Abstract
Acclimation of Chlamydomonas reinhardtii cells to low levels of singlet oxygen, produced either by photoreactive chemicals or high light treatment, induces a specific genetic response that strongly increases the tolerance of the algae to subsequent exposure to normally lethal singlet oxygen-producing conditions. The genetic response includes the increased expression of various oxidative stress response and detoxification genes, like the glutathione peroxidase homologous gene GPXH/GPX5 and the σ-class glutathione-S-transferase gene GSTS1. To identify components involved in the signal transduction and activation of the singlet oxygen-mediated response, a mutant selection was performed. This selection led to the isolation of the singlet oxygen resistant 1 (sor1) mutant, which is more tolerant to singlet oxygen-producing chemicals and shows a constitutively higher expression of GPXH and GSTS1. Map-based cloning revealed that the SOR1 gene encodes a basic leucine zipper transcription factor, which controls its own expression and the expression of a large number of oxidative stress response and detoxification genes. In the promoter region of many of these genes, a highly conserved 8-bp palindromic sequence element was found to be enriched. This element was essential for GSTS1 induction by increased levels of lipophilic reactive electrophile species (RES), suggesting that it functions as an electrophile response element (ERE). Furthermore, GSTS1 overexpression in sor1 requires the ERE, although it is unknown whether it occurs through direct binding of SOR1 to the ERE. RES can be formed after singlet oxygen-induced lipid peroxidation, indicating that RES-stimulated and SOR1-mediated responses of detoxification genes are part of the singlet oxygen-induced acclimation process in C. reinhardtii.
Keywords: photosynthesis, reactive oxygen species, retrograde signaling, oxylipin
In photosynthetic organisms, exposure to harsh environmental conditions, such as high light intensities or low temperature, can lower the photosynthetic efficiency and stimulate the formation of reactive oxygen species (ROS) like singlet oxygen (1O2) and superoxide anion radicals (1). To avoid ROS-induced damage, physiological acclimation processes are activated, many of which depend on the increased synthesis of nucleus-encoded proteins (2). Therefore, the expression of the corresponding genes is controlled by plastid signals responding to various factors such as the plastid gene expression activity, levels of chlorophyll precursors, the redox state of the photosynthetic electron transport chain, and the formation of ROS (recent reviews in refs. 3–5). Mutant analyses have revealed several plastid (e.g., GUN1–5 and EXECUTER1 and -2) or cytosolic proteins (ABI4, GLK, and PTM) that participate in the chloroplast to nucleus retrograde signaling (3, 6). However, the molecules transmitting the plastid signals of the chloroplast are still unknown.
Singlet oxygen is one ROS that can function as a plastid signal to activate nuclear gene expression (2). In Arabidopsis thaliana, 1O2 produced in the chloroplast of the conditional fluorescent (flu) mutant was shown to activate a programmed cell death (PCD) response (7). In Chlamydomonas reinhardtii, 1O2 produced during acclimation to high light intensities induces an acclimation response that involves increased expression of various nuclear defense proteins (8). This expression includes a thioredoxin peroxidase encoded by the glutathione peroxidase homologous gene GPXH/GPX5 (9), which was shown to be specifically induced by 1O2 (10, 11), and a σ-class glutathione-S-transferase (GSTS1) more generally induced by different ROS (8, 12). The high reactivity of 1O2 suggests that it acts close to its site of production (13). Thus, it was argued that, for induction of nuclear genes, 1O2 reaction products such as oxidized lipids might function as second messengers to transmit the signal out of the chloroplast (5, 14). In the A. thaliana flu mutant, fatty acid-derived oxidation products (oxylipins) were detected shortly after the dark–light shift, which stimulated 1O2 production (7). However, these oxylipins were formed enzymatically and resulted in the accumulation of the phytohormones 12-oxophytodienoic acid (OPDA) and jasmonic acid (JA) (15). These hormones are known to regulate various metabolic and developmental processes but are probably not specific second messengers of 1O2 signaling. Nevertheless, involvement of enzymatically formed oxylipins in 1O2 signaling has recently been shown using mutants of the 9-lipoxygenases initiating fatty acid oxidation in plants (16). However, nonenzymatic lipid oxidation products might function in 1O2-mediated signal transduction and activate redox-sensitive transcription factors by their strong electrophilic properties (5, 17). Interestingly, OPDA, itself a reactive electrophile species (RES), also stimulates the expression of a set of genes that are distinct from JA-induced genes and include many genes involved in oxidative stress response and detoxification of xenobiotics (18). In C. reinhardtii, fast GPXH induction by 1O2 produced by low rose bengal (RB) concentrations in the light was not affected by depleting lipophilic antioxidants, excluding fatty acid-derived RES as potential signal molecules for this sensitive 1O2 response (19). However, the GPXH gene is also induced by organic hydroperoxides through a slower-acting induction mechanism, indicating that both lipid peroxidation products and 1O2 activate the expression of the same defense gene, although with different kinetics (20). Thus, 1O2-derived RES might at least partially regulate the response of defense genes to 1O2.
Classical genetic approaches have provided valuable information about the regulation of oxidative stress responses in general and the response to 1O2 specifically in several reference systems. In Rhodobacter sphaeroides, mutants of the alternative σ-factor σE showed a reduced genetic response and were more sensitive to 1O2, suggesting that σE mediates the 1O2-specific induction of defense genes (21). In Saccharomyces cerevisiae, response to peroxide and 1O2 stress is regulated by the basic leucine zipper (bZIP) transcription factor YAP1, and yap1 mutants cannot acclimate to H2O2 and are sensitive to RB (22, 23). Screening for suppressor mutants of the PCD response in the A. thaliana flu mutant resulted in the identification of the executer1 and -2 mutants, in which the affected genes encode two chloroplast-localized proteins required for the induction of nuclear gene expression and PCD after 1O2 release (24, 25).
To identify components of the 1O2-signaling pathway in C. reinhardtii, mutants with an altered genetic response to 1O2-producing chemicals were isolated (26). However, none of these mutants was affected in any downstream signaling component. In the present study, we isolated RB-resistant mutants, one of which showed a strong constitutive expression of the GPXH and GSTS1 genes. Additional characterization of the mutant revealed a mutation in a gene encoding a putative bZIP transcription factor that regulates the expression of various oxidative stress and RES detoxification genes in response to RES signals.
Results
Isolation of Singlet Oxygen-Resistant Mutants.
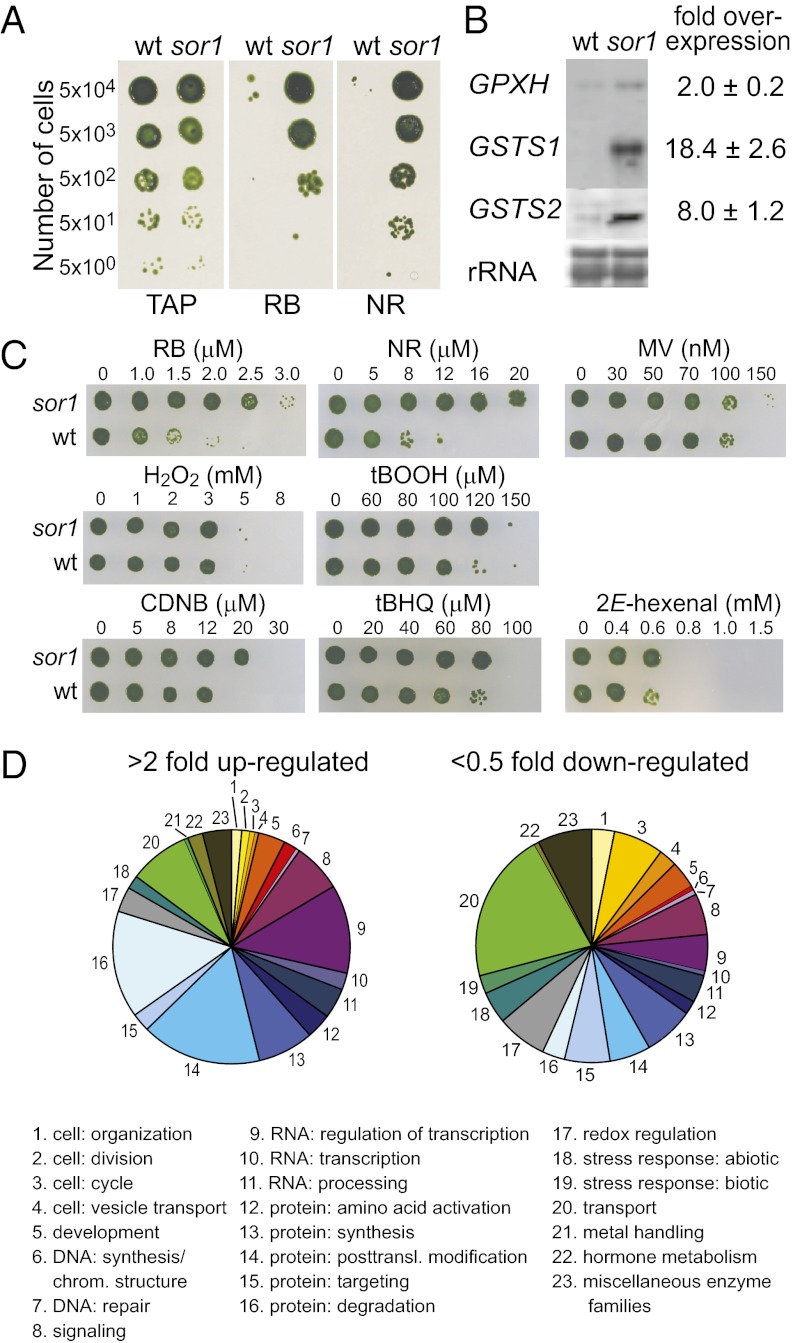
The green alga C. reinhardtii is able to increase its tolerance to 1O2 stress by activating an acclimation process during exposure to low levels of 1O2 that involves the specific induction of defense genes (8). To identify cellular components that trigger this acclimation process, we sought to isolate mutants showing constitutive acclimation and increased tolerance to 1O2. Therefore, the WT strain 4A+ was UV-mutagenized and plated onto agar plates containing a lethal concentration of the 1O2-photosensitizer RB. After an overnight recovery in the dark, plates were shifted to continuous light for 1 wk, at which point 40 RB-resistant clones were isolated. To confirm increased 1O2 resistance and identify mutants that are potentially affected in an upstream regulatory component of defense against 1O2, the 40 clones were rescreened for 1O2 tolerance using RB and another photosensitizer, neutral red (NR), and then tested for the expression of the putative target genes GPXH, GSTS1, and GSTS2. One of three mutants passing this second screen was called singlet oxygen resistant 1 (sor1), and it showed increased resistance to RB and NR and strong overexpression of 1O2-response genes (Fig. 1 A and B).
Fig. 1.
Phenotypic characterization of sor1. (A) sor1 was isolated as a 1O2-resistant mutant by growing on solid TAP medium containing lethal concentrations of the chemicals RB (2 μM) and NR (8 μM). (B) RNA gel blot analysis revealed that the sor1 mutant exhibits a constitutive higher expression of the 1O2-responsive genes GPXH, GSTS1, and GSTS2 compared with WT, which was confirmed quantitatively by qPCR. (C) Quantitative analyses of the sor1 resistance to various oxidative and electrophilic stress conditions were done in liquid cultures by growing cells with increasing concentration of individual chemical for 24 h and subsequent spotting on agar plates for recovery. (D) Distribution of annotated genes (496 up-regulated and 288 down-regulated) of 1,895 genes differently expressed at least twofold in sor1 compared with WT into different functional categories based on Mapman (28). Size of the slice for each functional group is based on the absolute number of up- or down-regulated genes annotated to each category.
GSTS1 and GSTS2 are both 1O2-response genes; however, their responses are less specific for 1O2 than GPXH induction, and they are also strongly induced by other oxidative stress conditions (12). Because sor1 exhibits a stronger overexpression of GSTS1 and GSTS2 than GPXH (Fig. 1B), we tested the resistance of sor1 to different ROS in a more quantitative manner. Cultures of sor1 and the WT were exposed to increasing concentrations of various ROS-producing chemicals in liquid cultures for 24 h to measure growth and then spotted on agar plates to analyze viability. In the absence of any chemical, there was no significant difference in the growth rates of the two strains (sor1: 1.91 ± 0.08 d−1; 4A+: 1.85 ± 0.11 d−1), showing that the metabolic cost for overexpression of the defense genes is probably low. In the presence of RB and NR, however, sor1 survived concentrations that were at least twofold higher than those concentrations survived by the WT strain, but sor1 did not show increased resistance to the superoxide radical-producing herbicide methyl viologen (MV) or H2O2 (Fig. 1C and Table 1). Interestingly, sor1 was slightly more tolerant to the organic tert-butylhydroperoxide (tBOOH). GSTs belong to the phase II detoxification system, which is involved in the removal of xenobiotics, including electrophiles like substituted quinones and plant oxylipins, from cells by conjugation and subsequent elimination (27). This finding prompted us to test resistance of sor1 to various RES; sor1 was found to be more resistant than the WT strain to the potential GST substrates 1-chloro-2,4-dinitrobenzene (CDNB) and 2E-hexenal, and also, it was found to be slightly more resistant to tert-butylhydroquinone (tBHQ).
Table 1.
Induction of a GSTS1-GLUC reporter construct in WT exposed to different chemical treatments and EC50 ± SEM of the chemicals in WT and sor1
| Chemical | Concentration | Fold induction (±SEM) |
EC50 (μM) |
||
| Light | Dark | WT | sor1 | ||
| Salt or osmotic stress | |||||
| NaCl | 200 mM | 1.8 ± 0.5 | n.d. | n.d. | n.d. |
| Sucrose | 600 mM | 1.5 ± 0.5 | n.d. | n.d. | n.d. |
| Photosynthesis inhibitor | |||||
| DCMU | 0.1 μM | 1.1 ± 0.2 | n.d. | n.d. | n.d. |
| DBMIB | 5 μM | 325 ± 120 | 230 ± 72 | 5.8 ± 0.7 | 5.7 ± 0.3 |
| ROS-generating chemicals | |||||
| H2O2 | 2 mM | 5.9 ± 2.4 | 1.0 ± 0.1 | 2,100 ± 205 | 1,977 ± 294 |
| MV | 0.1 μM | 1.1 ± 0.2 | 1.0 ± 0.1 | 0.056 ± 0.004 | 0.057 ± 0.002 |
| NR | 5 μM | 13.7 ± 2.0 | 1.0 ± 0.1 | 4.1 ± 0.4 | 7.9 ± 0.5* |
| RB | 1 μM | 4.2 ± 1.6 | 0.6 ± 0.2 | 1.3 ± 0.1 | 1.8 ± 0.1* |
| tBOOH | 100 μM | 30.3 ± 9.1 | 12.5 ± 2.1 | 111 ± 4 | 139 ± 4* |
| RES-generating chemicals | |||||
| p-Benzoquinone | 20 μM | 130 ± 8 | 53.2 ± 17.6 | 19.9 ± 0.4 | 30.5 ± 4.0† |
| tBHQ | 30 μM | 27.5 ± 6.8 | 40.3 ± 9.7 | 28.5 ± 1.0 | 30.8 ± 1.6 |
| CDNB | 10 μM | 125 ± 18 | 24.8 ± 6.6 | 8.7 ± 0.4 | 12.6 ± 0.8* |
| 2E-Hexenal | 500 μM | 232 ± 33 | 107 ± 16 | 471 ± 11 | 541 ± 18* |
| Phytohormones | |||||
| OPDA | 200 μM | 23.9 ± 5.6 | n.d. | n.d. | n.d. |
| JA | 200 μM | 1.0 ± 0.1 | n.d. | n.d. | n.d. |
n.d., not determined.
*Significant difference between sor1 and the WT resistance (P < 0.01).
†Significant difference between sor1 and the WT resistance (P < 0.05).
To identify more genes that are differentially regulated in sor1, a genome-wide expression analysis was performed. Overexpression of GPXH in sor1 was found to be higher during the light period of synchronously grown cultures. Therefore, total RNA was isolated from synchronously grown cultures of sor1 and WT 6 h after starting the light phase. Abundance of mRNAs was analyzed by RNA-Sequencing (RNA-Seq), and a total of 15,818 transcripts were identified, of which 1,287 were up- and 608 were down-regulated more than twofold in sor1 compared with the WT strain. Of the genes with known functions, the most strongly up-regulated genes in sor1 are a pyridoxamine 5′-phosphate oxidase required for vitamin B6 synthesis, an agmatine-iminohydrolase involved in a polyamine pathway, and GSTS1. For many other induced genes, it was only possible to annotate functions on the basis of Pfam protein domains. Additional analyses of cellular functions were done based on Mapman annotations (28), which revealed that the differently expressed genes encode for proteins with a broad range of cellular functions (Fig. 1D). However, only genes of a few functional groups, like cell division and transport, were slightly overrepresented in the fraction of overexpressed genes, whereas genes for vesicle transport were the only significant group in the down-regulated fraction (P < 0.05) (Table S1).
SOR1 Gene Encodes a Putative bZIP Transcription Factor.
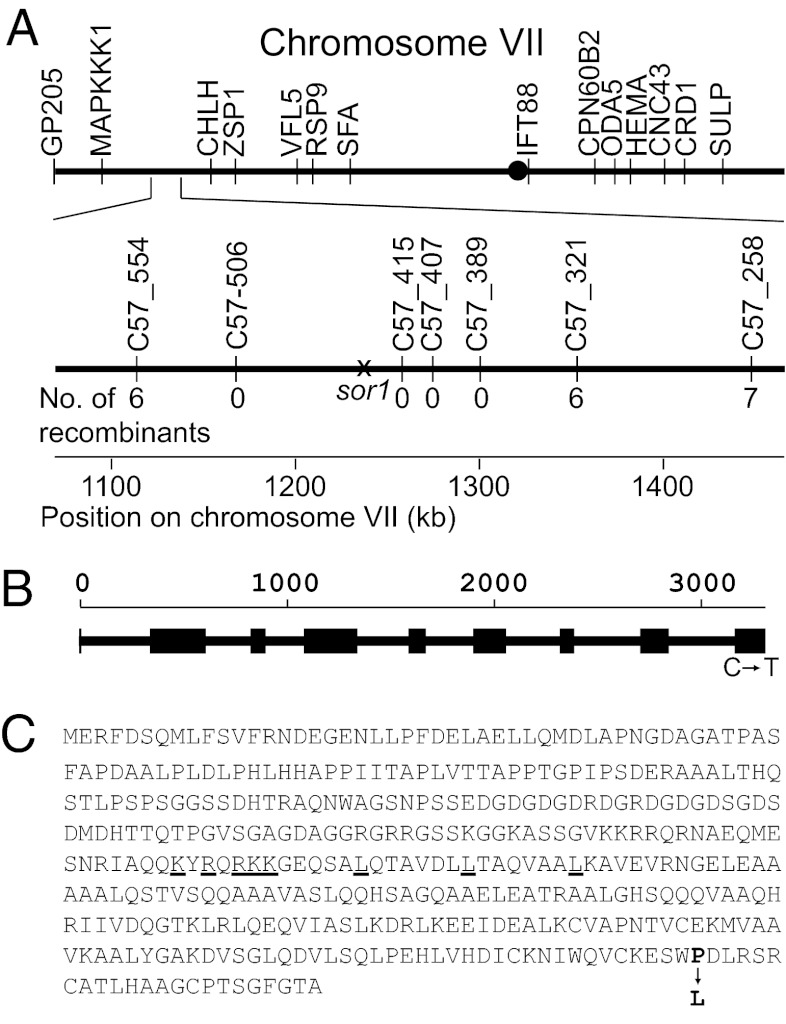
The sor1 mutant was created by UV mutagenesis and thus, was backcrossed four times to the WT to eliminate any undesired nuclear mutations. Segregation analysis of these backcrosses showed a clear 2:2 segregation of the resistance to RB and tBOOH in all tetrads tested, indicating that a single nuclear mutation is responsible for the sor1 phenotype (Fig. S1). To localize this mutation in the genome of C. reinhardtii, a map-based cloning approach was applied by crossing the mutant to the polymorphic strain S1-C5 and testing the progenies for segregation of the sor1 phenotype with different molecular markers (29). This process allowed an initial mapping of the sor1 mutation to the long arm of chromosome VII close to the chromosomal marker CHLH (Fig. 2A). For fine mapping, additional molecular markers were designed for the two strains, and analysis of a total of 214 progenies allowed localization of sor1 to a 200-kb region of the genome in which no additional recombination could be detected. This region was searched for candidate genes involved in regulation of gene expression. Six candidate genes were identified, including two transcription factors and four guanylyl cyclases. Sequencing of these genes revealed a C to T transition in the predicted coding region of one of two putative transcription factors (JGI-V4 protein ID 187531) (Fig. 2B). Sequencing of the corresponding transcript confirmed the computationally predicted model for the gene, which is composed of nine exons and eight introns, and positioned the mutation in the last exon, which is 3,263 bp downstream of the translation start codon. In this gene model, the sor1 mutation is predicted to cause a single amino acid change of proline 371 into a leucine near the C-terminal end of the 393-aa protein (Fig. 2C). Similarity searches for the SOR1 protein in common sequence databases revealed no homology to any known protein of other organisms, except for one putative ortholog (74% identity) in the genome of the closely related alga Volvox carteri. The protein was predicted to function as a transcription factor based on the identification of a putative bZIP DNA binding domain (E-value = 5.02 × 10−5), indicating that the sor1 phenotype could be caused by a mutation in a transcription factor.
Fig. 2.
Map-based cloning of SOR1. (A) Localization of the sor1 mutation to a 200-kb region on the long arm of chromosome VII. The position of established molecular markers as well as newly designed markers within the region of 1,100–1,500 kb chromosome VII and the number of recombinants for each marker are indicated. (B) Location of a C to T transition at position 3,263 bp relative to the translation start codon within the ninth exon of the 3,333-bp SOR1 gene. (C) Amino acid sequence (one letter code) of the corresponding SOR1 protein with the change of proline 371 into a leucine in the sor1 mutant indicated by an arrow. The positions of amino acids of the predicted bZIP DNA binding domain are underlined.
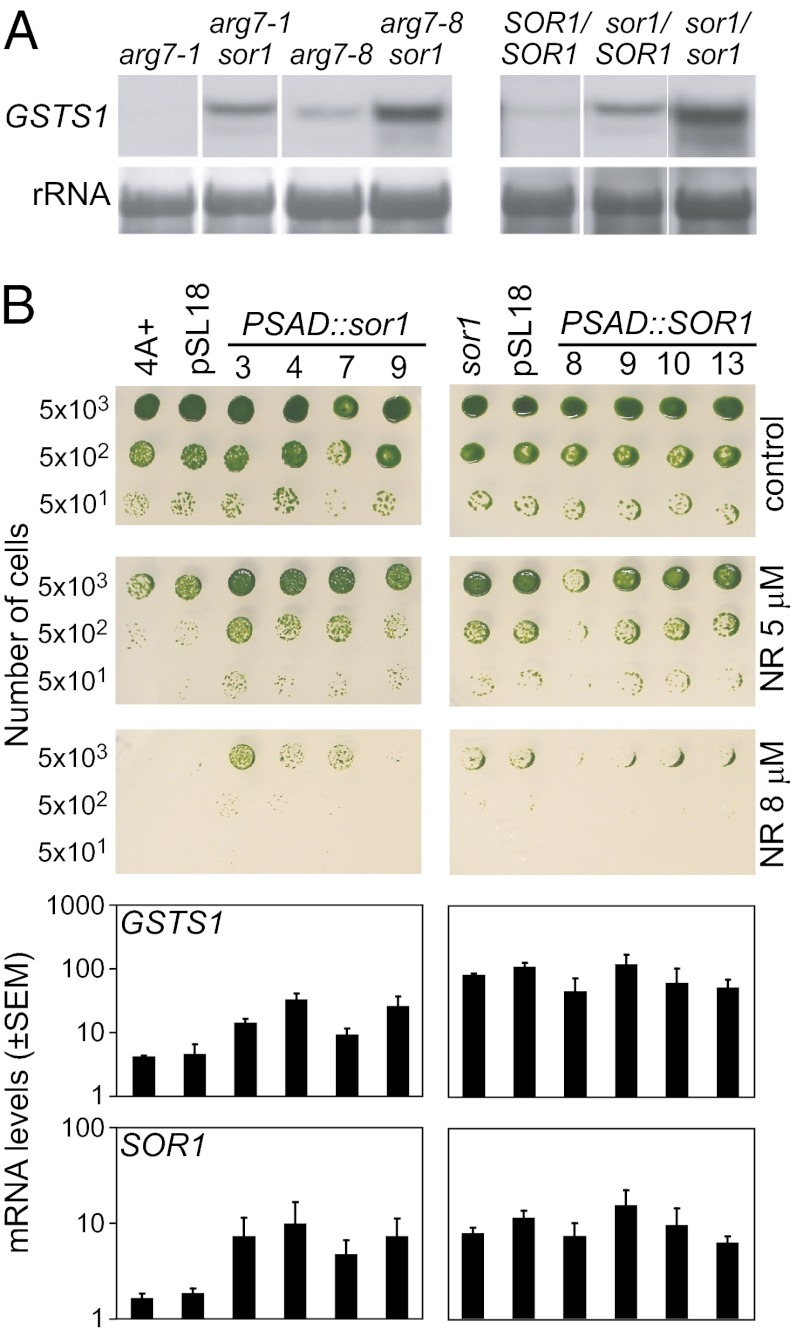
To confirm that the mutation identified in the putative bZIP transcription factor is responsible for the sor1 phenotype, complementation experiments were performed. However, sor1 might be a gain of function mutation, which would be dominant and hence, not be complemented by the WT allele. To test dominance of sor1, vegetative diploid C. reinhardtii strains were constructed. The heterozygous sor1/SOR1 diploid showed intermediate expression of GSTS1 compared with the sor1/sor1 and SOR1/SOR1 homozygote controls. This finding shows that the sor1 mutation is semidominant and most likely, a gain of function mutation (Fig. 3A).
Fig. 3.
Dominance and complementation of sor1. (A) The sor1 mutation was crossed into the arg7-1 and arg7-8 backgrounds, and vegetative diploids that are homozygous mutant (sor1/sor1), WT (SOR1/SOR1), or heterozygous (sor1/SOR1) were generated. Dominance of the mutation was tested by determining GSTS1 expression with RNA gel blot analysis using the amount of rRNA as a loading control. (B) Complementation of sor1. Phenotypes of four independent clones of WT (4A+) or sor1 transformed with constructs overexpressing either the mutant sor1 or WT SOR1 coding region with the PSAD promoter are shown. Because of the semidominant character of the mutated sor1 gene, 4A+ overexpressing sor1 became more resistance to NR on agar plates and exhibited higher expression of GSTS1 analyzed by qPCR compared with 4A+ or 4A+ transformed with the empty vector pSL18. However, when sor1 overexpressed the WT SOR1 protein, no complementation was detected for most transformants.
As a consequence of the semidominance of sor1, complementation analyses were performed by overexpressing either the sor1 mutant allele in the WT strain or the SOR1 WT gene in the sor1 mutant using the PSAD promoter (30). Transformants with the overexpression constructs were screened for their resistance to NR and tested for overexpression of the complementing sor1 or SOR1 transcript. Several clones of WT overexpressing the sor1 mutant gene were found to be more resistant to NR and express the GSTS1 gene at higher levels compared with the corresponding WT strain containing only the empty vector (Fig. 3B). This finding confirms that the mutated bZIP transcription factor is responsible for the sor1 phenotype and that sor1 is a gain of function mutation. Complementation of the sor1 mutant with the WT protein, however, resulted in only one clone with slightly reduced NR resistance and no significant reduction in GSTS1 expression compared with sor1 transformed with the empty vector. To our surprise, the SOR1 gene was, similar to GSTS1, already overexpressed in the sor1 mutant before transformation with any overexpression construct, suggesting that the gene autoregulates its own expression. This finding was confirmed by the RNA-Seq data showing that SOR1 mRNA abundance is 2.5-fold higher in the mutant compared with the WT strain.
Role of an 8-bp Palindromic Sequence Element in the Promoters of SOR1, GSTS1, and GPXH.
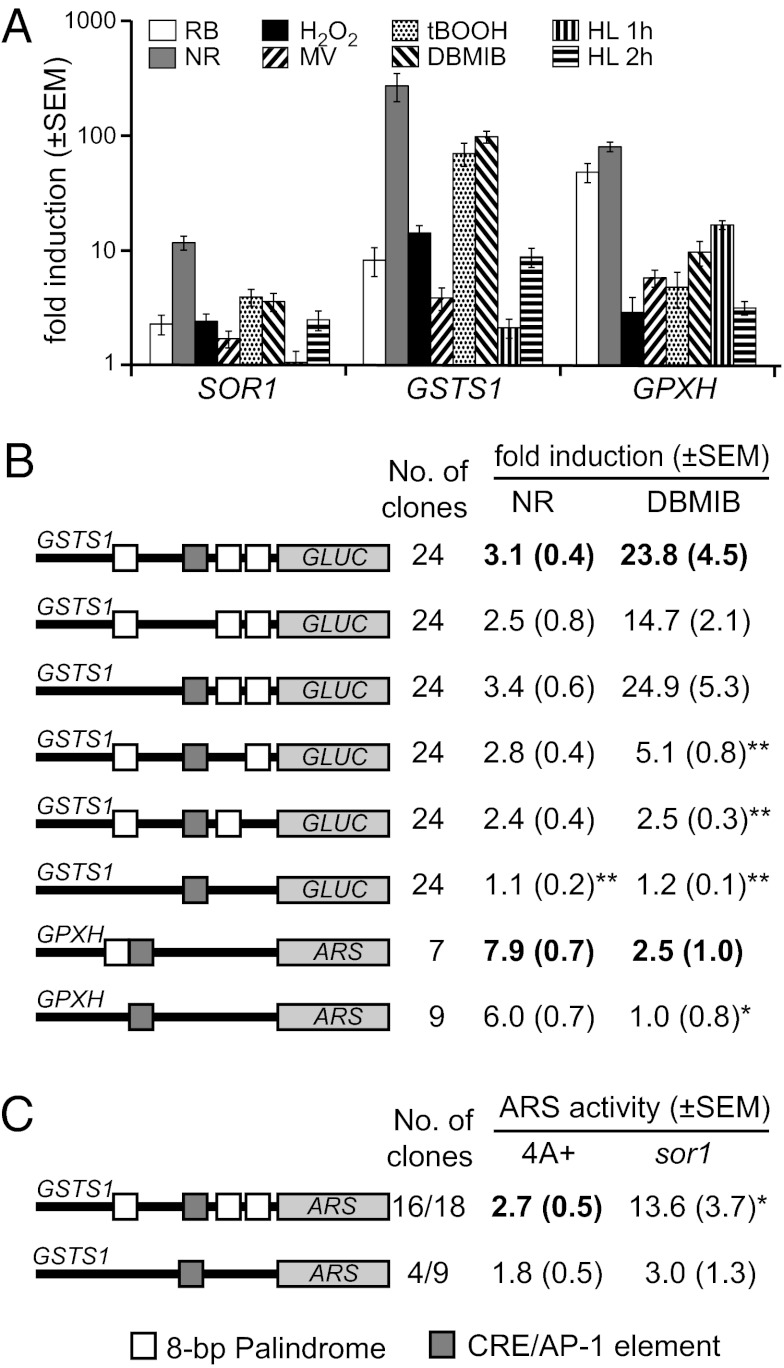
Overexpression of SOR1 in the sor1 mutant raised the question of whether the expression of SOR1 is similarly regulated compared with other genes that are up-regulated in the mutant. Therefore, we tested the induction of the SOR1 gene in the WT strain during exposure to various oxidative stress conditions known to stimulate the expression of GSTS1 and GPXH (12). SOR1 transcript levels indeed increased 11.5-fold after 1 h of NR treatment and were also up-regulated to a lower extent by RB, H2O2, MV, tBOOH, and the herbicide 2,5-dibromo-3-methyl-6-isopropyl-p-benzoquinone (DBMIB), which blocks the photosynthetic electron transport chain (Fig. 4A). Exposure to high light conditions, however, only stimulated SOR1 expression after prolonged exposure of at least 2 h. Although induction of SOR1 was generally lower than the induction of the GSTS1 and GPXH genes under the same condition, SOR1 showed a very similar response pattern to GSTS1, indicating that their expression might be controlled by similar regulatory mechanisms.
Fig. 4.
Induction mechanism of putative sor1 target genes. (A) Induction profiles of the SOR1, GSTS1, and GPXH genes after exposure to different oxidative stress conditions were compared: RB (1 μM), NR (3 μM), H2O2 (2 mM), MV (1 μM), tBOOH (100 μM), and DBMIB (2 μM) for 1 h and high light irradiation (HL; 2,000 μmol photons m−2 s−1) for 1 and 2 h. (B) Independent transformants containing different GSTS1-GLUC or GPXH-ARS reporter constructs were analyzed for the response of the constructs to either 3 μM NR or 2 μM DBMIB. To analyze the role of the putative regulatory elements CRE/AP1 (gray box) or the 8-bp palindrome (ERE; white boxes) in the response of the genes, various constructs with one or several mutated elements were tested for induction by the chemicals. (C) Average ARS activity of the indicated number of WT (4A+) or sor1 clones transformed either with the WT or palindrome-deleted GSTS1 reporter construct. Significant (*P < 0.05, **P < 0.01) differences to the responses of the WT constructs in the WT strain (bold letters) are indicated.
Expression of GPXH during different oxidative stress conditions was shown to be regulated by multiple mechanisms (20, 26). A promoter element homologous to the cAMP-responsive element (CRE) and activator protein 1 (AP-1) binding site in mammals was essential for the induction of a GPXH-arylsulfatase (ARS) reporter construct by 1O2 (11). Such a CRE/AP-1–like element was also found in the promoter region of the GSTS1 gene. To test a putative role of this element in the response of the GSTS1 gene to oxidative stress, reporter constructs with the GSTS1 promoter fused to the Gaussia luciferase (GLUC) gene adapted for C. reinhardtii were cloned (31). Although a high variability in expression and induction of the WT construct was detected in the different transformants, an average up-regulation of 23.8-fold could be measured during exposure to DBMIB, showing that the induction of GSTS1 by this chemical is caused by transcriptional activation (Fig. 4B). Interestingly, NR, which led to the highest GSTS1 transcript levels measured by quantitative PCR (qPCR), only weakly induced the expression of the reporter construct, indicating that this ROS stimulates GSTS1 expression by multiple regulatory mechanisms. Deletion of the CRE/AP-1–like element in the promoter of GSTS1 did not significantly reduce the response of the reporter construct to DBMIB or NR. Another putative regulatory element, consisting of an 8-bp palindromic core sequence (CAACGTTG), was found in the promoters of SOR1, GPXH, and GSTS1. GSTS1 contains three copies of the element within the first 330 bp upstream of the start codon. However, earlier investigations with reporter constructs containing a GPXH promoter with a mutated version of the palindrome revealed that this element was not required for GPXH induction by 1O2 (26). When the same construct was now tested for the response to DBMIB, the low but significant induction of the WT GPXH promoter was completely abolished in the mutated construct (Fig. 4B). We, therefore, deleted the three palindromes in the GSTS1 promoter, either individually or combined, and detected strong reduction of the response to DBMIB when removing either of the two proximal elements. No reduction was detected when the most distal element was deleted, showing that the distance of the palindrome to the transcription start site is important for activation. The triple mutant construct did not respond at all to DBMIB and NR, showing that these elements are required for activation of GSTS1 transcription by these chemicals.
We then investigated whether overexpression of GSTS1 in sor1 is caused by increased transcriptional activity and requires the 8-bp palindrome. Because of the very high expression variability of the same reporter construct containing the GLUC gene in different transformants, this reporter gene could not be used to compare difference in expression strength in sor1 and WT. ARS constructs, however, showed much less expression variability between clones (9). Thus, transformation of a GSTS1-ARS reporter construct into the sor1 strain resulted, on average, in a fivefold higher expression of the reporter construct than when the construct was introduced into the WT strain (Fig. 4C). However, ARS activity was not increased in transformants of sor1 containing the palindrome-deleted GSTS1-ARS reporter construct, which showed comparable average expression activity relative to transformants of the WT strain containing either construct. This finding indicates that overexpression of GSTS1 in sor1 is mediated through the same transcriptional activation mechanisms as DBMIB induction, both requiring the presence of the 8-bp palindrome in the promoter region. Furthermore, transcript half-lives are not changed in sor1, and therefore, a posttranscriptional mechanism cannot explain GSTS1 and SOR1 overexpression (Fig. S2).
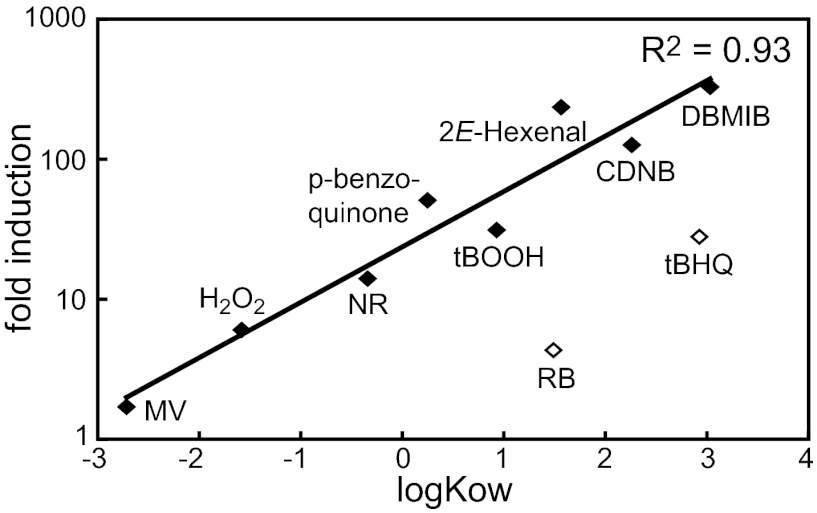
The different expression pattern of the GSTS1 WT gene and GSTS1-GLUC reporter construct by NR and DBMIB (Fig. 4) and the conclusion that multiple regulatory mechanisms might control GSTS1 expression raised the question about the strongest activation signal of GSTS1-GLUC transcription. Therefore, a selection of stress conditions was tested for potency to induce luciferase activity in one representative WT transformant. Nonoxidative stress conditions like salt stress or osmotic stress did not up-regulate the expression of the reporter construct (Table 1). The same was true for the photosynthetic electron transport inhibitor DCMU, whereas DBMIB again strongly stimulated expression by more than 300-fold. DBMIB is often used to study redox signaling by the reduced plastoquinol pool; however, a very strong induction of the GSTS1 reporter construct in the dark indicates that another chemical property of DBMIB is responsible for the genetic response. DBMIB is a p-benzoquinone-derivative, and this group of chemicals is known to be redox-active and electrophilic (32). We, thus, tested GSTS1-GLUC induction by a series of oxidative and electrophilic stress conditions. All of the chemicals that stimulate ROS production also induced the expression of the reporter construct. The strength of induction, however, was rather low compared with DBMIB and reached maximal levels of 30-fold by tBOOH treatment (Table 1). Stronger stimulation of expression was detected by the various RES, including p-benzoquinone, tBHQ, and CDNB. Endogenous electrophiles, such as reactive aldehydes, produced by lipid peroxidation are potential cellular signals to activate detoxification mechanisms (17, 33). Indeed, 2E-hexenal strongly activated GSTS1 transcription, and also, the electrophilic oxylipin OPDA induced the response, although to a lower extent. However, the OPDA-related nonelectrophilic hormone JA did not stimulate expression, indicating that, indeed, the electrophilic property of OPDA is responsible for the response. Furthermore, the GSTS1-GLUC induction strongly correlated with the lipophilic properties of the chemicals indicated by their octanol–water partitioning coefficient (Fig. 5).
Fig. 5.
Correlation of GSTS1-LUC induction by various ROS- or RES-forming chemicals with their putative membrane localization (lipophilicity) based on their octanol water partitioning coefficient logKOW (◆). The correlation coefficient was calculated excluding RB and tBHQ (◇) because of overestimation of their membrane localization by logKOW as a result of the negative charges (RB) or steric hindrance (tBHQ).
Comparison of the Genome-Wide Response in sor1 with Induction by ROS and RES.
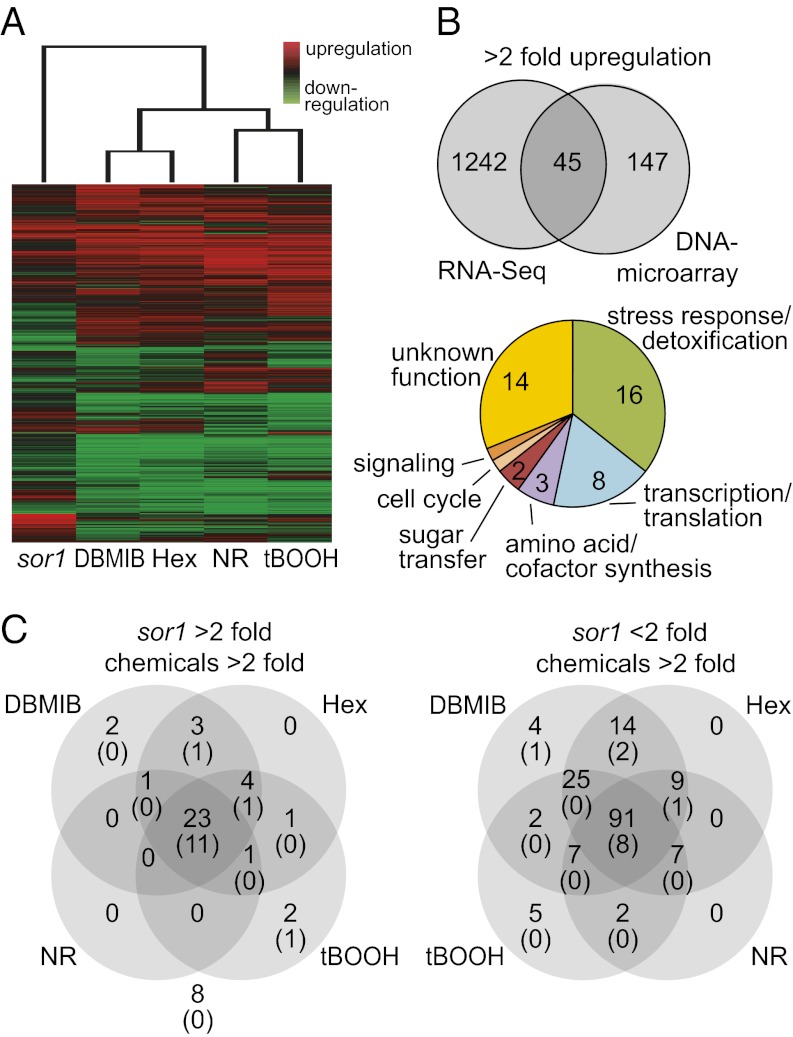
To compare the genetic response of sor1 with the induction profiles of oxidative and electrophilic stress conditions, genome-wide expression analyses were conducted with nonsynchronized cultures of sor1 and WT in the absence of chemicals or WT exposed to NR, tBOOH, DBMIB, or 2E-hexenal for 2 h. Expression levels after individual treatments were quantified with DNA microarrays enabling the assessment of 15,143 transcripts (Fig. 6A). Surprisingly, only 245 genes showed significantly (P < 0.05) different expression more than twofold (192 genes up and 53 genes down) in sor1 compared with WT in this experiment. This finding seems to contradict the much larger number of differently expressed genes found in the RNA-Seq experiments. However, this finding might reflect, on one hand, the lower sensitivity of DNA microarrays compared with sequencing-based expression analysis (34) and on the other hand, the consequence of the different growth conditions used for the experiment (i.e., synchronized vs. nonsynchronized cultures). Indeed, significant overrepresentation of genes involved in cell division among the sor1-overexpressed genes (Table S1) indicates that the sor1 mutant might be affected in the cell cycle, resulting in the strong differences in gene expression of synchronized cultures compared with WT. Still, comparison of the two expression analyses should allow filtering out of putative specific SOR1-responsive genes that are up-regulated in both conditions. This comparison revealed only 45 genes up-regulated more than twofold in both analyses, whereas the majority of genes might be induced in sor1 under certain growth conditions only (Fig. 6B). For 31 of 45 genes, a putative function could be annotated, and most of these genes seem to be either involved in stress response and detoxification (16 genes) or in the control of transcription and translation (8 genes) (Fig. 6B and Dataset S1A).
Fig. 6.
Global genetic response in sor1 and after ROS and RES treatment. (A) Total genome expression profiles analyzed by DNA microarrays in either the sor1 mutant or the WT strain exposed to one of four chemicals [DBMIB (2 μM), 2E-Hexenal (Hex; 0.3 mM), NR (3 μM), or tBOOH (100 μM)] for 2 h. Genes with significant differential expression (P < 0.001, 9,304 genes) in at least one of five conditions were clustered based on similar expression profiles and visualized in a color-based expression pattern. (B) Venn diagram showing the number of genes up-regulated more than twofold in sor1 determined by RNA-Seq in synchronized cultures and DNA microarrays in cultures exposed to continuous light (Right). The 45 genes overexpressed in both conditions were further analyzed for their putative function in cellular processes by homology searches and identification of functional domains (Lower). (C) Venn diagrams showing the distribution of the 45 genes consistently overexpressed twofold in sor1 (Left) and the 166 genes that are more than 20-fold induced by at least one of the chemical treatments but not in sor1 (Right) into different categories based on their more than twofold induction by the chemical treatments. The number of genes in each category (top number) and the number of genes that contains the 8-bp palindrome sequence (ERE) in their promoter regions (lower number in parentheses) are indicated.
Contrary to overexpression of genes in sor1 compared with WT, thousands of genes were induced by one or more of the chemical treatments. DBMIB, 2E-hexenal, and tBOOH induced 2,588, 2,421, and 2,637 genes, respectively, more than twofold (P < 0.05), whereas NR stimulated slightly fewer genes (1,757). Cluster analysis revealed that the induction profiles of DBMIB and 2E-hexenal were most similar (Fig. 6A) and that 1,685 of the genes induced more than twofold were in common. This finding is consistent with the view that DBMIB induces an electrophilic stress response and probably acts as an RES. Expression profiles of the two oxidative stress conditions NR and tBOOH were also more similar to each other than to either DBMIB or 2E-hexenal treatment. Still, the expression patterns of all four stress conditions correlated more strongly to each other than to the pattern of the sor1 mutant. This finding was caused by the rather low number of genes overexpressed in sor1 detected by the microarray experiments, and it showed that only a small part of the sor1 response correlated with the response to oxidative or electrophilic stress conditions. Still, most of the 45 genes up-regulated in both sor1 expression analyses, including all of the 16 stress response and detoxification genes as well as SOR1, were also induced more than twofold by several stress conditions (Fig. 6C and Dataset S1A).
All of the differently expressed genes were then searched for the presence of the 8-bp palindromic sequence element in a 2-kb promoter region upstream of their start codons. There are 411 promoters (2.6%) with at least one CAACGTTG element in the C. reinhardtii genome (of a total of 15,818 Augustus5 gene models). Using this frequency as background, we computed the significance for an overrepresentation of the 8-bp motif in different sets of promoters. For instance, 14 of the 45 genes overexpressed in sor1 contain at least one palindromic sequence (31%, hypergeometric P value = 2.2e-13). None of these 14 genes was in the group of 8 genes overexpressed in sor1 but not induced by any chemical (P value = 0.19), but 11 of these palindrome-containing genes belong to the set of 23 genes induced by all chemical stress conditions (48%, P value = 0) (Fig. 6C and Dataset S1A). One gene (protein ID 404979), encoding a protein of the DJ-1/PfpI family, had three copies of the palindrome like GSTS1. When the promoters of 166 genes strongly induced by chemical treatments (but not more than twofold in sor1) were screened, 12 genes contained the 8-bp palindrome, corresponding to a frequency of 7.2% (P value = 4.0e-04) (Dataset S1B); 5 of these 12 genes, a thiopurine-S-methyltransferase, a zinc-peptidase, a putative GST, GPXH, and a protein of unknown function (protein ID 194234) were overexpressed more than twofold in sor1 in at least one of two expression analyses and all chemical treatments, indicating that these genes might be similar to the group of genes induced in sor1 and by all chemicals. Interestingly, all but one of the 14 genes overexpressed in sor1 contained their palindrome within the first 410 bp upstream of the start codon, with a strong accumulation between positions −190 and −350 bp (10 genes) (Fig. S3). Palindromes of the 12 genes mainly induced by chemical treatments, however, were more equally distributed over the 2-kb regions, with 6 genes having their element farther upstream of the 410-bp region, which was often also associated with lower induction strength. This finding supports a correlation between the presence of the 8-bp palindrome in the proximal promoter region and a high expression of these genes in both sor1- and RES-treated cultures.
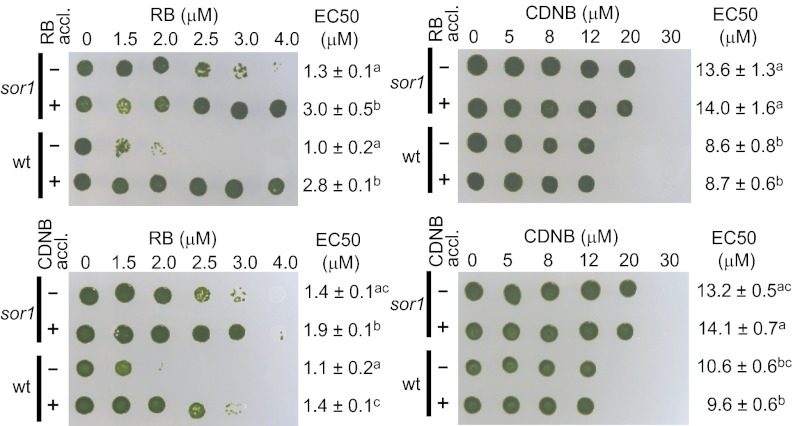
To connect the genetic response determined for 1O2, RES, and sor1 to increased tolerance of the algae to oxidative and electrophilic stress, cross-acclimation experiments were performed. Therefore, either sor1 or WT cells were preexposed to low levels of the 1O2-producing chemical RB or the RES CDNB and subsequently tested for resistance to high concentrations of the two chemicals (Fig. 7). The sor1 mutant and the WT strain both became more tolerant to RB when acclimated to either RB or CDNB for 2 h. However, neither of the pretreatments stimulated resistance to CDNB in the two strains, despite the fact that sor1 was more tolerant to the chemical than the WT strain.
Fig. 7.
Cross-acclimation experiments with the sor1 mutant and the WT strain preexposed to either 0.3 μM RB (+), 2 μM CDNB (+), or no chemical (−) for 2 h. After acclimation, cells were challenged with increasing concentrations of either of the two chemicals in liquid culture for 24 h and then spotted on agar plates to recover. EC50 ± SEM for growth inhibition was determined with four independent replicates, and significant differences (ANOVA, P < 0.05) are indicated by superscript letters, grouping nonsignificant different values with identical letters.
Discussion
Identification of SOR1 as a bZIP Transcription Factor Involved in Acclimation to Singlet Oxygen.
The response mechanisms of photosynthetic organisms to specific ROS, RES, and other redox signals have been extensively studied both in plants and algae (5, 35). Still, only few of the cellular components involved in the regulation of these responses are known. Here, we describe the isolation and characterization of a mutant, sor1, that exhibits constitutive high expression of various oxidative stress and RES detoxification genes; consequently, it is more tolerant to high levels of ROS and RES stresses. These phenotypes are caused by a single nucleotide mutation in the coding region of a putative bZIP transcription factor (protein ID 187531) causing a proline to leucine amino acid exchange near the C-terminal end of the mutant compared with WT protein. Except for the presence of the conserved bZIP DNA binding domain (N-x7-R-x9-L-x6-L-x6-L) (36), the SOR1 protein does not show any homology in its amino acid sequence to other members of this family, except for a putative ortholog present in the closely related species V. carteri and a paralogous sequence found in the C. reinhardtii genome (protein ID 157582). Phylogenetic analysis of plant and algal bZIP transcription factors revealed that SOR1 is one of seven putative bZIP proteins in C. reinhardtii, and it belongs to a group of algal-specific proteins with an unclear evolutionary relationship to other bZIP transcription factors of higher plants (37). SOR1 seems to be involved in the regulation of oxidative/electrophilic stress response genes, a cellular function found for various bZIP transcription factors in organisms from different phyla. The A. thaliana protein bZIP10 is activated by an ROS signal after pathogen infection, and it initiates a genetic response causing PCD (38). In yeast, the redox-sensitive transcription factor YAP1p is activated by ROS in a GPX3-dependent manner and stimulates the expression of more than 100 genes, including a large number of defense genes against ROS-induced stress (39). The mammalian Nrf2-Keap1 system is composed of the bZIP transcription factor Nrf2 and the redox sensitive negative regulator Keap1, which after activation by ROS and RES, releases Nrf2 to activate gene expression (40). Thus, induction of genes by reactive species seems to be a ubiquitous and evolutionarily conserved function of some bZIP transcription factors, and it seems also to be the cellular function of the SOR1 protein in C. reinhardtii.
The sor1 mutant was originally isolated in a screen for mutants with constitutive acclimation to 1O2 stress. However, the sor1 phenotype is not restricted to 1O2 resistance, and the mutant is also more tolerant to other ROS like tBOOH and various RES (Fig. 1C). This finding indicates that the SOR1 transcription factor might not be involved only in the regulation of an 1O2-specific acclimation process but rather, in a more general stress response mechanism. Although transcriptome analyses revealed large differences in the expression profiles of sor1 and different ROS and RES signals (Fig. 6A), all oxidative stress response and RES detoxification genes overexpressed in sor1 were also up-regulated more than twofold by almost all of the chemical treatments. Some of these genes might prevent ROS formation and accumulation, such as an alternative oxidase (41), a pyridoxamine 5′-phosphate oxidase involved in the synthesis of the ROS scavenger vitamin B6 (42), and a GDP-l-galactose phosphorylase involved in ascorbate synthesis (43). Other enzymes belong to the cellular detoxification systems, including GSTS1, GSTS2, a third GST (27), an ABC transporter, and three carbonyl reductases (one in the short-chain dehydrogenases family and two in the aldo/keto reductase family) (33, 44, 45). These members of the phases I, II, and III detoxification systems are involved in the activation, conjugation, and elimination, respectively, of exogenous and endogenous xenobiotics, including RES such as substituted quinones, α,β-unsaturated aldehydes, 2E-hexenal, and 4-hydroxy-2E-nonenal formed during lipid peroxidation (27, 44, 45). Such polyunsaturated fatty acid-derived RES can also be formed enzymatically during the synthesis of phytohormones like OPDA and dinor-OPDA (46). However, their formation is often strongly stimulated nonenzymatically by ROS-like 1O2 and hydroxyl radicals, leading to the accumulation of these toxic molecules (17, 33). A third group of genes overexpressed in sor1 encodes proteins that contain cysteines prone to oxidation to prevent oxidative damage, like a putative protein disulfide oxidoreductase, a methionine sulfoxide reductase (47), the glutaredoxin GRX2 (48), and two members of the DJ-1/PfpI superfamily (49). In humans, a connection between DJ-1 proteins and the ROS-induced Parkinson disease and cancer was shown. Thus, DJ-1 is considered to be a multifunctional oxidative stress response protein for which different functions, like ROS scavenging, stimulation of glutathione biosynthesis, and activation of Nrf2-dependent stress response, were suggested (49). Therefore, many of the putative SOR1 target genes are directly connected to the removal and detoxification of ROS, RES, and their reaction products.
Many Genes Overexpressed in sor1 Are Also Strongly Induced by Lipophilic Electrophiles Through an 8-bp Palindromic Regulatory Element.
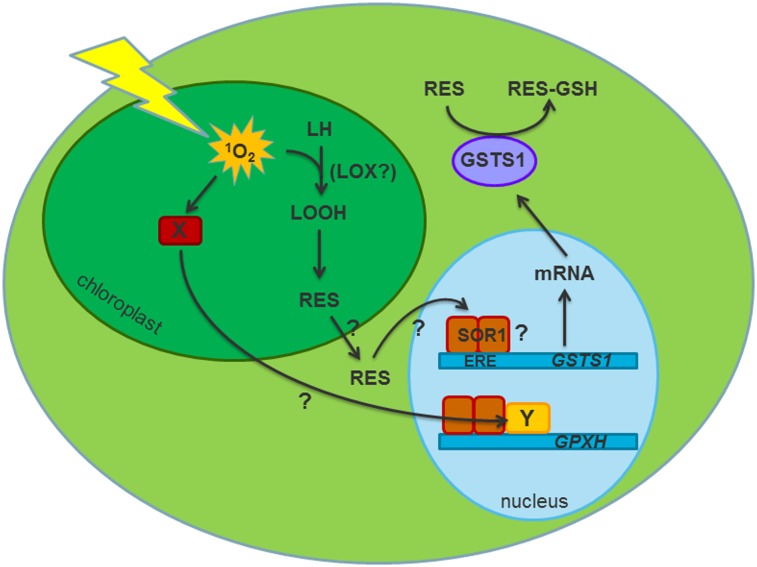
In addition to many defense genes, several regulatory genes involved in control of gene expression are overexpressed in sor1. These genes include SOR1 (protein ID 187531) itself as well as a SOR1 homologous gene (protein ID 157582) in C. reinhardtii, both of which were also induced by ROS and RES (Dataset S1A). Thus, SOR1 expression positively responds to the same signal as the overexpressed defense genes, which is mediated by its own gene product. This finding, together with the fact that sor1 is a gain of function mutation (Fig. 3A), shows that SOR1 likely encodes a positive regulator that is constitutively active in the sor1 mutant. However, increased expression of the SOR1 protein during stress treatment seems not to increase short-term induction of defense genes like GSTS1 and GSTS2, which was shown by the similar response of these genes to stress signals in the presence or absence of the protein synthesis inhibitor cycloheximide (Fig. S4). Still, coexpression of SOR1 and some defense genes is supported by the strong overrepresentation of an 8-bp palindromic sequence element (CAACGTTG) in the promoter region of these genes (Fig. 6C). The palindrome is essential for the induction of GSTS1 and GPXH by DBMIB and GSTS1 overexpression in sor1, showing that it is a functional cis-acting sequence directly involved in activation of transcription by RES and sor1 at least for the GSTS1 gene (Fig. 4). Furthermore, the effect of the palindromes on GSTS1 induction was lower for the more distal elements, showing that the distance from the start site influences the response. In agreement with that finding, we found strong accumulation (70% of genes) of the palindrome within the positions −70 and −340 bp of the promoters of induced genes (Fig. S3). Whereas expression of the GSTS1 WT gene was stimulated by NR and DBMIB to similar extents, the GSTS1-GLUC reporter construct responded much more strongly to RES than ROS-producing chemicals and positively correlated with the lipophilicity of the chemicals (Fig. 5 and Table 1). For many of the hydrophilic chemicals producing ROS (MV, RB, and NR) and high light treatment, increased induction was only observed after prolonged exposure, and this induction was accompanied by significant accumulation of the lipid peroxidation product malondialdehyde (Fig. 4A and Fig. S5) (20). This finding suggests that the signals for transcription activation of GSTS1 expression are probably not ROS themselves but rather, their reaction products with lipids. Thus, the 700-bp GSTS1 promoter fragment containing the three palindromes strongly responds to lipophilic RES signals, and the 8-bp palindrome is, therefore, designated as electrophile response element (ERE). The ERE shares strong functional similarities to the antioxidant response element and electrophile responsive element in mammals, which are activated by the redox active Keap1/Nrf2 regulatory complex (40). However, despite the functional similarities of these elements, the ERE core sequence is different from the mammalian antioxidant response element/electrophile responsive element consensus sequence, and it is also not related to any other known regulatory element, although it contains the ACGT core found in many bZIP binding motifs (36). Many of the ERE-containing genes belong to different groups of oxidative stress response and RES detoxification genes including two carbonyl reductases, two DJ1/Pfp1 genes, two GSTs, and several others (Dataset S1 A and B). Thus, the high number of stress response genes that contain an exact copy of the ERE in their proximal promoter regions and are induced by RES supports the hypothesis concerning its sequence and function.
Although the ERE was shown to be required for GSTS1 overexpression in sor1 (Fig. 4C), it is not clear whether the SOR1 protein directly binds to the element and stimulates transcription. Attempts to show direct binding of the SOR1 protein to the ERE element were, thus far, unsuccessful, and additional investigation is needed to test whether specific binding conditions, additional interaction partners, or heterodimer formation is required for transcriptional activation. Indeed, sequence analysis of the bZIP interaction domain of SOR1 revealed that putative repulsive interactions might hinder homodimer formation (50). However, not every gene that contains the ERE in its promoter is overexpressed in sor1, showing that the ERE is not sufficient to confer SOR1-dependent expression. Thus, for some genes, the ERE might function independently of SOR1 in one or more distinct signaling pathways.
SOR1, Singlet Oxygen Acclimation, and Response to RES.
How is SOR1 involved in the acclimation to 1O2? Quantitative comparison of 1O2 acclimation and sor1 resistance revealed that the acclimation response to 1O2 is stronger and more specific than the sor1 phenotype (Fig. 7 and Table 1) (8). This finding shows that, in C. reinhardtii, there is an 1O2-specific acclimation process that is SOR1-independent. However, an SOR1-dependent response to RES might at least partially contribute to the increased 1O2 tolerance during acclimation, which is supported by the increased cross-tolerance of CDNB-acclimated cultures to toxic RB concentrations (Fig. 7). A similar cross-tolerance of RES-acclimated cell cultures to 1O2 stress was shown in human skin cancer cells, which depended on the Keap1/Nrf2-induced gene expression (51). However, the fact that CDNB pretreatment in C. reinhardtii did not increase tolerance to CDNB itself suggests that pretreatment with this chemical resulted either in the response of fewer genes or a lower response than in sor1, which still was able to increase tolerance to RB. The same response to RES might be induced indirectly by 1O2 through the formation of α,β-unsaturated aldehydes during lipid peroxidation. This response would be in agreement with the general conclusion that oxidized lipids might function as more stable second messengers for 1O2 retrograde signaling to activate nuclear gene expression (13, 52). It was shown recently in A. thaliana that part of the 1O2 response was dependent on a functional oxygenation of fatty acids to form oxylipins (16). Similarly, enzymatically formed oxylipins affected the 1O2 response in the A. thaliana flu mutant, which accumulated increased levels of the electrophiles OPDA and dinor-OPDA (dnOPDA) and JA after dark–light shifts (15, 53). Although OPDA and JA seem not to directly act as second messengers for the 1O2-induced PCD process, they have been shown to contribute to the complex signaling cascade induced by 1O2 affecting the expression of some of the 1O2-induced genes (15). In the present study, a strong induction of GSTS1 was detected in response to OPDA but not JA, which is the nonelectrophilic product of OPDA, indicating that electrophilic oxylipins can be a signal to stimulate the RES-induced stress response in C. reinhardtii (Table 1). The specific induction of genes by OPDA but not JA was also shown in A. thaliana (18). Many of the induced genes were oxidative stress response and RES detoxification genes, suggesting that, in plants, OPDA plays an important role in the early response to oxidative and electrophilic stress conditions.
Other than RES signaling, another 1O2-specific response is likely to be involved in RB acclimation in C. reinhardtii. This 1O2-specific response includes the strong induction of GPXH and maybe other genes at low RB concentrations and during early high light response (8, 9, 20); however, it seems to not involve lipid-derived signals or the ERE (9, 19). The involvement of multiple signaling pathways in the response to 1O2 has also been suggested for A. thaliana (52). It has been suggested that such a redundancy in signaling processes helps to increase the robustness of the response, indicating its importance for the organism (54). In agreement, recent investigations to isolate mutants in the putative 1O2-specific signaling pathway were unsuccessful, supporting the robustness of a pathway that is either essential or redundant (26). SOR1 might even further increase this robustness through a positive feedback stimulation of SOR1 expression. It has been shown that 1O2 is the major ROS produced on high light stress in higher plants (55), and considering the presumed evolution of such a robust defense response mechanism in C. reinhardtii, it might also be the major ROS produced in green algae. In the present study, we identified SOR1 as one factor that can stimulate the tolerance of C. reinhardtii to high 1O2 formation by activating an RES-induced defense response, thereby contributing to the overall tolerance of this organism to deleterious photooxidative stress conditions.
Materials and Methods
Strains and Growth Conditions.
The WT strain 4A+, which is in a 137c strain background (56), was used to perform the mutant screen, the cell wall-deficient strain cw15 (CC-406) was used for transformation of reporter constructs, and the polymorphic strain S1-C5 (CC-1952) was used for mapping (29). All strains were grown mixotrophically in a Tris⋅acetate phosphate (TAP) medium (57) at 25 °C and 100 μmol photons m−2 s−1 photosynthetically active radiation either in liquid cultures on a rotary shaker (120 rpm) or on plates containing 1.5% agar. Experiments were usually performed by growing cultures mixotrophically until they reached a density of 2 × 106 cells mL−1. For UV mutagenesis, 20 mL culture were aliquoted into a sterile glass Petri dish (14-cm diameter) and exposed to 30–60 mJ cm−2 UV light in a UV Stratalinker 1800 (Stratagene). Cells were plated on TAP plates containing 4 μM RB, kept in the dark for 1 d to prevent light-activated DNA repair, and then shifted to 100 μmol photons m−2 s−1 to initiate selection for resistant clones. To test resistance of mutants after screening, 5 μL each 1:10 serial dilution of a concentrated cultures at 1 × 107 cells mL−1 were spotted onto plates containing either 2 μM RB or 8 μM NR and exposed to 100 μmol photons m−2 s−1 for several days. Quantitative evaluation of resistance to different stress conditions was done by transferring 1-mL aliquots of culture at 2 × 106 cells mL−1 into a 24-well culture plate and adding the individual chemical to the final concentrations indicated. Growth was analyzed by measuring OD at 750 nm after 0 and 24 h, and 5 μL each well were spotted on a TAP agar plate for recovery; EC50 values were determined by nonlinear fitting of a sigmoidal dose–response curve (four parameter logistic equation) with the program Prism version 4 from GraphPad Software.
RNA Isolation, Gel Blot Analysis, and Real-Time RT-qPCR.
To analyze gene expression, cultures were exposed to the treatment indicated, and cells were harvested by centrifugation. RNA was extracted using either the phenol/chloroform method described in Ledford et al. (8) or the RNeasy Mini Kit (Qiagen) as described by the manufacturer’s protocol with the following modifications. Cells were sonicated two times for 5 s in the RLT buffer on ice to break cells, and DNA was digested on column using the RNase-Free DNase Set (Qiagen). After purification, RNA was quantified with a NanoDrop2000 (Thermo Fisher Scientific), and quality was assessed on a 2100 Bioanalyzer (Agilent Technologies). RNA gel blot analyses of GPXH, GSTS1, and GSTS2 expression were performed as described before (8). Sequences of primers for qPCR were designed with the Primer Express software (Applied Biosystems) (Table S2), and qPCR reactions were performed on the ABI Prism 7500 Sequence Detection System using the SYBR Green technology (Applied Biosystems) as described before (9). Relative expression was calculated for each treatment compared with the untreated WT under the same growth conditions as an average with SE of three independent experiments.
RNA-Seq Experiments.
The cultures for RNA-Seq were grown in triplicates under 120 μmol photons m−2 s−1 in a 12-h dark–light cycle, and cells were collected 6 h after the beginning of the light cycle. RNA isolation and preparation and sequencing of the RNA-Seq libraries was done as described in the work by Castruita et al. (58) following the manufacturer’s instructions. In detail, RNA-Seq libraries of the three independent biological replicates of either the mutant and WT cultures were pooled and sequenced in one or two technical replicates, respectively, on an Illumina sequencer (Illumina). This process resulted in a total of 11,895,587 reads for the mutant and 5,143,241 or 5,260,441 reads for the WT library, with an average length of 36 bp per fragment. For individual library, between 84% and 88% of the fragments could be aligned to the C. reinhardtii genome, with 74–76% of the sequences providing a unique match. Additional data processing and statistical analyses were done as described in the work by Castruita et al. (58) selecting for genes differently expressed more than twofold between strains with a linear signal threshold level of six for the stronger expressed sample. All of the RNA-Seq data were submitted to the Gene Expression Omnibus database (http://www.ncbi.nlm.nih.gov/geo) under the accession no. GSE33548.
DNA Microarray Experiments.
Cultures of sor1 or 4A+ were exposed to the chemical treatments for 2 h before total RNA was isolated with the RNeasy Mini Kit (Qiagen), including the RNase-Free DNase Set (Qiagen). RNA was quantified with a NanoDrop2000 (Thermo Fisher Scientific), and quality was measured on the 2100 Bioanalyzer (Agilent Technologies). Microarray experiments were performed using a custom-made 4 × 44-K Chlamydomonas Whole Genome DNA Microarrays (Agilent Technologies) containing 15,143 specific probes designed based on the C. reinhardtii version 4 transcript models provided by the DOE Joint Genome Institute, with an average of three replicates for each probe. Labeling, purification, and hybridization was done according to the One-Color Microarray-Based Gene Expression Analysis system (Agilent Technologies) following the manufacturer’s protocol. Hybridized slides were scanned with the Agilent DNA Microarray scanner and quantified using the Agilent Feature Extraction software. Additional data analyses were performed with the R/Bioconductor software using background-corrected and quartile-normalized median signals. Significant differences of average signal intensities between conditions were determined by one-way ANOVA followed by Tukey’s pairwise comparison. Induction of significantly different expressed genes (P < 0.0001, false discovery rate = 0.000158) was calculated as log2 of the ratio between the sample and the untreated WT condition, excluding genes with high probability of cross-hybridization contaminations and minimal signal intensities below 10. Clustering was performed based on complete linkage of normalized and centered induction factors with Cluster 3.0, and the results were visualized in a color-based expression pattern using the TreeView 1.60 software (both designed by the Eisen Laboratory; http://rana.lbl.gov). The microarray data have been submitted to the Gene Expressiion Omnibus database (http://www.ncbi.nlm.nih.gov/geo) under accession number GSE30648.
Construction and Isolation of Vegetative Diploids.
C. reinhardtii is haploid, but vegetative diploids can be selected by crossing strains containing complementing alleles of the arg7 gene, arg7-1 and arg7-8. The ARG7 locus encodes argininosuccinate lyase, and arg7 mutants are arginine auxotrophs. Intragenic complementation of arg7-1 and arg7-8 allows diploids bearing both alleles to be selected by plating mated cells directly onto TAP plates without added arginine. Colonies isolated in this manner were then tested for the presence of both mating type alleles (plus and minus) by PCR.
Mapping.
To map the sor1 mutation on the C. reinhardtii genome, sor1 was crossed to the polymorphic strain S1-C5 mt- (CC-1952) (29), and the resulting zygospores were harvested and dissected as described in the work by Harris (57). A total of 214 progeny from independent tetrads was phenotyped based on RNA gel blot analysis of GSTS1 expression as described above. DNA was isolated as previously described, excluding the final CsCl purification step (56).
The molecular markers used in this study are listed in Table S3. CHLH, CPN60B2, and SFA markers were designed based on information in the work by Kathir et al. (29). All of the other markers in this study were created based on the JGI v2.0 C. reinhardtii nuclear genome sequence either by Qi Sun and David Stern (Boyce Thompson Institute for Plant Research, Ithaca, NY) or ourselves and were named based on their position on scaffold 57 in a kilobase scale (e.g., C57_389563 is C57_389). Whenever possible, S1-C5 EST sequences were used to design markers. Where S1-C5 sequence information was not available, primers were designed to hybridize to coding regions that flanked predicted introns and/or simple sequence repeats. When the amplified products from sor1 and S1-C5 did not differ obviously in size, PCR products from each parent were sequenced to find unique restriction sites that could be used to develop a cleaved amplified polymorphic sequence marker. All markers were run on 1.5% agarose gels.
Sequencing of candidate genes was done by amplifying overlapping fragments of 1 kb from genomic DNAs of sor1 and 4A+. For the SOR1 gene, each fragment was amplified and sequenced four to six times, and the identified nucleotide substitution in sor1 was confirmed to be present in a total of 48 progenies of a sor1×4A+ cross using a cleaved amplified polymorphic sequence marker (Table S3).
Cloning and Transformation of Plasmids and Reporter Assay.
Cloning of all plasmids is described in full details in SI Text. The GLUC reporter constructs were transformed into the cell wall-deficient strain CC-406. The ARS reporter construct and the pSL18-based overexpression constructs were transformed into 4A+ or sor1, respectively, following the protocol in the work by Kindle (59). The Streptomyces aminoglycoside 3′-phosphotransferase type VIII encoding gene (aphVIII) was used for selection of transformants on paromomycin in all cases.
Between 4 and 24 independent transformants expressing the reporter genes were selected and tested for strength of the expression either in different strains or after exposure to chemicals. To do testing, 1-mL cultures of strain CC-406 expressing the different GSTS1-GLUC constructs were grown to similar cell density in 24-well plates and exposed to increasing concentrations of the chemicals tested. After 1 h exposure, 20-μL samples were taken and frozen at −80 °C before luciferase activity was tested as described in the work by Shao and Bock (31). To test ARS activity of different strains, 1-mL cultures of each clone were grown to the same cell density, and samples of 150 μL were transferred into a transparent 96-well plate. Then, ARS activity was determined as described before (26), and significant difference was determined by one-way ANOVA followed by Tukey’s pairwise comparison using Prism version 4 from GraphPad Software.
Supplementary Material
Acknowledgments
We thank the Functional Genomics Center Zurich for technical support, especially Jelena Kühn Georgijevic and Hubert Rehrauer for their assistance in data generation. This work was supported by National Institute of General Medical Sciences Award R01GM071908 (to K.K.N.). The research at University of California at Los Angeles was supported by the Office of Science (Biological and Environmental Research), US Department of Energy, Cooperative Agreement DE-FC02-02ER63421.
Footnotes
The authors declare no conflict of interest.
Data deposition: The data reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, www.ncbi.nlm.nih.gov/geo [accession nos. GSE33548 (RNA sequence) and GSE30648 (DNA microarrays)].
This article is a PNAS Direct Submission.
See Author Summary on page 7611 (volume 109, number 20).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1116843109/-/DCSupplemental.
References
- 1.Krieger-Liszkay A. Singlet oxygen production in photosynthesis. J Exp Bot. 2005;56:337–346. doi: 10.1093/jxb/erh237. [DOI] [PubMed] [Google Scholar]
- 2.Li Z, Wakao S, Fischer BB, Niyogi KK. Sensing and responding to excess light. Annu Rev Plant Biol. 2009;60:239–260. doi: 10.1146/annurev.arplant.58.032806.103844. [DOI] [PubMed] [Google Scholar]
- 3.Inaba T, Yazu F, Ito-Inaba Y, Kakizaki T, Nakayama K. Retrograde signaling pathway from plastid to nucleus. Int Rev Cell Mol Biol. 2011;290:167–204. doi: 10.1016/B978-0-12-386037-8.00002-8. [DOI] [PubMed] [Google Scholar]
- 4.Chan KX, Crisp PA, Estavillo GM, Pogson BJ. Chloroplast-to-nucleus communication: Current knowledge, experimental strategies and relationship to drought stress signaling. Plant Signal Behav. 2010;5:1575–1582. doi: 10.4161/psb.5.12.13758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Galvez-Valdivieso G, Mullineaux PM. The role of reactive oxygen species in signalling from chloroplasts to the nucleus. Physiol Plant. 2010;138:430–439. doi: 10.1111/j.1399-3054.2009.01331.x. [DOI] [PubMed] [Google Scholar]
- 6.Sun X, et al. A chloroplast envelope-bound PHD transcription factor mediates chloroplast signals to the nucleus. Nat Commun. 2011;2:477. doi: 10.1038/ncomms1486. [DOI] [PubMed] [Google Scholar]
- 7.op den Camp RG, et al. Rapid induction of distinct stress responses after the release of singlet oxygen in Arabidopsis. Plant Cell. 2003;15:2320–2332. doi: 10.1105/tpc.014662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ledford HK, Chin BL, Niyogi KK. Acclimation to singlet oxygen stress in Chlamydomonas reinhardtii. Eukaryot Cell. 2007;6:919–930. doi: 10.1128/EC.00207-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fischer BB, et al. Function and regulation of the glutathione peroxidase homologous gene GPXH/GPX5 in Chlamydomonas reinhardtii. Plant Mol Biol. 2009;71:569–583. doi: 10.1007/s11103-009-9540-8. [DOI] [PubMed] [Google Scholar]
- 10.Fischer BB, Eggen RIL, Trebst A, Krieger-Liszkay A. The glutathione peroxidase homologous gene Gpxh in Chlamydomonas reinhardtii is upregulated by singlet oxygen produced in photosystem II. Planta. 2006;223:583–590. doi: 10.1007/s00425-005-0108-9. [DOI] [PubMed] [Google Scholar]
- 11.Leisinger U, et al. The glutathione peroxidase homologous gene from Chlamydomonas reinhardtii is transcriptionally up-regulated by singlet oxygen. Plant Mol Biol. 2001;46:395–408. doi: 10.1023/a:1010601424452. [DOI] [PubMed] [Google Scholar]
- 12.Fischer BB, Krieger-Liszkay A, Eggen RIL. Oxidative stress induced by the photosensitizers neutral red (type I) or rose bengal (type II) in the light causes different genetic responses in Chlamydomonas reinhardtii. Plant Sci. 2005;168:747–759. [Google Scholar]
- 13.Kochevar IE. Singlet oxygen signaling: From intimate to global. Sci STKE. 2004;2004:pe7. doi: 10.1126/stke.2212004pe7. [DOI] [PubMed] [Google Scholar]
- 14.Girotti AW, Kriska T. Role of lipid hydroperoxides in photo-oxidative stress signaling. Antioxid Redox Signal. 2004;6:301–310. doi: 10.1089/152308604322899369. [DOI] [PubMed] [Google Scholar]
- 15.Przybyla D, et al. Enzymatic, but not non-enzymatic, 1O2-mediated peroxidation of polyunsaturated fatty acids forms part of the EXECUTER1-dependent stress response program in the flu mutant of Arabidopsis thaliana. Plant J. 2008;54:236–248. doi: 10.1111/j.1365-313X.2008.03409.x. [DOI] [PubMed] [Google Scholar]
- 16.López MA, et al. Antagonistic role of 9-lipoxygenase-derived oxylipins and ethylene in the control of oxidative stress, lipid peroxidation and plant defence. Plant J. 2011;67:447–458. doi: 10.1111/j.1365-313X.2011.04608.x. [DOI] [PubMed] [Google Scholar]
- 17.Mueller MJ, Berger S. Reactive electrophilic oxylipins: Pattern recognition and signalling. Phytochemistry. 2009;70:1511–1521. doi: 10.1016/j.phytochem.2009.05.018. [DOI] [PubMed] [Google Scholar]
- 18.Taki N, et al. 12-oxo-phytodienoic acid triggers expression of a distinct set of genes and plays a role in wound-induced gene expression in Arabidopsis. Plant Physiol. 2005;139:1268–1283. doi: 10.1104/pp.105.067058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fischer BB, et al. Role of singlet oxygen in chloroplast to nucleus retrograde signaling in Chlamydomonas reinhardtii. FEBS Lett. 2007;581:5555–5560. doi: 10.1016/j.febslet.2007.11.003. [DOI] [PubMed] [Google Scholar]
- 20.Fischer BB, Dayer R, Wiesendanger M, Eggen RIL. Independent regulation of the GPXH gene expression by primary and secondary effects of high light stress in Chlamydomonas reinhardtii. Physiol Plant. 2007;130:195–206. [Google Scholar]
- 21.Anthony JR, Warczak KL, Donohue TJ. A transcriptional response to singlet oxygen, a toxic byproduct of photosynthesis. Proc Natl Acad Sci USA. 2005;102:6502–6507. doi: 10.1073/pnas.0502225102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Stephen DW, Rivers SL, Jamieson DJ. The role of the YAP1 and YAP2 genes in the regulation of the adaptive oxidative stress responses of Saccharomyces cerevisiae. Mol Microbiol. 1995;16:415–423. doi: 10.1111/j.1365-2958.1995.tb02407.x. [DOI] [PubMed] [Google Scholar]
- 23.Brombacher K, Fischer BB, Rüfenacht K, Eggen RIL. The role of Yap1p and Skn7p-mediated oxidative stress response in the defence of Saccharomyces cerevisiae against singlet oxygen. Yeast. 2006;23:741–750. doi: 10.1002/yea.1392. [DOI] [PubMed] [Google Scholar]
- 24.Wagner D, et al. The genetic basis of singlet oxygen-induced stress responses of Arabidopsis thaliana. Science. 2004;306:1183–1185. doi: 10.1126/science.1103178. [DOI] [PubMed] [Google Scholar]
- 25.Lee KP, Kim C, Landgraf F, Apel K. EXECUTER1- and EXECUTER2-dependent transfer of stress-related signals from the plastid to the nucleus of Arabidopsis thaliana. Proc Natl Acad Sci USA. 2007;104:10270–10275. doi: 10.1073/pnas.0702061104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fischer BB, Eggen RI, Niyogi KK. Characterization of singlet oxygen-accumulating mutants isolated in a screen for altered oxidative stress response in Chlamydomonas reinhardtii. BMC Plant Biol. 2010;10:279. doi: 10.1186/1471-2229-10-279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hayes JD, Flanagan JU, Jowsey IR. Glutathione transferases. Annu Rev Pharmacol Toxicol. 2005;45:51–88. doi: 10.1146/annurev.pharmtox.45.120403.095857. [DOI] [PubMed] [Google Scholar]
- 28.Thimm O, et al. MAPMAN: A user-driven tool to display genomics data sets onto diagrams of metabolic pathways and other biological processes. Plant J. 2004;37:914–939. doi: 10.1111/j.1365-313x.2004.02016.x. [DOI] [PubMed] [Google Scholar]
- 29.Kathir P, et al. Molecular map of the Chlamydomonas reinhardtii nuclear genome. Eukaryot Cell. 2003;2:362–379. doi: 10.1128/EC.2.2.362-379.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fischer N, Rochaix JD. The flanking regions of PsaD drive efficient gene expression in the nucleus of the green alga Chlamydomonas reinhardtii. Mol Genet Genomics. 2001;265:888–894. doi: 10.1007/s004380100485. [DOI] [PubMed] [Google Scholar]
- 31.Shao N, Bock R. A codon-optimized luciferase from Gaussia princeps facilitates the in vivo monitoring of gene expression in the model alga Chlamydomonas reinhardtii. Curr Genet. 2008;53:381–388. doi: 10.1007/s00294-008-0189-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Monks TJ, Hanzlik RP, Cohen GM, Ross D, Graham DG. Quinone chemistry and toxicity. Toxicol Appl Pharmacol. 1992;112:2–16. doi: 10.1016/0041-008x(92)90273-u. [DOI] [PubMed] [Google Scholar]
- 33.Farmer EE, Davoine C. Reactive electrophile species. Curr Opin Plant Biol. 2007;10:380–386. doi: 10.1016/j.pbi.2007.04.019. [DOI] [PubMed] [Google Scholar]
- 34.González-Ballester D, et al. RNA-seq analysis of sulfur-deprived Chlamydomonas cells reveals aspects of acclimation critical for cell survival. Plant Cell. 2010;22:2058–2084. doi: 10.1105/tpc.109.071167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Foyer CH, Noctor G. Redox regulation in photosynthetic organisms: Signaling, acclimation, and practical implications. Antioxid Redox Signal. 2009;11:861–905. doi: 10.1089/ars.2008.2177. [DOI] [PubMed] [Google Scholar]
- 36.Jakoby M, et al. bZIP transcription factors in Arabidopsis. Trends Plant Sci. 2002;7:106–111. doi: 10.1016/s1360-1385(01)02223-3. [DOI] [PubMed] [Google Scholar]
- 37.Corrêa LG, et al. The role of bZIP transcription factors in green plant evolution: Adaptive features emerging from four founder genes. PLoS One. 2008;3:e2944. doi: 10.1371/journal.pone.0002944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kaminaka H, et al. bZIP10-LSD1 antagonism modulates basal defense and cell death in Arabidopsis following infection. EMBO J. 2006;25:4400–4411. doi: 10.1038/sj.emboj.7601312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lee J, et al. Yap1 and Skn7 control two specialized oxidative stress response regulons in yeast. J Biol Chem. 1999;274:16040–16046. doi: 10.1074/jbc.274.23.16040. [DOI] [PubMed] [Google Scholar]
- 40.Itoh K, Mimura J, Yamamoto M. Discovery of the negative regulator of Nrf2, Keap1: A historical overview. Antioxid Redox Signal. 2010;13:1665–1678. doi: 10.1089/ars.2010.3222. [DOI] [PubMed] [Google Scholar]
- 41.Umbach AL, Fiorani F, Siedow JN. Characterization of transformed Arabidopsis with altered alternative oxidase levels and analysis of effects on reactive oxygen species in tissue. Plant Physiol. 2005;139:1806–1820. doi: 10.1104/pp.105.070763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bilski P, Li MY, Ehrenshaft M, Daub ME, Chignell CF. Vitamin B6 (pyridoxine) and its derivatives are efficient singlet oxygen quenchers and potential fungal antioxidants. Photochem Photobiol. 2000;71:129–134. doi: 10.1562/0031-8655(2000)071<0129:sipvbp>2.0.co;2. [DOI] [PubMed] [Google Scholar]
- 43.Linster CL, Clarke SG. L-Ascorbate biosynthesis in higher plants: The role of VTC2. Trends Plant Sci. 2008;13:567–573. doi: 10.1016/j.tplants.2008.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Oppermann U. Carbonyl reductases: The complex relationships of mammalian carbonyl- and quinone-reducing enzymes and their role in physiology. Annu Rev Pharmacol Toxicol. 2007;47:293–322. doi: 10.1146/annurev.pharmtox.47.120505.105316. [DOI] [PubMed] [Google Scholar]
- 45.Barski OA, Tipparaju SM, Bhatnagar A. The aldo-keto reductase superfamily and its role in drug metabolism and detoxification. Drug Metab Rev. 2008;40:553–624. doi: 10.1080/03602530802431439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Böttcher C, Pollmann S. Plant oxylipins: Plant responses to 12-oxo-phytodienoic acid are governed by its specific structural and functional properties. FEBS J. 2009;276:4693–4704. doi: 10.1111/j.1742-4658.2009.07195.x. [DOI] [PubMed] [Google Scholar]
- 47.Kim HY, Fomenko DE, Yoon YE, Gladyshev VN. Catalytic advantages provided by selenocysteine in methionine-S-sulfoxide reductases. Biochemistry. 2006;45:13697–13704. doi: 10.1021/bi0611614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lemaire SD. The glutaredoxin family in oxygenic photosynthetic organisms. Photosynth Res. 2004;79:305–318. doi: 10.1023/B:PRES.0000017174.60951.74. [DOI] [PubMed] [Google Scholar]
- 49.Wilson MA. The role of cysteine oxidation in DJ-1 function and dysfunction. Antioxid Redox Signal. 2011;15:111–122. doi: 10.1089/ars.2010.3481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Vinson C, Acharya A, Taparowsky EJ. Deciphering B-ZIP transcription factor interactions in vitro and in vivo. Biochim Biophys Acta. 2006;1759:4–12. doi: 10.1016/j.bbaexp.2005.12.005. [DOI] [PubMed] [Google Scholar]
- 51.Wondrak GT, et al. Cinnamoyl-based Nrf2-activators targeting human skin cell photo-oxidative stress. Free Radic Biol Med. 2008;45:385–395. doi: 10.1016/j.freeradbiomed.2008.04.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Baruah A, Simková K, Apel K, Laloi C. Arabidopsis mutants reveal multiple singlet oxygen signaling pathways involved in stress response and development. Plant Mol Biol. 2009;70:547–563. doi: 10.1007/s11103-009-9491-0. [DOI] [PubMed] [Google Scholar]
- 53.Danon A, Miersch O, Felix G, Camp RG, Apel K. Concurrent activation of cell death-regulating signaling pathways by singlet oxygen in Arabidopsis thaliana. Plant J. 2005;41:68–80. doi: 10.1111/j.1365-313X.2004.02276.x. [DOI] [PubMed] [Google Scholar]
- 54.Stelling J, Sauer U, Szallasi Z, Doyle FJ, 3rd, Doyle J. Robustness of cellular functions. Cell. 2004;118:675–685. doi: 10.1016/j.cell.2004.09.008. [DOI] [PubMed] [Google Scholar]
- 55.Triantaphylidès C, et al. Singlet oxygen is the major reactive oxygen species involved in photooxidative damage to plants. Plant Physiol. 2008;148:960–968. doi: 10.1104/pp.108.125690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Dent RM, Haglund CM, Chin BL, Kobayashi MC, Niyogi KK. Functional genomics of eukaryotic photosynthesis using insertional mutagenesis of Chlamydomonas reinhardtii. Plant Physiol. 2005;137:545–556. doi: 10.1104/pp.104.055244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Harris EH. The Chlamydomonas Sourcebook: A Comprehensive Guide to Biology and Laboratory Use. San Diego: Academic; 1989. [DOI] [PubMed] [Google Scholar]
- 58.Castruita M, et al. Systems biology approach in Chlamydomonas reveals connections between copper nutrition and multiple metabolic steps. Plant Cell. 2011;23:1273–1292. doi: 10.1105/tpc.111.084400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kindle KL. High-frequency nuclear transformation of Chlamydomonas reinhardtii. Proc Natl Acad Sci USA. 1990;87:1228–1232. doi: 10.1073/pnas.87.3.1228. [DOI] [PMC free article] [PubMed] [Google Scholar]