Abstract
Antibody-drug conjugates (ADCs) with biotin as a model cargo tethered to IgG1 mAbs via different linkers and conjugation methods were prepared and tested for thermostability and ability to bind target antigen and Fc receptor. Most conjugates demonstrated decreased thermostability relative to unconjugated antibody, based on DSC, with carbohydrate and amine coupled ADCs showing the least effect compared with thiol coupled conjugates. A strong correlation between biotin-load and loss of stability is observed with thiol conjugation to one IgG scaffold, but the stability of a second IgG scaffold is relatively insensitive to biotin load. The same correlation for amine coupling was less significant. Binding of antibody to antigen and Fc receptor was investigated using surface plasmon resonance. None of the conjugates exhibited altered antigen affinity. Fc receptor FcγIIb (CD32b) interactions were investigated using captured antibody conjugate. Protein G and Protein A, known inhibitors of Fc receptor (FcR) binding to IgG, were also used to extend the analysis of the impact of conjugation on Fc receptor binding. H10NPEG4 was the only conjugate to show significant negative impact to FcR binding, which is likely due to higher biotin-load compared with the other ADCs. The ADC aHISNLC and aHISTPEG8 demonstrated some loss in affinity for FcR, but to much lower extent. The general insensitivity of target binding and effector function of the IgG1 platform to conjugation highlight their utility. The observed changes in thermostability require consideration for the choice of conjugation chemistry, depending on the system being pursued and particular application of the conjugate.
Keywords: amine, carbohydrate, CD32b, Conjugate, DSC, Fc, linker, SPR, thermostability, thiol
The US Food and Drug Administration’s approval of brentuximab vedotin (AdcetrisTM) in August 2011 demonstrates the therapeutic potential of antibody-drug conjugates (ADCs) to treat many cancers. The therapeutic effects of ADCs can result from a complex combination of mechanisms, including anti-proliferative or cell-killing potential through delivery of cytotoxic agents, apoptotic signaling, antibody-dependent cell-mediated cytotoxicity (ADCC) and complement dependent cytotoxicity (CDC). The inherent specificity of ADCs, coupled with their long serum half-life and low immunogenicity have generated substantial interest and investment toward improving these drug delivery platforms.
The choice of linker that connects the drug to the antibody scaffold is a critical factor in determining the effectiveness of ADC therapy. There has been notable progress recently in linker technology and the range of chemical reagents available for coupling the antibody to the drug of interest.1 Several factors contribute to optimal linker function, including stability in vivo, immunogenicity, and efficiency of drug release from ADC. The linker should be sufficiently stable to allow the antibody to carry the toxic payload to the cell of interest and subsequently into the cell, where it must then release the active cytotoxic drug. This last step may be of critical importance, and it depends on the method of cellular uptake and internalization of the ADC, which in turn may change with linker properties.2,3 Furthermore, a linker should be chosen that induces no or minimal immunogenicity or off-target binding.
The site of conjugation must also be considered. Ideally, the site for conjugation must not interfere with any therapeutic function, nor significantly disrupt regions that may confer fold stability. The most common approach in preparing ADCs is to use heterobifunctional linkers. These consist of a spacer with chemically distinct reactive groups on either end that can couple to various functional groups on the respective antibody or drug molecule. This provides considerable control and flexibility in how one attaches the linker. There are several targets on the antibody available for conjugation. Three common strategies include thiol coupling to reduced cysteines, amine coupling to lysine residues, and coupling to oxidized sugar residues on glycosylated mAbs. In principle, each method offers advantages and disadvantages with regard to product heterogeneity, stability and potential impact on effector function. Because in some cases modification of antibody residues spatially distant from the CDR domains can affect antigen binding, it is reasonable to expect that conjugation to the different functional groups may have different functional affects.4
Since different IgG1s can in principle have different sensitivities to conjugation with drugs, it is important to determine whether the trends observed in effects of conjugation for one IgG1 can be generalized to others. In addition to variable linkers and coupling strategies, we compared two distinct IgG1 scaffolds, to determine if different Fab domains would differentially be affected by the different linkers. The IgG1s used here include anti-6xHis (aHIS), which is directed against his-tags and therefore can be used with varying antigens that vary in size or other property, and HyHEL-10 (H10) anti-hen lysozyme, which provides a well-characterized scaffold with well-understood interactions with its antigen. In effect, we constructed a matrix of linker, IgG1 scaffold, and conjugation chemistry to explore their effects on in vitro properties of ADCs.
Although the available literature suggests indirectly that the optimal choice of linker and conjugation site may vary with mAb idiotype, drug payload, conjugation site and antigen, no direct comparison of the effects of linker characteristics on functional properties for multiple mAbs and variable antigen has been reported in the public domain. General ‘rules’ about the advantages or disadvantages of specific linkers or conjugation methods are not well documented. Here we systematically evaluate the impact of linker and coupling chemistry on the properties of two model IgG1 antibodies (Fig. 1A). The results demonstrate some general trends concerning optimal linker strategies.
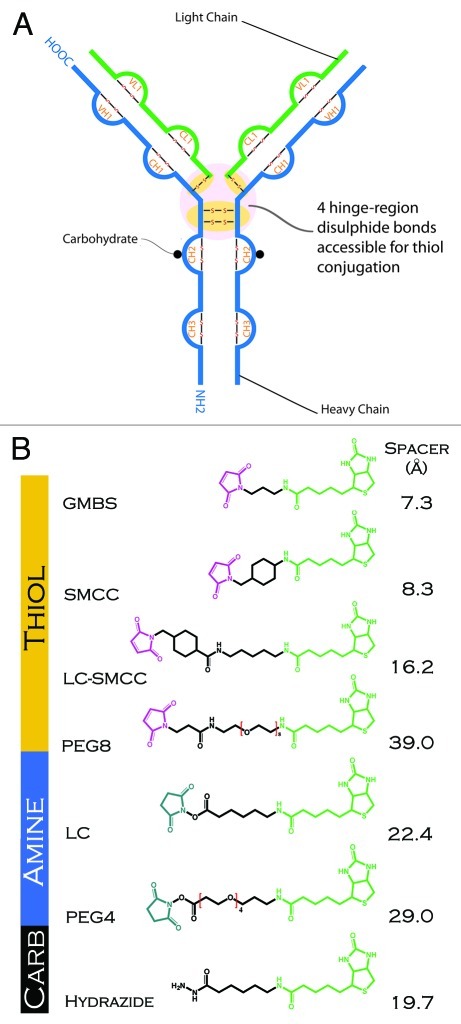
Figure 1. (A) Schematic representation of an IgG1 antibody showing sites of conjugation (thiol, amine and carbohydrate) used in generating the antibody-biotin conjugates. (B) Structure and length of linkers used in conjugating biotin (biocytin) to antibody. N-hydroxysuccinimide (blue), spacer arm (black), maleimide (violet), biotin (biocytin) (green).
Results
Overall strategy
To explore the effects of IgG1 structure, conjugation chemistry, and linker on ADC properties, we exploited two model IgG1 scaffolds, with multiple linkers of variable length and hydrophobicity. The linkers were used to attach the model ‘drug’ biotin to amines, reduced thiols, or oxidized carbohydrates on each IgG. In all cases, the form of biotin used for conjugation was biocytin, which allows for facile amine coupling of the biotin. Amine and carbohydrate linkers were purchased with this biocytin attached. For clarity, we refer to the group as biotin rather than biocytin in the remainder of this report. The structures of the linkers and biotin are shown in Figure 1B. For ADC identification, the subscripts ‘N’, ‘T’, and ‘C’ refer to coupling to amines, thiols, or carbohydrates respectively. Biotin was chosen as a model drug for several reasons. Biotin, while hydrophobic, has a relatively low propensity to form aggregates or to bind non-specifically to protein surfaces. It also provides a rapid method to verify conjugation via SPR, quantify the level of biotin load on the ADC (Table 1) and verify that the recognition of biotin by streptavidin is not compromised in the conjugates. All IgG-linker-biotin conjugates synthesized showed positive capture by the Biacore streptavidin-linked CAP sensor chip surface (data not shown). IgG1 idiotype monoclonal antibodies (mAbs), such as the mouse H10 and aHIS, are the most abundant and commonly used mAb platform for ADCs, and the IgG1 idiotype contains a total of 16 disulfide bonds including four interchain (2 heavy-heavy, two heavy-light) and 12 intrachain disulfide bonds (Fig. 1A). The four interchain disulfide bonds are accessible to reduction without unfolding the antibody. This gives a total of eight potential conjugation sites, assuming reduction of all four bonds. In practice, a mixture of thiol conjugates with different load (0, 2, 4, 6 and 8) is presumed. For amine coupling there are many surface accessible lysine residues distributed over a characteristic IgG1 structure. No attempt was made here to separate mixtures of antibodies loaded with different levels of biotin, and the average number of biotins per ADC were used as a measure of biotin load (Table 2).
Table 1. Impact of linker and conjugation chemistry on ADC thermal stability as measured by DSC.
| ADCa | Transition Temperature Tm (°C) |
|||
|---|---|---|---|---|
| Component 1 | Component 2 | Component 3 | Component 4 | |
| |
|
|
|
|
| H10 |
77.5 |
74.2 |
|
|
| H10 /w TCEP |
76.4 |
74.8 |
|
|
| H10NPEG4L |
76.4 |
73.3 |
|
|
| H10NPEG4H |
75.1 |
71.8 |
|
|
| H10TSMCC |
77.6 |
72.5 |
70.0 |
|
| H10TLC-SMCC |
78.2 |
72.4 |
69.0 |
|
| H10TGMBS |
77.4 |
72.9 |
66.8 |
|
| H10TPEG8 |
77.5 |
72.9 |
67.3 |
|
| H10C |
76.8 |
74.8 |
|
|
| aHIS |
78.9 |
71.2 |
67.5 |
61.7 |
| aHIS /w TCEP |
79.0 |
65.8 |
63.2 |
60.1 |
| aHISNPEG4 |
78.3 |
70.7 |
67.1 |
61.3 |
| aHISNLC |
78.1 |
70.6 |
67.0 |
61.1 |
| aHISTSMCC |
78.0 |
|
60.8 |
58.3 |
| aHISTLC-SMCC |
78.1 |
|
60.7 |
58.0 |
| aHISTGMBS |
78.2 |
|
60.7 |
58.0 |
| aHISTPEG8 | 77.7 | 60.5 | 57.9 | |
N, amine; T, thiol; C, Carbohydrate; L, low linker ratio; H, high linker ratio
Table 2. Impact of linker and conjugation chemistry on ADC binding to antigen.
| ADCa | KD (pM) | BTN:Abb | ||
|---|---|---|---|---|
| |
|
|
|
|
| H10 |
397 |
± 32.5 |
|
|
| H10NPEG4L |
546 |
± 80.4 |
6.4 |
± 0.3 |
| H10NPEG4H |
825 |
± 172.0 |
14.2 |
± 0.4 |
| H10TSMCC |
451 |
± 52.4 |
4.9 |
± 0.2 |
| H10TLC-SMCC |
297 |
± 38.6 |
4.9 |
± 0.8 |
| H10TGMBS |
376 |
± 63.3 |
6.6 |
± 0.3 |
| H10TPEG8 |
356 |
± 1.1 |
6.5 |
± 0.4 |
| H10C |
371 |
± 34.4 |
0.7 |
± 0.1 |
| |
|
|
|
|
| aHIS |
20.8 |
± 1.8 |
|
|
| aHISNPEG4 |
24.1 |
± 0.1 |
3.6 |
± 1.1 |
| aHISNLC |
21.8 |
± 0.3 |
3.7 |
± 0.9 |
| aHISTSMCC |
22.6 |
± 0.6 |
5.7 |
± 1.4 |
| aHISTLC-SMCC |
22.5 |
± 0.4 |
6.0 |
± 2.3 |
| aHISTGMBS |
22.8 |
± 2.4 |
4.9 |
± 0.6 |
| aHISTPEG8 | 23.3 | ± 2.8 | 7.7 | ± 0.04 |
N, amine; T, thiol; C, Carbohydrate; L, low linker ratio; H, high linker ratio. b. BTN, biotin; Ab, antibody
Thermal stability
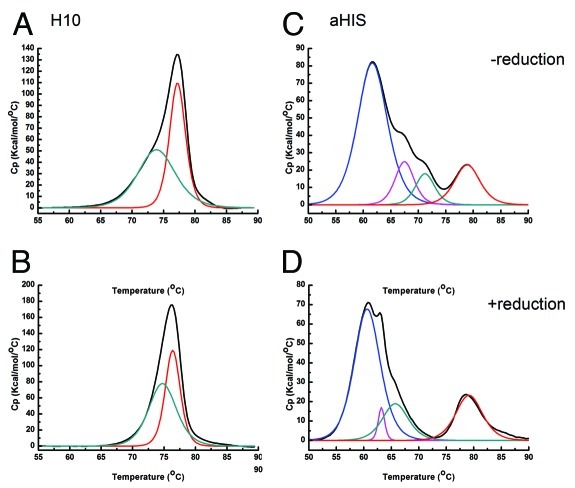
DSC was used to characterize each antibody and its corresponding ADCs to determine effects of linker and coupling strategy on thermal stability. Each DSC thermogram was fit to multiple components. A shift to lower Tm for a component is indicative of a loss in stability. Replicates for SD analysis are not available for each ADC. When evaluating significance of changes to Tm for a particular ADC from the naked antibody we used the conservative value of greater than 1°C as a minimum. In fact in the samples where a significant change in Tm is noted, the values reported are much greater than those characteristic for replicate samples with this instrument.5 The naked H10 antibody unfolds in two distinct but overlapping irreversible transitions (Fig. 2A), whereas the naked aHIS antibody unfolds with four distinct but overlapping irreversible transitions (Fig. 2C). IgG1 mAbs often exhibit two or more transitions in DSC thermograms due to the vastly different stabilities6 of the immunoglobin structural domains. The heterogeneity in drug-load with all ADC preparations is expected to cause some broadening of transitions seen in DSC thermograms. However, all DSC thermograms fit well to between two and four components using a non-two state model. In the absence of knowledge regarding the degree of heterogeneity in biotin load, a relative comparison of linkers and conjugation chemistries was pursued.
Figure 2. DSC thermograms and deconvoluted component transitions for the two unconjugated IgG1 antibodies in the presence and absence of TCEP. Left panels: (A) H10 without reducing agent (B) H10 with reducing agent; Right panels (C) aHIS without reducing agent (D) aHIS with reducing agent. Raw thermogram is shown in black. Individual transitions from non-two state fit are color coded as component 1 (red), component 2 (green), component 3 (magenta) and component 4 (blue).
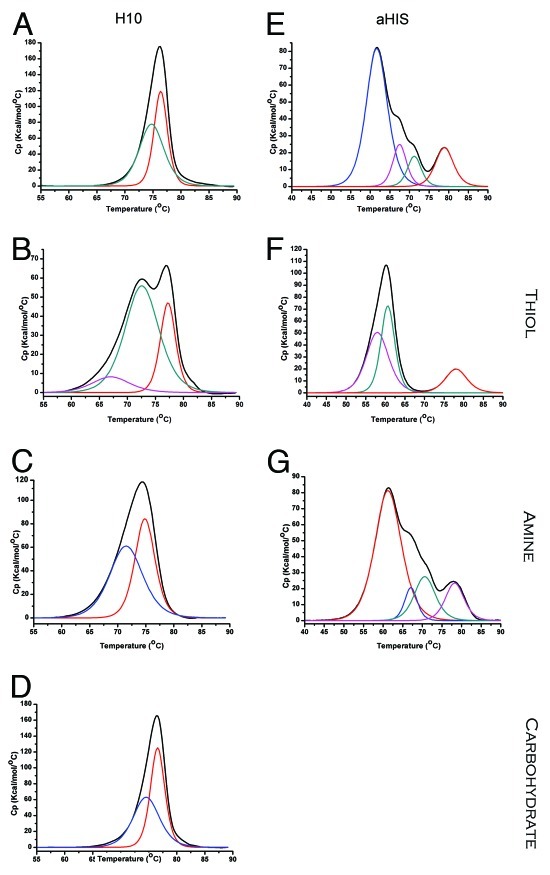
With H10 generated ADCs, the amine and carbohydrate conjugation chemistries did not alter the pattern of two component transitions seen with the naked antibody (Figs. 3A, C, D, Table 1). Amine conjugation to accessible surface lysine residues presents an additional strategy for conjugation of drugs, with the potential to modify stability of the antibody fold. There was a clear difference in stability observed with the amine coupled ADCs with a drop in Tm for both components 1 and 2. The greater the biotin load (Table 2) the greater the change in Tm. For the corresponding thiol conjugates (Fig. 3B; Table 1), a new component is observed with a much lower thermostability than the other two components. While component 1 does not change significantly in Tm for the thiol generated ADCs, the presence of this new lower temp transition results in ADC that are significantly less stable than the naked antibody, with H10TGMBS beginning to partially unfold at a Tm 7.4°C below that of any component in the naked antibody. This ADC was also the one with the highest biotin load.
Figure 3. DSC thermograms comparing ADCs prepared with different conjugation chemistries. Raw thermograms are shown in black. Individual transitions are color coded as: component 1 (red), component 2 (green), component 3 (magenta) and component 4 (blue). (A) H10, (B) H10TSMCC (C) H10NPEG4H, (D) H10C (E) aHIS (F) aHISTLC-SMCC (G) aHISNPEG4.
For the aHIS ADCs, the pattern of component transitions observed with DSC again differs depending on conjugation chemistry (Fig. 3E–G, Table 1). However, unlike H10 the number of visible components decreases from 4 with the naked antibody and amine-coupled ADCs to 3 for the thiol ADCs. It is unclear whether the lack of a component 2 for thiol-coupled ADCs reflects its superposition with another component in the thermogram that makes it indistinguishable. For amine aHIS ADCs, components 1–4 remain largely unaffected, even with higher biotin load (Table 2). For the thiol aHIS ADCs component 3 shows a large decline in Tm relative to the naked antibody of between 7.0–6.7°C which is comparable to the difference observed between components 2 and 3 for the thiol H10 ADCs. Component 4 showed a modest decline in Tm compared with the naked antibody, decreasing as much as 3.8°C for aHISTPEG8. So, in comparison to aHIS, the impact of thiol conjugation to thermostability was greater for H10 ADCs with comparable biotin load. The results for the shifts in low temperature DSC components with different model ADCs are summarized in Figure 4. In general, the results clearly demonstrate that different thiol-conjugates IgGs yield ADCs with significantly different sensitivity to drug load, even with the same linkers.
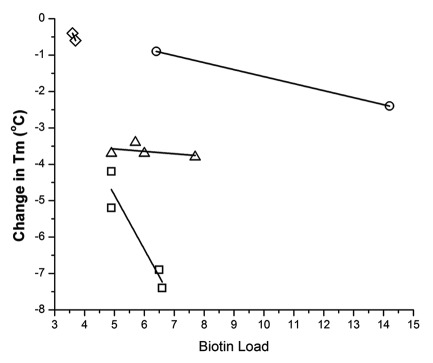
Figure 4. Correlation between biotin load and change in Tm for low-temperature components from DSC for ADC, grouped according to conjugation chemistry and/or antibody. (circle) amine-conjugated H10, (diamond) amine-conjugated aHIS, (triangle) thiol-conjugated aHIS and (square) thiol-conjugated H10.
For this work, we were interested in changes in thermostability that may be important for the manufacture and use of these ‘model’ therapeutics. Therefore, we focused on notable changes in the relative components with the lowest temperature transition. For thiol conjugates, H10 showed a strong correlation (R2 = 0.87) between higher biotin load and loss of stability. For aHIS, no correlation was observed for the corresponding thiol conjugates when comparing the lowest temperature transitions. However, among different DSC components there are similarities. This suggests that conjugation to free thiols located in the hinge region can differentially affect IgG1 scaffolds in ways that could have implications for their manufacture. With amine conjugates there were only two linkers used for each antibody. For amine H10 conjugates the correlation between biotin load and change in Tm for the low temperature component is less than that of thiol H10 conjugates. With amine aHIS conjugates the two Tm values are too close to confidently assess the correlation between biotin load and change in Tm. However, both values are below that observed with any of the thiol conjugates when those data sets are extrapolated to comparable biotin loads. What is clear from this data are that amine coupling has had less impact on thermostability compared with thiol conjugation for both antibodies. Also, for thiol conjugation the impact on thermostability with biotin load is very different among the two antibodies tested.
To determine the affect of reduction without chemical elaboration of the hinge region disulfides alone, both antibodies were unfolded in the presence of TCEP as the reducing agent. At the concentrations tested here (10 mM), TCEP had negligible impact on the DSC baseline. As stated previously, the H10 antibody has two characteristic component transitions (Fig. 2A) and the number of components, as well as the transition temperatures, did not change in the presence of this reducing agent (Fig. 2B). For the aHIS antibody, reduction of the hinge inter-chain disulfides with TCEP showed a slight negative affect on thermogram components 2, 3 and 4 with decreased thermostability (Fig. 2D). These data demonstrate that the effect of thiol reduction on stability can be IgG dependent, but the reduction of IgG by itself does not cause a significant decrease in thermal stability in either antibody used here.
In addition to thiol and amine conjugation strategies, we examined the effect of linking via carbohydrate. IgGs include N-linked oligosaccharides on two conserved asparagine residues (Asn297) in the CH2 domain of the Fc region and approximately 15–20% also have N-linked oligosaccharides in the variable region.7 Oxidation of these N-linked sugars offers a convenient means of conjugation of a drug or small molecule at a defined location, with potentially less of a destabilizing effect than hinge-region thiol conjugation. Among the conjugation strategies we tested, the carbohydrate linked ADCs had the smallest changes in Tm for DSC profile compared with the unconjugated antibody (Fig. 3D). This may be in part due to the much lower level of conjugation (Table 2). However, even taking this into account, the affect of conjugation on Tm was much less than that for the thiol or amine conjugated antibodies.
Antigen binding
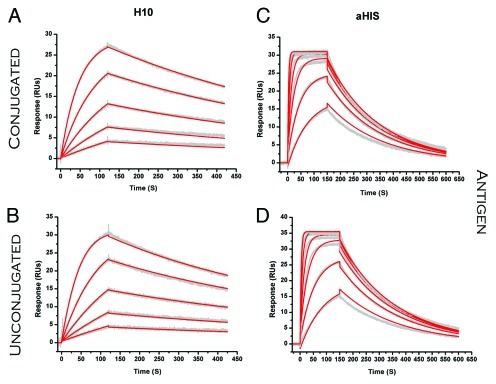
Because tight binding to its target is inherent in the strategy when using these drug delivery platforms, any effects on antigen binding due to conjugation are important. Such effects are possible, in principle, due to long range structural effects transmitted from conjugation sites to the antigen binding sites, or in the case of amine-conjugation potentially affecting hotspot lysine residues at the interface. Given the moderate role for disulfides in antibody stability (Fig. 2), we also studied whether conjugation to hinge-region disulfides affects antigen binding.8 Each individual antibody and its respective ADCs were tested for binding to antigen. Representative sensorgrams are shown in Figure 5; comparisons of KD were made by assessment of differences (> 2-fold difference from unconjugated antibody) with conjugation chemistry or linker. None of ADCs for either antibody were different than the unconjugated antibody (Table 2). For the mAbs tested in this study, there was no clear disadvantage to antigen binding for any of the linkers or chemistries investigated here. The amine-conjugated H10NPEG4H ADC was the one notable exception, and most likely reflects modification of a lysine residue at or near the paratope when the biotin load is sufficiently high.
Figure 5. SPR sensorgrams comparing binding of ADC (analyte) to antigen (ligand). (A) H10 binding to JQL (B). H10TPEG8 binding to JQL. (C) aHIS binding to 6xHis peptide (D). aHISNPEG4 binding to 6xHis peptide. Raw sensorgrams are shown in gray with results from fitting that data to a 1:1 binding model depicted in red.
Effector function and Fc binding
Effector functions mediated by binding of the Fc region of IgG1 antibodies to Fc receptors (FcR) can play an important role in the overall effectiveness of ADC therapies.9-11 Modifciation of residues directly involved or near the Fc recdeptor binding site, such as regions of glycosylation on antibodies, may have an important role in FcR binding and in the structural integrity.12,13 The potential therapeutic benefits of FcR activation or binding to other ‘FcR-like’ proteins make these interactions a target for optimization. All three conjugation strategies and linkers explored in this study could have direct or indirect effects on the structure and accessibility of the Fc region of the antibody. To test this, we captured the ADC to a SPR sensor surface on which native antigen was immobilized and then investigated binding of FcR to the captured ADC (Figs. 6, 7, 8). Effectively we began with a surface of ADC that was already bound to its corresponding antigen. This approach had several advantages over coupling of the antibody directly to the CM5 dextran surface of the chip or the Fc receptor. Because one of the approaches undertaken in conjugation involved amine coupling, the amine-sites available for coupling the ADCs to a CM5 sensor chip were much lower. Capturing the antibody on the SPR chip via its antigen also minimizes the possibility of interactions between the chip surface and the Fc region that could be confused with effects of N-linked biotin on the Fc region. The SPR data of FcR binding to ADC captured on the chip surface all fit very well to a one-site binding model. There was no statistically significant improvement of the fits to the data for a two-site binding model with non-equivalent binding sites, as observed by others.14 As noted above, one important distinction to other work using SPR in this area is that in our experiments the antibody/ADC was captured to the chip surface via the antigen rather than using direct amine coupling.
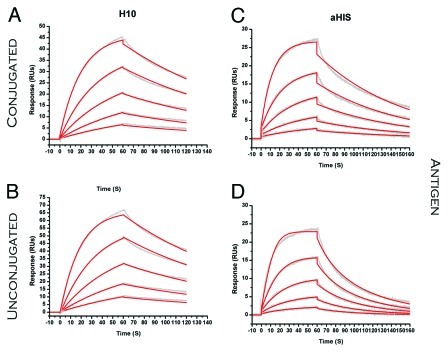
Figure 6. SPR sensorgrams comparing binding of ADC (ligand) to Protein G (analyte). (A) H10 (B). H10TPEG8 (C) aHIS (D). aHISNPEG4. Raw sensorgrams are shown in gray with results from fitting that data to a 1:1 binding model depicted in red.
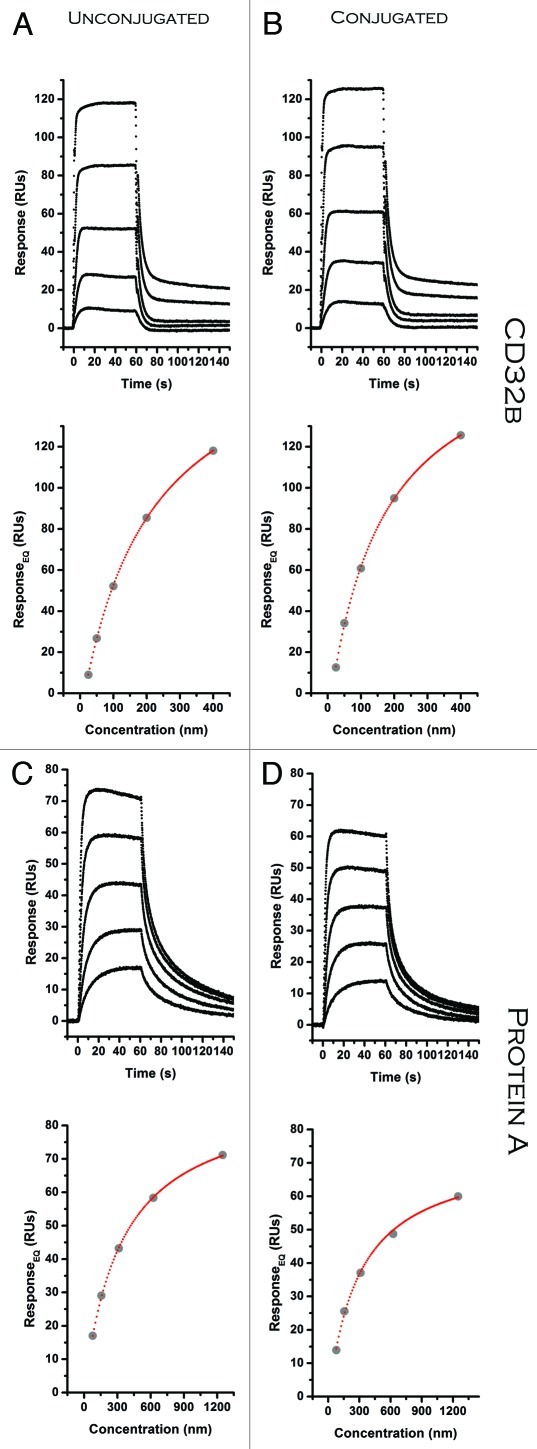
Figure 7. SPR equilibrium data comparing ADC (ligand) binding to CD32b and Protein A (analyte). (A) H10 (B). H10NPEG4L (C) H10 (D) H10NPEG4L. Raw sensorgrams are shown in black with corresponding results from fitting that data to a 1:1 binding model depicted in a binding isotherm below.
Figure 8. SPR equilibrium data comparing ADC (ligand) binding to CD32b and Protein A (analyte). (A) aHIS (B). aHISNLC (C) aHIS (D) aHISNPEG4. Raw sensorgrams are shown in black with corresponding results from fitting that data to a 1:1 binding model depicted in a binding isotherm below.
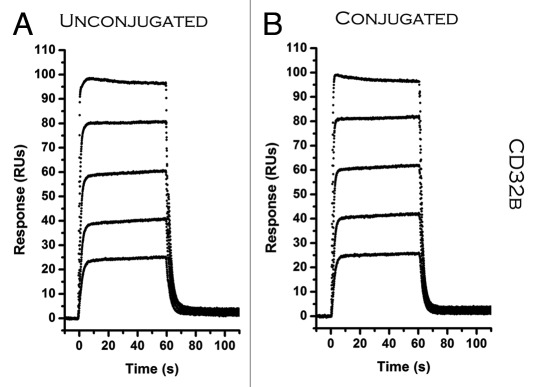
Some conjugation strategies were found to have a significant affect on binding to certain FcR-binding partners. For H10NPEG4H the affinity was reduced 17-fold for Protein G (Table 3). This may be due to coupling to lysine residues proximal to, or in direct contact with, Protein G binding residues. There was also a decrease in binding to Protein G observed with amine coupled aHISNLC. The aHISTPEG8 ADC was the only one to show some negative affect on binding with all FcR model proteins (CD32b,Protein A, and Protein G). For this conjugate, the higher ratio of biotin load may be one reason for this (Table 2). CD32b is an inhibitor for Fcγ receptors that are often involved in effector functions such as ADCC and C1q mediated complement activation. A reduction in expression of CD32b has been associated with enhanced antitumor activity for antibodies.15 These results suggest that amine coupling at high load-ratios can have a significant affect on binding to the Fc domain, and a similar evaluation would prove useful for any therapeutic ADC targeting effector function and using this conjugation chemistry. Other reports suggest a strong correlation with pegylation of antibodies and a reduction in FcR binding.16,17 We observed a large reduction, but only with one ADC binding to one particular FcR.
Table 3. Impact of linker and conjugation chemistry on ADC binding to Fc receptor.
| ADCa |
KD (nM) |
|||||
|---|---|---|---|---|---|---|
| CD32b | Protein G | Protein A | ||||
| H10 |
168.7 |
± 13.1 |
134.3 |
± 7.5 |
434.3 |
± 3.2 |
| H10NPEG4L |
150.3 |
± 10.8 |
324.3 |
± 17.1 |
510.0 |
± 5.2 |
| H10NPEG4H |
87.9 |
± 12.1 |
2314.0 |
± 175.0 |
334.0 |
± 5.2 |
| H10TSMCC |
134.0 |
± 10.5 |
124.3 |
± 18.9 |
495.3 |
± 10.4 |
| H10TLC-SMCC |
267.7 |
± 14.0 |
99.2 |
± 9.6 |
442.3 |
± 56.7 |
| H10TGMBS |
136.3 |
± 20.4 |
110.0 |
± 3.5 |
445.3 |
± 59.5 |
| H10TPEG8 |
227.3 |
± 10.6 |
132.3 |
± 18.6 |
454.3 |
± 55.4 |
| H10C |
124.7 |
± 6.4 |
90.5 |
± 18.2 |
454.7 |
± 34.5 |
| |
|
|
|
|
|
|
| aHIS |
203.0 |
± 9.5 |
119.6 |
± 28.1 |
309.6 |
± 28.6 |
| aHISNPEG4 |
163.4 |
± 20.5 |
137.0 |
± 25.1 |
361.6 |
± 73.4 |
| aHISNLC |
163.2 |
± 16.3 |
197.9 |
± 9.3 |
368.8 |
± 50.2 |
| aHISTSMCC |
138.8 |
± 27.9 |
157.3 |
± 36.3 |
165.9 |
± 39.4 |
| aHISTLC-SMCC |
139.2 |
± 25.6 |
163.3 |
± 8.4 |
164.8 |
± 36.7 |
| aHISTGMBS |
206.5 |
± 15.0 |
149.6 |
± 5.5 |
232.0 |
± 36.3 |
| aHISTPEG8 | 448.7 | ± 68.3 | 253.4 | ± 9.1 | 523.0 | ± 88.1 |
N, amine; T, thiol; C, Carb
Discussion
The lack of any significant change to antigen binding, or in most cases FcR binding, with any of the linkers or conjugation methods used, support the important role for antibodies as robust drug platforms for therapeutics. One attractive aspect to amine conjugation is the potential for increased drug load compared with using the fixed eight thiols presented by the IgG1 hinge disulfides. Because increased drug load on the ADC might be expected to improve therapeutic efficacy, the relatively mild perturbations to both Fc receptor binding and antigen binding observed in this work suggest that this advantage might be exploitable. However, in the case of thiol ADC, an increase in drug load does not necessarily correlate with improved therapeutic outcome.18 With amine coupling, these results suggest that an analysis of FcR binding may prove useful when comparing leads that may present differences in serum half-life.
Increasing circulating half-life has often been associated with improved therapeutic efficacy for ADCs. Correlation between protein stability and in vivo half-life has been known for some time.19,20 The more obvious correlation would be expected for changes in neonatal Fc receptor (FcRn) binding, which is a major factor in determining the rate of catabolism of IgGs not displaying target-mediated disposition, although there is some evidence that other factors may play a more significant role in serum half-life than affinity for FcRn.21 We chose the bacterial receptors Protein G and Protein A as surrogates for natural FcR since they are both known to overlap with FcR binding sites and compete for their binding.22,23 Our results suggest that binding to FcRn would not be expected to change with any of the conjugation chemistries or linkers investigated here. Therefore, the circulating half-life is not expected to change as a direct result of these modifications. However, catabolic mechanisms mediated by the cytosol are a possibility with any of these ADCs. Since terminal amine residues are a determining factor in that pathway, there is potential for influence on half-life in the case of amine coupling to the N-terminus of the antibody.
Biotin is small and somewhat water soluble whereas many chemotherapeutics are larger and more hydrophobic. Our results with biotinylating IgG1 antibodies demonstrate little or no impact on antigen/Fc receptor binding. With the exception of H10NPEG4H, these results are in contrast to several other reports which suggest that coupling of PEG molecules to an antibody can have a significant detrimental impact on antigen binding.24 The results with the H10NPEG4H ADC more likely reflect a difference in biotin load and location of conjugation on the antibody rather than the identity of the linker itself. The physiochemical characteristics and ultimate cellular disposition of the drug should also be considered when interpreting these results. Consideration of unintended consequences to secondary roles mediated through Fc receptor mediated binding would seem to warrant a similar analysis for ADCs currently being developed. There remains the possibility that a different small molecule, peptide, protein or nucleic acid bound to a different antibody via these methods might have a significant impact on antigen/Fc receptor binding. However, we tested two distinct IgG1 antibodies with completely different targets, and our results suggest a more general relationship in the case of smaller molecules. Clearly, many of the current chemotherapeutic drugs employed or explored for therapy differ significantly in size, charge and hydrophobicity from biotin. With much larger drug payloads like bacterial exotoxins we would expect that steric hindrance could play a significant role in modulating antigen/Fc receptor binding. While not a drug, there is clear evidence that with much larger PEG moieties used in the linker, the negative impact on antigen binding can be dramatic;17,25-27 In cases where the drug payload is highly hydrophobic and ‘sticky’, the chemical identity and length of the linker may influence the ability of those drug molecules to self-associate and promote aggregation.
While this report focuses on the relative thermal stability of conjugates, there has been much interest in investigating the stability of linkers in vivo. The work here demonstrates that linker identity has little or no impact on the inherent stability of the ADC construct, especially when compared with the relative impact of conjugation chemistry. Although outside the scope of this study, it would be interesting to compare conjugation chemistries known to impact thermal stability of an ADC with similar linkers and drugs to see if there is any correlation with the stability of a given linker to processing in vivo.
For all ADCs synthesized here, conjugation had some impact on thermostability of the molecule that could influence decisions for the manufacturing and storage for any particular ADC and may guide initial decisions on coupling strategies for therapeutic use based on possible correlations between thermal stability for the protein fold and serum half-life. Of the three functional groups investigated, thiol conjugation resulted in the greatest decreases in observed Tm as monitored by DSC. Furthermore, the degree of change in Tm is correlated with the number of biotin molecules linked to the antibody via thiols. The degree to which the Tm was independent of the length of the linker used or its hydrophobicity when the molecule being attached to the antibody is small and slightly hydrophobic. It remains to be seen if this relationship holds for much larger or more hydrophobic drugs such as exotoxins or auristatins. DSC showed clear differences in the number of components observed in thermograms for the two IgG1 antibodies and those differences also extended to how those component transitions were differentially impacted by conjugation chemistry. Perhaps most importantly, the results demonstrate the potential utility of DSC as a method to study comparability between ADC preparations.
Materials and Methods
Materials
Buffer salts and biocytin for ADC synthesis and purification were purchased from Sigma. Heterobifuctional linkers, both with and without biotin attached, T-gel, and Zeba columns were obtained from Thermo Scientific. Concentrated hybridoma supernatant containing mouse HyHEL-10 IgG1 (H10) whole antibody was provided by Dr. Sandi Smith-Gill, (National Cancer Institute). Japanese quail lysozyme was purified from egg white. Purified mouse monoclonal anti-6xHis IgG1 (aHIS) whole antibody was purchased from Invitrogen and dialyzed into PBS, pH 7.2. Hexa histidine peptides were obtained from AnaSpec. An engineered variant of apolipoprotein A1 (MSP1D1) was purified using the hepta-histidine tag via Ni-affinity chromatography as previously described.28 Reagents and consumables for DSC and SPR analysis were obtained from GE Health Sciences.
Purification of H10 antibody
Concentrated fractions of H10 antibody from hybridoma cultures were clarified by centrifugation at 50,000 × g for 20 min at 4°C. The supernatant was then applied to a T-gel column and purified according to the manufacturer’s instructions. Antibody fractions were concentrated via Millipore Amicon-Ultra 0.5 spin-ultrafiltration columns and then passed through a GE Superose 10/300 GL gel filtration column pre-equilibrated with HBS buffer (10 mM HEPES, 150 mM NaCl, 3 mM EDTA, pH 7.4). Purified antibody was stored in HBS at 4°C until further use.
Preparation of thiol conjugated ADCs
Stock solutions containing 5 μM H10 and aHIS were reduced with 10 mM TCEP in nitrogen purged phosphate buffer (10 mM sodium phosphate, 150 mM NaCl, 5 mM EDTA, pH 7.2) for 2 h at 37°C. A fresh solution of 30 mM SMCC (Fig. 1B) in DMSO was prepared and mixed with biocytin at a ratio of 1:20 in Phosphate Buffer for 2 h at 37°C. Initial attempts to reduce disulfide bonds in these antibodies to form thiol-linked conjugate using low ratios of TCEP:Ab29 proved unsuccessful. When using ratios below 5:1 no significant level of conjugate was observed. Considerably higher ratios of reducing agent:mAb were required (2000:1) at elevated temperatures, which is more consistent with those reported for other IgG1s.30 The reduced antibody was then buffer exchanged into HBS (PBS for aHIS) buffer using three Zeba desalting columns equilibrated with HBS buffer. Removal of TCEP reducing agent during this step prevented inhibition of the maleimide reaction.31 The reduced antibody was then mixed with 1/10 volume equivalent of the SMCC-biocytin reaction mixture and allowed to equilibrate at room temperature for 2.5 h in the dark. An excess of l-cysteine was then added to cap unreacted thiol groups and the mixture was allowed to incubate for 30 min at room temperature.
Preparation of Amine conjugated ADCs
NHS-PEG4-Biotin (Fig. 1B) was re-suspended in PBS and mixed with HyHEL-10 IgG in a final molar ratio of 20:1 and 40:1 and both reactions were incubated at room temperature for 2 h. For aHIS IgG, NHS-LC-Biotin (Fig. 1B) prepared in DMSO and NHS-PEG4-Biotin (Fig. 1B) were re-suspended in PBS and mixed with aHIS IgG in final molar ratio of 25:1 and incubated at room temperature for 30 min. In both cases, excess linker was removed with Zeba desalting columns.
Preparation of carbohydrate conjugated ADCs
Stock solutions of 50 μM H10 were oxidized in freshly prepared sodium periodate in PBS at room temperature in the dark for 2 h.32 Excess oxidizing agent was removed with Zeba desalting columns and the oxidized antibodies were immediately reacted with the hydrizide-biocytin linker (Fig. 1B) at a ratio of 1:20 for 2 h at room temperature.
Purification, determination of biotin-load and storage of ADCs
All ADC mixtures were further purified and buffer exchanged using a Superose 10/300 GL size exclusion column pre-equilibrated with HBS-E (10 mM HEPES, 150 mM NaCl, 3 mM EDTA) for H10 ADCs and PBS (10 mM sodium phosphate, 150 mM NaCl, pH 7.2) for aHIS ADCs at a flow rate of 0.55 mL/min, with UV detection at 280 nm. Protein fractions corresponding to the ADC peak (based on retention times of unconjugated antibody) were pooled and stored at 4°C until further use. Typical reaction yields after purification were between 30–55%. Degree of biotin conjugation on ADC was determined using the fluorescence biotin quantification kit from Thermo Scientific.
Stability analysis of conjugates using DSC
Differential scanning calorimetry was used to determine the affect of ligand and conjugation chemistry on the stability of all antibodies and ADCs. Samples were prepared immediately prior to analysis and any aggregates formed during storage were removed by size-exclusion chromatography in HBS-E buffer (10 mM HEPES, 150 mM NaCl, 3 mM EDTA, pH 7.4) or PBS for aHIS ADCs. Concentrations of antibody and conjugate used were 4–6 μM. All samples were analyzed using a Microcal VP-Capillary DSC equipped with autosampler and temperature controlled sample storage accessories. Unfolding was monitored between 10°C and 90°C (20°C and 90°C for aHIS ADCs) with a ramp rate of 1°C/min and a filtering period of 16 sec with no gain. H10 antibody was analyzed by DSC using ramp rates of 0.5°C/min and 1.5°C/min and there was no siginificant change in Tm or component profiles in the thermograms. Thermograms were normalized for concentration and then baseline corrected prior to fitting to a non-two-state unfolding model to determine transition melting temperature Tm for each component.
Determination of binding affinity for HEL antigen using SPR
SPR was used to compare effects of conjugation on ADC binding to its antigen. All experiments were conducted on a Biacore T100 with CM5 sensor chips. The antibodies were amine coupled to the sensor surface using 10 mM sodium acetate, pH 5.0 at a level that resulted in a maximum analyte response no higher than 40 response units (RUs). This, together with a flow rate of 100 µL/min, negated any significant mass transport effects for this very tight association. Several different concentrations of analyte diluted in HBS-EP running buffer (10 mM HEPES, 150 mM NaCl, 3 mM EDTA, 0.005% v/v P20) were injected for 120 sec followed by monitoring dissociation for 10 min. Raw sensorgrams were double reference subtracted using a reference flow cell with no ADC immobilized and by injections of running buffer. Corrected sensorgrams were fit globally to a 1:1 interaction model using the Biacore evaluation software. The quality of the nonlinear fit to this model was evaluated using coefficient of variance (CV) values and residuals. Unless otherwise stated in the figure captions, all values reported for apparent affinity KD are an average of three independent measurements made from separate dilutions of analyte stock. All experimental cycles were conducted at 25°C. Chip surfaces were regenerated after each cycle using 10 mM Glycine, pH 1.5.
Determination of binding affinity for 6xHis peptide antigen using SPR
ADCs were diluted into 10 mM sodium acetate buffer, pH 5.0 at a concentration of 10 µg/mL and immobilized on CM5 sensor surface chips using amine-coupling chemistry to give a maximum analyte response of approximately 40 response units (RUs). All binding analysis of antibodies with 6xHis peptides antigen were performed at 25°C in PBS (pH 7.2) as running buffer. Several concentrations of 6xHis peptides diluted in running buffer were injected for 150 sec at a flow rate of 50 µL/min followed by monitoring dissociation for 450 sec. Raw sensorgrams were double referenced by both subtracting a flow cell with no antibodies immobilized and subtracting a run of buffer only injection. The surface was regenerated with 10 mM glycine, pH 2.0 at a flow rate of 20 μL/min.
Determination of binding affinity for Fc receptor using SPR
Assessing the effect of conjugation on effector role function was done by first immobilizing the antigenic HEL ligand (a His-tagged apolipoprotein A1 variant for aHIS) onto a CM5 sensor surface onto which a high level of conjugate was captured. This configuration allows us to assess whether antigen bound ADC can still effectively bind Fc receptor. Between 3500–4000 RUs of capture ligand (antigen) was immobilized on each flow cell including the reference flow cell. To this was then captured approximately 500–1000 RUs of ADC diluted in SPR running buffer HBS-EP (PBS for aHIS). The level of captured ADC was chosen to give sufficient signal to provide good signal to noise for fitting while at the same time minimizing (for Protein G) the potential for mass transport limitations. Several concentrations of Fc receptor analyte were freshly prepared and injected over the antigen-captured ADC surface.
For Protein G, the protocol used was for kinetic measurements with a flow rate of 10 µL/min, an association time of 60 sec and a dissociation time of 60 sec (120 sec for aHIS). To assess potential mass-transport limitations tests with higher flow rates of 20 µL/min and 40 µL/min were conducted. No significantly change in on-rate was observed. Chip surfaces were regenerated after each cycle using 10 mM Glycine, pH 2.5. Analyte binding level was consistent among replicates, decreasing less than 5% after repeated regeneration cycles, and calculated rate used in determination of KD were highly reproducible from cycle to cycle. Raw sensorgrams were double referenced using a flow cell with no ligand immobilized and by injections of running buffer. Corrected sensorgrams were fit globally to a 1:1 interaction model using the Biacore evaluation software. Quality of the nonlinear fitting to this model was evaluated using CV values and the residuals. Values reported for apparent affinity KD are an average of three independent measurements made from separate dilutions of analyte stock.
Determination of binding affinity for CD32b and Protein A by equilibrium binding
For both CD32b binding and Protein A binding, the on rates and off rates were too fast to be measured with confidence using the T100. Therefore the apparent affinity was determined by measuring equilibrium binding levels at several different analyte concentrations in HBS-EP buffer. After correcting raw sensorgrams for the reference flow cell and buffer injections, equilibrium responses were plotted against analyte concentration using Biacore evaluation software and fit to a 1:1 association model using nonlinear Scatchard analysis. Each concentration was measured in triplicate and both methods were conducted using a sensor surface temperature of 25°C.
Acknowledgments
We wish to thank Sandi Smith-Gill from the National Cancer Institute for the kind gift of the mouse H10 IgG1 antibody. We would also like the thank John Sumida from the Bioanalytical Pharmacy Core, the Center for Intracellular Delivery Biologics and the Washington State Life Sciences Discovery Fund.
Glossary
Abbreviations:
- SPR
surface plasmon resonance
- DSC
differential scanning calorimetry
- FcγIIb
Fc gamma receptor IIb
- Tm
melting temperature
- ADC
antibody-drug conjugate
- MSP
membrane scaffold protein
- RU
response units
- SD
standard deviation
Footnotes
Previously published online: www.landesbioscience.com/journals/mabs/article/19449
References
- 1.Ojima I, Geng X, Wu X, Qu C, Borella CP, Xie H, et al. Tumor-specific novel taxoid-monoclonal antibody conjugates. J Med Chem. 2002;45:5620–3. doi: 10.1021/jm025540g. [DOI] [PubMed] [Google Scholar]
- 2.Smith LM, Nesterova A, Alley SC, Torgov MY, Carter PJ. Potent cytotoxicity of an auristatin-containing antibody-drug conjugate targeting melanoma cells expressing melanotransferrin/p97. Mol Cancer Ther. 2006;5:1474–82. doi: 10.1158/1535-7163.MCT-06-0026. [DOI] [PubMed] [Google Scholar]
- 3.Razani B, Woodman SE, Lisanti MP. Caveolae: from cell biology to animal physiology. Pharmacol Rev. 2002;54:431–67. doi: 10.1124/pr.54.3.431. [DOI] [PubMed] [Google Scholar]
- 4.Torres M, Casadevall A. The immunoglobulin constant region contributes to affinity and specificity. Trends Immunol. 2008;29:91–7. doi: 10.1016/j.it.2007.11.004. [DOI] [PubMed] [Google Scholar]
- 5.Wen J, Arthur K, Chemmalil L, Muzammil S, Gabrielson J, Jiang Y. Applications of differential scanning calorimetry for thermal stability analysis of proteins: qualification of DSC. J Pharm Sci. 2012;101:955–64. doi: 10.1002/jps.22820. [DOI] [PubMed] [Google Scholar]
- 6.Vermeer AW, Norde W. The thermal stability of immunoglobulin: unfolding and aggregation of a multi-domain protein. Biophys J. 2000;78:394–404. doi: 10.1016/S0006-3495(00)76602-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Holland M, Yagi H, Takahashi N, Kato K, Savage CO, Goodall DM, et al. Differential glycosylation of polyclonal IgG, IgG-Fc and IgG-Fab isolated from the sera of patients with ANCA-associated systemic vasculitis. Biochim Biophys Acta. 2006;1760:669–77. doi: 10.1016/j.bbagen.2005.11.021. [DOI] [PubMed] [Google Scholar]
- 8.Harris RJ. Heterogeneity of recombinant antibodies: linking structure to function. Dev Biol (Basel) 2005;122:117–27. [PubMed] [Google Scholar]
- 9.Sibéril S, Dutertre CA, Fridman WH, Teillaud JL. FcgammaR: The key to optimize therapeutic antibodies? Crit Rev Oncol Hematol. 2007;62:26–33. doi: 10.1016/j.critrevonc.2006.12.003. [DOI] [PubMed] [Google Scholar]
- 10.Natsume A, Niwa R, Satoh M. Improving effector functions of antibodies for cancer treatment: Enhancing ADCC and CDC. Drug Des Devel Ther. 2009;3:7–16. [PMC free article] [PubMed] [Google Scholar]
- 11.Moore GL, Chen H, Karki S, Lazar GA. Engineered Fc variant antibodies with enhanced ability to recruit complement and mediate effector functions. MAbs. 2010;2:181–9. doi: 10.4161/mabs.2.2.11158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Radaev S, Sun PD. Recognition of IgG by Fcgamma receptor. The role of Fc glycosylation and the binding of peptide inhibitors. J Biol Chem. 2001;276:16478–83. doi: 10.1074/jbc.M100351200. [DOI] [PubMed] [Google Scholar]
- 13.Krapp S, Mimura Y, Jefferis R, Huber R, Sondermann P. Structural analysis of human IgG-Fc glycoforms reveals a correlation between glycosylation and structural integrity. J Mol Biol. 2003;325:979–89. doi: 10.1016/S0022-2836(02)01250-0. [DOI] [PubMed] [Google Scholar]
- 14.Vaccaro C, Zhou J, Ober RJ, Ward ES. Engineering the Fc region of immunoglobulin G to modulate in vivo antibody levels. Nat Biotechnol. 2005;23:1283–8. doi: 10.1038/nbt1143. [DOI] [PubMed] [Google Scholar]
- 15.Clynes RA, Towers TL, Presta LG, Ravetch JV. Inhibitory Fc receptors modulate in vivo cytotoxicity against tumor targets. Nat Med. 2000;6:443–6. doi: 10.1038/74704. [DOI] [PubMed] [Google Scholar]
- 16.Cunningham-Rundles C, Zhuo Z, Griffith B, Keenan J. Biological activities of polyethylene-glycol immunoglobulin conjugates. Resistance to enzymatic degradation. J Immunol Methods. 1992;152:177–90. doi: 10.1016/0022-1759(92)90139-K. [DOI] [PubMed] [Google Scholar]
- 17.Anderson WL, Tomasi TB. Polymer modification of antibody to eliminate immune complex and Fc binding. J Immunol Methods. 1988;109:37–42. doi: 10.1016/0022-1759(88)90439-5. [DOI] [PubMed] [Google Scholar]
- 18.Hamblett KJ, Senter PD, Chace DF, Sun MM, Lenox J, Cerveny CG, et al. Effects of drug loading on the antitumor activity of a monoclonal antibody drug conjugate. Clin Cancer Res. 2004;10:7063–70. doi: 10.1158/1078-0432.CCR-04-0789. [DOI] [PubMed] [Google Scholar]
- 19.McLendon G, Radany E. Is protein turnover thermodynamically controlled? J Biol Chem. 1978;253:6335–7. [PubMed] [Google Scholar]
- 20.Fontana A, Zambonin M, Polverino de Laureto P, De Filippis V, Clementi A, Scaramella E. Probing the conformational state of apomyoglobin by limited proteolysis. J Mol Biol. 1997;266:223–30. doi: 10.1006/jmbi.1996.0787. [DOI] [PubMed] [Google Scholar]
- 21.Gurbaxani B, Dela Cruz LL, Chintalacharuvu K, Morrison SL. Analysis of a family of antibodies with different half-lives in mice fails to find a correlation between affinity for FcRn and serum half-life. Mol Immunol. 2006;43:1462–73. doi: 10.1016/j.molimm.2005.07.032. [DOI] [PubMed] [Google Scholar]
- 22.Sulica A, Medesan C, Laky M, Onică D, Sjöquist J, Ghetie V. Effect of protein A of Staphylococcus aureus on the binding of monomeric and polymeric IgG to Fc receptor-bearing cells. Immunology. 1979;38:173–9. [PMC free article] [PubMed] [Google Scholar]
- 23.Stone GC, Sjöbring U, Björck L, Sjöquist J, Barber CV, Nardella FA. The Fc binding site for streptococcal protein G is in the C gamma 2-C gamma 3 interface region of IgG and is related to the sites that bind staphylococcal protein A and human rheumatoid factors. J Immunol. 1989;143:565–70. [PubMed] [Google Scholar]
- 24.Chapman AP. PEGylated antibodies and antibody fragments for improved therapy: a review. Adv Drug Deliv Rev. 2002;54:531–45. doi: 10.1016/S0169-409X(02)00026-1. [DOI] [PubMed] [Google Scholar]
- 25.Kitamura K, Takahashi T, Yamaguchi T, Noguchi A, Noguchi A, Takashina K, et al. Chemical engineering of the monoclonal antibody A7 by polyethylene glycol for targeting cancer chemotherapy. Cancer Res. 1991;51:4310–5. [PubMed] [Google Scholar]
- 26.Suzuki T, Kanbara N, Tomono T, Hayashi N, Shinohara I. Physicochemical and biological properties of poly(ethylene glycol)-coupled immunoglobulin G. Biochim Biophys Acta. 1984;788:248–55. doi: 10.1016/0167-4838(84)90268-1. [DOI] [PubMed] [Google Scholar]
- 27.Takashina K, Kitamura K, Yamaguchi T, Noguchi A, Noguchi A, Tsurumi H, et al. Comparative pharmacokinetic properties of murine monoclonal antibody A7 modified with neocarzinostatin, dextran and polyethylene glycol. Jpn J Cancer Res. 1991;82:1145–50. doi: 10.1111/j.1349-7006.1991.tb01769.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ritchie TK, Grinkova YV, Bayburt TH, Denisov IG, Zolnerciks JK, Atkins WM, et al. Chapter 11 - Reconstitution of membrane proteins in phospholipid bilayer nanodiscs. Methods Enzymol. 2009;464:211–31. doi: 10.1016/S0076-6879(09)64011-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sun MM, Beam KS, Cerveny CG, Hamblett KJ, Blackmore RS, Torgov MY, et al. Reduction-alkylation strategies for the modification of specific monoclonal antibody disulfides. Bioconjug Chem. 2005;16:1282–90. doi: 10.1021/bc050201y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liu H, Chumsae C, Gaza-Bulseco G, Hurkmans K, Radziejewski CH. Ranking the susceptibility of disulfide bonds in human IgG1 antibodies by reduction, differential alkylation, and LC-MS analysis. Anal Chem. 2010;82:5219–26. doi: 10.1021/ac100575n. [DOI] [PubMed] [Google Scholar]
- 31.Getz EB, Xiao M, Chakrabarty T, Cooke R, Selvin PR. A comparison between the sulfhydryl reductants tris(2-carboxyethyl)phosphine and dithiothreitol for use in protein biochemistry. Anal Biochem. 1999;273:73–80. doi: 10.1006/abio.1999.4203. [DOI] [PubMed] [Google Scholar]
- 32.Hermanson GT. Bioconjugate techniques. Amsterdam; Boston: Elsevier Academic Press, 2008. [Google Scholar]