Abstract
Cullin/RING ubiquitin ligases (CRL) comprise the largest subfamily of ubiquitin ligases. CRLs are involved in cell cycle regulation, DNA replication, DNA damage response (DDR), development, immune response, transcriptional regulation, circadian rhythm, viral infection, and protein quality control. One of the main functions of CRLs is to regulate the DDR, a fundamental signaling cascade that maintains genome integrity. In this review, we will discuss the regulation of CRL ubiquitin ligases and their roles in control of the DDR.
Keywords: DNA damage, cullin, Cdc25A, claspin, Cdt1, cyclin D1, ubiquitin ligase, protein degradation
Introduction
Our genome is under constant attack by exogenous DNA insulting agents, such as UV light and chemical carcinogens, and by endogenous metabolic products, such as reactive oxidative species. In order to prevent genome instability, a hallmark of cancers, eukaryotic cells have evolved several protective mechanisms of the DNA damage response (DDR), including DNA replication and cell cycle arrest, and DNA repair initiation (Harper and Elledge, 2007; Negrini et al., 2010). During a DDR, cascades of phosphorylation events are initiated by a family of PI3-like family kinases, mainly an ataxia telangiectasia mutated (ATM), and ATM and Rad3-related (ATR) kinases (Harper and Elledge, 2007; Negrini et al., 2010). Recent studies, however, have indicated that ubiquitination also plays important roles in the DDR (Harper and Elledge, 2007; Messick and Greenberg, 2009; Negrini et al., 2010; Silverman et al., 2012).
Ubiquitin is an 8-kDa protein and can be covalently conjugated to other proteins via an isopeptide linkage between its C-terminal glycine and a primary amino group of its substrates, which is usually from a lysine side chain (Pickart, 2004; Hannah and Zhou, 2009). Ubiquitination is catalyzed by three enzymes, including a ubiquitin activating enzyme (E1), a ubiquitin conjugating enzyme (E2), and a ubiquitin ligase (E3) (Pickart, 2004; Hannah and Zhou, 2009). The ubiquitination machinery can conjugate a single ubiquitin to one lysine residue of substrates (called mono-ubiquitination), multiple single ubiquitin molecules to multiple lysine residues (called multi-ubiquitination), or multiple ubiquitins in a chain (called poly-ubiquitination) to substrates. Mono-ubiquitination and multi-ubiquitination play important roles in endocytosis, DNA repair, immune response, and transcriptional regulation, etc. Poly-ubiquitination via surface lysine residues of ubiquitin, such as Lysine-48 (K48) or Lysine-11 (K11) often leads to protein proteolysis through the 26S proteasome (Pickart, 2004; Pickart and Eddins, 2004). However, protein poly-ubiquitination does not necessarily drive protein turnover. Poly-ubiquitin chains via Lysine-63 residue (K63) of ubiquitin and linear poly-ubiquitin chains appear to play non-proteolytic functions and are involved in DNA repair, NF-κB activation, and Ras localization and signaling, etc. (Pickart, 2004; Jura et al., 2006; Yan et al., 2009, 2010; Xu et al., 2010).
The human genome contains two E1 with distinctive functions (Chiu et al., 2007; Jin et al., 2007; Pelzer et al., 2007; Lee et al., 2011), dozens of E2s and hundreds of E3 ligases which determine the specificity of substrates for ubiquitination (Pickart, 2004; Pickart and Eddins, 2004). Thus far, more than 600 ubiquitin ligases have been identified in the human genome, although many of them have not been linked to any substrates or biological activities. Based on their domain characteristics, ubiquitin ligases can be separated into two sub-groups, HECT domain E3 ligases and RING finger domain E3 ligases (Pickart, 2004). The cullin/RING ubiquitin ligases (CRL) comprise the largest subfamily of RING finger-containing E3s (Petroski and Deshaies, 2005; Sarikas et al., 2011). CRLs are modular ubiquitin ligases (Petroski and Deshaies, 2005; Sarikas et al., 2011). CRLs are involved in cell cycle regulation, DNA replication, DDR, development, immune response, transcriptional regulation, circadian rhythm, viral infection, and protein quality control (Petroski and Deshaies, 2005). This review will focus on the interplay between human CRL ubiquitin ligases and the DDR. We will discuss molecular features of CRLs and how they participate in the DDR by targeting DDR-regulatory proteins for ubiquitination.
Introduction of CRL Ubiquitin Ligases
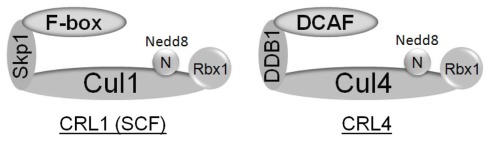
Cullin/RING ubiquitin ligases consist of multiple subunits (Petroski and Deshaies, 2005; Sarikas et al., 2011), including a RING finger protein (Rbx1 or Rbx2) that recruits an activated ubiquitin E2 enzyme, a scaffold subunit (cullin family proteins), and a substrate receptor that recognizes substrates. Many CRL ubiquitin ligases contain additional linker proteins, such as Skp1 in SCF (also called CRL1) and DDB1 in CRL4 (Figure 1). Recent studies indicate that Rbx1 associates with all cullins, except for Cul5, which associates specifically with Rbx2 (Kamura et al., 2004; Huang et al., 2009), suggesting distinct roles of the two Rbx homologs in human.
Figure 1.
Architecture of CRL1 (SCF) and CRL4 ubiquitin ligases. SCF ubiquitin ligases contain Cul1, Rbx1, Skp1, and F-box proteins which function as substrate receptors; whereas CRL4 has Cul4, Rbx1, DDB1, and DCAF proteins as substrate receptors. Both SCF and CRL4 can be activated by neddylation. F-box and DCAF proteins are substrate specificity factors.
The first CRL E3 ligase, named SCF complex (stands for Skp1/Cdc53 or cullin/F-box), was identified from budding yeast, Saccharomyces cerevisiae (Bai et al., 1996; Willems et al., 1996; Feldman et al., 1997; Skowyra et al., 1997; Verma et al., 1997). This finding initiated with studies of proteasome-dependent turnover of Sic1, an inhibitor of cyclins and cyclin-dependent kinases (Cdks) in budding yeast (Bai et al., 1996; Willems et al., 1996; Feldman et al., 1997; Skowyra et al., 1997; Verma et al., 1997). Degradation and ubiquitination of Sic1 at the G1/S transition were compromised in Cdc4, Cdc53 (Cul1 homolog in yeast), and Skp1 mutants in budding yeast, which suggested their common roles in Sic1 turnover (Bai et al., 1996; Feldman et al., 1997; Skowyra et al., 1997; Verma et al., 1997). The ubiquitination function of Cdc53 was observed in yeast Cln2 stability control as well (Willems et al., 1996). Biochemical evidence demonstrated that Cdc53 functions together with Cdc4 and Skp1 as a ubiquitin ligase (SCFCdc4) to catalyze the poly-ubiquitination of Sic1 both in vitro and in vivo (Feldman et al., 1997; Skowyra et al., 1997). Within the SCF complex, Cdc4, the F-box protein, is a substrate receptor that recognizes Sic1 within its WD-40 motif and interacts with Skp1 through its F-box domain. Skp1 is the linker protein that mediates association of Cdc4 with the scaffold cullin protein, Cdc53. Rbx1 (also called ROC1 or Hrt1 in yeast), a RING finger protein in the complex, was found to regulate the stability of yeast Sic1, human HIF1α, and other substrates (Lyapina et al., 1998; Kamura et al., 1999; Ohta et al., 1999; Seol et al., 1999; Skowyra et al., 1999; Tan et al., 1999). Structurally, Rbx1 binds to the C-terminal domain of yeast Cdc53 or human Cul1, and an E2 enzyme, Cdc34 (Zheng et al., 2002b; Zimmerman et al., 2010; Duda et al., 2011).
Regulation of CRL Ubiquitin Ligases
The activity of CRL ubiquitin ligases is regulated by NEDD8, a small ubiquitin-like protein (Deshaies et al., 2010). Like ubiquitin, NEDD8 can be conjugated to other proteins, especially cullins (Xirodimas et al., 2004; Watson et al., 2006; Jones et al., 2008). The conjugation of NEDD8, named neddylation, is catalyzed by NEDD8-specific E1, E2, and E3 (Dye and Schulman, 2007). The general consensus is that neddylation of cullins is required for activation of CRL ubiquitin ligases. However, untimely neddylation of cullins could drive destruction of substrate receptors via auto-ubiquitination (Cope and Deshaies, 2006). Therefore, the neddylation of cullins is counteracted by the deneddylation activity of a multifunctional protein complex, the COP9 signalosome (CSN) (Cope and Deshaies, 2003; Serino and Deng, 2003; Wei and Deng, 2003; Wei et al., 2008).
COP9 signalosome was initially found to be involved in plant photo morphogenesis and was later identified as a conserved complex in eukaryotes (Cope and Deshaies, 2003; Serino and Deng, 2003; Wei and Deng, 2003; Wei et al., 2008). CSN is comprised of eight subunits, which are CSN1–8 in order of decreasing molecular weight. The CSN complex participates in multiple biological events, including transcriptional regulation, cell division, and development, etc. (Tateishi et al., 2001; Lykke-Andersen et al., 2003; Panattoni et al., 2008). Part of the multi-functionality of CSN is linked with the neddylation system, with its isopeptidase activity to remove NEDD8 conjugation. This deneddylation activity is attributed to the metalloprotease motif of CSN5 but the whole CSN complex is required for the reaction (Cope et al., 2002). Indeed, conditional silencing of CSN5 in HEK293 cells increased the neddylation of cullins. Consequently, expression of multiple F-box proteins, but not the cullins, was decreased (Cope and Deshaies, 2006). The reduced expression of F-box proteins depends on Cul1 and the proteasome, further supporting an auto-ubiquitination and self-destruction mechanism (Cope and Deshaies, 2006). These data explain why accumulation of CRL substrates has been observed in cells where either CSN or the NEDD8 conjugation system is inactivated (Tateishi et al., 2001; Lykke-Andersen et al., 2003; Cope and Deshaies, 2006; Panattoni et al., 2008; Choo et al., 2011). It is clear that expression of some substrate receptors is not affected by neddylation, however (Cope and Deshaies, 2006).
Neddylation of cullins positively regulates the E3 ligase activity of CRLs by at least three mechanisms. First, neddylation of cullins enhances their interaction with ubiquitin-activated E2. It has been suggested that neddylation of Cul1 can increase the affinity between ubiquitin-activated E2 and Rbx1 (Kawakami et al., 2001). Using fluorescence resonance energy transfer (FRET) technology, Saha and Deshaies (2008) observed that neddylation of Cul1 can enhance Cdc34 binding to SCF ubiquitin ligase.
Second, neddylation positively regulates the ubiquitination activity of CRL ubiquitin ligases by enhancing ubiquitin transfer to substrates from the active E2 site and by positioning the active E2 site closer to ubiquitin accepting sites of substrates. Neddylation has been shown to stimulate the Cdc34-dependent ubiquitination activity of SCF by more than 10-fold (Saha and Deshaies, 2008). The κcat for ubiquitin transfer is enhanced by NEDD8 conjugation (Saha and Deshaies, 2008). According to the structural analysis of the SCFSkp2 ubiquitin ligase, Cul1 holds the Skp1–Skp2 and Rbx1 subunits more than 100 Å apart (Zheng et al., 2002b). A gap of 50 Å between the active cysteine of E2 and the leucine-rich repeat of Skp2 exists even after E2 joins SCFSkp2 (Zheng et al., 2002b). This gap was confirmed by additional SCF structural studies (Wu et al., 2003; Hao et al., 2007). In these scenarios, a gap of ∼50 Å is too large for E2 to transfer ubiquitin to its substrates. The neddylation of cullins, however, induces conformational changes in both the N-terminal and the C-terminal domains of cullins (Duda et al., 2008). These conformation changes make Rbx proteins acquire more flexibility to get access to their substrates. Such conformational rearrangements remain even after deneddylation, allowing both initiation and elongation of poly-ubiquitin chains.
Third, cullin neddylation prevents CRLs from associating with CAND1, a negative regulator of CRLs (Liu et al., 2002; Zheng et al., 2002a; Hwang et al., 2003; Min et al., 2003; Oshikawa et al., 2003; Goldenberg et al., 2004). The binding of CAND1 to cullins obscures their associations with their substrate receptors, which results in the inactivation of CRL E3 ligases. CAND1 only associates with non-neddylated cullins. Therefore, the popular model is that cullin neddylation favors the dissociation of CAND1 from CRLs. NEDD8-conjugated CRLs, which are free of CAND1, possess maximal E3 activity. After deneddylation by CSN, CAND1 is able to inhibit CRLs by binding to cullins. The next neddylation cycle can then prevent CAND1 from binding and CRL activity will resume. However, a recent study from the Harper group revealed that only a small fraction of cullins is associated with CAND1, whereas the majority of Cul1 proteins form complexes with F-box proteins, the substrate receptors (Bennett et al., 2010). Therefore, at least for the SCF ubiquitin ligases, the formation of SCF complexes is driven mainly by receptor binding.
Neddylation is catalyzed by its E3 ligase, Rbx (Huang et al., 2009). Recent studies indicated that the DCN family proteins function as additional E3 ligases to assist Rbx in cullin neddylation (Kurz et al., 2005, 2008; Ma et al., 2008; Meyer-Schaller et al., 2009). Wu et al. (2011) found that human DCN1 is modified by mono-ubiquitination, which drives nuclear export of DCN1. The biological impact of mono-ubiquitination of DCN1 under physiological condition remains to be determined, however. Interestingly, expression of DCN3L, a human homolog of yeast Dcn1p, can be enhanced by UV light (Ma et al., 2008), suggesting that the DNA damage signal may enhance neddylation of some, if not all, CRL ubiquitin ligases that are important for DDR.
The CRL family is one example of ubiquitin ligases that mediate many cellular signaling events (Petroski and Deshaies, 2005). In the following sections, we will discuss the roles of individual CRL ubiquitin ligases in control of DDR.
SCFβTRCP in DNA Damage Checkpoint Control
SCFβTRCP in DDR-induced Cdc25A ubiquitination and turnover
Cdc25A is a dual phosphatase that targets phosphorylated tyrosines and serines/threonines for de-phosphorylation (Karlsson-Rosenthal and Millar, 2006; Ray and Kiyokawa, 2007, 2008; Rudolph, 2007). Removal of inhibitory phosphate on tyrosine by Cdc25A activates cyclin/Cdk kinases, mainly cyclin E/Cdk2 and cyclin A/Cdk2, and enables the G1/S transition and mitotic entry. Cdc25A has two close homologs, Cdc25B and Cdc25C. It was proposed that Cdc25B initiates the activation of cyclin B/Cdk1 and Cdc25C completes this process in the nucleus during the G2/M transition (Boutros et al., 2006). However, both Cdc25B and Cdc25C are dispensable in mouse development, except for meiosis (Chen et al., 2001a; Ferguson et al., 2005). Cdc25B and Cdc25C double knockout mice are viable, but females are sterile, as are Cdc25B knockout females (Ferguson et al., 2005). In contrast, Cdc25A is essential for early embryo development (Ray et al., 2007a). In summary, Cdc25A is an essential gene for mouse development and is one of the major regulators of cell division (Molinari et al., 2000; Zhao et al., 2002; Ray and Kiyokawa, 2007, 2008). The Cdc25A protein level is tightly regulated via transcription and proteasome degradation in a cell cycle-dependent manner (Ray and Kiyokawa, 2007).
Over-expression of Cdc25A has been observed in multiple cancers (Kristjánsdóttir and Rudolph, 2004; Boutros et al., 2006). Studies in Cdc25A knockout mice indicated that Cdc25A is a rate-limiting oncogene that restricts tumorigenesis induced by the HER2/neu–RAS oncogenic pathway (Ray et al., 2007a). Consistent with this observation, MMTV-Cdc25A transgenic mice dramatically promote murine mammary tumorigenesis in cooperation with oncogenic RAS or neu (Ray et al., 2007b). Over-expression of Cdc25A in human cells can result in aberrant mitotic events, compromised DDR and destabilized chromosomes, such as chromosomal breaks at fragile sites (Cangi et al., 2008). Therefore, it is critical to maintain appropriate amounts of Cdc25A in human cells.
Cdc25A is an unstable protein and its protein levels are mediated by phosphorylation and the ubiquitin–proteasome pathway (Bernardi et al., 2000; Mailand et al., 2000, 2002; Falck et al., 2001; Donzelli et al., 2002, 2004; Shimuta et al., 2002; Zhao et al., 2002; Busino et al., 2003; Goloudina et al., 2003; Jin et al., 2003, 2008; Sørensen et al., 2003; Xiao et al., 2003; Kasahara et al., 2010). In response to DNA damage, Cdc25A is phosphorylated by the checkpoint kinase, Chk1, and subjected to proteasome-dependent degradation (Zhao et al., 2002; Goloudina et al., 2003; Jin et al., 2003; Sørensen et al., 2003; Xiao et al., 2003; Donzelli et al., 2004). The consequence of Cdc25A turnover is persistent Cdk phosphorylation and cell cycle arrest at S-phase upon DDR (Busino et al., 2003; Jin et al., 2003). IR-induced Cdc25A turnover in interphase was shown to depend on Cul1, suggesting the involvement of the SCF E3 ligase (Donzelli et al., 2002). In a screen to identify the substrate receptor of the SCF complex, the F-box proteins, βTRCP1 and βTRCP2, two homologs of human βTRCP, were found to be interacting with Cdc25A. Knockdown of both βTRCP1 and βTRCP2 by siRNA or expression of a dominant-negative βTRCP1 blocked DDR-induced Cdc25A proteolysis (Busino et al., 2003; Jin et al., 2003). In addition, depletion of both βTRCP1 and βTRCP2 results in hyperactive Cdk2 kinase activity and radio-resistant DNA synthesis, which are characteristics of a defective intra-S-phase checkpoint (Busino et al., 2003; Jin et al., 2003).
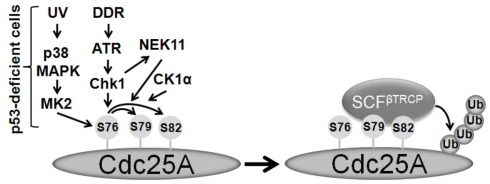
The recognition of ubiquitin substrates by F-box proteins often requires post-translational modification, usually phosphorylation, on target proteins (Petroski and Deshaies, 2005). Chk1 kinase activity is indispensible for in vitro ubiquitination of Cdc25A catalyzed by SCFβTRCP (Jin et al., 2003). Upon DDR, the Serine-296 (S296) site of Chk1 is autophosphorylated after it is phosphorylated at Serine-345 (S345) by ATR (Kasahara et al., 2010). Chk1 phosphorylated at S296 is further recognized by 14-3-3 gamma. 14-3-3 gamma not only tethers Chk1 in the nucleus, but also bridges the interaction between Chk1 and Cdc25A (Kasahara et al., 2010). Therefore, 14-3-3 gamma is important for Chk1-mediated Cdc25A turnover via the ubiquitin–proteasome pathway (Kasahara et al., 2010). Several Chk1 phosphorylation sites on Cdc25A were mapped, but only mutation on Serine-76 (S76) abolishes Cdc25A ubiquitination (Jin et al., 2003, 2008). Surprisingly, peptide scanning and mutagenesis revealed that S76 alone was not sufficient for direct binding with βTRCP (Jin et al., 2003). Adjacent to S76, there is a DSGFCLDSP sequence (residues 81–89) which resembles the classic phospho-degron motif in other βTRCP substrates (Cardozo and Pagano, 2004). Serine-88 is dispensable for SCFβTRCP-mediated Cdc25A ubiquitination (Jin et al., 2003). Although Aspartic acid-87 could mimic a phosphorylated-serine, it appears not to be involved in binding of Cdc25A to βTRCP (Jin et al., 2003). Phosphorylation of both Serine-79 (S79) and Serine-82 (S82) is required for Cdc25A ubiquitination (Jin et al., 2003). In addition, the phospho-S79 and phospho-S82 peptides are able to bind to βTRCP in vitro, indicating these two sites are central to where βTRCP recognizes Cdc25A (Jin et al., 2003). Chk1 can phosphorylate neither S79 nor S82, however. Therefore, it seems that active Chk1 phosphorylates S76 of Cdc25A in response to DNA damage, which creates a priming site for additional kinases to subsequently phosphorylate S79 and S82 residues. Phosphorylation of these residues recruits SCFβTRCP to ubiquitinate Cdc25A for turnover (Figure 2).
Figure 2.
Multiple kinases lead to the formation of phospho-degron of Cdc25A under DDR. Under DDR ATR activates Chk1 which then phosphorylates Cdc25A at S76 and turns on NEK11. Consequently, NEK11 phosphorylates Cdc25A at S82, whereas, CK1α phosphorylates both S79 and S82 of Cdc25A. In p53-deficient cells, p38MAPK and MK2 lead to the phosphorylation of S76 under UV damage. Upon the phospho-degron is created, Cdc25A is recognized and ubiquitinated by the SCFβTRCP ubiquitin ligase.
Besides Chk1, another checkpoint kinase Chk2 plays an important role in DDR-induced Cdc25A degradation (Falck et al., 2001). Chk2 has been found to phosphorylate Serine-124 (S124) of human Cdc25A, which is important for IR-induced Cdc25A turnover. Compared to S76, however, S124 phosphorylation appears to play a minor role in DDR-induced degradation of Cdc25A (Jin et al., 2003). Moreover, Chk2 can not phosphorylate S76 of Cdc25A in vitro efficiently (Jin et al., 2008). These data may explain why deletion of Chk2 in HCT116 cells has no impact on IR-induced Cdc25A turnover (Jin et al., 2008). This further emphasizes the primary importance of Chk1 phosphorylation at S76 in control of Cdc25A under DDR. The discrepancy in the role of Chk2 in DNA damage-induced Cdc25A degradation may depend on cell types and different DNA damage signals. Further studies are needed to resolve this issue.
In addition to Chk1 and Chk2, other protein kinases are involved in Cdc25A proteolysis in unsynchronized or stressed cells as well (Reinhardt et al., 2007; Kang et al., 2008; Melixetian et al., 2009; Honaker and Piwnica-Worms, 2010; Myer et al., 2011; Piao et al., 2011). In p53-deficient cells, Cdc25A proteolysis depends on p38 MAP kinase (p38MAPK), and MAPKAP Kinase-2 (MK2), but not Chk1 (Reinhardt et al., 2007). Apparently, Chk1 and MK2 recognize similar phosphorylation motifs, such as the one surrounding S76 of Cdc25A (Reinhardt et al., 2007). In this case, p38MAPK and MK2 mediate a third DNA damage checkpoint pathway for cell cycle arrest and survival after DNA damage, although an intact ATR–Chk1 pathway exists in p53-deficient cells (Reinhardt et al., 2007). Therefore, p38MAPK and MK2 are potential candidates for drug targets to kill cancer cells that are defective in p53 function. However, how p53 affects the selection of checkpoint pathways in Cdc25A proteolysis has yet to be determined.
Chk1 and SCFβTRCP mediate Cdc25A turnover in the S and G2 phases of cell division. At late G1-phase, Cdc25A stability is not controlled by Chk1, although Cdc25A ubiquitination is still SCFβTRCP-dependent. Kang et al. (2008) found that GSK3β phosphorylates S76 of Cdc25A at late G1-phase. Interestingly, S76 phosphorylation requires prior phosphorylation at Threonine-80 (T80) by PLK3 (Kang et al., 2008). A strong correlation between GSK3β inactivation and Cdc25A over-expression was observed in some human tumors, further supporting the negative role of GSK3β in Cdc25A production (Kang et al., 2008). However, under DDR, checkpoint kinases, such as Chk1 and MK2, are in charge of S76 phosphorylation of Cdc25A (Jin et al., 2003; Reinhardt et al., 2007).
S82 of Cdc25A is a primary phosphorylation site in the phospho-degron motif of Cdc25A. Several kinases have been reported to phosphorylate S82 to trigger Cdc25A turnover (Melixetian et al., 2009; Honaker and Piwnica-Worms, 2010; Piao et al., 2011). In a screen with a shRNA library, Melixetian et al. (2009) identified NIMA (never in mitosis gene A)-related kinase 11 (NEK11) as a gene that is involved in the G2/M checkpoint. Silencing NEK11 prevents ubiquitin-dependent proteolysis of Cdc25A, in both unsynchronized and DNA damaged cells. NEK11 phosphorylates S82 of Cdc25A during DDR. Moreover, NEK11 is a downstream effective kinase of the checkpoint pathway and is activated via phosphorylation at Serine-273 of NEK11 by Chk1. Thus far, there is no evidence to show whether NEK11 is capable of phosphorylating S79 of Cdc25A, although phosphorylation of S79 is important for S82 phosphorylation of Cdc25A and Cdc25A recognition by βTRCP. In a separate study, Honaker and Piwnica-Worms (2010) reported that casein kinase 1 alpha (CK1α) phosphorylates Cdc25A at both S79 and S82 during interphase and under genotoxic stress. In the same study, the authors claimed that NEK11 could not phosphorylate S82 of Cdc25A in vitro. However, it is unclear whether any priming kinase was included in the in vitro kinase assay. Paradoxically, Melixetian et al. (2009) found that NEK11 can phosphorylate S82 of Cdc25A in vitro, but the phosphorylation was independent of S76 phosphorylation. In contrast to the Honaker and Piwnica-Worms (2010) report, Piao et al. (2011) observed CK1ε directly phosphorylated S82 without any prior phosphorylation of Cdc25A, and silencing CK1ε stabilized cellular Cdc25A in HEK293 cells. These conflicting results reveal the complexity of phosphorylation-regulated Cdc25A proteolysis (Figure 2). They may also reflect the fact that Cdc25A stability is regulated by various signaling pathways and that different genetic background may select distinctive kinases to create a phospho-degron motif for Cdc25A. Further experiments are needed to clarify these discrepancies. Nevertheless, the consensus is that S76 is a priming site for generation of a phospho-degron sequence surrounding S79 and S82.
In addition to residues surrounding S82, other serine residues also contribute to DDR-induced Cdc25A degradation. PLK3 phosphorylates Serines-513 and -519 of Cdc25A in vitro (Myer et al., 2011). The role of PLK3 in Cdc25A stability was studied in PLK3 knockout mice where the G1/S checkpoint was found to be defective (Myer et al., 2011). PLK3-mediated Cdc25A stabilization did not translate into a significant increase in tumorigenesis in vivo (Myer et al., 2011), however, suggesting that PLK3-mediated Cdc25A turnover may not be a major pathway to control in vivo functions of Cdc25A.
SCFβTRCP in claspin ubiquitination and turnover during DDR recovery
While SCFβTRCP-mediated Cdc25A degradation is critical for the execution of the DNA damage checkpoint, SCFβTRCP-controlled Claspin turnover is important for checkpoint recovery. Claspin associates with the replication fork and is one of the major checkpoint mediators for Chk1 activation by ATR in response to replication stress (Chini and Chen, 2003; Kumagai and Dunphy, 2003; Lee et al., 2003; Lin et al., 2004). Claspin is a periodically synthesized protein and its expression peaks at S/G2 and is then degraded after cells transit into mitosis. In unsynchronized cells, the mitotic degradation of Claspin depends on SCFβTRCP, which binds to Claspin through a conserved DpSGxxpS phospho-degron motif (Cardozo and Pagano, 2004). Phosphorylation of these two serine residues is necessary for ubiquitination of Claspin. Alanine substitutions of these serine residues abolish the association of Claspin with SCFβTRCP and the poly-ubiquitination of Claspin (Mailand et al., 2006; Mamely et al., 2006; Peschiaroli et al., 2006). Apparently, PLK1 is the kinase that phosphorylates these serines, because PLK1 is necessary for in vitro ubiquitination and siRNA depletion of PLK1 suppresses mitotic Claspin degradation (Mailand et al., 2006; Mamely et al., 2006).
In G2, Claspin quickly accumulates after UV damage, suggesting a post-transcriptional regulatory mechanism. The level of Claspin correlates well with activation of Chk1, represented by phosphorylation of Serine-317 of Chk1 (Chini et al., 2006). Chk1 phosphorylates Threonine-916 of Claspin and is important to maintain Claspin protein stability (Bennett and Clarke, 2006; Chini and Chen, 2006; Chini et al., 2006). Although Chk1 affects Claspin turnover, apparently, the stability control of Clapsin is independent of ATR (Chini and Chen, 2006; Chini et al., 2006; Yang et al., 2008). Conversely, Claspin is also a stabilizer of Chk1 protein (Yang et al., 2008). During the recovery from replication stress, Chk1 activity is gradually diminished, concomitantly with the reduction of Claspin level. When serine residues in the phospho-degron of Claspin are mutated into alanine, Claspin is no longer a substrate of the SCFβTRCP ubiquitin ligase (Mailand et al., 2006; Mamely et al., 2006; Peschiaroli et al., 2006). Consequently, the activation of checkpoint pathway is extended and the percentage of mitotic cells is reduced (Mailand et al., 2006; Mamely et al., 2006; Peschiaroli et al., 2006). Therefore, SCFβTRCP-dependent turnover of Claspin is critical for cells to terminate the checkpoint pathway once the damage is repaired (Mailand et al., 2006; Mamely et al., 2006; Peschiaroli et al., 2006).
In summary, the SCFβTRCP ubiquitin ligase plays important roles in multiple steps of DDR via proteolytic control of multiple substrates which functions at different steps of DDR. Obviously, distinct protein kinases have been shown to create a conserved phospho-degron motif on different substrates. Nevertheless, as the substrate receptor, βTRCP is a key player to maintain genome integrity. Consistent with this, mutations of βTRCP1 have been identified in human cancer (He et al., 2005). The inhibitory role of βTRCP1 in tumorigenesis has been established (Kudo et al., 2004; Bhatia et al., 2011), suggesting that βTRCP1 is a bona fide tumor suppressor.
CRL4Cdt2 in DNA Damage Response
DNA replication is initiated from the sequential recruitment and assembly of regulatory proteins onto specialized chromosome regions known as DNA replication origins. During the late M-phase and G1-phase, pre-replicative complex (pre-RC) needs to be assembled onto origins. Origins then become “licensed” and can further recruit DNA polymerase for replication at S-phase (Dutta and Bell, 1997; Arias and Walter, 2007). The assembly of pre-RC is a stepwise process that starts with binding of origin recognition complex (ORC) to an origin. ORC then recruits Cdc6 and Cdt1, two DNA replication licensing factors which are required for loading of MCM complex. To prevent re-replication, where origins fire more than once during a single cell cycle, it is essential to dissemble the pre-RC complex immediately after replication is initiated (Dutta and Bell, 1997; Arias and Walter, 2007). In Xenopus and mammals, Cdt1 can be inactivated through binding to its inhibitor, Geminin, or via ubiquitination-mediated proteolysis (Wohlschlegel et al., 2000; Nishitani et al., 2001; Tada et al., 2001; Arias and Walter, 2005). Inhibition of Geminin or over-expression of Cdt1 stimulates DNA re-replication (Vaziri et al., 2003; Zhu et al., 2004; Takeda et al., 2005). Cdt1 transgenic mice develop thymic lymphoblastic lymphoma in the absence of p53 (Seo et al., 2005) and over-expression of Cdt1 and Cdc6 promotes malignant events by inducing genome instability and by abrogating antitumor barriers, suggesting their oncogenic functions (Liontos et al., 2007). Over-expression of Cdt1 and/or Cdc6 has been observed in certain cancers (Karakaidos et al., 2004). Therefore, DNA replication licensing machine could be a good therapeutic target for cancer cures.
CRL4Cdt2 in DDR-induced Cdt1 ubiquitination and turnover
In the unperturbed cell cycle, Cdt1 is degraded after replication is initiated at G1-/S-phase (Nishitani et al., 2001). Walter and colleagues demonstrated that Cdt1 degradation in Xenopus egg extract requires replication initiation (Arias and Walter, 2005). Depletion of Cdc45 or RPA, which are essential for origin firing, abolished the chromatin-associated Cdt1 ubiquitination and degradation (Arias and Walter, 2005). Two E3 ligases, SCFSkp2 and CRL4Cdt2, are identified to mediate cell cycle-dependent degradation of Cdt1 (Higa et al., 2003, 2006; Li et al., 2003; Zhong et al., 2003; Hu et al., 2004; Kondo et al., 2004; Arias and Walter, 2006; Hu and Xiong, 2006; Jin et al., 2006; Kim and Kipreos, 2006; Lovejoy et al., 2006; Nishitani et al., 2006; Ralph et al., 2006; Sansam et al., 2006; Senga et al., 2006). SCFSkp2 is the main one to control Cdt1 throughout the cell cycle in a Cdk-dependent manner, whereas CRL4Cdt2 is required for Cdt1 degradation in S-phase and during DDR.
In responding to UV or γ-irradiation, Cdt1 undergoes ubiquitination, and proteasome-dependent degradation, which presumably would prevent improper new origin firing before the damaged genome is repaired (Higa et al., 2003; Hu et al., 2004). The DDR-induced ubiquitination of Cdt1 is solely dependent on CRL4Cdt2, since the absence of Skp2 does not compromise Cdt1 degradation after UV. Deletion of Cdt2 in HeLa cells induced DNA re-replication and G2/M checkpoint activation, partly due to Cdt1 accumulation (Jin et al., 2006; Sansam et al., 2006). How CRL4Cdt2 recognizes and targets Cdt1 for ubiquitination has been intensively investigated. The degron signal recognized by CRL4Cdt2 is localized at the N-terminus of Cdt1 since the N-terminal fragment behaves like the full length Cdt1 in the aspect of UV-induced degradation. Truncation mutagenesis further narrows the degron region to be within the first 10 residues, which also encompass a PCNA-interacting motif (called PIP box). There are evidences indicating the essential role of PCNA in Cdt1 degradation (Arias and Walter, 2006; Hu and Xiong, 2006; Senga et al., 2006). Depletion of PCNA or mutation on the PIP box abolishes CRL4Cdt2-mediating Cdt1 ubiquitination and proteolysis in the unperturbed S-phase or UV damaged cells (Arias and Walter, 2006; Hu and Xiong, 2006; Senga et al., 2006). The Walter group further found that a TD motif inside the PIP box and a basic amino acid at four residues downstream of the PIP box of Cdt1 are important for Cdt1 to interact with PCNA strongly and to recruit CRL4Cdt2 to the Cdt1–PCNA complex (Havens and Walter, 2009). This observation was confirmed by two individual studies (Guarino et al., 2011; Michishita et al., 2011).
Although both UV and IR can trigger CRL4Cdt2-dependent Cdt1 degradation, the phosphorylation event of Cdt1 is only observed in UV damaged cells (Kondo et al., 2004). IR-induced degradation of Cdt1 is not dependent on either ATM or ATR checkpoint pathway, whereas UV-induced degradation is sensitive to the inhibitor of ATM and ATR, caffeine (Higa et al., 2003; Kondo et al., 2004). Although Cdt1 is phosphorylated after UV irradiation, none of the phosphorylation events are located at the N-terminus, where the degron motif is localized (Kondo et al., 2004; Senga et al., 2006). However, it is still unclear why caffeine, as the inhibitor of ATM and ATR, only affects UV-mediated Cdt1 proteolysis. How ATM and ATR regulate UV-controlled Cdt1 degradation is still unclear. Further lost-of-function investigations on ATM and ATR are necessary to validate their roles in UV-triggered Cdt1 proteolysis. Moreover, whether and how the phosphorylations of Cdt1 at its C-terminus are linked to Cdt1 ubiquitination and turnover are yet to be determined. Recent study from Cook’s group indicated that p38MAPK and c-Jun N-terminal kinase (JNK) can phosphorylate Cdt1 both during unperturbed G2 phase and under stress condition (Chandrasekaran et al., 2011). These results explained why Cdt1 becomes a stable protein when cells move into G2 and mitosis.
CRL4Cdt2 in DDR-induced p21 ubiquitination and turnover
CRL4Cdt2 is a key ubiquitin ligase that mediates ubiquitination of several important cell cycle regulators in cooperation with PCNA. The Cdk inhibitor, p21 (also called WAF1 or CIP1), is a substrate of CRL4Cdt2 in DNA replication licensing control. p21 is an inhibitor of Cdks whose kinase activity is required to drive Cdc6 out of the nucleus in order to prevent DNA replication relicensing (Kim et al., 2007). p21 binds to the cyclin/Cdk2 complex to suppress its kinase activity and prevents cells from entering into S-phase. In response to different stimuli, the transcription of p21 is activated by p53, through which cell cycle progression is arrested at the G1/S transition. In addition to functioning as a cell cycle regulator, p21 also forms a complex with PCNA and polymerase δ. Over-expression of p21 disrupts the PCNA–Fen1 interaction in vivo and potentially interferes with the DNA repair process (Chen et al., 1996).
Under low dose UV irradiation, p21 undergoes proteolysis, which could potentially promote DNA repair. Two E3 ligases, SCFSkp2 and CRL4Cdt2 are implicated in regulating p21 proteolysis under DNA damage conditions (Bornstein et al., 2003; Soria et al., 2006; Abbas et al., 2008; Kim et al., 2008; Nishitani et al., 2008). It has been shown that degradation of p21 after low dose UV irradiation (<40 J/m2) depends on ATR and Skp2. The interaction between Skp2 and p21 has been detected and is further increased after DNA damage (Bendjennat et al., 2003). Mutation of all lysine residues impaired UV-induced p21 degradation and cells exhibited defects in DNA repair. This implies that p21 ubiquitination promotes its degradation to prompt the accumulation of PCNA on chromatin for DNA repair process (Bendjennat et al., 2003).
p21, like Cdt1, has a PIP box motif for PCNA interaction. It was then speculated whether p21 degradation could be regulated like Cdt1, via CRL4Cdt2-mediated ubiquitination with PCNA as co-factor. Down-regulation of Cul4, DDB2, and Cdt2 in UV-irradiated p53 null HCT116 (to eliminate the signaling from p53) inhibited efficient p21 degradation (Abbas et al., 2008). As in the case for Cdt1, p21 ubiquitination by CRL4Cdt2 requires PCNA binding via the PIP box. The association of p21 and CRL4Cdt2 was detected by co-immunoprecipitation regardless of damage signaling. However, CRL4Cdt2 can only ubiquitinate p21 that possesses a phosphomimetic mutation at Serine-114, which is the target of the GSK3β kinase stimulated by ATR (Abbas et al., 2008).
The classic E2 enzyme for CRL ubiquitin ligases is Cdc34 (Petroski and Deshaies, 2005). A recent study, however, indicated that UbcH5 may function as a priming E2 to initiate ubiquitination of IκBα, a bona fide substrate of the SCFβTRCP ubiquitin ligase (Wu et al., 2010). Given the fact that CRLs share a common RING finger protein, Rbx, it is highly possible that the E2 priming step by UbcH5 may be common to poly-ubiquitination of many, if not all substrates of CRLs. Interestingly, the Dutta group found that UbcH8 mediates p21 ubiquitination, whereas the UBE2G family of E2s cooperate with CRL4Cdt2 to polyubiquitylate Cdt1 under DDR (Shibata et al., 2011). These data, if true, suggested that substrates also play important roles in E2 selection of CRL ubiquitin ligases. Because these experiments were done with single siRNA oligo, further studies are needed to confirm the roles of these E2 enzymes. UbcH8 lacks obvious features for poly-ubiquitin chain formation. Whether other E2s, such as UbcH5, can function together with UbcH8 in poly-ubiquitin chain generation on p21 remains to be determined. Moreover, like Cdc34, UBE2G E2s contain an acidic sequence that is required for poly-ubiquitin chain formation (Petroski and Deshaies, 2005). One important question is whether UBE2G, like Cdc34, needs assistance from another E2, such as UbcH5, as a priming subunit to boost its poly-ubiquitination capability. Another critical question is whether the CRL4Cdt2 ubiquitin ligase employs Cdc34, UbcH8, or UBE2G as E2 enzyme to conjugate poly-ubiquitin chains on Set8/PR-Set7, another critical substrate of CRL4Cdt2 under DDR (Abbas et al., 2010; Centore et al., 2010; Oda et al., 2010; Jørgensen et al., 2011). Understanding these questions is important to better comprehend protein poly-ubiquitination by CRL ubiquitin ligases under DDR.
CRL4DDB2 and CRL4CSA in DNA Repair Control
Nucleotide excision repair (NER) is a versatile repair mechanism to remove the damaged DNA lesions from the genome, including UV-induced cyclobutane pyrimidine dimers (CPD), 6–4 pyrimidine–pyrimidone photoproducts, and a variety of bulky adducts (Gillet and Schärer, 2006). It is a gap-filling process executed by DNA polymerase and ligase following the excision of a 24–32 DNA base oligonucleotide, where the lesion localizes. There are two sub-types of NER, global genome repair (GGR), and transcription-coupled repair (TCR), which differ in the mechanism of damage detection. GGR occurs throughout the whole genome, whereas TCR is restricted in the active transcribing regions. Mutations in the NER pathway are responsible for several rare inheritable diseases, such as xeroderma pigmentosum (XP) and Cockayne syndrome (Bootsma et al., 1998).
CRL4DDB2 in nucleotide excision repair
Xeroderma pigmentosum patients are hypersensitive to UV and are pre-disposed to skin cancer. Cells from XP patients are defective in repair of UV-induced DNA damage (Friedberg, 1995; Friedberg et al., 1995). In XP complementation group E (XP-E), the DDB2 gene is mutated. DDB2 encodes a WD-40 repeat protein that was originally identified as one subunit of a heterodimer with DDB1 (Dualan et al., 1995). The DDB1–DDB2 heterodimer binds to damaged DNA and is involved in NER. The crystal structure of DDB2–DDB1 dimer explains its high affinity toward UV-induced DNA lesion and its central role in NER. The hairpin motif of DDB2 inserts into the minor groove of a DNA duplex and thereby flips out the pyrimidine dimer to be recognized by the binding pocket of DDB2 (Scrima et al., 2008). The DNA lesion recognition ability of DDB2 is essential for subsequent recruitment of repair proteins to the damage site. Consistent with the clinical implication of DDB2 in human disease (XP-E), DDB2 knockout mice are susceptible to UV-induced skin tumorigenesis and develop spontaneous cancers at old age (Yoon et al., 2005).
DDB1 and DDB2 are two essential subunits of the CRL4DDB2 ubiquitin ligase where DDB2 functions as the substrate receptor (Groisman et al., 2003). Thus far, the known substrates for CRL4DDB2 are histones and XPC (Sugasawa et al., 2005; El-Mahdy et al., 2006; Kapetanaki et al., 2006; Wang et al., 2006; Guerrero-Santoro et al., 2008). CRL4DDB2 is not so active due to its association with the CSN (Groisman et al., 2003). However, UV radiation can trigger the translocation of CRL4DDB2 to chromatin. The recruitment of CRL4DDB2 is critical for orchestrating the repair machinery around the damage site. Chromatin-bound CRL4DDB2 is very active, because it does not associate with the CSN and its Cul4 subunit is neddylated (Groisman et al., 2003; Takedachi et al., 2010). Apparently, UV light sends signals to separate the CSN from CRL4DDB2, because the majority of CRL4DDB2 binds to chromatin after UV radiation (Groisman et al., 2003). Some DDB2 mutants from XP-E patients are defective in DDB1 binding (Jin et al., 2006; Takedachi et al., 2010). These mutants fail to recruit the other subunits of CRL4DDB2 to lesion DNA under DDR (Hwang et al., 1998; Shiyanov et al., 1999; Jin et al., 2006; Kapetanaki et al., 2006; Takedachi et al., 2010). As a result, XPC and histones are not ubiquitinated (Kapetanaki et al., 2006; Takedachi et al., 2010). This may explain how the DNA repair machine is impaired in some XP-E patients.
XPC is a central player in the NER pathway. XPC is necessary for the assembly of NER complex surrounding the lesion (Sugasawa, 2011), whereas its DNA binding affinity is less than that of DDB2. It has been shown that the poly-ubiquitination of XPC after UV treatment depends on the CRL4DDB2 ubiquitin ligase and that ubiquitination can augment the DNA binding affinity of XPC instead of targeting it to the proteasome for degradation (Batty et al., 2000; Sugasawa et al., 2005). One important question is why poly-ubiquitination of XPC by CRL4DDB2 does not lead to XPC proteolysis?
The poly-ubiquitination of DDB2 reduces its DNA binding ability and leads to its degradation (Chen et al., 2001b; Nag et al., 2001; Matsuda et al., 2005). Silencing of Cul4A was shown to stabilize DDB2, to prolong the retention of DDB2 at UV-induced lesions, and to impair the recruitment of XPC and the subsequent removal of CPD from the genome (El-Mahdy et al., 2006). Moreover, XPC ubiquitination may accelerate DDB2 turnover (Sugasawa et al., 2009). Therefore, it seems likely that a UV-induced DNA lesion is handed over from DDB2 to XPC for DNA repair and that this process depends on the poly-ubiquitination capability of the CRL4DDB2 ubiquitin ligase (Sugasawa et al., 2009). Whether XPC ubiquitination triggers DDB2 degradation via auto-ubiquitination is still debatable. However, a recent study from Arabidopsis suggested that DET1 is the substrate receptor of CRL4 that targets DDB2 for ubiquitination and degradation (Castells et al., 2011). Therefore, it will be interesting to see whether DET1 is responsible for UV- and Cul4A-mediated DDB2 ubiquitination and proteolysis in human cells. Using a chemical inhibitor, Zhao et al. (2008) demonstrated that p38MAPK is required for UV-induced DDB2 ubiquitination and proteolysis. The same group also found that DDB2 is phosphorylated by p38MAPK after UV treatment. However, the actual phosphorylation sites of DDB2 are yet to be identified. Moreover, it is still unclear whether phosphorylation of DDB2 under UV damage plays any critical roles in DDB2 ubiquitination and degradation. It is also unknown whether p38MAPK phosphorylates DDB2 directly or via its downstream kinases, such as MK2. Answering these questions will help understand the concise mechanism of NER under DDR.
In addition to XPC, histone proteins, H2A, H3, and H4, are conjugated with a single ubiquitin by CRL4DDB2 in response to UV irradiation. In XP-E patient-derived lymphoblastoid cells, UV-induced mono-ubiquitination of H2A is impaired (Kapetanaki et al., 2006). On one hand, some DDB2 mutants lose their functions as ubiquitin ligases, because they fail to interact with DDB1 (Jin et al., 2006; Kapetanaki et al., 2006). On the other hand, the K244E mutant of DDB2 contains only residual binding ability to DNA lesions (Takedachi et al., 2010). Although K244E maintains the ubiquitination activity of CRL4DDB2, it fails to ubiquitinate histones in nucleosome context (Takedachi et al., 2010). It has been proposed that ubiquitination of histone proteins by CRL4DDB2 loosens up the condensed chromatin structure into a relaxation state that is more accessible to the repair factors (Wang et al., 2006). However, inconsistent with this hypothesis, histone ubiquitination is not sufficient to destabilize the nucleosome in vitro (Takedachi et al., 2010). Therefore, one potential function of histone ubiquitination is to recruit other DNA repair factors to lesion DNA. One such factor could be XPA whose association with lesion DNA is enhanced by the ubiquitination activity of CRL4DDB2 (Takedachi et al., 2010). However, XPA does have an obvious ubiquitin-binding motif, suggesting that additional factors might be involved in the recruitment of XPA. CRLs usually conjugate poly-ubiquitin chains on their substrates (Petroski and Deshaies, 2005). How CRL4DDB2 is regulated to conjugate mono- versus poly-ubiquitin chains on its substrates remains enigmatic.
CRL4CSA in nucleotide excision repair
Besides CRL4DDB2, the CRL4CSA ubiquitin ligase is also actively involved in DNA repair (Groisman et al., 2003). In this complex, CSA is a substrate specific factor. CSA is also a WD-40 repeat protein and has 40% sequence homology with DDB2 from the second to the fifth WD-40 repeat. Similar to DDB2, CSA directly binds to DDB1 and is able to assemble an E3 ligase complex with Cul4 and Rbx1. In response to UV damage, CSA is found associating with the stalled RNA polymerase II (RNA pol II) (Groisman et al., 2003). Similar to CRL4DDB2, the ubiquitin ligase activity of CRL4CSA is inhibited due to the binding of CSN (Groisman et al., 2003). CSB, a DNA-dependent ATPase, is the only substrate identified for CRL4CSA thus far. CSB preferentially associates with elongating or stalled RNA pol II, and is potentially involved in the elongation step of RNA synthesis. Both CSA and CSB are mutated in Cockayne syndrome patients and involved in transcription-coupled DNA repair (Li et al., 2011). CSB fibroblasts exhibit hypersensitivity to UV irradiation and a defect in resumption of RNA synthesis after damage (Bregman et al., 1996; Balajee et al., 1997). Moreover, poly-ubiquitination and degradation of stalled RNA pol II in response to UV is also impaired in CSB cells (Bregman et al., 1996). CSB is a member of the SNF2-like family, which possesses chromatin-remodeling activity. CSB is believed to push away the stalled RNA pol II or to rearrange the chromatin structure so that the repair proteins can get access to the lesion (Groisman et al., 2006). The key function of CSB is to recruit histone acetyltransferase p300, NER proteins, and the CRL4CSA ubiquitin ligase to lesion DNA (van Gool et al., 1997; Fousteri et al., 2006); whereas, CSA is required to recruit XAB2, the nucleosomal binding protein HMGN1, and TFIIS in cooperation with CSB. If the lesion cannot be repaired, CSB then initiates poly-ubiquitination and degradation of stalled RNA pol II (Svejstrup, 2003). Three hours after damage, CSB itself is eliminated via CRL4CSA-mediated poly-ubiquitination and proteasome-dependent degradation (Groisman et al., 2006). The degradation of CSB is necessary for transcription to resume at a normal rate, and is coordinated with the dissociation of the CSN from the CRL4CSA ubiquitin ligase (Groisman et al., 2003), indicating that the ubiquitination function of CRL4CSA was inhibited during DDR, but activated after DNA repair was accomplished. However, how the CSN is released from the CRL4CSA ubiquitin ligase is still unclear.
In summary, CRL4CSA and CRL4DDB2 are two important ubiquitin ligases that not only function as ubiquitin ligases to ubiquitinate their cognate substrates, but also play distinct roles in recruiting critical DNA repair factors to lesion DNA upon DDR. Multiple mutations have been identified for DDB2 and CSA from human patients. Interestingly, most of these mutants fail to form ubiquitin ligases with other subunits of CRL4 (Jin et al., 2006). As a consequence, CRL4 ubiquitin ligases cannot be recruited to damage sites to assist DNA repair. Therefore, these disease-derived mutations all affect activities of CRL4CSA and CRL4DDB2 ubiquitin ligases, further supporting their fundamental roles in NER.
SCFFbxo4 and SCFFbxo31 in Cyclin D1 Ubiquitination
Cdk kinase activity is the master regulator that controls cell cycle progression. Cdk kinase activity can be modulated via phosphorylation, through binding to the inhibitors, or by association with cyclin co-factors. The level of cyclin proteins fluctuates during the cell cycle through periodic transcriptional activation or proteolysis. Cyclin D1 is expressed in G1-phase and its presence activates Cdk2 for entrance into S-phase. To prevent cells from entering into S-phase and propagating erroneous DNA, cyclin D1 is degraded via the ubiquitin–proteasome pathway after encountering genotoxic stress (Pagano et al., 1994; Diehl et al., 1997, 1998). Cyclin D1 is over-expressed in various malignant neoplasms, suggesting its role as an oncogene (Malumbres and Barbacid, 2001). MMTV-cyclin D1 transgenic mice develop mammary adenocarcinomas, indicating its oncogenic role in breast cancer (Wang et al., 1994; Hosokawa et al., 2001). Therefore, cyclin D1 expression is well controlled during cell division. Dysregulation of cyclin D1 can lead to genomic instability (Pontano et al., 2008). Cyclin D1 is an unstable protein. Its ubiquitination and degradation depends on phosphorylation of Threonine-286 (T286), which is mediated by GSK3β kinase (Diehl et al., 1997, 1998). Four F-box proteins, including Fbxo4, Fbxw8, Fbxo31, and Skp2, have been reported to recognize cyclin D1 and mediate its ubiquitination (Yu et al., 1998; Lin et al., 2006; Okabe et al., 2006; Barbash et al., 2009; Santra et al., 2009).
When cells progress into S-phase, the T286 on cyclin D1 is phosphorylated by GSK3β. Phosphorylated T286 and surrounding residues constitute the phospho-degron motif that is recognized by Fbxo4, a specificity factor in the SCFFbxo4 ubiquitin ligase (Diehl et al., 1997, 1998; Lin et al., 2006). Thus, phosphorylated cyclin D1 at T286 is an unstable protein that is subjected to poly-ubiquitination by SCFFbxo4 and then degradation by the 26S proteosome (Lin et al., 2006). Importantly, the ubiquitination of cyclin D1 by SCFFbxo4 requires a small heat-shock protein, αB crystallin (Lin et al., 2006). Over-expression of Fbxo4 and αB crystallin can stimulate cyclin D1 ubiquitination and accelerate its degradation, whereas knockdown of either of them can block cyclin D1 ubiquitination and increase its protein stability (Lin et al., 2006). In consistent with these observations, Fbxo4 and αB crystallin expression is reduced in subset of primary human tumors that overexpress cyclin D1 (Lin et al., 2006). In addition to Fbxo4, another F-box protein Fbxw8 was also identified as an E3 ligase for cyclin D1 in HCT116 cancer cell line. Fbxw8 can associate with either Cul1 or Cul7 to assemble an SCF or SCF-like complex, respectively (Okabe et al., 2006). Fbxw8 also recognizes the degron sequences surrounding phosphorylated T286. However, the phosphorylation of T286 in this case is mediated via the MAPK pathway rather than by GSK3β in the cell context examined (Okabe et al., 2006). The access of cyclin D1 to its cognate E3 ligase is spatially regulated. Similar to Fbxo4, majority of Fbxw8 is localized in the cytoplasm during G1- and S-phase, separating away from the nucleus localized cyclin D1. In S-phase, cyclin D1 is translocated into the cytoplasm, where it is now accessible to E3 ligase for subsequent ubiquitination and proteolysis. The phosphorylation of cyclin D1 by GSK3β at T286 is required for its nuclear export (Lin et al., 2006).
In contrast to Fbxo4 and Fbxw8, Fbxo31 is specifically dedicated to damage-induced degradation of cyclin D1 (Santra et al., 2009). In response to IR or other genotoxic stress, Fbxo31 is targeted by the checkpoint kinase ATM for phosphorylation, which results in the accumulation of Fbxo31. Fbxo31 binds to the same phospho-degron motif on cyclin D1 as Fbxo4 and Fbxw8 and mutation of T286 abolishes the interaction between cyclin D1 and all three F-box proteins. However, blocking of the MAPK pathway but not GSK3β prevents this damage-induced cyclin D1 degradation (Santra et al., 2009). It should be noted that the characterization of these four F-box proteins were done in different cell lines. Whether these four E3 ligases are involved in regulating cyclin D1 simultaneously or exclusively from each other remain to be determined. The Fbxo4 knockout mice develop various tumors whose cyclin D1 expression is enhanced (Vaites et al., 2011). However, a recent study in knockout mice of these ubiquitin ligases indicated that none of these four ubiquitin ligases is required for cyclin D1 proteolysis in mouse embryonic fibroblasts (Kanie et al., 2012), further supporting cell type-specific regulation of cyclin D1 stability. Therefore, it is intriguing that how cells choose among different ubiquitin ligases reacting to different stimuli for cyclin D1 destruction if they all recognize the same phospho-degron motif. Nevertheless, loss of heterozygosity of Fbxo31 has been reported in multiple cancers (Launonen et al., 2000; Lin et al., 2001; Härkönen et al., 2005; Kumar et al., 2005; Huang et al., 2010; Kogo et al., 2011). Inactive mutations of Fbxo4 are also identified in 15% of esophageal tumors (Barbash et al., 2008). These data support the idea that Fbxo31 and Fbxo4 are tumor suppressors.
Conclusion and Prospective
Here we discuss how CRL ubiquitin ligases mediate protein ubiquitination and turnover to enforce unidirectional signal transduction in the DDR pathway. Many of the substrate receptors of CRL are still uncharacterized. Growing evidence suggests that more CRL ubiquitin ligases will be found to mediate the DDR pathway and more DDR-related proteins will be identified as ubiquitin substrates of CRLs. Thus far, the main research focus has been on dissecting the roles of CRL1 (SCF) and CRL4 ubiquitin ligases in DDR. Emerging evidence suggests that other cullin-related ubiquitin ligases are also involved in DDR (Ribar et al., 2007; Yasukawa et al., 2008; Blackford et al., 2010). Further investigation is needed to explore the functions of cullin-related ubiquitin ligases other than CRL1 and CRL4 in DDR. Moreover, several groups have reported knockout mice models of Cul4A and suggest an important role of Cul4A in guarding the genome stability (Kopanja et al., 2009; Liu et al., 2009). The homolog of Cul4A, Cul4B shares significant sequence homology with Cul4A and has some redundant functions with Cul4A (Higa et al., 2003; Hu et al., 2004). Considering the distinct knockout phenotype of Cul4A in mice and cell lines (Nishitani et al., 2001; Karakaidos et al., 2004), it is conceivable that Cul4A and Cul4B play distinct roles in genome integrity maintenance via regulating protein stability of different substrates. However, Cul4B also plays significant roles in DDR (Kerzendorfer et al., 2010). In fact, degradation of p27 and p53 seems to depend on Cul4A solely (Banks et al., 2006; Higa et al., 2006). Therefore, more investigation is required to unravel the relationship between Cul4A and Cul4B and to advance our understanding of Cul4 as genome guardian.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Dr. Linda Guarino for critical reading of the manuscript. We apologize to those whose work could not be cited due to space limitation. Jianping Jin is a Pew Scholar and is supported by a grant (AU-1711) from the Welch Foundation.
References
- Abbas T., Shibata E., Park J., Jha S., Karnani N., Dutta A. (2010). CRL4Cdt2 regulates cell proliferation and histone gene expression by targeting PR-Set7/Set8 for degradation. Mol. Cell 40, 9–21 10.1016/j.molcel.2010.09.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abbas T., Sivaprasad U., Terai K., Amador V., Pagano M., Dutta A. (2008). PCNA-dependent regulation of p21 ubiquitination and degradation via the CRL4Cdt2 ubiquitin ligase complex. Genes Dev. 22, 2496–2506 10.1101/gad.1676108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arias E. E., Walter J. C. (2005). Replication-dependent destruction of Cdt1 limits DNA replication to a single round per cell cycle in Xenopus egg extracts. Genes Dev. 19, 114–126 10.1101/gad.1255805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arias E. E., Walter J. C. (2006). PCNA functions as a molecular platform to trigger Cdt1 destruction and prevent re-replication. Nat. Cell Biol. 8, 84–90 10.1038/ncb1346 [DOI] [PubMed] [Google Scholar]
- Arias E. E., Walter J. C. (2007). Strength in numbers: preventing rereplication via multiple mechanisms in eukaryotic cells. Genes Dev. 21, 497–518 10.1101/gad.1508907 [DOI] [PubMed] [Google Scholar]
- Bai C., Sen P., Hofmann K., Ma L., Goebl M., Harper J. W., Elledge S. J. (1996). SKP1 connects cell cycle regulators to the ubiquitin proteolysis machinery through a novel motif, the F-box. Cell 86, 263–274 10.1016/S0092-8674(00)80098-7 [DOI] [PubMed] [Google Scholar]
- Balajee A. S., May A., Dianov G. L., Friedberg E. C., Bohr V. A. (1997). Reduced RNA polymerase II transcription in intact and permeabilized Cockayne syndrome group B cells. Proc. Natl. Acad. Sci. U.S.A. 94, 4306–4311 10.1073/pnas.94.9.4306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banks D., Wu M., Higa L. A., Gavrilova N., Quan J., Ye T., Kobayashi R., Sun H., Zhang H. (2006). L2DTL/CDT2 and PCNA interact with p53 and regulate p53 polyubiquitination and protein stability through MDM2 and CUL4A/DDB1 complexes. Cell Cycle 5, 1719–1729 10.4161/cc.5.15.3150 [DOI] [PubMed] [Google Scholar]
- Barbash O., Egan E., Pontano L. L., Kosak J., Diehl J. A. (2009). Lysine 269 is essential for cyclin D1 ubiquitination by the SCFFbx4/alphaB-crystallin ligase and subsequent proteasome-dependent degradation. Oncogene 28, 4317–4325 10.1038/onc.2009.287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbash O., Zamfirova P., Lin D. I., Chen X., Yang K., Nakagawa H., Lu F., Rustgi A. K., Diehl J. A. (2008). Mutations in Fbx4 inhibit dimerization of the SCFFbx4 ligase and contribute to cyclin D1 overexpression in human cancer. Cancer Cell 14, 68–78 10.1016/j.ccr.2008.05.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batty D., Rapic’-Otrin V., Levine A. S., Wood R. D. (2000). Stable binding of human XPC complex to irradiated DNA confers strong discrimination for damaged sites. J. Mol. Biol. 300, 275–290 10.1006/jmbi.2000.3857 [DOI] [PubMed] [Google Scholar]
- Bendjennat M., Boulaire J., Jascur T., Brickner H., Barbier V., Sarasin A., Fotedar A., Fotedar R. (2003). UV irradiation triggers ubiquitin-dependent degradation of p21WAF1 to promote DNA repair. Cell 114, 599–610 10.1016/j.cell.2003.08.001 [DOI] [PubMed] [Google Scholar]
- Bennett E. J., Rush J., Gygi S. P., Harper J. W. (2010). Dynamics of cullin-RING ubiquitin ligase network revealed by systematic quantitative proteomics. Cell 143, 951–965 10.1016/j.cell.2010.11.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett L. N., Clarke P. R. (2006). Regulation of claspin degradation by the ubiquitin-proteosome pathway during the cell cycle and in response to ATR-dependent checkpoint activation. FEBS Lett. 580, 4176–4181 10.1016/j.febslet.2006.06.071 [DOI] [PubMed] [Google Scholar]
- Bernardi R., Liebermann D. A., Hoffman B. (2000). Cdc25A stability is controlled by the ubiquitin-proteasome pathway during cell cycle progression and terminal differentiation. Oncogene 19, 2447–2454 10.1038/sj.onc.1203564 [DOI] [PubMed] [Google Scholar]
- Bhatia N., Demmer T. A., Sharma A. K., Elcheva I., Spiegelman V. S. (2011). Role of β-TRCP ubiquitin ligase receptor in UVB mediated responses in skin. Arch. Biochem. Biophys. 508, 178–184 10.1016/j.abb.2011.01.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blackford A. N., Patel R. N., Forrester N. A., Theil K., Groitl P., Stewart G. S., Taylor A. M., Morgan I. M., Dobner T., Grand R. J., Turnell A. S. (2010). Adenovirus 12 E4orf6 inhibits ATR activation by promoting TOPBP1 degradation. Proc. Natl. Acad. Sci. U.S.A. 107, 12251–12256 10.1073/pnas.0914605107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bootsma D., Kreamer K. H., Cleaver J. E., Hoeijmakers J. H. (1998). “Nucleotide excision repair syndromes: xeroderma pigmento- sum, Cockayne syndrome, and trichothiodystrophy,” in The Genetic Basis of Human Cancer, eds Vogelsteain B., Kinzler K. W. (New York: McGraw-Hill; ), 254–257 [Google Scholar]
- Bornstein G., Bloom J., Sitry-Shevah D., Nakayama K., Pagano M., Hershko A. (2003). Role of the SCFSkp2 ubiquitin ligase in the degradation of p21Cip1 in S phase. J. Biol. Chem. 278, 25752–25757 10.1074/jbc.M301774200 [DOI] [PubMed] [Google Scholar]
- Boutros R., Dozier C., Ducommun B. (2006). The when and where of Cdc25 phosphatases. Curr. Opin. Cell Biol. 18, 185–191 10.1016/j.ceb.2006.02.003 [DOI] [PubMed] [Google Scholar]
- Bregman D. B., Halaban R., van Gool A. J., Henning K. A., Friedberg E. C., Warren S. L. (1996). UV-induced ubiquitination of RNA polymerase II: a novel modification deficient in Cockayne syndrome cells. Proc. Natl. Acad. Sci. U.S.A. 93, 11586–11590 10.1073/pnas.93.21.11586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Busino L., Donzelli M., Chiesa M., Guardavaccaro D., Ganoth D., Dorrello N. V., Hershko A., Pagano M., Draetta G. F. (2003). Degradation of Cdc25A by β-TRCP during S phase and in response to DNA damage. Nature 426, 87–91 10.1038/nature02082 [DOI] [PubMed] [Google Scholar]
- Cangi M. G., Piccinin S., Pecciarini L., Talarico A., Dal Cin. E., Grassi S., Grizzo A., Maestro R., Doglioni C. (2008). Constitutive overexpression of CDC25A in primary human mammary epithelial cells results in both defective DNA damage response and chromosomal breaks at fragile sites. Int. J. Cancer 123, 1466–1471 10.1002/ijc.23659 [DOI] [PubMed] [Google Scholar]
- Cardozo T., Pagano M. (2004). The SCF ubiquitin ligase: insights into a molecular machine. Nat. Rev. Mol. Cell Biol. 5, 739–751 10.1038/nrm1471 [DOI] [PubMed] [Google Scholar]
- Castells E., Molinier J., Benvenuto G., Bourbousse C., Zabulon G., Zalc A., Cazzaniga S., Genschik P., Barneche F., Bowler C. (2011). The conserved factor DE-ETIOLATED 1 cooperates with CUL4-DDB1DDB2 to maintain genome integrity upon UV stress. EMBO J. 30, 1162–1172 10.1038/emboj.2011.20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Centore R. C., Havens C. G., Manning A. L., Li J. M., Flynn R. L., Tse A., Jin J., Dyson N. J., Walter J. C., Zou L. (2010). CRL4Cdt2-mediated destruction of the histone methyltransferase Set8 prevents premature chromatin compaction in S phase. Mol. Cell 40, 22–33 10.1016/j.molcel.2010.09.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandrasekaran S., Tan T. X., Hall J. R., Cook J. G. (2011). Stress-stimulated mitogen-activated protein kinases control the stability and activity of the Cdt1 DNA replication licensing factor. Mol. Cell. Biol. 31, 4405–4416 10.1128/MCB.06163-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen M. S., Hurov J., White L. S., Woodford-Thomas T., Piwnica-Worms H. (2001a). Absence of apparent phenotype in mice lacking Cdc25C protein phosphatase. Mol. Cell. Biol. 21, 3853–3861 10.1128/MCB.21.22.7682-7695.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X., Zhang Y., Douglas L., Zhou P. (2001b). UV-damaged DNA-binding proteins are targets of CUL-4A-mediated ubiquitination and degradation. J. Biol. Chem. 276, 48175–48182 10.1074/jbc.M010501200 [DOI] [PubMed] [Google Scholar]
- Chen U., Chen S., Saha P., Dutta A. (1996). p21Cip1/Waf1 disrupts the recruitment of human Fen1 by proliferating-cell nuclear antigen into the DNA replication complex. Proc. Natl. Acad. Sci. U.S.A. 93, 11597–11602 10.1073/pnas.93.23.12992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chini C. C., Chen J. (2003). Human claspin is required for replication checkpoint control. J. Biol. Chem. 278, 30057–30062 10.1074/jbc.M301136200 [DOI] [PubMed] [Google Scholar]
- Chini C. C., Chen J. (2006). Repeated phosphopeptide motifs in human claspin are phosphorylated by Chk1 and mediate claspin function. J. Biol. Chem. 281, 33276–33282 10.1074/jbc.M604373200 [DOI] [PubMed] [Google Scholar]
- Chini C. C., Wood J., Chen J. (2006). Chk1 is required to maintain claspin stability. Oncogene 25, 4165–4171 10.1038/sj.onc.1209447 [DOI] [PubMed] [Google Scholar]
- Chiu Y. H., Sun Q., Chen Z. J. (2007). E1-L2 activates both ubiquitin and FAT10. Mol. Cell 27, 1014–1023 10.1016/j.molcel.2007.08.020 [DOI] [PubMed] [Google Scholar]
- Choo Y. Y., Boh B. K., Lou J. J., Eng J., Leck Y. C., Anders B., Smith P. G., Hagen T. (2011). Characterization of the role of COP9 signalosome in regulating cullin E3 ubiquitin ligase activity. Mol. Biol. Cell 22, 4706–4715 10.1091/mbc.E11-03-0251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cope G. A., Deshaies R. J. (2003). COP9 signalosome: a multifunctional regulator of SCF and other cullin-based ubiquitin ligases. Cell 114, 663–671 10.1016/S0092-8674(03)00722-0 [DOI] [PubMed] [Google Scholar]
- Cope G. A., Deshaies R. J. (2006). Targeted silencing of Jab1/Csn5 in human cells downregulates SCF activity through reduction of F-box protein levels. BMC Biochem. 7, 1. 10.1186/1471-2091-7-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cope G. A., Suh G. S., Aravind L., Schwarz S. E., Zipursky S. L., Koonin E. V., Deshaies R. J. (2002). Role of predicted metalloprotease motif of Jab1/Csn5 in cleavage of NEDD8 from Cul1. Science 298, 608–611 10.1126/science.1075901 [DOI] [PubMed] [Google Scholar]
- Deshaies R. J., Emberley E. D., Saha A. (2010). Control of cullin-ring ubiquitin ligase activity by NEDD8. Subcell. Biochem. 54, 41–56 10.1007/978-1-4419-6676-6_4 [DOI] [PubMed] [Google Scholar]
- Diehl J. A., Cheng M., Roussel M. F., Sherr C. J. (1998). Glycogen synthase kinase-3β regulates cyclin D1 proteolysis and subcellular localization. Genes Dev. 12, 3499–3511 10.1101/gad.12.22.3499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diehl J. A., Zindy F., Sherr C. J. (1997). Inhibition of cyclin D1 phosphorylation on threonine-286 prevents its rapid degradation via the ubiquitin-proteasome pathway. Genes Dev. 11, 957–972 10.1101/gad.11.8.957 [DOI] [PubMed] [Google Scholar]
- Donzelli M., Busino L., Chiesa M., Ganoth D., Hershko A., Draetta G. F. (2004). Hierarchical order of phosphorylation events commits Cdc25A to βTRCP-dependent degradation. Cell Cycle 3, 469–471 10.4161/cc.3.4.770 [DOI] [PubMed] [Google Scholar]
- Donzelli M., Squatrito M., Ganoth D., Hershko A., Pagano M., Draetta G. F. (2002). Dual mode of degradation of Cdc25A phosphatase. EMBO J. 21, 4875–4884 10.1093/emboj/cdf491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dualan R., Brody T., Keeney S., Nichols A. F., Admon A., Linn S. (1995). Chromosomal localization and cDNA cloning of the genes (DDB1 and DDB2) for the p127 and p48 subunits of a human damage-specific DNA binding protein. Genomics 29, 62–69 10.1006/geno.1995.1215 [DOI] [PubMed] [Google Scholar]
- Duda D. M., Borg L. A., Scott D. C., Hunt H. W., Hammel M., Schulman B. A. (2008). Structural insights into NEDD8 activation of cullin-RING ligases: conformational control of conjugation. Cell 134, 995–1006 10.1016/j.cell.2008.07.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duda D. M., Scott D. C., Calabrese M. F., Zimmerman E. S., Zheng N., Schulman B. A. (2011). Structural regulation of cullin-RING ubiquitin ligase complexes. Curr. Opin. Struct. Biol. 21, 257–264 10.1016/j.sbi.2011.01.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dutta A., Bell S. P. (1997). Initiation of DNA replication in eukaryotic cells. Annu. Rev. Cell Dev. Biol. 13, 293–332 10.1146/annurev.cellbio.13.1.293 [DOI] [PubMed] [Google Scholar]
- Dye B. T., Schulman B. A. (2007). Structural mechanisms underlying posttranslational modification by ubiquitin-like proteins. Annu. Rev. Biophys. Biomol. Struct. 36, 131–150 10.1146/annurev.biophys.36.040306.132820 [DOI] [PubMed] [Google Scholar]
- El-Mahdy M. A., Zhu Q., Wang Q. E., Wani G., Praetorius-Ibba M., Wani A. A. (2006). Cullin 4A-mediated proteolysis of DDB2 protein at DNA damage sites regulates in vivo lesion recognition by XPC. J. Biol. Chem. 281, 13404–13411 10.1074/jbc.M511834200 [DOI] [PubMed] [Google Scholar]
- Falck J., Mailand N., Syljuåsen R. G., Bartek J., Lukas J. (2001). The ATM-Chk2-Cdc25A checkpoint pathway guards against radioresistant DNA synthesis. Nature 410, 842–847 10.1038/35071124 [DOI] [PubMed] [Google Scholar]
- Feldman R. M., Correll C. C., Kaplan K. B., Deshaies R. J. (1997). A complex of Cdc4p, Skp1p, and Cdc53p/cullin catalyzes ubiquitination of the phosphorylated Cdk inhibitor Sic1p. Cell 91, 221–230 10.1016/S0092-8674(00)80404-3 [DOI] [PubMed] [Google Scholar]
- Ferguson A. M., White L. S., Donovan P. J., Piwnica-Worms H. (2005). Normal cell cycle and checkpoint responses in mice and cells lacking Cdc25B and Cdc25C protein phosphatases. Mol. Cell. Biol. 25, 2853–2860 10.1128/MCB.25.7.2853-2860.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fousteri M., Vermeulen W., van Zeeland A. A., Mullenders L. H. (2006). Cockayne syndrome A and B proteins differentially regulate recruitment of chromatin remodeling and repair factors to stalled RNA polymerase II in vivo. Mol. Cell 23, 471–482 10.1016/j.molcel.2006.06.029 [DOI] [PubMed] [Google Scholar]
- Friedberg E. C. (1995). Out of the shadows and into the light: the emergence of DNA repair. Trends Biochem. Sci. 20, 381. 10.1016/S0968-0004(00)89082-9 [DOI] [PubMed] [Google Scholar]
- Friedberg E. C., Walker G. C., Siede W. (1995). DNA Repair and Mutagenesis (Washington, DC: American Society of Microbiology; ). [Google Scholar]
- Gillet L. C., Schärer O. D. (2006). Molecular mechanisms of mammalian global genome nucleotide excision repair. Chem. Rev. 106, 253–276 10.1021/cr040483f [DOI] [PubMed] [Google Scholar]
- Goldenberg S. J., Cascio T. C., Shumway S. D., Garbutt K. C., Liu J., Xiong Y., Zheng N. (2004). Structure of the Cand1-Cul1-Roc1 complex reveals regulatory mechanisms for the assembly of the multisubunit cullin-dependent ubiquitin ligases. Cell 119, 517–528 10.1016/j.cell.2004.10.019 [DOI] [PubMed] [Google Scholar]
- Goloudina A., Yamaguchi H., Chervyakova D. B., Appella E., Fornace A. J., Jr., Bulavin D. V. (2003). Regulation of human Cdc25A stability by Serine 75 phosphorylation is not sufficient to activate a S phase checkpoint. Cell Cycle 2, 473–478 10.4161/cc.2.5.482 [DOI] [PubMed] [Google Scholar]
- Groisman R., Kuraoka I., Chevallier O., Gaye N., Magnaldo T., Tanaka K., Kisselev A. F., Harel-Bellan A., Nakatani Y. (2006). CSA-dependent degradation of CSB by the ubiquitin-proteasome pathway establishes a link between complementation factors of the Cockayne syndrome. Genes Dev. 20, 1429–1434 10.1101/gad.1438906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groisman R., Polanowska J., Kuraoka I., Sawada J., Saijo M., Drapkin R., Kisselev A. F., Tanaka K., Nakatani Y. (2003). The ubiquitin ligase activity in the DDB2 and CSA complexes is differentially regulated by the COP9 signalosome in response to DNA damage. Cell 113, 357–367 10.1016/S0092-8674(03)00316-7 [DOI] [PubMed] [Google Scholar]
- Guarino E., Shepherd M. E., Salguero I., Hua H., Deegan R. S., Kearsey S. E. (2011). Cdt1 proteolysis is promoted by dual PIP degrons and is modulated by PCNA ubiquitination. Nucleic Acids Res. 39, 5978–5990 10.1093/nar/gkr222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guerrero-Santoro J., Kapetanaki M. G., Hsieh C. L., Gorbachinsky I., Levine A. S., Rapic-Otrin V. (2008). The cullin 4B-based UV-damaged DNA-binding protein ligase binds to UV-damaged chromatin and ubiquitinates histone H2A. Cancer Res. 68, 5014–5022 10.1158/0008-5472.CAN-07-6162 [DOI] [PubMed] [Google Scholar]
- Hannah J., Zhou P. (2009). Regulation of DNA damage response pathways by the cullin-RING ubiquitin ligases. DNA Repair (Amst.) 8, 536–543 10.1016/j.dnarep.2009.01.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hao B., Oehlmann S., Sowa M. E., Harper J. W., Pavletich N. P. (2007). Structure of a Fbw7-Skp1-cyclin E complex: multisite-phosphorylated substrate recognition by SCF ubiquitin ligases. Mol. Cell 26, 131–143 10.1016/j.molcel.2007.02.022 [DOI] [PubMed] [Google Scholar]
- Härkönen P., Kyllönen A. P., Nordling S., Vihko P. (2005). Loss of heterozygosity in chromosomal region 16q24.3 associated with progression of prostate cancer. Prostate 62, 267–274 10.1002/pros.20147 [DOI] [PubMed] [Google Scholar]
- Harper J. W., Elledge S. J. (2007). The DNA damage response: ten years after. Mol. Cell 28, 739–745 10.1016/j.molcel.2007.11.015 [DOI] [PubMed] [Google Scholar]
- Havens C. G., Walter J. C. (2009). Docking of a specialized PIP Box onto chromatin-bound PCNA creates a degron for the ubiquitin ligase CRL4Cdt2. Mol. Cell 35, 93–104 10.1016/j.molcel.2009.05.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He N., Li C., Zhang X., Sheng T., Chi S., Chen K., Wang Q., Vertrees R., Logrono R., Xie J. (2005). Regulation of lung cancer cell growth and invasiveness by β-TRCP. Mol. Carcinog. 42, 18–28 10.1002/mc.20063 [DOI] [PubMed] [Google Scholar]
- Higa L. A., Banks D., Wu M., Kobayashi R., Sun H., Zhang H. (2006). L2DTL/CDT2 interacts with the CUL4/DDB1 complex and PCNA and regulates CDT1 proteolysis in response to DNA damage. Cell Cycle 5, 1675–1680 10.4161/cc.5.1.2266 [DOI] [PubMed] [Google Scholar]
- Higa L. A., Mihaylov I. S., Banks D. P., Zheng J., Zhang H. (2003). Radiation-mediated proteolysis of CDT1 by CUL4-ROC1 and CSN complexes constitutes a new checkpoint. Nat. Cell Biol. 5, 1008–1015 10.1038/ncb1061 [DOI] [PubMed] [Google Scholar]
- Honaker Y., Piwnica-Worms H. (2010). Casein kinase 1 functions as both penultimate and ultimate kinase in regulating Cdc25A destruction. Oncogene 29, 3324–3334 10.1038/onc.2010.96 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hosokawa Y., Papanikolaou A., Cardiff R. D., Yoshimoto K., Bernstein M., Wang T. C., Schmidt E. V., Arnold A. (2001). In vivo analysis of mammary and non-mammary tumorigenesis in MMTV-cyclin D1 transgenic mice deficient in p53. Transgenic Res. 10, 471–478 10.1023/A:1012064911751 [DOI] [PubMed] [Google Scholar]
- Hu J., McCall C. M., Ohta T., Xiong Y. (2004). Targeted ubiquitination of CDT1 by the DDB1-CUL4A-ROC1 ligase in response to DNA damage. Nat. Cell Biol. 6, 1003–1009 10.1038/ncb1172 [DOI] [PubMed] [Google Scholar]
- Hu J., Xiong Y. (2006). An evolutionarily conserved function of proliferating cell nuclear antigen for Cdt1 degradation by the Cul4-Ddb1 ubiquitin ligase in response to DNA damage. J. Biol. Chem. 281, 3753–3756 10.1074/jbc.M603682200 [DOI] [PubMed] [Google Scholar]
- Huang D. T., Ayrault O., Hunt H. W., Taherbhoy A. M., Duda D. M., Scott D. C., Borg L. A., Neale G., Murray P. J., Roussel M. F., Schulman B. A. (2009). E2-RING expansion of the NEDD8 cascade confers specificity to cullin modification. Mol. Cell 33, 483–495 10.1016/j.molcel.2009.01.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang H. L., Zheng W. L., Zhao R., Zhang B., Ma W. L. (2010). FBXO31 is down-regulated and may function as a tumor suppressor in hepatocellular carcinoma. Oncol. Rep. 24, 715–720 10.3892/or_00000981 [DOI] [PubMed] [Google Scholar]
- Hwang B. J., Toering S., Francke U., Chu G. (1998). p48 Activates a UV-damaged-DNA binding factor and is defective in xeroderma pigmentosum group E cells that lack binding activity. Mol. Cell. Biol. 18, 4391–4399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang J. W., Min K. W., Tamura T. A., Yoon J. B. (2003). TIP120A associates with unneddylated cullin 1 and regulates its neddylation. FEBS Lett. 541, 102–108 10.1016/S0014-5793(03)00321-1 [DOI] [PubMed] [Google Scholar]
- Jin J., Ang X. L., Ye X., Livingstone M., Harper J. W. (2008). Differential roles for checkpoint kinases in DNA damage-dependent degradation of the Cdc25A protein phosphatase. J. Biol. Chem. 283, 19322–19328 10.1074/jbc.M802785200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin J., Arias E. E., Chen J., Harper J. W., Walter J. C. (2006). A family of diverse Cul4-Ddb1-interacting proteins includes Cdt2, which is required for S phase destruction of the replication factor Cdt1. Mol. Cell 23, 709–721 10.1016/j.molcel.2006.08.010 [DOI] [PubMed] [Google Scholar]
- Jin J., Li X., Gygi S. P., Harper J. W. (2007). Dual E1 activation systems for ubiquitin differentially regulate E2 enzyme charging. Nature 447, 1135–1138 10.1038/nature05902 [DOI] [PubMed] [Google Scholar]
- Jin J., Shirogane T., Xu L., Nalepa G., Qin J., Elledge S. J., Harper J. W. (2003). SCFβTRCP links Chk1 signaling to degradation of the Cdc25A protein phosphatase. Genes Dev. 17, 3062–3074 10.1101/gad.1155703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones J., Wu K., Yang Y., Guerrero C., Nillegoda N., Pan Z. Q., Huang L. (2008). A targeted proteomic analysis of the ubiquitin-like modifier NEDD8 and associated proteins. J. Proteome Res. 7, 1274–1287 10.1021/pr700749v [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jørgensen S., Eskildsen M., Fugger K., Hansen L., Larsen M. S., Kousholt A. N., Syljuåsen R. G., Trelle M. B., Jensen O. N., Helin K., Sørensen C. S. (2011). SET8 is degraded via PCNA-coupled CRL4CDT2 ubiquitination in S phase and after UV irradiation. J. Cell Biol. 192, 43–54 10.1083/jcb.201009076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jura N., Scotto-Lavino E., Sobczyk A., Bar-Sagi D. (2006). Differential modification of Ras proteins by ubiquitination. Mol. Cell 21, 679–687 10.1016/j.molcel.2006.02.011 [DOI] [PubMed] [Google Scholar]
- Kamura T., Koepp D. M., Conrad M. N., Moreland R. J., Iliopoulos O., Lane W. S., Kaelin W. G., Jr., Elledge S. J., Conaway R. C., Harper J. W., Conaway J. W. (1999). Rbx1, a component of the VHL tumor suppressor complex and SCF ubiquitin ligase. Science 284, 657–661 10.1126/science.284.5414.657 [DOI] [PubMed] [Google Scholar]
- Kamura T., Maenaka K., Kotoshiba S., Matsumoto M., Kohda D., Conaway R. C., Conaway J. W., Nakayama K. I. (2004). VHL-box and SOCS-box domains determine binding specificity for Cul2-Rbx1 and Cul5-Rbx2 modules of ubiquitin ligases. Genes Dev. 18, 3055–3065 10.1101/gad.1252404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang T., Wei Y., Honaker Y., Yamaguchi H., Appella E., Hung M. C., Piwnica-Worms H. (2008). GSK-3 β targets Cdc25A for ubiquitin-mediated proteolysis, and GSK-3 β inactivation correlates with Cdc25A overproduction in human cancers. Cancer Cell 13, 36–47 10.1016/j.ccr.2007.12.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanie T., Onoyama I., Matsumoto A., Yamada M., Nakatsumi H., Tateishi Y., Yamamura S., Tsunematsu R., Matsumoto M., Nakayama K. I. (2012). Genetic reevaluation of the role of F-box proteins in cyclin D1 degradation. Mol. Cell. Biol. 32, 590–605 10.1128/MCB.06570-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapetanaki M. G., Guerrero-Santoro J., Bisi D. C., Hsieh C. L., Rapic-Otrin V., Levine A. S. (2006). The DDB1-CUL4ADDB2 ubiquitin ligase is deficient in xeroderma pigmentosum group E and targets histone H2A at UV-damaged DNA sites. Proc. Natl. Acad. Sci. U.S.A. 103, 2588–2593 10.1073/pnas.0511160103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karakaidos P., Taraviras S., Vassiliou L. V., Zacharatos P., Kastrinakis N. G., Kougiou D., Kouloukoussa M., Nishitani H., Papavassiliou A. G., Lygerou Z., Gorgoulis V. G. (2004). Overexpression of the replication licensing regulators hCdt1 and hCdc6 characterizes a subset of non-small-cell lung carcinomas: synergistic effect with mutant p53 on tumor growth and chromosomal instability – evidence of E2F-1 transcriptional control over hCdt1. Am. J. Pathol. 165, 1351–1365 10.1016/S0002-9440(10)63393-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karlsson-Rosenthal C., Millar J. B. (2006). Cdc25: mechanisms of checkpoint inhibition and recovery. Trends Cell Biol. 16, 285–292 10.1016/j.tcb.2006.04.002 [DOI] [PubMed] [Google Scholar]
- Kasahara K., Goto H., Enomoto M., Tomono Y., Kiyono T., Inagaki M. (2010). 14-3-3gamma mediates Cdc25A proteolysis to block premature mitotic entry after DNA damage. EMBO J. 29, 2802–2812 10.1038/emboj.2010.157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawakami T., Chiba T., Suzuki T., Iwai K., Yamanaka K., Minato N., Suzuki H., Shimbara N., Hidaka Y., Osaka F., Omata M., Tanaka K. (2001). NEDD8 recruits E2-ubiquitin to SCF E3 ligase. EMBO J. 20, 4003–4012 10.1093/emboj/20.15.4003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerzendorfer C., Whibley A., Carpenter G., Outwin E., Chiang S.-C., Turner G., Schwartz C., El-Khamisy S., Raymond F. L., O’Driscoll M. (2010). Mutations in cullin 4B result in a human syndrome associated with increased camptothecin-induced topoisomerase I-dependent DNA breaks. Hum. Mol. Genet. 19, 1324–1334 10.1093/hmg/ddq008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J., Feng H., Kipreos E. T. (2007). C. elegans CUL-4 prevents rereplication by promoting the nuclear export of CDC-6 via a CKI-1-dependent pathway. Curr. Biol. 17, 966–972 10.1016/j.cub.2007.09.040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y., Kipreos E. T. (2006). The Caenorhabditis elegans replication licensing factor CDT-1 is targeted for degradation by the CUL-4/DDB-1 complex. Mol. Cell. Biol. 27, 1394–1406 10.1128/MCB.00736-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y., Starostina N. G., Kipreos E. T. (2008). The CRL4Cdt2 ubiquitin ligase targets the degradation of p21Cip1 to control replication licensing. Genes Dev. 22, 2507–2519 10.1101/gad.1729508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kogo R., Mimori K., Tanaka F., Komune S., Mori M. (2011). FBXO31 determines poor prognosis in esophageal squamous cell carcinoma. Int. J. Oncol. 39, 155–159 [DOI] [PubMed] [Google Scholar]
- Kondo T., Kobayashi M., Tanaka J., Yokoyama A., Suzuki S., Kato N., Onozawa M., Chiba K., Hashino S., Imamura M., Minami Y., Minamino N., Asaka M. (2004). Rapid degradation of Cdt1 upon UV-induced DNA damage is mediated by SCFSkp2 complex. J. Biol. Chem. 279, 27315–27319 10.1074/jbc.M314023200 [DOI] [PubMed] [Google Scholar]
- Kopanja D., Stoyanova T., Okur M. N., Huang E., Bagchi S., Raychaudhuri P. (2009). Proliferation defects and genome instability in cells lacking Cul4A. Oncogene 28, 2456–2465 10.1038/onc.2009.86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kristjánsdóttir K., Rudolph J. (2004). Cdc25 phosphatases and cancer. Chem. Biol. 11, 1043–1051 10.1016/j.chembiol.2004.07.007 [DOI] [PubMed] [Google Scholar]
- Kudo Y., Guardavaccaro D., Santamaria P. G., Koyama-Nasu R., Latres E., Bronson R., Yamasaki L., Pagano M. (2004). Role of F-box protein βTRCP1 in mammary gland development and tumorigenesis. Mol. Cell. Biol. 24, 8184–8194 10.1128/MCB.24.10.4221-4228.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumagai A., Dunphy W. G. (2003). Repeated phosphopeptide motifs in claspin mediate the regulated binding of Chk1. Nat. Cell Biol. 5, 161–165 10.1038/ncb921 [DOI] [PubMed] [Google Scholar]
- Kumar R., Neilsen P. M., Crawford J., McKirdy R., Lee J., Powell J. A., Saif Z., Martin J. M., Lombaerts M., Cornelisse C. J., Cleton-Jansen A. M., Callen D. F. (2005). FBXO31 is the chromosome 16q24.3 senescence gene, a candidate breast tumor suppressor, and a component of an SCF complex. Cancer Res. 65, 11304–11313 10.1158/0008-5472.CAN-05-0189 [DOI] [PubMed] [Google Scholar]
- Kurz T., Chou Y. C., Willems A. R., Meyer-Schaller N., Hecht M. L., Tyers M., Peter M., Sicheri F. (2008). Dcn1 functions as a scaffold-type E3 ligase for cullin neddylation. Mol. Cell 29, 23–35 10.1016/j.molcel.2007.12.012 [DOI] [PubMed] [Google Scholar]
- Kurz T., Ozlü N., Rudolf F., O’Rourke S. M., Luke B., Hofmann K., Hyman A. A., Bowerman B., Peter M. (2005). The conserved protein DCN-1/Dcn1p is required for cullin neddylation in C. elegans and S. cerevisiae. Nature 435, 1257–1261 [DOI] [PubMed] [Google Scholar]
- Launonen V., Mannermaa A., Stenbäck F., Kosma V. M., Puistola U., Huusko P., Anttila M., Bloigu R., Saarikoski S., Kauppila A., Winqvist R. (2000). Loss of heterozygosity at chromosomes 3, 6, 8, 11, 16, and 17 in ovarian cancer: correlation to clinicopathological variables. Cancer Genet. Cytogenet. 122, 49–54 10.1016/S0165-4608(00)00279-X [DOI] [PubMed] [Google Scholar]
- Lee J., Kumagai A., Dunphy W. G. (2003). Claspin, a Chk1-regulatory protein, monitors DNA replication on chromatin independently of RPA, ATR, and Rad17. Mol. Cell 11, 329–340 10.1016/S1097-2765(03)00045-5 [DOI] [PubMed] [Google Scholar]
- Lee P. C., Sowa M. E., Gygi S. P., Harper J. W. (2011). Alternative ubiquitin activation/conjugation cascades interact with N-end rule ubiquitin ligases to control degradation of RGS proteins. Mol. Cell 43, 392–405 10.1016/j.molcel.2011.06.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J., Bhat A., Xiao W. (2011). Regulation of nucleotide excision repair through ubiquitination. Acta Biochim. Biophys. Sin. 43, 919–929 10.1093/abbs/gmr068 [DOI] [PubMed] [Google Scholar]
- Li X., Zhao Q., Liao R., Sun P., Wu X. (2003). The SCFSkp2 ubiquitin ligase complex interacts with the human replication licensing factor Cdt1 and regulates Cdt1 degradation. J. Biol. Chem. 278, 30854–30858 10.1074/jbc.C300251200 [DOI] [PubMed] [Google Scholar]
- Lin D. I., Barbash O., Kumar K. G., Weber J. D., Harper J. W., Klein-Szanto A. J., Rustgi A., Fuchs S. Y., Diehl J. A. (2006). Phosphorylation-dependent ubiquitination of cyclin D1 by the SCFFBX4-alphaB crystallin complex. Mol. Cell 24, 355–366 10.1016/j.molcel.2006.09.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin S. Y., Li K., Stewart G. S., Elledge S. J. (2004). Human claspin works with BRCA1 to both positively and negatively regulate cell proliferation. Proc. Natl. Acad. Sci. U.S.A. 101, 6484–6489 10.1073/pnas.0402496101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Y. W., Lee I. N., Chen C. H., Huang G. T., Lee H. S., Lee P. H., Lu F. J., Sheu J. C. (2001). Deletion mapping of chromosome 16q24 in hepatocellular carcinoma in Taiwan and mutational analysis of the 17-β-HSD gene localized to the region. Int. J. Cancer 93, 74–79 10.1002/ijc.1287 [DOI] [PubMed] [Google Scholar]
- Liontos M., Koutsami M., Sideridou M., Evangelou K., Kletsas D., Levy B., Kotsinas A., Nahum O., Zoumpourlis V., Kouloukoussa M., Lygerou Z., Taraviras S., Kittas C., Bartkova J., Papavassiliou A. G., Bartek J., Halazonetis T. D., Gorgoulis V. G. (2007). Deregulated overexpression of hCdt1 and hCdc6 promotes malignant behavior. Cancer Res. 67, 10899–10909 10.1158/0008-5472.CAN-07-2837 [DOI] [PubMed] [Google Scholar]
- Liu J., Furukawa M., Matsumoto T., Xiong Y. (2002). NEDD8 modification of CUL1 dissociates p120CAND1, an inhibitor of CUL1-SKP1 binding and SCF ligases. Mol. Cell 10, 1511–1518 10.1016/S1097-2765(02)00783-9 [DOI] [PubMed] [Google Scholar]
- Liu L., Lee S., Zhang J., Peters S. B., Hannah J., Zhang Y., Yin Y., Koff A., Ma L., Zhou P. (2009). CUL4A abrogation augments DNA damage response and protection against skin carcinogenesis. Mol. Cell 34, 4514–4560 10.1016/j.molcel.2009.04.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lovejoy C. A., Lock K., Yenamandra A., Cortez D. (2006). DDB1 maintains genome integrity through regulation of Cdt1. Mol. Cell. Biol. 26, 7977–7990 10.1128/MCB.00819-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyapina S. A., Correll C. C., Kipreos E. T., Deshaies R. J. (1998). Human CUL1 forms an evolutionarily conserved ubiquitin ligase complex (SCF) with SKP1 and an F-box protein. Proc. Natl. Acad. Sci. U.S.A. 95, 7451–7456 10.1073/pnas.95.13.7451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lykke-Andersen K., Schaefer L., Menon S., Deng X. W., Miller J. B., Wei N. (2003). Disruption of the COP9 signalosome Csn2 subunit in mice causes deficient cell proliferation, accumulation of p53 and cyclin E, and early embryonic death. Mol. Cell. Biol. 23, 6790–6797 10.1128/MCB.23.19.6790-6797.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma T., Shi T., Huang J., Wu L., Hu F., He P., Deng W., Gao P., Zhang Y., Song Q., Ma D., Qiu X. (2008). DCUN1D3, a novel UVC-responsive gene that is involved in cell cycle progression and cell growth. Cancer Sci. 99, 2128–2135 10.1111/j.1349-7006.2008.00836.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mailand N., Bekker-Jensen S., Bartek J., Lukas J. (2006). Destruction of claspin by SCFβTRCP restrains Chk1 activation and facilitates recovery from genotoxic stress. Mol. Cell 23, 307–318 10.1016/j.molcel.2006.06.016 [DOI] [PubMed] [Google Scholar]
- Mailand N., Falck J., Lukas C., Syljuâsen R. G., Welcker M., Bartek J., Lukas J. (2000). Rapid destruction of human Cdc25A in response to DNA damage. Science 288, 1425–1429 10.1126/science.288.5470.1425 [DOI] [PubMed] [Google Scholar]
- Mailand N., Podtelejnikov A. V., Groth A., Mann M., Bartek J., Lukas J. (2002). Regulation of G(2)/M events by Cdc25A through phosphorylation-dependent modulation of its stability. EMBO J. 21, 5911–5920 10.1093/emboj/cdf567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malumbres M., Barbacid M. (2001). To cycle or not to cycle: a critical decision in cancer. Nat. Rev. Cancer 1, 222–231 10.1038/35106065 [DOI] [PubMed] [Google Scholar]
- Mamely I., van Vugt M. A., Smits V. A., Semple J. I., Lemmens B., Perrakis A., Medema R. H., Freire R. (2006). Polo-like kinase-1 controls proteasome-dependent degradation of claspin during checkpoint recovery. Curr. Biol. 16, 1950–1955 10.1016/j.cub.2006.08.026 [DOI] [PubMed] [Google Scholar]
- Matsuda N., Azuma K., Saijo M., Iemura S., Hioki Y., Natsume T., Chiba T., Tanaka K., Tanaka K. (2005). DDB2, the xeroderma pigmentosum group E gene product, is directly ubiquitylated by cullin 4A-based ubiquitin ligase complex. DNA Repair (Amst.) 4, 537–545 10.1016/j.dnarep.2004.12.012 [DOI] [PubMed] [Google Scholar]
- Melixetian M., Klein D. K., Sørensen C. S., Helin K. (2009). NEK11 regulates CDC25A degradation and the IR-induced G2/M checkpoint. Nat. Cell Biol. 11, 1247–1253 10.1038/ncb1969 [DOI] [PubMed] [Google Scholar]
- Messick T. E., Greenberg R. A. (2009). The ubiquitin landscape at DNA double-strand breaks. J. Cell Biol. 187, 319–326 10.1083/jcb.200908074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer-Schaller N., Chou Y. C., Sumara I., Martin D. D., Kurz T., Katheder N., Hofmann K., Berthiaume L. G., Sicheri F., Peter M. (2009). The human Dcn1-like protein DCNL3 promotes Cul3 neddylation at membranes. Proc. Natl. Acad. Sci. U.S.A. 106, 12365–12370 10.1073/pnas.0812528106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michishita M., Morimoto A., Ishii T., Komori H., Shiomi Y., Higuchi Y., Nishitani H. (2011). Positively charged residues located downstream of PIP box, together with TD amino acids within PIP box, are important for CRL4Cdt2-mediated proteolysis. Genes Cells 16, 12–22 10.1111/j.1365-2443.2010.01464.x [DOI] [PubMed] [Google Scholar]
- Min K. W., Hwang J. W., Lee J. S., Park Y., Tamura T. A., Yoon J. B. (2003). TIP120A associates with cullins and modulates ubiquitin ligase activity. J. Biol. Chem. 278, 15905–15910 10.1074/jbc.M308493200 [DOI] [PubMed] [Google Scholar]
- Molinari M., Mercurio C., Dominguez J., Goubin F., Draetta G. F. (2000). Human Cdc25A inactivation in response to S phase inhibition and its role in preventing premature mitosis. EMBO Rep. 1, 71–79 10.1093/embo-reports/kvd018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myer D. L., Robbins S. B., Yin M., Boivin G. P., Liu Y., Greis K. D., Bahassi E. M., Stambrook P. J. (2011). Absence of polo-like kinase 3 in mice stabilizes Cdc25A after DNA damage but is not sufficient to produce tumors. Mutat. Res. 714, 1–10 10.1016/j.mrfmmm.2011.02.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nag A., Bondar T., Shiv S., Raychaudhuri P. (2001). The xeroderma pigmentosum group E gene product DDB2 is a specific target of cullin 4A in mammalian cells. Mol. Cell. Biol. 21, 6738–6747 10.1128/MCB.21.20.6738-6747.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Negrini S., Gorgoulis V. G., Halazonetis T. D. (2010). Genomic instability – an evolving hallmark of cancer. Nat. Rev. Mol. Cell Biol. 11, 220–228 10.1038/nrm2858 [DOI] [PubMed] [Google Scholar]
- Nishitani H., Shiomi Y., Iida H., Michishita M., Takami T., Tsurimoto T. (2008). Cdk inhibitor p21 is degraded by a proliferating cell nuclear antigen-coupled Cul4-DDB1Cdt2 pathway during S phase and after UV irradiation. J. Biol. Chem. 283, 29045–29052 10.1074/jbc.M806045200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishitani H., Sugimoto N., Roukos V., Nakanishi Y., Saijo M., Obuse C., Tsurimoto T., Nakayama K. I., Nakayama K., Fujita M., Lygerou Z., Nishimoto T. (2006). Two E3 ubiquitin ligases, SCF-Skp2 and DDB1-Cul4, target human Cdt1 for proteolysis. EMBO J. 25, 1126–1136 10.1038/sj.emboj.7601002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishitani H., Taraviras S., Lygerou Z., Nishimoto T. (2001). The human licensing factor for DNA replication Cdt1 accumulates in G1 and is destabilized after initiation of S-phase. J. Biol. Chem. 276, 44905–44911 10.1074/jbc.M105406200 [DOI] [PubMed] [Google Scholar]
- Oda H., Hübner M. R., Beck D. B., Vermeulen M., Hurwitz J., Spector D. L., Reinberg D. (2010). Regulation of the histone H4 monomethylase PR-Set7 by CRL4Cdt2-mediated PCNA-dependent degradation during DNA damage. Mol. Cell 40, 364–376 10.1016/j.molcel.2010.10.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohta T., Michel J. J., Schottelius A. J., Xiong Y. (1999). ROC1, a homolog of APC11, represents a family of cullin partners with an associated ubiquitin ligase activity. Mol. Cell 3, 535–541 10.1016/S1097-2765(00)80482-7 [DOI] [PubMed] [Google Scholar]
- Okabe H., Lee S. H., Phuchareon J., Albertson D. G., McCormick F., Tetsu O. (2006). A critical role for Fbxw8 and MAPK in cyclin D1 degradation and cancer cell proliferation. PLoS ONE 1, e128. 10.1371/journal.pone.0000128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oshikawa K., Matsumoto M., Yada M., Kamura T., Hatakeyama S., Nakayama K. I. (2003). Preferential interaction of TIP120A with Cul1 that is not modified by NEDD8 and not associated with Skp1. Biochem. Biophys. Res. Commun. 303, 1209–1216 10.1016/S0006-291X(03)00501-1 [DOI] [PubMed] [Google Scholar]
- Pagano M., Theodoras A. M., Tam S. W., Draetta G. F. (1994). Cyclin D1-mediated inhibition of repair and replicative DNA synthesis in human fibroblasts. Genes Dev. 8, 1627–1639 10.1101/gad.8.14.1627 [DOI] [PubMed] [Google Scholar]
- Panattoni M., Sanvito F., Basso V., Doglioni C., Casorati G., Montini E., Bender J. R., Mondino A., Pardi R. (2008). Targeted inactivation of the COP9 signalosome impairs multiple stages of T cell development. J. Exp. Med. 205, 465–477 10.1084/jem.20070725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelzer C., Kassner I., Matentzoglu K., Singh R. K., Wollscheid H. P., Scheffner M., Schmidtke G., Groettrup M. (2007). UBE1L2, a novel E1 enzyme specific for ubiquitin. J. Biol. Chem. 282, 23010–23014 10.1074/jbc.C700111200 [DOI] [PubMed] [Google Scholar]
- Peschiaroli A., Dorrello N. V., Guardavaccaro D., Venere M., Halazonetis T., Sherman N. E., Pagano M. (2006). SCFβTRCP-mediated degradation of claspin regulates recovery from the DNA replication checkpoint response. Mol. Cell 23, 319–329 10.1016/j.molcel.2006.06.013 [DOI] [PubMed] [Google Scholar]
- Petroski M. D., Deshaies R. J. (2005). Function and regulation of cullin-RING ubiquitin ligases. Nat. Rev. Mol. Cell Biol. 6, 9–20 10.1038/nrm1547 [DOI] [PubMed] [Google Scholar]
- Piao S., Lee S. J., Xu Y., Gwak J., Oh S., Park B. J., Ha N. C. (2011). CK1ε targets Cdc25A for ubiquitin-mediated proteolysis under normal conditions and in response to checkpoint activation. Cell Cycle 10, 531–537 10.4161/cc.10.3.14757 [DOI] [PubMed] [Google Scholar]
- Pickart C. M. (2004). Back to the future with ubiquitin. Cell 116, 181–190 10.1016/S0092-8674(03)01074-2 [DOI] [PubMed] [Google Scholar]
- Pickart C. M., Eddins M. J. (2004). Ubiquitin: structures, functions, mechanisms. Biochim. Biophys. Acta 1695, 55–72 10.1016/j.bbamcr.2004.09.019 [DOI] [PubMed] [Google Scholar]
- Pontano L. L., Aggarwal P., Barbash O., Brown E. J., Bassing C. H., Diehl J. A. (2008). Genotoxic stress-induced cyclin D1 phosphorylation and proteolysis are required for genomic stability. Mol. Cell. Biol. 28, 7245–7258 10.1128/MCB.01085-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ralph E., Boye E., Kearsey S. E. (2006). DNA damage induces Cdt1 proteolysis in fission yeast through a pathway dependent on Cdt2 and Ddb1. EMBO Rep. 7, 1134–1139 10.1038/sj.embor.7400827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ray D., Kiyokawa H. (2007). CDC25A levels determine the balance of proliferation and checkpoint response. Cell Cycle 6, 3039–3042 10.4161/cc.6.24.5104 [DOI] [PubMed] [Google Scholar]
- Ray D., Kiyokawa H. (2008). CDC25A phosphatase: a rate-limiting oncogene that determines genomic stability. Cancer Res. 68, 1251–1253 10.1158/0008-5472.CAN-07-5983 [DOI] [PubMed] [Google Scholar]
- Ray D., Terao Y., Nimbalkar D., Hirai H., Osmundson E. C., Zou X., Franks R., Christov K., Kiyokawa H. (2007a). Hemizygous disruption of Cdc25A inhibits cellular transformation and mammary tumorigenesis in mice. Cancer Res. 67, 6605–6611 10.1158/0008-5472.CAN-06-3065 [DOI] [PubMed] [Google Scholar]
- Ray D., Terao Y., Fuhrken P. G., Ma Z. Q., DeMayo F. J., Christov K., Heerema N. A., Franks R., Tsai S. Y., Papoutsakis E. T., Kiyokawa H. (2007b). Deregulated CDC25A expression promotes mammary tumorigenesis with genomic instability. Cancer Res. 67, 984–991 10.1158/0008-5472.CAN-06-3065 [DOI] [PubMed] [Google Scholar]
- Reinhardt H. C., Aslanian A. S., Lees J. A., Yaffe M. B. (2007). p53-deficient cells rely on ATM- and ATR-mediated checkpoint signaling through the p38MAPK/MK2 pathway for survival after DNA damage. Cancer Cell 11, 175–189 10.1016/j.ccr.2006.11.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ribar B., Prakash L., Prakash S. (2007). ELA1 and CUL3 are required along with ELC1 for RNA polymerase II polyubiquitination and degradation in DNA-damaged yeast cells. Mol. Cell. Biol. 27, 3211–3216 10.1128/MCB.00091-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudolph J. (2007). Cdc25 phosphatases: structure, specificity, and mechanism. Biochemistry 46, 3595–3604 10.1021/bi700026j [DOI] [PubMed] [Google Scholar]
- Saha A., Deshaies R. J. (2008). Multimodal activation of the ubiquitin ligase SCF by NEDD8 conjugation. Mol. Cell 32, 21–31 10.1016/j.molcel.2008.08.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sansam C. L., Shepard J. L., Lai K., Ianari A., Danielian P. S., Amsterdam A., Hopkins N., Lees J. A. (2006). DTL/CDT2 is essential for both CDT1 regulation and the early G2/M checkpoint. Genes Dev. 20, 3117–3129 10.1101/gad.1482106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santra M. K., Wajapeyee N., Green M. R. (2009). F-box protein FBXO31 mediates cyclin D1 degradation to induce G1 arrest after DNA damage. Nature 459, 722–725 10.1038/nature08011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarikas A., Hartmann T., Pan Z. Q. (2011). The cullin protein family. Genome Biol. 12, 220. 10.1186/gb-2011-12-4-220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scrima A., Konícková R., Czyzewski B. K., Kawasaki Y., Jeffrey P. D., Groisman R., Nakatani Y., Iwai S., Pavletich N. P., Thomä N. H. (2008). Structural basis of UV DNA-damage recognition by the DDB1-DDB2 complex. Cell 135, 1213–1223 10.1016/j.cell.2008.10.045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Senga T., Sivaprasad U., Zhu W., Park J. H., Arias E. E., Walter J. C., Dutta A. (2006). PCNA is a cofactor for Cdt1 degradation by CUL4/DDB1-mediated N-terminal ubiquitination. J. Biol. Chem. 281, 6246–6252 10.1074/jbc.M512705200 [DOI] [PubMed] [Google Scholar]
- Seo J., Chung Y. S., Sharma G. G., Moon E., Burack W. R., Pandita T. K., Choi K. (2005). Cdt1 transgenic mice develop lymphoblastic lymphoma in the absence of p53. Oncogene 24, 8176–8186 [DOI] [PubMed] [Google Scholar]
- Seol J. H., Feldman R. M., Zachariae W., Shevchenko A., Correll C. C., Lyapina S., Chi Y., Galova M., Claypool J., Sandmeyer S., Nasmyth K., Shevchenko A., Deshaies R. J. (1999). Cdc53/cullin and the essential Hrt1 RING-H2 subunit of SCF define a ubiquitin ligase module that activates the E2 enzyme Cdc34. Genes Dev. 13, 1614–1626 10.1101/gad.13.12.1614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serino G., Deng X. W. (2003). The COP9 signalosome: regulating plant development through the control of proteolysis. Annu. Rev. Plant Biol. 54, 165–182 10.1146/annurev.arplant.54.031902.134847 [DOI] [PubMed] [Google Scholar]
- Shibata E., Abbas T., Huang X., Wohlschlegel J. A., Dutta A. (2011). Selective ubiquitination of p21 and Cdt1 by UBCH8 and UBE2G ubiquitin-conjugating enzymes via the CRL4Cdt2 ubiquitin ligase complex. Mol. Cell. Biol. 31, 3136–3145 10.1128/MCB.05496-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimuta K., Nakajo N., Uto K., Hayano Y., Okazaki K., Sagata N. (2002). Chk1 is activated transiently and targets Cdc25A for degradation at the Xenopus midblastula transition. EMBO J. 21, 3694–3703 10.1093/emboj/cdf357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiyanov P., Hayes S. A., Donepudi M., Nichols A. F., Linn S., Slagle B. L., Raychaudhuri P. (1999). The naturally occurring mutants of DDB are impaired in stimulating nuclear import of the p125 subunit and E2F1-activated transcription. Mol. Cell. Biol. 19, 4935–4943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silverman J. S., Skaar J. R., Pagano M. (2012). SCF ubiquitin ligases in the maintenance of genome stability. Trends Biochem. Sci. 37, 66–73 10.1016/j.tibs.2011.10.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skowyra D., Craig K. L., Tyers M., Elledge S. J., Harper J. W. (1997). F-box proteins are receptors that recruit phosphorylated substrates to the SCF ubiquitin-ligase complex. Cell 91, 209–219 10.1016/S0092-8674(00)80403-1 [DOI] [PubMed] [Google Scholar]
- Skowyra D., Koepp D. M., Kamura T., Conrad M. N., Conaway R. C., Conaway J. W., Elledge S. J., Harper J. W. (1999). Reconstitution of G1 cyclin ubiquitination with complexes containing SCFGrr1 and Rbx1. Science 284, 662–665 10.1126/science.284.5414.662 [DOI] [PubMed] [Google Scholar]
- Sørensen C. S., Syljuåsen R. G., Falck J., Schroeder T., Rönnstrand L., Khanna K. K., Zhou B. B., Bartek J., Lukas J. (2003). Chk1 regulates the S phase checkpoint by coupling the physiological turnover and ionizing radiation-induced accelerated proteolysis of Cdc25A. Cancer Cell 3, 247–258 10.1016/S1535-6108(03)00048-5 [DOI] [PubMed] [Google Scholar]
- Soria G., Podhajcer O., Prives C., Gottifredi V. (2006). p21Cip1/WAF1 downregulation is required for efficient PCNA ubiquitination after UV irradiation. Oncogene 25, 2829–2838 10.1038/sj.onc.1209315 [DOI] [PubMed] [Google Scholar]
- Sugasawa K. (2011). Multiple DNA damage recognition factors involved in mammalian nucleotide excision repair. Biochemistry Mosc. 76, 16–23 10.1134/S0006297911010044 [DOI] [PubMed] [Google Scholar]
- Sugasawa K., Akagi J., Nishi R., Iwai S., Hanaoka F. (2009). Two-step recognition of DNA damage for mammalian nucleotide excision repair: directional binding of the XPC complex and DNA strand scanning. Mol. Cell 36, 642–653 10.1016/j.molcel.2009.09.035 [DOI] [PubMed] [Google Scholar]
- Sugasawa K., Okuda Y., Saijo M., Nishi R., Matsuda N., Chu G., Mori T., Iwai S., Tanaka K., Tanaka K., Hanaoka F. (2005). UV-induced ubiquitination of XPC protein mediated by UV-DDB-ubiquitin ligase complex. Cell 121, 387–400 10.1016/j.cell.2005.02.035 [DOI] [PubMed] [Google Scholar]
- Svejstrup J. Q. (2003). Rescue of arrested RNA polymerase II complexes. J. Cell Sci. 116, 447–451 10.1242/jcs.00271 [DOI] [PubMed] [Google Scholar]
- Tada S., Li A., Maiorano D., Méchali M., Blow J. J. (2001). Repression of origin assembly in metaphase depends on inhibition of RLF-B/Cdt1 by geminin. Nat. Cell Biol. 3, 107–113 10.1038/35074622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeda D. Y., Parvin J. D., Dutta A. (2005). Degradation of Cdt1 during S phase is Skp2-independent and is required for efficient progression of mammalian cells through S phase. J. Biol. Chem. 280, 23416–23423 10.1074/jbc.M501208200 [DOI] [PubMed] [Google Scholar]
- Takedachi A., Saijo M., Tanaka K. (2010). DDB2 complex-mediated ubiquitylation around DNA damage is oppositely regulated by XPC and Ku and contributes to the recruitment of XPA. Mol. Cell. Biol. 30, 2708–2723 10.1128/MCB.01460-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan P., Fuchs S. Y., Chen A., Wu K., Gomez C., Ronai Z., Pan Z. Q. (1999). Recruitment of a ROC1-CUL1 ubiquitin ligase by Skp1 and HOS to catalyze the ubiquitination of I kappa B alpha. Mol. Cell 3, 527–533 10.1016/S1097-2765(00)80481-5 [DOI] [PubMed] [Google Scholar]
- Tateishi K., Omata M., Tanaka K., Chiba T. (2001). The NEDD8 system is essential for cell cycle progression and morphogenetic pathway in mice. J. Cell Biol. 155, 571–579 10.1083/jcb.200104035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaites L. P., Lee E. K., Lian Z., Barbash O., Roy D., Wasik M., Klein-Szanto A. J., Rustgi A. K., Diehl J. A. (2011). The Fbx4 tumor suppressor regulates cyclin D1 accumulation and prevents neoplastic transformation. Mol. Cell. Biol. 31, 4513–4523 10.1128/MCB.05733-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Gool A. J., Citterio E., Rademakers S., van Os R., Vermeulen W., Constantinou A., Egly J. M., Bootsma D., Hoeijmakers J. H. (1997). The cockayne syndrome B protein, involved in transcription-coupled DNA repair, resides in an RNA polymerase II-containing complex. EMBO J. 16, 5955–5965 10.1093/emboj/16.14.4155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaziri C., Saxena S., Jeon Y., Lee C., Murata K., Machida Y., Wagle N., Hwang D. S., Dutta A. (2003). A p53-dependent checkpoint pathway prevents rereplication. Mol. Cell 11, 997–1008 10.1016/S1097-2765(03)00099-6 [DOI] [PubMed] [Google Scholar]
- Verma R., Feldman R. M., Deshaies R. J. (1997). SIC1 is ubiquitinated in vitro by a pathway that requires CDC4, CDC34, and cyclin/Cdk activities. Mol. Biol. Cell 8, 1427–1437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H., Zhai L., Xu J., Joo H. Y., Jackson S., Erdjument-Bromage H., Tempst P., Xiong Y., Zhang Y. (2006). Histone H3 and H4 ubiquitination by the CUL4-DDB-ROC1 ubiquitin ligase facilitates cellular response to DNA damage. Mol. Cell 22, 383–394 10.1016/j.molcel.2006.03.034 [DOI] [PubMed] [Google Scholar]
- Wang T. C., Cardiff R. D., Zukerberg L., Lees E., Arnold A., Schmidt E. V. (1994). Mammary hyperplasia and carcinoma in MMTV-cyclin D1 transgenic mice. Nature 369, 669–671 10.1038/369230a0 [DOI] [PubMed] [Google Scholar]
- Watson I. R., Blanch A., Lin D. C., Ohh M., Irwin M. S. (2006). Mdm2-mediated NEDD8 modification of TAp73 regulates its transactivation function. J. Biol. Chem. 281, 34096–34103 10.1074/jbc.M603654200 [DOI] [PubMed] [Google Scholar]
- Wei N., Deng X. W. (2003). The COP9 signalosome. Annu. Rev. Cell Dev. Biol. 19, 261–286 10.1146/annurev.cellbio.19.111301.112449 [DOI] [PubMed] [Google Scholar]
- Wei N., Serino G., Deng X. W. (2008). The COP9 signalosome: more than a protease. Trends Biochem. Sci. 33, 592–600 10.1016/j.tibs.2008.09.004 [DOI] [PubMed] [Google Scholar]
- Willems A. R., Lanker S., Patton E. E., Craig K. L., Nason T. F., Mathias N., Kobayashi R., Wittenberg C., Tyers M. (1996). Cdc53 targets phosphorylated G1 cyclins for degradation by the ubiquitin proteolytic pathway. Cell 86, 453–463 10.1016/S0092-8674(00)80118-X [DOI] [PubMed] [Google Scholar]
- Wohlschlegel J. A., Dwyer B. T., Dhar S. K., Cvetic C., Walter J. C., Dutta A. (2000). Inhibition of eukaryotic DNA replication by geminin binding to Cdt1. Science 290, 2309–2312 10.1126/science.290.5500.2309 [DOI] [PubMed] [Google Scholar]
- Wu G., Xu G., Schulman B. A., Jeffrey P. D., Harper J. W., Pavletich N. P. (2003). Structure of a β-TRCP1-Skp1-β-catenin complex: destruction motif binding and lysine specificity of the SCFβ-TRCP1 ubiquitin ligase. Mol. Cell 11, 1445–1456 10.1016/S1097-2765(03)00234-X [DOI] [PubMed] [Google Scholar]
- Wu K., Kovacev J., Pan Z. Q. (2010). Priming and extending: a UbcH5/Cdc34 E2 handoff mechanism for polyubiquitination on a SCF substrate. Mol. Cell 37, 784–796 10.1016/j.molcel.2010.02.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu K., Yan H., Fang L., Wang X., Pfleger C., Jiang X., Huang L., Pan Z. Q. (2011). Mono-ubiquitination drives nuclear export of the human DCN1-like protein hDCNL1. J. Biol. Chem. 286, 34060–34070 10.1074/jbc.M110.209130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao Z., Chen Z., Gunasekera A. H., Sowin T. J., Rosenberg S. H., Fesik S., Zhang H. (2003). Chk1 mediates S and G2 arrests through Cdc25A degradation in response to DNA-damaging agents. J. Biol. Chem. 278, 21767–21773 10.1074/jbc.M301596200 [DOI] [PubMed] [Google Scholar]
- Xirodimas D. P., Saville M. K., Bourdon J. C., Hay R. T., Lane D. P. (2004). Mdm2-mediated NEDD8 conjugation of p53 inhibits its transcriptional activity. Cell 118, 83–97 10.1016/j.cell.2004.06.016 [DOI] [PubMed] [Google Scholar]
- Xu L., Lubkov V., Taylor L. J., Bar-Sagi D. (2010). Feedback regulation of Ras signaling by Rabex-5-mediated ubiquitination. Curr. Biol. 20, 1372–1377 10.1016/j.cub.2010.09.043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan H., Chin M. L., Horvath E. A., Kane E. A., Pfleger CM. (2009). Impairment of ubiquitylation by mutation in Drosophila E1 promotes both cell-autonomous and non-cell-autonomous Ras-ERK activation in vivo. J. Cell Sci. 122, 1461–1470 10.1242/jcs.051904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan H., Jahanshahi M., Horvath E. A., Liu H. Y., Pfleger C. M. (2010). Rabex-5 ubiquitin ligase activity restricts Ras signaling to establish pathway homeostasis in Drosophila. Curr. Biol. 20, 1378–1382 10.1016/j.cub.2010.06.058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X. H., Shiotani B., Classon M., Zou L. (2008). Chk1 and claspin potentiate PCNA ubiquitination. Genes Dev. 22, 1147–1152 10.1101/gad.1682208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yasukawa T., Kamura T., Kitajima S., Conaway R. C., Conaway J. W., Aso T. (2008). Mammalian elongin a complex mediates DNA-damage-induced ubiquitination and degradation of Rpb1. EMBO J. 27, 3256–3266 10.1038/emboj.2008.249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon T., Chakrabortty A., Franks R., Valli T., Kiyokawa H., Raychaudhuri P. (2005). Tumor-prone phenotype of the DDB2-deficient mice. Oncogene 24, 469–478 10.1038/sj.onc.1208262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Z. K., Gervais J. L., Zhang H. (1998). Human CUL-1 associates with the SKP1/SKP2 complex and regulates p21CIP1/WAF1 and cyclin D proteins. Proc. Natl. Acad. Sci. U.S.A. 95, 11324–11329 10.1073/pnas.95.14.7993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao H., Watkins J. L., Piwnica-Worms H. (2002). Disruption of the checkpoint kinase 1/cell division cycle 25A pathway abrogates ionizing radiation-induced S and G2 checkpoints. Proc. Natl. Acad. Sci. U.S.A. 99, 14795–14800 10.1073/pnas.262533999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Q., Barakat B. M., Qin S., Ray A., El-Mahdy M. A., Wani G., Arafa E.-S., Mir S. N., Wang Q. E., Wani A. A. (2008). The p38 mitogen-activated protein kinase augments nucleotide excision repair by mediating DDB2 degradation and chromatin relaxation. J. Biol. Chem. 283, 32553–32561 10.1074/jbc.M803963200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng J., Yang X., Harrell J. M., Ryzhikov S., Shim E. H., Lykke-Andersen K., Wei N., Sun H., Kobayashi R., Zhang H. (2002a). CAND1 binds to unneddylated CUL1 and regulates the formation of SCF ubiquitin E3 ligase complex. Mol. Cell 10, 1519–1526 10.1016/S1097-2765(02)00784-0 [DOI] [PubMed] [Google Scholar]
- Zheng N., Schulman B. A., Song L., Miller J. J., Jeffrey P. D., Wang P., Chu C., Koepp D. M., Elledge S. J., Pagano M., Conaway R. C., Conaway J. W., Harper J. W., Pavletich N. P. (2002b). Structure of the Cul1-Rbx1-Skp1-F boxSkp2 SCF ubiquitin ligase complex. Nature 416, 703–709 10.1038/416703a [DOI] [PubMed] [Google Scholar]
- Zhong W., Feng H., Santiago F. E., Kipreos E. T. (2003). CUL-4 ubiquitin ligase maintains genome stability by restraining DNA-replication licensing. Nature 423, 885–889 10.1038/nature01747 [DOI] [PubMed] [Google Scholar]
- Zhu W., Chen Y., Dutta A. (2004). Rereplication by depletion of geminin is seen regardless of p53 status and activates a G2/M checkpoint. Mol. Cell. Biol. 24, 7140–7150 10.1128/MCB.24.11.4810-4823.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmerman E. S., Schulman B. A., Zheng N. (2010). Structural assembly of cullin-RING ubiquitin ligase complexes. Curr. Opin. Struct. Biol. 20, 714–721 10.1016/j.sbi.2010.08.010 [DOI] [PMC free article] [PubMed] [Google Scholar]