Abstract
The genus Agrobacterium is unique in its ability to conduct interkingdom genetic exchange. Virulent Agrobacterium strains transfer single-strand forms of T-DNA (T-strands) and several Virulence effector proteins through a bacterial type IV secretion system into plant host cells. T-strands must traverse the plant wall and plasma membrane, traffic through the cytoplasm, enter the nucleus, and ultimately target host chromatin for stable integration. Because any DNA sequence placed between T-DNA “borders” can be transferred to plants and integrated into the plant genome, the transfer and intracellular trafficking processes must be mediated by bacterial and host proteins that form complexes with T-strands. This review summarizes current knowledge of proteins that interact with T-strands in the plant cell, and discusses several models of T-complex (T-strand and associated proteins) trafficking. A detailed understanding of how these macromolecular complexes enter the host cell and traverse the plant cytoplasm will require development of novel technologies to follow molecules from their bacterial site of synthesis into the plant cell, and how these transferred molecules interact with host proteins and sub-cellular structures within the host cytoplasm and nucleus.
Keywords: Agrobacterium, T-DNA, virulence effector proteins, cytoplasmic trafficking, nuclear targeting
Introduction
Members of the genus Agrobacterium have the unique, natural ability to conduct horizontal genetic exchange between organisms of different phylogenetic kingdoms. Best known among Agrobacterium species is Agrobacterium tumefaciens, which causes the disease crown gall on a wide variety of dicotyledonous plants, as well as on some gymnosperms. Other pathogenic Agrobacterium species include A. rhizogenes, the causative agent for hairy root (root mat) disease, A. vitis, which causes tumors on grape and a few other species, and A. rubi, which causes cane gall disease (Kersters and De Ley, 1984; Farrand et al., 2003). The fundamental mechanism of pathogenesis is the same for each of these species: DNA transfer from the bacterium to the host plant leads to integration and expression of a portion of a large plasmid [Ti-(tumor inducing) or Ri-(root inducing) plasmid] originally extant in the bacterium. The region of DNA which is processed from these large plasmids is termed the T(transferred)-DNA region, and the transferred DNA is termed T-DNA. T-DNA is exported from Agrobacterium and enters the eukaryotic cell as a single-strand molecule called the T-strand. T-strands must traverse the host cell cytoplasm and enter the nucleus, where they eventually may integrate into the host genome. Plant species are the natural hosts for T-DNA transfer; however, animal and fungal cells can also participate as recipient hosts under laboratory conditions (Bundock et al., 1995; Piers et al., 1996; de Groot et al., 1998; Abuodeh et al., 2000; Kunik et al., 2001; Bulgakov et al., 2006; Lacroix et al., 2006a,b; Michielse et al., 2008).
Although scientists frequently consider Agrobacterium-mediated genetic transformation as T-DNA transfer, the molecular mechanism of transfer and intracellular T-strand movement depend on proteins likely associated with T-strands. This must be true because no specific T-DNA gene or DNA sequence is required for transformation: any sequence may be substituted between T-DNA borders for successful transformation to occur (Bevan et al., 1983; Fraley et al., 1983; Herrera-Estrella et al., 1983). It would appear that it is the various associated proteins, rather than T-DNA, that are acted upon to route T-DNA to its ultimate nuclear destination. Thus, it is probably more correct to think of T-DNA movement as protein, rather than DNA, trafficking. This review discusses this journey, and the responsible cellular components.
The Bacterial Players: Virulence Effector Proteins
As with many bacteria, Agrobacterium transfers several bacterial proteins into the host cell to achieve pathogenesis. These proteins are called Virulence effector proteins (or Vir proteins), and their export from Agrobacterium requires a type IV secretion system (T4SS). The Agrobacterium T4SS is composed of 11 VirB proteins and VirD4; their assembly and structure have been described in numerous studies (Christie et al., 2005; Fronzes et al., 2009).
VirD2
T-DNA transfer from Agrobacterium to host cells initiates with the induction of the virulence (vir) genes by plant-derived phenolic and sugar molecules (Stachel et al., 1985, 1986; Ankenbauer and Nester, 1990). Among the induced proteins is VirD2, an endonuclease that nicks the T-DNA region of Ti-/Ri-plasmids at border repeat sequences flanking T-DNA. During T-DNA processing, VirD2 covalently attaches to the 5′ end of the resulting T-strand (Herrera-Estrella et al., 1988; Ward and Barnes, 1988; Young and Nester, 1988; Durrenberger et al., 1989). VirD2 serves as a “pilot protein” to lead T-strands through the T4SS and into the host cell. Once in the host, VirD2 probably plays a major role in targeting T-strands to the nucleus. The C-terminal region of VirD2 contains a bipartite nuclear localization signal (NLS) sequence. This NLS (or even individual portions of the NLS) strongly targets affixed reporter proteins to the nucleus of plant, animal, and yeast cells (Herrera-Estrella et al., 1990; Howard et al., 1992; Tinland et al., 1992; Rossi et al., 1993; Citovsky et al., 1994; Mysore et al., 1998). A monopartite NLS in the N-terminal region of VirD2 can also target proteins to plant nuclei. However, this NLS probably does not function to target T-strands because it is located close to tyrosine 29, the amino acid to which T-strands link. Thus, this weaker NLS is likely occluded by T-DNA and does not appreciably contribute to T-strand nuclear targeting (Shurvinton et al., 1992; Vogel and Das, 1992).
Nuclear targeting of the VirD2/T-strand complex is likely mediated by importin α proteins. Importin α is an adaptor molecule that interacts with “classical” NLS motifs in cargo proteins and with the nuclear shuttle protein importin β. Under appropriate conditions, importin α/β/cargo protein complexes traverse through nuclear pores into the nucleoplasm, after which the complex dissociates; importins α and β cycle back to the cytoplasm where they can associate with additional cargoes for nuclear delivery (Terry et al., 2007).
Ballas and Citovsky (1997) initially described interaction of VirD2 with Arabidopsis thaliana AtKapα (subsequently renamed IMPa-1) in vitro and in yeast. Interaction required the VirD2 C-terminal bipartite NLS, and the complexes could be imported into nuclei of permeabilized yeast cells. Arabidopsis encodes nine importin α isoforms. Bhattacharjee et al. (2008) noted that VirD2 interacted with all tested importin α proteins in vitro, in yeast, and [using bimolecular fluorescence complementation (BiFC)] in plant cells. In plants, VirD2 complexed with IMPa-1, -4, -7, and -9 localized exclusively to nuclei, suggesting that various importin α isoforms could serve as adaptors for nuclear import of VirD2/T-strand complexes. However, a homozygous mutant of only impa4, but not of other tested IMP genes, was deficient in Agrobacterium-mediated transformation. Over-expression in the impa4 mutant background of any of seven tested IMP cDNAs could, however, reverse this phenotype. These results indicate that, although several importin α proteins may participate in transformation-associated events, IMPa-4 likely plays the major role.
VirD2 is a phosphoprotein (Bakó et al., 2003; Tao et al., 2004). Yeast two-hybrid analyses, using VirD2 as bait, have identified several additional plant proteins that interact with VirD2 and which may play roles in phosphorylation/dephosphorylation and VirD2/T-strand intracellular trafficking. Tao et al. (2004) discovered a tomato type 2C protein phosphatase (PP2C) that interacts with the C-terminal region of VirD2. Over-expression of this PP2C in tobacco protoplasts resulted in mis-localization of a GUS-VirD2 fusion peptide. Normally, the GUS-VirD2 protein localizes strongly to the nucleus. However, when the PP2C is over-expressed, the fusion protein fractionates between the nucleus and the cytoplasm. A likely VirD2 target site for the PP2C is serine394, which is located immediately upstream of the bipartite NLS. Indeed, alteration of this serine residue to alanine also resulted in a mutant VirD2 protein that did not efficiently target nuclei of protoplasts. Experiments are currently underway in the author’s laboratory to determine whether this serine-to-alanine mutation results in mis-localization of a VirD2–YFP fusion protein in roots of transgenic plants. Because roots are the natural targets for Agrobacterium infection, these results may indicate whether phosphorylation of VirD2 may potentiate its nuclear localization and, as a consequence, nuclear import of the VirD2/T-strand complexes.
VirD2 also interacts with and is phosphorylated by the cyclin-dependent kinase activating kinase CAK2Ms (Bakó et al., 2003). CAK2Ms also phosphorylates one of the RNA polymerase II large subunits, which in turn recruits a TATA-box binding protein important for transcription initiation. VirD2 interacts with this TATA-box binding protein, suggesting that VirD2 phosphorylation may play an additional role in targeting VirD2/T-strand complexes to chromatin. However, because T-DNA integrates randomly into the genome without regard to target site transcription (Kim et al., 2007; see below for a discussion of T-DNA chromatin targeting), the importance of these VirD2 protein interactions remains elusive.
Several authors have noted interaction of VirD2 with plant cyclophilins (Deng et al., 1998; Bakó et al., 2003). Cyclophilins may play a role in maintaining the correct conformation of proteins, and indeed, incubation of Arabidopsis and tobacco cells with the cyclophilin inhibitor Cyclosporin A resulted in decreased transformation. However, the recent finding by van Kregten et al. (2009) that the VirD2 cyclophilin-binding domain is not necessary for transformation suggests that cyclophilins may not be required for VirD2 function in the plant cell.
The role of VirD2 in T-DNA integration is not clear. Tinland et al. (1995) showed that a mutation in the “recombinase” domain of VirD2 (VirD2Rl29G) results in a protein that integrates T-DNA with normal frequency but with altered specificity of border use. Mysore et al. (1998) showed that a different mutation in VirD2 (a deletion/substitution in the ω domain) results in a protein that has normal integration specificity, but lower integration frequency. These latter results suggest that the ω domain may be involved in T-DNA integration. However, Bravo-Angel et al. (1998) showed that a deletion of this ω domain did not affect the efficiency or pattern of T-DNA integration, although T-DNA transfer was severely reduced. The difference in results described by Mysore et al. (1998) and Bravo-Angel et al. (1998) likely results from the different ω region mutations that the two groups used. Although the conflicting results from all of these experiments may be confusing with regard to the mechanistic role of VirD2, taken as a whole they indicate a role for VirD2 in T-DNA integration. The importance of VirD2 in T-DNA integration was further shown by Pelczar et al. (2004). These authors demonstrated that VirD2 affixed to the 5′ end of T-strands was important for maximal efficiency of mammalian cell transformation and precision of T-DNA integration by “artificial T-complexes” synthesized in vitro.
VirE2
VirE2 is a DNA binding protein that, in vitro, cooperatively forms complexes with any single-strand DNA sequence (Gietl et al., 1987; Christie et al., 1988; Citovsky et al., 1988, 1989; Das, 1988; Sen et al., 1989). T-DNA is transferred from Agrobacterium to host cells as a single-strand molecule (the T-strand), and therefore the original proposed function of VirE2 was to protect T-strands from nucleolytic degradation as they travel from the bacterium to the host cell (Tinland et al., 1994; Yusibov et al., 1994). Although an early publication indicated that VirE2 interacts with T-strands in Agrobacterium (Christie et al., 1988), this observation likely resulted from artifacts in the way the experiments were conducted. More recent experimental results indicate that VirE2 does not interact with T-strands in the bacterium (Cascales and Christie, 2004). Indeed, within Agrobacterium, VirE2 interacts with its chaperone VirE1 (Sundberg et al., 1996; Zhao et al., 2001). This interaction likely prevents VirE2 from forming complexes with T-strands in the bacterium. The importance of VirE2 in transformation occurs in the plant: plants expressing VirE2 could complement Agrobacterium virE2 mutants, that either lack a wild-type virE2 gene or make a form of VirE2 that cannot exit the bacterium, to full virulence (Citovsky et al., 1992; Simone et al., 2001).
VirE2 protects T-strands from nucleolytic degradation in the plant cell. Agrobacterium strains mutant in virE2 are not avirulent, but rather are severely attenuated in virulence. Yusibov et al. (1994) showed that T-strands transferred from virE2 mutant Agrobacterium accumulated to a much lesser extent than did T-strands delivered from wild-type bacteria. In addition, Rossi et al. (1996) showed that in the small number of tumors that did result from infection of plants with an Agrobacterium virE2 mutant, integrated T-DNA was often severely truncated from the 3′ end, which is the end not protected by VirD2. Taken together, these data suggest that VirE2 protein is important for maintaining T-strand integrity within the plant cell, and are consistent with the hypothesis that VirE2 coats and protects T-strands in vivo.
The role of VirE2 in nuclear targeting of T-strands remains controversial. Zupan et al. (1996) showed that fluorescently labeled single-strand DNA remained cytoplasmic when microinjected into Tradescantia cells. However, when the DNA was complexed with VirE2 prior to microinjection, fluorescence accumulated in the nucleus. Gelvin (1998) showed that an Agrobacterium strain that lacked VirE2 and the NLS region of VirD2 was avirulent on wild-type tobacco plants. However, this strain was virulent on tobacco plants that expressed VirE2. Because no known T-strand nuclear targeting signal was present in the mutant Agrobacterium strain, these data indicate that plant-produced VirE2 could interact with and target incoming T-strands to the nucleus, resulting in the production of tumors. Taken together, these experiments indicate that VirE2 plays a role in nuclear targeting of T-strands.
However, other reports suggested that VirE2 plays a minor, if any, role in T-strand nuclear targeting. Ziemienowicz et al. (2001) showed that VirE2-coated fluorescently labeled single-strand DNA molecules, 1 kb in length, did not accumulate in nuclei when introduced into permeabilized evacuolated tobacco BY-2 cells. However, when these VirE2-coated DNA molecules additionally contained VirD2 affixed to their 5′ ends, nuclear localization was rapid and efficient. Nuclear localization of these “artificial T-complexes” depended upon an intact C-terminal VirD2 NLS. These authors concluded that VirD2/T-strand molecules required the VirD2 NLS to target the complexes to the nucleus, but that VirE2 was required for nuclear entry. As described by others (Citovsky et al., 1989, 1997; Abu-Arish et al., 2004), they suggested that VirE2 altered the conformation of DNA molecules such that they could traverse the nuclear pores, but that once VirE2 interacted with DNA, its NLSs were not exposed and available to interact with nuclear import proteins. Thus, the role of VirE2 in nuclear targeting would be indirect: by interaction with importin α, VirD2 targets T-strands to nuclear pores, but nuclear import additionally requires VirE2 to shape T-strands to be able to traverse these pores.
The role of VirE2 in nuclear targeting of T-strands is also complicated by conflicting reports of the ability of VirE2 to interact with importin α. Ballas and Citovsky (1997) could not show interaction of VirE2 with the nuclear import adaptor AtKapα/IMPa-1 using a yeast two-hybrid system. However, Bhattacharjee et al. (2008) used yeast two-hybrid, in planta BiFC, and in vitro pull-down analyses to show that VirE2 could interact with multiple isoforms of importin α, and mapped domains of VirE2 responsible for this interaction. These latter authors went on to show that VirE2 complexed with single-strand DNA was still able to bind to importin α in vitro.
Additional confusion concerning the role of VirE2 in nuclear targeting of T-strands has arisen because of conflicting reports of its sub-cellular localization (Gelvin, 2010). Several studies indicated that VirE2, tagged at its N-terminus with reporter proteins, localized to the nucleus of transfected plant cells (Citovsky et al., 1992, 1994, 2004; Tzfira and Citovsky, 2001; Tzfira et al., 2001; Li et al., 2005). However, Bhattacharjee et al. (2008) demonstrated that such N-terminal tagging rendered VirE2, expressed in plant cells, unable to complement a virE2 mutant Agrobacterium strain for virulence, whereas C-terminally tagged VirE2 retained full activity in this assay. This and follow-up studies indicated that in most types of plant cells, VirE2 remains cytoplasmically localized, often forming perinuclear aggregates (Bhattacharjee et al., 2008; Grange et al., 2008; Lee et al., 2008; Gelvin, 2010). Similarly, VirE2 complexed with most investigated importin α isoforms, including AtKapa/IMPa-1, localizes within the cytoplasm of plant cells (Bhattacharjee et al., 2008; Lee et al., 2008). However, when VirE2 interacts with the Arabidopsis importin α isoform IMPa-4, the complex localizes to the nucleus of transfected plant cells (Bhattacharjee et al., 2008; Lee et al., 2008). Nuclear localization of VirE2/IMPa-4 complexes is particularly interesting in light of genetic data showing that mutation of Arabidopsis impa4, but not several other tested importin α genes, results in reduced transformation susceptibility of the roots of these mutant plants. VirD2 continues to localize to the nuclei of roots in impa4 mutants, suggesting that VirE2/IMPa-4 interactions may have “special” importance for nuclear import of T-strands.
An intriguing observation suggests that VirE2 may play yet another role in T-strand sub-cellular trafficking. Dumas et al. (2001) demonstrated that VirE2 could form gated channels that would allow single-strand DNA to traverse artificial “black membranes” in vitro. In plant cells, VirE2 localized within the cytoplasm, and may also accumulate in cell membranes (Grange et al., 2008). This latter study also showed that VirE2 could act as a molecular machine to impose a force against single-strand DNA. The authors argued that if single-strand DNA were traversing the plant cytoplasmic membrane after secretion from Agrobacterium, VirE2 within the plant cell could interact with the extra-membrane exposed portions of DNA and “pull” these molecules into the plant cell.
The apparent contradictions in the literature regarding the sub-cellular localization of VirE2 may be explained by observations of Djamei et al. (2007). This study investigated the sub-cellular localization of the protein VirE2 Interacting Protein 1 (VIP1). VIP1 plays an important role in Agrobacterium-mediated transformation. Mutation of vip1 reduces transformation susceptibility, whereas over-expression increases susceptibility (Tzfira et al., 2001, 2002; Li et al., 2005). Because VIP1 interacts with both VirE2 and with IMPa-1, VIP1 has been described as a molecular “intermediary” that mediates interactions between proteins that form the hypothetical T-complex with T-strands and the nuclear import apparatus (Tzfira et al., 2001; Li et al., 2005; Lacroix et al., 2006b). Djamei et al. (2007) showed that VIP1 can localize in both the cytoplasm and the nucleus. Nuclear localization is effected by phosphorylation of VIP1 on serine-79. Mutation of this amino acid to alanine favors cytoplasmic localization, whereas substitution with an aspartate residue drives VIP1 to the nucleus. VIP1 phosphorylation, which can be triggered by pathogen attack, also mediates transformation susceptibility. The phosphorylation status of VIP1 may therefore result in nuclear targeting of a VIP1–VirE2 complex and, thus, T-strand trafficking to the nucleus. This model remains to be tested. What is clear, however, is that sub-cellular localization of VirE2 depends on the cellular environment. Indeed, the sub-cellular localization of a VirE2–YFP fusion protein in transgenic Arabidopsis differs with the cell type: VirE2 localizes in perinuclear aggregates in root and leaf mesophyll cells, but in the nucleus of leaf trichomes (Gelvin, 2010).
“Non-essential” virulence effector proteins
Three additional A. tumefaciens Virulence effector proteins, VirE3, VirD5, and VirF, are secreted into plant host cells. Initial experiments, using mutant Agrobacterium strains with transposon insertions in the genes encoding these proteins, indicated that they were either “quantitative” or “host-range” factors that influenced the extent of transformation of different plant species (Stachel and Nester, 1986; Lin and Kado, 1993; Regensburg-Tuink and Hooykaas, 1993; Kalogeraki et al., 2000). Thus, because of their seeming lack of importance for virulence, the elucidation of the roles these proteins play in transformation occurred only recently.
As mentioned above, the plant protein VIP1 is important for efficient transformation. However, VIP1 is not an abundant protein, and its scarcity may render some plant species only weakly susceptible. VIP1 is a novel form of bZIP transcription factor that regulates expression of many plant defense genes (Pitzschke et al., 2009a,b). Because of its nuclear localization in plants, its ability to “auto-activate” reporter genes when bound to a Gal4 DNA binding domain in yeast, and its interaction in yeast and in vitro with the plant TFIIB general transcription factor pBrp, Garcia-Rodriguez et al. (2006) suggested that VirE3 may also be a plant transcription factor. VirE3 interacts with VirE2, and can complement a vip1 mutant for both nuclear import of VirE2 and for transformation susceptibility (Lacroix et al., 2005). These authors suggested that VirE3 can substitute for VIP1 in those plants where VIP1 may be limiting. Thus, Agrobacterium appears to have a “back-up” system of Virulence effector proteins that mimics the activity of plant proteins important for transformation.
VirF was initially described as a “host-range” transformation factor. Many plants are efficiently transformed by virF mutant Agrobacterium strains. However, virF mutant bacterial strains, or nopaline-type strains which lack virF, show weak virulence on species such as Nicotiana glauca and tomato (Regensburg-Tuink and Hooykaas, 1993). Transgenic plants expressing virF are efficiently transformed by these Agrobacterium strains, demonstrating that VirF functions in the plant host. In yeast, VirF interacts with Skp1 protein, a part of the SCF complex which “tags” proteins with ubiquitin and targets them for destruction by the 26S proteosome. Indeed, VirF is a F-box protein. Thus, VirF may be involved in targeted proteolysis of proteins, such as certain Virulence effector proteins, that must be stripped from T-strands prior to or during T-DNA integration. Tzfira et al. (2004) showed that VirF can mediate targeted proteolysis of both VirE2 and VIP1 in yeast and in planta. Inhibition of proteasomal function by the drug MG132 resulted in decreased transformation, suggesting that protein degradation is required for efficient transformation.
As with the situation with VirE3 and VIP1, VirF has a functional ortholog in plants. The plant-encoded F-box protein VBF (VIP1-binding F-box protein) can supply VirF activity to Agrobacterium strains either lacking or mutant in virF (Zaltsman et al., 2010). VBF is induced by Agrobacterium infection, and VBF protein can destabilize VirE2 and VIP1 in a manner consistent with that of VirF. Anti-sense VBF transgenic plants show lower susceptibility to Agrobacterium-mediated transformation, whereas expression of a VBF–T4SS signal fusion protein in Agrobacterium increases virulence of a virF mutant Agrobacterium strain. Thus again, Agrobacterium uses a back-up system, VirF, to substitute for a plant functional ortholog.
As a mediator of VirE2 and VIP1 degradation, VirF is an important virulence factor for some plant species. However, VirF is itself unstable in the plant. Magori and Citovsky (2011) recently showed that VirD5, another Agrobacterium Virulence effector protein, interacts with and stabilizes VirF in plants. Thus, Agrobacterium secretes effector proteins that both carry out functions missing in some plant species, and functions to protect the activity of other Virulence effector proteins.
The Agrobacterium rhizogenes effector protein galls
All A. tumefaciens, and many A. rhizogenes, strains encode three virE operon genes. As discussed above, VirE2 plays a major role in virulence, and VirE3 is also important for transformation of some plant species in which VIP1 activity is low. It thus came as a surprise that some highly virulent A. rhizogenes strains lack virE1 and virE2 from their Ri-plasmids (and from their chromosomes as well; Moriguchi et al., 2001). Rather, these strains contain the gene GALLS. GALLS can complement a virE2 mutant Agrobacterium strain to full virulence (Hodges et al., 2004), although other than a NLS domain, it does not in any way resemble VirE2 (Hodges et al., 2006). GALLS is an exported Virulence effector protein (Hodges et al., 2006), and can be made in two forms: GALLS-FL (full-length) and GALLS-CT (C-terminal). GALLS-CT, the most abundant of the two GALLS proteins, derives from the C-terminal portion of GALLS-FL, and is generated by translation initiation from within the GALLS coding region rather than by proteolytic cleavage of the GALLS-FL protein (Hodges et al., 2009). On most tested plant species, both GALLS-FL and GALLS-CT are required for full virulence. GALLS-CT lacks a NLS and, unlike GALLS-FL which localizes to the nucleus, GALLS-CT remains cytoplasmic. The two GALLS isoforms interact in plant cells with themselves and with each other. Interestingly, GALLS-FL also interacts with IMPa-4 and with VirD2 (Hodges et al., 2009). These data suggest that GALLS may aid in stabilization and/or nuclear import of VirD2/T-strand complexes.
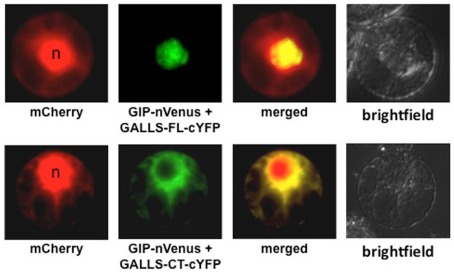
As an important Virulence effector protein for some A. rhizogenes strains, we conducted a search for plant proteins that interact with GALLS. Using yeast two-hybrid, plant BiFC, and in vitro pull-down analyses, we recently characterized interaction of GALLS with several members of the Arabidopsis LSH family of proteins. Interaction of GALLS-FL with GIP (GALLS interacting protein, one of the LSH family members) occurs predominantly in the nucleus, whereas interaction of GALLS-CT with GIP occurs in the cytoplasm (Figure 1; Y. Wang, L.-Y. Lee, L. Hodges, W. Ream, and S. B. Gelvin, unpublished). The LSH protein family may be transcription factors: several members of this family, as Gal4 DNA binding domain fusion proteins, auto-activate reporter genes in yeast. Because GALLS proteins interact with LSH protein family members, we tested Arabidopsis T-DNA insertion mutants in several LSH genes. No single gene disruption affected transformation susceptibility, suggesting that functional redundancy among these family members may obscure their role in transformation. However, over-expression of some, but not all, LSH family members in transgenic plants resulted in substantially increased transformation susceptibility of the derived transgenic lines. Most interesting, over-expression of these family members resulted in low-level transformation susceptibility to Agrobacterium strains lacking both virE2 and GALLS. No transformation was observed using these Agrobacterium strains and wild-type Arabidopsis plants. These data suggest that LSH family members may act in transformation at some step downstream of where VirE2 or GALLS proteins normally function.
Figure 1.
Interaction of GIP (GALLS Interacting Protein) with GALLS-FL and GALLS-CT. Tobacco BY-2 protoplasts were transfected with cDNAs encoding mCherry, nVenus-tagged GIP (one of the LSH protein family members), and either GALLS-FL or GALLS-CT containing a cYFP tag. The cells were imaged by epifluorescence microscopy 18 h later. mCherry marks both the nucleus and the cytoplasm. GIP-nVenus interacts with GALLS-FL-cYFP (upper row) or with GALLS-CT-cYFP (lower row) to restore fluorescence using BiFC. n Indicates the nucleus. Data courtesy of Dr. Yaling Wang.
Role of Agrobacterium and Plant Proteins in Targeting T-strands to Chromatin
Ultimately, T-strands (or double-strand DNA synthesized from T-strands in the nucleus) must integrate into plant nuclear DNA to establish stable transformation. Although the mechanism of T-DNA integration remains unknown, several important players involved in the process have been identified.
Numerous early reports suggested that T-DNA preferentially integrates into gene-rich, transcribed, A/T-rich, and/or promoter regions of genes (Koncz et al., 1989; Brunaud et al., 2002; Szabados et al., 2002; Alonso et al., 2003; Chen et al., 2003; Sallaud et al., 2004; Schneeberger et al., 2005). These analyses were conducted on large populations of Arabidopsis or rice transgenic plants mutagenized by T-DNA insertions. However, plants in these mutant populations that contain T-DNA insertions were selected by expression of antibiotic or herbicide resistance genes. If T-DNA had integrated into a region of chromatin that silenced the selection marker, many of these plants would have been discarded. Kim et al. (2007) analyzed T-DNA/plant DNA junction sequences from plant cells that had not been selected for the expression of antibiotic resistance transgenes. Their analysis indicated that T-DNA does not prefer any particular chromatin environment for integration. T-DNA was found, proportionally to the sequences of the Arabidopsis genome, in gene-rich and gene-poor regions, highly repetitive DNA, centromeres, and telomeres. Integration sites were not preferentially transcribed or methylated. The randomness of T-DNA integration suggests that general chromatin factors (such as histones, etc.) rather than other proteins (such as specific transcription factors) may be chromatin targets for T-strand/protein complexes. Indeed, the importance of particular histone proteins and chromatin modifying proteins in T-DNA integration has been well-established (Mysore et al., 2000; Yi et al., 2002, 2006; Zhu et al., 2003; Anand et al., 2007b; Crane and Gelvin, 2007; Tenea et al., 2009).
The importance of histones in T-DNA integration was further inferred from a study of VirE2 interacting protein 2 (VIP2). Because vip2 mutant Arabidopsis or VIGS-silenced N. benthamiana plants display normal transient Agrobacterium-mediated transformation, but are deficient in stable transformation, VIP2 was postulated to be important for T-DNA integration but not T-DNA transfer (Anand et al., 2007a). VIP2 is a NOT-domain protein that is hypothesized to be a transcriptional activator. Mutation of AtVIP2 results in decreased levels of histone mRNA, explaining its importance in T-DNA integration.
Several reports have confirmed that proteins important for Agrobacterium-mediated transformation target specific chromatin proteins. Loyter et al. (2005) showed that VIP1 could interact with each of the core histone proteins (H2A, H2B, H3, and H4) in vitro, and with H2A in planta. These results were confirmed by Li et al. (2005), and further extended by Lacroix et al. (2008), who showed that VIP1 could bind to purified mononucleosomes in vitro. Binding to nucleosomes depended upon the presence of both VIP1 and VirE2, and VirF could also form part of this complex.
How Do T-strand/Vir Protein/Plant Protein Complexes Traffic through the Cell?
The mechanism by which T-strands, complexed with bacterial and plant proteins, move through the cell is unknown. One possibility is that cytoskeletal structures, and the molecular motors that traffic macromolecules, protein complexes, and organelles along them, may be involved. Salman et al. (2005) showed that ssDNA–VirE2 complexes could migrate along microtubules in cell-free Xenopus oocyte extracts, that migration depended upon an intact VirE2 NLS, and that dynein motor proteins are important for movement. Zhu et al. (2003) presented a compendium of Arabidopsis mutants that are resistant to Agrobacterium transformation (rat mutants). Among these was a kinesin mutant.
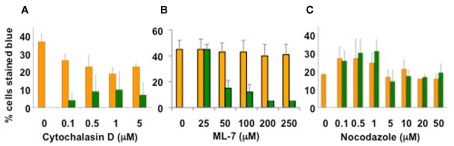
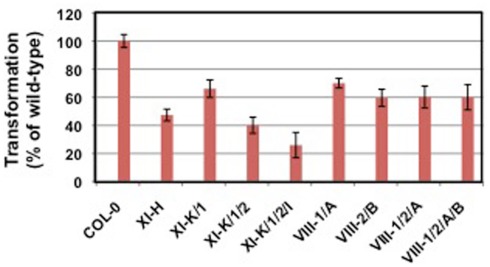
Unpublished data suggest that the actin cytoskeleton may be involved in Agrobacterium-mediated transformation. Arabidopsis lines mutant for actin-2, which is expressed in roots, are resistant to root transformation; complementation of act2 mutants with a wild-type ACT2 cDNA restored transformation susceptibility. Tobacco BY-2 cells treated with the actin microfilament inhibitor cytochalasin D or the myosin light chain kinase inhibitor ML-7 showed reduced transformation susceptibility, whereas cells treated with the microtubule inhibitor nocodazole retained full virulence (Figure 2; P. Rao and S. B. Gelvin, unpublished). We have recently screened numerous Arabidopsis myosin single- and multiple-mutant plants for transformation susceptibility. Several mutant plants showed reduced transformation competence using both transient and stable transformation assays (Figure 3; Y. Yu and S. B. Gelvin, unpublished). Preliminary data indicate that the sub-cellular localization of some fluorescently tagged Virulence effector proteins is altered when these proteins are expressed in leaf protoplasts from these mutant plants (L.-Y. Lee and S. B. Gelvin, unpublished). However, the true in vivo importance of the actin cytoskeleton in Virulence protein trafficking awaits characterization of transgenic myosin mutant Arabidopsis plants expressing tagged Virulence effector proteins.
Figure 2.
Treatment of tobacco cells with actin cytoskeleton inhibitors decreases Agrobacterium-mediated transient transformation. Tobacco BY-2 cells were treated for 1 h with the indicated chemicals (A) Cytochalasin D, (B) ML-7, (C) Nocodazole, or DMSO solvent, prior to infection by A. tumefaciens At849 at a ratio of 1000 bacteria/plant cell. After 48 h, the cells were washed and stained for GUS activity with X-gluc. Orange bars, solvent treatment; green bars, chemical treatment. Data courtesy of Dr. Praveen Rao.
Figure 3.
Transformation susceptibility of myosin mutant Arabidopsis plants. Root segments of wild-type (Col-0) and mutant plants were infected with the tumorigenic strain A. tumefaciens A208 at a concentration of 106 cfu/ml. Tumors were scored 30 days later. Data courtesy of Yanjun Yu.
Models and Future Directions: Novel Approaches to Understand T-strand/Protein Trafficking
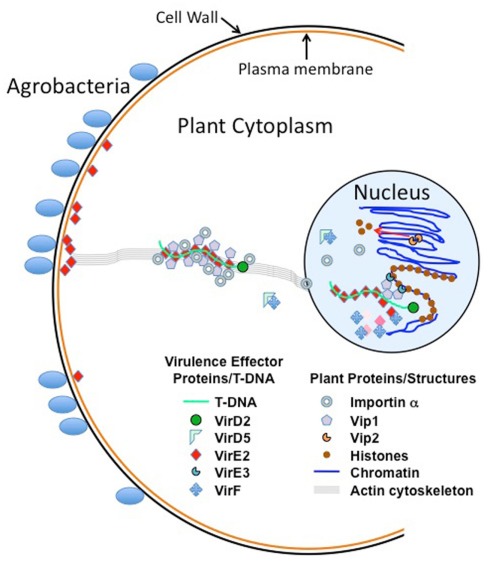
Figure 4 presents a model of T-DNA movement through the plant cell, and the proteins and structures with which it interacts. Although there are abundant data characterizing interactions of Agrobacterium Virulence effector proteins among themselves and with plant proteins, most of these studies were conducted either in vitro or by over-expressing test proteins in plant cells. Thus, the “true” levels of these proteins in the plant are likely obscured, and over-expression may force interactions to occur that may not normally occur, or saturate sub-cellular trafficking routes, resulting in mis-localization of proteins and protein complexes. In addition, the synthesis of bacterial proteins in plants, or their introduction into plant cells by artificial methods (e.g., electroporation, permeabilization of plant membranes, etc.) results in proteins that have not entered the plant cell through the “normal” channels used during Agrobacterium infection. For example, VirE2 molecules synthesized in planta, or introduced by microinjection, bypass the plant plasma membrane. However, VirE2 (and the other Virulence effector proteins) are normally delivered through the Agrobacterium type IV secretion system and through the plant plasma membrane. One model of T-DNA transfer posits that VirE2 may sit in the plant plasma membrane, form a channel through which T-strands pass, and “pick up” T-strands as they enter the plant cell (Duckely and Hohn, 2003). Thus, past analyses would have missed this important step. Most Vir–Vir and Vir–plant protein interaction experiments have been conducted in the absence of T-strands. It is possible that proteins complexed with single-strand DNA will interact with other proteins, or with other cellular components, differently than they would in isolation from DNA. Finally, although extensive in vitro biochemical and genetic data strongly suggest that T-complexes (i.e., T-strands with VirD2 covalently linked to the 5′ end, and coated with VirE2) exist, no such molecules have yet been identified or characterized following a normal infection. Thus, this author refers to these as “hypothetical T-complexes.”
Figure 4.
Model of T-DNA movement through the plant cell, and the proteins and structures with which it interacts. This model assumes that the T-strand/VirD2/VirE2 T-complex exists in a plant cell as hypothesized. Sub-cellular movement of T-complexes and/or Virulence effector proteins may utilize the actin cytoskeleton, as shown.
Virtually all protein localization and protein-protein interaction experiments have been conducted using transient expression assays, and in tissues other than root, the target for this soil phytopathogen. Thus, it would be useful to use root tissue to express proteins for interaction studies. Although this can be done transiently, stably transformed plants provide opportunities to visualize protein sub-cellular localization in numerous different tissues and cell types. In order to mitigate over-expression artifacts, plant proteins should be expressed from native promoters. Experiments in which Agrobacterium Virulence effector proteins are expressed in plants should be conducted with “low-level” promoters, rather than the strong cauliflower mosaic virus (CaMV) 35S promoter usually employed. To increase sensitivity, highly fluorescent autofluorescent protein derivatives could be used, such as Venus instead of YFP or GFP.
A major concern about understanding Agrobacterium T-DNA and Virulence protein transfer is that current methodologies bypass the normal way in which these molecules enter the plant cell. It would be useful to express “tagged” Virulence effector proteins in Agrobacterium, and follow the tag as these proteins journey to and through the plant cell. However, Agrobacterium effector proteins cannot be tagged at their C-termini, as this blocks the type IV secretion signal (Vergunst et al., 2000, 2003, 2005; Hodges et al., 2006). In addition, tagging proteins at their N-termini frequently disrupts transport and/or function, either in the bacterium or in the plant (Zhou and Christie, 1999; Bhattacharjee et al., 2008; van Kregten et al., 2009). We have recently generated VirD2, VirE2, and GALLS constructions with full or partial YFP-tags internally placed within the Virulence proteins. In many instances, these proteins maintain full or substantial activity, as measured by virulence assays in which the internally tagged proteins substitute for wild-type proteins (Table 1). We shall be using full-length YFP-tagged Virulence effector proteins to follow their passage from Agrobacterium to the plant cell. In addition, we shall be “pairing” partial YFP-tagged Virulence effector proteins, made in Agrobacterium, with interacting plant proteins tagged with the cognate YFP-tags in BiFC experiments to determine the site of interaction of these proteins, made/introduced at normal levels, in the plant cell.
Table 1.
Effect of internally tagging Virulence effector proteins upon virulence.
| Virulence effector protein | Autofluorescent protein taga | Virulence relative to wild-type protein (%)b |
|---|---|---|
| VirD2 | None | 100 |
| Venus | 0–2 | |
| mCherry | 10–20 | |
| nVenus | 42–80 | |
| VirE2 | None | 100 |
| Venus | 0 | |
| mCherry | 0 | |
| nVenus | 0 | |
| cCFP | 44–100 | |
| GALLS-FL | None | 100 |
| YFP | 45–76 | |
| cCFP | 62–100 |
aFor any particular Virulence effector protein, the different autofluorescent protein, or protein fragment, was inserted into the same position in the effector protein.
bRange of numbers represents several experiments performed with bacterial inoculum at 107 and 108 cfu/ml.
Finally, identification of “hypothetical” T-complexes formed in plant cells by T-strand and Virulence effector proteins secreted from Agrobacterium could confirm extant models of T-strand sub-cellular trafficking, and allow biochemical characterization of bacterial and host proteins comprising these mobile complexes.
Conflict of Interest Statement
The author declares that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The author thanks Dr. Lan-Ying Lee for critical reading of the manuscript. Work in the author’s laboratory was supported by the US National Science Foundation, the Department of Energy, the Biotechnology Research and Development Corporation, the Corporation for Plant Biotechnology Research, and Dow AgroSciences.
References
- Abu-Arish A., Frenkiel-Krispin D., Fricke T., Tzfira T., Citovsky V., Wolf S. G., Elbaum M. (2004). Three-dimensional reconstruction of Agrobacterium VirE2 protein with single-stranded DNA. J. Biol. Chem. 279, 25359–25363 10.1074/jbc.M401804200 [DOI] [PubMed] [Google Scholar]
- Abuodeh R. O., Orbach M. J., Mandel M. A., Das A., Galgiani J. N. (2000). Genetic transformation of Coccidioides immitis facilitated by Agrobacterium tumefaciens. J. Infect. Dis. 181, 2106–2110 10.1086/315525 [DOI] [PubMed] [Google Scholar]
- Alonso J. M., Stepanova A. N., Leisse T. J., Kim C. J., Chen H., Shinn P., Stevenson D. K., Zimmerman J., Barajas P., Cheuk R., Gadrinab C., Heller C., Jeske A., Koesema E., Meyers C. C., Parker H., Prednis L., Ansari Y., Choy N., Deen H., Geralt M., Hazari N., Hom E., Karnes M., Mulholland C., Ndubaku R., Schmidt I., Guzman P., Aguilar-Henonin L., Schmid M., Weigel D., Carter D. E., Marchand T., Risseeuw E., Brogden D., Zeko A., Crosby W. L., Berry C. C., Ecker J. R. (2003). Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301, 653–657 10.1126/science.1086391 [DOI] [PubMed] [Google Scholar]
- Anand A., Krichevsky A., Schomack S., Lahaye T., Tzfira T., Tang Y., Citovsky V., Mysore K. S. (2007a). Arabidopsis VirE2 interacting protein2 is required for Agrobacterium T-DNA integration in plants. Plant Cell 19, 1695–1708 10.1105/tpc.106.042903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anand A., Vaghchhipawala Z., Ryu C.-M., Kang L., Wang K., del-Pozo O., Martin G. B., Mysore K. S. (2007b). Identification and characterization of plant genes involved in Agrobacterium-mediated plant transformation by virus-induced gene silencing. Mol. Plant Microbe Interact. 20, 41–52 10.1094/MPMI-20-0041 [DOI] [PubMed] [Google Scholar]
- Ankenbauer R. G., Nester E. W. (1990). Sugar-mediated induction of Agrobacterium tumefaciens virulence genes: structural specificity and activities of monosaccharides. J. Bacteriol. 172, 6442–6446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bakó L., Umeda M., Tiburcio A. F., Schell J., Koncz C. (2003). The VirD2 pilot protein of Agrobacterium-transferred DNA interacts with the TATA box-binding protein and a nuclear protein kinase in plants. Proc. Natl. Acad. Sci. U.S.A. 100, 10108–10113 10.1073/pnas.1733208100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ballas N., Citovsky V. (1997). Nuclear localization signal binding protein from Arabidopsis mediates nuclear import of Agrobacterium VirD2 protein. Proc. Natl. Acad. Sci. U.S.A. 94, 10723–10728 10.1073/pnas.94.20.10723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bevan M. W., Flavell R. B., Chilton M.-D. (1983). A chimeric antibiotic resistance gene as a selectable marker for plant cell transformation. Nature 304, 184–187 10.1038/304184a0 [DOI] [PubMed] [Google Scholar]
- Bhattacharjee S., Lee L.-Y., Oltmanns H., Cao H., Veena, Cuperus J., Gelvin S. B. (2008). AtImpa-4, an Arabidopsis importin α isoform, is preferentially involved in Agrobacterium-mediated plant transformation. Plant Cell 20, 2661–2680 10.1105/tpc.108.060467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bravo-Angel A. M., Hohn B., Tinland B. (1998). The omega sequence of VirD2 is important but not essential for efficient transfer of T-DNA by Agrobacterium tumefaciens. Mol. Plant Microbe Interact. 11, 57–63 10.1094/MPMI.1998.11.1.57 [DOI] [PubMed] [Google Scholar]
- Brunaud V., Balzergue S., Dubreucq B., Aubourg S., Samson F., Chauvin S., Bechtold N., Cruaud C., DeRose R., Pelletier G., Lepiniec L., Caboche M., Lecharny A. (2002). T-DNA integraton into the Arabidopsis genome depends on sequences of pre-insertion sites. EMBO Rep. 3, 1152–1157 10.1093/embo-reports/kvf237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bulgakov V. P., Kisselev K. V., Yakovlev K. V., Zhuravlev Y. N., Gontcharov A. A., Odintsova N. A. (2006). Agrobacterium-mediated transformation of sea urchin embryos. Biotechnol. J. 1, 454–461 [DOI] [PubMed] [Google Scholar]
- Bundock P., den Dulk-Ras A., Beijersbergen A., Hooykaas P. J. J. (1995). Trans-kingdom T-DNA transfer from Agrobacterium tumefaciens to Saccharomyces cerevisiae. EMBO J. 14, 3206–3214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cascales E., Christie P. J. (2004). Definition of a bacterial type IV secretion pathway for a DNA substrate. Science 304, 1170–1173 10.1126/science.1095211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen S., Jin W., Wang M., Zhang F., Zhou J., Jia Q., Wu Y., Liu F., Wu P. (2003). Distribution and characterization of over 1000 T-DNA tags in rice genome. Plant J. 36, 105–113 10.1046/j.1365-313X.2003.01902.x [DOI] [PubMed] [Google Scholar]
- Christie P. J., Atmakuri K., Krishnamoorthy V., Jakubowski S., Cascales E. (2005). Biogenesis, architecture, and function of bacterial type IV secretion systems. Annu. Rev. Microbiol. 59, 451–485 10.1146/annurev.micro.58.030603.123630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christie P. J., Ward J. E., Winans S. C., Nester E. W. (1988). The Agrobacterium tumefaciens virE2 gene product is a single-stranded-DNA-binding protein that associates with T-DNA. J. Bacteriol. 170, 2659–2667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Citovsky V., De Vos G., Zambryski P. (1988). Single-stranded DNA binding protein encoded by the virE locus of Agrobacterium tumefaciens. Science 240, 501–504 10.1126/science.240.4851.501 [DOI] [PubMed] [Google Scholar]
- Citovsky V., Guralnick B., Simon M. N., Wall J. S. (1997). The molecular structure of Agrobacterium VirE2-single stranded DNA complexes involved in nuclear import. J. Mol. Biol. 271, 718–727 10.1006/jmbi.1997.1230 [DOI] [PubMed] [Google Scholar]
- Citovsky V., Kapelnikov A., Oliel S., Zakai N., Rojas M. R., Gilbertson R. L., Tzfira T., Loyter A. (2004). Protein interactions involved in nuclear import of the Agrobacterim VirE2 protein in vivo and in vitro. J. Biol. Chem. 279, 29528–29533 10.1074/jbc.M403159200 [DOI] [PubMed] [Google Scholar]
- Citovsky V., Warnick D., Zambryski P. (1994). Nuclear import of Agrobacterium VirD2 and VirE2 proteins in maize and tobacco. Proc. Natl. Acad. Sci. U.S.A. 91, 3210–3214 10.1073/pnas.91.8.3210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Citovsky V., Wong M. L., Zambryski P. (1989). Cooperative interaction of Agrobacterium VirE2 protein with single-stranded DNA: implications for the T-DNA transfer process. Proc. Natl. Acad. Sci. U.S.A. 86, 1193–1197 10.1073/pnas.86.4.1193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Citovsky V., Zupan J., Warnick D., Zambryski P. (1992). Nuclear localization of Agrobacterium VirE2 protein in plant cells. Science 256, 1802–1805 10.1126/science.1615325 [DOI] [PubMed] [Google Scholar]
- Crane Y. M., Gelvin S. B. (2007). RNAi-mediated gene silencing reveals involvement of Arabidopsis chromatin-related genes in Agrobacterium-mediated root transformation. Proc. Natl. Acad. Sci. U.S.A. 104, 15156–15161 10.1073/pnas.0706986104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das A. (1988). Agrobacterium tumefaciens virE operon encodes a single-stranded DNA-binding protein. Proc. Natl. Acad. Sci. U.S.A. 85, 2909–2913 10.1073/pnas.85.9.2909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Groot M. J. A., Bundock P., Hooykaas P. J. J., Beijersbergen A. G. M. (1998). Agrobacterium tumefaciens-mediated transformation of filamentous fungi. Nat. Biotechnol. 16, 839–842 [DOI] [PubMed] [Google Scholar]
- Deng W., Chen L., Wood D. W., Metcalfe T., Liang X., Gordon M. P., Comai L., Nester E. W. (1998). Agrobacterium VirD2 protein interacts with plant host cyclophilins. Proc. Natl. Acad. Sci. U.S.A. 95, 7040–7045 10.1073/pnas.95.12.7040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Djamei A., Pitzschke A., Nakagami H., Rajh I., Hirt H. (2007). Trojan horse strategy in Agrobacterium transformation: abusing MAPK defense signaling. Science 318, 453–456 10.1126/science.1148110 [DOI] [PubMed] [Google Scholar]
- Duckely M., Hohn B. (2003). The VirE2 protein of Agrobacterium tumefaciens: the Yin and Yang of T-DNA transfer. FEMS Microbiol. Lett. 223, 1–6 10.1016/S0378-1097(03)00246-5 [DOI] [PubMed] [Google Scholar]
- Dumas F., Duckely M., Pelczar P., Van Gelder P., Hohn B. (2001). An Agrobacterium VirE2 channel for transferred-DNA transport into plant cells. Proc. Natl. Acad. Sci. U.S.A. 98, 485–490 10.1073/pnas.011477898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durrenberger F., Crameri A., Hohn B., Koukolikova-Nicola Z. (1989). Covalently bound VirD2 protein of Agrobacterium tumefaciens protects the T-DNA from exonucleolytic degradation. Proc. Natl. Acad. Sci. U.S.A. 86, 9154–9158 10.1073/pnas.86.23.9154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farrand S. K., van Berkum P. B., Oger J. (2003). Agrobacterium is a definable genus of the family Rhizobiaceae. Int. J. Syst. Evol. Microbiol. 53, 1681–1687 10.1099/ijs.0.02445-0 [DOI] [PubMed] [Google Scholar]
- Fraley R. T., Rogers S. G., Horsch R. B., Sanders P. R., Flick J. S., Adams S. P., Bittner M. L., Brand L. A., Fink C. L., Fry J. S., Galluppi G. R., Goldberg S. B., Hoffmann N. L., Woo S. C. (1983). Expression of bacterial genes in plant cells. Proc. Natl. Acad. Sci. U.S.A. 80, 4803–4807 10.1073/pnas.80.15.4803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fronzes R., Christie P. J., Waksman g. (2009). The structural biology of type IV secretion systems. Nat. Rev. Microbiol. 7, 703–714 10.1038/nrmicro2218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Rodriguez F. M., Schrammeijer B., Hooykaas P. J. J. (2006). The Agrobacterium VirE3 effector protein: a potential plant transcriptional activator. Nucleic Acids Res. 34, 6496–6504 10.1093/nar/gkl877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gelvin S. B. (1998). Agrobacterium VirE2 proteins can form a complex with T strands in the plant cytoplasm. J. Bacteriol. 180, 4300–4302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gelvin S. B. (2010). Finding a way to the nucleus. Curr. Opin. Microbiol. 13, 53–58 10.1016/j.mib.2009.11.003 [DOI] [PubMed] [Google Scholar]
- Gietl C., Koukolikova-Nicola Z., Hohn B. (1987). Mobilization of T-DNA from Agrobacterium to plant cells involves a protein that binds single-stranded DNA. Proc. Natl. Acad. Sci. U.S.A. 84, 9006–9010 10.1073/pnas.84.24.9006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grange W., Duckely M., Husale S., Jacob S., Engel A., Hegner M. (2008). VirE2: a unique ssDNA-compacting molecular machine. PLoS Biol. 6, e44. 10.1371/journal.pbio.0060044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrera-Estrella A., Chen Z.-M., Van Montagu M., Wang K. (1988). VirD proteins of Agrobacterium tumefaciens are required for the formation of a covalent DNA-protein complex at the 5′ terminus of T-strand molecules. EMBO J. 7, 4055–4062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrera-Estrella A., Van Montagu M., Wang K. (1990). A bacterial peptide acting as a plant nuclear targeting signal: the amino-terminal portion of Agrobacterium VirD2 protein directs a β-galactosidase fusion protein into tobacco nuclei. Proc. Natl. Acad. Sci. U.S.A. 87, 9534–9537 10.1073/pnas.87.24.9534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrera-Estrella L., DeBlock M., Messens E., Hernalsteens J.-P., Van Montagu M., Schell J. (1983). Chimeric genes as dominant selectable markers in plant cells. EMBO J. 2, 987–996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodges L., Cuperus J., Ream W. (2004). Agrobacterium rhizogenes GALLS protein substitutes for Agobacterium tumefaciens single-stranded DNA-binding protein VirE2. J. Bacteriol. 186, 3065–3077 10.1128/JB.186.10.3065-3077.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodges L. D., Lee L.-Y., McNett H., Gelvin S. B., Ream W. (2009). The Agrobacterium rhizogenes GALLS gene encodes two secreted proteins required for genetic transformation of plants. J. Bacteriol. 191, 355–364 10.1128/JB.01018-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodges L. D., Vergunst A. C., Neal-McKinney J., den Dulk-Ras A., Moyer D. M., Hooykaas P. J. J., Ream W. (2006). Agrobacterium rhizogenes GALLS protein contains domains for ATP binding, nuclear localization, and type IV secretion. J. Bacteriol. 188, 8222–8230 10.1128/JB.00747-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howard E. A., Zupan J. R., Citovsky V., Zambryski P. C. (1992). The VirD2 protein of A. tumefaciens contains a C-terminal bipartite nuclear localization signal: implications for nuclear uptake of DNA in plant cells. Cell 68, 109–118 10.1016/0092-8674(92)90210-4 [DOI] [PubMed] [Google Scholar]
- Kalogeraki V. S., Zhu J., Stryker J. L., Winans S. C. (2000). The right end of the vir region of an octopine-type Ti plasmid contains four new members of the vir regulon that are not essential for pathogenesis. J. Bacteriol. 182, 1774–1778 10.1128/JB.182.6.1774-1778.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kersters K., De Ley L. (1984). “Genus III. Agrobacterium Conn 1942, 359AL,” in Bergey’s Manual of Systematic Bacteriology, Vol. 1, eds Krieg N. R., Holt J. G. (Baltimore: Williams & Wilkins; ), 244–254 [Google Scholar]
- Kim S.-I., Veena, Gelvin S. B. (2007). Genome-wide analysis of Agrobacterium T-DNA integration sites in the Arabidopsis genome generated under non-selective conditions. Plant J. 51, 779–791 10.1111/j.1365-313X.2007.03183.x [DOI] [PubMed] [Google Scholar]
- Koncz C., Martini N., Mayerhofer R., Koncz-Kalman Z., Korber H., Redei G. P., Schell J. (1989). High-frequency T-DNA-mediated gene tagging in plants. Proc. Natl. Acad. Sci. U.S.A. 86, 8467–8471 10.1073/pnas.86.21.8467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunik T., Tzfira T., Kapulnik Y., Gafni Y., Dingwall C., Citovsky V. (2001). Genetic transformation of HeLa cells by Agrobacterium. Proc. Natl. Acad. Sci. U.S.A. 98, 1871–1876 10.1073/pnas.041327598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lacroix B., Loyter A., Citovsky V. (2008). Association of the Agrobacterium T-DNA-protein complex with plant nucleosomes. Proc. Natl. Acad. Sci. U.S.A. 105, 15429–15434 10.1073/pnas.0805641105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lacroix B., Tzfira T., Vainstein A., Citovsky V. (2006a). A case of promiscuity: Agrobacterium’s endless hunt for new partners. Trends Genet. 22, 29–37 10.1016/j.tig.2005.10.004 [DOI] [PubMed] [Google Scholar]
- Lacroix B., Li J., Tzfira T., Citovsky V. (2006b). Will you let me use your nucleus? How Agrobacterium gets its T-DNA expressed in the host plant cell. Can. J. Physiol. Pharmacol. 84, 333–345 10.1139/y05-108 [DOI] [PubMed] [Google Scholar]
- Lacroix B., Vaidya M., Tzfira T., Citovsky V. (2005). The VirE3 protein of Agrobacterium mimics a host cell function required for plant genetic transformation. EMBO J. 24, 428–437 10.1038/sj.emboj.7600524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee L.-Y., Fang M.-J., Kuang L.-Y., Gelvin S. B. (2008). Vectors for multi-color bimolecular fluorescence complementation to investigate protein-protein interactions in living plant cells. Plant Methods 4, 24. 10.1186/1746-4811-4-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J., Krichevsky A., Vaidya M., Tzfira T., Citovsky V. (2005). Uncoupling of the functions of the Arabidopsis VIP1 protein in transient and stable plant genetic transformation by Agrobacterium. Proc. Natl. Acad. Sci. U.S.A. 102, 5733–5738 10.1073/pnas.0504799102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin T.-S., Kado C. I. (1993). The virD4 gene is required for virulence while virD3 and orf5 are not required for virulence of Agrobacterium tumefaciens. Mol. Microbiol. 9, 803. 10.1111/j.1365-2958.1993.tb01739.x [DOI] [PubMed] [Google Scholar]
- Loyter A., Rosenbluh J., Zakai N., Li J., Kozlovsky S. V., Tzfira T., Citovsky V. (2005). The plant VirE2 interacting protein 1. A molecular link between the Agrobacterium T-complex and the host cell chromatin? Plant Physiol. 138, 1318–1321 10.1104/pp.105.062547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magori S., Citovsky V. (2011). Agrobacterium counteracts host-induced degradation of its effector F-box protein. Sci. Signal. 4, ra69. 10.1126/scisignal.2002124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michielse C. B., Hooykaas P. J. J., van den Hondel C. A. M. J. J., Ram A. F. J. (2008). Agrobacterium-mediated transformation of the filamentous fungus Aspergillus awamori. Nat Protoc 3, 1671–1678 10.1038/nprot.2008.154 [DOI] [PubMed] [Google Scholar]
- Moriguchi K., Maeda Y., Satou M., Hardayani N. S. N., Kataoka M., Tanaka N., Yoshida K. (2001). The complete nucleotide sequence of a plant root-inducing (Ri) plasmid indicates its chimeric structure and evolutionary relationship between tumor-inducing (Ti) and symbiotic (Sym) plasmids in Rhizobiaceae. J. Mol. Biol. 307, 771–784 10.1006/jmbi.2001.4488 [DOI] [PubMed] [Google Scholar]
- Mysore K. S., Bassuner B., Deng X.-B., Darbinian N. S., Motchoulski A., Ream W., Gelvin S. B. (1998). Role of the Agrobacterium tumefaciens VirD2 protein in T-DNA transfer and integration. Mol. Plant Microbe Interact. 11, 668–683 10.1094/MPMI.1998.11.7.668 [DOI] [PubMed] [Google Scholar]
- Mysore K. S., Nam J., Gelvin S. B. (2000). An Arabidopsis histone H2A mutant is deficient in Agrobacterium T-DNA integration. Proc. Natl. Acad. Sci. U.S.A. 97, 948–953 10.1073/pnas.97.2.948 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelczar P., Kalck V., Gomez D., Hohn B. (2004). Agrobacterium proteins VirD2 and VirE2 mediate precise integration of synthetic T-DNA complexes in mammalian cells. EMBO Rep. 5, 632–637 10.1038/sj.embor.7400165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piers K. L., Heath J. D., Liang X., Stephens K. M., Nester E. W. (1996). Agrobacterium tumefaciens-mediated transformation of yeast. Proc. Natl. Acad. Sci. U.S.A. 93, 1613–1618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pitzschke A., Schikora A., Hirt H. (2009a). MAPK cascade signaling networks in plant defense. Curr. Opin. Plant Biol. 12, 421–426 10.1016/j.pbi.2009.06.008 [DOI] [PubMed] [Google Scholar]
- Pitzschke A., Djamei A., Teige M., Hirt H. (2009b). VIP1 response elements mediate mitogen-activated protein kinase 3-induced stress gene expression. Proc. Natl. Acad. Sci. U.S.A. 106, 18414–18419 10.1073/pnas.0905599106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Regensburg-Tuink A. J. G., Hooykaas P. J. J. (1993). Transgenic N. glauca plants expressing bacterial virulence gene virF are converted into hosts for nopaline strains of A. tumefaciens. Nature 363, 69–71 10.1038/363069a0 [DOI] [PubMed] [Google Scholar]
- Rossi L., Hohn B., Tinland B. (1993). The VirD2 protein of Agrobacterium tumefaciens carries nuclear localization signals important for transfer of T-DNA to plant. Mol. Gen. Genet. 239, 345–353 10.1007/BF00276932 [DOI] [PubMed] [Google Scholar]
- Rossi L., Hohn B., Tinland B. (1996). Integration of complete transferred DNA units is dependent on the activity of virulence E2 protein of Agrobacterium tumefaciens. Proc. Natl. Acad. Sci. U.S.A. 93, 126–130 10.1073/pnas.93.1.126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sallaud C., Gay C., Larmande P., Bes M., Piffanelli P., Piegu B., Droc G., Regad F., Bourgeois E., Meynard D., Perin C., Sabau X., Ghesquiere A., Glaszmann J. C., Delseny M., Guiderdoni E. (2004). High throughput T-DNA insertion mutagenesis in rice: a first step towards in silico reverse genetics. Plant J. 39, 450–464 10.1111/j.1365-313X.2004.02145.x [DOI] [PubMed] [Google Scholar]
- Salman H., Abu-Arish A., Oliel S., Loyter A., Klafter J., Granek R., Elbaum M. (2005). Nuclear localization signal peptides induce molecular delivery along microtubules. Biophys. J. 89, 2134–2145 10.1529/biophysj.105.060160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneeberger R. G., Zhang K., Tatarinova T., Troukhan M., Kwok S. F., Drais J., Klinger K., Orejudos F., Macy K., Bhakta A., Burns J., Subramanian G., Donson J., Flavell R., Feldmann K. A. (2005). Agrobacterium T-DNA integration in Arabidopsis is correlated with DNA sequence compositions that occur frequently in gene promoter regions. Funct. Integr. Genomics 5, 240–253 10.1007/s10142-005-0138-1 [DOI] [PubMed] [Google Scholar]
- Sen P., Pazour G. J., Anderson D., Das A. (1989). Cooperative binding of Agrobacterium tumefaciens VirE2 protein to single-stranded DNA. J. Bacteriol. 171, 2573–2580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shurvinton C. E., Hodges L., Ream W. (1992). A nuclear localization signal and the C-terminal omega sequence in the Agrobacterium tumefaciens VirD2 endonuclease are important for tumor formation. Proc. Natl. Acad. Sci. U.S.A. 89, 11837–11841 10.1073/pnas.89.24.11837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simone M., McCullen C. A., Stahl L. E., Binns A. N. (2001). The carboxy-terminus of VirE2 from Agrobacterium tumefaciens is required for its transport to host cells by the virB-encoded type IV transport system. Mol. Microbiol. 41, 1283–1293 10.1046/j.1365-2958.2001.02582.x [DOI] [PubMed] [Google Scholar]
- Stachel S. E., Messens E., Van Montagu M., Zambryski P. (1985). Identification of the signal molecules produced by wounded plant cells that activate T-DNA transfer in Agrobacterium tumefaciens. Nature 318, 624–629 10.1038/318024a0 [DOI] [Google Scholar]
- Stachel S. E., Nester E. W. (1986). The genetic and transcriptional organization of the vir region of the A6 Ti plasmid of Agrobacterium tumefaciens. EMBO J. 5, 1445–1454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stachel S. E., Nester E. W., Zambryski P. C. (1986). A plant cell factor induces Agrobacterium tumefaciens vir gene expression. Proc. Natl. Acad. Sci. U.S.A. 83, 379–383 10.1073/pnas.83.2.379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sundberg C., Meek L., Carroll K., Das A., Ream W. (1996). VirE1 protein mediates export of the single-stranded DNA-binding protein VirE2 from Agrobacterium tumefaciens into plant cells. J. Bacteriol. 178, 1207–1212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szabados L., Kovacs I., Oberschall A., Abraham E., Kerekes I., Zsigmond L., Nagy R., Alvarado M., Krasovskaja I., Gal M., Berente A., Redei G. P., Ben Haim A., Koncz C. (2002). Distribution of 1000 sequenced T-DNA tags in the Arabidopsis genome. Plant J. 32, 233–242 10.1046/j.1365-313X.2002.01417.x [DOI] [PubMed] [Google Scholar]
- Tao Y., Rao P. K., Bhattacharjee S., Gelvin S. B. (2004). Expression of plant protein phosphatase 2C interferes with nuclear import of the Agrobacterium T-complex protein VirD2. Proc. Natl. Acad. Sci. U.S.A. 101, 5164–5169 10.1073/pnas.0300084101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tenea G. N., Spantzel J., Lee L.-Y., Zhu Y., Lin K., Johnson S. J., Gelvin S. B. (2009). Overexpression of several Arabidopsis histone genes increases Agrobacterium-mediated transformation and transgene expression in plants. Plant Cell 21, 3350–3367 10.1105/tpc.109.070607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terry L. J., Shows E. B., Wente S. R. (2007). Crossing the nuclear envelope: hierarchical regulation of nucleocytoplasmic transport. Science 318, 1412–1416 10.1126/science.1145147 [DOI] [PubMed] [Google Scholar]
- Tinland B., Hohn B., Puchta H. (1994). Agrobacterium tumefaciens transfers single-stranded transferred DNA (T-DNA) into the plant cell nucleus. Proc. Natl. Acad. Sci. U.S.A. 91, 8000–8004 10.1073/pnas.91.17.8000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tinland B., Koukolikova-Nicola Z., Hall M. N., Hohn B. (1992). The T-DNA-linked VirD2 protein contains two distinct functional nuclear localization signals. Proc. Natl. Acad. Sci. U.S.A. 89, 7442–7446 10.1073/pnas.89.16.7442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tinland B., Schoumacher F., Gloeckler V., Bravo-Angel A. M., Hohn B. (1995). The Agrobacterium tumefaciens virulence D2 protein is responsible for precise integration of T-DNA into the plant genome. EMBO J. 14, 3585–3595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tzfira T., Citovsky V. (2001). Comparison between nuclear localization of nopaline- and octopine-specific Agrobacterium VirE2 proteins in plant, yeast and mammalian cells. Mol. Plant Pathol. 2, 171–176 10.1046/j.1364-3703.2001.00065.x [DOI] [PubMed] [Google Scholar]
- Tzfira T., Vaidya M., Citovsky V. (2001). VIP1, an Arabidopsis protein that interacts with Agrobacterium VirE2, is involved in VirE2 nuclear import and Agrobacterium infectivity. EMBO J. 20, 3596–3607 10.1093/emboj/20.13.3596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tzfira T., Vaidya M., Citovsky V. (2002). Increasing plant susceptibility to Agrobacterium infection by over-expression of the Arabidopsis nuclear protein VIP1. Proc. Natl. Acad. Sci. U.S.A. 99, 10435–10440 10.1073/pnas.162304099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tzfira T., Vaidya M., Citovsky V. (2004). Involvement of targeted proteolysis in plant genetic transformation by Agrobacterium. Nature 431, 87–92 10.1038/nature02857 [DOI] [PubMed] [Google Scholar]
- van Kregten M., Lindhout B. I., Hooykaas P. J. J., van der Zaal B. J. (2009). Agrobacterium-mediated T-DNA transfer and integration by minimal VirD2 consisting of the relaxase domain and a type IV secretion system translocation signal. Mol. Plant Microbe Interact. 22, 1356–1365 [DOI] [PubMed] [Google Scholar]
- Vergunst A. C., Schrammeijer B., den Dulk-Ras A., de Vlaam C. M. T., Regensburg-Tuink T. J. G., Hooykaas P. J. J. (2000). VirB/D4-dependent protein translocation from Agrobacterium into plant cells. Science 290, 979–982 10.1126/science.290.5493.979 [DOI] [PubMed] [Google Scholar]
- Vergunst A. C., van Lier M. C. M., den Duld-Ras A., Hooykaas P. J. J. (2003). Recognition of the Agrobacterium tumefaciens VirE2 translocation signal by the VirB/D4 transport system does not require VirE1. Plant Physiol. 133, 978–988 10.1104/pp.103.029223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vergunst A. C., van Lier M. C. M., den Dulk-Ras A., Stuve T. A. G., Ouwehand A., Hooykaas P. J. J. (2005). Positive charge is an important feature of the C-terminal transport signal of the VirB/D4-translocated proteins of Agrobacterium. Proc. Natl. Acad. Sci. U.S.A. 102, 832–837 10.1073/pnas.0406241102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogel A. M., Das A. (1992). Mutational analysis of Agrobacterium tumefaciens virD2: tyrosine 29 is essential for endonuclease activity. J. Bacteriol. 174, 303–308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward E. R., Barnes W. M. (1988). VirD2 protein of Agrobacterium tumefaciens very tightly linked to the 5′ end of T-strand DNA. Science 242, 927–930 [Google Scholar]
- Yi H., Mysore K. S., Gelvin S. (2002). Expression of the Arabidopsis histone H2A-1 gene correlates with susceptibility to Agrobacterium transformation. Plant J. 32, 285–298 10.1046/j.1365-313X.2002.01425.x [DOI] [PubMed] [Google Scholar]
- Yi H., Sardesai N., Fujinuma T., Chan C.-W., Veena, Gelvin S. B. (2006). Constitutive expression exposes functional redundancy between the Arabidopsis histone H2A gene HTA1 and other H2A gene family members. Plant Cell 18, 1575–1589 10.1105/tpc.105.039719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young C., Nester E. W. (1988). Association of the VirD2 protein with the 5′ end of T strands in Agrobacterium tumefaciens. J. Bacteriol. 170, 3367–3374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yusibov V. M., Steck T. R., Gupta V., Gelvin S. B. (1994). Association of single-stranded transferred DNA from Agrobacterium tumefaciens with tobacco cells. Proc. Natl. Acad. Sci. U.S.A. 91, 2994–2998 10.1073/pnas.91.8.2994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaltsman A., Krichevsky A., Loyter A., Citovsky V. (2010). Agrobacterium induces expression of a host F-box protein required for tumorigenicity. Cell Host Microbe 7, 197–209 10.1016/j.chom.2010.02.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Z., Sagulenko E., Ding Z., Christie P. J. (2001). Activities of virE1 and the VirE1 secretion chaperone in export of the multifunctional VirE2 effector via an Agrobacterium type IV secretion pathway. J. Bacteriol. 183, 3855–3865 10.1128/JB.183.13.3855-3865.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou X.-R., Christie P. J. (1999). Mutagenesis of the Agrobacterium VirE2 single-stranded DNA-binding protein identifies regions required for self-association and interaction with VirE1 and a permissive site for hybrid protein construction. J. Bacteriol. 181, 4342–4352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Y., Nam J., Humara J. M., Mysore K. S., Lee L.-Y., Cao H., Valentine L., Li J., Kaiser A. D., Kopecky A. L., Hwang H.-H., Bhattacharjee S., Rao P. K., Tzfira T., Rajagopal J., Yi H., Veena, Yadav B. S., Crane Y. M., Lin K., Larcher Y., Gelvin M. J. K., Knue M., Ramos-Oliva C., Zhao X., Davis S. J., Kim S.-I., Ranjith-Kumar C. T., Choi Y.-J., Hallan V. K., Chattopadhyay S., Sui X., Ziemienowicz A., Matthysse A. G., Citovsky V., Hohn B., Gelvin S. B. (2003). Identification of Arabidopsis rat mutants. Plant Physiol. 132, 494–505 10.1104/pp.102.018101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziemienowicz A., Merkle T., Schoumacher F., Hohn B., Rossi L. (2001). Import of Agrobacterium T-DNA into plant nuclei: two distinct functions of VirD2 and VirE2 proteins. Plant Cell 13, 369–383 10.1105/tpc.13.2.369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zupan J. R., Citovsky V., Zambryski P. (1996). Agrobacterium VirE2 protein mediates nuclear uptake of single-stranded DNA in plant cells. Proc. Natl. Acad. Sci. U.S.A. 93, 2392–2397 10.1073/pnas.93.6.2392 [DOI] [PMC free article] [PubMed] [Google Scholar]