Abstract
The engulfment of a photoautotrophic cyanobacterium by a primitive mitochondria-bearing eukaryote traces back to more than 1.2 billion years ago. This single endosymbiotic event not only provided the early petroalgae with the metabolic capacity to perform oxygenic photosynthesis, but also introduced a plethora of other metabolic routes ranging from fatty acids and amino acids biosynthesis, nitrogen and sulfur assimilation to secondary compounds synthesis. This implicated the integration and coordination of the newly acquired metabolic entity with the host metabolism. The interface between the host cytosol and the plastidic stroma became of crucial importance in sorting precursors and products between the plastid and other cellular compartments. The plastid envelope membranes fulfill different tasks: they perform important metabolic functions, as they are involved in the synthesis of carotenoids, chlorophylls, and galactolipids. In addition, since most genes of cyanobacterial origin have been transferred to the nucleus, plastidial proteins encoded by nuclear genes are post-translationally transported across the envelopes through the TIC–TOC import machinery. Most importantly, chloroplasts supply the photoautotrophic cell with photosynthates in form of reduced carbon. The innermost bilayer of the plastidic envelope represents the permeability barrier for the metabolites involved in the carbon cycle and is literally stuffed with transporter proteins facilitating their transfer. The intracellular metabolite transporters consist of polytopic proteins containing membrane spans usually in the number of four or more α-helices. Phylogenetic analyses revealed that connecting the plastid with the host metabolism was mainly a process driven by the host cell. In Arabidopsis, 58% of the metabolite transporters are of host origin, whereas only 12% are attributable to the cyanobacterial endosymbiont. This review focuses on the metabolite transporters of the inner envelope membrane of plastids, in particular the electrochemical potential-driven class of transporters. Recent advances in elucidating the plastidial complement of metabolite transporters are provided, with an update on phylogenetic relationship of selected proteins.
Keywords: endosymbiosis, envelope membrane, translocator
Introduction
Oxygenic photosynthesis is the process by which plants are able to convert the solar energy into stable chemical bonds. By trapping the ephemeral energy contained in the photon, photosynthetic organisms are able to form and stabilize chemical bonds.
In the oxygenic photosynthesis reduced carbon is formed, using electrons extracted from water. The energy stored in the C–H bonds can be released during respiration, where the electrons are transferred back to oxygen, leading to the formation of water. In this way photosynthesis and respiration define a water–oxygen cycle, thereby re-distributing the solar energy to the whole biosphere (Hohmann-Marriott and Blankenship, 2011).
Long before oxygenic photosynthesis arose, the solar energy was trapped by non-oxygenic organisms, using hydrogen, ferrous iron, and hydrogen sulfide as the main electron source. Some of today’s bacteria still use this source of electrons instead of water. These are the purple non-sulfur bacteria (e.g., Rhodopseudomonas), which perform cyclic electron flow around a PSII-like protein complex, and the green sulfur bacteria (e.g., Chlorobium), which possess a PSI-like reaction center able to extract electrons from hydrogen sulfide and transferring them to NAD+ via linear electron transport (Allen and Martin, 2007; Hohmann-Marriott and Blankenship, 2011). Different hypotheses exist for explaining the events that gave rise to the oxygen-evolving protocyanobacterium, possessing both types of reaction centers and using water as electron donor. One states that lateral gene transfer occurred between a purple non-sulfur bacterium with a quinone-based (Type II) reaction center and a green sulfur bacterium with a FeS-based (Type I) reaction center, while in an alternate hypothesis the structural similarity of the two photosystems indicates that they might have evolved from a single ancestor via gene duplication and divergence (Xiong et al., 2000; Mulkidjanian et al., 2006; Allen and Martin, 2007).
Whatever model best explains the integration of the two photosystems, the key in evolving oxygenic photosynthesis is the development of the water-splitting oxygen-evolving complex (OEC), containing four Mn atoms and a Ca atom capable of sequentially removing four electrons from two water molecules. This process ultimately releases molecular oxygen as by-product. The accumulation of O2 converted the atmosphere into an oxidizing one, permitting the development of more complex life forms that utilize oxygen as electron acceptor during the aerobic respiration.
This all posed the bases for the next level of complexity: endosymbiotic associations. Eukaryotic origin is defined as being concomitant with the acquisition of mitochondria, likely an α-proteobacterium, either by a nucleus-bearing amitochondriate cell, or by a prokaryotic host cell, later evolving eukaryote-specific features (Embley and Martin, 2006).
Later on the engulfment of a photoautotrophic cyanobacterium by a primitive mitochondriate eukaryote established the lineage of the Archaeplastida, also known as the Plantae (Gould et al., 2008; Keeling, 2010). All plastids are believed to have originated after a single endosymbiotic event, more than 1.2 billion years ago (Björn, 2009). One exception is provided by Paulinella chromatophora, which represents a case of ongoing primary endosymbiosis between an ameba and a cyanobacterium-like endosymbiont (Figure 1; Kies, 1974). Soon after the primary endosymbiosis, three major autotrophic lineages arose: the glaucophytes, the green algae, and the red algae (Figure 1; Adl et al., 2005).
Figure 1.
Plastid evolution in photosynthetic eukaryotes. The uptake of a cyanobacterium resulted in a photosynthetic plantae ancestor which subsequently diverged in the three lineages containing primary plastids: the chlorophytes (including the land plants), the rhodophytes, and the glaucophytes. The subsequent secondary endosymbioses of green and red algae engulfed by different hosts resulted in the euglenophyta and chlorarachniophyta (greens) and in the possibly monophyletic chromalveolates (reds). Paulinella chromatophora represents an independent primary endosymbiosis in progress.
Glaucophytes are a small group of unicellular, chlorophyll a containing freshwater algae consisting of four genera with at least seven species. They were the first lineage branching off and contain a plastid (muroplast) with a thin peptidoglycan wall between the two envelope membranes (Martin and Herrmann, 1998; Steiner and Löffelhardt, 2002; Sato et al., 2009). The peptidoglycan layer is believed to be a relic of the cyanobacterial cell wall, thus supporting the endosymbiotic theory of plastid origin. Other similarities to the cyanobacterial progenitors are the maintenance of phycobilisomes as light-harvesting antenna and a carboxysomal-like carbon-concentrating mechanism (Burey et al., 2007). Red algae (rhodophytes) also possess plastids with phycobilisomes but lack any remnant of the peptidoglycan wall and carboxysomes. In addition, they possess membrane-intrinsic light-harvesting complexes (LHCs) similar to those of the chloroplasts of the Viridiplantae associated with the PSI, thus representing a transitional state between cyanobacteria and chloroplasts (Wolfe et al., 1994). Rhodophyta have furthermore contributed to at least six other algal lineages by secondary endosymbiosis, as indicated by the presence of up to four membranes surrounding their plastids. These include members from the chromalveolates supergroup: the cryptophytes (still retaining a remnant nucleus, the nucleomorph), the haptophytes, the plastid-containing heterokontophyta (e.g., diatoms), and the alveolates. (Figure 1; Reyes-Prieto et al., 2007; Archibald, 2009).
The green lineage (chlorophyta) is represented by the green algae and the land plants. This is the most derived lineage. In contrast to glaucophytes and red algae (collectively termed biliphytes) their photosynthetic plastids lack the phycobilisomes; they possess instead chlorophyll b-containing LHCs associated with both photosystems as well as other accessory pigments (Keeling, 2010). A secondary endosymbiotic event involving one member of the green algae has been proposed to have occurred once, leading to the Cabozoa which include the euglenophyta and the chlorarachniophyta (Figure 1).
Even if the presence of the plastid is often associated with the capability to perform photosynthesis, a diverse number of lineages reduced the plastids metabolic capability to the point where photosynthesis was lost. These include for example the red algal-derived apicoplast of the malaria parasite Plasmodium (which still retains important anabolic pathways providing the host cell with fatty acids, isoprene units, and haem; Ralph et al., 2004) and secondary endosymbionts containing plastids of green origin occurring in the excavata, such as the colorless euglenozoa.
Primary plastids are delimited by two membranes, whose composition reflects their bacterial origin: besides the already mentioned peptidoglycan layer, the outer envelope membrane contains galactolipids and β-barrel proteins (Jarvis et al., 2000; Schleiff et al., 2003). However, envelope membranes were subjected to extensive modifications: whereas the inner envelope derives from the cyanobacterial plasma membrane, the outer envelope is of chimeric origin, with the outer leaflet containing lipids derived from the host endomembrane system (Cavalier-Smith, 2000).
The events occurring at the interface between the endosymbiont and the host were of crucial importance for the establishment of the plastid. Insertion of host protein translocators into the envelope was necessary to take full advantage of the newly acquired metabolic entity. Besides the photosynthetic reactions that lead to carbon fixation, endosymbiosis introduced into the petroalgae a set of metabolic pathways, including nitrogen and sulfur assimilation, the biosynthesis of amino acids, fatty acids, vitamins, hormones, and a plethora of secondary compounds (Weber and Flügge, 2002; Weber et al., 2006; Weber and Fischer, 2007). Metabolite translocators were therefore of utmost importance for integrating and coordinating the flux of precursors and products between the plastid and other cellular compartments. The metabolite traffic between plastids and other cellular compartments is predominantly facilitated by antiporters embedded in the inner membrane. The choice of antiporter systems ensures that the exchange of a substrate on one side of the membrane is always accompanied by the presence of another counter-exchange substrate on the trans-side (Flügge, 1995). This antiport transport mechanism is common to other subcellular compartments such as mitochondria, peroxisomes, and the endomembrane system, and is important to link different metabolite pools subjecting them to a strict control. In the context of endosymbiosis, the regulated exchange of metabolites would have been of crucial importance in maintaining the metabolic homeostasis between the cyanobacterium and the host (Weber and Linka, 2011).
The outer envelope contains porin-like channels and has long been considered as not acting as a permeability barrier, proposing that solute movement across the outer envelope membrane is limited solely by size (Flügge, 2000). The discovery of low-affinity, high-specific channels in the outer envelope points to a more active role of the outer envelope, even if their regulatory role in vivo is still a matter of debate (Soll et al., 2000; Bölter and Soll, 2001; Duy et al., 2007). The reader is referred to recent reviews describing the outer envelope proteins (e.g., Duy et al., 2007; Inoue, 2011).
This review focuses on the metabolite transporters of the inner envelope membrane of plastids, in particular the electrochemical potential-driven class of transporters. Recent advances in elucidating the plastidial complement of metabolite transporters are provided, with an update on phylogenetic relationship of selected proteins.
Plastidic Phosphate Translocators
The plastidic phosphate translocator (pPT) family comprises four members of the drug/metabolite transporters (DMT) superfamily (TC 2.A.7; Saier et al., 2006, 2009). These antiporters catalyze the exchange of phosphorylated C3-, C5-, and C6-compounds for inorganic phosphate (Pi). The homo-exchange guarantees a balance in the phosphate content of the stroma and the cytosol and ensures a constant provision of phosphate to sustain ATP synthesis (Weber et al., 2005).
Phylogenetic analyses revealed that pPTs are monophyletic and originated from an existing endomembrane translocator (Weber et al., 2006). The major plant plastid translocators co-localize in branches containing homologs from red and green algae, supporting their monophyly and an origin of the translocators in their common ancestor (Weber et al., 2006). pPTs are more closely related to the nucleotide sugar transporters (NSTs) than to other families of the DMT superfamily (Knappe et al., 2003a).
This is in agreement with the need of the petroalgae to export the adenosine-diphosphoglucose (ADP-Glc) produced by the cyanobiont into the host cytosol. The first metabolic connection between the two compartments would thus have been through an ADP-Glc translocator. Indeed, it was recently shown that members of the NSTs possess an innate ability to transport ADP-Glc, filling the link between the carbon metabolism of endosymbiont and host cell (Colleoni et al., 2010). The establishment of a carbon flux was thus a host-driven process, by recruiting a translocator of its endomembrane system and redirecting it to the endosymbiont, later on evolving into the pPT family of transporters (Weber et al., 2006; Tyra et al., 2007; Colleoni et al., 2010). This hypothesis, however, awaits support from analyzing the plastid permeome of a member of the third lineage of the Archaeplastida, the glaucophyta. The host contribution to the establishment of the endosymbiosis was further supported by a work in which transporter proteins broadly distributed in the plantae kingdom were subjected to phylogenetic analysis: out of 83 proteins scrutinized, 58% were shown to be of host origin (Tyra et al., 2007).
The triose phosphate/phosphate translocator
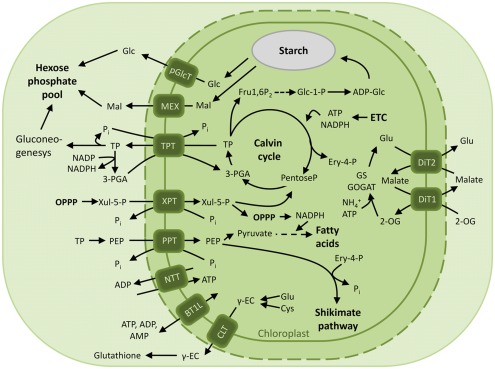
The plastid provides the cell with reduced carbon compounds by the assimilation of carbon dioxide during photosynthesis. In plants the photoassimilates are either exported in their phosphorylated form by members of the pPT family to fuel sucrose biosynthesis in the cytosol, or they are retained in the plastid to drive starch synthesis (Figure 2). The daily path for carbon export occurs in form of triose phosphates (TP) via the triose phosphate/phosphate translocator (TPT). TPT accepts TPs (dihydroxyacetone phosphate, DHAP, and glyceraldehyde 3-phosphate) as well as 3-phosphoglycerate (3-PGA) as substrates (Table 1; Fliege et al., 1978; Flügge and Heldt, 1984). In addition to its role in allocating the carbon between the stroma and the cytosol, the TPT has the ability to export reducing equivalents to the cytosol. The TP/3-PGA shuttle exports TPs, which are then converted by the glyceraldehyde phosphate dehydrogenase to 3-PGA, reducing one molecule of NADP+ to NADPH, required for biosynthetic processes. The 3-PGA is then transported back to the chloroplast stroma by the TPT and can re-enter the Calvin–Benson cycle (Figure 2; Flügge and Heldt, 1984).
Figure 2.
Overview of the characterized chloroplast envelope metabolite transporters. Transport processes of carbon, nitrogen compounds and energy across the envelope membrane of photosynthetic plastids in green plants are depicted. Abbreviations: 2-OG, 2-oxoglutarate; ADP-Glc, ADP-glucose; BT1L, BT1-like transporter; CLT, CRT-like transporter; Cys, cysteine; Ery 4-P, Erythrose 4-phosphate; ETC, electron transport chain; Fru1,6P2, fructose-1,6-bisphosphate; Γ-EC, Γ-glutamylcysteine; Glc, glucose; Glc 1-P, glucose 1-phosphate; Glc 6-P, glucose 6-phosphate; Glu, glutamine; GPT, glucose 6-phosphate/phosphate translocator; Mal, maltose; MEX, maltose exporter; NTT, nucleoside triphosphate transporter; OPPP, oxidative pentose phosphate pathway; 3-PGA, 3-phosphoglyceric acid; PEP, phosphoenolpyruvate; pGlcT, plastidic glucose transporter; Pi, inorganic phosphate; PPT, PEP/phosphate translocator; TP, triose phosphate; TPT, triose phosphate/phosphate translocator; XPT, xylulose 5-phosphate/phosphate translocator; Xul 5-P, xylulose 5-phosphate.
Table 1.
Phosphate translocators of the plastidic inner envelope membrane.
| TPT (triose phosphate/phosphate translocator) | PPT (phospho-enol-pyruvate/phosphate translocator) | GPT (glucose-6- phosphate/phosphate translocator) | XPT (xylulose-5- phosphate/phosphate translocator | ||
|---|---|---|---|---|---|
| AGI number | At5g17630 | At5g33320, At3g01550 | At5g54800, At1g61800 | At5g17630 | |
| TC number | TC 2.A.7.9 | ||||
| Substrates | Green plants | TP, 3-PGA | PEP | Glc-6-P, TP, 3-PGA | TP, Xul-5-P, Ru-5-P, Ery-4-P |
| Red algae | TP | PEP | n.d. | – | |
| Glaucophytes | – | – | – | – | |
| Apicomplexas cryptomonads | TP, PEP | – | – | – | |
| Kinetic constant | Substrate | ||||
| Km (app) | Phosphate | 1.0 | 0.8 | 1.1 | 1.0 |
| Ki (app) | TrioseP | 1.0 | 8.0 | 0.6 | 0.4 |
| Ki (app) | 3-PGA | 1.0 | 4.6 | 1.8 | 10.6 |
| Ki (app) | PEP | 3.3 | 0.3 | 2.9 | 2.1 |
| Ki (app) | Glc-6-P | >50 | >50 | 1.1 | 6.6 |
| Ki (app) | Ery-4-P | 3.3 | |||
| Ki (app) | Xul-5-P | 0.8 | |||
| Ki (app) | Ru-5-P | 3.5 | |||
Apparent Km (phosphate) and Ki values expressed in millimolar for various phosphorylated metabolites. Transport activities of the recombinant translocators from green plants were measured in a reconstituted system using proteoliposomes pre-loaded with inorganic phosphate (Fischer et al., 1997; Kammerer et al., 1998; Eicks et al., 2002).
In the red alga Galdieria sulphuraria the TPT shows narrower substrate specificity, being able to transport TPs but not 3-PGA (Figure 3B). Moreover, the GsTPT displays a three-fold higher affinity to Pi and the substrate DHAP has a two-fold higher affinity to the Pi binding site than its counterpart from the chlorophytes (Table 1; Linka et al., 2008a). While in organisms descending from the green lineage the products of the carbon fixation are stored in insoluble starch granules inside the chloroplasts, rhodophytes accumulate a starch-like polymer in the cytosol through the use of UDP-glucose, called floridean starch, resembling amylopectin in its structure (Ball and Morell, 2003; Patron and Keeling, 2005). The heteroside floridoside is synthesized from UDP-Gal and glyceraldehyde 3-phosphate (Gly-3-P) in the cytosol as well and represents the main soluble pool of fixed carbon in analogy with sucrose in chlorophytes (Figure 3B; Viola et al., 2001). This means that the partitioning of the photoassimilates in red algae occurs exclusively in the cytosol, with GsTPT acting only as TP-exporter. This also explains the higher affinity of GsTPT for its substrates, enabling the export of TPs even at low concentrations (Linka et al., 2008a).
Figure 3.
Characterized and putative metabolite transporters of the glaucophaye Cyanophora paradoxa (A) and the rhodophyte Galdieria sulphuraria (B). Proposed model of carbon export from C. paradoxa based on uptake experiments with isolated cyanelles. The identity of the transporter responsible for the TPT activity is not elucidated yet (A). Characterized metabolite transporters of G. sulphuraria and their substrate specificities. The GsTPT, unlike for the green plant chloroplasts, is unable to transport 3-PGA pointing to the presence of an alternate reduction shuttle in the red algae (B). Abbreviations: ADP-Glc, ADP-glucose; ETC, electron transport chain; Glc 1-P, glucose 1-phosphate; Glc 6-P, glucose 6-phosphate; GPT, glucose 6-phosphate/phosphate translocator; NTT, nucleoside triphosphate transporter; OPPP, oxidative pentose phosphate pathway; 3-PGA, 3-phosphoglyceric acid; PEP, phosphoenolpyruvate; Pi, inorganic phosphate; PPT, PEP/phosphate translocator; TP, triose phosphate; TPT, triose phosphate/phosphate translocator; UDP-Glc, UDP-glucose; UDP-Gal, UDP-galactose.
In addition to its ability to grow heterotrophically, G. sulphuraria can grow on more than 50 different carbon sources (Gross and Schnarrenberger, 1995; Barbier et al., 2005). Unlike land plants, where TPT is expressed in photosynthetic active tissues, GsTPT is also expressed in heterotrophically grown cells (Schulz et al., 1993; Linka et al., 2008a). Whereas plants provide heterotrophic plastids with glucose 6-phosphate (Glc6P) through the Glucose 6-phosphate/phosphate translocator (GPT), red algae do not appear to have pentose phosphate or hexose phosphate translocators (Oesterhelt et al., 2007).
Since the GsTPT is unable to transport 3-PGA, its role in the abovementioned NADPH shuttle can be excluded. While in plants the enzyme fructose-1,6-bisphosphatase is redox regulated and inactivated by oxidation, the G. sulphuraria homolog lacks redox modulation (Reichert et al., 2003). In this way the TPs can be converted into hexose phosphates in heterotrophic growth conditions and in the dark by the plastidic FBPase, ensuring the production of reduction equivalents (Reichert et al., 2003; Oesterhelt et al., 2007).
The expression pattern of GsTPT thus reflects the route for carbon import into heterotrophically grown cells with GsTPT as unique mediator of the carbon flux between plastid and cytosol (Linka et al., 2008a).
The lineage of the glaucophytes is best studied in the freshwater alga Cyanophora paradoxa (Löffelhardt et al., 1997). As for the red algae, glaucophytes synthesize and store the starch in the cytosol through an UDP-glucose pathway (Plancke et al., 2008). Transport assays with isolated cyanelles demonstrated the activity of a translocator transferring phosphate, DHAP, and 3-PGA, in analogy with chloroplasts of higher plants (Figure 3A; Schlichting and Bothe, 1993). It is expected that the ongoing C. paradoxa genome sequencing project will soon provide clues as to whether a TPT ortholog also exists in glaucophytes.
The plastids of organisms derived by secondary endosymbiosis can have up to four envelope membranes, like the apicoplast of the malaria parasite Plasmodium falciparum. The apicoplast is no longer able to perform photosynthetic carbon fixation, but retained the capacity to synthesize fatty acids, heme, iron sulfur clusters, and isoprene subunits (Ralph et al., 2004). The compartmentalization of these biosynthetic pathways poses as prerequisite the net import of carbon skeletons, ATP, and reducing equivalents. Indeed, P. falciparum has two TPTs, PfoTPT, and PfiTPT, previously shown to reside in the outermost and innermost membranes of the apicoplast (Figure 4A; Mullin et al., 2006).They both have a substrate preference for DHAP, 3-PGA, and phosphoenolpyruvate (PEP), suggesting that they operate in tandem (Table 1; Lim et al., 2010). The discovery of PEP as substrate of a member of the TPTs is unprecedented and provides a plausible strategy to solve the needs of the apicoplast anabolism. PEP can be converted to pyruvate yielding one molecule of ATP by substrate-level phosphorylation. Pyruvate can then be channeled to fatty acid biosynthesis further producing NADH (Lim et al., 2010). It still remains unclear how the C3 compounds cross the two internal membranes. In another apicomplexan, Toxoplasma gondii, a pfoTPT homolog (TgAPT1) has been proposed to reside in more than one membrane, providing a solution to the issue of transport (Karnataki et al., 2007).
Figure 4.
Substrates of the triose phosphate/phosphate translocators (TPTs) and nucleoside triphosphate transporters (NTTs) of the apicoplast of Plasmodium falciparum (A), of the diatom plastids from Thalassiosira pseudonana and Phaedactylum tricornutum (B), and of the crytomonad plastid from Guillardia theta (C). Overview of the metabolite transport processes for the characterized transporters of the chromalveolates. The presence of four envelope membranes reflects their secondary endosymbiotic origin. The TPT of the apicoplast of P. falciparum has a broader substrate specificity compared to the green counterpart, accepting also PEP (A). Diatom plastids are not able to synthesize nucleotides and therefore they depend on the import of nucleotides from the cytosol. They possess NTTs which catalyze a uniport mode of transport as in the chlamydial intracellular parasites from where these transporters were acquired (B). The cryptomonads contain a less reduced secondary endosymbiont, still harboring a vestigial nucleus in the periplasmic compartment, the former algal cytoplasm, where the starch is synthesized. The day and night path of carbon metabolism are regulated by differential expression of TPT genes whose products localize in different envelopes (C). Abbreviations: (d)NTP, (deoxy)nucleotide triphosphate; G3P, glycerol 3-phosphate; PEP, phosphoenolpyruvate; PfiTPT, P. falciparum innermost envelope TPT; PfoTPT, P. falciparum outermost envelope TPT; 3-PGA, 3-phosphoglycerate; Pi, inorganic phosphate; TP, triose phosphate; UDP-Glc, UDP-glucose.
The cryptomonad Guillardia theta harbors a complex plastid which retained a vestigial nucleus (nucleomorph) from the engulfed red alga. The nucleomorph resides in the periplastid compartment, the former algal cytoplasm. This intermembrane space is also the site of starch accumulation (Haferkamp et al., 2006). The genome of G. theta encodes for two putative TPTs, able to import Pi in counter-exchange to DHAP but not 3-PGA (Table 1). Interestingly, the genes encoding these phosphate translocators (TPT1 and TPT2) are regulated in opposite directions. TPT1 transcripts accumulate during the night phase, whereas TPT2 accumulates during the light phase (Haferkamp et al., 2006). It has been proposed that that this expression pattern reflects the metabolic involvement of the corresponding carrier proteins, if TPT2 resides in the innermost membrane and favors the export of TPs in the intermembrane space during the day. On the other hand, TPT1 could reside in the first or second outermost membrane and function as exporter of the starch degradation products during the night (Figure 4C; Haferkamp et al., 2006).
The glucose 6-phosphate/phosphate translocator
In contrast to photosynthetic plastids which are able to synthesize carbon skeletons and reducing equivalents, heterotrophic plastids of sink tissues rely on the import of photosynthates. The phosphorylated C3 sugar phosphates synthesized in the stroma of photosynthetically active chloroplasts are exported to the cytosol through the TPT and converted into sucrose, which is in turn loaded into the phloem and redirected to sink tissues such as tubers, fruits, and seeds. Here sucrose is cleaved by the action of invertase or sucrose synthase and converted into hexose phosphates. The primary hexose phosphate, glucose 6-phosphate, is ultimately imported into the plastid through the (GPT; Figure 5).
Figure 5.
Overview of the characterized metabolite transporters of the heterotrophic plastid. Non-photosynthetic plastids rely on the import of photosynthates to drive the metabolic reactions in the sink tissues. Glucose 6-phosphate is transported by GPT to provide heterotrophic plastids with substrates further used to produce reducing equivalents and storage compounds. In maize, the substrate for starch synthesis, ADP-glucose, is produced in the cytoplasm and shuttled to the seeds by the brittle-1 transporter. Abbreviations: ADP-Glc, ADP-glucose; BT1, Brittle-1 ADP-glucose carrier; Glc 1-P, glucose 1-phosphate; Glc 6-P, glucose 6-phosphate; GPT, glucose 6-phosphate/phosphate translocator; Mal, maltose; NTT, nucleoside triphosphate transporter; OPPP, oxidative pentose phosphate pathway; 3-PGA, 3-phosphoglyceric acid; PEP, phosphoenolpyruvate; Pi, inorganic phosphate; PPi, pyrophosphate; PPT, PEP/phosphate translocator; XPT, xylulose 5-phosphate/phosphate translocator; Xul 5-P, xylulose 5-phosphate.
Glucose 6-phosphate/phosphate translocator is preferentially expressed in non-green tissues and mediates the import of glucose 6-P (Glc6P) as well as TPs and 3-PGA (Table 1; Kammerer et al., 1998). Glc6P is further channeled into the starch biosynthetic pathway or into the oxidative pentose phosphate pathway (OPPP), producing reduction equivalents required for diverse anabolic pathways (Figure 5; Flügge et al., 2011). The importance of GPT in plant metabolism is demonstrated by the lethal phenotype resulting after disruption of the AtGPT1 homolog of Arabidopsis thaliana (Niewiadomski et al., 2005). Mutant plants are severely impaired in the gametogenesis, due to the lack of NADPH produced by the OPPP to sustain fatty acid biosynthesis. Shortage of Glc6P as substrate for the OPPP results in reduced formation of lipid bodies and non-physiological cell death (Niewiadomski et al., 2005). In a recent report the GPT2 ortholog of oil palm (Elaeis guineensis Jacq) could be associated with a function in fatty acid biosynthesis and oil accumulation in the mesocarp (Bourgis et al., 2011). Oil palm can accumulate up to 90% oil in its mesocarp. This is achieved through a high flux of glycolytic intermediates providing pyruvate for oil synthesis. Up-regulation of GPT2 and PPT supplies glycolytic substrates and intermediates to the plastid by transport from the cytosol, funneling carbon compounds toward pyruvate to sustain the high demand of substrates for fatty acid biosynthesis during mesocarp ripening (Bourgis et al., 2011).
Reconstituted G. sulphuraria membranes showed high activity of phosphate/phosphate and phosphate/DHAP counter-exchange, whereas 3-PGA, 2-PGA, PEP, and Glc 6-P transport rates did not vary from that of the negative controls (Weber et al., 2004). Indeed, uptake experiments with reconstituted liposomes containing a recombinant protein with significant homology to higher plant GPT demonstrated that Glc6P is not a relevant substrate and suggests that G. sulphuraria and likely red algae in general do not possess a plastidic hexose-P importer (Linka et al., 2008a). As mentioned above, GPT supplies heterotrophic plastids of green plants due to the absence of FBPase activity. Since in G. sulphuraria the FBPase is not redox regulated, import of TPs mediated by GsTPT could ensure the supply of hexose-P for NADPH production during the night or under heterotrophic growth conditions.
The xylulose 5-phosphate/phosphate translocator
The XPT of Arabidopsis was the last member of plant pPT to be identified and is able to transport inorganic phosphate, TPs, 3-P-glycerate, xylulose 5-P (Xul 5-P), and, to a lesser extent, erythrose 4-P (Ery 4-P) and ribulose 5-P (Ru 5-P; Table 1; Eicks et al., 2002). The XPT protein displays high homology to the GPT, and most likely derived from the latter by retrotranscription and genome insertion, as suggested by the lack of introns in the XPT gene (Knappe et al., 2003a). Phylogenetic analysis of the phosphate transporters of the red algae G. sulphuraria and Cyanidioschyzon merolae showed that members of this family are present which localize within the branch of the GPT/XPT group of transporters (Weber et al., 2006).
The proposed function of XPT is to import pentose phosphates into the stroma in the form of Xyl 5-P to be further metabolized in the Calvin cycle and the OPPP (Figures 2 and 5). A further function of the XPT would be the supply of carbon skeletons intermediates withdrawn from the Calvin cycle and the OPPP and channeled into other biosynthetic pathways such as the synthesis of nucleotides and the shikimate pathway requiring Rib 5-P and Ery 4-P, respectively (Eicks et al., 2002).
The phosphoenolpyruvate/phosphate translocator
The phosphoenolpyruvate/phosphate translocator (PPT) mediates the import of cytosolic PEP into the plastid stroma in exchange with Pi (Table 1; Fischer et al., 1997).
Since the chloroplasts and most plastids of non-green tissues lack the enzymes required to form PEP from TPs via the glycolytic pathway, PEP has to be imported from the cytosol (Stitt and Aprees, 1979; Borchert et al., 1993). The energy-rich compound PEP serves as substrate for the shikimate pathway, whose starting point is the condensation of one molecule of PEP and one of Ery 4-P (Figure 2). The shikimate pathway is exclusively localized within the plastid and is involved in the biosynthesis of aromatic amino acids and of a variety of secondary compounds such as lignin, alkaloids, and flavonoids (Herrmann, 1995; Herrmann and Weaver, 1999). PEP can also be metabolized to pyruvate and acetyl-CoA which serves as precursor for the biosynthesis of fatty acids, as already mentioned for the GPT (Figures 2 and 4; Rajasekharan and Nachiappan, 2010; Bourgis et al., 2011).
Arabidopsis contains two PPT genes (PPT1 and PPT2) which differ in their expression pattern (Knappe et al., 2003b). PPT1 is expressed in the leaf vasculature and in some root tissues. The abolishment of its activity in the cue1 mutant causes a reticulate leaf phenotype with abnormally developed mesophyll cells (MC) (Streatfield et al., 1999). The expression pattern of PPT1 and the observation that the biosynthesis of aromatic amino acids is not severely affected in the cue1 mutant led to the suggestion that it might be involved in the development of the MCs trough a not yet identified signaling molecule likely derived from the shikimate pathway. AtPPT1 is presumably involved in the synthesis of secondary metabolites that regulate leaf development, whereas AtPPT2 which is ubiquitously expressed in leaves could have a more housekeeping function in supplying the plastidic shikimate pathway with PEP (Knappe et al., 2003b; Voll et al., 2003).
Another way to provide plastids with PEP is from plastidic pyruvate by pyruvate, orthophosphate dikinase (PPDK). This reaction is an essential step in C4- and CAM-plants where PEP is exported to the cytosol of MCs and serves as substrate for the PEP-carboxylase during CO2 fixation (Bräutigam et al., 2008). In C4 and CAM-plants PPT is therefore an exporter of PEP (Weber and Von Caemmerer, 2010).
In the red alga G. sulphuraria, preliminary uptake experiments with isolated total membranes did not show pronounced PPT activity (Weber et al., 2004). However, phylogenetic analysis revealed the presence of a candidate ortholog in the red algal genome, suggesting the presence of a potential coding capability for PPT (Weber et al., 2006). Indeed, the recombinant protein was functionally characterized in reconstituted liposome membranes and displayed the signature of PPT activity (Linka et al., 2008a). As its green counterpart, the G. sulphuraria PPT is highly specific for Pi and PEP (Table 1).
PEP synthesis inside the rhodoplast is assumed to be unlikely since the isozymes of both routes for synthesizing PEP in the plastid (i.e., the glycolytic pathway through phosphoglyceromutase and enolase, and the conversion of pyruvate to PEP by the PPDK) do not possess obvious subcellular targeting sequences and are predicted to be cytosolic (Linka et al., 2008a). Since fatty acid biosynthesis and the shikimate pathway in red algae are predicted to be similar to those of the green lineage of Archaeplastida, it is assumed that conversion of TP to PEP is not possible in G. sulphuraria, thus depending on PEP import from the cytosol to drive PEP-depended reactions in the stroma (Figure 3B; Weber et al., 2004; Richards et al., 2006; Linka et al., 2008a).
The glutathione transporter
Recently, a new transporter member of the DMT superfamily has been characterized as glutathione transporter in Arabidopsis. In plants, glutathione is in the center of an antioxidant network that buffers the reactive oxygen species (ROS) originating as by-product of the aerobic energy metabolism. Glutathione is a tripeptide enzymatically synthesized by the glutamate–cysteine ligase (GSH1) and the glutathione synthase (GSH2). In Arabidopsis, GSH1 is plastid localized, while GSH2 is targeted to both plastid and cytosol (Meister, 1995; Wachter et al., 2005). Consequently, the pathway intermediate, Γ-glutamylcysteine (Γ-EC), must be exported from the plastid to allow for cytosolic GSH biosynthesis (Figure 2). In a mutant screening designed to select mutants resistant to l-buthionine-SR-sulfoximine (BSO), an inhibitor of GSH1, the gene At5g19380, encoding a predicted membrane transport protein, was identified as responsible for the revertant phenotype (Maughan et al., 2010). The gene was found to be related to the chloroquine resistance transporter (PfCRT) of the malaria parasite P. falciparum and was consequently designated CRT-like transporter1 (CLT1). Arabidopsis mutants lacking CLT1 are heavy metal-sensitive, GSH-deficient, and hypersensitive to Phytophthora infection. CLT1 is a member of a small gene family comprising three members in Arabidopsis and all localize to the plastid envelope. Expression of the transporter in Xenopus oocytes confirmed that it can mediate GSH uptake (Maughan et al., 2010). Homology search revealed related genes in higher plants, in the moss Physcomitrella patens, in green algae and in parasitic protozoans, suggesting an ancient ancestry for this transporter.
The ATP:ADP Antiporter Family
Members of the ATP:ADP Antiporter (AAA) Family (TC 2.A.12), also called nucleoside triphosphate transporters (NTT) catalyze a counter-exchange mode of transport and may be distantly related to members of the major facilitator superfamily (MFS; TC 2.A.1). They are not related to the mitochondrial ATP/ADP exchangers of the mitochondrial carrier family (MCF; TC 2.A.5), which pump ATP out of mitochondria in accordance with the polarity of the mitochondrial membrane potential. In addition, the plant mitochondrial adenylate translocator is highly sensitive to specific inhibitors, whereas the plastidic ATP/ADP translocator is only slightly sensitive to them (Vignais et al., 1976; Ardila et al., 1993; Schunemann et al., 1993; Winkler and Neuhaus, 1999).
In prokaryotes these transporters enable “energy parasitism,” as in Rickettsia and Chlamydia (Neuhaus et al., 1997). In addition, putative nucleotide transport proteins with weak sequence similarities to the NTT family of transporters have been identified in the nuclear genome of the obligate intracellular parasite Encephalitozoon cuniculi (Katinka et al., 2001). The bacterial nucleotide carriers have conspicuous sequence similarity to the NTT proteins from chloroplasts of higher plants (Neuhaus et al., 1997).
In plants, the main function proposed for the NTT family of transporters is to supply ATP-dependent reactions in non-photosynthetic plastids. Amyloplasts are heterotrophic plastids of sink tissues and contain various anabolic pathways, like the biosyntheses of starch and fatty acids or nitrogen assimilation. During starch biosynthesis in potato tubers, glucose 1-phosphate (Glc1P) is converted to ADP-Glc by the AGPase, a reaction which is ATP-dependent and relies on the supply of cytosolic ATP by NTT (Figure 5). This is corroborated by transgenic plants where repression of the potato NTT leads to decreased tuber yield and starch contents (Tjaden et al., 1998). In contrast to heterotrophic plastids, the role of NTTs in chloroplasts is not obvious. Rather than exporting ATP out of the photosynthetically active plastid during the day, NTT import ATP during the night or when photosynthetic ATP biosynthesis is not sufficient to fuel anabolic reactions requiring ATP (Reinhold et al., 2007).
Arabidopsis knock-out mutants demonstrated the involvement of NTT and ATP import in the assembly of magnesium chelatase, an enzyme involved in chlorophyll biosynthesis (Kobayashi et al., 2008). Plants repressing NTTs display necrotic lesions caused by photo-oxidation due to an accumulation of high amounts of the phototoxic chlorophyll biosynthesis intermediate protoporphyrin IX, which induces ROS production and photooxidative damage. Furthermore, the severity of the stress depended on the size of the plastidic starch pool, with plants grown in short day conditions being more affected (Reinhold et al., 2007). It has been proposed that the sensitivity to photooxidative stress in plants with limited plastidic ATP supply may link the appearance of the light harvesting complexes in the green lineage with the return of the storage polysaccharide to the plastid (Deschamps et al., 2008b). In plants, the main pathway of starch breakdown does not allow for the direct generation of ATP within the plastid. However, hexose phosphates can be generated in the stroma by plastidial starch phosphorylase and further metabolized within the plastid to generate the required ATP pools, thereby circumventing protoporphyrin IX-induced oxidative stress (Deschamps et al., 2008b; Rathore et al., 2009). The authors propose that a sufficient supply of ATP was a potentially important protective innovation necessary for “safe” proliferation of LHCs.
The glaucophyte C. paradoxa also possesses a NTT, although the activity for ATP/ADP translocation could not be demonstrated in isolated muroplasts (Schlichting et al., 1990; Linka et al., 2003). In C. paradoxa, both fatty acids and starch biosynthesis are cytosol-localized, indicating that ATP import into the muroplast is not required for these reactions (Figure 3A; Ma et al., 2001; Plancke et al., 2008). Functional expression and reconstitution into membranes of the glaucophyte NTT ortholog will be required to elucidate its physiological role and demonstrate whether the protein is able to mediate ATP/ADP exchange.
The red algae C. merolae and G. sulphuraria contain each one gene coding for NTT (Tyra et al., 2007). In Galdieria, similarly to the plant NTT, the ortholog transporter is able to exchange ATP for ADP (Linka et al., 2003). In red algae the synthesis of storage carbon is confined in the cytoplasmic compartment, however, other anabolic reactions such as fatty acid biosynthesis require energy supply (Viola et al., 2001). The synthesis of fatty acids in G. sulphuraria has been postulated to occur in the plastid, as indicated by the presence of the subunits of the plastidic acetyl-coA carboxylase, an ATP-dependent enzyme (Weber et al., 2004). As mentioned before, the occurrence of a glycolytic pathway in the plastids of red algae is unlikely and ATP import would be required to drive fatty acid biosynthesis (Figure 3B; Linka et al., 2008a).
In contrast to higher plants, the secondary derived plastids of the diatoms are not able to synthesize nucleotides. Plants export newly synthesized nucleotides through a member of the MCF, an adenine nucleotide uniporter (Leroch et al., 2005). In diatoms, the synthesis of nucleotides is confined to the cytosol, implying the existence of an import mechanism. Thalassiosira pseudonana and Phaeodactylum tricornutum possess diverse members of the NTTs mediating the import of purine and pyrimidine nucleotides (Ast et al., 2009). In these organisms the NTTs are therefore involved in the nucleotide supply to maintain organellar metabolism (such as nucleic acids synthesis) rather than energy provision. Diatoms possess therefore plastidial NTT proteins with biochemical properties similar to those of some bacterial intracellular parasites, which catalyze nucleotide transport in a unidirectional mode, a feature not observed for any eukaryotic NTT (Figure 4B; Tjaden et al., 1999). This role in providing the plastid with externally synthesized nucleotides is similar to that of metabolically impaired intracellular bacteria such as Chlamydia which depend on host-derived metabolites. The functional relatedness of Chlamydia and plant NTTs reflects their common origin. The prokaryote transporters with the highest sequence similarity to plant NTT proteins occur in Chlamydia relatives and phylogenetic analyses of plant, Chlamydiae and Rickettsiae NTTs suggest that the translocator was transferred from Chlamydia to plants (Schmitz-Esser et al., 2004). The Arabidopsis AtNTT1 and AtNTT2 ATP/ADP translocases were shown to be monophyletic with the chlamydial translocators (Tyra et al., 2007). Moreover, free-living cyanobacteria are devoid of genes encoding for NTTs. Although Chlamydiae are not found in plants, an unexpected number of chlamydial genes display a high degree of similarity to their plant homologs. An analysis of bacteria–eukaryote protein similarities revealed that, beyond the well explainable abundance in cyanobacterial and α-proteobacterial sequences, a notable number of plant-like genes were found in the Chlamydiaceae genomes and, most intriguingly, the vast majority of these genes contain a plastid-targeting signal (Brinkman et al., 2002; Horn et al., 2004). A phylogenetic analysis of the C. merolae genome aimed at searching for genes that are evolutionary related to Chlamydia retrieved 21 genes which are broadly present in primary photosynthetic eukaryotes. The direction of transfer is proposed to be from the bacteria to the plants, since the genes are predominantly distributed in bacteria and the cyanobacterial homologs form a well supported group distinct from the chlamydial homologs (Huang and Gogarten, 2007). Genes of chlamydial origin are mainly restricted to photosynthetic eukaryotes and non-photosynthetic plastid-bearing lineages, an observation that supports an association between Chlamydiae and the plant ancestor. A deeper phylogenomic analysis of 17 plant genomes extended the list to 55 proteins of chlamydial origin, two-thirds of which are of plastid function (Moustafa et al., 2008). The scenario evolving from these comparative genomics surveys suggests a three-way partnership between the host, the cyanobacterial endosymbiont and an environmental Chlamydia. The chlamydial contribution may have consisted in facilitating the establishment of the cyanobacterial endosymbiont by providing the necessary transport systems required for the equilibration of their metabolism, like the ADP/ATP translocator needed for the energy flux into the organelle. Later on, when the organelle was fully established in the host cell, the bacterial parasite became dispensable and was eventually lost.
Chlamydia could have thus given a contribution in the early steps of endosymbiosis by providing the petroalgae with the genetic tools necessary to establish a favorable interaction.
Mitochondrial Carrier Family
Members of the MC family (TC 2.A.29) catalyze the passage of hydrophobic compounds across the inner mitochondrial membrane and preferentially catalyze the exchange of one solute acting as antiporters (Haferkamp, 2007; Palmieri et al., 2011). Members of the MCF are involved in transporting keto acids, amino acids, nucleotides, inorganic ions, and co-factors across the mitochondrial inner membrane. Although most are found in mitochondria, recently members of the MCF were found in peroxisomes, in hydrogenosomes of anaerobic fungi, and chloroplasts (Picault et al., 2004; Haferkamp, 2007; Linka et al., 2008b). In Arabidopsis, 58 genes encode for MCF proteins (Picault et al., 2004; Palmieri et al., 2011). The number in other sequenced plant genomes varies from 37 to 125, thus being larger than that of Saccharomyces cerevisiae, which encodes for 35 MCs and comparable with that of humans which contain 53 members (Palmieri et al., 1996; Palmieri, 2004).
Members of the MCF located in the plastid membrane represent transporters which are of host origin and were likely relocated to the envelope to substitute the pre-existing cyanobacterial transporters with the endomembrane-derived host transporters (Tyra et al., 2007). No homologs of mitochondrial carriers have ever been found in prokaryotes or archaea, meaning that MCFs are a later addition during the development of the eukaryotic cell (Kunji, 2004). In plants plastids, they mediate the transport of different substrates like folates, S-adenosylmethionine, NAD, ADP-glucose or ATP, ADP, and AMP (Haferkamp, 2007).
The ADP-glucose transporter brittle-1
Besides the abovementioned NTT transporter, plastids contain another type of nucleotide transporter, the ADP-glucose transporter Brittle-1 (BT1), first identified in maize endosperm (Sullivan et al., 1991). The brittle1 mutant was named after the reduced starch content in the endosperm, resulting in collapsed, brittle kernels (Mangelsdorf and Jones, 1926). In the cereals endosperm, ADP-glucose pyrophosphorylase is localized in the cytosol (Denyer et al., 1996). The enzyme generates ADP-glucose, which must be imported into the plastid to drive starch biosynthesis. BT1 is localized to the inner envelope membrane of the amyloplast and transports ADP-glucose in antiport with the ADP released during starch biosynthesis, ensuring the balance of adenylates between stroma and cytoplasm (Figure 5; Sullivan and Kaneko, 1995; Kirchberger et al., 2007).
Another maize Brittle transporter protein, Brittle-1-2, and homologs of these transporters are found in other plants (Kirchberger et al., 2007). Interestingly, two types of Brittle-1 transporters can be identified: the BT1 from maize endosperm is present only in monocotyledonous plants and catalyzes ADP-glucose-ADP antiport, whereas the BT1-2 isoform has homologs also in dicotyledonous plants and transports nucleotides but not ADP-glucose (Leroch et al., 2005; Kirchberger et al., 2008). The StBT1 from potato and the AtBT1 from Arabidopsis locate to the inner plastid envelope and transport AMP, ADP, and ATP. Moreover, they show a unidirectional mode of transport rather than antiport (Figure 2; Leroch et al., 2005; Kirchberger et al., 2008).
Transporters involved in the one-carbon metabolism
Several cellular processes such as the synthesis of amino acids, purines, secondary metabolites, and the photorespiratory pathway depend upon the supply or removal of one-carbon units by tetrahydrofolate (THF) or S-adenosylmethionine (SAM).
Tetrahydrofolate and its derivatives are a family of co-factors that are essential for all cellular one-carbon transfer reactions: they are required for the synthesis of purines, thymidylate, methionine, pantothenate, and the interconversion of serine and glycine (Hanson and Roje, 2001). Folates are tripartite molecules composed of a pterin, a p-aminobenzoic acid (pABA), and a glutamate chain with a variable number of glutamate moieties.
In plants, folate synthesis presents a complex spatial organization, involving three subcellular compartments: the cytosol, the plastid, and the mitochondria.
In Arabidopsis, one member of the MCF was identified as folate carrier of the plastid envelope (Bedhomme et al., 2005). The AtFOLT1 protein is the closest homolog of the human mitochondrial folate transporter. Functional complementation of a glycine auxotroph Chinese hamster ovary (CHO) cell line deficient for the transport of folates into mitochondria could identify the folate carriers both in human and Arabidopsis (Titus and Moran, 2000; Bedhomme et al., 2005). The expression of AtFOLT1 in E. coli confers to the bacteria the ability to uptake exogenous folate, whereas the disruption of the gene did not led to a deficient folate supply to the plastid, since wild-type levels of folates were detected in the chloroplasts and the enzymatic capacity to catalyze folate-dependent reactions was preserved (Bedhomme et al., 2005). This suggests that alternative routes for folate uptake into plastids are present.
SAM serves as methyl-group donor and it is exclusively synthesized in the plant cytosol. Consequently, it has to be imported into plastids and mitochondria where it acts as substrate for the methyltransferases. The product resulting from the transmethylation reaction, S-adenosylhomocysteine (SAHC) has to be in turn exported to undergo re-methylation in the cytosol. The carrier that catalyzes the counter exchange of SAM for SAHC has been recently identified in Arabidopsis (Bouvier et al., 2006; Palmieri et al., 2006). The SAM transporter (SAMT) exhibits a broad expression in various plant tissues, is localized to the plastid envelope membrane and, interestingly, the complete sequence contains additional information for mitochondrial targeting (Palmieri et al., 2006).
Other Transporters
The dicarboxylate translocators
For plants, nitrogen is available in the form of nitrate, which is sequentially reduced to nitrite in the cytosol and finally to ammonia in the chloroplast stroma. Ammonia is further assimilated into nitrogen compounds via the plastidic glutamine synthetase/glutamate synthetase (GS/GOGAT) cycle. Glutamate serves as a universal organic nitrogen donor for the biosynthesis of a vast variety nitrogen-containing compounds. Assimilation of ammonia in the plastids involves two distinct dicarboxylate translocators (DiTs) with partially overlapping substrate specificities, the 2-oxoglutarate/malate-translocator (DiT1), and the glutamate/malate-translocator (DiT2). DiT1 imports the precursor of ammonia assimilation, 2-oxoglutarate, in exchange with malate; DiT2 exports the end product of ammonia assimilation, glutamate, to the cytosol, importing back the malate (Figure 2; Weber et al., 1995; Weber and Flügge, 2002; Renne et al., 2003). These transporters play also an important role in recycling the ammonia lost during photorespiration by the mitochondrial glycine decarboxylase, as indicated by the photorespiratory phenotype displayed by Arabidopsis plants lacking DiT2 and tobacco DiT1 antisense plants (Renne et al., 2003; Linka and Weber, 2005; Schneidereit et al., 2006). DiT1 also displays a high affinity for oxaloacetate (OAA) and has been proposed as the long-sought OAA/malate valve (Taniguchi et al., 2002). The malate valve functions as redox sink to dissipate excess of electrons and protect the photosynthetic apparatus from photoinhibition by exporting reducing equivalents thus balancing the stromal ATP/NADPH ratio (Heineke et al., 1991; Scheibe, 2004; Scheibe et al., 2005). The role as malate valve for DiT1 was recently proven by mutant analysis: CO2-dependent O2 evolution assays showed that cytosolic oxaloacetate is efficiently transported into chloroplasts mainly by DiT1 and O2 evolution is not enhanced by addition of OAA in the mutant, supporting the absence of additional oxaloacetate transporters (Kinoshita et al., 2011).
Dicarboxylate translocators belong to the Divalent Anion:Na+ Symporter (DASS) Family (TC 2.A.47) which transport organic di- and tri-carboxylates of the Krebs Cycle as well as dicarboxylate amino acid, inorganic sulfate, and phosphate. These proteins are found in Gram-negative bacteria, cyanobacteria, archaea, plant chloroplasts, yeast, and animals. DiTs are present only in the green lineage of plants and in bacteria, but not in red algae or glaucophytes. Phylogenetic surveys indicate that the dicarboxylate transporters are of chlamydial origin, reinforcing the hypothesis on the importance these intracellular parasites had in the establishment of endosymbiosis (Tyra et al., 2007). In isolated C. paradoxa muroplasts, transport activities for glutamine and 2-oxoglutarate could be measured (Kloos et al., 1993). In uptake experiments with isolated cyanelles by silicone oil filtering it could be shown that glutamate poorly penetrated into cyanelles, whereas glutamine was enriched in the plastid fraction by 1.7 fold within 10 min. Glutamine uptake proceeded in two phases and was stimulated by 2-oxoglutarate (Kloos et al., 1993). Ammonia formed by nitrite reduction inside the cyanelles is incorporated by GS into glutamine and then exported jointly with oxoglutarate (Kloos et al., 1993). The export of glutamine appears to be the route for providing the host cell with fixed nitrogen, a situation that encounters a parallel in the nitrogen-fixing filamentous cyanobacteria. In cyanobacteria however, the transporter seems not to discriminate between glutamine and glutamate, indicating that the glutamine export route is unique to C. paradoxa (Chapman and Meeks, 1983; Flores and Muropastor, 1988; Kloos et al., 1993).
The maltose transporter
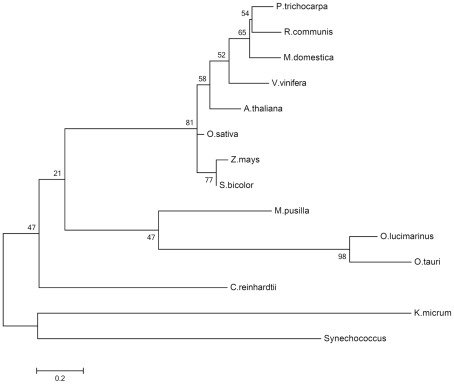
In the dark, the starch which has accumulated during the day is degraded to supply sucrose biosynthesis. Maltose and glucose are the main products of starch degradation, with maltose being the most prominent sugar exported to the cytosol at night or during increased sink demand (Figure 2; Weise et al., 2004). The maltose transporter was identified by a screening of Arabidopsis mutants displaying a maltose excess (MEX) phenotype (Niittyla et al., 2004). MEX is unrelated to any other membrane transporter and belongs to a family represented solely by the A. thaliana member (TC 2.A.84). The maltose exporter is found only in the genomes of chloroplastida and is absent in red algae, glaucophytes, and photosynthetic organisms derived by secondary endosymbiosis. The presence of MEX orthologs in the dinoflagellate Karlodinium micrum and in Synechococcus likely is the result of independent horizontal gene transfers (Figure 6; Tyra et al., 2007). The pathway of starch synthesis and degradation is presumed to have been cytosolic in the common ancestor of the three Archaeplastida lineages (Deschamps et al., 2008a). MEX appears therefore to be a plant-specific transporter, evolved as a consequence of the relocation of the starch metabolism to the stroma of the green algal ancestor (Tyra et al., 2007; Deschamps et al., 2008a).
Figure 6.
Molecular phylogenetic analysis of the MEX1 proteins. The evolutionary history was inferred by using the maximum likelihood method based on the WAG model (Whelan and Goldman, 2001). Sequence alignment was performed with MUSCLE included in MEGA5 (Tamura et al., 2011). Alignment quality was assessed by GUIDANCE giving an alignment score of 0.783. The bootstrap consensus tree inferred from 500 replicates is taken to represent the evolutionary history of the taxa analyzed. The percentage of replicate trees in which the associated taxa clustered together in the bootstrap test (500 replicates) are shown next to the branches. A discrete gamma distribution was used to model evolutionary rate differences among sites (five categories, +G, parameter = 3.3094). The rate variation model allowed for some sites to be evolutionarily invariable ([+I], 5.4532% sites). The tree is drawn to scale, with branch lengths measured in the number of substitutions per site. The analysis involved 14 amino acid sequences. All positions containing gaps and missing data were eliminated. There were a total of 173 positions in the final dataset. Evolutionary analyses were conducted in MEGA5 (Tamura et al., 2011).
The plastidic glucose transporter
Starch breakdown initiates by the action of the phosphorylating enzymes glucan, water dikinase (GWD) and phosphoglucan, water dikinase (PDW) and, after involvement of a debranching enzyme, results in the release linear glucans (Smith et al., 2005). β-amylases catalyze the production of maltose from linear glucans, and, to a lesser extent, maltotriose which is further metabolized to glucose by the disproportionating enzyme DPE1 (Critchley et al., 2001; Smith et al., 2005). A transporter involved in carbon mobilization mediating glucose export has been initially characterized and two decades later cloned and identified (Schafer et al., 1977; Weber et al., 2000). The glucose transporter (pGlcT) mediates the export of glucose released during the onset of starch degradation and has additional and still unknown roles suggested by its expression in some heterotrophic tissues that do not contain starch (Butowt et al., 2003). pGlcT contributes to the export of photoassimilates from chloroplasts, even though it is not the major route for the export of starch degradation products (Figure 2). The pglct-1/mex1 double knock-out Arabidopsis line displays a severe growth retardation phenotype suggesting that the effects of pGlcT loss on photoassimilate partitioning and plant growth are observable when the major route for exporting starch degradation products is blocked (Hahn et al., 2011). pGlcT belongs to the (MFS, TC 2.A.1) a very old, large and diverse superfamily whose members catalyze uniport, solute:cation symport and/or solute:H+ or solute:solute antiport (Law et al., 2008). This superfamily is ubiquitous in all kingdoms of life, however, close relatives of pGlcT are found only within the Chloroplastida and, as for the maltose exporter, it reflects an acquisition by the green lineage imposed by the compartmentalization of the starch metabolism within the chloroplast (Tyra et al., 2007).
The plastidic bile Acid:Na symporter
Glucosinolates (GSLs) are sulfur- and nitrogen-containing plant secondary metabolites derived from amino acids and sugar. They are mainly present in the Brassicaceae family, including Arabidopsis, and have repellent activity against herbivore insects and pathogens (Fahey et al., 2001).
The biosynthesis of methionine-derived glucosinolates involves both the chloroplast and the cytosolic compartments and starts in the cytosol where Met is transaminated by the aminotransferase BCAT4. The resulting 2-keto acid, 4-methylthio-2-oxobutanoate (MTOB) is imported into the chloroplast where it undergoes repeated condensations with acetyl-CoA and subsequent decarboxylations to the corresponding α-keto acid. The chain-elongated keto acid is aminated to the amino acid and transported back to the cytosol for the following core structure synthesis (Fahey et al., 2001; Knill et al., 2008; Nour-Eldin and Halkier, 2009). Thus, the methionine derivatives must be transported at least twice across the chloroplast membranes. The first transporter involved in the biosynthesis of Met-derived glucosinolates was recently identified as member of the Bile Acid:Na Symporter (BASS) Family (TC 2.A.28) due to significant sequence similarities to mammalian sodium-coupled bile acid transporters (Gigolashvili et al., 2009; Sawada et al., 2009). The BAT5 protein localizes to the plastid and loss-of-function mutants defective in BAT5 function contained strongly reduced levels of aliphatic glucosinolates. Feeding experiments suggest that BAT5 transports MTOB and chain-elongated 2-keto acids across the chloroplast envelope membrane, indicating a role for BAT5 in the transport of GS intermediates (Gigolashvili et al., 2009; Sawada et al., 2009).
Homologs of the sodium-coupled bile acid transporters are also found in the algal genomes, however no data about their localization or function are to date available.
The plastidic folate carrier
In addition to the plastidic folate carrier belonging to the MCF, a second folate carrier was identified which belongs to the folate-biopterin transporter (FBT) Family (TC 2.A.71). The FBT family includes functionally characterized members from protozoa and homologs are found in cyanobacteria (Synechocystis) and plants (A. thaliana). The Synechocystis and Arabidopsis FBTs were identified based on their homology to the folate transporter of the trypanosomatid protozoa Leishmania (Klaus et al., 2005). These transporters are able to complement E. coli mutants impaired in folate uptake or production; in particular they are capable of transporting the monoglutamyl form of folate (Klaus et al., 2005). The plant FBT likely originated by endosymbiotic gene transfer (EGT) from the captured cyanobacterium and contributes to a redundant folate uptake system involving also the abovementioned host-derived FOLT1 transporter (Tyra et al., 2007).
The pyruvate carrier
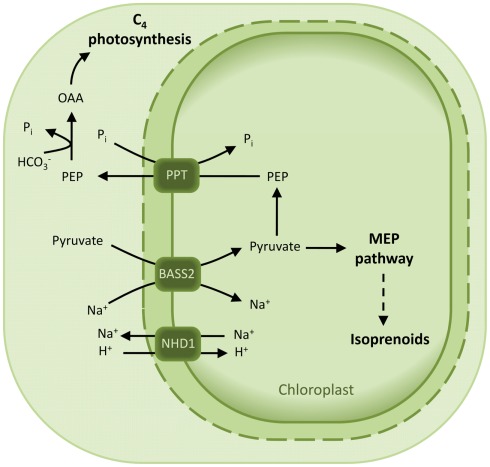
Plants that perform C4 photosynthesis require a massive flux of metabolites across the membranes of their MC and bundle sheat cells (BSC). In particular, pyruvate is formed in the chloroplasts of the BSC by decarboxylation of the C4 acid malate and has to be transported back to the MC chloroplasts to regenerate the CO2 acceptor PEP (Bräutigam and Weber, 2011). Pyruvate transport in C4 plants has been shown to occur either in a proton- or sodium-dependent manner (Ohnishi and Kanai, 1987, 1990). Accordingly, a putative plastidic proton:sodium symporter (NHD1) is 16-fold up-regulated in the C4 species Cleome gynandra with respect to the C3 relative Cleome spinosa (Bräutigam et al., 2011). It is proposed that NDH1 functions in exporting sodium out of the chloroplast to sustain a gradient necessary for pyruvate import. In addition, a strong up-regulation of a bile acid:sodium cotransporter was observed in C. gyandra (Bräutigam et al., 2011) and in C4 Flaveria species (Gowik et al., 2011). This cotransporter is a member of the BASS Family (TC 2.A.28) and is not found in maize, a species which displays a proton-dependent transport of pyruvate (Bräutigam et al., 2008). The biochemical function NDH1 and BASS2 was recently demonstrated by expression in an heterologous system of BASS2 alone and in presence of NDH1 (Furumoto et al., 2011). Indeed, these proteins represent a two-translocator system, consisting of the sodium:pyruvate cotransporter BASS2 and the sodium:proton antiporter NHD1 (Figure 7; Furumoto et al., 2011).
Figure 7.
Model for the pyruvate uptake into the plastid. The two-translocator system responsible for pyruvate uptake into the plastid of certain C4 species is based on a sodium:pyruvate cotransporter (BASS2) and a sodium:proton antiporter (NHD1). NHD1 is able to establish a sodium gradient across the envelope membrane, which in turn drives pyruvate import by the cotransporter BASS2. Abbreviations: BASS2, BILE ACID:SODIUM SYMPORTER FAMILY PROTEIN 2; NHD1, sodium:proton antiporter; OAA, oxaloacetate; PEP, phosphoenolpyruvate; PPT, PEP/phosphate translocator.
Conclusion
The transport of organic compounds across the cyanobacterial membrane was of crucial importance for establishing endosymbiosis and insertion of protein translocators into the envelope was necessary to take full advantage of the newly acquired metabolic entity. The pPTs were most likely pioneers in settling a connection between the host and the cyanobiont in order to tap the photosynthates, as suggested by their ubiquitous distribution among the plastid-bearing organisms. The majority of the plastid translocators, including the PTs, are of host origin, indicating that establishment of the plastid was a host-driven process. However, evidence is accumulating which implies a small but substantial contribution from the intracellular parasite Chlamydia. The requirement of three organisms in the early steps of endosymbiosis could explain why this event occurred only once during evolution. Understanding the metabolic connection between plastid and cytosol is also crucial for future attempts to generate synthetic organelles, such as synthetic plastids that harbor tailor-made metabolic capacities and that are introduced into plastid-free recipient cells (Weber and Osteryoung, 2010).
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
- Adl S. M., Simpson A. G., Farmer M. A., Andersen R. A., Anderson O. R., Barta J. R., Bowser S. S., Brugerolle G., Fensome R. A., Fredericq S., James T. Y., Karpov S., Kugrens P., Krug J., Lane C. E., Lewis L. A., Lodge J., Lynn D. H., Mann D. G., Mccourt R. M., Mendoza L., Moestrup O., Mozley-Standridge S. E., Nerad T. A., Shearer C. A., Smirnov A. V., Spiegel F. W., Taylor M. F. (2005). The new higher level classification of eukaryotes with emphasis on the taxonomy of protists. J. Eukaryot. Microbiol. 52, 399–451 10.1111/j.1550-7408.2005.00053.x [DOI] [PubMed] [Google Scholar]
- Allen J. F., Martin W. (2007). Evolutionary biology: out of thin air. Nature 445, 610–612 10.1038/445610a [DOI] [PubMed] [Google Scholar]
- Archibald J. M. (2009). The puzzle of plastid evolution. Curr. Biol. 19, R81–R88 10.1016/j.cub.2008.11.067 [DOI] [PubMed] [Google Scholar]
- Ardila F., Pozuetaromero J., Akazawa T. (1993). Adenylate uptake by proplastids from cultured-cells of tobacco (nicotiana-tabacum-L Cv-By2) indicates that an adenylate translocator is present in all types of plastid. Plant Cell Physiol. 34, 237–242 [Google Scholar]
- Ast M., Gruber A., Schmitz-Esser S., Neuhaus H. E., Kroth P. G., Horn M., Haferkamp I. (2009). Diatom plastids depend on nucleotide import from the cytosol. Proc. Natl. Acad. Sci. U.S.A. 106, 3621–3626 10.1073/pnas.0808862106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ball S. G., Morell M. K. (2003). From bacterial glycogen to starch: understanding the biogenesis of the plant starch granule. Annu. Rev. Plant Biol. 54, 207–233 10.1146/annurev.arplant.54.031902.134927 [DOI] [PubMed] [Google Scholar]
- Barbier G., Oesterhelt C., Larson M. D., Halgren R. G., Wilkerson C., Garavito R. M., Benning C., Weber A. P. (2005). Comparative genomics of two closely related unicellular thermo-acidophilic red algae, Galdieria sulphuraria and Cyanidioschyzon merolae, reveals the molecular basis of the metabolic flexibility of Galdieria sulphuraria and significant differences in carbohydrate metabolism of both algae. Plant Physiol. 137, 460–474 10.1104/pp.104.051169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bedhomme M., Hoffmann M., Mccarthy E. A., Gambonnet B., Moran R. G., Rebeille F., Ravanel S. (2005). Folate metabolism in plants – an Arabidopsis homolog of the mammalian mitochondrial folate transporter mediates folate import into chloroplasts. J. Biol. Chem. 280, 34823–34831 10.1074/jbc.M506045200 [DOI] [PubMed] [Google Scholar]
- Björn L. O. (2009). The evolution of photosynthesis and chloroplasts. Curr. Sci. 96, 1466–1474 [Google Scholar]
- Bölter B., Soll J. (2001). Ion channels in the outer membranes of chloroplasts and mitochondria: open doors or regulated gates? EMBO J. 20, 935–940 10.1093/emboj/20.5.935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borchert S., Harborth J., Schunemann D., Hoferichter P., Heldt H. W. (1993). Studies of the enzymic capacities and transport properties of Pea Root Plastids. Plant Physiol. 101, 303–312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourgis F., Kilaru A., Cao X., Ngando-Ebongue G. F., Drira N., Ohlrogge J. B., Arondel V. (2011). Comparative transcriptome and metabolite analysis of oil palm and date palm mesocarp that differ dramatically in carbon partitioning. Proc. Natl. Acad. Sci. U.S.A. 108, 12527–12532 10.1073/pnas.1106502108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouvier F., Linka N., Isner J. C., Mutterer J., Weber A. P. M., Camara B. (2006). Arabidopsis SAMT1 defines a plastid transporter regulating plastid biogenesis and plant development. Plant Cell 18, 3088–3105 10.1105/tpc.105.040741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bräutigam A., Hoffmann-Benning S., Weber A. P. (2008). Comparative proteomics of chloroplast envelopes from C3 and C4 plants reveals specific adaptations of the plastid envelope to C4 photosynthesis and candidate proteins required for maintaining C4 metabolite fluxes. Plant Physiol. 148, 568–579 10.1104/pp.108.121012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bräutigam A., Kajala K., Wullenweber J., Sommer M., Gagneul D., Weber K. L., Carr K. M., Gowik U., Mass J., Lercher M. J., Westhoff P., Hibberd J. M., Weber A. P. (2011). An mRNA blueprint for C4 photosynthesis derived from comparative transcriptomics of closely related C3 and C4 species. Plant Physiol. 155, 142–156 10.1104/pp.110.159442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bräutigam A., Weber A. P. M. (2011). “Transport processes: connecting the reactions of C4 photosynthesis,” in C4 Photosynthesis and Related CO2 Concentrating Mechanisms, eds Raghavendra A. S., Sage R. F. (Berlin: Springer; ), 199–219 [Google Scholar]
- Brinkman F. S., Blanchard J. L., Cherkasov A., Av-Gay Y., Brunham R. C., Fernandez R. C., Finlay B. B., Otto S. P., Ouellette B. F., Keeling P. J., Rose A. M., Hancock R. E., Jones S. J., Greberg H. (2002). Evidence that plant-like genes in Chlamydia species reflect an ancestral relationship between Chlamydiaceae, cyanobacteria, and the chloroplast. Genome Res. 12, 1159–1167 10.1101/gr.341802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burey S. C., Poroyko V., Ergen Z. N., Fathi-Nejad S., Schüller C., Ohnishi N., Fukuzawa H., Bohnert H. J., Löffelhardt W. (2007). Acclimation to low [CO2] by an inorganic carbon-concentrating mechanism in Cyanophora paradoxa. Plant Cell Environ. 30, 1422–1435 10.1111/j.1365-3040.2007.01715.x [DOI] [PubMed] [Google Scholar]
- Butowt R., Granot D., Rodriguez-Garcia M. I. (2003). A putative plastidic glucose translocator is expressed in heterotrophic tissues that do not contain starch, during olive (Olea europea L.) fruit ripening. Plant Cell Physiol. 44, 1152–1161 10.1093/pcp/pcg149 [DOI] [PubMed] [Google Scholar]
- Cavalier-Smith T. (2000). Membrane heredity and early chloroplast evolution. Trends Plant Sci. 5, 174–182 10.1016/S1360-1385(00)01598-3 [DOI] [PubMed] [Google Scholar]
- Chapman J. S., Meeks J. C. (1983). Glutamine and glutamate transport by Anabaena-variabilis. J. Bacteriol. 156, 122–129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colleoni C., Linka M., Deschamps P., Handford M. G., Dupree P., Weber A. P., Ball S. G. (2010). Phylogenetic and biochemical evidence supports the recruitment of an ADP-glucose translocator for the export of photosynthate during plastid endosymbiosis. Mol. Biol. Evol. 27, 2691–2701 10.1093/molbev/msq158 [DOI] [PubMed] [Google Scholar]
- Critchley J. H., Zeeman S. C., Takaha T., Smith A. M., Smith S. M. (2001). A critical role for disproportionating enzyme in starch breakdown is revealed by a knock-out mutation in Arabidopsis. Plant J. 26, 89–100 10.1046/j.1365-313x.2001.01012.x [DOI] [PubMed] [Google Scholar]
- Denyer K., Dunlap F., Thorbjornsen T., Keeling P., Smith A. M. (1996). The major form of ADP-glucose pyrophosphorylase in maize endosperm is extra-plastidial. Plant Physiol. 112, 779–785 10.1104/pp.112.2.779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deschamps P., Haferkamp I., D’Hulst C., Neuhaus H. E., Ball S. G. (2008a). The relocation of starch metabolism to chloroplasts: when, why and how. Trends Plant Sci. 13, 574–582 10.1016/j.tplants.2008.08.009 [DOI] [PubMed] [Google Scholar]
- Deschamps P., Moreau H., Worden A. Z., Dauvillee D., Ball S. G. (2008b). Early gene duplication within chloroplastida and its correspondence with relocation of starch metabolism to chloroplasts. Genetics 178, 2373–2387 10.1534/genetics.108.087205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duy D., Soll J., Philippar K. (2007). Solute channels of the outer membrane: from bacteria to chloroplasts. Biol. Chem. 388, 879–889 10.1515/BC.2007.120 [DOI] [PubMed] [Google Scholar]
- Eicks M., Maurino V., Knappe S., Flügge U.-I., Fischer K. (2002). The plastidic pentose phosphate translocator represents a link between the cytosolic and the plastidic pentose phosphate pathways in plants. Plant Physiol. 128, 512–522 10.1104/pp.010576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Embley T. M., Martin W. (2006). Eukaryotic evolution, changes and challenges. Nature 440, 623–630 10.1038/nature04546 [DOI] [PubMed] [Google Scholar]
- Fahey J. W., Zalcmann A. T., Talalay P. (2001). The chemical diversity and distribution of glucosinolates and isothiocyanates among plants. Phytochemistry 56, 5–51 10.1016/S0031-9422(00)00316-2 [DOI] [PubMed] [Google Scholar]
- Fischer K., Kammerer B., Gutensohn M., Arbinger B., Weber A., Häusler R. E., Flügge U. I. (1997). A new class of plastidic phosphate translocators: a putative link between primary and secondary metabolism by the phosphoenolpyruvate/phosphate antiporter. Plant Cell 9, 453–462 10.1105/tpc.9.3.453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fliege R., Flügge U. I., Werdan K., Heldt H. W. (1978). Specific transport of inorganic phosphate, 3-phosphoglycerate and triosephosphates across the inner membrane of the envelope in spinach chloroplasts. Biochim. Biophys. Acta 502, 232–247 10.1016/0005-2728(78)90045-2 [DOI] [PubMed] [Google Scholar]
- Flores E., Muropastor M. I. (1988). Uptake of glutamine and glutamate by the dinitrogen-fixing cyanobacterium anabaena Sp Pcc7120. FEMS Microbiol. Lett. 56, 127–130 10.1111/j.1574-6968.1988.tb03163.x [DOI] [Google Scholar]
- Flügge U. I. (1995). Phosphate translocation in the regulation of photosynthesis. J. Exp. Bot. 46, 1317–1323 [Google Scholar]
- Flügge U. I. (2000). Transport in and out of plastids: does the outer envelope membrane control the flow? Trends Plant Sci. 5, 135–137 10.1016/S1360-1385(00)01578-8 [DOI] [PubMed] [Google Scholar]
- Flügge U. I., Heldt H. W. (1984). The phosphate-triose phosphate-phosphoglycerate translocator of the chloroplast. Trends Biochem. Sci. 9, 530–533 10.1016/0968-0004(84)90283-4 [DOI] [Google Scholar]
- Flügge U.-I., Häusler R. E., Ludewig F., Gierth M. (2011). The role of transporters in supplying energy to plant plastids. J. Exp. Bot. 62, 2381–2392 10.1093/jxb/erq361 [DOI] [PubMed] [Google Scholar]
- Furumoto T., Yamaguchi T., Ohshima-Ichie Y., Nakamura M., Tsuchida-Iwata Y., Shimamura M., Ohnishi J., Hata S., Gowik U., Westhoff P., Bräutigam A., Weber A. P. M., Izui K. (2011). A plastidial sodium-dependent pyruvate transporter. Nature 476, 472–475 10.1038/nature10250 [DOI] [PubMed] [Google Scholar]
- Gigolashvili T., Yatusevich R., Rollwitz I., Humphry M., Gershenzon J., Flügge U. I. (2009). The plastidic bile acid transporter 5 is required for the biosynthesis of methionine-derived glucosinolates in Arabidopsis thaliana. Plant Cell 21, 1813–1829 10.1105/tpc.109.066399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gould S. B., Waller R. F., Mcfadden G. I. (2008). Plastid evolution. Annu. Rev. Plant Biol. 59, 491–517 10.1146/annurev.arplant.59.032607.092915 [DOI] [PubMed] [Google Scholar]
- Gowik U., Bräutigam A., Weber K. L., Weber A. P. M., Westhoff P. (2011). Evolution of C4 photosynthesis in the genus Flaveria - how many and which genes does it take to make C4? Plant Cell 23, 2087–2105 10.1105/tpc.111.086264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gross W., Schnarrenberger C. (1995). Heterotrophic growth of two strains of the acido-thermophilic red alga Galdieria sulphuraria. Plant Cell Physiol. 36, 633 [Google Scholar]
- Haferkamp I. (2007). The diverse members of the mitochondrial carrier family in plants. FEBS Lett. 581, 2375–2379 10.1016/j.febslet.2007.02.020 [DOI] [PubMed] [Google Scholar]
- Haferkamp I., Deschamps P., Ast M., Jeblick W., Maier U., Ball S., Neuhaus H. E. (2006). Molecular and biochemical analysis of periplastidial starch metabolism in the cryptophyte Guillardia theta. Eukaryot. Cell 5, 964–971 10.1128/EC.00381-05 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hahn T. R., Cho M. H., Lim H., Shin D. H., Jeon J. S., Bhoo S. H., Park Y. I. (2011). Role of the plastidic glucose translocator in the export of starch degradation products from the chloroplasts in Arabidopsis thaliana. New Phytol. 190, 101–112 10.1111/j.1469-8137.2010.03580.x [DOI] [PubMed] [Google Scholar]
- Hanson A. D., Roje S. (2001). One-carbon metabolism in higher plants. Annu. Rev. Plant Physiol. Plant Mol. Biol. 52, 119–137 10.1146/annurev.arplant.52.1.119 [DOI] [PubMed] [Google Scholar]
- Heineke D., Riens B., Grosse H., Hoferichter P., Peter U., Flügge U. I., Heldt H. W. (1991). Redox transfer across the inner chloroplast envelope membrane. Plant Physiol. 95, 1131–1137 10.1104/pp.95.4.1131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrmann K. M. (1995). The Shikimate pathway: early steps in the biosynthesis of aromatic compounds. Plant Cell 7, 907–919 10.1105/tpc.7.7.907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrmann K. M., Weaver L. M. (1999). The Shikimate pathway. Annu. Rev. Plant Physiol. Plant Mol. Biol. 50, 473–503 10.1146/annurev.arplant.50.1.473 [DOI] [PubMed] [Google Scholar]
- Hohmann-Marriott M. F., Blankenship R. E. (2011). Evolution of photosynthesis. Annu. Rev. Plant Biol. 62, 515–548 10.1146/annurev-arplant-042110-103811 [DOI] [PubMed] [Google Scholar]
- Horn M., Collingro A., Schmitz-Esser S., Beier C. L., Purkhold U., Fartmann B., Brandt P., Nyakatura G. J., Droege M., Frishman D., Rattei T., Mewes H. W., Wagner M. (2004). Illuminating the evolutionary history of Chlamydiae. Science 304, 728–730 10.1126/science.1096330 [DOI] [PubMed] [Google Scholar]
- Huang J. L., Gogarten J. P. (2007). Did an ancient chlamydial endosymbiosis facilitate the establishment of primary plastids? Genome Biol. 8, R99. 10.1186/gb-2007-8-6-r99 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue K. (2011). Emerging roles of the chloroplast outer envelope membrane. Trends Plant Sci. 10.1016/j.tplants.2011.06.005 [DOI] [PubMed] [Google Scholar]
- Jarvis P., Dörmann P., Peto C. A., Lutes J., Benning C., Chory J. (2000). Galactolipid deficiency and abnormal chloroplast development in the Arabidopsis MGD synthase 1 mutant. Proc. Natl. Acad. Sci. U.S.A. 97, 8175–8179 10.1073/pnas.100132197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kammerer B., Fischer K., Hilpert B., Schubert S., Gutensohn M., Weber A., Flügge U. I. (1998). Molecular characterization of a carbon transporter in plastids from heterotrophic tissues: the glucose 6-phosphate/phosphate antiporter. Plant Cell 10, 105–117 10.1105/tpc.10.1.105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karnataki A., Derocher A., Coppens I., Nash C., Feagin J. E., Parsons M. (2007). Cell cycle-regulated vesicular trafficking of Toxoplasma APT1, a protein localized to multiple apicoplast membranes. Mol. Microbiol. 63, 1653–1668 10.1111/j.1365-2958.2007.05619.x [DOI] [PubMed] [Google Scholar]
- Katinka M. D., Duprat S., Cornillot E., Metenier G., Thomarat F., Prensier G., Barbe V., Peyretaillade E., Brottier P., Wincker P., Delbac F., El Alaoui H., Peyret P., Saurin W., Gouy M., Weissenbach J., Vivares C. P. (2001). Genome sequence and gene compaction of the eukaryote parasite Encephalitozoon cuniculi. Nature 414, 450–453 10.1038/35106579 [DOI] [PubMed] [Google Scholar]
- Keeling P. J. (2010). The endosymbiotic origin, diversification and fate of plastids. Philos. Trans. R. Soc. Lond. B Biol. Sci. 365, 729–748 10.1098/rstb.2009.0103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kies L. (1974). Electron microscopical investigations on Paulinella chromatophora Lauterborn, a thecamoeba containing blue-green endosymbioints (cyanelles). Protoplasma 80, 69–89 [DOI] [PubMed] [Google Scholar]
- Kinoshita H., Nagasaki J., Yoshikawa N., Yamamoto A., Takito S., Kawasaki M., Sugiyama T., Miyake H., Weber A. P. M., Taniguchi M. (2011). The chloroplastic 2-oxoglutarate/malate transporter has dual function as the malate valve and in carbon/nitrogen metabolism. Plant J. 65, 15–26 10.1111/j.1365-313X.2010.04397.x [DOI] [PubMed] [Google Scholar]
- Kirchberger S., Leroch M., Huynen M. A., Wahl M., Neuhaus H. E., Tjaden J. (2007). Molecular and biochemical analysis of the plastidic ADP-glucose transporter (ZmBT1) from Zea mays. J. Biol. Chem. 282, 22481–22491 10.1074/jbc.M702484200 [DOI] [PubMed] [Google Scholar]
- Kirchberger S., Tjaden J., Neuhaus H. E. (2008). Characterization of the Arabidopsis Brittle1 transport protein and impact of reduced activity on plant metabolism. Plant J. 56, 51–63 10.1111/j.1365-313X.2008.03583.x [DOI] [PubMed] [Google Scholar]
- Klaus S. M., Kunji E. R., Bozzo G. G., Noiriel A., De La Garza R. D., Basset G. J., Ravanel S., Rebeille F., Gregory J. F., Hanson A. D. (2005). Higher plant plastids and cyanobacteria have folate carriers related to those of trypanosomatids. J. Biol. Chem. 280, 38457–38463 10.1074/jbc.M507432200 [DOI] [PubMed] [Google Scholar]
- Kloos K., Schlichting R., Zimmer W., Bothe H. (1993). Glutamine and glutamate transport in cyanophora-paradoxa. Bot. Acta 106, 435–440 [Google Scholar]
- Knappe S., Flügge U. I., Fischer K. (2003a). Analysis of the plastidic phosphate translocator gene family in Arabidopsis and identification of new phosphate translocator-homologous transporters, classified by their putative substrate-binding site. Plant Physiol. 131, 1178–1190 10.1104/pp.016519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knappe S., Lottgert T., Schneider A., Voll L., Flügge U. I., Fischer K. (2003b). Characterization of two functional phosphoenolpyruvate/phosphate translocator (PPT) genes in Arabidopsis-AtPPT1 may be involved in the provision of signals for correct mesophyll development. Plant J. 36, 411–420 10.1046/j.1365-313X.2003.01888.x [DOI] [PubMed] [Google Scholar]
- Knill T., Schuster J., Reichelt M., Gershenzon J., Binder S. (2008). Arabidopsis branched-chain aminotransferase 3 functions in both amino acid and glucosinolate biosynthesis. Plant Physiol. 146, 1028–1039 10.1104/pp.107.111609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi K., Mochizuki N., Yoshimura N., Motohashi K., Hisabori T., Masuda T. (2008). Functional analysis of Arabidopsis thaliana isoforms of the Mg-chelatase CHLI subunit. Photochem. Photobiol. Sci. 7, 1188–1195 10.1039/b802604c [DOI] [PubMed] [Google Scholar]
- Kunji E. R. (2004). The role and structure of mitochondrial carriers. FEBS Lett. 564, 239–244 10.1016/S0014-5793(04)00242-X [DOI] [PubMed] [Google Scholar]
- Law C. J., Maloney P. C., Wang D. N. (2008). Ins and outs of major facilitator superfamily, antiporters. Annu. Rev. Microbiol. 62, 289–305 10.1146/annurev.micro.61.080706.093329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leroch M., Kirchberger S., Haferkamp I., Wahl M., Neuhaus H. E., Tjaden J. (2005). Identification and characterization of a novel plastidic adenine nucleotide uniporter from Solanum tuberosum. J. Biol. Chem. 280, 17992–18000 10.1074/jbc.M412462200 [DOI] [PubMed] [Google Scholar]
- Lim L., Linka M., Mullin K. A., Weber A. P., Mcfadden G. I. (2010). The carbon and energy sources of the non-photosynthetic plastid in the malaria parasite. FEBS Lett. 584, 549–554 10.1016/j.febslet.2010.04.006 [DOI] [PubMed] [Google Scholar]
- Linka M., Jamai A., Weber A. P. M. (2008a). Functional characterization of the plastidic phosphate translocator gene family from the thermo-acidophilic red alga Galdieria sulphuraria reveals specific adaptations of primary carbon partitioning in green plants and red algae. Plant Physiol. 148, 1487–1496 10.1104/pp.108.129478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linka N., Theodoulou F. L., Haslam R. P., Linka M., Napier J. A., Neuhaus H. E., Weber A. P. M. (2008b). Peroxisomal ATP import is essential for seedling development in Arabidopsis thaliana. Plant Cell 20, 3241–3257 10.1105/tpc.108.062042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linka M., Weber A. P. (2005). Shuffling ammonia between mitochondria and plastids during photorespiration. Trends Plant Sci. 10, 461–465 10.1016/j.tplants.2005.08.002 [DOI] [PubMed] [Google Scholar]
- Linka N., Hurka H., Lang B. F., Burger G., Winkler H. H., Stamme C., Urbany C., Seil I., Kusch J., Neuhaus H. E. (2003). Phylogenetic relationships of non-mitochondrial nucleotide transport proteins in bacteria and eukaryotes. Gene 306, 27–35 10.1016/S0378-1119(03)00429-3 [DOI] [PubMed] [Google Scholar]
- Löffelhardt W., Bohnert H. J., Bryant D. A. (1997). The cyanelles of Cyanophora paradoxa. CRC Crit. Rev. Plant Sci. 16, 393–413 10.1080/713608152 [DOI] [Google Scholar]
- Ma Y., Jakowitsch J., Maier T. L., Bayer M. G., Muller N. E., Schenk H. E., Löffelhardt W. (2001). ATP citrate lyase in the glaucocystophyte alga Cyanophora paradoxa is a cytosolic enzyme: characterisation of the gene for the large subunit at the cDNA and genomic levels. Mol. Genet. Genomics 266, 231–238 10.1007/s004380100545 [DOI] [PubMed] [Google Scholar]
- Mangelsdorf P. C., Jones D. F. (1926). The expression of mendelian factors in the gametophyte of maize. Genetics 11, 423–455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin W., Herrmann R. G. (1998). Gene transfer from organelles to the nucleus: how much, what happens, and why? Plant Physiol. 118, 9–17 10.1104/pp.118.1.9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maughan S. C., Pasternak M., Cairns N., Kiddle G., Brach T., Jarvis R., Haas F., Nieuwland J., Lim B., Muller C., Salcedo-Sora E., Kruse C., Orsel M., Hell R., Miller A. J., Bray P., Foyer C. H., Murray J. A., Meyer A. J., Cobbett C. S. (2010). Plant homologs of the Plasmodium falciparum chloroquine-resistance transporter, PfCRT, are required for glutathione homeostasis and stress responses. Proc. Natl. Acad. Sci. U.S.A. 107, 2331–2336 10.1073/pnas.0913689107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meister A. (1995). Glutathione biosynthesis and its inhibition. Biothiols Pt B 252, 26–30 [DOI] [PubMed] [Google Scholar]
- Moustafa A., Reyes-Prieto A., Bhattacharya D. (2008). Chlamydiae has contributed at least 55 genes to plantae with predominantly plastid functions. PLoS ONE 3, e2205. 10.1371/journal.pone.0002205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulkidjanian A. Y., Koonin E. V., Makarova K. S., Mekhedov S. L., Sorokin A., Wolf Y. I., Dufresne A., Partensky F., Burd H., Kaznadzey D., Haselkorn R., Galperin M. Y. (2006). The cyanobacterial genome core and the origin of photosynthesis. Proc. Natl. Acad. Sci. U.S.A. 103, 13126–13131 10.1073/pnas.0605709103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullin K. A., Lim L., Ralph S. A., Spurck T. P., Handman E., Mcfadden G. I. (2006). Membrane transporters in the relict plastid of malaria parasites. Proc. Natl. Acad. Sci. U.S.A. 103, 9572–9577 10.1073/pnas.0602293103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neuhaus H. E., Thom E., Mohlmann T., Steup M., Kampfenkel K. (1997). Characterization of a novel eukaryotic ATP/ADP translocator located in the plastid envelope of Arabidopsis thaliana L. Plant J. 11, 73–82 10.1046/j.1365-313X.1997.11010073.x [DOI] [PubMed] [Google Scholar]
- Niewiadomski P., Knappe S., Geimer S., Fischer K., Schulz B., Unte U. S., Rosso M. G., Ache P., Flügge U. I., Schneider A. (2005). The Arabidopsis plastidic glucose 6-phosphate/phosphate translocator GPT1 is essential for pollen maturation and embryo sac development. Plant Cell 17, 760–775 10.1105/tpc.104.029124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niittyla T., Messerli G., Trevisan M., Chen J., Smith A. M., Zeeman S. C. (2004). A previously unknown maltose transporter essential for starch degradation in leaves. Science 303, 87–89 10.1126/science.1091811 [DOI] [PubMed] [Google Scholar]
- Nour-Eldin H. H., Halkier B. A. (2009). Piecing together the transport pathway of aliphatic glucosinolates. Phytochem. Rev. 8, 53–67 10.1007/s11101-008-9110-8 [DOI] [Google Scholar]
- Oesterhelt C., Klocke S., Holtgrefe S., Linke V., Weber A. P. M., Scheibe R. (2007). Redox regulation of chloroplast enzymes in Galdieria sulphuraria in view of eukaryotic evolution. Plant Cell Physiol. 48, 1359–1373 10.1093/pcp/pcm108 [DOI] [PubMed] [Google Scholar]
- Ohnishi J., Kanai R. (1990). Pyruvate uptake induced by a Ph jump in mesophyll chloroplasts of maize and sorghum, Nadp-malic enzyme type-C4 species. FEBS Lett. 269, 122–124 10.1016/0014-5793(90)81134-A [DOI] [PubMed] [Google Scholar]
- Ohnishi J. I., Kanai R. (1987). Na+-induced uptake of pyruvate into mesophyll chloroplasts of a C4 plant, Panicum miliaceum. FEBS Lett. 219, 347–350 10.1016/0014-5793(87)80250-8 [DOI] [Google Scholar]
- Palmieri F. (2004). The mitochondrial transporter family (SLC25): physiological and pathological implications. Pflugers Arch. 447, 689–709 10.1007/s00424-003-1099-7 [DOI] [PubMed] [Google Scholar]
- Palmieri F., Palmieri L., Arrigoni R., Blanco E., Carrari F., Zanor M. I., Studart-Guimaraes C., Fernie A. R. (2006). Molecular identification of an Arabidopsis S-adenosylmethionine transporter. Analysis of organ distribution, bacterial expression, reconstitution into liposomes, and functional characterization. Plant Physiol. 142, 855–865 10.1104/pp.106.086975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmieri F., Pierri C. L., De Grassi A., Nunes-Nesi A., Fernie A. R. (2011). Evolution, structure and function of mitochondrial carriers: a review with new insights. Plant J. 66, 161–181 10.1111/j.1365-313X.2011.04516.x [DOI] [PubMed] [Google Scholar]
- Palmieri L., Palmieri F., Runswick M. J., Walker J. E. (1996). Identification by bacterial expression and functional reconstitution of the yeast genomic sequence encoding the mitochondrial dicarboxylate carrier protein. FEBS Lett. 399, 299–302 10.1016/S0014-5793(96)01350-6 [DOI] [PubMed] [Google Scholar]
- Patron N. J., Keeling P. J. (2005). Common evolutionary origin of starch biosynthetic enzymes in green and red algae. J. Phycol. 41, 1131–1141 10.1111/j.1529-8817.2005.00135.x [DOI] [Google Scholar]
- Picault N., Hodges M., Paimieri L., Palmieri F. (2004). The growing family of mitochondrial carriers in Arabidopsis. Trends Plant Sci. 9, 138–146 10.1016/j.tplants.2004.01.007 [DOI] [PubMed] [Google Scholar]
- Plancke C., Colleoni C., Deschamps P., Dauvillee D., Nakamura Y., Haebel S., Ritte G., Steup M., Buleon A., Putaux J. L., Dupeyre D., D’Hulst C., Ral J. P., Löffelhardt W., Ball S. G. (2008). Pathway of cytosolic starch synthesis in the model glaucophyte Cyanophora paradoxa. Eukaryot. Cell 7, 247–257 10.1128/EC.00373-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajasekharan R., Nachiappan V. (2010). Fatty acid biosynthesis and regulation in plants. Plant Dev. Biol. Biotechnol. Perspect. 105–115 [Google Scholar]
- Ralph S. A., Van Dooren G. G., Waller R. F., Crawford M. J., Fraunholz M. J., Foth B. J., Tonkin C. J., Roos D. S., Mcfadden G. I. (2004). Tropical infectious diseases: metabolic maps and functions of the Plasmodium falciparum apicoplast. Nat. Rev. Microbiol. 2, 203–216 10.1038/nrmicro843 [DOI] [PubMed] [Google Scholar]
- Rathore R. S., Garg N., Garg S., Kumar A. (2009). Starch phosphorylase: role in starch metabolism and biotechnological applications. Crit. Rev. Biotechnol. 29, 214–224 10.1080/07388550902926063 [DOI] [PubMed] [Google Scholar]
- Reichert A., Dennes A., Vetter S., Scheibe R. (2003). Chloroplast fructose 1,6-bisphosphatase with changed redox modulation: comparison of the Galdieria enzyme with cysteine mutants from spinach. Biochim. Biophys. Acta 1645, 212–217 [DOI] [PubMed] [Google Scholar]
- Reinhold T., Alawady A., Grimm B., Beran K. C., Jahns P., Conrath U., Bauer J., Reiser J., Melzer M., Jeblick W., Neuhaus H. E. (2007). Limitation of nocturnal import of ATP into Arabidopsis chloroplasts leads to photooxidative damage. Plant J. 50, 293–304 10.1111/j.1365-313X.2007.03049.x [DOI] [PubMed] [Google Scholar]
- Renne P., Dressen U., Hebbeker U., Hille D., Flügge U. I., Westhoff P., Weber A. P. M. (2003). The Arabidopsis mutant dct is deficient in the plastidic glutamate/malate translocator DiT2. Plant J. 35, 316–331 10.1046/j.1365-313X.2003.01806.x [DOI] [PubMed] [Google Scholar]
- Reyes-Prieto A., Weber A. P. M., Bhattacharya D. (2007). The origin and establishment of the plastid in algae and plants. Annu. Rev. Genet. 41, 147–168 10.1146/annurev.genet.41.110306.130134 [DOI] [PubMed] [Google Scholar]
- Richards T. A., Dacks J. B., Campbell S. A., Blanchard J. L., Foster P. G., Mcleod R., Roberts C. W. (2006). Evolutionary origins of the eukaryotic shikimate pathway: gene fusions, horizontal gene transfer, and endosymbiotic replacements. Eukaryot. Cell 5, 1517–1531 10.1128/EC.00106-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saier M. H., Tran C. V., Barabote R. D. (2006). TCDB: the transporter classification database for membrane transport protein analyses and information. Nucleic Acids Res. 34, D181–D186 10.1093/nar/gkj001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saier M. H., Yen M. R., Noto K., Tamang D. G., Elkan C. (2009). The transporter classification database: recent advances. Nucleic Acids Res. 37, D274–D278 10.1093/nar/gkn862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato M., Mogi Y., Nishikawa T., Miyamura S., Nagumo T., Kawano S. (2009). The dynamic surface of dividing cyanelles and ultrastructure of the region directly below the surface in Cyanophora paradoxa. Planta 229, 781–791 10.1007/s00425-008-0872-4 [DOI] [PubMed] [Google Scholar]
- Sawada Y., Toyooka K., Kuwahara A., Sakata A., Nagano M., Saito K., Hirai M. Y. (2009). Arabidopsis bile acid:sodium symporter family protein 5 is involved in methionine-derived glucosinolate biosynthesis. Plant Cell Physiol. 50, 1579–1586 10.1093/pcp/pcp058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schafer G., Heber U., Heldt H. W. (1977). Glucose transport into spinach chloroplasts. Plant Physiol. 60, 286. 10.1104/pp.60.2.286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheibe R. (2004). Malate valves to balance cellular energy supply. Physiol. Plant 120, 21–26 10.1111/j.0031-9317.2004.0222.x [DOI] [PubMed] [Google Scholar]
- Scheibe R., Backhausen J. E., Emmerlich V., Holtgrefe S. (2005). Strategies to maintain redox homeostasis during photosynthesis under changing conditions. J. Exp. Bot. 56, 1481–1489 10.1093/jxb/eri181 [DOI] [PubMed] [Google Scholar]
- Schleiff E., Eichacker L. A., Eckart K., Becker T., Mirus O., Stahl T., Soll J. (2003). Prediction of the plant beta-barrel proteome: a case study of the chloroplast outer envelope. Protein Sci. 12, 748–759 10.1110/ps.0237503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlichting R., Bothe H. (1993). The cyanelles (organelles of a low evolutionary scale) possess a phosphate-translocator and a glucose-carrier in cyanophora-paradoxa. Bot. Acta 106, 428–434 [Google Scholar]
- Schlichting R., Zimmer W., Bothe H. (1990). Exchange of metabolites in cyanophora-paradoxa and its cyanelles. Bot. Acta 103, 392–398 [Google Scholar]
- Schmitz-Esser S., Linka N., Collingro A., Beier C. L., Neuhaus H. E., Wagner M., Horn M. (2004). ATP/ADP translocases: a common feature of obligate intracellular amoebal symbionts related to Chlamydiae and Rickettsiae. J. Bacteriol. 186, 683–691 10.1128/JB.186.3.683-691.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneidereit J., Häusler R. E., Fiene G., Kaiser W. M., Weber A. P. M. (2006). Antisense repression reveals a crucial role of the plastidic 2-oxoglutarate/malate translocator DiT1 at the interface between carbon and nitrogen metabolism. Plant J. 45, 206–224 10.1111/j.1365-313X.2005.02594.x [DOI] [PubMed] [Google Scholar]
- Schulz B., Frommer W. B., Flügge U. I., Hummel S., Fischer K., Willmitzer L. (1993). Expression of the triose phosphate translocator gene from potato is light-dependent and restricted to green tissues. Mol. Gen. Genet. 238, 357–361 10.1007/BF00291994 [DOI] [PubMed] [Google Scholar]
- Schunemann D., Borchert S., Flügge U. I., Heldt H. W. (1993). Adp/Atp translocator from pea root plastids – comparison with translocators from spinach-chloroplasts and pea leaf mitochondria. Plant Physiol. 103, 131–137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith A. M., Zeeman S. C., Smith S. M. (2005). Starch degradation. Annu. Rev. Plant Biol. 56, 73–98 10.1146/annurev.arplant.56.032604.144257 [DOI] [PubMed] [Google Scholar]
- Soll J., Bölter B., Wagner R., Hinnah S. C. (2000). …response: the chloroplast outer envelope: a molecular sieve? Trends Plant Sci. 5, 137–138 10.1016/S1360-1385(00)01579-X [DOI] [PubMed] [Google Scholar]
- Steiner J. M., Löffelhardt W. (2002). Protein import into cyanelles. Trends Plant Sci. 7, 72–77 10.1016/S1360-1385(01)02179-3 [DOI] [PubMed] [Google Scholar]
- Stitt M., Aprees T. (1979). Capacities of pea-chloroplasts to catalyze the oxidative pentose-phosphate pathway and glycolysis. Phytochemistry 18, 1905–1911 10.1016/S0031-9422(00)82700-4 [DOI] [Google Scholar]
- Streatfield S. J., Weber A., Kinsman E. A., Häusler R. E., Li J. M., Post-Beittenmiller D., Kaiser W. M., Pyke K. A., Flügge U. I., Chory J. (1999). The phosphoenolpyruvate/phosphate translocator is required for phenolic metabolism, palisade cell development, and plastid-dependent nuclear gene expression. Plant Cell 11, 1609–1621 10.1105/tpc.11.9.1609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan T. D., Kaneko Y. (1995). The maize brittle1 gene encodes amyloplast membrane polypeptides. Planta 196, 477–484 10.1007/BF00203647 [DOI] [PubMed] [Google Scholar]
- Sullivan T. D., Strelow L. I., Illingworth C. A., Phillips R. L., Nelson O. E. (1991). Analysis of maize brittle-1 alleles and a defective suppressor-mutator-induced mutable allele. Plant Cell 3, 1337–1348 10.1105/tpc.3.12.1337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K., Peterson D., Peterson N., Stecher G., Nei M., Kumar S. (2011). MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 10.1093/molbev/msr121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taniguchi M., Taniguchi Y., Kawasaki M., Takeda S., Kato T., Sato S., Tahata S., Miyake H., Sugiyama T. (2002). Identifying and characterizing plastidic 2-oxoglutarate/malate and dicarboxylate transporters in Arabidopsis thaliana. Plant Cell Physiol. 43, 706–717 10.1093/pcp/pcf109 [DOI] [PubMed] [Google Scholar]
- Titus S. A., Moran R. G. (2000). Retrovirally mediated complementation of the glyB phenotype – cloning of a human gene encoding the carrier for entry of folates into mitochondria. J. Biol. Chem. 275, 36811–36817 10.1074/jbc.M005163200 [DOI] [PubMed] [Google Scholar]
- Tjaden J., Mohlmann T., Kampfenkel K., Henrichs G., Neuhaus H. E. (1998). Altered plastidic ATP/ADP-transporter activity influences potato (Solanum tuberosum L.) tuber morphology, yield and composition of tuber starch. Plant J. 16, 531–540 10.1046/j.1365-313x.1998.00317.x [DOI] [Google Scholar]
- Tjaden J., Winkler H. H., Schwoppe C., Van Der Laan M., Mohlmann T., Neuhaus H. E. (1999). Two nucleotide transport proteins in Chlamydia trachomatis, one for net nucleoside triphosphate uptake and the other for transport of energy. J. Bacteriol. 181, 1196–1202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyra H. M., Linka M., Weber A. P. M., Bhattacharya D. (2007). Host origin of plastid solute transporters in the first photosynthetic eukaryotes. Genome Biol. 8, R212. 10.1186/gb-2007-8-10-r212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vignais P. V., Douce R., Lauquin G. J., Vignais P. M. (1976). Binding of radioactively labeled carboxyatractyloside, atractyloside and bongkrekic acid to Adp translocator of potato mitochondria. Biochim. Biophys. Acta 440, 688–696 10.1016/0005-2728(76)90051-7 [DOI] [PubMed] [Google Scholar]
- Viola R., Nyvall P., Pedersen M. (2001). The unique features of starch metabolism in red algae. Proc. Biol. Sci. 268, 1417–1422 10.1098/rspb.2001.1644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voll L., Häusler R. E., Hecker R., Weber A., Weissenbock G., Fiene G., Waffenschmidt S., Flügge U. I. (2003). The phenotype of the Arabidopsis cue1 mutant is not simply caused by a general restriction of the shikimate pathway. Plant J. 36, 301–317 10.1046/j.1365-313X.2003.01889.x [DOI] [PubMed] [Google Scholar]
- Wachter A., Wolf S., Steininger H., Bogs J., Rausch T. (2005). Differential targeting of GSH1 and GSH2 is achieved by multiple transcription initiation: implications for the compartmentation of glutathione biosynthesis in the Brassicaceae. Plant J. 41, 15–30 10.1111/j.1365-313X.2004.02269.x [DOI] [PubMed] [Google Scholar]
- Weber A., Flügge U.-I. (2002). Interaction of cytosolic and plastidic nitrogen metabolism in plants. J. Exp. Bot. 53, 865–874 10.1093/jexbot/53.370.865 [DOI] [PubMed] [Google Scholar]
- Weber A., Menzlaff E., Arbinger B., Gutensohn M., Eckerskorn C., Flügge U. I. (1995). The 2-oxoglutarate malate translocator of chloroplast envelope membranes – molecular-cloning of a transporter containing a 12-helix motif and expression of the functional protein in yeast-cells. Biochemistry 34, 2621–2627 10.1021/bi00008a028 [DOI] [PubMed] [Google Scholar]
- Weber A., Servaites J. C., Geiger D. R., Kofler H., Hille D., Groner F., Hebbeker U., Flügge U. I. (2000). Identification, purification, and molecular cloning of a putative plastidic glucose translocator. Plant Cell 12, 787–801 10.1105/tpc.12.5.787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber A. P. M., Fischer K. (2007). Making the connections-the crucial role of metabolite transporters at the interface between chloroplast and cytosol. FEBS Lett. 581, 2215–2222 10.1016/j.febslet.2007.02.010 [DOI] [PubMed] [Google Scholar]
- Weber A. P. M., Linka M., Bhattacharya D. (2006). Single, ancient origin of a plastid metabolite translocator family in plantae from an endomembrane-derived ancestor. Eukaryot. Cell 5, 609–612 10.1128/EC.5.3.609-612.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber A. P. M., Linka N. (2011). Connecting the plastid: transporters of the plastid envelope and their role in linking plastidial with cytosolic metabolism. Annu. Rev. Plant Biol. 62, 53–77 10.1146/annurev-arplant-042110-103903 [DOI] [PubMed] [Google Scholar]
- Weber A. P. M., Oesterhelt C., Gross W., Bräutigam A., Imboden L. A., Krassovskaya I., Linka N., Truchina J., Schneidereit J., Voll H., Voll L. M., Zimmermann M., Jamai A., Riekhof W. R., Yu B., Garavito R. M., Benning C. (2004). EST-analysis of the thermo-acidophilic red microalga Galdieria sulphuraria reveals potential for lipid A biosynthesis and unveils the pathway of carbon export from rhodoplasts. Plant Mol. Biol. 55, 17–32 10.1007/s11103-004-0376-y [DOI] [PubMed] [Google Scholar]
- Weber A. P. M., Osteryoung K. W. (2010). From endosymbiosis to synthetic photosynthetic life. Plant Physiol. 154, 593–597 10.1104/pp.110.161216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber A. P. M., Schwacke R., Flügge U.-I. (2005). Solute transporters of the plastid envelope membrane. Annu. Rev. Plant Biol. 56, 133–164 10.1146/annurev.arplant.56.032604.144201 [DOI] [PubMed] [Google Scholar]
- Weber A. P. M., Von Caemmerer S. (2010). Plastid transport and metabolism of C3 and C4 plants – comparative analysis and possible biotechnological exploitation. Curr. Opin. Plant Biol. 13, 257–265 10.1016/j.pbi.2010.01.007 [DOI] [PubMed] [Google Scholar]
- Weise S. E., Weber A. P. M., Sharkey T. D. (2004). Maltose is the major form of carbon exported from the chloroplast at night. Planta 218, 474–482 10.1007/s00425-003-1128-y [DOI] [PubMed] [Google Scholar]
- Whelan S., Goldman N. (2001). A general empirical model of protein evolution derived from multiple protein families using a maximum-likelihood approach. Mol. Biol. Evol. 18, 691–699 [DOI] [PubMed] [Google Scholar]
- Winkler H. H., Neuhaus H. E. (1999). Non-mitochondrial ATP transport. Trends Biochem. Sci. 24, 64–68 10.1016/S0968-0004(98)01334-6 [DOI] [PubMed] [Google Scholar]
- Wolfe G. R., Cunningham F. X., Durnford D., Green B. R., Gantt E. (1994). Evidence for a common origin of chloroplasts with light-harvesting complexes of different pigmentation. Nature 367, 566–568 10.1038/367566a0 [DOI] [Google Scholar]
- Xiong J., Fischer W. M., Inoue K., Nakahara M., Bauer C. E. (2000). Molecular evidence for the early evolution of photosynthesis. Science 289, 1724–1730 10.1126/science.289.5485.1724 [DOI] [PubMed] [Google Scholar]