Abstract
The Env protein of murine leukemia virus matures by two cleavage events. First, cellular furin separates the receptor binding surface (SU) subunit from the fusion-active transmembrane (TM) subunit and then, in the newly assembled particle, the viral protease removes a 16-residue peptide, the R-peptide from the endodomain of the TM. Both cleavage events are required to prime the Env for receptor-triggered activation. Cryoelectron microscopy (cryo-EM) analyses have shown that the mature Env forms an open cage-like structure composed of three SU–TM complexes, where the TM subunits formed separated Env legs. Here we have studied the structure of the R-peptide precursor Env by cryo-EM. TM cleavage in Moloney murine leukemia virus was inhibited by amprenavir, and the Envs were solubilized in Triton X-100 and isolated by sedimentation in a sucrose gradient. We found that the legs of the R-peptide Env were held together by trimeric interactions at the very bottom of the Env. This suggested that the R-peptide ties the TM legs together and that this prevents the activation of the TM for fusion. The model was supported by further cryo-EM studies using an R-peptide Env mutant that was fusion-competent despite an uncleaved R-peptide. The Env legs of this mutant were found to be separated, like in the mature Env. This shows that it is the TM leg separation, normally caused by R-peptide cleavage, that primes the Env for receptor triggering.
Keywords: three-dimensional structure, retrovirus, spike protein
The spike protein Env on the surface of the retrovirus murine leukemia virus (MLV) matures by two proteolytic cleavage events mediated by cellular furin and the viral protease (1–3). Env is composed of an 80-kDa transmembrane precursor protein in the rough endoplasmic reticulum of the infected cell (4, 5). Here it trimerizes before it is routed to the cell surface for assembly with the internal Gag and GagPol precursors into virus particles in a budding process (6). The furin cleavage of Env occurs in the trans-Golgi, and it separates the surface (SU) subunit from the transmembrane (TM) (Pr15E) subunit. This cleavage also releases the viral fusion peptide at the N-terminal end of the TM. The viral protease cleavage occurs after virus assembly in the newly formed particle. It removes a 16-residue-long peptide, the R-peptide from the C terminus of the TM, forming p15E (7). Not until this second cleavage is completed is Env primed for receptor-mediated triggering into further conformations that can direct virus entry through fusion of the viral membrane with the cell membrane (8, 9). The viral protease also cleaves the Gag and GagPol precursors into their mature proteins and enzymes.
The structure of the MLV Env has been studied by cryoelectron microscopy (cryo-EM) both in intact particles and as solubilized trimers (10, 11). It reveals a remarkably open structure with separated legs. The protomer of the solubilized Env is formed from three protrusions—upper, middle, and lower. Together, they encage a large central cavity, with the top protrusions forming the roof and upper sides and the two other protrusions forming the rest of the sides and the legs at the bottom. The SU has been shown to consist of a receptor-binding N-terminal domain (RBD) and a C-terminal domain (12). The atomic structure of the RBD has been solved and fits into the top protrusion, suggesting that the middle one represents the C-terminal SU domain and that the lower, forming the separated Env legs, represents the TM subunit (11, 13).
Receptor binding has been suggested to trigger a cascade of conformational signals from RBD to TM via the C-terminal SU domain (14, 15). The SU and TM are coupled by a disulfide bond, and activation requires its isomerization (16). This is possible because the SU cysteine of the disulfide is part of an isomerization-active CXXC motif in the C-terminal SU domain. Thus, upon Env triggering, the RBD transmits a signal to the C-terminal SU domain that activates the free thiol of the CXXC motif to attack the intersubunit disulfide and rearrange it into a disulfide isomer within the motif. If an alkylator is present during Env triggering, the isomerization-active thiol becomes modified before it can attack the intersubunit disulfide and the Env will be arrested in its activation process at an intermediate stage, the isomerization-arrested stage (IAS). This form of the Env has also been studied by cryo-EM, and the structure shows that activation is initiated by a clockwise rotation of the RBDs, opening up the closed roof of the Env (11). Most likely, the rotation triggers the disulfide isomerization. This might then allow the TM to extend and reach the cell membrane with its fusion peptide through the hole in the Env roof and, further, in a subsequent step, to approach the viral and cell membranes for fusion through backfolding.
In this work, we have posed the question of how the precursor structures of the Env prevent triggering. To this end, we have studied the structure of the R-peptide precursor Env. Previously it has been shown that Env, which is produced in cells expressing only the Env gene, retains the R-peptide and cannot support cell–cell fusion (8, 9). Fusion was possible only if the R-peptide was deleted. Similarly, virus, where the R-peptide cleavage has been inhibited, cannot fuse with cells (17). It was suggested that the R-peptide ties the TM subunits together in the endodomain and that this prevents conformational changes in the Env that are necessary for fusion. Indeed, the endodomain with the R-peptide has been modeled as a trimeric coiled coil (18). This model has been supported by mutational studies using Moloney (Mo) MLV. In particular, an L651A mutation, just downstream of the R-peptide cleavage site, which should violate the heptad repeat of hydrophobic residues supporting coiled-coil formation, has been shown to rescue the inhibited fusion capacity of cell-associated Env (19, 20). Here we have produced wild-type and L651A Env mutant Mo-MLV in the presence of an inhibitor of the viral protease, at concentrations that preferentially inhibit R-peptide cleavage. The Envs with the R-peptide retained have been solubilized and the structures were analyzed by cryo-EM. We found that the splayed TM legs of the mature Env are tied together in the R-peptide containing endodomain of the wild type, but not of the mutant. This shows that the R-peptide mediates a trimerization interaction, possibly a trimeric coiled coil, in the endodomain of the Env. Furthermore, the data indicate that R-peptide cleavage removes the trimer interaction and primes the Env for receptor triggering by splaying the TM legs.
Results
Three-Dimensional Structure of the R-Peptide Env.
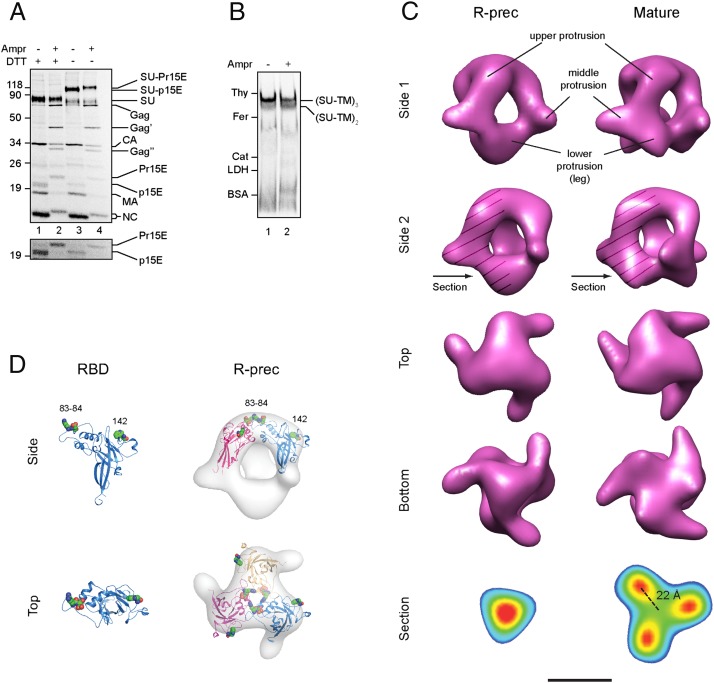
Mo-MLV was produced in the transformed MOV-3 cell line. Cleavage of the R-peptide was inhibited by including 0.8 μM viral protease inhibitor amprenavir in the growth medium, and the virus was isolated by centrifugation in a sucrose step gradient. Using [35S]Cys-labeled virus, we first made a biochemical characterization of the virus. Mo-MLV produced in the absence of the inhibitor was used as control. Reducing SDS/PAGE showed that the virus produced in the presence of the inhibitor carried the R-peptide containing Pr15E form of the TM subunit (Fig. 1A, lane 2). This migrated more slowly than the R-peptide free p15E form of TM present in the control virus (Fig. 1A, lane 1). As shown by high contrast of the TM region of the gel, the inhibition of R-peptide cleavage appears virtually complete. The identities of the p15E and Pr15E bands have earlier been confirmed by immunoprecipitation (21). The PAGE analysis also shows the SU subunit and the Gag-cleavage products capsid (CA), matrix (MA), and nucleocapsid (NC), and, in the case of the R-peptide virus, some intact Gag and Gag-cleavage intermediates (Gag′ and Gag′′). Nonreducing SDS/PAGE showed the disulfide-linked SU–TM complexes in both preparations together with some apparently isomerized SU and Pr15E/p15E (Fig. 1A, lanes 3 and 4). The complex with the retained R-peptide (SU–Pr15E) migrated slightly more slowly than the one with the R-peptide cleaved (SU–p15E). In blue native (BN)/PAGE, both complexes migrated predominantly as apparent trimers (Fig. 1B, lanes 1 and 2). Thus, we could confirm earlier results demonstrating that low levels of amprenavir preferentially inhibited R-peptide cleavage, leaving Gag cleavages more functional (21). Further, the Env subunits of the R-peptide virus formed disulfide-linked complexes that were noncovalently associated into trimeric Envs, like in the control virus.
Fig. 1.
Structure of the R-peptide precursor Env. (A) Analyses by SDS/PAGE. [35S]Cys-labeled virus was produced in the presence or absence of amprenavir to inhibit or allow R-peptide cleavage. Viral proteins of R-peptide virus (lanes 2 and 4, Ampr+) and mature virus (lanes 1 and 3, Ampr−) were analyzed by reducing (lanes 1 and 2, DTT+) and nonreducing (lanes 3 and 4, DTT−) SDS/PAGE. Viral proteins, their precursors, and disulfide-linked complexes are indicated. The migrations of standard proteins are shown (Left). The figure represents a phosphorimage. The TM region of the PAGE analysis is also shown at higher contrast below. (B) Analyses by BN/PAGE. [35S]Cys-labeled R-peptide (lane 2) and mature (lane 1) virus were solubilized in cold Triton X-100 and analyzed by BN/PAGE. The complete Envs, namely trimers of the SU–TM complexes, and incomplete ones, namely dimers of the SU–TM complexes, are indicated. The migrations of standard proteins are shown (Left). (C) A comparison of the surface-rendered 3D reconstruction of the R-peptide Env structure (R-prec) with that of the mature Env (mature) as determined previously. Envs were solubilized from unlabeled R-peptide virus and isolated by gradient centrifugation for structure reconstruction by cryo-EM. Two side views, one top and one bottom, are shown. One protomeric unit is hatched in side view 2, and protrusions are indicated in side view 1. Additionally, virtual sections of the bottom regions of both reconstructions, color-coded from blue to red according to the EM density gradient, are shown (Bottom). The sections are perpendicular to the threefold axis of the Env and their positions are indicated by arrows in side view 2 panels. In the section of the mature Env, the distance of the TM high-density nodes from the threefold axis is indicated. (Scale bar, 50 Å.) (D) Fitting of the atomic structure of RBD into the R-peptide Env. The modeled atomic structure of Mo-MLV RBD (Left) fits into the upper protrusions of the R-peptide Env (Right). The RBD structure is shown as a cartoon representation, whereas the Env reconstruction has been surface-rendered at full volume. Atoms of Arg83, Asp84, and Trp142 in the two receptor binding sites of RBD are shown as spheres, and residues are numbered. Two RBDs (pink and blue) have been fitted into the side view, and three (pink, blue, and yellow) are fitted into the top view of the Env.
For the structural analyses, we used unlabeled R-peptide virus. The Envs were solubilized by Triton X-100 in the cold and isolated by centrifugation in a continuous sucrose gradient. This separated the trimeric Envs from dimeric and monomeric forms of the SU–TM complexes and from other viral constituents. The separation was monitored using BN/PAGE and silver staining. The trimeric Envs were analyzed by cryo-EM, and the 3D structure was reconstructed using the EMAN single-particle approach. The structure, at a resolution of 21 Å, is shown in different projections in Fig. 1C (Left). It is compared with the structure of the mature Env with the R-peptide cleaved, which we have determined earlier (Fig. 1C, Right) (11). The R-peptide Env showed a similar open cage-like structure as the mature one. The protomeric unit was composed of three protrusions, of which the top and the middle formed the roof and the sides of the cage, like in the mature Env. However, the bottom protrusion did not form the separated Env legs typical for the mature virus, but rather converged into a common central body. The atomic structure of the RBD of Friend-MLV has been determined and used to model the structure of the Mo-MLV RBD (11). It is shaped like a bent finger and has earlier been fitted into the upper protrusion of the mature Env. It shows a similar unique fitting into the upper protrusion of the R-peptide Env, exposing its two patches of receptor-binding residues in the roof center and periphery (Fig. 1D) (22). This suggested that the middle protrusion and the common central body at the bottom were composed of the C-terminal SU domain and the TM subunits, respectively. The middle protrusion appeared slightly smaller in the R-peptide Env compared with the mature one. We concluded that the R-peptide ties the bottom protrusions, namely the TM legs, together by trimeric interactions.
This is particularly evident when examining the density distribution in the two Env forms. Fig. S1 (Left and Center) shows the precursor and mature Env structure reconstructions surface-rendered at full volume and at two increased-density thresholds. Fig. 1C (Bottom) shows virtual sections of the common central body and the leg regions of respective reconstructions, color-coded according to EM density. These analyses demonstrate a single central TM density node in the bottom of the precursor Env and three separated ones in that of the mature Env. To give a quantitative value of the change in TM organization upon R-peptide cleavage, we measured the distance of the separated TM density nodes of the mature Env from the threefold axis, which is 22 Å (indicated in Fig. 1C, Bottom Right).
Structure of the R-Peptide Env in Its Isomerization-Arrested Intermediate State.
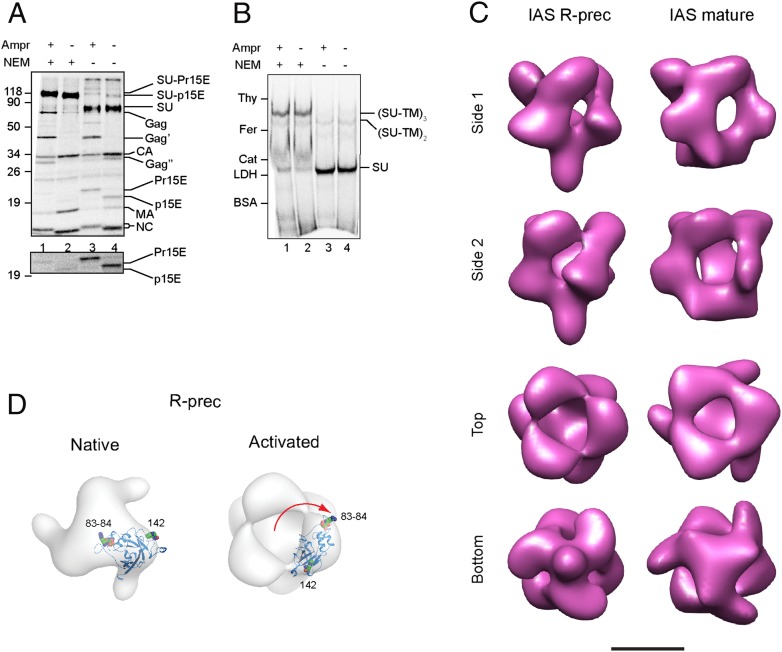
Earlier, we showed that the R-peptide Env could be triggered in vitro by Ca2+ depletion and in vivo by the receptor on the cell surface, but that the activation process was slower than that of the mature Env and did not proceed to completion (17). To find out how the R-peptide impedes Env activation, we studied the structure of the R-peptide Env in its isomerization-arrested state. To this end, we incubated R-peptide virus in EDTA- and Triton X-100–containing buffer at 37 °C in the presence of the thiol alkylator N-ethylmaleimide (NEM). Ca2+ depletion triggers Env activation, which, however, will be arrested at a step before isomerization (IAS) because of alkylation of the active thiol in the CXXC motif of the SU. To monitor the IAS formation, we used [35S]Cys-labeled virus and nonreducing SDS/PAGE. R-peptide virus incubated in the absence of alkylator showed completely isomerized, free SU and Pr15E subunits, whereas the subunits remained covalently bound as SU–TM complexes in the alkylated virus (Fig. 2A, lane 3, NEM− and lane 1, NEM+). Alkylated and nonalkylated control virus, produced in the absence of amprenavir and therefore carrying the mature Env, showed a corresponding band pattern, but with a slightly faster migrating SU–TM complex and with an R-peptide cleaved TM, that is, p15E instead of Pr15E (Fig. 2A, lanes 2 and 4). BN/PAGE analyses showed that the SU–TM complexes were associated into trimeric Envs in their alkylated but not in their nonalkylated form, both in the R-peptide and the control virus (Fig. 2B).
Fig. 2.
Structure of the R-peptide precursor Env in its activation intermediate form (IAS). (A) Analyses of viral proteins by nonreducing SDS/PAGE. [35S]Cys-labeled R-peptide virus was solubilized in the presence of EDTA and alkylator to generate the IAS form of the R-peptide Env (lane 1) or without alkylator to generate the fully isomerized and activated form (lane 3). Virus made in the absence of amprenavir and therefore with R-peptide–cleaved Envs was used as control (lanes 2 and 4). A high-contrast picture of the TM region of the gel is shown below. (B) BN/PAGE of corresponding samples. (C) A comparison of the surface-rendered 3D reconstruction of the IAS R-peptide Env structure (Left) with that of the IAS form of the mature Env (Right) as determined previously. IAS R-peptide Envs were solubilized from unlabeled R-peptide virus and isolated by gradient centrifugation for structure reconstruction by cryo-EM. (Scale bar, 50 Å.) (D) Fitting of the atomic structure of RBD into the IAS R-peptide Env (Right) compared with that into the native R-peptide Env (Left). Residues of RBD in receptor binding sites are indicated as in Fig. 1D. The red arrow indicates RBD rotation during activation.
The IAS Envs of unlabeled R-peptide virus were isolated by centrifugation and analyzed by cryo-EM and subsequent image processing. The structure, at a resolution of 22 Å, is shown in Fig. 2C (Left). It is compared with the structure of the earlier-determined IAS form of the mature Env (Fig. 2C, Right) (11). The protomeric units of the R-peptide IAS Env still encaged a central cavity, but the three protrusions had changed their forms and/or their locations. In the top of the Env, a central opening appeared in the roof (Fig. 2C, Left, second panel from the bottom). Fitting of the atomic structure of the RBD into the top protrusion suggested that the opening was generated by a 110° clockwise rotation of the RBD from its position in the native R-peptide Env (Fig. 2D). This roof opening through RBD rotation was very reminiscent of that occurring during IAS formation of the mature Env (Fig. 2C, Right). Further, the middle projection had become less pronounced in the R-peptide IAS Env compared with its native form (Fig. 1C, Left). Finally, the bottom projections were almost totally melded together, forming an elongated central body (Fig. 2C, Left). We concluded that the R-peptide Env underwent initial activation events in the top, that is, the opening of the roof through RBD rotation like the mature Env, whereas the TM subunits in the bottom seemed to join together even more by extending their threefold interactions.
Structure of the R-Peptide L651A Mutant Env.
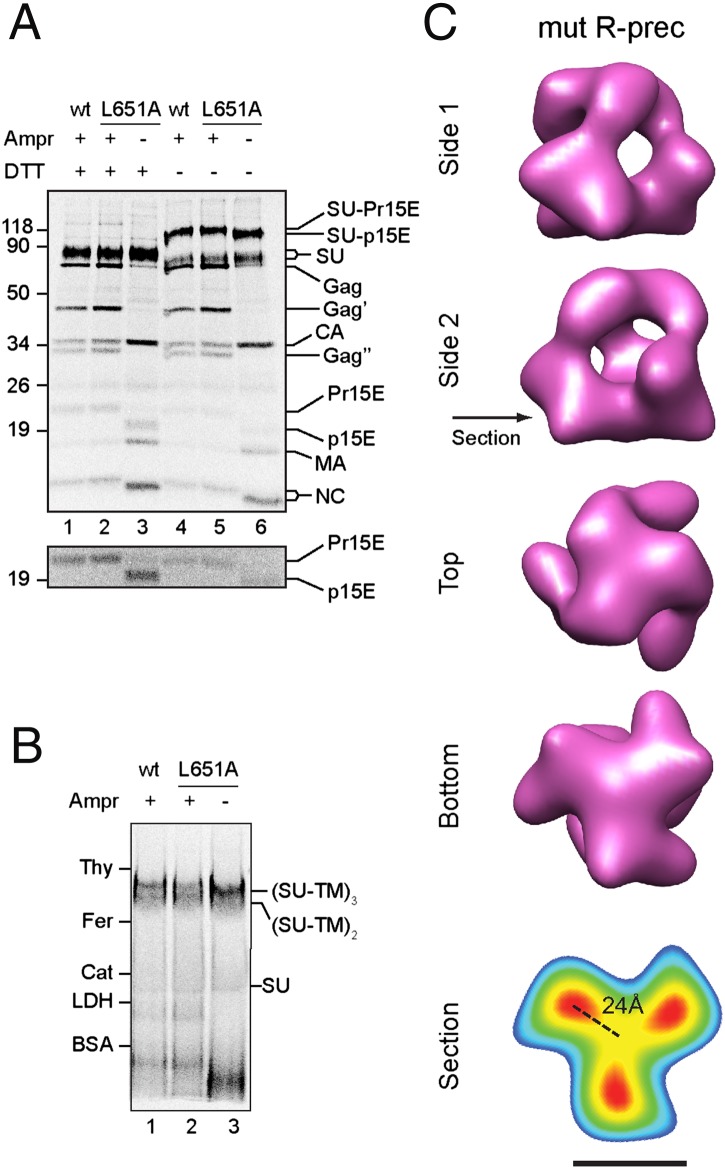
We were interested to find out whether the R-peptide L651A mutant Env with rescued fusion activity had separated TM legs like the mature Env. This would support the hypothesis that the R-peptide inhibited Env activation by tethering the TM endodomains together by coiled-coil formation. Therefore, we introduced the mutation into the proviral DNA clone of the Mo-MLV and produced the mutant virus in transfected 293T cells in the presence and absence of 0.8 μM amprenavir. Nonreducing/reducing SDS/PAGE of [35S]Cys-labeled virus showed that the mutant Env was incorporated into the virus similarly to WT (Fig. 3A, lanes 1 and 2), the SU and TM subunits were disulfide-linked (Fig. 3A, lanes 4–6), and amprenavir preferentially inhibited R-peptide cleavage (Fig. 3A, lane 2). When produced without amprenavir, the R-peptide of the mutant was cleaved normally (Fig. 3A, lane 3). Furthermore, BN/PAGE showed that the R-peptide L651A Env was migrating like an SU–TM trimer (Fig. 3B). To study the structure of the R-peptide mutant Env, it was isolated as before and analyzed by cryo-EM. Despite the presence of the R-peptide, the mutant Env did not tie the TM legs together, as in the WT R-peptide Env, but had separated TM legs like the mature Env (Fig. 3C, resolution 22 Å). This is most clearly seen when examining the mutant Env reconstruction at increased-density thresholds (Fig. S1, Right). The extent of leg separation (24 Å) is indicated in the section analysis of the leg region (Fig. 3C, Bottom). We concluded that the L651A mutation prevented the formation of the trimeric TM interaction, probably a coiled coil, which was induced by the WT R-peptide. It allowed the TM subunits to splay into a peripheral location instead. Thus, it is the peripheral relocation of the TM subunits or their loss of trimeric interaction and not the R-peptide cleavage per se that appeared to be required for the fusion function of the Env.
Fig. 3.
Structure of the R-peptide L651A mutant Env. (A) Analyses by SDS/PAGE. [35S]Cys-labeled mutant and WT virus were produced in the presence and absence of amprenavir to inhibit and allow R-peptide cleavage. Viral proteins were analyzed by reducing (lanes 1–3) and nonreducing (lanes 4–6) SDS/PAGE. A high-contrast picture of the TM region of the gel is shown below. (B) Analyses by BN/PAGE. [35S]Cys-labeled R-peptide mutant virus (lanes 2 and 3) and mature WT virus (lane 1) were solubilized by Triton X-100 in the cold and analyzed by BN/PAGE. (C) Surface-rendered 3D reconstruction of the R-peptide mutant Env structure. R-peptide mutant Envs were solubilized from unlabeled R-peptide mutant virus and isolated by gradient centrifugation for structure reconstruction by cryo-EM. Additionally, a virtual section of the bottom region of the reconstruction, color-coded according to EM densities, is shown (Bottom). The position of the section is indicated by an arrow in side view 2. (Scale bar, 50 Å.)
Discussion
Our earlier cryo-EM study of the mature Mo-MLV Env revealed an open cage-like structure, where the TM subunits seemed to form separated Env legs (11). A tripod Env was also found in another study of the Mo-MLV (10). In this latter work, cryo-EM tomography of intact particles was used to determine the Env structure, and not the cryo-EM single-particle approach of solubilized and purified Envs we have used. In the present study, we show that the TM legs of the R-peptide precursor of the Env are tied together into a central body at the Env bottom. This suggests that the R-peptide mediates trimerization of the C-terminal region of the TM subunits. Furthermore, the results show that R-peptide cleavage causes a relocation of the TM subunits from the central body in the R-peptide precursor to a splayed, peripheral location in the mature Env. Thus, the leg separation appears to be the major determinant for the final priming of the Env to be triggered by the receptor. The structural alteration in the Env endodomain seems also to transmit changes into the ectodomain. Although in our structure reconstructions we were unable to find significant differences between the ectodomains of the precursor and the mature Env, biochemical mapping using antibodies and biotinylation have shown that the R-peptide cleavage is associated with ectodomain changes (20).
According to the prevailing model, the receptor triggers changes in the SU subunit, including isomerization of the intersubunit disulfide (14–16). As a result, the TM subunits can refold, probably first into the putative extended form that can reach the target cell membrane with its N-terminal fusion peptide, and then into the backfolded form, the formation of which is thought to drag the viral and cell membranes together for fusion. The model is supported by the atomic structure of the TM ectodomain (23). This showed a trimer of TM hairpins, where the N-terminal parts formed a central coiled coil upon which the C-terminal parts were folded in an antiparallel orientation. A possible scenario for a successful fusion is that the TM subunits are first released to interact with the target membrane with their fusion peptides, and then form the coiled coil in their N-terminal part, and finally start to approach the membranes by backfolding the C-terminal parts into the grooves of the coiled coil. The latter reaction might require that the transmembrane part of the TM is free to rotate and move laterally in the plane of the membrane (24). If this is the case, then tying the endodomains of the TM together by the R-peptide should be an efficient way to prevent the backfolding step of Env activation in the R-peptide precursor Env. In this manner, premature fusion activation would be avoided. Further, the R-peptide cleavage would appear to be an elegant mechanism to prime the Env for receptor triggering. In the splayed conformation, the C-terminal parts of the TM should have the freedom to backfold once the extended form with the N-terminal coiled coil has formed.
Earlier, we showed that the R-peptide Env was able to undergo early activation steps, although at a reduced rate (17). In particular, it isomerized its intersubunit disulfide when triggered in vitro by Ca2+ depletion. Here we studied the structure of the in vitro activated intermediate form of the R-peptide Env (IAS). We found that the roof of the cage-like Env indeed had opened up, apparently by a similar clockwise rotation of the RBDs as found during the activation of the mature Env. This opening probably gives space for the putative extended form of the TM subunits to reach the target cell membrane with its fusion peptides. However, like in the case of the WT IAS Env, such an extended form of TM was not visualized, possibly because it is not stable without interacting with the target membrane. Instead, the N-terminal part of the TM might have reoriented toward the Env bottom, where the fusion peptide can interact with the detergent surrounding the transmembrane segments. It is conceivable that such reoriented N-terminal parts of the TM subunits also can form a coiled coil by their helical regions and that the C-terminal segments can at least to some extent interact with it. This proposed structure might correspond to the elongated central body in the bottom of the IAS form of the R-peptide Env.
Most of the ∼32-residue-long endodomains of the TM subunits, including their C-terminal 16-residue-long R-peptides, have been modeled as trimeric coiled coils (18). This has been suggested to inhibit the receptor triggering of the R-peptide Env precursor. The model was tested by analyzing the fusion phenotype of an Env with a mutation of Leu651 into Ala (19, 20). Leu651 is positioned just behind the Leu649/Val650 cleavage site and is predicted to be one key residue in the heptad repeat of hydrophobic residues specifying the trimeric coiled-coil structure. The fusion phenotype was characterized in cells expressing only the Env gene. This resulted in the accumulation of Envs with uncleaved R-peptide on the cell surface. Such cells were unable to fuse with receptor-carrying cells unless the gene region for the R-peptide was deleted (8, 9). Importantly, L651A Envs, which carried the R-peptide were almost as efficient in cell–cell fusion as the Envs with the R-peptide deletion. Thus, this phenotype of the mutant supports the notion that the R-peptide forms a trimeric coiled coil that inhibits receptor-induced Env triggering. In the present study, we determined the cryo-EM structure of the L651A Env and found that, despite the presence of the R-peptide it showed a separation of the TM legs similar to the mature form of the Env without the R-peptide. This showed that a separation of the TM legs, with or without the presence of the R-peptide is associated with a priming of the Env for receptor-triggered activation.
An evident future study is to elucidate the structure of the furin precursor. Earlier, we found that the more glycosylated form, gp90, of the Env precursor gp80 migrated significantly more slowly in SDS/PAGE after furin cleavage into a disulfide-linked SU–TM complex (25). The change in migration was not due to glycosylation but possibly to a structural change. One possibility is that the furin precursor protomers do not form the open cage-like structure but a much more closed one, which might not even allow the RBD to undergo initial activation by rotation. This should be possible to find out by determining the cryo-EM structure of the furin precursor Env.
Materials and Methods
Cells and Reagents.
293T cells and Mo-MLV–producing MOV-3 cells were maintained as described (21). MOV-3 cells represent Mo-MLV–transformed 3T3 cells (G. Schmidt, GSF-National Research Center for Environment and Health, Neuherberg, Germany). The viral protease inhibitor amprenavir was from the AIDS Research and Reference Reagent Program, Division of AIDS, National Institute of Allergy and Infectious Diseases, National Institutes of Health.
Construction of the L651A Env Virus Mutant.
The codon TTG for Leu at codon position 651 of the Env gene was changed to GCG for Ala by fusion PCR using the pNCA plasmid containing the Mo-MLV provirus (26).
Production and Isolation of Virus.
Virus was produced either in MOV-3 cells (WT only) or in 293T cells that had been transfected with corresponding proviral DNA (21). Amprenavir (0.8 μM) was included in the growth medium to completely inhibit R-peptide cleavage. [35S]Cys-labeled WT virus and mutant virus were prepared by labeling 293T cells with [35S]Cys for 24 h, starting at 24 h posttransfection, as described (21). Virus in the culture medium was isolated by sedimentation in a 20%/50% (wt/wt) sucrose step gradient in HNC buffer (50 mM Hepes, 100 mM NaCl, 1.8 mM CaCl2, pH 7.4) (21). The virus was recovered from the 20%/50% sucrose interphase.
PAGE.
The virus was incubated in HNC buffer containing 0.3% Triton X-100 on ice for 30 min to solubilize the Envs in their native form. For generation of the activated IAS intermediate form of the Envs, the virus was solubilized in HN buffer (50 mM Hepes, 100 mM NaCl, pH 7.4) containing 0.3% of Triton X-100, 10 mM EDTA, and 10 mM NEM at 37 °C for 30 min. SDS/PAGE sample buffer containing 5 mM NEM was added, and the samples were analyzed by reducing and nonreducing 13% SDS/PAGE using prestained proteins with lot-specific molecular masses of 118, 90, 50, 34, 26, and 19 kDa (Fisher Scientific) as markers (27). Radioactive proteins were visualized by phosphorimaging. Alternatively, the solubilized virus was analyzed by 4–10% BN/PAGE (28, 29). Standard proteins were thyroglobulin (Thy; 669 kDa), ferritin (Fer; 440 kDa), catalase (Cat; 232 kDa), lactate dehydrogenase (LDH; 140 kDa), and BSA (66 kDa).
Isolation of Envs.
Solubilized Envs, either in their native form or in their activated intermediate form (IAS), were isolated by sedimentation in a linear 5–20% sucrose gradient in HNC buffer containing 0.05% Triton X-100, superimposed by a 0–0.2% glutaraldehyde gradient (30). Trimers of disulfide-linked SU–TM complexes were identified in gradient fractions by BN/PAGE and silver staining and used for EM analyses.
Cryo-EM and Image Processing.
Isolated Envs were adsorbed to grids coated with a holey carbon film, blotted, and plunge-frozen in ethane using an FEI Vitrobot. Micrographs were recorded with a JEOL 2100F transmission electron microscope at 200 kV as described (30). In brief, particles were selected and visually verified in boxer, and corrected for the contrast transfer function using ctfit, both in EMAN 1 (http://blake.bcm.edu/emanwiki/EMAN). The particle set was loaded in EMAN 2, and reference-free class averages were generated using e2refined2d. These showed C3 symmetry, which was then imposed throughout the reconstruction. A set of 10 initial models was generated in e2initialmodel and tested through the iterative processes of alignment, classification, averaging, and creation of a new 3D model for 12 cycles. At this stage, the majority of the initial models had converged into a similar structure. One of those was chosen for refinement until no further improvement could be achieved. Complete galleries of the class averages and projections of the final 3D reconstructions of the native form of the R-peptide Env, the IAS form of the R-peptide Env, and the native form of the R-peptide L651A Env mutant are shown in Figs. S2–S4. There was good agreement between the projections and the corresponding class averages. As shown in Fig. S5 A–C, there was also good representation of particle orientations in the reconstructions. The resolution of the final 3D reconstruction was estimated from a Fourier shell correlation curve at the point where the curve dropped below 0.5. Information below that resolution was truncated from the reconstruction by low-pass filtering. The resolutions obtained were 21–22 Å for the reconstructions (Fig. S6).
Molecular Modeling.
The modeled structure of the RBD of Mo-MLV was first manually fitted into the native or IAS form of the R-peptide Env (11). The fitting was then refined using the Fit in map module in UCSF Chimera (http://www.cgl.ucsf.edu/chimera/). The visualization was done in UCSF Chimera and PyMOL (http://www.pymol.org/).
Supplementary Material
Acknowledgments
We thank Drs. Hans Hebert and Philip J. B. Koeck for helpful discussions. Swedish Science Foundation Grant 2778 and Swedish Cancer Foundation Grant 0525 (to H.G.) and European Union FP7-People-ITN-2008 Marie Curie Actions Project Virus Entry 235649 supported this work.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The electron microscopy density maps reported in this paper have been deposited in the Electron Microscopy Data Bank (EMDB) (accession nos. EMD-2065, EMD-2066, and EMD-2067).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1118125109/-/DCSupplemental.
References
- 1.Hallenberger S, et al. Inhibition of furin-mediated cleavage activation of HIV-1 glycoprotein gp160. Nature. 1992;360:358–361. doi: 10.1038/360358a0. [DOI] [PubMed] [Google Scholar]
- 2.Green N, et al. Sequence-specific antibodies show that maturation of Moloney leukemia virus envelope polyprotein involves removal of a COOH-terminal peptide. Proc Natl Acad Sci USA. 1981;78:6023–6027. doi: 10.1073/pnas.78.10.6023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Schultz A, Rein A. Maturation of murine leukemia virus env proteins in the absence of other viral proteins. Virology. 1985;145:335–339. doi: 10.1016/0042-6822(85)90168-0. [DOI] [PubMed] [Google Scholar]
- 4.Shapiro SZ, Strand M, August JT. High molecular weight precursor polypeptides to structural proteins of Rauscher murine leukemia virus. J Mol Biol. 1976;107:459–477. doi: 10.1016/s0022-2836(76)80078-2. [DOI] [PubMed] [Google Scholar]
- 5.Ng VL, Wood TG, Arlinghaus RB. Processing of the env gene products of Moloney murine leukaemia virus. J Gen Virol. 1982;59:329–343. doi: 10.1099/0022-1317-59-2-329. [DOI] [PubMed] [Google Scholar]
- 6.Hunter E. Viral entry and receptors. In: Coffin JM, Hughes SH, Varmus HE, editors. Retroviruses. Cold Spring Harbor, NY: Cold Spring Harbor Lab Press; 1997. pp. 71–119. [PubMed] [Google Scholar]
- 7.Henderson LE, Sowder R, Copeland TD, Smythers G, Oroszlan S. Quantitative separation of murine leukemia virus proteins by reversed-phase high-pressure liquid chromatography reveals newly described gag and env cleavage products. J Virol. 1984;52:492–500. doi: 10.1128/jvi.52.2.492-500.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ragheb JA, Anderson WF. pH-independent murine leukemia virus ecotropic envelope-mediated cell fusion: Implications for the role of the R peptide and p12E TM in viral entry. J Virol. 1994;68:3220–3231. doi: 10.1128/jvi.68.5.3220-3231.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rein A, Mirro J, Haynes JG, Ernst SM, Nagashima K. Function of the cytoplasmic domain of a retroviral transmembrane protein: p15E-p2E cleavage activates the membrane fusion capability of the murine leukemia virus Env protein. J Virol. 1994;68:1773–1781. doi: 10.1128/jvi.68.3.1773-1781.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Förster F, Medalia O, Zauberman N, Baumeister W, Fass D. Retrovirus envelope protein complex structure in situ studied by cryo-electron tomography. Proc Natl Acad Sci USA. 2005;102:4729–4734. doi: 10.1073/pnas.0409178102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wu SR, et al. Turning of the receptor-binding domains opens up the murine leukaemia virus Env for membrane fusion. EMBO J. 2008;27:2799–2808. doi: 10.1038/emboj.2008.187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pinter A, Honnen WJ, Tung JS, O’Donnell PV, Hämmerling U. Structural domains of endogenous murine leukemia virus gp70s containing specific antigenic determinants defined by monoclonal antibodies. Virology. 1982;116:499–516. doi: 10.1016/0042-6822(82)90143-x. [DOI] [PubMed] [Google Scholar]
- 13.Fass D, et al. Structure of a murine leukemia virus receptor-binding glycoprotein at 2.0 angstrom resolution. Science. 1997;277:1662–1666. doi: 10.1126/science.277.5332.1662. [DOI] [PubMed] [Google Scholar]
- 14.Lavillette D, Boson B, Russell SJ, Cosset FL. Activation of membrane fusion by murine leukemia viruses is controlled in cis or in trans by interactions between the receptor-binding domain and a conserved disulfide loop of the carboxy terminus of the surface glycoprotein. J Virol. 2001;75:3685–3695. doi: 10.1128/JVI.75.8.3685-3695.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Barnett AL, Davey RA, Cunningham JM. Modular organization of the Friend murine leukemia virus envelope protein underlies the mechanism of infection. Proc Natl Acad Sci USA. 2001;98:4113–4118. doi: 10.1073/pnas.071432398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wallin M, Ekstrom M, Garoff H. Isomerization of the intersubunit disulphide-bond in Env controls retrovirus fusion. EMBO J. 2004;23:54–65. doi: 10.1038/sj.emboj.7600012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Löving R, Li K, Wallin M, Sjöberg M, Garoff H. R-peptide cleavage potentiates fusion-controlling isomerization of the intersubunit disulfide in Moloney murine leukemia virus Env. J Virol. 2008;82:2594–2597. doi: 10.1128/JVI.02039-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Taylor GM, Sanders DA. Structural criteria for regulation of membrane fusion and virion incorporation by the murine leukemia virus TM cytoplasmic domain. Virology. 2003;312:295–305. doi: 10.1016/s0042-6822(03)00297-6. [DOI] [PubMed] [Google Scholar]
- 19.Yang C, Compans RW. Analysis of the murine leukemia virus R peptide: Delineation of the molecular determinants which are important for its fusion inhibition activity. J Virol. 1997;71:8490–8496. doi: 10.1128/jvi.71.11.8490-8496.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Aguilar HC, Anderson WF, Cannon PM. Cytoplasmic tail of Moloney murine leukemia virus envelope protein influences the conformation of the extracellular domain: Implications for mechanism of action of the R peptide. J Virol. 2003;77:1281–1291. doi: 10.1128/JVI.77.2.1281-1291.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Löving R, Kronqvist M, Sjöberg M, Garoff H. Cooperative cleavage of the R peptide in the Env trimer of Moloney murine leukemia virus facilitates its maturation for fusion competence. J Virol. 2011;85:3262–3269. doi: 10.1128/JVI.02500-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zavorotinskaya T, Albritton LM. A hydrophobic patch in ecotropic murine leukemia virus envelope protein is the putative binding site for a critical tyrosine residue on the cellular receptor. J Virol. 1999;73:10164–10172. doi: 10.1128/jvi.73.12.10164-10172.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fass D, Harrison SC, Kim PS. Retrovirus envelope domain at 1.7 angstrom resolution. Nat Struct Biol. 1996;3:465–469. doi: 10.1038/nsb0596-465. [DOI] [PubMed] [Google Scholar]
- 24.Waning DL, Russell CJ, Jardetzky TS, Lamb RA. Activation of a paramyxovirus fusion protein is modulated by inside-out signaling from the cytoplasmic tail. Proc Natl Acad Sci USA. 2004;101:9217–9222. doi: 10.1073/pnas.0403339101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sjöberg M, Wallin M, Lindqvist B, Garoff H. Furin cleavage potentiates the membrane fusion-controlling intersubunit disulfide bond isomerization activity of leukemia virus Env. J Virol. 2006;80:5540–5551. doi: 10.1128/JVI.01851-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Colicelli J, Goff SP. Sequence and spacing requirements of a retrovirus integration site. J Mol Biol. 1988;199(1):47–59. doi: 10.1016/0022-2836(88)90378-6. [DOI] [PubMed] [Google Scholar]
- 27.Opstelten DJ, Wallin M, Garoff H. Moloney murine leukemia virus envelope protein subunits, gp70 and Pr15E, form a stable disulfide-linked complex. J Virol. 1998;72:6537–6545. doi: 10.1128/jvi.72.8.6537-6545.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schägger H, von Jagow G. Blue native electrophoresis for isolation of membrane protein complexes in enzymatically active form. Anal Biochem. 1991;199:223–231. doi: 10.1016/0003-2697(91)90094-a. [DOI] [PubMed] [Google Scholar]
- 29.Sjöberg M, Lindqvist B, Garoff H. Stabilization of TM trimer interactions during activation of Moloney murine leukemia virus Env. J Virol. 2008;82:2358–2366. doi: 10.1128/JVI.01931-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wu SR, et al. Single-particle cryoelectron microscopy analysis reveals the HIV-1 spike as a tripod structure. Proc Natl Acad Sci USA. 2010;107:18844–18849. doi: 10.1073/pnas.1007227107. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.