Abstract
Organelles are surrounded by membranes with a distinct lipid and protein composition. While it is well established that lipids affect protein functioning and vice versa, it has been only recently suggested that elevated membrane protein concentrations may affect the shape and organization of membranes. We therefore analyzed the effects of high chloroplast envelope protein concentrations on membrane structures using an in vivo approach with protoplasts. Transient expression of outer envelope proteins or protein domains such as CHUP1-TM–GFP, outer envelope protein of 7 kDa–GFP, or outer envelope protein of 24 kDa–GFP at high levels led to the formation of punctate, circular, and tubular membrane protrusions. Expression of inner membrane proteins such as translocase of inner chloroplast membrane 20, isoform II (Tic20-II)–GFP led to membrane protrusions including invaginations. Using increasing amounts of DNA for transfection, we could show that the frequency, size, and intensity of these protrusions increased with protein concentration. The membrane deformations were absent after cycloheximide treatment. Co-expression of CHUP1-TM–Cherry and Tic20-II–GFP led to membrane protrusions of various shapes and sizes including some stromule-like structures, for which several functions have been proposed. Interestingly, some structures seemed to contain both proteins, while others seem to contain one protein exclusively, indicating that outer and inner envelope dynamics might be regulated independently. While it was more difficult to investigate the effects of high expression levels of membrane proteins on mitochondrial membrane shapes using confocal imaging, it was striking that the expression of the outer membrane protein Tom20 led to more elongate mitochondria. We discuss that the effect of protein concentrations on membrane structure is possibly caused by an imbalance in the lipid to protein ratio and may be involved in a signaling pathway regulating membrane biogenesis. Finally, the observed phenomenon provides a valuable experimental approach to investigate the relationship between lipid synthesis and membrane protein expression in future studies.
Keywords: membrane proteins, chloroplast envelopes, membrane structure, organelle structure, lipid to protein ratio
Introduction
Membranes not only confine cells and organelles, they are important gateways regulating and facilitating the exchange of proteins, lipids, and metabolites between different compartments. The lipidaceous and proteinaceous components of membranes exhibit many well-known functional interrelations (Lee, 2003; Marsh, 2008; van Meer et al., 2008), for example, specific lipids have been shown to modulate membrane protein function and topology (Schleiff et al., 2001; Lee, 2004). It is therefore not surprising that membranes of organelles such as chloroplasts exhibit defined lipid compositions and specific protein to lipid ratios (Benning, 2009). However, the composition of membranes of these organelles is not static and can change in response to certain environmental conditions such as light or temperature (Uemura et al., 1995; Okawa et al., 2008; Burgos et al., 2011).
Membranes not only exhibit plasticity with regard to their composition but also their morphology. A fairly well-studied phenomenon is the vesicle budding and fusion event of the endomembrane system (Melser et al., 2011). Membrane dynamics have also been observed to various degrees for membranes of chloroplasts and mitochondria. For example, in plastids, dynamic tubular extrusions of the inner and outer envelopes, termed stromules, have been described (Gray et al., 2001; Kwok and Hanson, 2004b). The study of these fragile and highly dynamic membrane structures was made possible by fluorescently labeled proteins of the outer envelope or the stroma (Kohler et al., 1997). The exact shape and frequency of stromules depends on the tissue, plastid type and environmental conditions (Holzinger et al., 2007; Hanson and Sattarzadeh, 2008, 2011; Itoh and Fujiwara, 2010; Shaw and Gray, 2011). It had been speculated that since stromules increase the surface area of plastids they might facilitate the interaction with other plastids, mitochondria, and peroxisomes (Kwok and Hanson, 2004a,b; Holzinger et al., 2007; Lutz and Engel, 2007; Hanson and Sattarzadeh, 2008, 2011; Itoh and Fujiwara, 2010; Shaw and Gray, 2011). Recently it was shown that the dynamic behavior of stromules also coincides with the dynamics of the endoplasmic reticulum, possibly allowing the exchange of metabolites at sites of contact (Schattat et al., 2011).
Mitochondria exhibit even more dynamic behavior as visualized by fusing GFP to N-terminal mitochondrial targeting sequences. In mesophyll cells mitochondria are mainly spherical or elongate, often in close proximity to chloroplasts and do move in response to changes in light intensity (Islam et al., 2009). In epidermal or root cells, mitochondria change their shape from spherical to elongate to branched within seconds, frequently undergoing fusion and fission and rapidly changing their location within a cell (Logan and Leaver, 2000; Logan, 2006).
There is mounting evidence that an increase of the protein concentration in membranes of the ER (Wright et al., 1988; Gong et al., 1996) or the plasma membrane of various organisms (Arechaga et al., 2000; Lefman et al., 2004) can influence membrane dynamics, e.g., via the formation of membrane proliferations which accommodate the increased levels of protein. Similarly, it was recently observed that expression of outer or inner envelope proteins of chloroplasts at high levels can lead to alterations in the shape of the respective membrane (Fourrier et al., 2008; Schmidt von Braun and Schleiff, 2008; Singh et al., 2008). We therefore investigated the potential effects of overexpression of outer and inner membrane proteins of endosymbiotically derived organelles on membrane morphology.
Materials and Methods
Construct generation
For the construction of atTic20-II and atOEP7 GFP fusion proteins atTIC20-II (At2g47840) and atOEP7 (AT3g52420) were amplified from cDNA and cloned into the plant expression vector pML94–myc–GFP (Bionda et al., 2010). psOEP24 was amplified from cDNA and cloned into pAVA (Sommer et al., 2011). The coding sequence for YC3.60 (kind gift from A. Miyawaki, RIKEN, Tokyo) was cloned into pML94–myc–GFP (Bionda et al., 2010). CHUP1-TM (AT3G25690, aa 1–57; Schmidt von Braun and Schleiff, 2008) was amplified from cDNA and cloned N-terminally to YC3.60 via KpnI and BcuI. CHUP1-TM was cut out from pML94–CHUP1-TM–YC3.60 via BcuI and KpnI and placed into pML94–Cherry to yield pML94–CHUP1-TM–Cherry. Tom20-TM (P3 domain, AT3G27070, aa 156–188; Perry et al., 2006) was amplified from cDNA and cloned C-terminally to YC3.60 via NheI and BamHI. The Cherry–SKL, precursor of the alternative oxidase (pAOX), chloroplast inner membrane localized rhomboid-like protein (cpRhomboid), mitochondrial-targeted rhomboid-like protein (mtRhomboid), precursor of small subunit (pSSU), and pNTT1 constructs were described before (Kmiec-Wisniewska et al., 2008; Bionda et al., 2010). Ferredoxin 2 (AT1g60950) was amplified from Arabidopsis thaliana cDNA and cloned into plant expression vector pML94–myc–GFP via KpnI and BcuI.
Protoplast transformation, GFP visualization, and Western Blotting
Isolation and transformation of A. thaliana Col0 protoplasts (used in Figures 1 and 3–5) was previously described (Mishra et al., 2002 and Sommer et al., 2011). Eight to 10 h post-transfection, protoplasts were analyzed using a Leica SP5 confocal laser scanning microscope. GFP was excited at 488 nm and the emission was recorded at 495–525 nm. YFP and chlorophyll a were excited at 514 nm and recorded at 530–570 or 660–710 nm. Cherry was excited at 561 nm and recorded at 595–615 nm. Mitochondria were stained with MitoTracker® CMTMRos at a concentration of 200 nM for 30 min. MitoTracker fluorescence was excited at 514 nm and the emission recorded at 575–610 nm. For western blotting 105 protoplasts were harvested for each time point, spun down for 1 min at 20,000 × g and analyzed via SDS-PAGE. Western blots were preformed using mouse monoclonal α-GFP (Roche Mannheim), α-Actin (SigmaAldrich), or rabbit polyclonal α-Tic40 (gift of J. Froehlich, Plant Research Lab, MSU, MI, USA) antibodies.
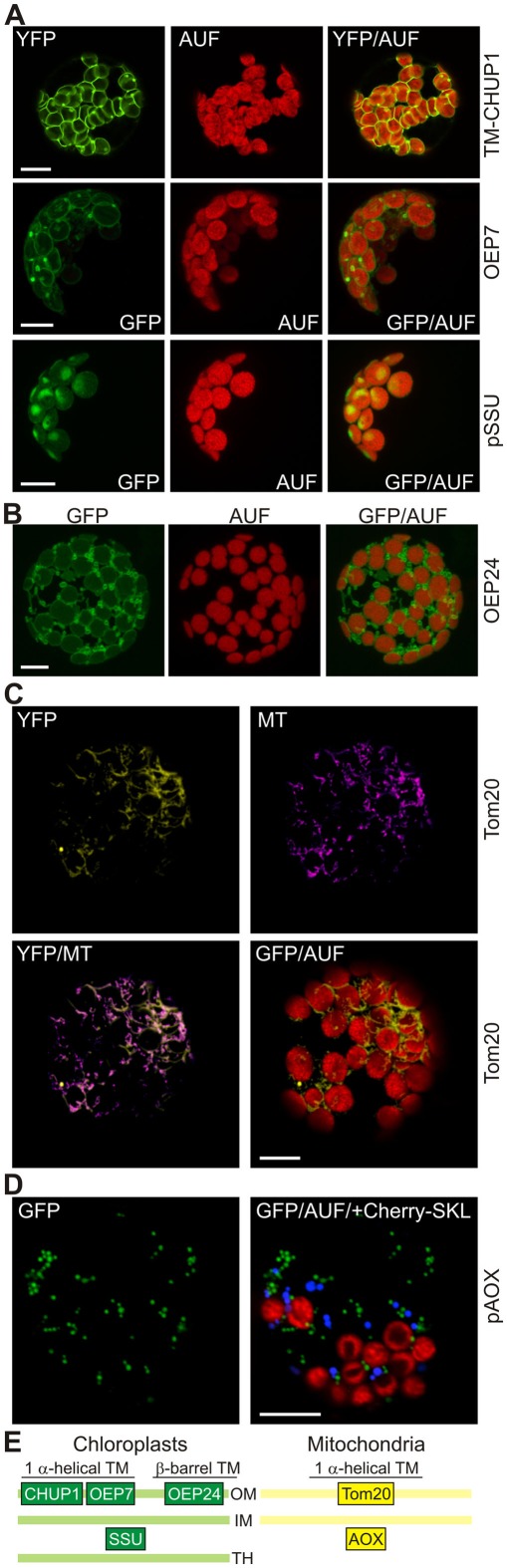
Figure 1.
High expression levels of outer membrane proteins lead to changes in chloroplast and mitochondrial membrane morphology. (A) The transmembrane (TM) domain of the chloroplast outer envelope protein CHUP1, full length outer envelope protein 7 (OEP7), or the small subunit of RUBISCO (pSSU) were fused to the N-terminus of GFP and expressed in protoplasts under control of a 35S promoter. GFP fluorescence (GFP), chlorophyll autofluorescence (AUF), and the overlay (GFP/AUF) of representative protoplasts are shown. The CHUP1 and pSSU images are single optical sections, OEP7 images are maximum projections of optical sections. (B) The β-barrel-shaped OEP24, fused to GFP was expressed in A. thaliana mesophyll protoplasts under control of the 35S promoter. GFP fluorescence (GFP), autofluorescence (AUF), and the overlay (GFP/AUF) of a representative protoplast are shown. Images are maximum projections of optical sections. (C) The transmembrane domain of the mitochondrial protein Tom20 was fused to YFP and expressed in protoplasts under control of a 35S promoter. YFP fluorescence, MitoTracker (MT) staining, and the overlays of the YFP fluorescence with MT or the chlorophyll autofluorescence (AUF) of representative protoplasts are shown. Images are single optical sections. (D) pAOX fused to GFP and Cherry with an SKL at the C-terminus to induce peroxisomal targeting were co-expressed in protoplasts under control of a 35S promoter. GFP fluorescence (GFP), chlorophyll autofluorescence (AUF), and the overlay of GFP/AUF and the Cherry–SKL signal (blue) of a representative protoplast are shown. Images are single optical sections. Scale bars in (A–D) represent 10 μm. (E) The localization of the proteins used in this figure is shown as a schematic (OM, outer membrane; IM, inner membrane; TH, thylakoid membrane).
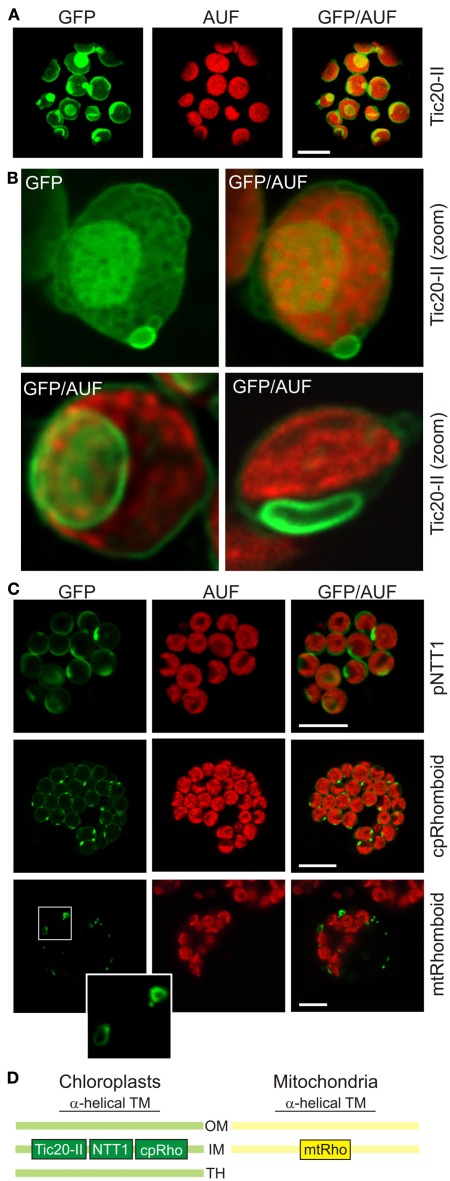
Figure 3.
High expression levels of inner membrane proteins lead to changes in chloroplast and mitochondrial membrane morphology. (A) The chloroplast inner envelope protein atTic20-II was fused to GFP and expressed in A. thaliana mesophyll protoplasts under control of a 35S promoter. GFP fluorescence (GFP), chlorophyll autofluorescence (AUF), and overlays of GFP/AUF of representative protoplasts are shown. Images are single optical sections. (B) Representative chloroplasts with vesicle-like extrusions (top) or intrusions (bottom) of atTic20-II–GFP protoplasts are shown. Images either show GFP only or the overlay (GFP/AUF). Images are single optical sections. (C) The chloroplast inner envelope protein pNTT1 and the chloroplast and mitochondrial cpRhomboid–GFP or mtRhomboid–GFP (Table 1) were expressed in A. thaliana mesophyll protoplasts under control of a 35S promoter. GFP fluorescence (GFP), chlorophyll autofluorescence (AUF), and overlays of GFP/AUF of representative protoplasts are shown. Images are maximum projections of optical sections. The mitochondrial structures framed are enlarged. Scale bars in (A,C) represent 10 μm. (D) The localization of the proteins used in this figure is shown schematically (OM, outer membrane; IM, inner membrane; TH, thylakoid membrane).
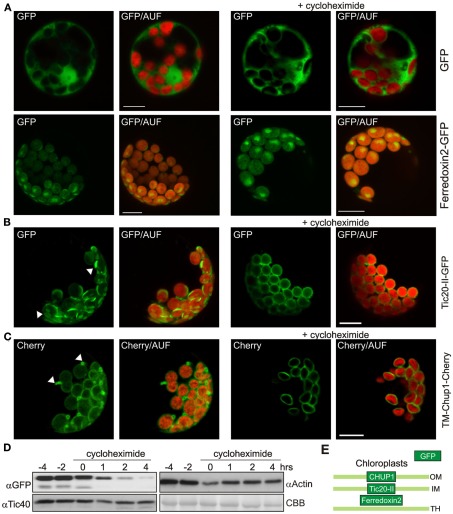
Figure 5.
The deformation of the membranes is reversible. (A) GFP and the stromal atFerredoxin 2 fused to GFP were expressed in A. thaliana mesophyll protoplasts under control of a 35S promoter (left panels). Two hour prior to analysis one set was treated with cycloheximide (right panels). GFP fluorescence (GFP), and overlays of GFP and chlorophyll autofluorescence (GFP/AUF) of representative protoplasts are shown. Images are maximum projections of optical sections. (B) The inner envelope protein atTic20-II fused to GFP was expressed in A. thaliana mesophyll protoplasts under control of a 35S promoter (left panels). Two hour prior to analysis one set was treated with cycloheximide (right panels). GFP fluorescence (GFP), and overlays of GFP and chlorophyll autofluorescence (GFP/AUF) of representative protoplasts are shown. Images are maximum projections of optical sections. (C) The transmembrane domain of the outer envelope protein CHUP1 (TM-CHUP1) was fused to Cherry and expressed in A. thaliana mesophyll protoplasts under control of a 35S promoter (left panels). Two hour prior to analysis one set was treated with cycloheximide (right panels). Cherry fluorescence (cherry), and overlays of cherry and chlorophyll autofluorescence (Cherry/AUF) of representative protoplasts are shown. Images are maximum projections of optical sections. In (B,C), white arrowheads point to membrane deformations in the absence of cycloheximide. Scale bars represent 10 μm. (D) Western blot analysis of protoplasts overexpressing atTic20-II–GFP before and after addition of cycloheximide (at 0 h). Coomassie brilliant blue stained RUBISCO (large subunit) as loading control (CBB). (E) The localization of the proteins used in this figure is shown schematically (OM, outer membrane; IM, inner membrane; TH, thylakoid membrane).
Protoplast treatment with cycloheximide
Transformed protoplasts were incubated overnight for 8–10 h at 23°C and subsequently treated with cycloheximide at a concentration of 5 μg/ml and further incubated for 2–3 h at 20°C in the dark.
Results
Elevated levels of outer membrane proteins alter the membrane structure
While analyzing which part of the sequence of the chloroplast unusual positioning protein 1 (CHUP1; Table 1) is important for its localization to the outer envelope of the chloroplast, we noticed that expression of 35S::CHUP1–GFP frequently led to the formation of membrane protrusions of the outer envelope to which CHUP1 is also localized (Schmidt von Braun and Schleiff, 2008). However, since full length CHUP1 is known to interact with actin filaments (Schmidt von Braun and Schleiff, 2008), which per se might alter the structure of the outer envelope, we determined the effects of high expression levels of the transmembrane (TM) domain of CHUP1–GFP on the structure of outer envelope membranes of chloroplasts. Interestingly we observed the same membrane protrusions (Figure 1A, top), as seen for the full length CHUP1–GFP constructs when transforming wild type protoplasts. This suggests that the alteration of the outer envelope structure is related to the increased concentration of the envelope protein rather than the specific activity of CHUP1. To further test this hypothesis we transiently overexpressed the bitopic outer envelope protein of 7 kDa (OEP7) as a GFP construct under the control of the constitutive 35S promoter in wild type protoplasts. We confirmed that OEP7 localizes to the chloroplast envelope (Figure 1A, middle; Schleiff et al., 2001) resulting in punctate protrusions similar to those seen in protoplasts expressing CHUP1-TM–GFP. We confirmed that the GFP-labeled protrusions were caused by the expression of outer envelope proteins, because expression of pSSU the small subunit of RUBISCO fused to GFP (pSSU–GFP) did not lead to an alteration of the envelope structure (Figure 1A, bottom) as previously established (Lee et al., 2006).
Table 1.
The proteins used in this study.
| Protein | Property | Construct | Reference |
|---|---|---|---|
| CHUP1 | Bitopic α-helical COEa protein | CHUP1–Cherry | Schmidt von Braun and Schleiff (2008) |
| TM-CHUP1-YC3.60 | |||
| cpRhomboid | Bitopic α-helical CIEb protein | cpRhomboid–GFP | Kmiec-Wisniewska et al. (2008) |
| Ferredoxin 2 | Stromal protein | Ferredoxin 2–GFP | Hanke et al. (2004) |
| mtRhomboid | Bitopic α-helical MIMc protein | mtRhomboid–GFP | Kmiec-Wisniewska et al. (2008) |
| OEP7 | Bitopic α-helical COE protein | OEP7–GFP | Schleiff et al. (2001) |
| OEP24 | β-Barrel COE protein | OEP24–GFP(S11) | Pohlmeyer et al. (1998) |
| pAOX | Matrix protein | pAOX–GFP | Carrie et al. (2009) |
| pNTT1 | Politopic α-helical CIE protein | pNTT1–GFP | Bionda et al. (2010) |
| pSSU | Stromal protein | pSSU–GFP | Bionda et al. (2010) |
| Tic20-II | Politopic α-helical CIE protein | Tic20-II–GFP | Machettira et al. (2011) |
| Tom20 | Bitopic α-helical MOMd protein | Tom20–YC3.60 | Perry et al. (2006) |
aCOE, chloroplast outer envelope; bCIE, chloroplast inner envelope; cMIM, mitochondrial inner membrane; dMOM, mitochondrial outer membrane.
Both TM-CHUP1 and OEP7 are helical in structure, and therefore we tested the effects of high expression levels of an outer membrane protein with β-barrel shape on membrane morphology, namely the outer envelope protein of 24 kDa (OEP24; Table 1). In this case we used a self-assembling GFP system (Sommer et al., 2011) in which only the 11th beta strand of GFP (S11) is linked to the protein of choice while the strands 1–10 of GFP (S1–10) are expressed in the cytosol (Sommer et al., 2011). Again we frequently observed vesicle-like structures and even tubular protrusions of the outer envelope membrane (Figure 1B). Similar results are obtained by (Breuers et al., 2012), when overexpressing A. thaliana Toc64-III or LACS9 fused to GFP in N. benthamiana protoplasts.
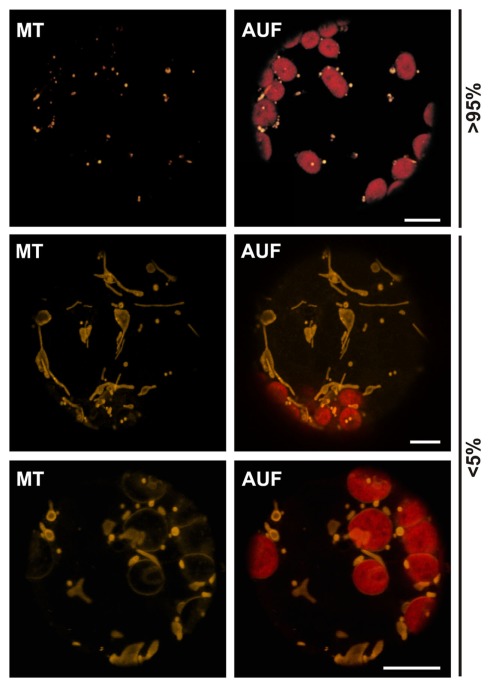
To test if the same holds true for proteins in the outer membrane of mitochondria we expressed 35S::Tom20–YC3.60 (translocase of outer mitochondrial membrane protein of 20 kDa; Perry et al., 2006) in wild type protoplasts and analyzed the YFP fluorescence (Table 1; Figure 1C). In general spherical or slightly elongated mitochondria are observed in protoplasts (Daschner et al., 2001; Matthes et al., 2007), which can be visualized by expressing 35S::pAOX–GFP, a precursor of a matrix protein tightly bound to the inner mitochondrial membrane (pAOX; Figure 1D; Kuhn et al., 2009). The mitochondria are generally even smaller than peroxisomes as shown by targeting of Cherry to the peroxisomes by fusion to the C-terminal peroxisomal targeting signal composed of the amino acids SKL. In contrast, we observed mitochondria with a drastically different morphology in more than 80% of the protoplast expressing 35S::Tom20–YC3.60. The mitochondria are characterized by an increased presence of tubular structures. To confirm the mitochondrial localization of Tom20-YC3.60 we imaged mitochondria by analyzing YFP fluorescence and MitoTracker staining simultaneously (Figure 1C), the latter of which mainly accumulates in the mitochondrial matrix (Presley et al., 2003). We not only observed overlapping signals for most parts, but also found structures that were only visible by either MitoTracker or YFP. This prompted us to investigate the mitochondrial structure in protoplasts stained with MitoTracker only (Figure 2). In general we observed the punctuated staining as expected from the pAOX–GFP images, however, in very rare cases (<5%) we observed staining of tubular structures even surrounding the chloroplasts as well (Figure 2). Thus, expression of Tom20-YC3.60 either induced the formation of the observed mitochondrial structures, or assuming equilibrium between tubular and individual mitochondria, it at least shifted the equilibrium towards tubulization. Overall, we can conclude that high expression levels of both helical and β-barrel-shaped outer membrane proteins of chloroplasts and mitochondria resulted in significant alterations of the respective membrane structures.
Figure 2.
Morphological differences of plant mitochondria. Untransfected wild type protoplasts from A. thaliana stained with MitoTracker reveal the different morphologies of plant mitochondria. Images are single optical sections. MitoTracker staining (MT), chlorophyll autofluorescence (AUF). Scale bars represent 10 μm.
Manipulation of inner membrane protein levels alters the membrane structure
To test if high expression levels of inner membrane proteins of chloroplasts and mitochondria also cause changes in membrane morphology, we transformed wild type protoplasts with different GFP constructs under the control of a 35S promoter (Table 1). For example, the expression of the inner chloroplast envelope membrane translocase of 20kDa, isoform II (Tic20-II) labeled, as expected, the circumference of the chloroplasts when imaged using chlorophyll autofluorescence (Kasmati et al., 2011). In addition, we observed regions of intense fluorescence, vesicle-like protrusions in approximately 60–80% of all transfected cells (Figure 3A). These invaginations appeared to be sometimes composed of multiple layers of membranes (Figure 3B). Thus, the morphology of these structures is remarkably different from the ones observed for outer membrane proteins (Figure 1A). A similar phenomenon, namely the invaginations of the inner chloroplast envelope membrane, was reported for Tic40 (Schmidt von Braun and Schleiff, 2008; Singh et al., 2008; Breuers et al., 2012). Since both Tic40 and Tic20 are components of the protein translocation system, we analyzed whether changes in membrane structure can be observed for inner membrane proteins with other functions as well. Expression of a chloroplast inner membrane localized nucleotide transporter protein 1 (NTT1) or a cpRhomboid (Kmiec-Wisniewska et al., 2008) fused to GFP in protoplasts led to the expected localization of the protein (Figure 3C). In addition, protrusions with enhanced fluorescence intensities were observed in approximately 50–70% of all transfected cells. The same vesicle-like protrusions are recently observed when overexpressing TPT, the triosephosphate carrier in the inner envelope membrane, AtAPG1, the membrane-associated protein catalyzing the methylation of demethylplastoquinol to plastoquinone-9 or AtLrgB, a highly abundant protein of unknown molecular function in tobacco protoplasts (Breuers et al., 2012 and references therein). Therefore, as shown for outer membrane structures, the protrusions and invaginations of the membrane are not dependent on the protein function, but only on elevated protein concentrations.
We then tested whether high expression levels of inner membrane proteins of mitochondria also lead to correct targeting plus altered membrane morphologies. We used a mtRhomboid protein fused to GFP (Figure 3C, bottom). Some rhomboid–GFP-labeled mitochondria (Kmiec-Wisniewska et al., 2008) exhibited the expected round to oval shape just as those labeled with pAOX–GFP (Figure 1D). In addition, we observed some mitochondria with bulb-like protrusions in approximately 30% of all transfected cells. Hence high expression levels of a mitochondrial inner membrane protein also led to alterations of the mitochondrial membrane morphology. However, the morphological changes were never as pronounced as those observed when expressing high levels of GFP-labeled mitochondrial outer membrane proteins.
Changes of membrane morphology are protein concentration dependent
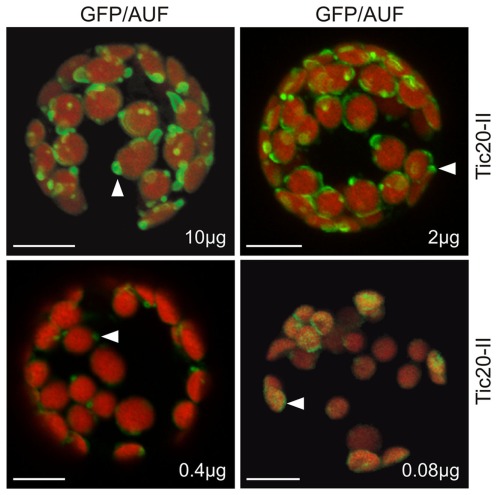
We investigated in more detail the relationship between protein concentration and membrane structure to confirm that the observed membrane alterations were indeed concentration dependent. It is known that protein levels can be manipulated by changing the amount of plasmid DNA used for protoplast transformation while keeping the final amount of DNA constant by making up the difference with empty vector (Mishra et al., 2002). When we transformed protoplasts with increasing amounts of Tic20-II–GFP plasmid DNA we observed a gradual increase in the frequency and intensity of membrane protrusions with increasing protein concentrations (Figure 4). This confirms that membrane deformation is directly related to the protein content.
Figure 4.
The alteration of membrane morphology is concentration dependent. Various concentrations (10, 2, 0.4, and 0.08 μg) of atTic20-II–GFP plasmid DNA adjusted to a total of 10 μg DNA with empty vector were used to transfect A. thaliana mesophyll protoplasts. Arrowheads point to examples of changes in membrane morphology.
To test whether the deformation is a short-term reaction during insertion of the proteins or whether these structures are reversible, we inhibited the protein synthesis for 2 h by adding cycloheximide 8 h post transfection. Cycloheximide is a frequently used drug to inhibit protein synthesis. The treatment with cycloheximide did not lead to a significant reduction of the fluorescence of cytoplasmically expressed GFP (Figure 5A, top right panel) or the stroma targeted Ferredoxin 2–GFP fusion protein (Figure 5A, bottom right panel). Analyzing the protoplasts transformed with Tic20-II after cycloheximide treatment revealed an even distribution of the GFP signal surrounding the autofluorescence of the chlorophyll (Figure 5B, right panel), which stands in contrast to the observation in the absence of cycloheximide (Figure 5B, left panel). The analysis of the GFP fluorescence distribution when fused to TM-CHUP1 confirms this observation. In the absence of cycloheximide we observed extrusions (Figure 5C, left panel), while after cycloheximide treatment no extrusions were found (Figure 5C, right panel). These observations together with the remaining GFP fluorescence intensity after cycloheximide treatment strongly suggests that the deformation of the inner envelope is reversible and enforced by massive insertion of the membrane proteins into the envelopes. To follow the fate of the protein level after cycloheximide treatment we performed western blots with antibodies either against GFP (to detect Tic20-II–GFP) or Tic40 (as control for the steady state level of inner membrane proteins, as well as against actin; Figure 5D). It can be clearly seen, that the Tic20-II–GFP levels are drastically decreased after cycloheximide treatment, whereas changes in the amounts of Tic40 and actin are only moderate. Thus, the disappearance of the membrane protrusions is probably a consequence of the decreasing protein levels.
Simultaneous expression of outer and inner envelope proteins causes a range of changes in membrane morphology
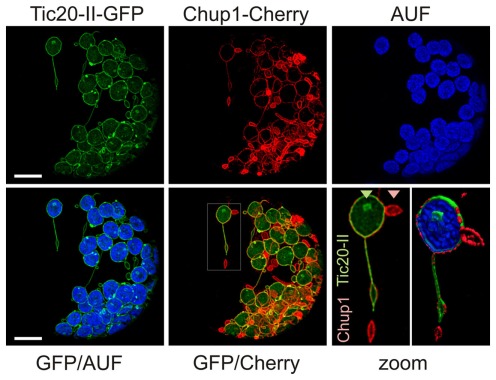
We analyzed the change of chloroplast membrane structure in response to the overexpression of both an inner and an outer envelope protein to determine whether the deformation of the two membranes is coordinated. We transformed protoplasts with TM-CHUP1–mCherry and Tic20-II–GFP and observed even more extreme changes in membrane morphology than when protoplasts were transformed with one construct only (Figure 6 versus Figures 1 and 3). These changes in membrane structure become distinctly visible in scans through the confocal XYZ stacks of the recorded chloroplast/protoplast sections and 3D projections thereof (Videos S1–S5 in Supplementary Material). Here the intrusions and extrusions of the inner and outer envelope membranes can be seen in direct relation to the chloroplast thylakoids (arrow heads). In addition to individual circular and tubular protrusions (green and red arrow heads in Videos S1 and S2 in Supplementary Material) we sometimes observed long, tubular protrusions reminiscent of stromules (Hanson and Sattarzadeh, 2011; Figure 6 and Video S2 in Supplementary Material, white arrow head). These structures have a diameter of less than 1 μm and various lengths (Figure 6, inset), both in the range previously reported for stromules (Kwok and Hanson, 2004c; Natesan et al., 2005; Hanson and Sattarzadeh, 2011). However, stromules are highly dynamic with an extension rate of 0.2 μm s−1 and an even faster retraction rate of 1.2 μm s−1 (Gunning, 2005). The structures observed here do not show such rapid dynamics as no drastic alterations of the protrusions were observed within 30 s (Figure 6, inset). In general, we observed some structures, in which fluorescence of both proteins could be observed, while for others we only observed signals of one of the two fluorophores (only TM-CHUP1–mCherry as highlighted in the zoomed section; Figure 6, zoom). However, it remains to be tested whether outer membrane proliferations indeed represent structures without any inner membrane proteins.
Figure 6.
Co-expression of outer and inner chloroplast membrane proteins at high levels induces many types of membrane alterations including stromule-like structures. A. thaliana mesophyll protoplasts were co-transfected with atTic20-II–GFP (inner envelope) and TM-CHUP1–Cherry (outer envelope; see Figure 5E). GFP fluorescence (GFP), Cherry fluorescence (Cherry), chlorophyll autofluorescence (AUF) or overlays of GFP/AUF and GFP/Cherry of a representative protoplast are shown. Images are maximum projections of optical sections. The scale bar represents 10 μm. The framed region in the GFP/Cherry overlay is zoomed in bottom right (scale bare 2 μm). The triangles point to labeling with GFP or Cherry only. Scale bars represent 10 μm.
Discussion
GFP and its derivatives has been a powerful tool to study the in vivo localization of fusion proteins and has provided great insights into questions ranging from the identification of sequences that are important for proper protein localization and functioning (Aihara et al., 2008; Millar et al., 2009) to the nature of dynamic changes in protein localization (Sakamoto and Briggs, 2002). Protein labeling has allowed for the dynamic imaging of organelles, cytoskeletal elements and various membranous structures (Ehrhardt, 2003). For example, fusion of GFP to random cDNA led to the identification of Arabidopsis lines in which dynamic changes in organelles and novel structures could be imaged (Cutler et al., 2000). In addition, targeted fusion of targeting peptides or sequences of proteins with well-defined localization to GFP has allowed for the visualization of highly dynamic organelles such as mitochondria (Niwa et al., 1999) or structures such as stromules (Kohler et al., 1997; Hanson and Sattarzadeh, 2011).
Our results show that even at high expression levels of outer and inner membrane proteins of chloroplasts and mitochondria the general expected localization of these well-studied proteins was observed (Figures 1 and 3). However, the membranes in many but not all protoplasts formed protrusions regardless of the function of the proteins or the structure of the membrane anchor. When inner envelope proteins were overexpressed we even observed invaginations in some cases (Figure 3). There is evidence from other protein–GFP constructs that high levels of proteins can lead to the formation of unusual membrane protrusions. For example, the protein SFR2, which plays an important role in the freezing tolerance of A. thaliana and localizes to the outer envelope membrane, is also found in protrusions similar to those that we observed for outer envelope proteins even though the construct was expressed in mutant protoplasts (Fourrier et al., 2008).
Chloroplast stromules – storage place for imported outer envelope proteins?
We observed membrane protrusions for both helical and barrel-shaped proteins. It had been shown before that helical peptides can induce non-bilayer phases even at low concentrations (Lewis et al., 2007), suggesting that processes such as membrane deformation and the formation of vesicles and tubules may be under the control of intramembrane proteins (Marsh, 2008; van Meer et al., 2008). For example, in tobacco it has been shown that special integral membrane proteins are involved in the restriction of tubular ER structures (namely reticulons; see Sparkes et al., 2010). Invaginations of the membrane were also observed when the inner membrane protein Tic40 was overexpressed in tobacco protoplasts (Singh et al., 2008). Furthermore, membrane dynamics of inner and outer envelope membranes seem to be regulated independent of each other (Figures 1 and 3–5; Videos S1 and S2 in Supplementary Material) as expression of Tic40 only led to changes in inner membrane morphology and to the upregulation of other inner membrane proteins (Singh et al., 2008). Interestingly, these membrane outgrowths were folded in several layers as shown by electron microscopy and were accompanied by the upregulation of the expression of other inner membrane proteins (Singh et al., 2008).
When both inner and outer envelope proteins were expressed at high levels we observed a range of different protrusions including some that were stromule-like (stromules; see Natesan et al., 2005; Hanson and Sattarzadeh, 2008 and references therein). Mesophyll chloroplasts usually have – if at all – few and relatively small stromules, in contrast to leucoplasts. The biological significance of stromules is not established, but it has been speculated that they help to increase the surface area of different plastid types especially those with relatively small plastid bodies. Stromules are envisioned as platforms for the interaction with other plastids, mitochondria, and the endoplasmic reticulum or in general as structures that increase the uptake or release capacity of chloroplasts for proteins and solutes (Natesan et al., 2005). It is not clear how stromules are formed, if proteins are pushing from the main plastid body or if external cytoskeletal elements are pulling (Hanson and Sattarzadeh, 2008), but it was interesting to see that similar structures could come into being by simultaneously overexpressing both inner and outer envelope proteins. However, we did not observe a dynamic behavior of the protrusions described in here in the same time range as seen for stromules (Kwok and Hanson, 2004c; Natesan et al., 2005; Hanson and Sattarzadeh, 2011), indicating that they are different in nature than true stromules. Nevertheless, our data suggest the observed structures might be viewed as “storage compartments” after massive import of membrane proteins, but the question remains, why the extrusions are locally restricted.
Mitochondrial membrane protein expression results in a fission disruption like “phenotype”
The effects of overexpression of proteins on membrane structure were more pronounced in chloroplasts than in mitochondria which could be due to several reasons. Mitochondria are much smaller in size and much more dynamic in shape than chloroplasts, making alterations in membrane structure more difficult to pinpoint using confocal imaging. We saw the most pronounced effects on mitochondrial structure when overexpressing Tom20 (Figure 1C), which led to an abundance of elongated mitochondria in protoplasts. Interestingly mitochondria of similar shapes were seen in a dominant-negative mutant of ADL1C[K48C], a dynamin-like protein that plays a crucial role in mitochondrial fission (Jin et al., 2003). Since the shape of mitochondria is influenced by the balance between fission and fusion (Sesaki and Jensen, 2001; Palmer et al., 2011), it is not surprising that fission mutants exhibit altered mitochondrial forms. There are two classes of fission proteins, one that localizes to the outer membrane and another that localizes to the inner membrane (Jin et al., 2003). Evidence suggests that in A. thaliana BIGYIN is inserted in the outer membrane and interacts with cytoplasmic proteins such as ELM1, DRP3A, and DRP3B during fission (Arimura et al., 2008). It is not clear how the high concentrations of Tom20-II–GFP would interfere with fission, but most likely they would prevent the binding or proper localization of factors influencing the fission of the outer membrane. The overexpression of mtRhomboid resulted in a few mitochondria with a bulb-like structure (Figure 3C). Mitochondria with similar structures were seen in protoplasts of bigyin1 mutants in which fission was disrupted (Scott et al., 2006).
Protein to lipid ratio as determinant of membrane structure
The formation of membrane protrusions in our and other studies was concentration dependent but independent of protein function (Wright et al., 1988; Gong et al., 1996; Arechaga et al., 2000; Lefman et al., 2004; Schmidt von Braun and Schleiff, 2008; Singh et al., 2008). Hence the formation of membrane protrusions under these conditions might reflect a compensatory mechanism of membranes to handle the insertion of the high amounts of proteins while keeping the ratio of proteins to lipids in most of the membranes constant. In line with such notion, 2 h after inhibition of protein synthesis the deformation of the membrane disappeared (Figure 5). Furthermore, the formation of membrane tubules can be induced by injecting large amounts of proteins into vesicles (Domanov and Kinnunen, 2006). This suggests that the lipid–peptide interaction alone without the involvement of cytoskeletal elements can lead to the modulation of the membrane structure. Interestingly even extreme alterations in membrane structure did not seem to affect the growth or fertility of the organism tested (Wright et al., 1988; Singh et al., 2008). It has been proposed that a regulatory network exists that synchronizes lipid synthesis and membrane protein expression. It is not clear what triggers these coordinated membrane alterations and the coordination between chloroplasts and nucleus but it might be a phase transition induced by alteration of the lipid to protein ratio (e.g., Jähnig et al., 1982; Vigh et al., 1998). Accordingly the degradation of overexpressed proteins leads to the subsequent disappearance of the membrane protrusions (Figure 5). The targeted expression of envelope proteins at various concentrations may be exploited as a tool to investigate the regulatory network that controls the lipid to protein ratio in various membranous environments. The latter will be of major importance for an understanding of the regulation of membrane biogenesis in general.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Videos S1–S5 for this article can be found online at http://www.frontiersin.org/technical_advances_in_plant_science/10.3389/fpls.2011.00118/abstract
Videos S1–S5 show a single chloroplast (Videos S1 and S2) or a section (Videos S3–S5) of an Arabidopsis thaliana mesophyll protoplast overexpressing atTic20-II–GFP (inner envelope, green) and TM-CHUP1–Cherry (outer envelope, red; see also Figure 6).
Scan through a confocal image stack in Z-axis showing the fluorescence of the GFP (green) and Cherry (red) fusion proteins in relation to the chloroplast autofluorescence (in blue).
Three hundred sixty degrees rotation of the single chloroplast of Video S1 along the X-axis after deconvolution and 3D reconstruction. The extrusions of the outer and the intrusions of the inner membrane are clearly visible. Extrusions of the outer envelope are marked with red, intrusions of the inner with green arrow heads. White arrow heads point to the stromule-like tubular extrusions of both membranes.
Scan through a confocal image stack in Z-axis of a protoplast section as in Video S1.
Ninety degrees rotation of S3 round the X- (Video S4) or the Y-axis (Video S5). Arrowheads point to proliferations of the outer membrane. GFP was excited at 488 nm and the emission was recorded at 495–525 nm using a Leica SP5 microscope with a HCX PL APO CS 40 × 1.25 NA 1.25 oil objective. Imaris ×64 6.2.1 software (Bitplane Inc., CT, USA) was used for video processing. Cherry was excited at 561 nm and recorded at 595–615 nm. GFP is in green, Cherry in red, chloroplast autofluorescence is in blue (Videos S2, S4, and S5). Three hundred sixty degrees rotations of Video S1 or S3 after deconvolution of the data using Auto Quant X Version ×2.1.1 with AutoDeblur Gold GWF (Media Cybernetics Inc., MD, USA), respectively. Any fairly recent version of the Windows MediaPlayer is capable of playing the supplied video files. Other players and/or operating systems might require the installation of the Microsoft MPEG-4 codec version 3 (MP43) or any other compatible MPEG-4 codec (e.g., www.xvid.org).
Acknowledgments
We thank Klaus Dieter Scharf for helpful discussions while designing the experimental setup, Beata Kmiec-Wisniewska (University of Wroclaw) for providing us the Rhomboid expression plasmids. The work was supported by grants from Deutsche Forschungsgemeinschaft (SFB807-P17), the Center of Membrane Proteomics Frankfurt (CMP), the Cluster of Excellence “Macromolecular Complexes” and Volkswagenstiftung to Enrico Schleiff.
References
- Aihara Y., Tabata R., Suzuki T., Shimazaki K., Nagatani A. (2008). Molecular basis of the functional specificities of phototropin 1 and 2. Plant J. 56, 364–375 10.1111/j.1365-313X.2008.03605.x [DOI] [PubMed] [Google Scholar]
- Arechaga I., Miroux B., Karrasch S., Huijbregts R., De Kruijff B., Runswick M. J., Walker J. E. (2000). Characterisation of new intracellular membranes in Escherichia coli accompanying large scale over-production of the β subunit of F(1)F(o) ATP synthase. FEBS Lett. 482, 215–219 10.1016/S0014-5793(00)02054-8 [DOI] [PubMed] [Google Scholar]
- Arimura S., Fujimoto M., Doniwa Y., Kadoya N., Nakazono M., Sakamoto W., Tsutsumi N. (2008). Arabidopsis ELONGATED MITOCHONDRIA1 is required for localization of DYNAMIN- RELATED PROTEIN3A to mitochondrial fission sites. Plant Cell 20, 1555–1566 10.1105/tpc.108.058578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benning C. (2009). Mechanisms of lipid transport involved in organelle biogenesis in plant cells. Annu. Rev. Cell Dev. Biol. 25, 71–91 10.1146/annurev.cellbio.042308.113414 [DOI] [PubMed] [Google Scholar]
- Bionda T., Tillmann B., Simm S., Beilstein K., Ruprecht M., Schleiff E. (2010). Chloroplast import signals: the length requirement for translocation in vitro and in vivo. J. Mol. Biol. 402, 510–523 10.1016/j.jmb.2010.07.052 [DOI] [PubMed] [Google Scholar]
- Breuers F. K. H., Braeutigam A., Geimer S., Welzel U. Y., Stefano G., Renna L., Brandizzi F., Weber A. P. M. (2012). Dynamic remodeling of the plastid envelope membranes – a tool for chloroplast envelope in vivo localizations. Front. Plant Sci. 10.3389/fpls.2012.00007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burgos A., Szymanski J., Seiwert B., Degenkolbe T., Hannah M. A., Giavalisco P., Willmitzer L. (2011). Analysis of short-term changes in the Arabidopsis thaliana glycerolipidome in response to temperature and light. Plant J. 66, 656–668 10.1111/j.1365-313X.2011.04531.x [DOI] [PubMed] [Google Scholar]
- Carrie C., Kuhn K., Murcha M. W., Duncan O., Small I. D., O’toole N., Whelan J. (2009). Approaches to defining dual-targeted proteins in Arabidopsis. Plant J. 57, 1128–1139 10.1111/j.1365-313X.2008.03745.x [DOI] [PubMed] [Google Scholar]
- Cutler S. R., Ehrhardt D. W., Griffitts J. S., Somerville C. R. (2000). Random GFP::cDNA fusions enable visualization of subcellular structures in cells of Arabidopsis at a high frequency. Proc. Natl. Acad. Sci. U.S.A. 97, 3718–3723 10.1073/pnas.97.7.3718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daschner K., Couee I., Binder S. (2001). The mitochondrial isovaleryl-coenzyme a dehydrogenase of Arabidopsis oxidizes intermediates of leucine and valine catabolism. Plant Physiol. 126, 601–612 10.1104/pp.126.2.601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Domanov Y. A., Kinnunen P. K. (2006). Antimicrobial peptides temporins B and L induce formation of tubular lipid protrusions from supported phospholipid bilayers. Biophys. J. 91, 4427–4439 10.1529/biophysj.106.091702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehrhardt D. (2003). GFP technology for live cell imaging. Curr. Opin. Plant Biol. 6, 622–628 10.1016/j.pbi.2003.09.014 [DOI] [PubMed] [Google Scholar]
- Fourrier N., Bedard J., Lopez-Juez E., Barbrook A., Bowyer J., Jarvis P., Warren G., Thorlby G. (2008). A role for SENSITIVE TO FREEZING2 in protecting chloroplasts against freeze-induced damage in Arabidopsis. Plant J. 55, 734–745 10.1111/j.1365-313X.2008.03549.x [DOI] [PubMed] [Google Scholar]
- Gong F. C., Giddings T. H., Meehl J. B., Staehelin L. A., Galbraith D. W. (1996). Z-membranes: artificial organelles for overexpressing recombinant integral membrane proteins. Proc. Natl. Acad. Sci. U.S.A. 93, 2219–2223 10.1073/pnas.93.3.1340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray J. C., Sullivan J. A., Hibberd J. M., Hansen M. R. (2001). Stromules: mobile protrusions and interconnections between plastids. Plant Biol. 3, 223–233 10.1055/s-2001-15204 [DOI] [Google Scholar]
- Gunning B. E. (2005). Plastid stromules: video microscopy of their outgrowth, retraction, tensioning, anchoring, branching, bridging, and tip-shedding. Protoplasma 225, 33–42 10.1007/s00709-004-0073-3 [DOI] [PubMed] [Google Scholar]
- Hanke G. T., Kimata-Ariga Y., Taniguchi I., Hase T. (2004). A post genomic characterization of Arabidopsis ferredoxins. Plant Physiol. 134, 255–264 10.1104/pp.103.032755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanson M. R., Sattarzadeh A. (2008). Dynamic morphology of plastids and stromules in angiosperm plants. Plant Cell Environ. 31, 646–657 10.1111/j.1365-3040.2007.01768.x [DOI] [PubMed] [Google Scholar]
- Hanson M. R., Sattarzadeh A. (2011). Stromules: recent insights into a long neglected feature of plastid morphology and function. Plant Physiol. 155, 1486–1492 10.1104/pp.110.170852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holzinger A., Buchner O., Lutz C., Hanson M. R. (2007). Temperature-sensitive formation of chloroplast protrusions and stromules in mesophyll cells of Arabidopsis thaliana. Protoplasma 230, 23–30 10.1007/s00709-006-0222-y [DOI] [PubMed] [Google Scholar]
- Islam M. S., Niwa Y., Takagi S. (2009). Light-dependent intracellular positioning of mitochondria in Arabidopsis thaliana mesophyll cells. Plant Cell Physiol. 50, 1032–1040 10.1093/pcp/pcp066 [DOI] [PubMed] [Google Scholar]
- Itoh R. D., Fujiwara M. T. (2010). Regulation of leucoplast morphology in roots. Interorganellar signaling from mitochondria? Plant Signal. Behav. 5, 856–859 10.4161/psb.5.7.11893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jähnig F., Vogel H., Best L. (1982). Unifying description of the effect of membrane proteins on lipid order. Verification for the melittin/dimyristoylphosphatidylcholine system. Biochemistry 21, 6790–6798 10.1021/bi00269a027 [DOI] [PubMed] [Google Scholar]
- Jin J. B., Bae H., Kim S. J., Jin Y. H., Goh C. H., Kim D. H., Lee Y. J., Tse Y. C., Jiang L., Hwang I. (2003). The Arabidopsis dynamin-like proteins ADL1C and ADL1E play a critical role in mitochondrial morphogenesis. Plant Cell 15, 2357–2369 10.1105/tpc.015222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasmati A. R., Topel M., Patel R., Murtaza G., Jarvis P. (2011). Molecular and genetic analyses of Tic20 homologues in Arabidopsis thaliana chloroplasts. Plant J. 66, 877–889 10.1111/j.1365-313X.2011.04551.x [DOI] [PubMed] [Google Scholar]
- Kmiec-Wisniewska B., Krumpe K., Urantowka A., Sakamoto W., Pratje E., Janska H. (2008). Plant mitochondrial rhomboid, AtRBL12, has different substrate specificity from its yeast counterpart. Plant Mol. Biol. 68, 159–171 10.1007/s11103-008-9359-8 [DOI] [PubMed] [Google Scholar]
- Kohler R. H., Cao J., Zipfel W. R., Webb W. W., Hanson M. R. (1997). Exchange of protein molecules through connections between higher plant plastids. Science 276, 2039–2042 10.1126/science.276.5321.2039 [DOI] [PubMed] [Google Scholar]
- Kuhn S., Bussemer J., Chigri F., Vothknecht U. C. (2009). Calcium depletion and calmodulin inhibition affect the import of nuclear-encoded proteins into plant mitochondria. Plant J. 58, 694–705 10.1111/j.1365-313X.2009.03810.x [DOI] [PubMed] [Google Scholar]
- Kwok E. Y., Hanson M. R. (2004a). GFP-labelled Rubisco and aspartate aminotransferase are present in plastid stromules and traffic between plastids. J. Exp. Bot. 55, 595–604 10.1093/jxb/erh062 [DOI] [PubMed] [Google Scholar]
- Kwok E. Y., Hanson M. R. (2004b). In vivo analysis of interactions between GFP-labeled microfilaments and plastid stromules. BMC Plant Biol. 4, 2. 10.1186/1471-2229-4-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwok E. Y., Hanson M. R. (2004c). Stromules and the dynamic nature of plastid morphology. J. Microsc. 214, 124–137 10.1111/j.0022-2720.2004.01317.x [DOI] [PubMed] [Google Scholar]
- Lee A. G. (2003). Lipid-protein interactions in biological membranes: a structural perspective. Biochim. Biophys. Acta 1612, 1–40 10.1016/S0005-2736(03)00056-7 [DOI] [PubMed] [Google Scholar]
- Lee A. G. (2004). How lipids affect the activities of integral membrane proteins. Biochim. Biophys. Acta 1666, 62–87 10.1016/j.bbamem.2004.05.012 [DOI] [PubMed] [Google Scholar]
- Lee D. W., Lee S., Lee G. J., Lee K. H., Kim S., Cheong G. W., Hwang I. (2006). Functional characterization of sequence motifs in the transit peptide of Arabidopsis small subunit of rubisco. Plant Physiol. 140, 466–483 10.1104/pp.105.074575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lefman J., Zhang P., Hirai T., Weis R. M., Juliani J., Bliss D., Kessel M., Bos E., Peters P. J., Subramaniam S. (2004). Three-dimensional electron microscopic imaging of membrane invaginations in Escherichia coli overproducing the chemotaxis receptor Tsr. J. Bacteriol. 186, 5052–5061 10.1128/JB.186.15.5052-5061.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis R. N., Liu F., Krivanek R., Rybar P., Hianik T., Flach C. R., Mendelsohn R., Chen Y., Mant C. T., Hodges R. S., McElhaney R. N. (2007). Studies of the minimum hydrophobicity of alpha-helical peptides required to maintain a stable transmembrane association with phospholipid bilayer membranes. Biochemistry 46, 1042–1054 10.1021/bi061891b [DOI] [PMC free article] [PubMed] [Google Scholar]
- Logan D. C. (2006). Plant mitochondrial dynamics. Biochim. Biophys. Acta 1763, 430–441 10.1016/j.bbamcr.2006.01.003 [DOI] [PubMed] [Google Scholar]
- Logan D. C., Leaver C. J. (2000). Mitochondria-targeted GFP highlights the heterogeneity of mitochondrial shape, size and movement within living plant cells. J. Exp. Bot. 51, 865–871 10.1093/jexbot/51.346.865 [DOI] [PubMed] [Google Scholar]
- Lutz C., Engel L. (2007). Changes in chloroplast ultrastructure in some high-alpine plants: adaptation to metabolic demands and climate? Protoplasma 231, 183–192 10.1007/s00709-007-0249-8 [DOI] [PubMed] [Google Scholar]
- Machettira A. B., Gross L. E., Sommer M. S., Weis B. L. M., Englich G., Tripp J., Schleiff E. (2011). The localization of Tic20 proteins in Arabidopsis thaliana is not restricted to the inner envelope membrane of chloroplasts. Plant Mol. Biol. 77, 381–390 10.1007/s11103-011-9818-5 [DOI] [PubMed] [Google Scholar]
- Marsh D. (2008). Protein modulation of lipids, and vice-versa, in membranes. Biochim. Biophys. Acta 1778, 1545–1575 10.1016/j.bbamem.2008.01.015 [DOI] [PubMed] [Google Scholar]
- Matthes A., Schmidt-Gattung S., Kohler D., Forner J., Wildum S., Raabe M., Urlaub H., Binder S. (2007). Two DEAD-box proteins may be part of RNA-dependent high-molecular-mass protein complexes in Arabidopsis mitochondria. Plant Physiol. 145, 1637–1646 10.1104/pp.107.108076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melser S., Molino D., Batailler B., Peypelut M., Laloi M., Wattelet-Boyer V., Bellec Y., Faure J. D., Moreau P. (2011). Links between lipid homeostasis, organelle morphodynamics and protein trafficking in eukaryotic and plant secretory pathways. Plant Cell Rep. 30, 177–193 10.1007/s00299-011-1037-7 [DOI] [PubMed] [Google Scholar]
- Millar A. H., Carrie C., Pogson B., Whelan J. (2009). Exploring the function-location nexus: using multiple lines of evidence in defining the subcellular location of plant proteins. Plant Cell 21, 1625–1631 10.1105/tpc.109.066019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mishra S. K., Tripp J., Winkelhaus S., Tschiersch B., Theres K., Nover L., Scharf K. D. (2002). In the complex family of heat stress transcription factors, HsfA1 has a unique role as master regulator of thermotolerance in tomato. Genes Dev. 16, 1555–1567 10.1101/gad.228802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Natesan S. K., Sullivan J. A., Gray J. C. (2005). Stromules: a characteristic cell-specific feature of plastid morphology. J. Exp. Bot. 56, 787–797 10.1093/jxb/eri088 [DOI] [PubMed] [Google Scholar]
- Niwa Y., Hirano T., Yoshimoto K., Shimizu M., Kobayashi H. (1999). Non-invasive quantitative detection and applications of non-toxic, S65T-type green fluorescent protein in living plants. Plant J. 18, 455–463 10.1046/j.1365-313X.1999.00464.x [DOI] [PubMed] [Google Scholar]
- Okawa K., Nakayama K., Kakizaki T., Yamashita T., Inaba T. (2008). Identification and characterization of Cor413im proteins as novel components of the chloroplast inner envelope. Plant Cell Environ. 31, 1470–1483 10.1111/j.1365-3040.2008.01854.x [DOI] [PubMed] [Google Scholar]
- Palmer C. S., Osellame L. D., Stojanovski D., Ryan M. T. (2011). The regulation of mitochondrial morphology: intricate mechanisms and dynamic machinery. Cell. Signal. 23, 1534–1545 10.1016/j.cellsig.2011.05.021 [DOI] [PubMed] [Google Scholar]
- Perry A. J., Hulett J. M., Likic V. A., Lithgow T., Gooley P. R. (2006). Convergent evolution of receptors for protein import into mitochondria. Curr. Biol. 16, 221–229 10.1016/j.cub.2005.12.034 [DOI] [PubMed] [Google Scholar]
- Pohlmeyer K., Soll J., Grimm R., Hill K., Wagner R. (1998). A high-conductance solute channel in the chloroplastic outer envelope from Pea. Plant Cell 10, 1207–1216 10.2307/3870722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Presley A. D., Fuller K. M., Arriaga E. A. (2003). MitoTracker green labeling of mitochondrial proteins and their subsequent analysis by capillary electrophoresis with laser-induced fluorescence detection. J. Chromatogr. B Analyt. Technol. Biomed. Life Sci. 793, 141–150 10.1016/S1570-0232(03)00371-4 [DOI] [PubMed] [Google Scholar]
- Sakamoto K., Briggs W. R. (2002). Cellular and subcellular localization of phototropin 1. Plant Cell 14, 1723–1735 10.1105/tpc.003293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schattat M., Kiah Barton K., Mathur J. (2011). Correlated behavior implicates stromules in increasing the interactive surface between plastids and ER tubules. Plant Signal. Behav. 6, 715–718 10.4161/psb.6.5.15085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schleiff E., Tien R., Salomon M., Soll J. (2001). Lipid composition of outer leaflet of chloroplast outer envelope determines topology of OEP7. Mol. Biol. Cell 12, 4090–4102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt von Braun S., Schleiff E. (2008). The chloroplast outer membrane protein CHUP1 interacts with actin and profilin. Planta 227, 1151–1159 10.1007/s00425-007-0688-7 [DOI] [PubMed] [Google Scholar]
- Scott I., Tobin A. K., Logan D. C. (2006). BIGYIN, an orthologue of human and yeast FIS1 genes functions in the control of mitochondrial size and number in Arabidopsis thaliana. J. Exp. Bot. 57, 1275–1280 10.1093/jxb/erj096 [DOI] [PubMed] [Google Scholar]
- Sesaki H., Jensen R. E. (2001). UGO1 encodes an outer membrane protein required for mitochondrial fusion. J. Cell Biol. 152, 1123–1134 10.1083/jcb.152.6.1123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw D. J., Gray J. C. (2011). Visualisation of stromules in transgenic wheat expressing a plastid-targeted yellow fluorescent protein. Planta 233, 961–970 10.1007/s00425-011-1351-x [DOI] [PubMed] [Google Scholar]
- Singh N. D., Li M., Lee S. B., Schnell D., Daniell H. (2008). Arabidopsis Tic40 expression in tobacco chloroplasts results in massive proliferation of the inner envelope membrane and upregulation of associated proteins. Plant Cell 20, 3405–3417 10.1105/tpc.108.063172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sommer M. S., Daum B., Gross L. E., Weis B. L., Mirus O., Abram L., Maier U. G., Kuhlbrandt W., Schleiff E. (2011). Chloroplast Omp85 proteins change orientation during evolution. Proc. Natl. Acad. Sci. U.S.A. 108, 13841–13846 10.1073/pnas.1108626108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sparkes I., Tolley N., Aller I., Svozil J., Osterrieder A., Botchway S., Mueller C., Frigerio L., Hawes C. (2010). Five Arabidopsis reticulon isoforms share endoplasmic reticulum location, topology, and membrane-shaping properties. Plant Cell 22, 1333–1343 10.1105/tpc.110.074385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uemura M., Joseph R. A., Steponkus P. L. (1995). Cold acclimation of Arabidopsis thaliana (effect on plasma membrane lipid composition and freeze-induced lesions). Plant Physiol. 109, 15–30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Meer G., Voelker D. R., Feigenson G. W. (2008). Membrane lipids: where they are and how they behave. Nat. Rev. Mol. Cell Biol. 9, 112–124 10.1038/nrm2330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vigh L., Maresca B., Harwood J. L. (1998). Does the membrane’s physical state control the expression of heat shock and other genes? Trends Biochem. Sci. 23, 369–374 10.1016/S0968-0004(98)01279-1 [DOI] [PubMed] [Google Scholar]
- Wright R., Basson M., D’ari L., Rine J. (1988). Increased amounts of HMG-CoA reductase induce “karmellae”: a proliferation of stacked membrane pairs surrounding the yeast nucleus. J. Cell Biol. 107, 101–114 10.1083/jcb.107.1.101 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Scan through a confocal image stack in Z-axis showing the fluorescence of the GFP (green) and Cherry (red) fusion proteins in relation to the chloroplast autofluorescence (in blue).
Three hundred sixty degrees rotation of the single chloroplast of Video S1 along the X-axis after deconvolution and 3D reconstruction. The extrusions of the outer and the intrusions of the inner membrane are clearly visible. Extrusions of the outer envelope are marked with red, intrusions of the inner with green arrow heads. White arrow heads point to the stromule-like tubular extrusions of both membranes.
Scan through a confocal image stack in Z-axis of a protoplast section as in Video S1.
Ninety degrees rotation of S3 round the X- (Video S4) or the Y-axis (Video S5). Arrowheads point to proliferations of the outer membrane. GFP was excited at 488 nm and the emission was recorded at 495–525 nm using a Leica SP5 microscope with a HCX PL APO CS 40 × 1.25 NA 1.25 oil objective. Imaris ×64 6.2.1 software (Bitplane Inc., CT, USA) was used for video processing. Cherry was excited at 561 nm and recorded at 595–615 nm. GFP is in green, Cherry in red, chloroplast autofluorescence is in blue (Videos S2, S4, and S5). Three hundred sixty degrees rotations of Video S1 or S3 after deconvolution of the data using Auto Quant X Version ×2.1.1 with AutoDeblur Gold GWF (Media Cybernetics Inc., MD, USA), respectively. Any fairly recent version of the Windows MediaPlayer is capable of playing the supplied video files. Other players and/or operating systems might require the installation of the Microsoft MPEG-4 codec version 3 (MP43) or any other compatible MPEG-4 codec (e.g., www.xvid.org).