doi: 10.1126/science.1164382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yarden Y, Sliwkowski MX. Untangling the ErbB signalling network. Nat Rev Mol Cell Biol. 2001 Feb;2(2):127–137. doi: 10.1038/35052073. [DOI] [PubMed] [Google Scholar]
- 7.Furnari FB, Fenton T, Bachoo RM, Mukasa A, Stommel JM, Stegh A, et al. Malignant astrocytic glioma: genetics, biology, and paths to treatment. Genes Dev. 2007 Nov 1;21(21):2683–2710. doi: 10.1101/gad.1596707. [DOI] [PubMed] [Google Scholar]
- 8.Pao W, Chmielecki J. Rational, biologically based treatment of EGFR-mutant non-small-cell lung cancer. Nat Rev Cancer. 2010 Nov;10(11):760–774. doi: 10.1038/nrc2947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brandes AA, Franceschi E, Tosoni A, Hegi ME, Stupp R. Epidermal growth factor receptor inhibitors in neuro-oncology: hopes and disappointments. Clin Cancer Res. 2008 Feb 15;14(4):957–960. doi: 10.1158/1078-0432.CCR-07-1810. [DOI] [PubMed] [Google Scholar]
- 10.Stommel JM, Kimmelman AC, Ying H, Nabioullin R, Ponugoti AH, Wiedemeyer R, et al. Coactivation of receptor tyrosine kinases affects the response of tumor cells to targeted therapies. Science. 2007 Oct 12;318(5848):287–290. doi: 10.1126/science.1142946. [DOI] [PubMed] [Google Scholar]
- 11.Inda MM, Bonavia R, Mukasa A, Narita Y, Sah DW, Vandenberg S, et al. Tumor heterogeneity is an active process maintained by a mutant EGFR-induced cytokine circuit in glioblastoma. Genes Dev. 2010 Aug 15;24(16):1731–1745. doi: 10.1101/gad.1890510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sharma SV, Bell DW, Settleman J, Haber DA. Epidermal growth factor receptor mutations in lung cancer. Nat Rev Cancer. 2007 Mar;7(3):169–181. doi: 10.1038/nrc2088. [DOI] [PubMed] [Google Scholar]
- 13.Lee JC, Vivanco I, Beroukhim R, Huang JH, Feng WL, Debiasi RM, et al. Epidermal Growth Factor Receptor Activation in Glioblastoma through Novel Missense Mutations in the Extracellular Domain. PLoS Med. 2006 Dec 19;3(12):e485. doi: 10.1371/journal.pmed.0030485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Batra SK, Castelino-Prabhu S, Wikstrand CJ, Zhu X, Humphrey PA, Friedman HS, et al. Epidermal growth factor ligand-independent, unregulated, cell-transforming potential of a naturally occurring human mutant EGFRvIII gene. Cell Growth Differ. 1995 Oct;6(10):1251–1259. [PubMed] [Google Scholar]
- 15.Greulich H, Chen TH, Feng W, Janne PA, Alvarez JV, Zappaterra M, et al. Oncogenic Transformation by Inhibitor-Sensitive and -Resistant EGFR Mutants. PLoS Med. 2005 Oct 4;2(11):e313. doi: 10.1371/journal.pmed.0020313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sonoda Y, Ozawa T, Hirose Y, Aldape KD, McMahon M, Berger MS, et al. Formation of intracranial tumors by genetically modified human astrocytes defines four pathways critical in the development of human anaplastic astrocytoma. Cancer Res. 2001 Jul 1;61(13):4956–4960. [PubMed] [Google Scholar]
- 17.Vivanco I, Rohle D, Versele M, Iwanami A, Kuga D, Oldrini B, et al. The phosphatase and tensin homolog regulates epidermal growth factor receptor (EGFR) inhibitor response by targeting EGFR for degradation. Proc Natl Acad Sci U S A. 2010 Apr 6;107(14):6459–6464. doi: 10.1073/pnas.0911188107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hynes NE, Lane HA. ERBB receptors and cancer: the complexity of targeted inhibitors. Nat Rev Cancer. 2005 May;5(5):341–354. doi: 10.1038/nrc1609. [DOI] [PubMed] [Google Scholar]
- 19.Weihua Z, Tsan R, Huang WC, Wu Q, Chiu CH, Fidler IJ, et al. Survival of cancer cells is maintained by EGFR independent of its kinase activity. Cancer Cell. 2008 May;13(5):385–393. doi: 10.1016/j.ccr.2008.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rabindran SK, Discafani CM, Rosfjord EC, Baxter M, Floyd MB, Golas J, et al. Antitumor activity of HKI-272, an orally active, irreversible inhibitor of the HER-2 tyrosine kinase. Cancer Res. 2004 Jun 1;64(11):3958–3965. doi: 10.1158/0008-5472.CAN-03-2868. [DOI] [PubMed] [Google Scholar]
- 21.Fry DW, Bridges AJ, Denny WA, Doherty A, Greis KD, Hicks JL, et al. Specific, irreversible inactivation of the epidermal growth factor receptor and erbB2, by a new class of tyrosine kinase inhibitor. Proc Natl Acad Sci U S A. 1998 Sep 29;95(20):12022–12027. doi: 10.1073/pnas.95.20.12022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Slichenmyer WJ, Elliott WL, Fry DW. CI-1033, a pan-erbB tyrosine kinase inhibitor. Semin Oncol. 2001 Oct;28(5) Suppl 16:80–85. doi: 10.1016/s0093-7754(01)90285-4. [DOI] [PubMed] [Google Scholar]
- 23.Lynch TJ, Bell DW, Sordella R, Gurubhagavatula S, Okimoto RA, Brannigan BW, et al. Activating mutations in the epidermal growth factor receptor underlying responsiveness of non-small-cell lung cancer to gefitinib. N Engl J Med. 2004 May 20;350(21):2129–2139. doi: 10.1056/NEJMoa040938. [DOI] [PubMed] [Google Scholar]
- 24.Carey KD, Garton AJ, Romero MS, Kahler J, Thomson S, Ross S, et al. Kinetic analysis of epidermal growth factor receptor somatic mutant proteins shows increased sensitivity to the epidermal growth factor receptor tyrosine kinase inhibitor, erlotinib. Cancer Res. 2006 Aug 15;66(16):8163–8171. doi: 10.1158/0008-5472.CAN-06-0453. [DOI] [PubMed] [Google Scholar]
- 25.Paez JG, Janne PA, Lee JC, Tracy S, Greulich H, Gabriel S, et al. EGFR mutations in lung cancer: correlation with clinical response to gefitinib therapy. Science. 2004 Jun 4;304(5676):1497–1500. doi: 10.1126/science.1099314. [DOI] [PubMed] [Google Scholar]
- 26.Pao W, Miller V, Zakowski M, Doherty J, Politi K, Sarkaria I, et al. EGF receptor gene mutations are common in lung cancers from "never smokers" and are associated with sensitivity of tumors to gefitinib and erlotinib. Proc Natl Acad Sci U S A. 2004 Sep 7;101(36):13306–13311. doi: 10.1073/pnas.0405220101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jura N, Zhang X, Endres NF, Seeliger MA, Schindler T, Kuriyan J. Catalytic control in the EGF receptor and its connection to general kinase regulatory mechanisms. Mol Cell. 2011 Apr 8;42(1):9–22. doi: 10.1016/j.molcel.2011.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lemmon MA, Schlessinger J. Cell signaling by receptor tyrosine kinases. Cell. 2010 Jun 25;141(7):1117–1134. doi: 10.1016/j.cell.2010.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wood ER, Truesdale AT, McDonald OB, Yuan D, Hassell A, Dickerson SH, et al. A unique structure for epidermal growth factor receptor bound to GW572016 (Lapatinib): relationships among protein conformation, inhibitor off-rate, and receptor activity in tumor cells. Cancer Res. 2004 Sep 15;64(18):6652–6659. doi: 10.1158/0008-5472.CAN-04-1168. [DOI] [PubMed] [Google Scholar]
- 30.Stamos J, Sliwkowski MX, Eigenbrot C. Structure of the epidermal growth factor receptor kinase domain alone and in complex with a 4-anilinoquinazoline inhibitor. J Biol Chem. 2002 Nov 29;277(48):46265–46272. doi: 10.1074/jbc.M207135200. [DOI] [PubMed] [Google Scholar]
- 31.Yun CH, Mengwasser KE, Toms AV, Woo MS, Greulich H, Wong KK, et al. The T790M mutation in EGFR kinase causes drug resistance by increasing the affinity for ATP. Proc Natl Acad Sci U S A. 2008 Jan 28; doi: 10.1073/pnas.0709662105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pruss RM, Herschman HR. Variants of 3T3 cells lacking mitogenic response to epidermal growth factor. Proc Natl Acad Sci U S A. 1977 Sep;74(9):3918–3921. doi: 10.1073/pnas.74.9.3918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Di Fiore PP, Pierce JH, Fleming TP, Hazan R, Ullrich A, King CR, et al. Overexpression of the human EGF receptor confers an EGF-dependent transformed phenotype to NIH 3T3 cells. Cell. 1987 Dec 24;51(6):1063–1070. doi: 10.1016/0092-8674(87)90592-7. [DOI] [PubMed] [Google Scholar]
- 34.Gilmer TM, Cable L, Alligood K, Rusnak D, Spehar G, Gallagher KT, et al. Impact of common epidermal growth factor receptor and HER2 variants on receptor activity and inhibition by lapatinib. Cancer Res. 2008 Jan 15;68(2):571–579. doi: 10.1158/0008-5472.CAN-07-2404. [DOI] [PubMed] [Google Scholar]
- 35.Lassman AB, Rossi MR, Raizer JJ, Abrey LE, Lieberman FS, Grefe CN, et al. Molecular study of malignant gliomas treated with epidermal growth factor receptor inhibitors: tissue analysis from North American Brain Tumor Consortium Trials 01–03 and 00–01. Clin Cancer Res. 2005 Nov 1;11(21):7841–7850. doi: 10.1158/1078-0432.CCR-05-0421. [DOI] [PubMed] [Google Scholar]
- 36.Hegi ME, Diserens AC, Bady P, Kamoshima Y, Kouwenhoven MC, Delorenzi M, et al. Pathway analysis of glioblastoma tissue after preoperative treatment with the EGFR tyrosine kinase inhibitor gefitinib--a phase II trial. Mol Cancer Ther. 2011 Jun;10(6):1102–1112. doi: 10.1158/1535-7163.MCT-11-0048. [DOI] [PubMed] [Google Scholar]
- 37.Burris HA, 3rd, Taylor CW, Jones SF, Koch KM, Versola MJ, Arya N, et al. A phase I and pharmacokinetic study of oral lapatinib administered once or twice daily in patients with solid malignancies. Clin Cancer Res. 2009 Nov 1;15(21):6702–6708. doi: 10.1158/1078-0432.CCR-09-0369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rusnak DW, Affleck K, Cockerill SG, Stubberfield C, Harris R, Page M, et al. The characterization of novel, dual ErbB-2/EGFR, tyrosine kinase inhibitors: potential therapy for cancer. Cancer Res. 2001 Oct 1;61(19):7196–7203. [PubMed] [Google Scholar]
- 39.Bigner SH, Humphrey PA, Wong AJ, Vogelstein B, Mark J, Friedman HS, et al. Characterization of the epidermal growth factor receptor in human glioma cell lines and xenografts. Cancer Res. 1990 Dec 15;50(24):8017–8022. [PubMed] [Google Scholar]
- 40.Karaman MW, Herrgard S, Treiber DK, Gallant P, Atteridge CE, Campbell BT, et al. A quantitative analysis of kinase inhibitor selectivity. Nat Biotechnol. 2008 Jan;26(1):127–132. doi: 10.1038/nbt1358. [DOI] [PubMed] [Google Scholar]
- 41.Shah NP, Kasap C, Weier C, Balbas M, Nicoll JM, Bleickardt E, et al. Transient potent BCR-ABL inhibition is sufficient to commit chronic myeloid leukemia cells irreversibly to apoptosis. Cancer Cell. 2008 Dec 9;14(6):485–493. doi: 10.1016/j.ccr.2008.11.001. [DOI] [PubMed] [Google Scholar]
- 42.Amin DN, Sergina N, Ahuja D, McMahon M, Blair JA, Wang D, et al. Resiliency and vulnerability in the HER2-HER3 tumorigenic driver. Sci Transl Med. 2010 Jan 27;2(16):16ra7. doi: 10.1126/scitranslmed.3000389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ciardiello F, Tortora G. EGFR antagonists in cancer treatment. N Engl J Med. 2008 Mar 13;358(11):1160–1174. doi: 10.1056/NEJMra0707704. [DOI] [PubMed] [Google Scholar]
- 44.Eck MJ, Yun CH. Structural and mechanistic underpinnings of the differential drug sensitivity of EGFR mutations in non-small cell lung cancer. Biochim Biophys Acta. 2010 Mar;1804(3):559–566. doi: 10.1016/j.bbapap.2009.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Baselga J. Targeting tyrosine kinases in cancer: the second wave. Science. 2006 May 26;312(5777):1175–1178. doi: 10.1126/science.1125951. [DOI] [PubMed] [Google Scholar]
- 46.Mi LZ, Lu C, Li Z, Nishida N, Walz T, Springer TA. Simultaneous visualization of the extracellular and cytoplasmic domains of the epidermal growth factor receptor. Nat Struct Mol Biol. 2011 Sep;18(9):984–989. doi: 10.1038/nsmb.2092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Thiessen B, Stewart C, Tsao M, Kamel-Reid S, Schaiquevich P, Mason W, et al. A phase I/II trial of GW572016 (lapatinib) in recurrent glioblastoma multiforme: clinical outcomes, pharmacokinetics and molecular correlation. Cancer Chemother Pharmacol. 2010 Jan;65(2):353–361. doi: 10.1007/s00280-009-1041-6. [DOI] [PubMed] [Google Scholar]
- 48.Gerber DE, Minna JD. ALK inhibition for non-small cell lung cancer: from discovery to therapy in record time. Cancer Cell. Dec 14;18(6):548–551. doi: 10.1016/j.ccr.2010.11.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bollag G, Hirth P, Tsai J, Zhang J, Ibrahim PN, Cho H, et al. Clinical efficacy of a RAF inhibitor needs broad target blockade in BRAF-mutant melanoma. Nature. 2010 Sep 30;467(7315):596–599. doi: 10.1038/nature09454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chien AJ, Illi JA, Ko AH, Korn WM, Fong L, Chen LM, et al. A phase I study of a 2-day lapatinib chemosensitization pulse preceding nanoparticle albumin-bound Paclitaxel for advanced solid malignancies. Clin Cancer Res. 2009 Sep 1;15(17):5569–5575. doi: 10.1158/1078-0432.CCR-09-0522. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.