Abstract
Receptor-like kinases (RLK) are among the largest gene families encoded by plant genomes. Common structural features of plant RLKs are an extracellular ligand binding domain, a membrane spanning domain, and an intracellular protein kinase domain. The largest subfamily of plant RLKs is characterized by extracellular leucine-rich repeat (LRR-RLK) structures that are known biochemical modules for mediating ligand binding and protein–protein interactions. In the frame of the Arabidopsis Functional Genomics Network initiative of the German Research Foundation (DFG) we have conducted a comprehensive survey for and functional characterization of LRR-RLKs potentially implicated in Arabidopsis thaliana immunity to microbial infection. Arabidopsis gene expression patterns suggested an important role of this class of proteins in biotic stress adaptation. Detailed biochemical and physiological characterization of the brassinosteroid insensitive 1-associated receptor kinase 1 (BAK1) revealed brassinolide-independent roles of this protein in plant immunity, in addition to its well-established function in plant development. The LRR-RLK BAK1 has further been shown to form heteromeric complexes with various other LRR-RLKs in a ligand-dependent manner, suggesting a role as adapter or co-receptor in plant receptor complexes. Here, we review the current status of BAK1 and BAK1-interacting LRR-RLKs in plant immunity.
Keywords: plant innate immunity, LRR-RLKs, receptor complexes, BAK1
Introduction
Plant receptor-like kinases (RLKs) belong to the monophyletic interleukin-1 receptor-associated kinase (IRAK) or RLK/Pelle family (Shiu et al., 2004). 600 family members make this family one of the largest in Arabidopsis thaliana (Lehti-Shiu et al., 2009). Likewise, sequencing of the genomes of rice, poplar, soybean, or potato has revealed the presence of large RLK families in these plants. RLKs are commonly built of N-terminal ligand binding domains and C-terminal serine/threonine protein kinase domains (Morillo and Tax, 2006). Many RLKs are located in the plasma membrane. In these cases, transmembrane domains flanked by extra- and intra-cellular juxtamembrane domains separate the ligand sensor/protein interaction domains and protein kinase domains. The current mechanistic mode of action of RLK proteins comprises ligand binding-induced conformational switches within the RLK proteins that trigger downstream signaling events subsequently activating a signal-specific cellular program (Chinchilla et al., 2009). As numerous RLKs share conserved structural features within the extracellular domains, they can be grouped into protein subfamilies (Shiu et al., 2004). A. thaliana encodes ∼235 RLKs with extracytoplasmic leucine-rich repeat (LRR) domains (LRR-RLK), which is the largest RLK subfamily in this plant (Lehti-Shiu et al., 2009). Forward and reverse genetic approaches have revealed various physiological functions of Arabidopsis LRR-RLKs (Morillo and Tax, 2006). Brassinosteroid insensitive 1 (BRI1), the receptor for the plant steroid hormone, brassinolide (BL), constitutes one of the best studied plant LRR-RLK, and was shown to regulate stem elongation, vascular differentiation, seed size, fertility, flowering time, and senescence (Li et al., 2002; Nam and Li, 2002; Wang et al., 2005) in a hormone-dependent manner. Binding of brassinolide to an island domain that folds back between LRR repeat 21 and 22 was suggested to provide a docking platform for the formation of heteromeric complexes with another LRR-RLK, BAK1 (BRI1-associated receptor kinase 1). Another subset of plant LRR-RLKs has been shown to function as pattern recognition receptors mediating the recognition of microbial surface structures (pathogen or microbe-associated molecular patterns, PAMPs/MAMPs) and plant innate immunity to microbial infection (Nürnberger and Kemmerling, 2006). For example, Arabidopsis FLS2 (Flagellin Sensing 2) and EFR (EF-Tu Receptor) sense bacterial flagellin and elongation factor EF-Tu, and thereby confer basal immunity to microbial pathogens displaying the respective cognate ligand (Gomez-Gomez and Boller, 2000; Zipfel et al., 2006). The large number of LRR-RLK that are encoded by the A. thaliana genome and the proven role of FLS2 in plant immunity have prompted us to undertake a systematic survey for additional LRR-RLKs that mediate plant–pathogen encounters (Kemmerling et al., 2007).
A Transcriptomics-Based Search for LRR-RLKs with Putative Roles in Plant Defense to Microbial Infection
Responses to abiotic or biotic stimuli in plants have often been reported to be associated with increased accumulation of transcripts encoding proteins that are required for initiation and/or execution of adaptive physiological programs. A preferred technology to identify such proteins is a genome-based analysis of stimulus-induced alterations within the entire transcriptome of a whole organism, an organ, individual cell layers, or even single cells (Kilian et al., 2007). The AtGenExpress initiative funded by the German Research Foundation (DFG) aimed at establishing transcriptome patterns for various developmental stages, tissues, and adaptive responses to abiotic and biotic stresses. Contributing partner labs agreed upon using the same A. thaliana ecotype Col-0 seed stock, comparable conditions for plant growth and procedures for plant treatment, such as infections that were representative of and informative for the respective plant research communities. Likewise, procedures for data handling, normalization of raw data and statistical analyses followed standards shared by all participating groups.
Within the frame of this program, we analyzed microbial infection or microbial pattern-induced transcript accumulation in infected or infiltrated Arabidopsis Col-0 leaves. Experiments conducted included treatments with bacterial patterns flagellin (flg22; Felix et al., 1999), lipopolysaccharide (LPS; Dow et al., 2000), or HrpZ (Lee et al., 2001), the Phytophthora parasitica-derived protein NLP (Fellbrich et al., 2002; Qutob et al., 2006; Ottmann et al., 2009). In addition, plants were infected with different strains of the bacterial pathogen, Pseudomonas syringae pv. tomato (virulent strain DC3000; avirulent strain DC3000 AvrRpm1; non-pathogenic strain DC3000 hrcC−) or P. syringae pv. phaseolicola (Qutob et al., 2006; Kemmerling et al., 2007). Experimental details and information on protocols and materials used are found at The Arabidopsis Information Resource (TAIR; www.arabidopsis.org).
The ATH1 gene array (Affymetrix©) contains oligonucleotide sequences representing 221 LRR-RLK encoding Arabidopsis genes. We have analyzed transcript accumulation of these genes for the experimental set-ups as described above. Table 1 shows those LRR-RLK genes of which expression was enhanced by the respective treatments. In total, a subset of 49 LRR-RLK genes showed significantly increased (more than twofold) transcript accumulation upon one or more treatments. It is important to note, that individual LRR-RLK genes did not show stimulus-specific gene expressions, but were expressed upon various treatments (Postel et al., 2010). This is consistent with the view that plants mount generic defenses against infection rather than microbe or pattern-specific immune responses. Among those LRR-RLKs found to be expressed upon microbial infection or treatment were a number of those that are indeed implicated in plant immunity. These genes encode for example the pattern recognition receptors FLS2 and EFR, as well as the flagellin-induced LRR-RLK FRK1 (He et al., 2006; Zipfel et al., 2006; Table 1). Strikingly, numerous LRR-RLKs encoding proteins previously implicated in developmental programs were found to be expressed upon infection, suggesting a hitherto undetected role of these proteins in plant immunity in addition to their well-established role in plant development. Such proteins comprise members of the somatic embryogenesis receptor kinase family (SERK1, SERK2, SERK3/BAK1, SERK4/BKK1; Albrecht et al., 2008), HAESA (has a role in floral organ abscission; Jinn et al., 2000), and brassinolide receptors BRL1, BRL3 (Cano-Delgado et al., 2004). Altogether, identification of receptor kinases implicated in brassinolide perception and signaling raised the question whether or not the plant hormone, brassinolide, is required for plant immunity.
Table 1.
Microarray expression analysis of Arabidopsis LRR-RLK genes.
| AGI code | Name | Fold induction | AGI code | Name | Fold induction |
|---|---|---|---|---|---|
| At1g05700 | 9.8 | At2g19190 | FRK1 | 102.8 | |
| At1g09970 | RLK7/IKU2 | 6.9 | At2g24130 | 2 | |
| At1g12460 | 2.1 | At2g31880 | SOBIR1 | 3.7 | |
| At1g14390 | 2.1 | At3g02880 | 3.7 | ||
| At1g16670 | 3.7 | At3g09010 | 6.3 | ||
| At1g17750 | PEPR2 | 5.7 | At3g13380 | BRL3 | 4.8 |
| At1g34210 | SERK2 | 2.5 | At3g28450 | 2.4 | |
| At1g34420 | 3.2 | At3g47580 | 2.9 | ||
| At1g35710 | 3.5 | At4g08850 | 4.4 | ||
| At1g51790 | 9.3 | At4g10390 | 4.2 | ||
| At1g51800 | IOS1 | 24.8 | At4g28490 | HAESA | 3.5 |
| At1g51820 | 32.9 | At4g33430 | BAK1 | 3.6 | |
| At1g51850 | 263.7 | At4g39270 | 7.2 | ||
| At1g51860 | 149.7 | At5g01950 | 2.9 | ||
| At1g51890 | 30.4 | At5g15730 | 3.5 | ||
| At1g53430 | 3.6 | At5g20480 | EFR | 30.3 | |
| At1g55610 | BRL1 | 2.3 | At5g25930 | 10.5 | |
| At1g56120 | 6.7 | At5g42440 | 4.2 | ||
| At1g66830 | 9.9 | At5g46330 | FLS2 | 2.2 | |
| At1g69270 | RPK1 | 2.6 | At5g48380 | BIR1 | 3.1 |
| At1g71830 | SERK1 | 3.1 | At5g53320 | 8.6 | |
| At1g73080 | PEPR1 | 4.1 | At5g54590 | 4.6 | |
| At1g74360 | 34.1 | At5g59680 | 13.8 | ||
| At2g02220 | PSKR1 | 10.9 | At5g65240 | 2.5 | |
| At2g13790 | SERK4 | 3.2 |
Maximum fold inductions of 49 LRR-RLK genes induced after PAMP or pathogen treatment.
BAK1 Controls the Extent of Necrotic Cell Death in Infected Plants in a Brassinolide-Independent Manner
Brassinolides (BL) regulate plant growth, differentiation, and development (Vert et al., 2005). Hormone binding to the ectopic domain of BRI1 results in the establishment of a docking site for heteromeric complex formation with BAK1 (Hothorn et al., 2011; She et al., 2011). Infection-induced transcript accumulation of four members of the SERK family initiated analyses on a potential role of BAK1 in Arabidopsis innate immunity to bacterial infection. Reverse genetic approaches using various bak1 loss-of-function alleles in infection experiments with the virulent bacterial pathogen, P. syringae pv. tomato strain DC3000, or the fungal pathogen, Alternaria brassicicola, yielded substantially enhanced disease symptom formation on the mutants relative to those observed in wild-type plants (Kemmerling et al., 2007). Symptom development in bak1 mutants was characterized by extensive spreading of necrotic areas beyond the sites of infection. Growth rates of fungal pathogens A. brassicicola or Botrytis cinerea, but not of various bacterial (P. syringae pv. tomato) strains were increased in bak1 mutants. These findings suggested that accelerated cell death facilitated growth of necrotrophic pathogens, but not of (hemi)biotrophic pathogens (Kemmerling et al., 2007). A pathophysiological investigation of the bak1bkk1 (serk3serk4) double mutant revealed increased levels of necrosis and susceptibility to fungal infection, suggesting partial functional redundancy of these two proteins in plant immunity to microbial infection (He et al., 2007; Albrecht et al., 2008). In sum, these findings demonstrate a role of BAK1 in controlling infection-induced cell death.
BAK1 gene expression in infected plants suggested a role as first messenger of the plant hormone BL in evoking plant immunity and cell death control (Kemmerling et al., 2007; Chinchilla et al., 2009; Postel et al., 2010). However, treatment with BL prior to infection assays did not complement the susceptibility to fungal infection observed in bak1 mutants. Importantly, growth defects in bak1 mutants that are due to deficiencies in BL sensing were rescued by BL treatment. Moreover, other Arabidopsis genotypes deficient in BL biosynthesis or BL sensitivity were not more susceptible to fungal infection. Likewise, transcriptome responses to BL treatment and bacterial or fungal infection were very dissimilar (Kemmerling et al., 2007). Altogether, these findings indicate a BL-independent function of BAK1 in plant immunity programs that is separable from the BL-dependent activity of BAK1 in plant development.
BAK1 is Required for the Functionality of Plant Pattern Recognition Receptors of the LRR-RLK Type
Activation of plant basal immunity to host-adapted (virulent) pathogens as well as plant immunity to host non-adapted (non-virulent) microbes is based upon recognition of microbe-derived molecular surface structures (patterns) through plant pattern recognition receptors (Nürnberger et al., 2004; Jones and Dangl, 2006; Boller and Felix, 2009; Postel and Kemmerling, 2009). Two major types of plant pattern recognition receptors have been identified, including LRR-RLKs and RLKs with ectopic domains composed of bacterial lysin-motifs (LysM-RLK). Typical pattern recognition receptors of the LRR-RLK type comprise Arabidopsis sensors for bacterial flagellin (FLS2) or bacterial elongation factor Tu (EFR). Our genome-wide survey for LRR-RLKs of which expression is triggered by P. syringae-derived flagellin (flg22; Felix et al., 1999) revealed that flg22 treatment indeed resulted in increased levels of BAK1 transcripts. Since bak1 mutants showed wild-type levels of flg22 binding, a role of BAK1 in flagellin binding was ruled out (Chinchilla et al., 2007). However, subsequent analyses of flagellin-induced cellular responses revealed partial impairment of all responses in different bak1 genotypes. Thus, BAK1 is an element of flagellin-induced signaling subsequently leading to the activation of inducible plant immune responses. Notably, when exogenously administered, BL did not rescue flagellin response deficiencies in bak1 genotypes which is in agreement with the aforementioned BL-independent role of BAK1 in plant immunity-associated programs, such as basal immunity or infection-induced cell death (Chinchilla et al., 2007, 2009).
Ligand-induced interaction of BRI1 and BAK1 in brassinolide-dependent plant development can serve as a paradigm for the co-operation of heteromeric LRR-RLKs. Indeed, flg22 treatment of Arabidopsis seedlings resulted within a few seconds in heteromeric complex formation between the two LRR-RLKs (Chinchilla et al., 2007). It was further shown that BAK1 kinase activity was required for flg22-inducible plant responses, and that BAK1 likely phosphorylated itself as well as FLS2 within 15 s upon flg22 treatment (Schulze et al., 2010; Schwessinger et al., 2011). Likewise, EF-Tu responses were reduced in bak1 genotypes (Chinchilla et al., 2007), and the EF-Tu receptor EFR and BAK1 were shown to interact physically in a ligand-dependent manner (Schwessinger et al., 2011). Altogether, these findings suggest that BAK1 functions as an adapter protein in complexes with various LRR-RLKs (BRI1, FLS2, EFR) and that ligand-specific heteromerization of BAK1 with different LRR-RLKs gives rise to the activation of a ligand-specific plant response (Chinchilla et al., 2009; Postel et al., 2010). Likewise, SERK4/BKK1, a close homolog of BAK1 (Albrecht et al., 2008) has recently been shown to act in tandem with BAK1 thereby facilitating full activation of plant immunity to infection (Roux et al., 2011). Phosphorylation by BAK1 of U-Box E3-ubiquitin ligases PUB12 and PUB13 activates proteasome-dependent degradation of FLS2, suggesting that BAK1 may not only be involved in pattern signaling, but may also determine temporary desensitization of the system and PRR turnover (Lu et al., 2011).
Recently, a novel bak1 mutant allele was isolated that carries a point mutation in the kinase domain (Schwessinger et al., 2011). This mutant allele was impaired in PAMP responsiveness, but lacked the cell death phenotypes and BL-specific deficiencies in plant growth. The mechanistic basis of the observed functional differences is currently unclear. Nevertheless, this novel mutant allele will prove valuable in future attempts to unravel how signal specificity is maintained in three different BAK1-dependent physiological programs.
LysM-domain RLKs mediate the recognition of microbial patterns including fungal chitin (Miya et al., 2007) or bacterial peptidoglycan (our unpublished findings) and immunity to fungal or bacterial infection, respectively. However, chitin or peptidoglycan-inducible responses were unaltered in bak1 genotypes relative to those observed in wild-type lines, suggesting that plant pattern recognition receptors of the LysM-RLK type do not require BAK1 activity.
BAK1 is a Target of Immune Suppression by Bacterial Effectors
PAMP-triggered immunity (PTI) is an efficient layer of immunity to infection by host non-adapted pathogens. In turn, host-adapted pathogens have acquired means to strive on host plants by suppressing PTI (Nürnberger et al., 2004). Bacterial, fungal, or oomycete phytopathogens produce multiple effector proteins many of which are translocated to the host cells to suppress host immunity. P. syringae pv. tomato produces two effectors AvrPto and AvrPtoB, both of which have been shown to suppress PTI triggered by bacterial flagellin (He et al., 2006). Elucidation of the 3D-structure revealed that the tertiary structure of AvrPto is similar to that of protein kinase inhibitors (Xing et al., 2007). Shan et al. (2008) therefore suggested that interruption of PTI might already occur at the level of pattern sensing through FLS2/BAK1 interaction. Over-expression of AvrPto and subsequent co-immunoprecipitation experiments showed interaction of the effector with FLS2 and BAK1, suggesting that PTI was suppressed through interference with the receptor complex (Xiang et al., 2008). Newer studies of this group show that FLS2 but not BAK1 might be the direct target of AvrPto (Xiang et al., 2011). Another study provided evidence that AvrPto interacted preferentially with the kinase domain of BAK1, but not with that of FLS2 (Shan et al., 2008). The same study also showed that AvrPto interfered with flagellin-induced heterodimerization of BAK1 and FLS2. While the structural requirements of the RLKs for interaction with the bacterial effector AvrPto remain elusive, these findings indicate that one mechanism by which bacterial effector-mediated suppression of basal immunity is brought about is the interference with pattern recognition receptor complex function. It remains to be shown whether interaction of AvrPto with the adapter LRR-RLK BAK1, which interacts with several pattern recognition receptors (FLS2, EFR, and likely others) and therefore constitutes a key node in PTI activation, is the preferred immunosuppressive mode of action of this effector.
BIR1 is a Negative Regulator of Immunity-Associated BAK1 Function
As constitutive activation of BAK1-dependent immune responses is likely detrimental to plant growth, negative regulatory control mechanisms for plant immune programs have been suggested (Chinchilla et al., 2009; Mazzotta and Kemmerling, 2011). A screen for BAK1-interacting proteins has been performed that revealed BAK1-interacting RLK (BIR1) as a potential interaction partner of BAK1 in planta (Gao et al., 2009). Phenotypes associated with loss of BIR1 included spontaneous cell death that was dependent on salicylic acid and thereby resembled hypersensitive cell death (HR) observed during effector-triggered immunity (ETI). Double mutants with known mutants in the ETI signaling cascade as eds1, pad4, and ndr1 show partial suppression of the bir1 cell death phenotype supporting its involvement in ETI control. Indeed, bir1-1 plants showed enhanced resistance to virulent races of the biotrophic oomycete, Hyaloperonospora arabidopsidis, suggesting that BAK1 serves a negative regulatory role in ETI. The authors of this study suggested a model in which BAK1/BIR1 complexes guard ETI immune receptors in the resting state (at times when there is no infection) thereby preventing untimely activation of ETI and hypersensitive cell death. Upon infection with an avirulent pathogen, manipulation by microbial effectors of unknown host plant targets leads to a release of immune receptors from the BAK1/BIR1 dock thereby activating HR and subsequent execution of ETI (Gao et al., 2009). Very recently, an Arabidopsis calcium-dependent phospholipid-binding protein (BON1) was identified that interacts physically with BIR1 and BAK1 in planta and, as BIR1, is phosphorylated by BAK1 in vitro (Wang et al., 2011). bon1 mutant phenotypes enhanced bir1 mutant cell death and ETI phenotypes, indicating that BIR1, BON1, and BAK1 might form a tripartite complex as a negative control element for the activation of ETI.
BAK1 Interacts with LRR-RLKs AtPEPR1 and AtPEPR2
Our genome-wide transcriptomics-based survey for LRR-RLKs revealed that PAMP treatment and bacterial infection resulted in accumulation of transcripts encoding the LRR-RLK, AtPEPR1, and its close homolog AtPEPR2 (Table 1). Both constitute functionally redundant LRR-RLKs that mediate recognition of plant peptides AtPEP1 and AtPEP2 (Huffaker et al., 2006; Yamaguchi et al., 2006, 2010; Krol et al., 2010). AtPEPs are plant-derived peptides that are produced upon wounding, PAMP treatment, or microbial infection (Yamaguchi and Huffaker, 2011), and AtPEP1 treatment of Arabidopsis plants triggers innate immune responses and enhanced resistance to Pythium irregulare infection (Huffaker et al., 2006; Yamaguchi et al., 2006). AtPEPR1 and AtPEPR2 were shown to interact physically with BAK1 (Postel et al., 2010; Schulze et al., 2010). Likewise, AtPEP1 and AtPEP2-inducible responses did not only require AtPEPR1 and AtPEPR2, but also BAK1 (Krol et al., 2010; Schulze et al., 2010) suggesting again a role of BAK1 as an adapter protein in LRR-RLK receptor function. The AtPEPR1/AtPEPR2/BAK1 receptor complex is therefore likely to function mechanistically similar to the FLS2/BAK1 or EFR/BAK1 complexes in plant immunity. A major difference between these complexes is that the latter are assembled upon perception of a microbial pattern, whereas the AtPEPR1/AtPEPR2/BAK1 receptor complex is formed upon perception of plant-derived AtPEP1 and AtPEP2. These peptides are thought to represent so-called danger signals that are produced upon host damage inflicted by bacterial infection (Boller and Felix, 2009). Physiologically, these signals are believed to potentiate PAMP-inducible plant responses (Huffaker and Ryan, 2007; Yamaguchi and Huffaker, 2011) thereby resembling animal cytokines mediating activation of animal innate immune responses, such as inflammasome activation.
Conclusion
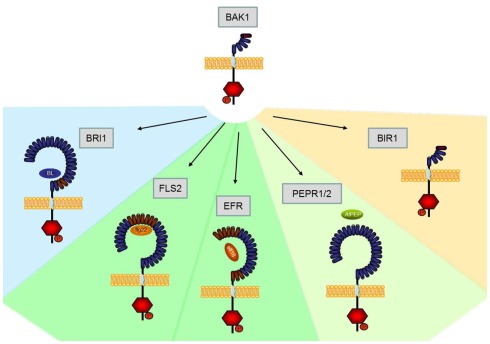
Transcriptome-based identification of receptor kinases potentially implicated in different physiological programs (development, innate immunity) has paved the way for the functional analysis of plant receptor proteins with dual functions. For example, research over the past several years has provided ample evidence that BAK1 serves both brassinolide-dependent roles in plant development and brassinolide-independent functions in plant immunity. Mechanistically, functionality of BAK1 is brought about by heteromeric complex formation with other LRR-RLKs that mediate regulatory activities of BAK1 in different physiological programs. Figure 1 provides an overview of the interactions of BAK1 and its implications in plant development and immunity. Importantly, BAK1 can function as both positive and negative regulatory component. For example, positive regulatory functions of BAK1 comprise stimulus-induced complex formations with LRR-RLKs BRI1, FLS2, EFR, or AtPEPR1/AtPEPR2. In turn, negative regulatory functions of BAK1 comprise constitutive interactions with LRR-RLKs, such as BIR1. Importantly, BAK1 appears to be a control element of virtually all elements of the plant immune system comprising PTI, ETI, and the control of infection-induced necrotic cell death. One of the major challenges of the future will be to unravel at the molecular level how signal specificity within BAK1-dependent physiological programs is determined.
Figure 1.
Schematic overview of Arabidopsis LRR-RLKs interacting with BAK1. While currently BRI1 is the only known BAK1-interacting LRR-RLK involved in developmental processes (blue area), several other RLKs are involved in plant innate immunity (PTI, green area; ETI orange area).
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This work was supported by the Deutsche Forschungsgemeinschaft (DFG Nu 70/4-1, 70/4-2, 70/4-3, Nu/Ke 70/7-1, SFB446).
References
- Albrecht C., Russinova E., Kemmerling B., Kwaaitaal M., de Vries S. C. (2008). Arabidopsis somatic embryogenesis receptor kinase proteins serve brassinosteroid-dependent and -independent signaling pathways. Plant Physiol. 148, 611–619 10.1104/pp.108.123216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boller T., Felix G. (2009). A renaissance of elicitors: perception of microbe-associated molecular patterns and danger signals by pattern-recognition receptors. Annu. Rev. Plant Biol. 60, 379–406 10.1146/annurev.arplant.57.032905.105346 [DOI] [PubMed] [Google Scholar]
- Cano-Delgado A., Yin Y., Yu C., Vafeados D., Mora-Garcia S., Cheng J. C., Nam K. H., Li J., Chory J. (2004). BRL1 and BRL3 are novel brassinosteroid receptors that function in vascular differentiation in Arabidopsis. Development 131, 5341–5351 10.1242/dev.01403 [DOI] [PubMed] [Google Scholar]
- Chinchilla D., Shan L., He P., de Vries S., Kemmerling B. (2009). One for all: the receptor-associated kinase BAK1. Trends Plant Sci. 14, 535–541 10.1016/j.tplants.2009.08.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chinchilla D., Zipfel C., Robatzek S., Kemmerling B., Nürnberger T., Jones J. D., Felix G., Boller T. (2007). A flagellin-induced complex of the receptor FLS2 and BAK1 initiates plant defence. Nature 448, 497–500 10.1038/nature05999 [DOI] [PubMed] [Google Scholar]
- Dow M., Newman M.-A., von Roepenack E. (2000). The induction and modulation of plant defense responses by bacterial lipopolysaccharides. Annu. Rev. Phytopathol. 38, 241–261 10.1146/annurev.phyto.38.1.241 [DOI] [PubMed] [Google Scholar]
- Felix G., Duran J. D., Volko S., Boller T. (1999). Plants have a sensitive perception system for the most conserved domain of bacterial flagellin. Plant J. 18, 265–276 10.1046/j.1365-313X.1999.00265.x [DOI] [PubMed] [Google Scholar]
- Fellbrich G., Romanski A., Varet A., Blume B., Brunner F., Engelhardt S., Felix G., Kemmerling B., Krzymowska M., Nürnberger T. (2002). NPP1, a Phytophthora-associated trigger of plant defense in parsley and Arabidopsis. Plant J. 32, 375–390 10.1046/j.1365-313X.2002.01454.x [DOI] [PubMed] [Google Scholar]
- Gao M., Wang X., Wang D., Xu F., Ding X., Zhang Z., Bi D., Cheng Y. T., Chen S., Li X., Zhang Y. (2009). Regulation of cell death and innate immunity by two receptor-like kinases in Arabidopsis. Cell Host Microbe 6, 34–44 10.1016/j.chom.2009.05.019 [DOI] [PubMed] [Google Scholar]
- Gomez-Gomez L., Boller T. (2000). FLS2: an LRR receptor-like kinase involved in the perception of the bacterial elicitor flagellin in Arabidopsis. Mol. Cell 5, 1003–1011 10.1016/S1097-2765(00)80265-8 [DOI] [PubMed] [Google Scholar]
- He K., Gou X., Yuan T., Lin H., Asami T., Yoshida S., Russell S. D., Li J. (2007). BAK1 and BKK1 Regulate brassinosteroid-dependent growth and brassinosteroid-independent cell-death pathways. Curr. Biol. 17, 1109–1115 10.1016/j.cub.2007.05.036 [DOI] [PubMed] [Google Scholar]
- He P., Shan L., Lin N.-C., Martin G., Kemmerling B., Nürnberger T., Sheen J. (2006). Specific bacterial suppressors of MAMP signaling upstream of MAPKKK in Arabidopsis innate immunity. Cell 125, 563–575 10.1016/j.cell.2006.03.045 [DOI] [PubMed] [Google Scholar]
- Hothorn M., Belkhadir Y., Dreux M., Dabi T., Noel J. P., Wilson I. A., Chory J. (2011). Structural basis of steroid hormone perception by the receptor kinase BRI1. Nature 474, 467–471 10.1038/nature10153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huffaker A., Pearce G., Ryan C. A. (2006). An endogenous peptide signal in Arabidopsis activates components of the innate immune response. Proc. Natl. Acad. Sci. U.S.A. 103, 10098–10103 10.1073/pnas.0603727103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huffaker A., Ryan C. A. (2007). Endogenous peptide defense signals in Arabidopsis differentially amplify signaling for the innate immune response. Proc. Natl. Acad. Sci. U.S.A. 104, 10732–10736 10.1073/pnas.0703343104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jinn T. L., Stone J. M., Walker J. C. (2000). HAESA, an Arabidopsis leucine-rich repeat receptor kinase, controls floral organ abscission. Genes Dev. 14, 108–117 [PMC free article] [PubMed] [Google Scholar]
- Jones J. D., Dangl J. L. (2006). The plant immune system. Nature 444, 323–329 10.1038/nature05286 [DOI] [PubMed] [Google Scholar]
- Kemmerling B., Schwedt A., Rodriguez P., Mazzotta S., Frank M., Qamar S. A., Mengiste T., Betsuyaku S., Parker J. E., Mussig C., Thomma B. P., Albrecht C., de Vries S. C., Hirt H., Nürnberger T. (2007). The BRI1-associated kinase 1, BAK1, has a brassinolide-independent role in plant cell-death control. Curr. Biol. 17, 1116–1122 10.1016/j.cub.2007.05.046 [DOI] [PubMed] [Google Scholar]
- Kilian J., Whitehead D., Horak J., Wanke D., Weinl S., Batistic O., D’Angelo C., Bornberg-Bauer E., Kudla J., Harter K. (2007). The AtGenExpress global stress expression data set: protocols, evaluation and model data analysis of UV-B light, drought and cold stress responses. Plant J. 50, 347–363 10.1111/j.1365-313X.2007.03052.x [DOI] [PubMed] [Google Scholar]
- Krol E., Mentzel T., Chinchilla D., Boller T., Felix G., Kemmerling B., Postel S., Arents M., Jeworutzki E., Al-Rasheid K. A., Becker D., Hedrich R. (2010). Perception of the Arabidopsis danger signal peptide 1 involves the pattern recognition receptor AtPEPR1 and its close homologue AtPEPR2. J. Biol. Chem. 285, 13471–13479 10.1074/jbc.M109.097394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J., Klüsener B., Tsiamis G., Stevens C., Neyt C., Tampakaki A. P., Panopoulos N. J., Noller J., Weiler E. W., Cornelis G. R., Mansfield J. W., Nürnberger T. (2001). HrpZ(Psph) from the plant pathogen Pseudomonas syringae pv. phaseolicola binds to lipid bilayers and forms an ion-conducting pore in vitro. Proc. Natl. Acad. Sci. U.S.A. 98, 289–294 10.1073/pnas.121064698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehti-Shiu M. D., Zou C., Hanada K., Shiu S. H. (2009). Evolutionary history and stress regulation of plant receptor-like kinase/pelle genes. Plant Physiol. 150, 12–26 10.1104/pp.108.134353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J., Wen J., Lease K. A., Doke J. T., Tax F. E., Walker J. C. (2002). BAK1, an Arabidopsis LRR receptor-like protein kinase, interacts with BRI1 and modulates brassinosteroid signaling. Cell 110, 213–222 10.1016/S0092-8674(02)00812-7 [DOI] [PubMed] [Google Scholar]
- Lu D., Lin W., Gao X., Wu S., Cheng C., Avila J., Heese A., Devarenne T. P., He P., Shan L. (2011). Direct ubiquitination of pattern recognition receptor FLS2 attenuates plant innate immunity. Science 332, 1439–1442 10.1126/science.1199098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazzotta S., Kemmerling B. (2011). Pattern recognition in plant innate immunity. J. Plant Pathol. 93, 7–17 [Google Scholar]
- Miya A., Albert P., Shinya T., Desaki Y., Ichimura K., Shirasu K., Narusaka Y., Kawakami N., Kaku H., Shibuya N. (2007). CERK1, a LysM receptor kinase, is essential for chitin elicitor signaling in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 104, 19613–19618 10.1073/pnas.0705147104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morillo S. A., Tax F. E. (2006). Functional analysis of receptor-like kinases in monocots and dicots. Curr. Opin. Plant Biol. 9, 460–469 10.1016/j.pbi.2006.07.009 [DOI] [PubMed] [Google Scholar]
- Nam K. H., Li J. (2002). BRI1/BAK1, a receptor kinase pair mediating brassinosteroid signaling. Cell 110, 203–212 10.1016/S0092-8674(02)00814-0 [DOI] [PubMed] [Google Scholar]
- Nürnberger T., Brunner F., Kemmerling B., Piater L. (2004). Innate immunity in plants and animals: striking similarities and obvious differences. Immunol. Rev. 198, 249–266 10.1111/j.0105-2896.2004.0119.x [DOI] [PubMed] [Google Scholar]
- Nürnberger T., Kemmerling B. (2006). Receptor protein kinases – pattern recognition receptors in plant immunity. Trends Plant Sci. 11, 519–522 10.1016/j.tplants.2006.09.005 [DOI] [PubMed] [Google Scholar]
- Ottmann C., Luberacki B., Kufner I., Koch W., Brunner F., Weyand M., Mattinen L., Pirhonen M., Anderluh G., Seitz H. U., Nürnberger T., Oecking C. (2009). A common toxin fold mediates microbial attack and plant defense. Proc. Natl. Acad. Sci. U.S.A. 106, 10359–10364 10.1073/pnas.0907587106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Postel S., Kemmerling B. (2009). Plant systems for recognition of pathogen-associated molecular patterns. Semin. Cell Dev. Biol. 20, 1025–1031 10.1016/j.semcdb.2009.06.002 [DOI] [PubMed] [Google Scholar]
- Postel S., Küfner I., Beuter C., Mazzotta S., Schwedt A., Borlotti A., Halter T., Kemmerling B., Nürnberger T. (2010). The multifunctional leucine-rich repeat receptor kinase BAK1 is implicated in Arabidopsis development and immunity. Eur. J. Cell Biol. 89, 169–174 10.1016/j.ejcb.2009.11.001 [DOI] [PubMed] [Google Scholar]
- Qutob D., Kemmerling B., Brunner F., Küfner I., Engelhardt S., Gust A. A., Luberacki B., Seitz H. U., Stahl D., Rauhut T., Glawischnig E., Schween G., Lacombe B., Watanabe N., Lam E., Schlichting R., Scheel D., Nau K., Dodt G., Hubert D., Gijzen M., Nürnberger T. (2006). Phytotoxicity and innate immune responses induced by Nep1-like proteins. Plant Cell 18, 3721–3744 10.1105/tpc.106.044180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roux M., Schwessinger B., Albrecht C., Chinchilla D., Jones A., Holton N., Malinovsky F. G., Tör M., de Vries S., Zipfel C. (2011). The Arabidopsis leucine rich-repeat receptor-like kinases BAK1/SERK3 and BKK1/SERK4 are required for innate immunity to hemibiotrophic and biotrophic pathogens. Plant Cell 23, 2240–2255 10.1105/tpc.111.084301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulze B., Mentzel T., Jehle A. K., Mueller K., Beeler S., Boller T., Felix G., Chinchilla D. (2010). Rapid heteromerization and phosphorylation of ligand-activated plant transmembrane receptors and their associated kinase BAK1. J. Biol. Chem. 285, 9444–9451 10.1074/jbc.M109.096842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwessinger B., Roux M., Kadota Y., Ntoukakis V., Sklenar J., Jones A., Zipfel C. (2011). Phosphorylation-dependent differential regulation of plant growth, cell death, and innate immunity by the regulatory receptor-like kinase BAK1. PLoS Genet. 7, e1002046. 10.1371/journal.pgen.1002046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shan L., He P., Li J., Heese A., Peck S. C., Nürnberger T., Martin G. B., Sheen J. (2008). Bacterial effectors target the common signaling partner BAK1 to disrupt multiple MAMP receptor-signaling complexes and impede plant immunity. Cell Host Microbe 4, 17–27 10.1016/j.chom.2008.05.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- She J., Han Z., Kim T. W., Wang J., Cheng W., Chang J., Shi S., Yang M., Wang Z. Y., Chai J. (2011). Structural insight into brassinosteroid perception by BRI1. Nature 474, 472–476 10.1038/nature10178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiu S. H., Karlowski W. M., Pan R., Tzeng Y. H., Mayer K. F., Li W. H. (2004). Comparative analysis of the receptor-like kinase family in Arabidopsis and rice. Plant Cell 16, 1220–1234 10.1105/tpc.020834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vert G., Nemhauser J. L., Geldner N., Hong F., Chory J. (2005). Molecular mechanisms of steroid hormone signaling in plants. Annu. Rev. Cell Dev. Biol. 21, 177–201 10.1146/annurev.cellbio.21.090704.151241 [DOI] [PubMed] [Google Scholar]
- Wang X., Goshe M. B., Soderblom E. J., Phinney B. S., Kuchar J. A., Li J., Asami T., Yoshida S., Huber S. C., Clouse S. D. (2005). Identification and functional analysis of in vivo phosphorylation sites of the Arabidopsis brassinosteroid-insensitive1 receptor kinase. Plant Cell 17, 1685–1703 10.1105/tpc.105.033076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z., Meng P., Zhang X., Ren D., Yang S. (2011). BON1 interacts with the protein kinases BIR1 and BAK1 in modulation of temperature-dependent plant growth and cell death in Arabidopsis. Plant J. 67, 1081–1093 10.1111/j.1365-313X.2011.04655.x [DOI] [PubMed] [Google Scholar]
- Xiang T., Zong N., Zhang J., Chen J., Chen M., Zhou J. M. (2011). BAK1 is not a target of the Pseudomonas syringae effector AvrPto. Mol. Plant Microbe Interact. 24, 100–107 10.1094/MPMI-04-10-0096 [DOI] [PubMed] [Google Scholar]
- Xiang T., Zong N., Zou Y., Wu Y., Zhang J., Xing W., Li Y., Tang X., Zhu L., Chai J., Zhou J. M. (2008). Pseudomonas syringae effector AvrPto blocks innate immunity by targeting receptor kinases. Curr. Biol. 18, 74–80 10.1016/j.cub.2007.12.020 [DOI] [PubMed] [Google Scholar]
- Xing W., Zou Y., Liu Q., Liu J., Luo X., Huang Q., Chen S., Zhu L., Bi R., Hao Q., Wu J. W., Zhou J. M., Chai J. (2007). The structural basis for activation of plant immunity by bacterial effector protein AvrPto. Nature 449, 243–247 10.1038/nature06109 [DOI] [PubMed] [Google Scholar]
- Yamaguchi Y., Huffaker A. (2011). Endogenous peptide elicitors in higher plants. Curr. Opin. Plant Biol. 14, 351–357 10.1016/j.pbi.2011.05.001 [DOI] [PubMed] [Google Scholar]
- Yamaguchi Y., Huffaker A., Bryan A. C., Tax F. E., Ryan C. A. (2010). PEPR2 is a second receptor for the Pep1 and Pep2 peptides and contributes to defense responses in Arabidopsis. Plant Cell 22, 508–522 10.1105/tpc.109.068874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi Y., Pearce G., Ryan C. A. (2006). The cell surface leucine-rich repeat receptor for AtPep1, an endogenous peptide elicitor in Arabidopsis, is functional in transgenic tobacco cells. Proc. Natl. Acad. Sci. U.S.A. 103, 10104–10109 10.1073/pnas.0603729103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zipfel C., Kunze K., Chinchilla D., Caniard A., Jones J. D. G., Boller T., Felix G. (2006). Perception of the bacterial PAMP EF-Tu by the Arabidopsis receptor kinase EFR restricts Agrobacterium-mediated transformation. Cell 125, 749–760 10.1016/j.cell.2006.03.037 [DOI] [PubMed] [Google Scholar]