Abstract
Aberrant DNA methylation patterns have been linked to molecular and cellular alterations in the aging brain. Caloric restriction (CR) and upregulation of antioxidants have been proposed as interventions to prevent or delay age-related brain pathology. Previously, we have shown in large cohorts of aging mice, that age-related increases in DNA methyltransferase 3a (Dnmt3a) immunoreactivity in the mouse hippocampus were attenuated by CR, but not by overexpression of superoxide dismutase 1 (SOD1). Here, we investigated age-related alterations of 5-methylcytidine (5-mC), a marker of DNA methylation levels, in a hippocampal subregion-specific manner. Examination of 5-mC immunoreactivity in 12- and 24-month-old wild type (WT) mice on control diet, mice overexpressing SOD1 on control diet, wild type mice on CR, and SOD1 mice on CR, indicated an age-related increase in 5-mC immunoreactivity in the hippocampal dentate gyrus, CA3, and CA1–2 regions, which was prevented by CR but not by SOD1 overexpression. Moreover, positive correlations between 5-mC and Dnmt3a immunoreactivity were observed in the CA3 and CA1–2. These findings suggest a crucial role for DNA methylation in hippocampal aging and in the mediation of the beneficial effects of CR on aging.
Keywords: Aging, Epigenesis, Epigenetics, DNA methylation, 5-methylcytidine (5-mC), Caloric restriction, Antioxidants, Superoxide dismutase (SOD), Hippocampus
1. Introduction
Aging of the brain is associated with various molecular and morphological alterations that can lead to cognitive decline and increased risk for the development of neurodegenerative diseases. Brain shrinkage, neuronal loss, increased DNA damage, vascular dysfunction, and changes in numbers and morphology of dendritic spines have all been linked with the selective vulnerability to aging of hippocampal and neocortical circuits (Dickstein et al., 2007, 2010; Luebke et al., 2010; Rutten et al., 2003, 2007). Recently, the epigenetic mechanism of DNA methylation, has been linked to mediation of the age-related alterations in gene expression and with memory formation (Calvanese et al., 2009; Chouliaras et al., 2010a; Day and Sweatt, 2010; Fraga and Esteller, 2007; Levenson et al., 2006; Liu et al., 2009; Miller and Sweatt, 2007; Murgatroyd et al., 2010; Penner et al., 2010a). Furthermore, manipulations of DNA methyltransferases (Dnmts) in mice have been shown to affect memory processing (Feng et al., 2010; Levenson et al., 2006). We recently reported an age-related increase in Dnmt3a levels in the mouse hippocampus, which was prevented by caloric restriction (CR) (Chouliaras et al., 2011). CR has been previously shown to increase lifespan, prevent age-related alterations, and delay disease onset in many species (Adams et al., 2008; Anderson et al., 2009; Bordone and Guarente, 2005; Colman et al., 2009; Levenson and Rich, 2007; Maswood et al., 2004; Mattson et al., 2002; Mouton et al., 2009; Patel et al., 2005; Sohal and Weindruch, 1996), possibly acting via altering epigenetic mechanisms. Besides CR, upregulation of the endogenous human antioxidant Cu/Zn-superoxide dismutase 1 (SOD1) has been shown to reduce oxidative damage to DNA and prevent neurodegeneration after brain injury in rodents (Borg and London, 2002; Cadet et al., 1994; Cardozo-Pelaez et al., 1998; Chan et al., 1994; Rutten et al., 2002), while oxidative stress has also been described to impact on DNA methylation.
Cohorts of wild type and transgenic mice overexpressing SOD1 were generated, fed with either a control diet or a diet restricted in calories, which were examined at 12 and 24 months of age. Recently, we reported reductions in hippocampal volumes associated with aging and CR (Rutten et al., 2010), as well as increased hippocampal Dnmt3a immunoreactivity (IR) with aging that was prevented by caloric restriction (Chouliaras et al., 2011). In the present study, levels of DNA methylation in situ, by immunohistochemical analysis of 5-mC, were examined in a hippocampal subregion-specific analysis aiming to investigate the effects of aging, CR, and overexpression of SOD1 on DNA methylation levels and test whether levels of 5-mC correlate with Dnmt3a levels.
2. Methods
2.1. Animals
Wild type (WT) C57Bl6J and transgenic mice overexpressing normal human SOD1 were used, derived from a total cohort of 240 male mice. Both types of animals were assigned to receive either a control diet (CD) or a calorie restricted diet. Details on the generation and characterization of the cohort, preparation of the diets, and weight and survival curves of the animals have been described previously (Rutten et al., 2010; Weindruch et al., 1986). Briefly, the cohort was generated from 4 breeder pairs of female WT C57Bl6J and male transgenic hemizygous for SOD1 on a C57Bl6J background (carrying 7 copies of the entire human SOD1 sequence, in order to achieve increased expression and enzyme activity in brain and other tissues; Epstein et al., 1987; Przedborski et al., 1992). The animals were assigned to the 2 diet groups after weaning and for their entire life. The reduction of caloric intake in the CR groups was approximately 50%, while the CD was approximately 85% of the ad libitum consumption, in order to monitor the caloric intake and ensure that all the food pellets would be consumed. The detailed compositions, feeding patterns, and the weight and survival curves have been reported by (Rutten et al., 2010). All experiments were carried on the Central Animal Facilities, Maastricht University, Maastricht, The Netherlands, and the animals were maintained under standard temperature and humidity conditions on a 12-hour light-dark circle, housed individually, with water available ad libitum, under specified pathogen-free conditions. All experiments were approved by the Animals Ethics Board of Maastricht University.
2.2. Experimental design
Four groups were generated, based on the genotype and diet of the animals: (1) WT mice on control diet (WT-CD), (2) SOD1 mice on control diet (SOD1-CD), (3) WT mice on caloric restriction (WT-CR), and (4) SOD1 mice on caloric restriction (SOD1-CR). Six animals from each group were euthanized at the age of 12 months and another 6 animals per group at 24 months of age for histological analyses.
2.3. Tissue processing
The animals were deeply anesthetized and transcardially perfused with 20 mL tyrode solution and 20 mL of fixative solution (4% parafolmaldehyde, 0.9% NaCl) followed by 30 mL of a second fixative solution (8% parafolmaldehyde, 0.9% NaCl, 1% acetic acid). After removal, the brains were post-fixed at 4 °C for 24 hours in the 8% parafolmaldehyde, without the acetic acid. Consequently, the brains were hemisected in the midsagittal line, cryoprotected in sucrose solution (10%, 20%, and finally 30% sucrose in Tris-HCl buffer, 2 × 12 hours per solution at 4 °C) and embedded in Tissue Tek® (Sakura Finetec Europe, Zoeterwoude, The Netherlands). Then, the left brain halves were quickly frozen and stored at −80 °C, until they were cut serially in 30-µm thick free-floating coronal sections using a cryostat (type HM 500 OMV, Microm, Walldorf, Germany), yielding 10 subseries of every tenth section, and stored until further histological processing. The right brain halves were not used in the present study.
2.4. Immunohistochemical detection of 5-mC
Using standard immunohistochemical procedures that were previously described (Chouliaras et al., 2011), with a mouse monoclonal anti-5-mC (dilution 1:500; GenWay Biotech, San Diego, CA, USA) as primary antibody and a donkey antimouse biotine (dilution 1:200; Jackson, Westgrove, PA, USA) as a secondary antibody, a series of sections containing the hippocampus were stained. In order to visualize the reaction product, the sections were incubated in 3,3′-diaminobenzidine tetrahydrochloride (DAB) solution (1:1 3,3′-diaminobenzidine tetrahydrochloride: Tris-HCl, 0.3% H2O2) (Sigma, Uithoorn, The Netherlands) for 10 minutes. Control experiments, omitting the primary antibody or using an anti-5-mC primary antibody from another company (Santa Cruz, CA, USA) confirmed the specificity of immunoreactivity (data not shown).
2.5. Immunofluorescent labeling of 5-mC and DAPI
After incubation with the primary and secondary antibodies (see above), the sections were incubated with streptavidin Alexa Fluor 594 conjugate (Invitrogen, Eugene, OR, USA) and counterstained with 4′,6-diamidino-2-phenylindole dihydrochloride (DAPI; Sigma Aldrich, Zwijndrecht, The Netherlands).
2.6. Analysis of 5-mC IR
2.6.1. Mean intensity and surface area of 5-mC IR
Gray value measurements and percentage of surface area below (i.e., darker as compared with) the calculated threshold (see below) were used for the assessment of 5-mC IR. As illustrated using the white boxes in Fig. 1A, 4 images from the granule cell layer of the dentate gyrus (DG), 2 images from the CA3 cell layer, and 2 images from the CA1–2 regions were taken in 4 selected sections per series, at 4 bregma levels (−1.58, −1.82, −3.40, and −3.52) according to Franklin and Paxinos (1997). Thus, a total of 32 images per animal were analyzed. The images were taken using the 40× objective, with a digital camera (f-view; Olympus, Tokyo, Japan) connected to an Olympus AX70 brightfield microscope (analySIS; Imaging System, Münster, Germany). The pattern of the staining revealed densely stained small particles, reflecting hypermethylated DNA fragments (Brown et al., 2008; Hernandez-Blazquez et al., 2000). Threshold values were set in order to correct for background signal (Strackx et al., 2008). Surface area was defined as the percentage of the area delineated with a gray value below the background threshold. Further, the surface area measurements were corrected for total hippocampal volume differences (Rutten et al., 2010), simply by multiplying the surface area by the volume. However, the surface area measurements, despite the performed corrections, might still be hampered by hippocampal volume differences. Therefore, in addition, mean gray values were calculated in order to further replicate the results. The mean gray value was defined as the sum of gray values of all pixels below the calculated threshold in the selected area divided by the number of all pixels, and such measurement is independent of volume or even cell number differences. Results on gray values are represented as mean intensity, i.e., the inverse mean gray value, with higher intensities representing increased IR.
Fig. 1.
5-Methylcytidine (5-mC) immunoreactivity (IR) in mouse hippocampus. Representative image of a hippocampal section stained for 5-mC (bregma level −1.58). The white boxes indicate the places where the high-magnification images were taken for the analysis (i.e., 4 images per section for dentate gyrus (DG), 2 images per sections for CA3, and 2 images per section for CA1–2). Scale bar = 165 µm (see text for more details).
2.6.2. Imaging of 5-mC and DAPI
Colocalization of 5-mC and DAPI was demonstrated after double immunofluorescence (Fig. 2A–F). Image stacks of 16-µm thick and consisting of 80 confocal images (0.2 µm apart) of double-labeled cell particles were made with a 100× objective (Olympus UPlanSApo; numerical aperture [NA] = 1.40) and the SI-SD system (MBF Bioscience, Williston, VT, USA). The system consisted of a modified Olympus BX51 fluorescence microscope with customized spinning disk unit (DSU; Olympus), computer-controlled excitation and emission filter wheels (Olympus), 3-axis high-accuracy computer-controlled stepping motor specimen stage (4 × 4 Grid Encoded Stage, Ludl Electronic Products, Hawthorne, NY, USA), linear z-axis position encoder (Ludl), ultra-high sensitivity monochrome electron multiplier CCD camera (1000 × 1000 pixels, C9100-02, Hamamatsu Photonics, Hamamatsu City, Japan) and controlling software (StereoInvestigator; MBF BioScience, Williston, VT, USA).
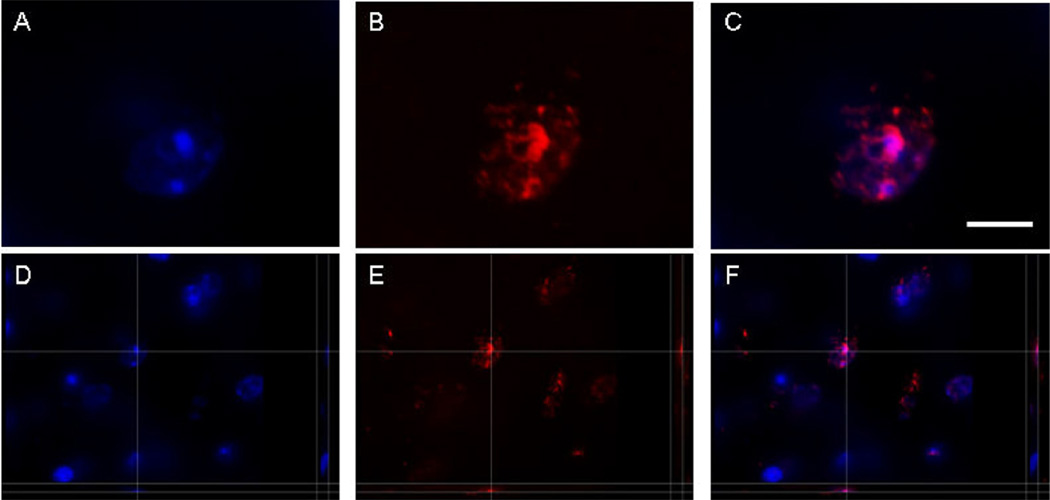
Fig. 2.
5-Methylcytidine (5-mC) and 4′,6-diamidino-2-phenylindole dihydrochloride (DAPI) colocalization. Representative images of a nucleus of a hippocampal cell in a 12-month-old mouse showing fluorescent labeling of DAPI (blue) in (A) and (D), and 5-mC (red) in (B) and (E), at the corresponding location and focal plane, with merged pictures in (C) and (F), showing colocalization (pink). All images represent 1 stack taken with the SI-SD system (see text for more details). Images (D–F) show 3-dimensional reconstructions. Scale bar = 3 µm. Projections and minor corrections in intensity and contrast were made with the Imaris software program (Bitplane AG, Zurich, Switzerland).
2.7. Correlation analysis
Dnmt3a IR was assessed previously in sections from the same mouse brains that were used in the present study. Pearson’s correlation coefficient (rp) was calculated for gray values when correlating Dnmt3a IR with 5-mC IR per hippocampal subregion per animal.
2.8. Statistical analysis
All data are presented as mean and standard error of the mean. The general linear model univariate analysis of variance was used for comparisons between groups, accounting for the main and interactive effects of age, genotype, and diet. Statistical significance was set at an α level of 0.05. Pair-wise comparisons were performed with a Bonferroni post hoc test. All statistical calculations were performed using the Statistical Package for the Social Sciences, (SPSS 16, SPSS, Inc., Chicago, IL, USA). Graphs were built in GraphPad Prism (Version 4, GraphPad Software, San Diego, CA, USA).
3. Results
3.1. Qualitative analysis of 5-mC IR
Nuclear IR of 5-mC was observed in cells throughout the hippocampus (see Fig. 3). Qualitative microscopic inspection of the stained section indicated an evident and profound increase in 5-mC IR occurring with aging, which seemed to be attenuated in animals fed with CR (Fig. 3). Quantitative analyses were carried out to confirm these findings. Further, colocalization with DAPI showed qualitative verification of the nuclear signal (see Fig. 2). The densely-stained 5-mC-immunoreactive particles colocalized with the DAPI signal (Fig. 2C).
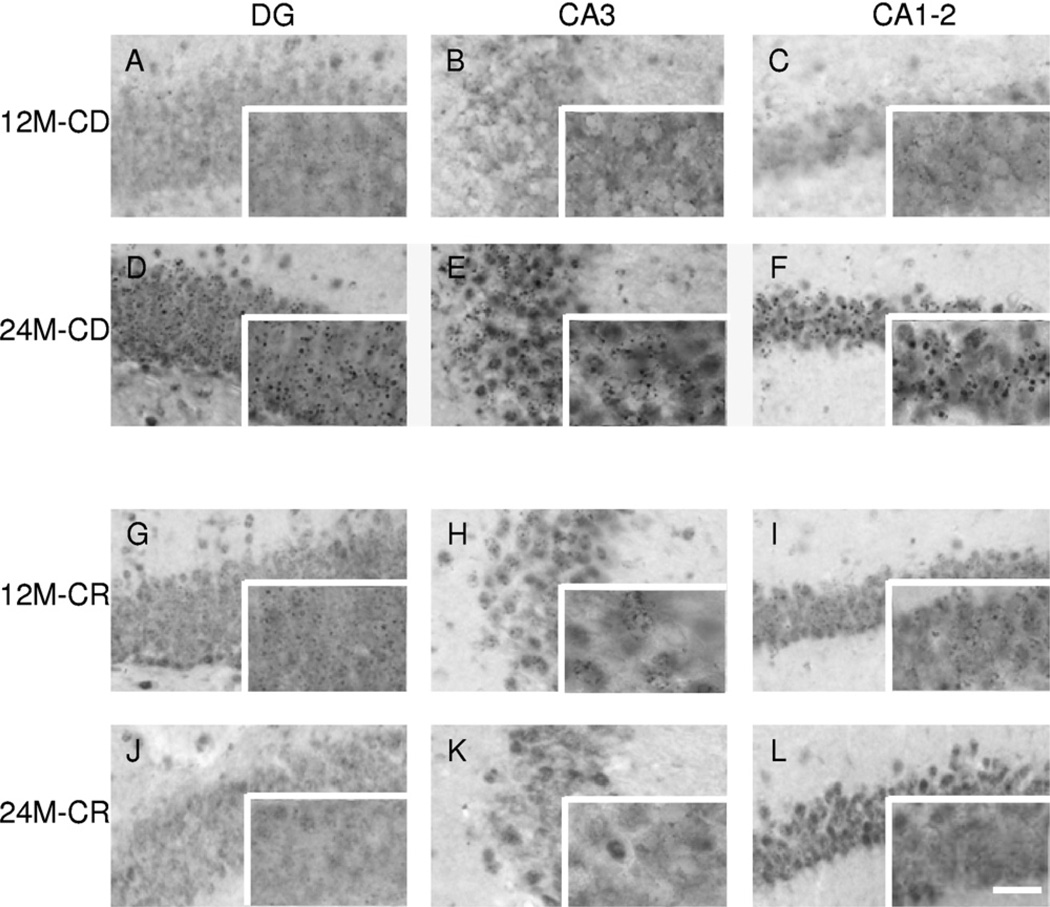
Fig. 3.
Representative images of 5-methylcytidine (5-mC) immunoreactivity (IR). High-magnification images of 5-mC IR. Representative images of the hippocampal regions dentate gyrus (DG), CA3, and CA1–2 are presented for a 12-month-old control diet (CD) mouse in (A), (B), and (C), for a 24-month-old CD mouse in (D), (E), and (F), for a 12-month-old caloric restriction (CR) mouse in (G), (H), and (I), and for a 24-month-old CR mouse in (J), (K), and (L). Note: An evident increase in 5-mC IR from 12 to 24 months in CD mice, which is not seen in CR mice. The full size images were taken with a 40× objective while the images in the bottom right box of each micrograph with a 100× objective. Scale bar = 20 µm in images (A–L) and 50 µm in the high-magnification images in the bottom right boxes.
3.2. 5-mC intensity
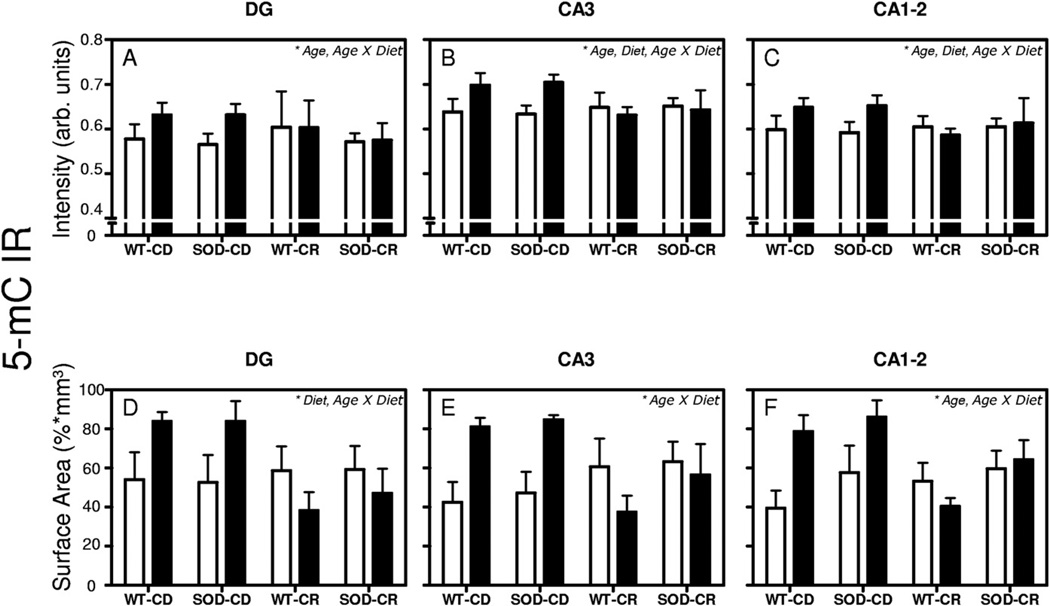
Mean intensity of 5-mC IR (Fig. 4A–C) was increased with aging from 12- to 24-month-old mice in all investigated regions (p = 0.028 for DG; p = 0,004 for CA3 and p = 0.009 for CA1–2) and decreased by CR diet only in the CA3 (p = 0.005) and CA1–2 (p = 0.002) regions. Interestingly, age × diet interactions were also observed for all hippocampal subregions investigated (p = 0.037 for DG; p < 0.001 for CA3; and p = 0.002 for CA1–2). Pairwise post hoc comparisons using Bonferroni correction in the CA3 region showed a significant increase of 5-mC IR in 24-month-old WT-CD mice as compared with 12-month-old WT-CD mice (+ 9.3%, p =0.024) and similarly for SOD1-CD mice (24- vs. 12-month-old, + 11.2%; p=0.005). On the contrary, a decrease of 5-mC IR was observed when comparing 24-month-old WT-CR to 24-month-old WT-CD mice (−10.6%, p = 0.005) and 24-month-old SOD1-CR to 24-month-old SOD1-CD mice (−9.6%, p = 0.034). In the CA1–2 region post hoc comparisons showed a decrease in 5-mC IR in 24-month-old WT-CR mice when compared with 24-month-old WT-CD mice (−10.6%, p = 0.029). No specific effects of the SOD1 genotype were observed. Thus, we observed an age-related increase of 5-mC IR in the CD groups but not in the CR groups.
Fig. 4.
5-Methylcytidine (5-mC) intensity and surface area. Mean and standard error of the mean of mean intensity value measurements of 5-mC immunoreactivity (IR) (A–F). Pooled data from the 4 groups of 12-month-old (white bars) and 24-month-old mice (black bars) are presented separately for dentate gyrus (DG) (A and D), CA3 (B and E), and CA1–2 (C and F). Surface area occupied by 5-mC IR measurements (D–F) were corrected for volume differences. The significant effects observed of each analysis are noted in the top right corner of each graph. p < 0.05 was considered as statistically significant.
3.3. 5-mC surface area
In the same way (Fig. 4D–F), diet (CR vs. CD) decreased the surface area of 5-mC IR in the DG (p = 0.038) and age (24 months vs. 12 months) increased it in the CA1–2 region (p = 0.041). Furthermore, significant age × diet interactions were observed in all hippocampal subregions (p = 0.007 for DG; p = 0.01 for CA3, and p = 0.01 for CA1–2), pointing to an age-related increase in the CD groups that was not observed in the CR groups. No effects of the SOD1 genotype were observed. Furthermore, pair-wise post hoc comparisons with Bonferroni correction showed no significant differences among individual groups.
3.4. 5-mC and Dnmt3a correlation analysis
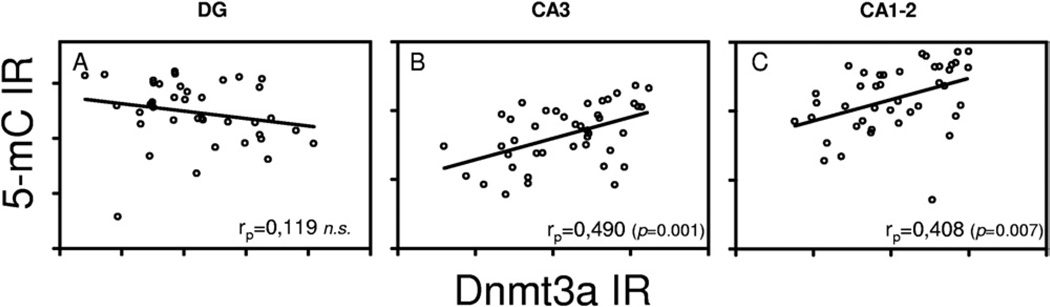
Previously, we observed, in the same cohort of mice, age-related changes of Dnmt3a IR that were attenuated by CR (Chouliaras et al., 2011). Thus, correlation analysis between 5-mC and Dnmt3a gray values was carried out, in order to examine to what extent the observed changes in 5-mC IR may be mediated, at least in part, by Dnmt3a. Positive linear correlation was observed in the CA3 (rp = 0.490, p = 0.001) and CA1–2 (rp = 0.408, p = 0.007) regions, but not in the DG (Fig. 5A–C).
Fig. 5.
5-Methylcytidine (5-mC) and DNA methyltransferase 3a (Dnmt3a) correlation analysis. Correlation analysis of 5-mC immunoreactivity (IR) and Dnmt3a IR for the dentate gyrus (DG), CA3 and CA1–2. Significant Spearman’s correlation coefficients are noted in the bottom right of each graph.
4. Discussion
Qualitative and quantitative assessment of 5-mC IR revealed densely-stained hypermethylated regions with nuclear localization, which were more pronounced in brains of 24-month-old animals when compared with 12-month-old mice. The age-related increase in 5-mC IR observed in CD animals was not observed in CR animals, suggesting that CR prevented these age-related alterations. In contrast, mice overexpressing SOD1 did not show any differences as compared with WT mice. Positive correlation with previously reported Dnmt3a IR analysis showed that the changes in 5-mC IR in CA3 and CA1–2 are associated with alterations in Dnmt3a.
4.1. Age-related increase in 5-mC
DNA methylation is a dynamic process subject to environmental influences and the aging process (Day and Sweatt, 2010; McGowan et al., 2009; Meaney and Ferguson-Smith, 2010; Murgatroyd et al., 2009; Rutten and Mill, 2009; Zhang et al., 2010). For instance, it has been shown that DNA methylation differences between monozygotic twins increase with age (Fraga et al., 2005). Furthermore, aberrant methylation patterns have been described in aging, such as global hypomethylation as well as hypermethylation of CpG islands, i.e. genomic regions rich in nucleotide sequences of cytosine (C) followed by guanine (G) (Christensen et al., 2009; Murgatroyd et al., 2010; Siegmund et al., 2007), whereas in the rodent hippocampus, age-associated DNA methylation changes of the memory-related Arc gene have been reported (Penner et al., 2010b). In addition, a global loss of 5-mC and methylation stabilizing factors, such as Dnmt1 and methyl-CpG binding protein 2 (MeCP2), has been observed in brains of Alzheimer’s disease (AD) patients when compared with aged-matched controls (Mastroeni et al., 2010). Similar findings were observed in a case report of a monozygotic twin pair discordant for AD (Mastroeni et al., 2009). Various other studies have shown epigenetic dysregulation in AD, pointing toward a shift from normal aging to AD, possibly related to stochastic factors, sustainable effects of environmental exposures, as well as gene × environment interactions (Avramopoulos et al., 2011; Cao et al., 2010; Chouliaras et al., 2010a, 2010b; Fuso and Scarpa, 2011; Lahiri et al., 2009; Mastroeni et al., 2011; Wang et al., 2008). As the 5-mC IR mainly reflects DNA methylation in CpG-rich regions, our findings are compatible with the previously reported age-associated hypermethylation of CpG islands, within the brain and other tissues (Christensen et al., 2009; Fraga and Esteller, 2007; Fraga et al., 2007; Siegmund et al., 2007). Furthermore, increased CpG methylation with aging has been reported in the frontal cortex, temporal cortex, pons, and cerebellum of human brains, with the hypermethylated loci mainly situated in genes related to DNA binding and transcriptional regulation (Hernandez et al., 2011). Moreover, the functional impact of the observed increased hippocampal levels of 5-mC with aging is thus not yet clear, but may contribute to age-related changes in gene expression and their regional specificity (Berchtold et al., 2008; Haberman et al., 2009; Penner et al., 2010b).
4.2. Correlation of Dnmt3a and 5-mC
Previously, we reported an age-related increase of Dnmt3a IR that was also attenuated by CR (Chouliaras et al., 2011). Dnmt3a is a de novo methyltransferase, implicated in learning, memory, and associated synaptic plasticity and neurogenesis. The observed positive correlation between Dnmt3a and 5-mC IR suggests that Dnmt3a could be a mediator of increased 5-mC IR in the CA1–2 and CA3 regions.
4.3. Prevention of age-related increase in 5-mC by caloric restriction
The age-related increase in 5-mC IR observed in the mouse hippocampus was prevented by CR. While an increase in 5-mC IR was found in 24-month-old CD mice as compared with the 12-month-old animals, CR mice did not show such age-related differences. The mechanism of this effect is not yet fully clear. So far, it is known that dietary and nutritional factors can alter DNA methylation patterns by affecting the 1 carbon metabolism, the main regulator of the methylation potential (Sugden, 2006). For instance, it has been shown that dietary manipulations targeting the 1 carbon metabolism may impact on methylation of various genes that have been linked to neurodegeneration (Fuso et al., 2011; Scarpa et al., 2006). Furthermore, a wealth of evidence suggests that the beneficial effects of CR are mediated by sirtuins, molecules with histone deacetylase properties (Blander and Guarente, 2004; Bordone and Guarente, 2005; Chen and Guarente, 2007; Chen et al., 2005, 2008; Dillin and Kelly, 2007) and that regulation of chromatin via histone alterations is intrinsically intertwined with the regulation of DNA methylation (Dong et al., 2007; Kishikawa et al., 2002). Thus, it is reasonable to speculate that CR induces both changes in DNA methylation as well as chromatin modifications and that these epigenetic changes are causally involved in the beneficial effects on life span.
4.4. No effects of SOD1 overexpression
Mice overexpressing SOD1 did not show any differences as compared with WT mice. Such a finding is in line with our previous reports of no effects of SOD1 on brain volumes (Rutten et al., 2010) and Dnmt3a (Chouliaras et al., 2011). Although the role of oxidative damage in aging has been well documented, its possible reduction by SOD1 did not show any effect in relation to 5-mC IR or previously reported Dnmt3a IR. Either 5-mC IR is not directly related to changes in oxidative stress, or such changes are not reflected in the hippocampus, or the levels of SOD1 and other antioxidants in this model were not sufficient enough to attenuate the effects of aging (Epstein et al., 1987; Przedborski et al., 1992).
4.5. Strengths and limitations
Besides the major strengths and limitations of the study design, as discussed previously (Chouliaras et al., 2011; Rutten et al., 2010), one of the major strengths of the present study is that the standard and controlled conditions under which the animals were kept minimize the effects of variations in the environment and point to effects of stochastic factors during life. Furthermore, in contrast to DNA extracted from tissue homogenates, the in situ DNA methylation detection by using a specific 5-mC antibody does not affect the integrity of the region of interest, allowing the identification of neuron- and region-specific differences. Conversely, a possible limitation of using such an antibody is the simultaneous detection of non-CpG methylation, which might serve a distinct role (Lister et al., 2009). Nevertheless, while non-CpG methylation is abundant in stem cells, its presence in postmitotic cells is very low (Lister et al., 2009; Ramsahoye et al., 2000). Thus, considering the relatively low proportion of dividing cells compared with differentiated neurons in the hippocampus, it is unlikely that non-CpG methylation contributes significantly to the present results.
4.6. Future directions
While the present study focused on relative measures of global 5-mC IR within cells, it would be very attractive to extend these with analyses of methylation profiles of genes using whole-genome interrogation or targeted hypothesis-based approaches. Unfortunately, the current experimental design did not allow the isolation of mRNA, which prevented us from examining whether the changes in 5-mC IR have functional consequences for gene expression. Although numerous studies have already reported the effects of aging and CR on gene expression (Lee et al., 2000; Park and Prolla, 2005; Prolla, 2002; Prolla and Mattson, 2001; Verbitsky et al., 2004; Weindruch et al., 2002), it would be highly interesting to study both epigenetic phenomena as well as their functional impact on gene expression in the same sample. Moreover, the role of other epigenetic modifications of the DNA (e.g., DNA hydroxymethylation) (Kriaucionis and Heintz, 2009; Tahiliani et al., 2009), and chromatin (e.g., histone modifications) awaits further clarification. The current analysis focused on levels of 5-mC per se and did not assess age-related changes in DNA hydroxymethylation. Interestingly, recent studies have suggested that 5-hydroxymethylcytosine (5-hmC) is an exclusive product of 5-mC (Williams et al., 2011), and is associated with chromatin opening and transcriptional activation (Jin et al., 2011; Valinluck et al., 2004; Williams et al., 2011). Moreover, it has been shown that DNA hydroxymethylation serves as an intermediate step in the process of active DNA demethylation in the brain (Guo et al., 2011). As for DNA methylation, changes in DNA hydroxymethylation may play a pivotal role in the aging process. Another interesting candidate for further investigations is repetitive element methylation. Recent studies have suggested altered repetitive element methylation in peripheral lymphocytes in association with advanced age and AD, namely a decrease in Alu methylation with age and an increase in long interspersed element-1 (LINE-1) methylation in AD (Bollati et al., 2009, 2011).
Altogether, these first observational epigenetic studies on the aging brain may serve as an important basis for future experimental animal research investigating the effects of interventions that alter DNA methylation on age-related molecular and cellular processes in the brain and on age-related functional decline. Such studies will be necessary to allow further interpretation on the causal relationships between the observed age-related changes in 5-mC and age-related functional decline.
In conclusion, the current findings reveal changes of DNA methylation in the hippocampus with aging, in a region-specific manner, and suggest that CR has a beneficial effect on the life span through impacting on DNA methylation.
Acknowledgements
Funds have been provided by the Internationale Stichting Alzheimer Onderzoek (ISAO), grant no. 09552, and the Netherlands Organization for Scientific Research (NWO, Veni Award 916.11.086) to B.P.F. Rutten, by the ISAO, grant no. 07551, to D.L.A. van den Hove and by a Marie Curie Host Fellowship Grant MC-EST 020589 EURON to L. Chouliaras. P.R. Hof is supported by NIH grant P50 AG05138. Figure 2 was generated with an SI-SD system (MBF Bioscience), which was obtained by NWO grant no. 91106003.
Footnotes
Disclosure statement
The authors declare no actual or potential conflicts of interest relating to this work.
All experiments were conducted in compliance with the Principles of Laboratory Animal Care (NIH publication no. 86-23, revised 1985) and the Dutch animal protection law, considering international standards. The experimental protocols were approved by the Animal Subjects Ethics Committee of The Maastricht University.
References
- Adams MM, Shi L, Linville MC, Forbes ME, Long AB, Bennett C, Newton IG, Carter CS, Sonntag WE, Riddle DR, Brunso-Bechtold JK. Caloric restriction and age affect synaptic proteins in hippocampal CA3 and spatial learning ability. Exp. Neurol. 2008;211:141–149. doi: 10.1016/j.expneurol.2008.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson RM, Shanmuganayagam D, Weindruch R. Caloric restriction and aging: studies in mice and monkeys. Toxicol. Pathol. 2009;37:47–51. doi: 10.1177/0192623308329476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avramopoulos D, Szymanski M, Wang R, Bassett S. Gene expression reveals overlap between normal aging and Alzheimer’s disease genes. Neurobiol. Aging. 2011:7. doi: 10.1016/j.neurobiolaging.2010.04.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berchtold NC, Cribbs DH, Coleman PD, Rogers J, Head E, Kim R, Beach T, Miller C, Troncoso J, Trojanowski JQ, Zielke HR, Cotman CW. Gene expression changes in the course of normal brain aging are sexually dimorphic. Proc. Natl. Acad. Sci. U. S. A. 2008;105:15605–15610. doi: 10.1073/pnas.0806883105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blander G, Guarente L. The Sir2 family of protein deacetylases. Annu. Rev. Biochem. 2004;73:417–435. doi: 10.1146/annurev.biochem.73.011303.073651. [DOI] [PubMed] [Google Scholar]
- Bollati V, Galimberti D, Pergoli L, Dalla Valle E, Barretta F, Cortini F, Scarpini E, Bertazzi PA, Baccarelli A. DNA methylation in repetitive elements and Alzheimer disease. Brain Behav. Immun. 2011 doi: 10.1016/j.bbi.2011.01.017. in press; [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bollati V, Schwartz J, Wright R, Litonjua A, Tarantini L, Suh H, Sparrow D, Vokonas P, Baccarelli A. Decline in genomic DNA methylation through aging in a cohort of elderly subjects. Mech. Ageing Dev. 2009;130:234–239. doi: 10.1016/j.mad.2008.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bordone L, Guarente L. Calorie restriction, SIRT1 and metabolism: understanding longevity. Nat. Rev. Mol. Cell Biol. 2005;6:298–305. doi: 10.1038/nrm1616. [DOI] [PubMed] [Google Scholar]
- Borg J, London J. Copper/zinc superoxide dismutase overexpression promotes survival of cortical neurons exposed to neurotoxins in vitro. J. Neurosci. Res. 2002;70:180–189. doi: 10.1002/jnr.10404. [DOI] [PubMed] [Google Scholar]
- Brown SE, Weaver IC, Meaney MJ, Szyf M. Regional-specific global cytosine methylation and DNA methyltransferase expression in the adult rat hippocampus. Neurosci. Lett. 2008;440:49–53. doi: 10.1016/j.neulet.2008.05.028. [DOI] [PubMed] [Google Scholar]
- Cadet JL, Sheng P, Ali S, Rothman R, Carlson E, Epstein C. Attenuation of methamphetamine-induced neurotoxicity in copper/zinc superoxide dismutase transgenic mice. J. Neurochem. 1994;62:380–383. doi: 10.1046/j.1471-4159.1994.62010380.x. [DOI] [PubMed] [Google Scholar]
- Calvanese V, Lara E, Kahn A, Fraga MF. The role of epigenetics in aging and age-related diseases. Ageing Res. Rev. 2009;8:268–276. doi: 10.1016/j.arr.2009.03.004. [DOI] [PubMed] [Google Scholar]
- Cao K, Chen-Plotkin AS, Plotkin JB, Wang LS. Age-correlated gene expression in normal and neurodegenerative human brain tissues. PLoS One. 2010;5 doi: 10.1371/journal.pone.0013098. pii: e13098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cardozo-Pelaez F, Song S, Parthasarathy A, Epstein CJ, Sanchez-Ramos J. Attenuation of age-dependent oxidative damage to DNA and protein in brainstem of Tg Cu/Zn SOD mice. Neurobiol. Aging. 1998;19:311–316. doi: 10.1016/s0197-4580(98)00067-0. [DOI] [PubMed] [Google Scholar]
- Chan PH, Epstein CJ, Kinouchi H, Kamii H, Imaizumi S, Yang G, Chen SF, Gafni J, Carlson E. SOD-1 transgenic mice as a model for studies of neuroprotection in stroke and brain trauma. Ann. N. Y. Acad. Sci. 1994;738:93–103. doi: 10.1111/j.1749-6632.1994.tb21794.x. [DOI] [PubMed] [Google Scholar]
- Chen D, Bruno J, Easlon E, Lin SJ, Cheng HL, Alt FW, Guarente L. Tissue-specific regulation of SIRT1 by calorie restriction. Genes Dev. 2008;22:1753–1757. doi: 10.1101/gad.1650608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen D, Guarente L. SIR2: a potential target for calorie restriction mimetics. Trends Mol. Med. 2007;13:64–71. doi: 10.1016/j.molmed.2006.12.004. [DOI] [PubMed] [Google Scholar]
- Chen D, Steele AD, Lindquist S, Guarente L. Increase in activity during calorie restriction requires Sirt1. Science. 2005;310:1641. doi: 10.1126/science.1118357. [DOI] [PubMed] [Google Scholar]
- Chouliaras L, Rutten BP, Kenis G, Peerbooms O, Visser PJ, Verhey F, van Os J, Steinbusch HW, van den Hove DL. Epigenetic regulation in the pathophysiology of Alzheimer’s disease. Prog. Neurobiol. 2010a;90:498–510. doi: 10.1016/j.pneurobio.2010.01.002. [DOI] [PubMed] [Google Scholar]
- Chouliaras L, Sierksma AS, Kenis G, Prickaerts J, Lemmens MA, Brasnjevic I, van Donkelaar EL, Martinez-Martinez P, Losen M, De Baets MH, Kholod N, van Leeuwen F, Hof PR, van Os J, Steinbusch HW, van den Hove DL, Rutten BP. Geneenvironment interaction research and transgenic mouse models of Alzheimer’s disease. Int. J. Alzheimers Dis. 2010b;2010 doi: 10.4061/2010/859101. pii: 859101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chouliaras L, van den Hove DL, Kenis G, Dela Cruz J, Lemmens MA, van Os J, Steinbusch HW, Schmitz C, Rutten BP. Caloric restriction attenuates age-related changes of DNA methyltransferase 3a in mouse hippocampus. Brain Behav. Immun. 2011;25:616–623. doi: 10.1016/j.bbi.2010.11.016. [DOI] [PubMed] [Google Scholar]
- Christensen BC, Houseman EA, Marsit CJ, Zheng S, Wrensch MR, Wiemels JL, Nelson HH, Karagas MR, Padbury JF, Bueno R, Sugarbaker DJ, Yeh RF, Wiencke JK, Kelsey KT. Aging and environmental exposures alter tissue-specific DNA methylation dependent upon CpG island context. PLoS Genet. 2009;5 doi: 10.1371/journal.pgen.1000602. e1000602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colman RJ, Anderson RM, Johnson SC, Kastman EK, Kosmatka KJ, Beasley TM, Allison DB, Cruzen C, Simmons HA, Kemnitz JW, Weindruch R. Caloric restriction delays disease onset and mortality in rhesus monkeys. Science. 2009;325:201–204. doi: 10.1126/science.1173635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Day JJ, Sweatt JD. DNA methylation and memory formation. Nat. Neurosci. 2010;13:1319–1323. doi: 10.1038/nn.2666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickstein DL, Kabaso D, Rocher AB, Luebke JI, Wearne SL, Hof PR. Changes in the structural complexity of the aged brain. Aging Cell. 2007;6:275–284. doi: 10.1111/j.1474-9726.2007.00289.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickstein DL, Walsh J, Brautigam H, Stockton SD, Jr., Gandy S, Hof PR. Role of vascular risk factors and vascular dysfunction in Alzheimer’s disease. Mt. Sinai J. Med. 2010;77:82–102. doi: 10.1002/msj.20155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dillin A, Kelly JW. Medicine. The yin-yang of sirtuins. Science. 2007;317:461–462. doi: 10.1126/science.1146585. [DOI] [PubMed] [Google Scholar]
- Dong E, Guidotti A, Grayson DR, Costa E. Histone hyperacetylation induces demethylation of reelin and 67-kDa glutamic acid decarboxylase promoters. Proc. Natl. Acad. Sci. U. S. A. 2007;104:4676–4681. doi: 10.1073/pnas.0700529104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Epstein CJ, Avraham KB, Lovett M, Smith S, Elroy-Stein O, Rotman G, Bry C, Groner Y. Transgenic mice with increased Cu/Zn-superoxide dismutase activity: animal model of dosage effects in Down syndrome. Proc. Natl. Acad. Sci. U. S. A. 1987;84:8044–8048. doi: 10.1073/pnas.84.22.8044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng J, Zhou Y, Campbell SL, Le T, Li E, Sweatt JD, Silva AJ, Fan G. Dnmt1 and Dnmt3a maintain DNA methylation and regulate synaptic function in adult forebrain neurons. Nat. Neurosci. 2010;13:423–430. doi: 10.1038/nn.2514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraga MF, Agrelo R, Esteller M. Cross-talk between aging and cancer: the epigenetic language. Ann. N. Y. Acad. Sci. 2007;1100:60–74. doi: 10.1196/annals.1395.005. [DOI] [PubMed] [Google Scholar]
- Fraga MF, Ballestar E, Paz MF, Ropero S, Setien F, Ballestar ML, Heine-Suñer D, Cigudosa JC, Urioste M, Benitez J, Boix-Chornet M, Sanchez-Aguilera A, Ling C, Carlsson E, Poulsen P, Vaag A, Stephan Z, Spector TD, Wu YZ, Plass C, Esteller M. Epigenetic differences arise during the lifetime of monozygotic twins. Proc. Natl. Acad. Sci. U. S. A. 2005;102:10604–10609. doi: 10.1073/pnas.0500398102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraga MF, Esteller M. Epigenetics and aging: the targets and the marks. Trends Genet. 2007;23:413–418. doi: 10.1016/j.tig.2007.05.008. [DOI] [PubMed] [Google Scholar]
- Franklin KB, Paxinos G. The Mouse Brain in Stereotaxic Coordinates. San Diego: Academic Press; 1997. [Google Scholar]
- Fuso A, Nicolia V, Pasqualato A, Fiorenza MT, Cavallaro RA, Scarpa S. Changes in Presenilin 1 gene methylation pattern in diet-induced B vitamin deficiency. Neurobiol. Aging. 2011;32:187–199. doi: 10.1016/j.neurobiolaging.2009.02.013. [DOI] [PubMed] [Google Scholar]
- Fuso A, Scarpa S. One-carbon metabolism and Alzheimer’s disease: is it all a methylation matter? Neurobiol. Aging. 2011;32:1192–1195. doi: 10.1016/j.neurobiolaging.2011.01.012. [DOI] [PubMed] [Google Scholar]
- Guo JU, Su Y, Zhong C, Ming GL, Song H. Hydroxylation of 5-methylcytosine by TET1 promotes active DNA demethylation in the adult brain. Cell. 2011;145:423–434. doi: 10.1016/j.cell.2011.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haberman RP, Colantuoni C, Stocker AM, Schmidt AC, Pedersen JT, Gallagher M. Prominent hippocampal CA3 gene expression profile in neurocognitive aging. Neurobiol. Aging. 2009 doi: 10.1016/j.neurobiolaging.2009.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernandez DG, Nalls MA, Gibbs JR, Arepalli S, van der Brug M, Chong S, Moore M, Longo DL, Cookson MR, Traynor BJ, Singleton AB. Distinct DNA methylation changes highly correlated with chronological age in the human brain. Hum. Mol. Genet. 2011;20:1164–1172. doi: 10.1093/hmg/ddq561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernandez-Blazquez FJ, Habib M, Dumollard JM, Barthelemy C, Benchaib M, de Capoa A, Niveleau A. Evaluation of global DNA hypomethylation in human colon cancer tissues by immunohistochemistry and image analysis. Gut. 2000;47:689–693. doi: 10.1136/gut.47.5.689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin SG, Wu X, Li AX, Pfeifer GP, Benchaib M. Genomic mapping of 5-hydroxymethylcytosine in the human brain. Nucleic Acids Res. 2011 doi: 10.1093/nar/gkr120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kishikawa S, Ugai H, Murata T, Yokoyama KK. Roles of histone acetylation in the Dnmt1 gene expression. Nucleic Acids Res. Suppl. 2002:209–210. doi: 10.1093/nass/2.1.209. [DOI] [PubMed] [Google Scholar]
- Kriaucionis S, Heintz N. The nuclear DNA base 5-hydroxymethylcytosine is present in Purkinje neurons and the brain. Science. 2009;324:929–930. doi: 10.1126/science.1169786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lahiri DK, Maloney B, Zawia NH. The LEARn model: an epigenetic explanation for idiopathic neurobiological diseases. Mol. Psychiatry. 2009;14:992–1003. doi: 10.1038/mp.2009.82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee CK, Weindruch R, Prolla TA. Gene-expression profile of the ageing brain in mice. Nat. Genet. 2000;25:294–297. doi: 10.1038/77046. [DOI] [PubMed] [Google Scholar]
- Levenson CW, Rich NJ. Eat less, live longer? New insights into the role of caloric restriction in the brain. Nutr. Rev. 2007;65:412–415. doi: 10.1111/j.1753-4887.2007.tb00319.x. [DOI] [PubMed] [Google Scholar]
- Levenson JM, Roth TL, Lubin FD, Miller CA, Huang IC, Desai P, Malone LM, Sweatt JD. Evidence that DNA (cytosine-5) methyltransferase regulates synaptic plasticity in the hippocampus. J. Biol. Chem. 2006;281:15763–15773. doi: 10.1074/jbc.M511767200. [DOI] [PubMed] [Google Scholar]
- Lister R, Pelizzola M, Dowen RH, Hawkins RD, Hon G, Tonti-Filippini J, Nery JR, Lee L, Ye Z, Ngo QM, Edsall L, Antosiewicz-Bourget J, Stewart R, Ruotti V, Millar AH, Thomson JA, Ren B, Ecker JR. Human DNA methylomes at base resolution show widespread epigenomic differences. Nature. 2009;462:315–322. doi: 10.1038/nature08514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L, van Groen T, Kadish I, Tollefsbol TO. DNA methylation impacts on learning and memory in aging. Neurobiol. Aging. 2009;30:549–560. doi: 10.1016/j.neurobiolaging.2007.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luebke JI, Weaver CM, Rocher AB, Rodriguez A, Crimins JL, Dickstein DL, Wearne SL, Hof PR. Dendritic vulnerability in neurodegenerative disease: insights from analyses of cortical pyramidal neurons in transgenic mouse models. Brain Struct. Funct. 2010;214:181–199. doi: 10.1007/s00429-010-0244-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mastroeni D, Grover A, Delvaux E, Whiteside C, Coleman PD, Rogers J. Epigenetic changes in Alzheimer’s disease: decrements in DNA methylation. Neurobiol. Aging. 2010;31:2025–2037. doi: 10.1016/j.neurobiolaging.2008.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mastroeni D, Grover A, Delvaux E, Whiteside C, Coleman PD, Rogers J. Epigenetic changes in Alzheimer’s disease: decrements in DNA methylation. Neurobiol. Aging. 2011;31:2025–2037. doi: 10.1016/j.neurobiolaging.2008.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mastroeni D, McKee A, Grover A, Rogers J, Coleman PD. Epigenetic differences in cortical neurons from a pair of monozygotic twins discordant for Alzheimer’s disease. PLoS One. 2009;4:e6617. doi: 10.1371/journal.pone.0006617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maswood N, Young J, Tilmont E, Zhang Z, Gash DM, Gerhardt GA, Grondin R, Roth GS, Mattison J, Lane MA, Carson RE, Cohen RM, Mouton PR, Quigley C, Mattson MP, Ingram DK. Caloric restriction increases neurotrophic factor levels and attenuates neurochemical and behavioral deficits in a primate model of Parkinson’s disease. Proc. Natl. Acad. Sci. U. S. A. 2004;101:18171–18176. doi: 10.1073/pnas.0405831102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattson MP, Duan W, Chan SL, Cheng A, Haughey N, Gary DS, Guo Z, Lee J, Furukawa K. Neuroprotective and neurorestorative signal transduction mechanisms in brain aging: modification by genes, diet and behavior. Neurobiol. Aging. 2002;23:695–705. doi: 10.1016/s0197-4580(02)00025-8. [DOI] [PubMed] [Google Scholar]
- McGowan PO, Sasaki A, D’Alessio AC, Dymov S, Labonté B, Szyf M, Turecki G, Meaney MJ. Epigenetic regulation of the glucocorticoid receptor in human brain associates with childhood abuse. Nat. Neurosci. 2009;12:342–348. doi: 10.1038/nn.2270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meaney MJ, Ferguson-Smith AC. Epigenetic regulation of the neural transcriptome: the meaning of the marks. Nat. Neurosci. 2010;13:1313–1318. doi: 10.1038/nn1110-1313. [DOI] [PubMed] [Google Scholar]
- Miller CA, Sweatt JD. Covalent modification of DNA regulates memory formation. Neuron. 2007;53:857–869. doi: 10.1016/j.neuron.2007.02.022. [DOI] [PubMed] [Google Scholar]
- Mouton PR, Chachich ME, Quigley C, Spangler E, Ingram DK. Caloric restriction attenuates amyloid deposition in middle-aged dtg APP/PS1 mice. Neurosci. Lett. 2009;464:184–187. doi: 10.1016/j.neulet.2009.08.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murgatroyd C, Patchev AV, Wu Y, Micale V, Bockmühl Y, Fischer D, Holsboer F, Wotjak CT, Almeida OF, Spengler D. Dynamic DNA methylation programs persistent adverse effects of early-life stress. Nat. Neurosci. 2009;12:1559–1566. doi: 10.1038/nn.2436. [DOI] [PubMed] [Google Scholar]
- Murgatroyd C, Wu Y, Bockmuhl Y, Spengler D. The Janus face of DNA methylation in aging. Aging [Albany] 2010;2:107–110. doi: 10.18632/aging.100124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park SK, Prolla TA. Lessons learned from gene expression profile studies of aging and caloric restriction. Ageing Res. Rev. 2005;4:55–65. doi: 10.1016/j.arr.2004.09.003. [DOI] [PubMed] [Google Scholar]
- Patel NV, Gordon MN, Connor KE, Good RA, Engelman RW, Mason J, Morgan DG, Morgan TE, Finch CE. Caloric restriction attenuates Abeta-deposition in Alzheimer transgenic models. Neurobiol. Aging. 2005;26:995–1000. doi: 10.1016/j.neurobiolaging.2004.09.014. [DOI] [PubMed] [Google Scholar]
- Penner MR, Roth TL, Barnes CA, Sweatt JD. An epigenetic hypothesis of aging-related cognitive dysfunction. Front. Aging Neurosci. 2010a;2:9. doi: 10.3389/fnagi.2010.00009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penner MR, Roth TL, Chawla MK, Hoang LT, Roth ED, Lubin FD, Sweatt JD, Worley PF, Barnes CA. Age-related changes in Arc transcription and DNA methylation within the hippocampus. Neurobiol. Aging. 2010b doi: 10.1016/j.neurobiolaging.2010.01.009. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prolla TA. DNA microarray analysis of the aging brain. Chem. Sens. 2002;27:299–306. doi: 10.1093/chemse/27.3.299. [DOI] [PubMed] [Google Scholar]
- Prolla TA, Mattson MP. Molecular mechanisms of brain aging and neurodegenerative disorders: lessons from dietary restriction. Trends Neurosci. 2001;24:S21–S31. doi: 10.1016/s0166-2236(00)01957-3. [DOI] [PubMed] [Google Scholar]
- Przedborski S, Jackson-Lewis V, Kostic V, Carlson E, Epstein CJ, Cadet JL. Superoxide dismutase, catalase, and glutathione peroxidase activities in copper/zinc-superoxide dismutase transgenic mice. J. Neurochem. 1992;58:1760–1767. doi: 10.1111/j.1471-4159.1992.tb10051.x. [DOI] [PubMed] [Google Scholar]
- Ramsahoye BH, Biniszkiewicz D, Lyko F, Clark V, Bird AP, Jaenisch R. Non-CpG methylation is prevalent in embryonic stem cells and may be mediated by DNA methyltransferase 3a. Proc. Natl. Acad. Sci. U. S. A. 2000;97:5237–5242. doi: 10.1073/pnas.97.10.5237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rutten BP, Brasnjevic I, Steinbusch HW, Schmitz C. Caloric restriction and aging but not overexpression of SOD1 affect hippocampal volumes in mice. Mech. Ageing Dev. 2010;131:574–579. doi: 10.1016/j.mad.2010.08.002. [DOI] [PubMed] [Google Scholar]
- Rutten BP, Korr H, Steinbusch HW, Schmitz C. The aging brain: less neurons could be better. Mech. Ageing Dev. 2003;124:349–355. doi: 10.1016/s0047-6374(03)00002-2. [DOI] [PubMed] [Google Scholar]
- Rutten BP, Mill J. Epigenetic mediation of environmental influences in major psychotic disorders. Schizophr. Bull. 2009;35:1045–1056. doi: 10.1093/schbul/sbp104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rutten BP, Schmitz C, Gerlach OH, Oyen HM, de Mesquita EB, Steinbusch HW, Korr H. The aging brain: accumulation of DNA damage or neuron loss? Neurobiol. Aging. 2007;28:91–98. doi: 10.1016/j.neurobiolaging.2005.10.019. [DOI] [PubMed] [Google Scholar]
- Rutten BP, Steinbusch HW, Korr H, Schmitz C. Antioxidants and Alzheimer’s disease: from bench to bedside (and back again) Curr. Opin. Clin. Nutr. Metab. Care. 2002;5:645–651. doi: 10.1097/00075197-200211000-00006. [DOI] [PubMed] [Google Scholar]
- Scarpa S, Cavallaro RA, D’Anselmi F, Fuso A. Gene silencing through methylation: an epigenetic intervention on Alzheimer disease. J. Alzheimers Dis. 2006;9:407–414. doi: 10.3233/jad-2006-9406. [DOI] [PubMed] [Google Scholar]
- Siegmund KD, Connor CM, Campan M, Long TI, Weisenberger DJ, Biniszkiewicz D, Jaenisch R, Laird PW, Akbarian S. DNA methylation in the human cerebral cortex is dynamically regulated throughout the life span and involves differentiated neurons. PLoS One. 2007;2:e895. doi: 10.1371/journal.pone.0000895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sohal RS, Weindruch R. Oxidative stress, caloric restriction, and aging. Science. 1996;273:59–63. doi: 10.1126/science.273.5271.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strackx E, van den Hove DL, Steinbusch HP, Prickaerts J, Vles JS, Blanco CE, Steinbusch HW, Gavilanes AW. Fetal asphyxia leads to a decrease in dorsal raphe serotonergic neurons. Dev. Neurosci. 2008;30:358–366. doi: 10.1159/000155218. [DOI] [PubMed] [Google Scholar]
- Sugden C. One-carbon metabolism in psychiatric illness. Nutr. Res. Rev. 2006;19:117–136. doi: 10.1079/NRR2006119. [DOI] [PubMed] [Google Scholar]
- Tahiliani M, Koh KP, Shen Y, Pastor WA, Bandukwala H, Brudno Y, Agarwal S, Iyer LM, Liu DR, Aravind L, Rao A. Conversion of 5-methylcytosine to 5-hydroxymethylcytosine in mammalian DNA by MLL partner TET1. Science. 2009;324:930–935. doi: 10.1126/science.1170116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valinluck V, Tsai HH, Rogstad DK, Burdzy A, Bird A, Sowers LC. Oxidative damage to methyl-CpG sequences inhibits the binding of the methyl-CpG binding domain (MBD) of methyl-CpG binding protein 2 (MeCP2) Nucleic Acids Res. 2004;32:4100–4108. doi: 10.1093/nar/gkh739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verbitsky M, Yonan AL, Malleret G, Kandel ER, Gilliam TC, Pavlidis P. Altered hippocampal transcript profile accompanies an age-related spatial memory deficit in mice. Learn. Mem. 2004;11:253–260. doi: 10.1101/lm.68204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang SC, Oelze B, Schumacher A. Age-specific epigenetic drift in late-onset Alzheimer’s disease. PLoS ONE. 2008;3:e2698. doi: 10.1371/journal.pone.0002698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weindruch R, Kayo T, Lee CK, Prolla TA. Gene expression profiling of aging using DNA microarrays. Mech. Ageing Dev. 2002;123:177–193. doi: 10.1016/s0047-6374(01)00344-x. [DOI] [PubMed] [Google Scholar]
- Weindruch R, Walford RL, Fligiel S, Guthrie D. The retardation of aging in mice by dietary restriction: longevity, cancer, immunity and lifetime energy intake. J. Nutr. 1986;116:641–654. doi: 10.1093/jn/116.4.641. [DOI] [PubMed] [Google Scholar]
- Williams K, Christensen J, Pedersen MT, Johansen JV, Cloos PA, Rappsilber J, Helin K. TET1 and hydroxymethylcytosine in transcription and DNA methylation fidelity. Nature. 2011;473:343–348. doi: 10.1038/nature10066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang TY, Hellstrom IC, Bagot RC, Wen X, Diorio J, Meaney MJ. Maternal care and DNA methylation of a glutamic acid decarboxylase 1 promoter in rat hippocampus. J. Neurosci. 2010;30:13130–13137. doi: 10.1523/JNEUROSCI.1039-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]