Abstract
A hallmark of human cancer is heterogeneity, reflecting the complex series of changes resulting in the activation of oncogenes coupled with inactivation of tumor suppressor genes. Breast cancer is no exception and indeed, many studies have revealed considerable complexity and heterogeneity in the population of primary breast tumors and substantial changes in a recurrent breast tumor that has acquired drug resistance. We have made use of a Myc-inducible transgenic mouse model of breast cancer in which elimination of Myc activity following tumor development initially leads to a regression of a subset of tumors generally followed by de novo Myc-independent growth. We have observed that tumors that grow independent of MYC expression have gene profiles and histologies that are distinct from the primary tumors and have acquired features resembling epithelial-mesenchymal transition (EMT) and “tumor initiating” cells. Analyses of the genetic pathways underlying these histological changes revealed a strong correlation between activation of the Ras, TGFβ, and TNFα pathways with Myc-independent growth. Collectively, the data reveal genetic alterations that underlie an escape from Myc-dependent growth and tumor progression that may parallel what occurs in human cancers as they acquire drug resistance.
Keywords: mouse, mammary, EMT, MYC, cancer
Introduction
Human cancer, including breast cancer, is characterized by genetic complexity reflecting the acquisition of multiple mutations, amplifications, deletions, and gene arrangements over a period of time. Recent genome-scale studies of copy number variation and large-scale DNA sequencing efforts have provided direct evidence for this complexity (Ding et al., 2008; Mullighan et al., 2007; Sjoblom et al., 2006; Weir et al., 2007). This heterogeneity is manifested in the primary tumors that exhibit considerable patient to patient variation that determines disease outcome and response to therapies as well as in the recurrent tumors that may additionally acquire the ability to metastasize to distant organs. An ability to model the complexity that gives rise to tumor heterogeneity is thus essential to understanding the oncogenic process and would enable the development of additional therapeutics to target cancers that evade conventional therapy.
Mouse models genetically engineered to explore the concept of oncogene addiction have guided our understanding of the genetic complexity underlying the progression of cancers. Prior work using transgenic mouse models that conditionally overexpress the MYC oncogene has shown that inactivation of MYC can be sufficient to induce sustained tumor regression. Particularly in lymphomas, islet cell tumors and skin tumors, withdrawal of MYC expression resulted in rapid tumor cell elimination through apoptosis (Pelengaris et al., 2002). Similarly, MYC inactivation in osteogenic sarcoma resulted in terminal differentiation of tumor cells into mature bone cells resulting in cells that no longer have tumorgenic potential (Jain et al., 2002). While tumor initiation and progression in these systems appeared to be dependent on a single oncogenic event, more recent mouse models of MYC-induced epithelial cancers have exhibited a more complex situation apparently reflecting the action of multiple oncogenic events (Boxer et al., 2004; D’Cruz et al., 2001; Moody et al., 2005; Shachaf et al., 2004). In a majority of these studies, inactivation of the oncogene resulted in regression of only a subset of the tumors. Tumors that initially regressed, showing a dependence on a specific oncogenic pathway, generally recurred exhibiting acquired resistance to pathway inhibition similar to what is observed during the course of human cancers. Additional studies employing models that combine inducible Myc and Ras oncogenes have described further complexity with evidence of a hierarchy of oncogene dependence (Podsypanina et al., 2008). The evident heterogeneity observed in these transgenic mice that conditionally overexpress an oncogene or oncogenes with respect to dependency on the initiating event suggests that these models can serve to reveal the complexities of the oncogenic process.
A particularly powerful approach to the study of genomic complexities that underlie these complex phenotypes has been the use of whole genome expression analyses. Our previous work has made use of patterns of gene expression, as well as patterns of pathway activity revealed using signatures of pathway activation, to identify heterogeneity of tumors within mouse models. We have further explored this heterogeneity, now in the context of the genomic alterations that occur to allow tumors to lose dependence on the initial oncogene activating event similar to how human cancers may acquire drug resistance.
Results
Genomic Analysis of Primary and Myc-independent Tumors
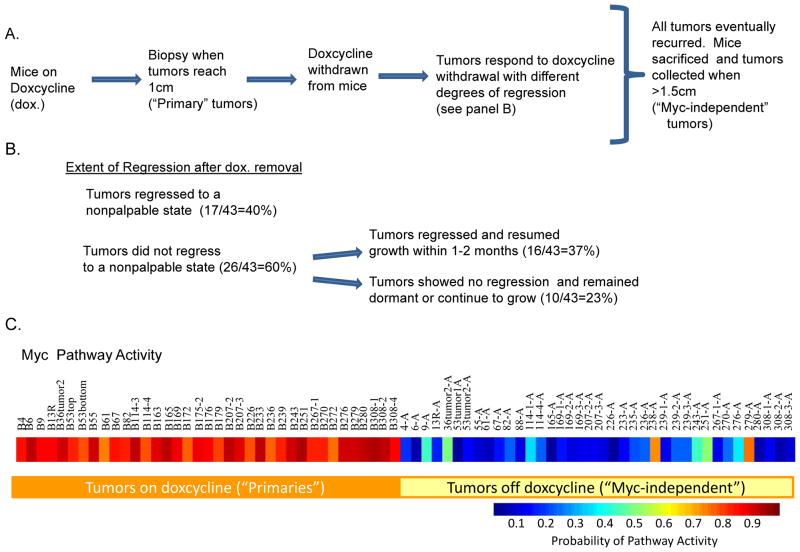
The transgenic line MMTV-rtTA (MTB) carries the reverse tetracycline-dependent transcriptional activator, rtTA, under the control of the mouse mammary tumor virus (MMTV). When mated to mice carrying the c-MYC transgene fused to a tetracycline-dependent promoter (TetO-Myc )(TOM), bitransgenic animals (MTB/TOM) are produced in which Myc is induced in the presence of doxycycline, resulting in the development of adenocarcinomas with 100% penetrance. Tumors that develop are heterogeneneous since previous work has shown that upon withdrawal of doxycycline, approximately half of the adenocarcinomas regressed while the remaining half persisted independent of MYC expression(Boxer et al., 2004).
We induced 35 mice with doxycycline and consistent with previous observations, chronically induced MTB/TOM developed mammary tumors with a mean latency of 21.8 weeks (Boxer et al., 2004; D’Cruz et al., 2001). From these 35 mice, we observed a total of 43 tumors. Upon withdrawal of doxycycline, 17 out of 43 (40%) tumors regressed to a nonpalpable state while the remaining 26 tumors (60%) showed variable degrees of regression but did not reach a nonpalpable state (Figure 1B). Tumors that regressed to a nonpalpable state generally recurred within 1–6 months and mice were euthanized when tumors reached >1cm3. Among the tumors that did not reach a nonpalpable state, 16/43 (37%) tumors showed variable degrees of regression (from >90% regression where the tumor was barely palpable to less than 10% reduction in tumor volume) and resumed growth within 1–2 months. The remaining 10/43 tumors (23%) showed no signs of regression and either remained dormant or continued to grow.
Figure 1.
A. Flow diagram describes when tumors were collected in MTB/TOM mice. Genome-scale expression of the “primary” and “Myc-independent” tumors was assessed on Affymetrix Mouse 430A.2 arrays. B. Approximately 40% of tumors regressed to a nonpalpable state while 60% of tumors never reached a nonpalpable state after removal of Myc expression. Among the 60% of tumors that never reached a nonpalpable state, 37% showed some extent of regression but quickly resumed growth while 23% showed no signs of regression and either remained dormant or continued to grow. C. MYC pathway activity in tumor samples growing in the presence of doxycycline and tumor samples growing after withdrawal of doxycycline.
As an approach to the analysis of the genomic changes that might underlie the differential outcome following elimination of Myc, we utilized genome-scale gene expression analysis. We assessed gene expression profiles on Affymetrix 430A.2 gene expression arrays of 37 out of 43 tumors collected when the mice were administered doxycycline (hereon referred to as the “primary” tumors) and 38 tumors that recurred or continued to grow after doxycycline withdrawal (hereon referred to as the “Myc-independent” tumors) (Figure 1A). As a starting point of our analyses, we first confirmed that MYC expression was tightly controlled by doxycycline, by comparing MYC pathway activity using a Myc gene expression signature that had previously been generated in our lab and validated on independent mouse data sets(Bild et al., 2006; Gatza et al.; Huang et al., 2003). The advantage of utilizing gene expression signatures is the ability to assess gene patterns represented by a collection of genes related to MYC expression thus offering an assessment of MYC activity rather than just MYC expression. Pathway analysis of the primary tumors showed high MYC activity when mice were still on doxycycline, as shown in Figure 1C. MYC pathway activity was diminished in recurrent tumors collected when the mice were off doxycycline with the exception of two tumors (238A and 279A). Not surprisingly, both of these tumors did not regress to a nonpalpable state following removal of doxycycline, consistent with continued MYC function. Although we have not looked further into the underlying cause of constitutive high MYC activity in these tumors, Podsypanina et al had previously reported the acquisition of spontaneous somatic mutations in the rtTA transgene cDNA resulting in the enhanced ability of the rtTA to bind and constitutively activate the tetO promoter (Podsypanina et al., 2008). We speculate that this may be a possible explanation for the high MYC activity we observe in these tumors.
Numerous studies have documented the synergistic actions of MYC and mutant Kras2 in mammary tumorgenesis (Andrechek et al., 2009; Boxer et al., 2004; D’Cruz et al., 2001; Podsypanina et al., 2008). In principle, Ras activation could be an explanation for the development of Myc independence in a fraction of the tumors. To address this possibility, we sequenced the mutational hot spots in Kras2 (codons 12, 13, and 61 of exons 1 and 2). Sequence analyses reveal mutations in Kras2 in 49% of total tumors analyzed (Table 1). Among tumors that regressed to a nonpalpable state 35% of tumors harbored Kras mutations while tumors that regressed but were still palpable and tumors that were either dormant or continued growing after removal of Myc harbored K-ras mutations in 44% and 80% of tumors, respectively. A majority of the mutations were observed in codon 12 with the exception of two tumors with mutations in codon 61. The amino acid changes in the mutations varied with a majority of them being a glycine to aspartic acid mutation. Mutations in Kras2 that were observed in the primary tumor samples were also observed in the recurring tumors thus confirming that the tumors represent bona fide recurrences rather than de novo neoplasms. Four of the primary tumors (B114-4, B169, B179 and B233) that did not initially have mutations in Kras2 eventually acquired mutations in Kras2 when the tumors were collected upon necropsy. Our results suggest that although Kras mutation status initially plays a role in Myc-independent growth, additional genetic alterations have to also be involved since all tumors eventually thrive independent of MYC expression.
Table 1.
Kras mutation status in MTB/TOM mouse mammary tumor samples.
| Kras mutation status | % of tumors with Kras mutations | |
|---|---|---|
| Tumors regressed to a nonpalpable state(17/43) | 6/17 * | 35% |
| Tumors regressed but were still palpable(16/43) | 7/16 | 44% |
| Tumors were dormant or continued growing (10/43) | 8/10 | 80% |
One sample not determined
Cluster Analysis Segregate Mouse Mammary Tumors that Escape Dependence on Myc
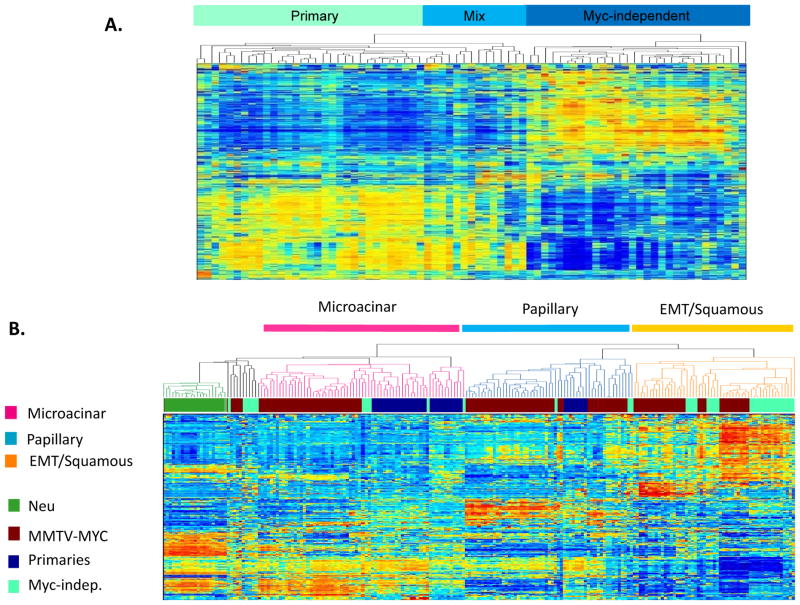
The Myc-induced mammary tumors from the MTB/TOM mouse model, although initiated by one oncogenic event, showed variable responses to withdrawal of Myc expression likely reflecting additional alterations acquired in the tumors. To determine whether differential responses to Myc withdrawal is reflected in specific gene expression patterns dictating tumor outcome, we performed an unsupervised clustering of the microarray data we obtained from all the mouse mammary tumors. As shown in Figure 2A, there was a very clear distinction in gene expression patterns between the primary tumors initiated by Myc compared to tumors that recur or persist in the absence of Myc transgene expression (Figure 2A).
Figure 2.
A. Unsupervised hierarchical clustering of MTB/TOM mammary tumors. The primary and Myc-independent tumors are separated into two different groups as denoted by the labels at the bottom. The middle “mixed” group contains both primary and Myc-independent tumors. B. Unsupervised clustering of mouse mammary tumors from the MTB/TOM model together with the MMTV-MYC model that constitutively expressed MYC. The “primary” samples cluster primarily with the papillary and microacinar group while the “recurrent” samples extend to all groups with most of them clustering with the EMT squamous group. The red bars represent groups of MTB/TOM tumors comprised of the following: Group I- one recurrent tumor, 82A; Group II-mix of primary tumors (B13R, B61, and B172) and recurring tumors (36-2A and 238A); Group III- primaries and two recurrent samples (9A and 279A); Group IV- recurrent samples.
To further characterize the mammary tumors we obtained from the MTB/TOM mouse model, we performed an unsupervised clustering again with tumors we previously characterized from a mouse model that constitutively expressed Myc (MMTV-MYC). Our prior work has described considerable heterogeneity in tumors from this MMTV-Myc mouse model as distinct gene expression patterns that correlated with distinct histological subtypes (Andrechek et al., 2009). Mammary tumors with papillary and microacinar histological types clustered in a group together apart from the EMT/squamous tumors. An EMT tumor signature that was generated from this data was shown to identify human breast tumors that were likely to metastasize. As shown in Figure 2B, cluster analysis revealed a majority of the MTB/TOM primary tumors clustered most closely with the previously characterized microacinar tumors while the Myc-independent tumors primarily clustered with the previously characterized EMT and squamous tumors.
Analysis of EMT Markers in Myc-independent Tumors
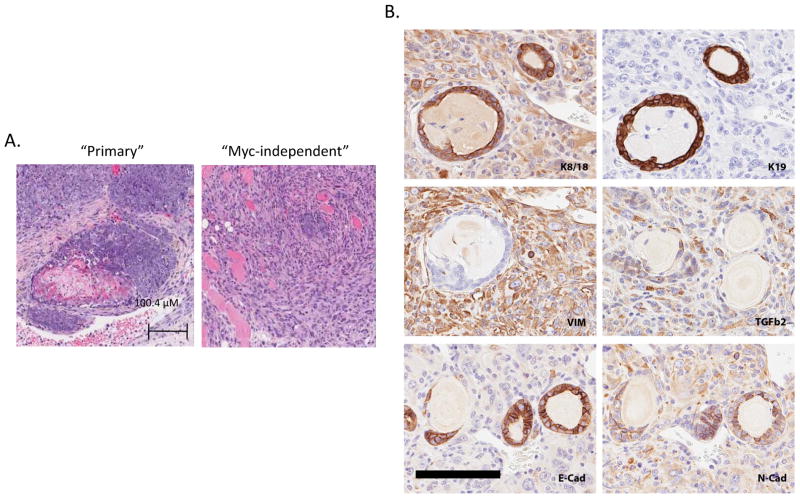
Previous studies with Her2/neu-induced mammary tumors have observed that tumors recurring independent of Her2/neu transgene expression generally exhibited EMT type histologies(Moody et al., 2005). The unsupervised clustering analysis presented in Figure 2 panel B provides evidence that the Myc independent tumors are genetically distinct from the primary tumors and cluster with tumors that exhibit an EMT/squamous phenotype. To further confirm that the distinction in gene patterns reflects acquired genetic heterogeneity that allow tumors to become more resilient and escape Myc dependence rather than mere differences resulting from the absence of a Myc signature, we set out to investigate evidence for EMT in the MTB/TOM tumors. Histological analysis of hematoxylin and eosin stained mouse mammary tumor samples from the MTB/TOM mouse revealed the primary samples to be predominantly of the microacinar and large blue cell type while the Myc-independent samples were a mixture of large blue cell, squamous, papillary and spindle cell type that are often characteristic of EMT. Although not all of the primary samples were examined histologically, the squamous and EMT-like histology pattern was not observed among the primary tumor samples we examined (Supplementary Table I and Figure 3A).
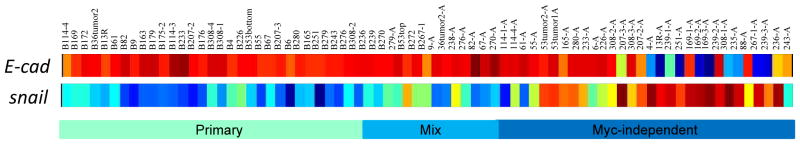
Figure 3.
Histological differences are observed between primary tumors initiated by MYC and tumors that have escaped MYC-dependence. A. Hemotoxylin and eosin staining of a sample histology of a “primary” tumor with the corresponding tumor Myc-independent tumor is shown. B. Heat map showing microarray probe expression of E-cadherin, fibronectin and vimentin. Downregulation of E-cadherin and upregualtion of fibronectin and vimentin are observed in the Myc-independent tumors. C. Expression of EMT markers from microarray expression analysis. E-cadherin and cyotokeratin 18 expression is downregulated and expression of fibronectin and vimentin are upregulated.
To further investigate the EMT-like nature of these tumor samples, we first examined expression of several documented markers of EMT including E-cadherin, vimentin, and fibronectin from our microarray data (Thiery, 2002). Consistent with the EMT nature of these Myc-independent tumors, we observed increased expression of vimentin and fibronectin expression using probes on Affymetrix arrays (Figure 3B). Downregulation of E-cadherin was also observed in a subset of the Myc-independent tumors. Immunohistochemistry by diaminobenzidine (DAB) staining on selected samples confirmed the expression of the EMT markers from the microarray analysis and additionally demononstrated a reduction in cytokeratin 18 (Figure 3C). Collectively, our results thus far show significant histological and genetic changes in the mouse mammary tumors as they escape MYC dependence and these changes are indicative of a transition to a more mesenchymal phenotype.
Activation of Ras, TGFβ and TNFα pathways
To further define and elucidate genetic differences between the primary and Myc-independent tumors, we used human signatures of cell signaling pathway. These signatures represent the activation of a cell signaling pathways in the form of a pattern of gene expression unique to that circumstance and can be quantitated and assessed in other biological samples. We previously validated and applied the Myc and Ras pathway signatures on various mouse datasets (Andrechek et al., 2009; Bild et al., 2006; Huang et al., 2003). We now extend this analysis for the TGFβ and TNFα pathway signatures using various mouse data sets (see Supplementary Figure I) and applied the TGFβ, Ras and TNFα gene signatures to the primary and Myc-independent MTB/TOM tumors. Our analyses showed the Myc-independent tumors have a higher probability of activation of these pathways versus the primary tumors (Figure 4A). The numerical values representing predicted pathway probabilities with the upper and lower limits of the predicted values are provided in the Supplementary Tables. We additionally confirmed activation of these pathways by immunohistochemisty staining with antibodies to TNFα and phospho-Smad3 as an indication of TGFβ activation Figure 4B.
Figure 4. Analysis of pathway activity in the MTB/TOM mammary tumors.
A. Heat maps showing TGFβ, TNFα, and Ras pathway activity in primary and Myc-independent MTB/TOM mouse mammary tumors. Numerical values for predicted pathway probabilities with upper and lower limits for all pathway predictions and training data are provided in the Supplementary Tables. B. Immunohistochemical staining to confirm activation of TNFalpha, and TGFbeta. C. Graph showing correlation between predicted pathway probabilities of Ras vs. TGFbeta; TNFalpha vs. TGFbeta; and Ras vs TNFalpha.
An abundance of work has noted an association of TGFβ, TNFα and Ras activation with EMT-like features. We determined whether there was any correlation between activation of these pathways. A Pearson correlation test showed a positive correlation between these pathways that showed to be statistically significant by a two-tailed T-test (Figure 4C).
MYC-independent growth has features of cancer initiating cells
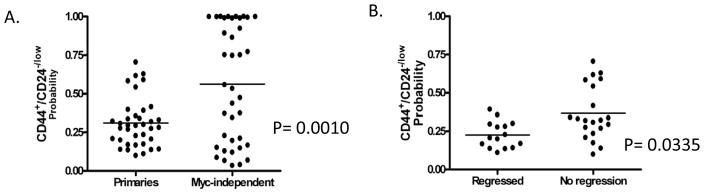
A variety of studies has proposed a role for so-called tumor-initiating cells as a component of the underlying mechanism of tumor recurrence and the acquisition of drug resistance. These cells generally display characteristics of EMT, show mammosphere-forming and higher tumor seeding capabilities, and are identified by expression of cell surface markers (CD44+/CD24−/low) (Al-Hajj et al., 2003; Creighton et al., 2009; Diehn et al., 2009). These “tumor-initiating” cells are thought to be resistant to therapy and may therefore contribute to cancer relapse. Given that the MTB/TOM mouse model studied here recapitulates some features of tumor recurrence and resistance comparable to human cancers, we sought to explore the extent to which the recurrent tumors were comparable to “tumor initiating” cells previously characterized from human breast cancers. For this analysis, we utilized data from human breast cancers that have been flow-sorted for expression of the CD44+/CD24−/low cell surface marker (GSE7513) (Al-Hajj et al., 2003). We generated a gene expression signature and validated this signature on an independent data set (GSE6883; Supplementary Figure II). We then examined the probability of this CD44+/CD24−/low gene signature in the primary and Myc-independent mouse mammary tumors. As shown in Figure 5A, tumors that have escaped MYC dependence had a statistically significant higher probability of exhibiting the CD44+/CD24−/low gene signature. Additionally, an analysis of our primary tumors showed a higher probability of the CD44+/CD24−/low gene signature in tumors that regressed to a nonpalpable state versus tumors that do not regress to a nonpalpable state in response to initial Myc removal (Figure 5B). Collectively, these results showed MYC-independent growth is generally associated with a higher CD44+/CD24−/low gene signature probability and suggests that MYC-independent growth may be acquired through mechanisms reminiscent of how human tumors recur and acquire drug resistance.
Figure 5.
C and D. The probability of the CD44+/CD24−/low gene expression signature generated in “A” was assessed in our primary and recurrent samples.
Discussion
Perhaps the largest challenge facing the effective treatment of cancer, including breast cancer, is the substantial heterogeneity and complexity that is evident in these disease states. Breast cancer is not one disease but rather a multitude of disorders with distinct etiologies. This can be seen in the variation evident in primary tumors that defines response to therapies as well as differential prognosis. It can also be seen in the recurrence of disease following initial therapy that most often is then associated with mortality. Elucidating the heterogeneity that permit tumor cells to escape blockade of a dominant oncogenic pathway, survive in a latent state and eventually reestablish malignant growth is thus of fundamental importance to making progress towards the goal of effective treatments and improved disease outcomes. The ability to use a mouse genetic model as a means to study these events, particularly the distinctions amongst primary tumors that define response to therapy and subsequent disease progression, is a key opportunity to better understand the human disease. We suggest that the observations we describe here, that provide clear evidence for heterogeneity and complexity within a mouse model reflecting events seen in the human disease, is one such opportunity.
Although the mouse mammary tumors studied here were initiated by a single oncogenic event, it is nevertheless clear that there is substantial heterogeneity in the primary tumors that determined the response to Myc withdrawal and the likelihood for progression to the Myc-independent state. In reality, the withdrawal of Myc expression as enabled in this model is a mimic for a Myc-specific drug. As such, it is apparent that while some of the tumors are indeed more reliant on the action of Myc and thus their growth is subdued by the inhibition of Myc function, others are not. This variation, that determines the outcome of the ‘Myc therapy’, mirrors similar situations in the treatment of human breast cancer where for instance there is variation in the response to Herceptin within the Her2 positive population of breast tumors. Although Ras activating mutations might be partly responsible for this variation, given the previous evidence for the coincidence of these mutations in the MMTV Myc model, it is apparent that this is perhaps not sufficient to explain the variability in outcome in a small subset of tumors that do not harbor ras mutations or pathway activity.
Prior work by Moody et al has shown an EMT phenotype to be associated with recurrence of Neu-induced mouse mammary tumors (Moody et al., 2002). In light of these observations together with our results, we suggest that the acquisition of an EMT phenotype in the mouse model is not specific to a particular oncogene initiating event but is a general event associated with more resilient tumors at a later stage of tumor progression. Our work provides novelty in that we have observed significant gene profile differences including activation of the TGFβ, TNFα, and RAS signaling pathways associated with an escape from oncogene dependent growth in the MTB/TOM mouse model. Consistent with this observation, an abundance of literature has reported the TGFβ, TNFα, and RAS signaling pathways to have a prometastatic role associated with an EMT phenotype during the later stages of tumorigenesis. (Acloque et al., 2008; Singh and Settleman, 2010; Thiery, 2002; Thiery et al., 2009). Our observations reported here suggest the MTB/TOM mouse to be a good model system for investigating future therapeutic opportunities in targeting a combination of these signaling pathways in more advanced human cancers.
Recent work has connected EMT to the emergence of cancer stem cells (Mani et al., 2008). Consistent with EMT-like features and the propensity for more aggressive tumor growth, a higher probability of the CD44+/CD24−/low gene signature was associated with MYC-independent growth. Tumorgenic breast cancer cells that express high levels of CD44 and low CD24 in humans have been proposed to be resistant to chemotherapy and are thought to be responsible for cancer relapse (Creighton et al., 2009). We emphasize that although these results do not suggest that our recurrent tumors are cancer-initiating cells, these observations do suggest that these tumors share properties of EMT and are indicative of cells that are more robust and resistant to cell death. Primary samples that did not regress after withdrawal of MYC expression were more likely to exhibit characteristics of the CD44+/CD24−/low phenotype. These results suggest CD44+/CD24−/low features may be a good predictor of the extent to which a tumor has escaped MYC dependence in this mouse model and further suggests this model to be good system for developing combination therapeutic strategies to target cells that are resistant to the ablation of one oncogenic pathway. Furthermore, studies by Creighton et al. have alluded to the similarities between CD44+/CD24−/low and mammosphere-forming cells to human breast cancers of the “claudin low” type, previously characterized by Herschkowitz et al (Herschkowitz et al., 2007). These findings highlight the value of further defining the potential of this MTB/TOM mouse model in studying the molecular complexity underlying specific groups of human cancers. .
Finally, an area of research that is becoming increasingly more intriguing is the mechanisms underlying the phenomenon of oncogene addiction and how some tumors escape this addiction. What is evident in the studies we present here is the degree of heterogeneity acquired in tumors that evolve to thrive independent of expression from the oncogene-initiating event. The value in this determination is the extent to which the heterogeneity itself is a model of human disease and thus provides an opportunity for further dissection of this heterogeneity with the goal of working towards personalized therapeutics in human cancer.
Materials and Methods
Animals
Animal use and husbandry was in accordance with institutional and federal guidelines. Bitransgenic animals were induced by administering doxycycline (2mg/ml;Sigma) in their drinking water which was replaced weekly. Animals were monitored for tumor growth weekly.
Tissue collection
A biopsy was taken when mammary tumors were ~1cm. Mice with recurring tumors or tumors that did not regressed were euthanized when tumors were ~1.5–2 cm. Tumor tissues collected for RNA extraction were flash frozen in liquid nitrogen. Tumor tissues collected for histology were fixed in formalin and then were processed for routine histology.
Kras2 mutation analysis
cDNA was generated and PCR amplified for sequencing using Titanium one step RT-PCR (Clontech). DNA fragments from Kras2 were gel purified and sequence analysis was performed by Duke DNA Sequencing Facility.
Microarray analysis
Flash frozen RNA samples from mouse tissue were purified using the RNeasy Mini Kit (Qiagen) after roto-stator homogenization and submitted to the Duke Microarray facility for hybridization to Affymetrix Mouse Genome 430A 2.0 array platforms. The resulting .CEL files were normalized by Robust Multi-Array (RMA) or MicroArray Suite (MAS5) using Affymetrix Expression Console. MAS5 normalized files were additionally log2 transformed. The RAW .CEL files have been deposited into the Gene Expression Omnibus (GEO) database under the accession number GSE22406.
Unsupervised Cluster Analysis
Unsupervised clustering was performed with the publicly available Cluster 3.0 software. Using the SD function under the “filter” tab, genes were filtered down to approximately 1000 genes. Under the “adjusted” tab the option to center genes and center arrays was checked. Under the “hierarchical” tab, the option to cluster genes and arrays were checked and average linkage was chosen. The results were visualized with JavaTreeView. Matlab was then used to generate a color heat map of the results.
Statistical Analysis of Pathway Predictions
The statistical methods for pathway analysis have been thoroughly described but are reiterated here for clarification (Bild et al., 2006; Huang et al., 2003; West et al., 2001; Gatza et al.). The following explanation is from Gatza et al. In our pathway analysis, a signature represents a group of genes that collectively exhibit a consistent pattern of expression. This signature enables a distinction between two phenotypes. A metagene representing a group of genes that collectively demonstrate a consistent pattern of expression for a specific phenotype is identified from the training data (Phenotype A versus Phenotype B). Each signature summarizes its constituent genes as a single expression profile and is derived from the first principal component of that gene set. This factor corresponds to the largest singular value as determined by singular value decomposition (SVD). Bayesian methods are then used to estimate binary probability regression models based on a given set of expression vectors (values across metagenes) derived from the training data. Application of these models to an independent validation dataset enables the evaluation of predictive probabilities of each of the two phenotypic states for each sample in the validation dataset. In these analyses, gene selection and identification is based solely on the training data. Metagene values are computed using the principal components of the training data ensuring reproducibility of the signature irrespective of the composition of the validation dataset. Bayesian fitting of binary probability regression models to the training data enable assessment of the relevance of the metagene signature in within-sample classification as well as estimation and uncertainty assessment for the binary regression weights. This results in the mapping of metagenes to probabilities of relative pathway status. Evaluation of independent tumor or cell line samples results in the prediction of relative pathway status generating estimated relative probabilities, and associated measures of uncertainty, of activation or deregulation for each sample in the validation dataset.
To ensure that over-fitting does not occur in the generation of each signature, a leave-one-out cross validation was performed for each set of training data to examine the stability and predictive capabilities of our model. In this analysis, each sample is left out, one at a time, of the dataset and the model was refitted (both the metagene factors and the partitions used) using the remaining samples. The software for this analysis can be downloaded from http://www.duke.edu/~dinbarry/BINREG/.
Pathway Predictions for MTB/TOM Mouse Mammary Tumors
The procedures for the generation of training data followed by predictions of pathway activity are described below.
I. Pathway Analysis of TNF alpha activity
Generation of TNFα training data: The TNFα training data was generated using datasets from GEO (GSE2838 and GSE2639). RAW.CEL files were Robust Multi-array Average (RMA) normalized using Affymetrix Expression Console Version 1.1. This file is provided in the Supplementary Information(SI) as “tnfa_train__rma”. In the “tnfa_train__rma” file, “0” represents untreated conditions and the columns designated as “1” represents cells treated with TNFα. The file was loaded onto BINREG2 which can be downloaded at http://www.duke.edu/~dinbarry/BINREG/ for binary regression analysis. The following parameters were chosen in BINREG2: 80 gene/ 3 metagenes/ Shift scale norm/ no quantile norm/ data is already logged/ 1000 burn in / 5000 iterations. The metagene scores for the training data is included in the SI as “Predicted Pathway Probabilities-Training.TNFalpha”
Converting Mouse Affymetrix probe IDs to Human probe IDs for Analysis: Mouse 430A 2.0 mouse probe IDs from the MTB/TOM data and Mouse 430 probIDs from GEO-GSE19272 were converted to human U133 probed IDs using Chip Comparer http://chipcomparer.genome.duke.edu/ and File Merger http://filemerger.genome.duke.edu/.
Validation of TNFα signature on publicly available mouse data: The RAW .CEL files representing primary murine hepatocytes mock treated or treated with TNFalpha were downloaded from GEO with the gene accession number GSE19272. The data was RMA normalized using Affymetrix Expression Console Version 1.1. Mouse Affymetrix probe IDs were converted to human Affymetrix probe IDs as described above. Analysis in BINREG2 was performed using the parameters described above to generate the training data. The result of this validation set is reported by BINREG2 as an output file denoted as validation.txt. Columns of output are: 1) the column number in input file, 2) phenotype call, 3) average probability, 4) and 5) upper and lower limits of the credible interval, 6) metagene score. The results are provided in the Supplementary Information as the “Predicted Pathway Probabilities-GSE19272.TNFalpha.validation” file. The average probabilities were then used to generate a heat map in Matlab.
Predicting TNFα pathway activity in MTB/TOM tumors: The data was RMA normalized using Affymetrix Expression Console Version 1.1. Mouse Affymetrix probe IDs were converted to human Affymetrix probe IDs as described above. Analysis in BINREG2 was performed using the parameters described above to generate the training data. The result of this validation set is reported by BINREG2 as an output file denoted as validation.txt. Columns of output are: 1) the column number in input file, 2) phenotype call, 3) average probability, 4) and 5) upper and lower limits of the credible interval, 6) metagene score. The results are provided in the Supplementary Information as the “Predicted Pathway Probabilities-MTBTOM.TNFalpha” file The average probabilities were then used to generate a heat map in Matlab.
II. Pathway Analysis of TGFβ pathway activity
Generation of TGFβ training data: The TGFβ training data was generated from mock or TGFβ treated lung cancer cells. Gene expression profiles were assessed by Affymetrix U133 arrays and RAW.CEL files were Robust Multi-array Average (RMA) normalized using Affymetrix Expression Console Version 1.1. This file is provided in the Supplementary Information as “tgfb_train_rma”. In the “tgfb_train_rma” file, “0” represents mock treated and the columns designated as “1” represents cells treated with TGFβ. The file was loaded onto BINREG2 which can be downloaded at http://www.duke.edu/~dinbarry/BINREG/ for binary regression analysis. The following parameters were chosen in BINREG2: 125 gene/ 3 metagenes/ Shift scale norm/ no quantile norm/ data is already logged/ 1000 burn in / 5000 iterations. The metagene scores for the training data is included in the SI as “Prediceted Pathway Probabilities-Training.TGFbeta”
Converting Mouse Affymetrix probe IDs to Human probe IDs for Analysis: Mouse 430A 2.0 mouse probe IDs from the MTB/TOM data and Mouse 430 probIDs from GEO-GSE13986 were converted to human U133 probed IDs using Chip Comparer http://chipcomparer.genome.duke.edu/ and File Merger http://filemerger.genome.duke.edu/.
Validation of TGFβ signature on publicly available mouse data: The RAW .CEL files representing gene expression profiles from control mice or mice induced to express TGFβ were downloaded from the data set with the gene accession number GSE13986. The data was RMA normalized using Affymetrix Expression Console Version 1.1. Mouse Affymetrix probe IDs were converted to human Affymetrix probe IDs as described above. Analysis in BINREG2 was performed using the parameter described above to generate the training data. The result of this validation set is reported by Binreg as an output file denoted as validation.txt. Columns of output are: 1) the column number in input file, 2) phenotype call, 3) average probability, 4) and 5) upper and lower limits of the credible interval, 6) metagene score. The results are provided in the Supplementary Information as the “Predicted Pathway Probabilities-GSE13986. TGFbeta.validation” file. The average probabilities were then used to generate a heat map in Matlab.
Predicting TGFβ pathway activity in MTB/TOM tumors: The data was RMA normalized using Affymetrix Expression Console Version 1.1. Mouse Affymetrix probe IDs were converted to human Affymetrix probe IDs as described above. Analysis in BINREG2 was performed using the parameters described above to generate the training data. The result of this validation set is reported by BINREG2 as an output file denoted as validation.txt. Columns of output are: 1) the column number in input file, 2) phenotype call, 3) average probability, 4) and 5) upper and lower limits of the credible interval, 6) metagene score. The results are provided in the Supplementary Information as the “Predicted Pathway Probabilities-MTBTOM.TGFbeta” file. The average probabilities were then used to generate a heat map in Matlab.
III. Pathway Analysis of Ras pathway activity
Generation of RAS training data: The RAS training data was generated from HMECs infected with an adenovirus expressing GFP or RAS. Gene expression profiles were assessed by Affymetrix U133 arrays and RAW.CEL files were MAS5 normalized using Affymetrix Expression Console Version 1.1 and log2 transformed. This file is provided in the Supplementary Information as “ras_train_mas5log2”. In the “ras_train_mas5log2” file, “0” represents cells infected with GFP-adenovirus and the columns designated as “1” represents cells expressing RAS-adenovirus. The file was loaded onto BINREG2 which can be downloaded at http://www.duke.edu/~dinbarry/BINREG/ for binary regression analysis. The following parameters were chosen in BINREG2: 350 gene/ 2 metagenes/Shift scale norm/ quantile norm/ data is already logged/ 1000 burn in / 5000 iterations. The metagene scores for the training data is included in the SI as “Predicted Pathway Probabilities-Training.Ras”
Converting Mouse Affymetrix probe IDs to Human probe IDs for Analysis: Mouse 430A 2.0 mouse probe IDs from the MTB/TOM data and Mouse 430 probIDs from GEO-GSE13986 were converted to human U133 probed IDs using Chip Comparer http://chipcomparer.genome.duke.edu/ and File Merger http://filemerger.genome.duke.edu/.
Validation of RAS signature on publicly available mouse data: The RAW .CEL files representing gene expression profiles from control mice or mice induced to express RAS were downloaded from the data set with the gene accession number GSE13986. The data was RMA normalized using Affymetrix Expression Console Version 1.1. Mouse Affymetrix probe IDs were converted to human Affymetrix probe IDs as described above. Analysis in BINREG2 was performed using the parameter described above to generate the training data. The result of this validation set is reported by Binreg as an output file denoted as validation.txt. Columns of output are: 1) the column number in input file, 2) phenotype call, 3) average probability, 4) and 5) upper and lower limits of the credible interval, 6) metagene score. The results are provided in the Supplementary Information as the “Predicted Pathway Probabilities-GSE13986. Ras.validation” file. The average probabilities were then used to generate a heat map in Matlab.
Predicting RAS pathway activity in MTB/TOM tumors: The data was RMA normalized using Affymetrix Expression Console Version 1.1. Mouse Affymetrix probe IDs were converted to human Affymetrix probe IDs as described above. Analysis in BINREG2 was performed using the parameters described above to generate the training data. The result of this validation set is reported by BINREG2 as an output file denoted as validation.txt. Columns of output are: 1) the column number in input file, 2) phenotype call, 3) average probability, 4) and 5) upper and lower limits of the credible interval, 6) metagene score. The results are provided in the Supplementary Information as the “Predicted Pathway Probabilities-MTBTOM.Ras” file. The average probabilities were then used to generate a heat map in Matlab.
IV. Analysis of Myc Pathway Activity
Prediction of Myc pathway activity was performed as described above for prediction of probabilities for TGFβ, TNFα and Ras pathway activity. The Myc training data published in Huang et al, Bild et al, and Gatza et al was used (Bild et al., 2006; Gatza et al.; Huang et al., 2003). Gene expression profiles were previously assessed by Affymetrix U133 arrays and RAW.CEL files were MAS5 normalized using Affymetrix Expression Console Version 1.1 and log2 transformed. The training data is provided in SI as “myc_train_mas5log2”. Given that the Myc training data has already been validated on mouse samples previously, we did not validate this training data on additional mouse data here. Mouse probe IDs from the 430A.2 microarray chip were converted to human U133 probe IDs using Chip Comparer and File Merger and described above and the probability of Myc pathway activity was assessed using BINREG2 using the following parameters: 500 gene/ 2 metagenes/Shift scale norm/ quantile norm/ data is already logged/ 1000 burn in / 5000 iterations.
Generation of a CD44+/CD24−/low Gene Signature and Analysis of Data
Generation of CD44+/CD24−/low training data: The CD44+/CD24−/low training data was generated from GEO dataset GSE7513. RAW.CEL files were RMA normalized using Affymetrix Expression Console Version 1.1. This file is provided in the Supplementary Information as “CD44.CD24_train_RMA”. In the “CD44.CD24_train_RMA” file, “0” represents unsorted cells and the columns designated as “1” represents CD44+/CD24−/sorted cells. The file was loaded onto BINREG2 which can be downloaded at http://www.duke.edu/~dinbarry/BINREG/ for binary regression analysis. The following parameters were chosen in BINREG2: 400 genes/ 3 metagenes /1000 burn in /5000 iterations. The metagene scores for the training data is included in the SI as “Predicted Pathway Probabilities-Training.CD44+CD24−(GSE7513).”
Converting Mouse Affymetrix probe IDs to Human probe IDs for Analysis and merging of datasets: Mouse 430A 2.0 mouse probe IDs from the MTB/TOM data were converted to human U133 probed IDs using Chip Comparer http://chipcomparer.genome.duke.edu/ and File Merger http://filemerger.genome.duke.edu/. The GSE7513, GSE6883 and MTB/TOM data were then normalized again and merged using Bayesian Factor Regression Model (BFRM) (Carvalho et al., 2008; Lucas et al., 2009)to remove technical variation between data sets.
Validation of CD44+/CD24−/low signature on publicly available mouse data: The RAW .CEL files representing sorted and unsorted cells were downloaded from the data set with the gene accession number GSE6883. The data was RMA normalized using Affymetrix Expression Console Version 1.1 and merged and normalized to GSE7513 as described above. The result of this validation set is reported by BINREG2 as an output file denoted as validation.txt. Columns of output are: 1) the column number in input file, 2) phenotype call, 3) average probability, 4) and 5) upper and lower limits of the credible interval, 6) metagene score. The results are provided in the Supplementary Information as the “Predicted Pathway Probabilities-GSE6883.CD44+CD24−.validation” file.
Predicting CD44+/CD24−/low pathway activity in MTB/TOM tumors: The data was RMA normalized using Affymetrix Expression Console Version 1.1. Mouse Affymetrix probe IDs were converted to human Affymetrix probe IDs as described above. Analysis in BINREG2 was performed using the parameters described above to generate the training data. The result of this validation set is reported by BINREG2 as an output file denoted as validation.txt. Columns of output are: 1) the column number in input file, 2) phenotype call, 3) average probability, 4) and 5) upper and lower limits of the credible interval, 6) metagene score. The results are provided in the Supplementary Information as the “Predicted Pathway Probabilities-MTBTOM.CD44+CD24−(GSE7513)” and “Predicted Pathway Probabilities-MTBTOM(primaries_only).CD44+CD24−(GSE7513)” file.
Comparison of Predicted CD44+/CD24−/low probabilities: Predicted probabilities were graphed on Graph Pad Prism 4.0. A two-tailed T-test was used to statistical significance of the comparisons between “Primaries” vs. “Myc-independent” samples and “Regressed” vs. “No Regression” as shown in Figure 5.
Correlation Analysis of Signaling Pathways
A Pearson correlation analysis between signaling pathways were performed using GraphPad Prism version 4.00 for Windows. Predicted pathway probabilities were enter into GraphPad followed by selection of the “Analyze”, “Built-in analysis”, and “Correlation” options, sequentially. A Pearson correlation and a two-tailed P value were then selected for the analysis and a “r squared” value representing the coefficient of determination was reported. The data was graphed in Excel.
Immunohistochemistry
Mouse mammary tissues were sectioned and paraffin embedded by Duke Pathology Department. H&E staining Immunohistochemical staining of tissues for EMT markers were performed using the Vector M.O.M Immunodetection Peroxidase Kit. In brief, paraffin embedded mouse mammary tissues were deparaffinized and hydrated using standard procedures. Antigen unmasking was performed by heating slides in a citrate buffer solution (10mM Citric Acid, pH 6.0, 0.05% Tween 20). Endogenous peroxidase activity was blocked in 3% hydrogen peroxide and Avidin/Biotin blocking was performed using Vector Avidin/Biotin Blocking Kit (cat. No. SP-2001). Immunohistochemistry staining was performed using the Vector M.O.M Immunodetection Peroxidase Kit following manufacturer’s instructions. Diaminobenzidine (DAB) was used as a substrate and haemotoxylin was used as a counterstain. Slides were mounted and viewed on a confocal microscope. The following antibodies were used: E-cadhering G-10 (sc-8426), Cytokeratin18 RGE53 (sc-32324), Vimentin RV202 (sc-32322), pSmad3 (sc-130218) and TNFα (sc-52746) were from Santa Cruz. Fibronectin (610077) was from BD Transduction Laboratories.
Supplementary Material
Supplementary Figure I. Pathway activity was assessed for NMuMG cells treated with a control or TGFbeta (GSE13986); Murine primary hepatocytes treated with a control or TNFalpha (GSE 19272); and Mouse mammary tissue uninduced or induced for transgenic Ras expression (GSE13986) using training data generated in panel A. Differential activation of the oncogenic pathways examined is observed between the “primary” and “recurrent” tumors. The middle “mixed” group contains both “primary” and “recurrent” tumors.
Supplementary Figure II. A. Heat map of genes used to generate a gene signature representing CD44+/CD24−/low cells. B. CD44+/CD24−/low pathway probabilities were assessed for CD44+/CD24−/low -sorted tumor cells from an independent data set (GSE 6883).
Acknowledgments
The MTB/TOM mouse was a kind gift from Lewis A. Chodosh. We thank Kenichiro Fujiwara for assistance in animal husbandry and Rachel E. Rempel for helpful advice. We are grateful to Kaye Culler for her assistance with the manuscript.
This work was supported by grants to JRN (RO1-CA104663, and U54-CA112952) from the National Cancer Institute. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Cancer Institute or the National Institutes of Health.
Footnotes
Potential Conflicts of Interest
Joseph R. Nevins:
Expression Analysis - ownership interest
Millenium Pharmaceuticals and Qiagen - member of SAB
Literature Cited
- Acloque H, Thiery JP, Nieto MA. The physiology and pathology of the EMT. Meeting on the epithelial-mesenchymal transition. EMBO Rep. 2008;9:322–326. doi: 10.1038/embor.2008.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al-Hajj M, Wicha MS, Benito-Hernandez A, Morrison SJ, Clarke MF. Prospective identification of tumorigenic breast cancer cells. Proc Natl Acad Sci U S A. 2003;100:3983–3988. doi: 10.1073/pnas.0530291100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrechek ER, Cardiff RD, Chang JT, Gatza ML, Acharya CR, Potti A, Nevins JR. Genetic heterogeneity of Myc-induced mammary tumors reflecting diverse phenotypes including metastatic potential. Proc Natl Acad Sci U S A. 2009;106:16387–16392. doi: 10.1073/pnas.0901250106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bild AH, Yao G, Chang JT, Wang Q, Potti A, Chasse D, Joshi MB, Harpole D, Lancaster JM, Berchuck A, et al. Oncogenic pathway signatures in human cancers as a guide to targeted therapies. Nature. 2006;439:353–357. doi: 10.1038/nature04296. [DOI] [PubMed] [Google Scholar]
- Boxer RB, Jang JW, Sintasath L, Chodosh LA. Lack of sustained regression of c-MYC-induced mammary adenocarcinomas following brief or prolonged MYC inactivation. Cancer Cell. 2004;6:577–586. doi: 10.1016/j.ccr.2004.10.013. [DOI] [PubMed] [Google Scholar]
- Carvalho CM, Chang J, Lucas JE, Nevins JR, Wang Q, West M. High-Dimensional Sparse Factor Modeling: Applications in Gene Expression Genomics. J Am Stat Assoc. 2008;103:1438–1456. doi: 10.1198/016214508000000869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Creighton CJ, Li X, Landis M, Dixon JM, Neumeister VM, Sjolund A, Rimm DL, Wong H, Rodriguez A, Herschkowitz JI, et al. Residual breast cancers after conventional therapy display mesenchymal as well as tumor-initiating features. Proc Natl Acad Sci U S A. 2009;106:13820–13825. doi: 10.1073/pnas.0905718106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Cruz CM, Gunther EJ, Boxer RB, Hartman JL, Sintasath L, Moody SE, Cox JD, Ha SI, Belka GK, Golant A, et al. c-MYC induces mammary tumorigenesis by means of a preferred pathway involving spontaneous Kras2 mutations. Nat Med. 2001;7:235–239. doi: 10.1038/84691. [DOI] [PubMed] [Google Scholar]
- Diehn M, Cho RW, Clarke MF. Therapeutic implications of the cancer stem cell hypothesis. Semin Radiat Oncol. 2009;19:78–86. doi: 10.1016/j.semradonc.2008.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding L, Getz G, Wheeler DA, Mardis ER, McLellan MD, Cibulskis K, Sougnez C, Greulich H, Muzny DM, Morgan MB, et al. Somatic mutations affect key pathways in lung adenocarcinoma. Nature. 2008;455:1069–1075. doi: 10.1038/nature07423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gatza ML, Lucas JE, Barry WT, Kim JW, Wang Q, Crawford MD, Datto MB, Kelley M, Mathey-Prevot B, Potti A, Nevins JR. A pathway-based classification of human breast cancer. Proc Natl Acad Sci U S A. 107:6994–6999. doi: 10.1073/pnas.0912708107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herschkowitz JI, Simin K, Weigman VJ, Mikaelian I, Usary J, Hu Z, Rasmussen KE, Jones LP, Assefnia S, Chandrasekharan S, et al. Identification of conserved gene expression features between murine mammary carcinoma models and human breast tumors. Genome Biol. 2007;8:R76. doi: 10.1186/gb-2007-8-5-r76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang E, Ishida S, Pittman J, Dressman H, Bild A, Kloos M, D’Amico M, Pestell RG, West M, Nevins JR. Gene expression phenotypic models that predict the activity of oncogenic pathways. Nat Genet. 2003;34:226–230. doi: 10.1038/ng1167. [DOI] [PubMed] [Google Scholar]
- Jain M, Arvanitis C, Chu K, Dewey W, Leonhardt E, Trinh M, Sundberg CD, Bishop JM, Felsher DW. Sustained loss of a neoplastic phenotype by brief inactivation of MYC. Science. 2002;297:102–104. doi: 10.1126/science.1071489. [DOI] [PubMed] [Google Scholar]
- Lucas J, Carvalho C, West M. A bayesian analysis strategy for cross-study translation of gene expression biomarkers. Stat Appl Genet Mol Biol. 2009;8:Article 11. doi: 10.2202/1544-6115.1436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mani SA, Guo W, Liao MJ, Eaton EN, Ayyanan A, Zhou AY, Brooks M, Reinhard F, Zhang CC, Shipitsin M, et al. The epithelial-mesenchymal transition generates cells with properties of stem cells. Cell. 2008;133:704–715. doi: 10.1016/j.cell.2008.03.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moody SE, Perez D, Pan TC, Sarkisian CJ, Portocarrero CP, Sterner CJ, Notorfrancesco KL, Cardiff RD, Chodosh LA. The transcriptional repressor Snail promotes mammary tumor recurrence. Cancer Cell. 2005;8:197–209. doi: 10.1016/j.ccr.2005.07.009. [DOI] [PubMed] [Google Scholar]
- Moody SE, Sarkisian CJ, Hahn KT, Gunther EJ, Pickup S, Dugan KD, Innocent N, Cardiff RD, Schnall MD, Chodosh LA. Conditional activation of Neu in the mammary epithelium of transgenic mice results in reversible pulmonary metastasis. Cancer Cell. 2002;2:451–461. doi: 10.1016/s1535-6108(02)00212-x. [DOI] [PubMed] [Google Scholar]
- Mullighan CG, Goorha S, Radtke I, Miller CB, Coustan-Smith E, Dalton JD, Girtman K, Mathew S, Ma J, Pounds SB, et al. Genome-wide analysis of genetic alterations in acute lymphoblastic leukaemia. Nature. 2007;446:758–764. doi: 10.1038/nature05690. [DOI] [PubMed] [Google Scholar]
- Pelengaris S, Khan M, Evan GI. Suppression of Myc-induced apoptosis in beta cells exposes multiple oncogenic properties of Myc and triggers carcinogenic progression. Cell. 2002;109:321–334. doi: 10.1016/s0092-8674(02)00738-9. [DOI] [PubMed] [Google Scholar]
- Podsypanina K, Politi K, Beverly LJ, Varmus HE. Oncogene cooperation in tumor maintenance and tumor recurrence in mouse mammary tumors induced by Myc and mutant Kras. Proc Natl Acad Sci U S A. 2008;105:5242–5247. doi: 10.1073/pnas.0801197105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shachaf CM, Kopelman AM, Arvanitis C, Karlsson A, Beer S, Mandl S, Bachmann MH, Borowsky AD, Ruebner B, Cardiff RD, et al. MYC inactivation uncovers pluripotent differentiation and tumour dormancy in hepatocellular cancer. Nature. 2004;431:1112–1117. doi: 10.1038/nature03043. [DOI] [PubMed] [Google Scholar]
- Singh A, Settleman J. EMT, cancer stem cells and drug resistance: an emerging axis of evil in the war on cancer. Oncogene. 2010;29:4741–4751. doi: 10.1038/onc.2010.215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sjoblom T, Jones S, Wood LD, Parsons DW, Lin J, Barber TD, Mandelker D, Leary RJ, Ptak J, Silliman N, et al. The consensus coding sequences of human breast and colorectal cancers. Science. 2006;314:268–274. doi: 10.1126/science.1133427. [DOI] [PubMed] [Google Scholar]
- Thiery JP. Epithelial-mesenchymal transitions in tumour progression. Nat Rev Cancer. 2002;2:442–454. doi: 10.1038/nrc822. [DOI] [PubMed] [Google Scholar]
- Thiery JP, Acloque H, Huang RY, Nieto MA. Epithelial-mesenchymal transitions in development and disease. Cell. 2009;139:871–890. doi: 10.1016/j.cell.2009.11.007. [DOI] [PubMed] [Google Scholar]
- Weir BA, Woo MS, Getz G, Perner S, Ding L, Beroukhim R, Lin WM, Province MA, Kraja A, Johnson LA, et al. Characterizing the cancer genome in lung adenocarcinoma. Nature. 2007;450:893–898. doi: 10.1038/nature06358. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure I. Pathway activity was assessed for NMuMG cells treated with a control or TGFbeta (GSE13986); Murine primary hepatocytes treated with a control or TNFalpha (GSE 19272); and Mouse mammary tissue uninduced or induced for transgenic Ras expression (GSE13986) using training data generated in panel A. Differential activation of the oncogenic pathways examined is observed between the “primary” and “recurrent” tumors. The middle “mixed” group contains both “primary” and “recurrent” tumors.
Supplementary Figure II. A. Heat map of genes used to generate a gene signature representing CD44+/CD24−/low cells. B. CD44+/CD24−/low pathway probabilities were assessed for CD44+/CD24−/low -sorted tumor cells from an independent data set (GSE 6883).