Abstract
Recombineering, a recently developed technique for efficient genetic manipulation of bacteria, is facilitated by phage-derived recombination proteins and has the advantage of using DNA substrates with short regions of homology. This system was first developed in E. coli but has since been adapted for use in other bacteria. It is now widely used in a number of different systems for a variety of purposes, and the construction of chromosomal gene knockouts, deletions, insertions, point mutations, as well as in vivo cloning, mutagenesis of bacterial artificial chromosomes and phasmids, and the construction of genomic libraries has been reported. However, these methods also can be effectively applied to the genetic modification of bacteriophage genomes, in both their prophage and lytically growing states. The ever-growing collection of fully sequenced bacteriophages raises more questions than they answer, including the unknown functions of vast numbers of genes with no known homologs and of unknown function. Recombineering of phage genomes is central to addressing these questions, enabling the simple construction of mutants, determination of gene essentiality, and elucidation of gene function. In turn, advances in our understanding of phage genomics should present similar recombineering tools for dissecting a multitude of other genetically naïve bacterial systems.
Keywords: bacteriophage, BRED, recombineering
Introduction
Bacteriophages are extremely abundant, and it has been estimated that the number of phage particles in the biosphere is approximately 1031.1 These phages are a source of high genetic diversity and are replete with genetic novelty.2 They have played key roles in the development of bacterial genetics and can provide effective solutions for the development of systems to manipulate genetically naïve bacteria.3 Genetic modification of phages can facilitate the study of these viruses and, in consequence, the development of new tools for bacterial manipulation.
In contrast to the range of tools that have been described for targeted mutagenesis of bacterial chromosomes, fewer methods have been implemented for targeted mutagenesis of lytically growing phages, because high frequency events are required in the absence of a selectable system.
Historically, construction of mutations in bacteriophages was accomplished by general mutagenesis using UV irradiation or chemical compounds that can generate DNA damage.
A homologous recombination approach has also been used to make gene deletions or recombinant phages,4,5 but the frequency is low and screening to find the desired mutant can be tedious and time consuming.
Recently, a new in vivo technology to introduce genetic changes in bacterial genomes has been developed. This technique—named “recombineering”—refers to the engineering of recombinant DNA by homologous recombination.6-8 Recombineering employs recombination systems encoded by bacteriophages to enhance the frequency of homologous recombination, allowing the construction of chromosomal gene knock outs, deletions, insertions, point mutations, in vivo cloning, mutagenesis of bacterial artificial chromosomes, phasmids and genomic libraries.6-15
The increased availability of bacterial genome sequences has facilitated the use of recombineering, because it is only possible to recombineer an organism whose sequence is known.
The first systems described employed recombination functions encoded by bacteriophage lambda and the Rac prophage, and were originally designed for modification of E. coli. Subsequently, these were successfully employed in other Gram-negative bacteria, such as Salmonella and Shigella,16,17 and further adapted for application to other more divergent bacteria, like the insect endosymbiont Sodalis glossinidius and the promising subject for biotechnological exploitation, Pantoea ananatis.18,19
Double-stranded DNA recombineering in E. coli using the lambda Red system involves three phage-encoded proteins, Exo, Beta and Gam. Exo is an exonuclease that degrades one strand of DNA from a double-stranded end to generate a ssDNA substrate. Beta is a DNA pairing enzyme that anneals the recombineering substrate to its chromosomal target. Gam inhibits the E.coli RecBCD and SbcD enzymes, preventing degradation of the double-stranded DNA (dsDNA) substrate.20-23 For optimal dsDNA recombineering, RecBCD should be inactivated, either by mutation or by lambda gam.6,8,24 The original model proposed that after degradation by Exo, a 3′ single stranded DNA tail is exposed to which Beta can bind.8 Homologous recombination then occurs, following the strand invasion or DNA annealing model.25-28 Recently, a new model to explain the mechanism of dsDNA recombination mediated by lambda phage proteins has been proposed.29,30 In this alternative model, lambda exonuclease entirely degrades one strand, while leaving the other strand intact as single-stranded DNA. This single-stranded intermediate then recombines via β recombinase catalyzed annealing at the replication fork.
The Rac prophage encodes only RecE and RecT, which are functionally equivalent to lambda Exo and Beta, respectively.12,31
Beta belongs to a large family of ssDNA annealing proteins (SSAPs),32-34 and only this protein is necessary for single-stranded oligonucleotide recombination,7 used for the creation of point mutations and deletions. A strand bias in recombination levels has been observed when comparing two complementary ssDNAs. Higher frequencies are obtained when using an oligonucleotide that anneals to the DNA strand undergoing discontinuous synthesis (lagging strand). As a result of the replication process, transient regions of ssDNA may be accessible for Beta-mediated annealing of the oligo. The increased recombination efficiency of the ‘‘lagging strand’’ oligos may reflect the increased availability of single-stranded regions during lagging vs. leading strand synthesis.7,12,35
Regulated expression of the bacteriophage recombination system typically is required for recombineering. These functions can be integrated into the chromosome or contained within an extrachromosomal plasmid. A lambda lysogen with a defective prophage can be used to express the Red proteins from the strong lambda pL promoter.6 The recombineering functions have also been transferred to a variety of different plasmids,36 including constructs in which the functions are under control of the arabinose promoter pBAD.14 Some plasmids carry a temperature-sensitive (ts) origin of replication, so they can be cured from the cell after recombination. The prophage constructs contain the ts cI857 allele of the phage lambda repressor allowing protein expression to be controlled by temperature. At low temperature (30°C), recombineering functions are strongly repressed, and expression occurs after a shift to 42°C for only 15 min.
The efficiency of recombineering in E. coli and the possibility of using DNA segments with short regions of homology (approx. 50 bp) to the target sequences has led to the search for proteins with similarity to the lambda/Rac encoded proteins in other phages or bacteria, especially because adapting the lambda/Rac systems for its use in other organisms, particularly those that are Gram-positive could not give optimal results.37-39
The complete genome sequences of more than 80 mycobacteriophage genomes40-45 has provided us with thousands of unique genes, making this an ideal reservoir in which to search for novel recombineering functions. Recently, RecE and RecT homologs—the products of genes 60 and 61, respectively—were identified in mycobacteriophage Che9c, the expression of which substantially enhances mycobacterial recombination frequencies. These proteins have been utilized to develop an efficient recombineering system, facilitating the construction of gene-knockouts and point mutations in the hard to manipulate and pathogenic bacterium Mycobacterium tuberculosis, as well as in fast-growing related species like Mycobacterium smegmatis, which is routinely use as a model system.46,47
Mycobacterial strains constructed for recombineering contain an extra chromosomal plasmid in which the phage recombination genes are under the control of the inducible acetamidase promoter.48 Similar to what has been shown in E. coli, dsDNA recombineering in the mycobacteria requires both an exonuclease (gp60) and its associated recombinase (gp61),39 while recombination using ssDNA substrates requires only the recombinase.47 Targeted gene replacement mutants are engineered using linear dsDNA allelic exchange substrates (AESs), containing regions of homology upstream and downstream of the target gene flanking a cassette for antibiotic resistance, which are electroporated into either M. smegmatis or M. tuberculosis recombineering strains.39 Substrates for ssDNA recombineering are short oligonucleotides—a minimum length of 50 bases is recommended—encoding the desired mutation, and this technique provides a simple and efficient method for constructing point mutations in mycobacterial genomes.47 The overall efficiency of ssDNA recombineering is substantially higher than with dsDNA substrates for both chromosomal and plasmid targets. However, optimal efficiencies are only obtained when using oligonucleotides that target, and anneal to, the DNA strand undergoing discontinuous synthesis (lagging strand template).47 These can display efficiencies that are up to 10,000-fold higher than oligonucleotides targeting the leading strand, which far exceeds the 2–50 fold strand biases observed in the λ Red system.7,35 It is not clear why the biases are so much larger in mycobacteria than in E. coli, but this may reflect fundamental differences in the DNA replication systems and/or in how gp61 interacts with the replication machinery and supports the necessity of finding recombineering systems specific for each bacteria or related bacterial group.
Recombineering systems based on the mycobacteriophage Che9c-encoded proteins have provided new approaches to mycobacterial mutagenesis and greatly expanded the genetic toolbox available to study the pathogenic mycobacteria.
Datta et al.37 have also identified and characterized genes that are similar to Beta or RecT from Gram-positive and other Gram-negative bacteria and their phages. Nine genes, including Che9c gp61, were expressed in E. coli under control of lambda pL, and protein activity was assayed using an oligo recombination system.49 Seven of these nine genes were adjacent to a known or putative exonuclease gene. In the E. coli system, all the ssDNA binding proteins tested were able to catalyze oligo-mediated recombination but with variable efficiency; Che9c gp61 recombination levels were approximately 1000-fold lower than those observed with lambda β. When present with their canonical exonucleases, three of the four recombinase-exonuclease pairs tested were also able to carry out low level recombination with linear dsDNA, but not when coupled to lambda Exo; presumably because a physical and specific coupling is required between the cognate exonuclease and single strand annealing proteins. Thus, the exonuclease seems to be more species-specific, whereas the recombinase can function in diverse hosts.
After failure to observe lambda Red-mediated recombination in Pseudomonas syringae, and based on the hypothesis that recombinases found in, or associated with, particular species would be more likely to function in close relatives than distantly species,32,50,51 Swingle et al.38used a combination of bioinformatic tools to identify proteins homologous to RecE/RecT in Pseudomas syringae pv syringae B728a. The so-called recTEPSY genes are located in a 72 kb region that has been horizontally acquired in Pseudomas syringae pv syringae B728a. This region contains 86 annotated open reading frames (ORFs), the majority of which encode hypothetical proteins. Thirteen ORFs in this locus are annotated as phage-related proteins, and it is likely this region corresponds to a prophage or a remnant of a prophage containing the recombination functions. Depending on the oligo concentration, up to a 450-fold difference in recombination frequency between the RecTPSY expression and vector control strains was observed. Additionally, dsDNA recombination was observed when RecTPSY was assisted by RecEPSY.
Recombineering can be used to target the bacterial chromosome or extrachromosomal replicating molecules. As a corollary to this, it also can be extended for efficient and straightforward modification of bacteriophage genomes. Here, we will discuss the generation of E. coli phage mutants using recombineering with lambda Red proteins, as well as the construction of mycobacteriophage mutants using BRED (Bacteriophage Recombineering of Electroporated DNA).52
Recombineering Prophages
One relatively straightforward approach to engineering phage genomes is to use dsDNA recombineering to modify an integrated prophage, effectively treating it as any other chromosomal locus. There are, however, several limitations and disadvantages. First, this approach is restricted to temperate phages. Moreover, it is useful for these to be inducible phages, such that lytically growing particles of the constructed mutant can be generated. While this is often not a concern for well-characterized systems such as phage lambda, it may be generally so. Second, this approach typically requires the use of a selectable marker to recover the desired mutant, and introduction of a marker such as a drug resistance gene can both introduce genetic polarity, and increase the size of the phage genome beyond a packaging limit. These effects can be minimized by using a second event—such as by using site-specific recombination—to remove the selectable marker, but this then requires multiple sequential manipulations. Strategies to modify an integrated prophage are described in more detail below.
Recombineering E. coli Phages
E. coli phage mutants have played key roles in the study of phage biology,53-57 but the construction and/or screening of the desired mutation can be tedious and time consuming. Recombineering has simplified mutant phage construction, by increasing the frequency of recombination events in the cell, enabling the engineering of lytically growing phages. This should be applicable to all or most phages that can be propagated in the laboratory and avoids the need for genetic selection and the associated problems with genetic polarity. Two main approaches to recombineering lytically growing phages have been described, one using phage lambda as a model system, and the second using mycobacteriophages as a model system.
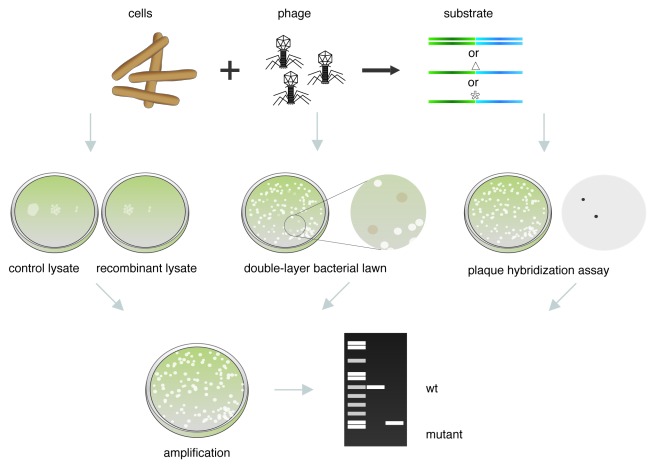
A method for engineering phage lambda was originally described by Oppenheim et al.58-60 In general, this involves infecting cells with phage, preparing competent cells, introducing the dsDNA or ssDNA substrates by electroporation and analyzing the resulting plaques for the presence of the mutation. A scheme of the general strategy used to modify phages infecting E. coli is shown in Figure 1. Because this method relies on physical screening to identify the desired mutants, it is critical that not only the recombination frequency is high, but that the electroporation efficiency is also very high (which is true for E. coli), such that a very high proportion of cells take up the dsDNA or ssDNA substrate.
Figure 1.E. coli phage recombineering. Schematic representation of the strategy used to construct and screen phage mutants by recombineering in E. coli. If the phage to be modified is not already present as a prophage in the genome, the cells are infected with phage; subsequently, recombination functions are expressed, and cells are made electrocompetent. The substrate is introduced into the infected cells by electroporation (dsDNA, ssDNA containing a deletion Δ, or a point mutation *), and cultures are recovered to allow phage assembly. The recombinant lysate is then screened for the desired mutant. Plating properties of the recombinant lysate can be compared with a control lysate obtained using the same protocol but without addition of the substrate. The double-layer bacterial lawn technique can be used when a strain that supports amplification of the mutant phage (that cannot replicate in the isogenic strain) is available. Alternatively, the desired mutant can be screened by plaque hybridization using a probe that can only hybridize to the mutant but not to the wt phage. After the mutant phage is identified and purified, a stock can be prepared, and the presence of the desired mutation corroborated by PCR or DNA sequencing.
Although most of the strains carrying the defective lamboid prophage expressing β, exo and gam are W3110 or MG1655 derivatives, the prophage can be moved to other backgrounds by P1 transduction,6,61,62 or the desired strain can be transformed with a plasmid expressing the recombination functions.36 For that reason, with some modifications, this protocol originally developed for lambda phage, can be adapted to make mutations in most E. coli phages. Alternatively, an intact prophage can serve as both the source of the recombination functions and the target for recombineering. In this case, not only must the recombination functions be present, the prophage must also be inducible in order to recover phage particles after recombineering.
In both cases, cells are grown to exponential phase. If a lytically growing phage is going to be modified, the cells are infected at a multiplicity of infection (MOI) of 1–3 and left standing to allow the phage adsorb (adsorption conditions for each phage must be taken in consideration for this step). This is not necessary if the phage is already present as a prophage in the cells.
In the next step, expression of Beta, Exo and Gam (when it is present) is induced for a short period of time. Conditions for induction have to be determined on the basis of the selected strain (temperature shift, addition of the inducer, etc.). The cells are then gently washed, and the appropriate DNA substrate is introduced by electroporation. When modifying an intact prophage, full induction time should only be used if the prophage is going to be induced and a recombinant lysate prepared. The progeny phage must then be screened for the mutation. Alternatively, the prophage may be targeted using a selectable marker, such as a drug resistance cassette, and the recombinant bacterial colonies identified using selection (see below).
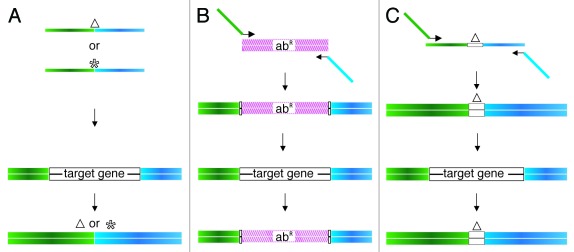
A single-stranded oligo or a dsDNA PCR product can be used as substrate for bacteriophage recombineering (Fig. 2). When a ssDNA substrate is used (for base substitution or small deletions or insertions), these are typically about 70 bases in length and are designed such that the alteration to be introduced is contained in the center of the oligo. An 80-nucleotide oligo was successfully used to generate a 326 bp deletion of the cII gene in lambda.60 In contrast to what has been seen with oligo-mediated recombination in bacteria, both complementary oligos were equally efficient in promoting recombination, perhaps reflecting the rolling circle replication employed by this phage.
Figure 2. Type of substrates used in phage recombineering. (A) ssDNA oligos (70 nt) containing either a small deletion (Δ) or a point mutation (*) in the center, and which will anneal to the lagging strand template during DNA replication. (B) When the mutation is going to be constructed in a prophage, the target gene can be replaced by an antibiotic resistance marker (abR). In this case, primers that amplify the resistance marker and contain about 50 bases with homology to each side of the target gene are used to generate the dsDNA substrate. (C) Schematic representation of the strategy used to increase the homology to the target by PCR. A 200 bp substrate can be obtained from a 100-mer oligonucleotide with 50 bases of homology to each side of the deletion and two 72-mers. In (A?C) each, blue and green rectangles indicate the homology regions to or flanking the target gene.
When a dsDNA substrate is used, a PCR product with 50 base pairs of homology (at each end) to a target in the phage chromosome is created following the strategy outlined in Figure 2.
It should be noted that drug resistance markers can only be used if selection is going to be done in the lysogen, since these markers cannot be employed in lytically replicating phages. Care should be taken if a plasmid is utilized as the template in the PCR reaction used to generate the substrate, because any residual plasmid in the reaction products will transform the cells efficiently, generating undesirable background. Digestion of the PCR reaction with DpnI (this will not cut the PCR product but will cleave the plasmid if it was purified from a Dam+ host) can be used to reduce the residual plasmid.
Oppenheim et al.60 used this system to precisely replace the rexA and rexB genes in lambda with a gene conferring ampicillin resistance. The substrate to target the lambda chromosome was generated as described in Figure 2. In this case, a phage lysate was grown from the electroporation mix and used to form lysogens in naïve E. coli. AmpR lysogens were selected, and the replacement of the rexAB genes was confirmed by PCR.
If, as an alternative, recombinant lysogens of a complete prophage (carrying for example, a drug resistance marker) are going to be selected, the electroporation mixture can be spread on a filter that is placed on top of a plate of rich nonselective medium. After incubation for a few hours the filter is transferred to a plate containing the selective drug. After overnight growth, recombinants cells will give rise to colonies, and prophages can be excised to recover mutant phage particles.
When a lytically propagated phage genome is the target, the recombinant lysate has to be plated for direct selection of the plaques or for further screening. Preliminarily, dilutions of the control and recombinant lysates can be plated on lawns of a bacterial indicator to compare plating properties of both lysates (Fig. 1). If available, a selective indicator strain should be used. For example, if an amber mutation is introduced in an essential gene, the double-layer bacterial lawn technique63 can be used to identify phages that carry the mutation. In this case, a double-layer containing two different indicator strains is utilized to plate the recombinant lysate. The lower layer contains the restrictive host where only the wild-type (wt) phage can multiply, and the top layer contains a suppressor strain that supports growth of both the mutant and wt phage. Amber mutants can only lyse the suppressor strain and thus form “cloudy” plaques because the non-suppresor strain can grow to confluence (Fig. 1).
If phage mutants cannot be selected or distinguished visually, a plaque hybridization assay can be used to detect the mutant phage derivatives (Fig. 1). In order to identify the mutant, at least a 15 bp DNA sequence that is present in the recombinant phage, but not in the original phage, is needed to use as a target for the probe.5 To corroborate the presence of the desired mutation, an isolated plaque can be resuspended in buffer and used directly as a template in a PCR reaction, using two primers that flank the deletion/insertion.15,52 An amplicon of differential size is expected in the mutant as compared with the parent phage. MAMA-PCR (mismatch amplification mutation assay PCR) can also be used to screen for point mutations.64 If a point mutation that alters a restriction site was generated, the base change can be identified after restriction of the amplified PCR product and visualization by gel electrophoresis. Finally, a lysate of the corroborated recombinant phage can be prepared and used for further studies.
Construction of Mycobacteriophage Mutants by Recombineering
Mycobacteriophages—viruses that infect mycobacteria—have been instrumental in overcoming the many challenges of performing genetic studies in the human pathogen, M. tuberculosis. These formed the basis for the development of shuttle phasmids,65 and have been used for transposon mutagenesis,66 reporter gene delivery,67-69 and specialized transduction,70 as well as for the construction of integration-proficient vectors, enabling the stable introduction of foreign genes into this bacterium.42,43,71-73 However, although mycobacteriophages have been instrumental in the development of techniques for mycobacterial mutant construction, genetic tools enabling mutagenesis of the phages themselves have been lacking.
In general, constructing mutant derivatives of bacteriophages is more challenging than manipulating the bacterial host chromosome, mostly because antibiotic selection is not useful in lytically propagated viruses. To circumvent this, mycobacterial recombineering could be employed for the manipulation of prophages, which are integrated into the host chromosome, in a manner similar to that described for the manipulation of lambda prophages above. Unfortunately, relatively few mycobacteriophages form stable lysogens, significantly limiting the use of this strategy. Until recently, the most powerful method for manipulating lytic mycobacteriophages was through the use of shuttle phasmids74—phage-plasmid hybrids that can be packaged into phage λ particles and propagated in E. coli. In one example, this strategy was used to make defined mutations in the tape measure gene of a shuttle phasmid derivative of mycobacteriophage TM4 by performing the mutagenesis in E. coli (by λ Red recombineering) and recovering the mutant phage in M. smegmatis.15 In spite of this, and the obvious advantages of this technique, use of mycobacteriophage phasmids remains limited. This is due, in part, to the multiple steps required to construct the phasmids, but is primarily a function of the relatively small packaging capacity (less than ~50 kb) of λ capsids, as compared with the large average size of mycobacteriophage genomes (~70 kb).40,41 Thus, most mycobacteriophage genomes are too large for shuttle phasmid construction, and this approach is not broadly applicable for functional genomic studies on this phage population.
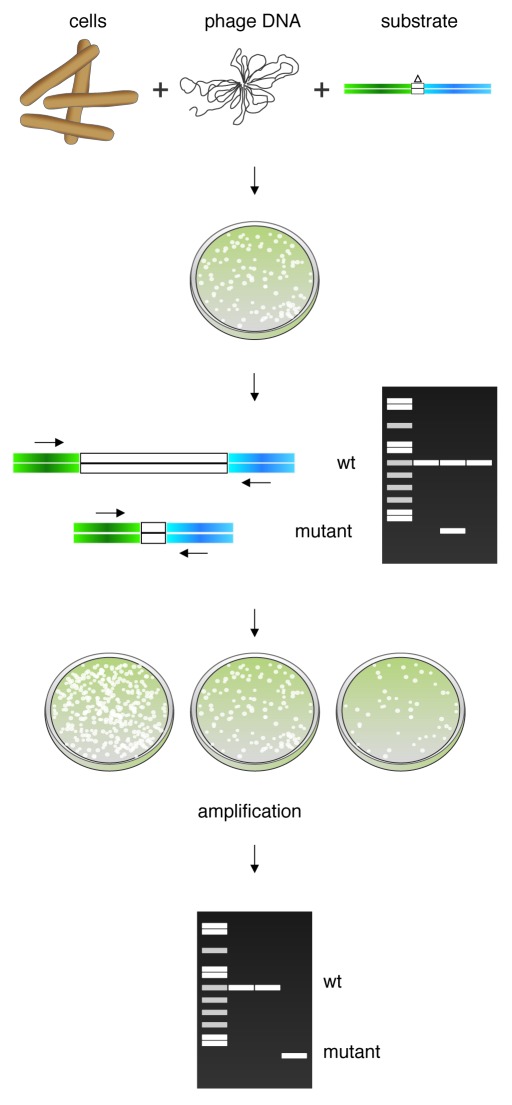
In contrast, mycobacterial recombineering systems offer a new, potentially powerful approach to constructing mutants of any lytically growing mycobacteriophage, using a method termed Bacteriophage Recombineering of Electroporated DNA (BRED),52 the general scheme of which is illustrated in Figure 3. A critical aspect of this technique is based on the observation that, in mycobacteria, recombineering frequencies appear to be primarily limited by relatively poor DNA uptake efficiencies75; that is, even for electrocompetent mycobacteria, only 1 out of 1000 viable cells can productively take up DNA. This presents a challenge when constructing mutations for which there is no selection, such as chromosomal point mutations with no known phenotype, as 99.9% colonies recovered will arise from cells that were unable to take up the recombineering substrate.47 The issue is circumvented in ssDNA recombineering by employing a counter-selection strategy, in which a selectable plasmid (or a second oligonucleotide that incorporates a selectable point mutation) is co-transformed with the ssDNA substrate.6,7 This effectively selects against non-competent cells in the population, and the transformants can then be screened for the mutation using MAMA-PCR,10,64 which selectively amplifies the mutant allele. In this way, non-selectable mutations can be easily recovered due to the high efficiency of ssDNA recombineering.
Figure 3. Mycobacteriophage recombineering. Schematic representation of the strategy used to construct phage mutants in Mycobacterium smegmatis by recombineering. M. smegmatis cells inducibly expressing the recombination functions are co-electroporated with phage DNA and the recombineering substrate; these are mixed with M. smegmatis plating cells and plated to obtain individual plaques. Primary plaques are screened by PCR with flanking primers to detect those containing mutant phage that is present at some frequency in combination with the wt phage. Once identified in this way, a positive plaque containing the mutant allele will be diluted and plated. If a non-essential gene was targeted, the mutant phage can be easily identified by PCR. Essentiality of the target gene can be inferred if the mutant allele is not detected in a lysate of pooled secondary plaques.
The technique described in the previous section for the recombineering of an E. coli phage, such as λ, involves infecting with the phage to be targeted, inducing the recombineering functions, and electroporating with the DNA substrate;60 however, the inefficiency of DNA uptake in mycobacteria precludes this approach. Therefore, in BRED, we have adopted an alternative approach derived from the co-selection methods described above.52 In this procedure, an M. smegmatis strain carrying a recombineering plasmid is induced with acetamide, and electrocompetent cells are prepared. This electroporation is performed with both the genomic DNA of the phage to be manipulated and a PCR-generated or synthetic DNA substrate that targets this phage (Fig. 2). After a brief recovery period, the reactions are mixed with additional plating cells of M. smegmatis and plated as in an infectious center assay (i.e., prior to lysis, such as plaques are generated from infected cells, not from viral particles). In this way, only cells that have taken up phage DNA will give rise to plaques within the bacterial lawn, effectively selecting against those that are non-transformable; on average, about 100 plaques are obtained from 50–100 ng of phage DNA in such an experiment. All of these primary plaques will contain wild-type phage DNA, and a subset (5–50%) will also contain the mutant allele,52 which can be readily identified by PCR screening. Provided the mutation does not ablate a region that is essential for phage growth, purified mutant phage derivatives can be isolated by plating serial dilutions of the mixed plaques, followed by purification and PCR analysis of the resulting secondary plaques. The frequency with which the pure mutants are recovered can vary significantly depending on factors such as the amount of mutant allele present in the primary plaque and the phenotype of the mutant (e.g., it will be recovered at a lower frequency if it has a reduced burst size as compared with wild-type). Often, analysis of 12–18 secondary plaques is sufficient to identify a homogenously mutant strain, but in other cases, it has been necessary to screen secondary plaques in pools of 5–10 in order to recover the mutant. In these cases, once a pool containing the mutant is identified, a third plating is necessary to completely purify the mutant. This pooling strategy essentially enriches for a mutation that might have been present at very low levels in the initial primary plaque.
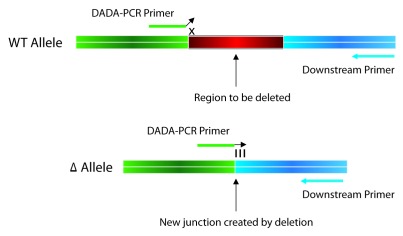
To date, the most common application for BRED has been the construction of unmarked gene deletions; however, this technique can also be used to efficiently engineer in-frame internal deletions, small insertions, point mutations and gene replacements in the phage genome, none of which require selection for mutant recovery52 (Hatfull Lab, unpublished data). When constructing a deletion or insertion mutant, a 200 bp dsDNA substrate, having 100 bp of homology flanking the region to be deleted or inserted is recommended, as a higher proportion of primary plaques contain the desired mutant allele when using dsDNA vs. ssDNA substrates, and larger substrates (200 bp) vs. shorter ones (100 bp). Insertions are typically screened by PCR using one primer that anneals within the inserted region and another that anneals to a downstream region in the phage chromosome. For deletions, primers that flank the deleted region are used; when the mutant allele is present in a primary plaque, these produce a smaller PCR product that is most often easily visible on an agarose gel. In some cases—usually when the mutants are somewhat defective—mutant alleles are present at low frequencies in the primary plaques and cannot be detected by flanking primers. For these situations, we have adapted the MAMA-PCR strategy described above and developed a technique termed Deletion Amplification Detection Assay (DADA) PCR.52 This employs a DADA-PCR primer that anneals across the new junction created by the deletion at its 3′ end and another that anneals to the flanking DNA, either up- or downstream of this. The DADA-PCR primer is designed such that in the mutant allele, the primer will anneal perfectly across the deletion site (this is the site created by the joining of the two DNA ends that flank the deleted region), but in the wild-type phage, the terminal one to three 3′ nucleotides (depending on the primer design) will not base pair with the DNA template (Fig. 4). A high fidelity DNA polymerase will have difficulty synthesizing through this mismatch, and thus, under stringent conditions, the wild-type allele is not amplified.
Figure 4. Deletion Amplification Assay (DADA) PCR. DADA-PCR is used to specifically detect the presence of a gene deletion allele that is present at a low frequency or is otherwise difficult to detect in a mixed population of mutant and WT phage. The DADA-PCR primer is designed such that the 3′ end anneals across the new junction created by the joining of the two DNA ends flanking the deleted region. When this primer anneals to the WT allele, this creates mismatch (shown an ‘x’ in the figure) through which a high fidelity DNA polymerase cannot synthesize under stringent PCR conditions. However, the DADA-PCR primer is complementary to the deletion allele, and with an appropriate down- (or up-) stream primer, will only generate a product if the mutant allele is present.
One application of BRED in which the DADA-PCR detection strategy has been quite useful is in the deletion of genes that are essential—or suspected to be essential—for phage growth. We have been able to delete a number of essential genes in mycobacteriophage Giles (such as the lysin A)52 (Hatfull lab, unpublished data) and other phages by constructing a complementation strain of M. smegmatis for mutant isolation, which expresses the phage gene to be deleted. In these cases, it is often necessary to use DADA-PCR to detect primary plaques containing the mutant allele. These can be recovered on wild-type bacteria, presumably due to the presence of wild-type helper phage in the primary plaque. Once such a plaque is recovered, essentiality can be inferred if the mutant allele is not detected in lysates prepared from a re-plating of these mixed plaques on wild-type cells; that is, the individual mutant particles could not give rise to plaques in the secondary plating. The mixed primary plaque must then be serially diluted and plated on the complementation strain. Secondary plaques are picked and patched in duplicate on wild-type and complementing bacteria, and a mutant phage can be identified as one that is only able to infect and kill the complementing M. smegmatis. This strategy provides a powerful and effective way to screen for gene essentially, allowing us to determine which of the many unique genes found in the mycobacteriophages are required for lytic growth.
Another type of genetic manipulation for which BRED has proved successful is the engineering of point mutations, and for this, ssDNA synthetic substrates are employed.52 Although an extensive length dependence analysis has not been performed, we find that oligonucleotides that are at least 71 nt in length are sufficient to efficiently recover the mutation (Marinelli, unpublished data). Based on the observations from mycobacterial ssDNA recombineering47; however, it is possible that shorter substrates might also work well. Importantly, due to the nature of phage DNA replication, when constructing point mutations, oligonucleotides targeting both strands are used and are transformed together with the phage genomic DNA.52 The primary and secondary plaques are screened for the mutant allele using MAMA-PCR and purified using the same techniques outlined above.
Finally, we have shown that BRED can be used for the construction of gene replacement mutants within the phage genome.52 As mentioned above, antibiotic resistance cassettes are not useful in lytic phages; however, numerous other applications—such as the construction of fluorescent reporter phages67-69—could be envisioned for which it would be desirable to insert a foreign gene into a phage genome. The fairly strict packaging size constraints that exist for dsDNA bacteriophages would necessitate inserting a gene such as egfp (enhanced green fluorescent protein) in place of a similarly sized non-essential phage gene. For this type of BRED, we use dsDNA substrates, synthesized by PCR, that contain the cassette of choice flanked by 100 bp homologous to either end of the target gene that is being replaced. Such mutations are easily detected using a primer pair in which one anneals within the newly inserted gene and the other anneals to a region up- or downstream within the phage genome.
To date, BRED has been successfully used on nine different mycobacteriophages52,76-78 (Hatfull Lab, unpublished data), and it is likely that the full utility of these approaches for mycobacteriophage mutagenesis has yet to be realized. For example, the coupling of efficient point mutagenesis with mycobacterial nonsense suppressor strains79 could provide a relatively simple method for making conditional lethal mutants in mycobacteriophage genomes. It might also be possible to utilize this technique for the manipulation of phages that infect other species of mycobacteria or those infecting other closely related bacteria.
Perspectives
Coordinated efforts spanning the last decade have yielded a vast collection of fully sequenced bacteriophages. About 600 fully sequenced and annotated bacteriophage genomes are available from the NCBI database, revealing a genetically diverse population of architecturally mosaic genomes. This immense reservoir of genetic information has created numerous opportunities for functional genomic studies aimed at uncovering novel functions within this phage population.
Just for Mycobacteriophages alone, an enormous number of predicted open reading frames of unknown function have been found, which make up over 3,000 phamilies of unique genes having no detectable sequence similarity to known proteins.41-45,73,80 An increasing interest in the hunt for phage proteins that can promote recombination functions has been particularly driven by the number of bacteriophage genome sequences available.50 These functions can potentially be used to develop species-specific recombineering systems in a diverse suite of organisms.
Bacteriophage recombineering has shown to be a highly effective approach for the construction of gene deletions, small insertions, gene replacements, and point mutations in lytically replicating phages.52,60 This technique will, for the first time, allow us to perform broad functional genomic studies on bacteriophages, potentially revealing new and exciting features of this diverse phage population.
Here we have described the modification of E. coli phages and mycobacteriophages. However, with slight modifications to these protocols, and providing the appropriate recombination systems are found, these methods can potentially be adapted for use with many other phages. Any new bacteriophage isolated can be a possible source for new recombination proteins. For this purpose, genetic manipulation of these phages could be very useful and recombineering could become the perfect ally to accomplish this objective. Thus, bacteriophage recombineering will not only facilitate the study of orphan genes and provide insights into phage biology, but also could help to uncover new and valuable instruments for the manipulation of genetically naive bacteria.
Acknowledgments
This work was supported by US National Institutes of Health grant 1R03TW008926-01, ANPCyT-PICT2009-0095, CONICET-PIP2011-0222.
Footnotes
Previously published online: www.landesbioscience.com/journals/bacteriophage/article/18778
References
- 1.Hendrix RW, Lawrence JG, Hatfull GF, Casjens S. The origins and ongoing evolution of viruses. Trends Microbiol. 2000;8:504–8. doi: 10.1016/S0966-842X(00)01863-1. [DOI] [PubMed] [Google Scholar]
- 2.Brüssow H, Hendrix RW. Phage Genomics: Small Is Beautiful. Cell. 2002;108:13–6. doi: 10.1016/s0092-8674(01)00637-7. [DOI] [PubMed] [Google Scholar]
- 3.Jacobs WR Jr. Mycobacterium tuberculosis: A Once Genetically Intractable Organism. In: Hatfull GF, Jacobs Jr. WR, eds. Molecular Genetics of the Mycobacteria. Washington, DC: ASM Press, 2000:1-16. [Google Scholar]
- 4.Sarkis GJ, Jacobs WR, Hatfulll GF. L5 luciferase reporter mycobacteriophages: a sensitive tool for the detection and assay of live mycobacteria. Mol Microbiol. 1995;15:1055–67. doi: 10.1111/j.1365-2958.1995.tb02281.x. [DOI] [PubMed] [Google Scholar]
- 5.Tanji Y, Furukawa C, Na S-H, Hijikata T, Miyanaga K, Unno H. Escherichia coli detection by GFP-labeled lysozyme-inactivated T4 bacteriophage. J Biotechnol. 2004;114:11–20. doi: 10.1016/j.jbiotec.2004.05.011. [DOI] [PubMed] [Google Scholar]
- 6.Yu D, Ellis HM, Lee EC, Jenkins NA, Copeland NG, Court DL. An efficient recombination system for chromosome engineering in Escherichia coli. Proc Natl Acad Sci USA. 2000;97:5978–83. doi: 10.1073/pnas.100127597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ellis HM, Yu D, DiTizio T, Court DL. High efficiency mutagenesis, repair, and engineering of chromosomal DNA using single-stranded oligonucleotides. Proc Natl Acad Sci USA. 2001;98:6742–6. doi: 10.1073/pnas.121164898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Court DL, Sawitzke JA, Thomason LC. Genetic engineering using homologous recombination. Annu Rev Genet. 2002;36:361–88. doi: 10.1146/annurev.genet.36.061102.093104. [DOI] [PubMed] [Google Scholar]
- 9.Copeland NG, Jenkins NA, Court DL. Recombineering: a powerful new tool for mouse functional genomics. Nat Rev Genet. 2001;2:769–79. doi: 10.1038/35093556. [DOI] [PubMed] [Google Scholar]
- 10.Swaminathan S, Ellis HM, Waters LS, Yu D, Lee EC, Court DL, et al. Rapid engineering of bacterial artificial chromosomes using oligonucleotides. Genesis. 2001;29:14–21. doi: 10.1002/1526-968X(200101)29:1<14::AID-GENE1001>3.0.CO;2-X. [DOI] [PubMed] [Google Scholar]
- 11.Sarov M, Schneider S, Pozniakovski A, Roguev A, Ernst S, Zhang Y, et al. A recombineering pipeline for functional genomics applied to Caenorhabditis elegans. Nat Methods. 2006;3:839–44. doi: 10.1038/nmeth933. [DOI] [PubMed] [Google Scholar]
- 12.Zhang Y, Buchholz F, Muyrers JP, Stewart AF. A new logic for DNA engineering using recombination in Escherichia coli. Nat Genet. 1998;20:123–8. doi: 10.1038/2417. [DOI] [PubMed] [Google Scholar]
- 13.Muyrers JP, Zhang Y, Testa G, Stewart AF. Rapid modification of bacterial artificial chromosomes by ET-recombination. Nucleic Acids Res. 1999;27:1555–7. doi: 10.1093/nar/27.6.1555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Datsenko KA, Wanner BL. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc Natl Acad Sci USA. 2000;97:6640–5. doi: 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Piuri M, Hatfull GF. A peptidoglycan hydrolase motif within the mycobacteriophage TM4 tape measure protein promotes efficient infection of stationary phase cells. Mol Microbiol. 2006;62:1569–85. doi: 10.1111/j.1365-2958.2006.05473.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ranallo RT, Barnoy S, Thakkar S, Urick T, Venkatesan MM. Developing live Shigella vaccines using λ Red recombineering. FEMS Immunol Med Microbiol. 2006;47:462–9. doi: 10.1111/j.1574-695X.2006.00118.x. [DOI] [PubMed] [Google Scholar]
- 17.Swingle B, Markel E, Costantino N, Bubunenko MG, Cartinhour S, Court DL. Oligonucleotide recombination in Gram-negative bacteria. Mol Microbiol. 2010;75:138–48. doi: 10.1111/j.1365-2958.2009.06976.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pontes MH, Dale C. Lambda red-mediated genetic modification of the insect endosymbiont Sodalis glossinidius. Appl Environ Microbiol. 2011;77:1918–20. doi: 10.1128/AEM.02166-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Katashkina JI, Hara Y, Golubeva LI, Andreeva IG, Kuvaeva TM, Mashko SV. Use of the lambda Red-recombineering method for genetic engineering of Pantoea ananatis. BMC Mol Biol. 2009;10:34. doi: 10.1186/1471-2199-10-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kulkarni SK, Stahl FW. Interaction between the sbcC gene of Escherichia coli and the gam gene of phage lambda. Genetics. 1989;123:249–53. doi: 10.1093/genetics/123.2.249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Murphy KC. The lambda Gam protein inhibits RecBCD binding to dsDNA ends. J Mol Biol. 2007;371:19–24. doi: 10.1016/j.jmb.2007.05.085. [DOI] [PubMed] [Google Scholar]
- 22.Marsić N, Roje S, Stojiljkovic I, Salaj-Smic E, Trgovcevic Z. In vivo studies on the interaction of RecBCD enzyme and lambda Gam protein. J Bacteriol. 1993;175:4738–43. doi: 10.1128/jb.175.15.4738-4743.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Court R, Cook N, Saikrishnan K, Wigley D. The crystal structure of lambda-Gam protein suggests a model for RecBCD inhibition. J Mol Biol. 2007;371:25–33. doi: 10.1016/j.jmb.2007.05.037. [DOI] [PubMed] [Google Scholar]
- 24.Yu D, Sawitzke JA, Ellis H, Court DL. Recombineering with overlapping single-stranded DNA oligonucleotides: testing a recombination intermediate. Proc Natl Acad Sci USA. 2003;100:7207–12. doi: 10.1073/pnas.1232375100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Stahl MM, Thomason L, Poteete AR, Tarkowski T, Kuzminov A, Stahl FW. Annealing vs. invasion in phage lambda recombination. Genetics. 1997;147:961–77. doi: 10.1093/genetics/147.3.961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Langston LD, Symington LS. Gene targeting in yeast is initiated by two independent strand invasions. Proc Natl Acad Sci USA. 2004;101:15392–7. doi: 10.1073/pnas.0403748101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pâques F, Haber JE. Multiple pathways of recombination induced by double-strand breaks in Saccharomyces cerevisiae. Microbiol Mol Biol Rev. 1999;63:349–404. doi: 10.1128/mmbr.63.2.349-404.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Smith GR. Homologous recombination near and far from DNA breaks: alternative roles and contrasting views. Annu Rev Genet. 2001;35:243–74. doi: 10.1146/annurev.genet.35.102401.090509. [DOI] [PubMed] [Google Scholar]
- 29.Maresca M, Erler A, Fu J, Friedrich A, Zhang Y, Stewart AF. Single-stranded heteroduplex intermediates in lambda Red homologous recombination. BMC Mol Biol. 2010;11:54. doi: 10.1186/1471-2199-11-54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mosberg JA, Lajoie MJ, Church GM. Lambda red recombineering in Escherichia coli occurs through a fully single-stranded intermediate. Genetics. 2010;186:791–9. doi: 10.1534/genetics.110.120782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gottesman MM, Gottesman ME, Gottesman S, Gellert M. Characterization of bacteriophage lambda reverse as an Escherichia coli phage carrying a unique set of host-derived recombination functions. J Mol Biol. 1974;88:471–87. doi: 10.1016/0022-2836(74)90496-3. [DOI] [PubMed] [Google Scholar]
- 32.Iyer LM, Koonin EV, Aravind L. Classification and evolutionary history of the single-strand annealing proteins, RecT, Redbeta, ERF and RAD52. BMC Genomics. 2002;3:8. doi: 10.1186/1471-2164-3-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hall SD, Kolodner RD. Homologous pairing and strand exchange promoted by the Escherichia coli RecT protein. Proc Natl Acad Sci USA. 1994;91:3205–9. doi: 10.1073/pnas.91.8.3205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Noirot P, Kolodner RD. DNA strand invasion promoted by Escherichia coli RecT protein. J Biol Chem. 1998;273:12274–80. doi: 10.1074/jbc.273.20.12274. [DOI] [PubMed] [Google Scholar]
- 35.Zhang Y, Muyrers JP, Rientjes J, Stewart AF. Phage annealing proteins promote oligonucleotide-directed mutagenesis in Escherichia coli and mouse ES cells. BMC Mol Biol. 2003;4:1. doi: 10.1186/1471-2199-4-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Datta S, Costantino N, Court DL. A set of recombineering plasmids for gram-negative bacteria. Gene. 2006;379:109–15. doi: 10.1016/j.gene.2006.04.018. [DOI] [PubMed] [Google Scholar]
- 37.Datta S, Costantino N, Zhou X, Court DL. Identification and analysis of recombineering functions from Gram-negative and Gram-positive bacteria and their phages. Proc Natl Acad Sci USA. 2008;105:1626–31. doi: 10.1073/pnas.0709089105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Swingle B, Bao Z, Markel E, Chambers A, Cartinhour S. Recombineering using RecTE from Pseudomonas syringae. Appl Environ Microbiol. 2010;76:4960–8. doi: 10.1128/AEM.00911-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.van Kessel JC, Hatfull GF. Recombineering in Mycobacterium tuberculosis. Nat Methods. 2007;4:147–52. doi: 10.1038/nmeth996. [DOI] [PubMed] [Google Scholar]
- 40.Hatfull GF, Jacobs-Sera D, Lawrence JG, Pope WH, Russell DA, Ko CC, et al. Comparative genomic analysis of 60 Mycobacteriophage genomes: genome clustering, gene acquisition, and gene size. J Mol Biol. 2010;397:119–43. doi: 10.1016/j.jmb.2010.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pope WH, Jacobs-Sera D, Russell DA, Peebles CL, Al-Atrache Z, Alcoser TA, et al. Expanding the diversity of mycobacteriophages: insights into genome architecture and evolution. PLoS ONE. 2011;6:e16329. doi: 10.1371/journal.pone.0016329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Morris P, Marinelli LJ, Jacobs-Sera D, Hendrix RW, Hatfull GF. Genomic characterization of mycobacteriophage Giles: evidence for phage acquisition of host DNA by illegitimate recombination. J Bacteriol. 2008;190:2172–82. doi: 10.1128/JB.01657-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pham TT, Jacobs-Sera D, Pedulla ML, Hendrix RW, Hatfull GF. Comparative genomic analysis of mycobacteriophage Tweety: evolutionary insights and construction of compatible site-specific integration vectors for mycobacteria. Microbiology. 2007;153:2711–23. doi: 10.1099/mic.0.2007/008904-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hatfull GF, Pedulla ML, Jacobs-Sera D, Cichon PM, Foley A, Ford ME, et al. Exploring the mycobacteriophage metaproteome: phage genomics as an educational platform. PLoS Genet. 2006;2:e92. doi: 10.1371/journal.pgen.0020092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pedulla ML, Ford ME, Houtz JM, Karthikeyan T, Wadsworth C, Lewis JA, et al. Origins of highly mosaic mycobacteriophage genomes. Cell. 2003;113:171–82. doi: 10.1016/S0092-8674(03)00233-2. [DOI] [PubMed] [Google Scholar]
- 46.van Kessel JC, Hatfull GF. Mycobacterial recombineering. Methods Mol Biol. 2008;435:203–15. doi: 10.1007/978-1-59745-232-8_15. [DOI] [PubMed] [Google Scholar]
- 47.van Kessel JC, Hatfull GF. Efficient point mutagenesis in mycobacteria using single-stranded DNA recombineering: characterization of antimycobacterial drug targets. Mol Microbiol. 2008;67:1094–107. doi: 10.1111/j.1365-2958.2008.06109.x. [DOI] [PubMed] [Google Scholar]
- 48.Parish T, Mahenthiralingam E, Draper P, Davis EO, Colston MJ. Regulation of the inducible acetamidase gene of Mycobacterium smegmatis. Microbiology. 1997;143:2267–76. doi: 10.1099/00221287-143-7-2267. [DOI] [PubMed] [Google Scholar]
- 49.Costantino N, Court DL. Enhanced levels of lambda Red-mediated recombinants in mismatch repair mutants. Proc Natl Acad Sci USA. 2003;100:15748–53. doi: 10.1073/pnas.2434959100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lopes A, Amarir-Bouhram J, Faure G, Petit MA, Guerois R. Detection of novel recombinases in bacteriophage genomes unveils Rad52, Rad51 and Gp2.5 remote homologs. Nucleic Acids Res. 2010;38:3952–62. doi: 10.1093/nar/gkq096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Marchler-Bauer A, Anderson JB, Chitsaz F, Derbyshire MK, DeWeese-Scott C, Fong JH, et al. CDD: specific functional annotation with the Conserved Domain Database. Nucleic Acids Res. 2009;37:D205–10. doi: 10.1093/nar/gkn845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Marinelli LJ, Piuri M, Swigonova Z, Balachandran A, Oldfield LM, van Kessel JC, et al. BRED: a simple and powerful tool for constructing mutant and recombinant bacteriophage genomes. PLoS ONE. 2008;3:e3957. doi: 10.1371/journal.pone.0003957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hsiao CL, Black LW. DNA packaging and the pathway of bacteriophage T4 head assembly. Proc Natl Acad Sci USA. 1977;74:3652–6. doi: 10.1073/pnas.74.9.3652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Völker TA, Kuhn A, Showe MK, Bickle TA. Gene 67, a new, essential bacteriophage T4 head gene codes for a prehead core component, PIP. II. The construction in vitro of unconditionally lethal mutants and their maintenance. J Mol Biol. 1982;161:491–504. doi: 10.1016/0022-2836(82)90403-X. [DOI] [PubMed] [Google Scholar]
- 55.Müller UR. Enzymatic construction and selection of bacteriophage G4 mutants with modifications of a DNA secondary structure in the J-F intercistronic region. J Virol. 1983;48:170–9. doi: 10.1128/jvi.48.1.170-179.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yagil E, Dorgai L, Weisberg RA. Identifying determinants of recombination specificity: construction and characterization of chimeric bacteriophage integrases. J Mol Biol. 1995;252:163–77. doi: 10.1006/jmbi.1995.0485. [DOI] [PubMed] [Google Scholar]
- 57.Katsura I, Hendrix RW. Length determination in bacteriophage lambda tails. Cell. 1984;39:691–8. doi: 10.1016/0092-8674(84)90476-8. [DOI] [PubMed] [Google Scholar]
- 58.Thomason L, Court DL, Bubunenko M, Costantino N, Wilson H, Datta S, et al. Recombineering: genetic engineering in bacteria using homologous recombination. Curr Protoc Mol Biol 2007; Chapter 1:Unit 1 16. [DOI] [PubMed] [Google Scholar]
- 59.Thomason LC, Oppenheim AB, Court DL. Modifying bacteriophage lambda with recombineering. Methods Mol Biol. 2009;501:239–51. doi: 10.1007/978-1-60327-164-6_21. [DOI] [PubMed] [Google Scholar]
- 60.Oppenheim AB, Rattray AJ, Bubunenko M, Thomason LC, Court DL. In vivo recombineering of bacteriophage lambda by PCR fragments and single-strand oligonucleotides. Virology. 2004;319:185–9. doi: 10.1016/j.virol.2003.11.007. [DOI] [PubMed] [Google Scholar]
- 61.Moore SD. Assembling new Escherichia coli strains by transduction using phage P1. Methods Mol Biol. 2011;765:155–69. doi: 10.1007/978-1-61779-197-0_10. [DOI] [PubMed] [Google Scholar]
- 62.Thomason LC, Costantino N, Court DLE. coli genome manipulation by P1 transduction. Curr Protoc Mol Biol 2007; Chapter 1:Unit 1 17. [DOI] [PubMed] [Google Scholar]
- 63.Campbell A. The bacteriophage lambda. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press, 1971. [Google Scholar]
- 64.Cha RS, Zarbl H, Keohavong P, Thilly WG. Mismatch amplification mutation assay (MAMA): application to the c-H-ras gene. PCR Methods Appl. 1992;2:14–20. doi: 10.1101/gr.2.1.14. [DOI] [PubMed] [Google Scholar]
- 65.Jacobs WR, Jr., Tuckman M, Bloom BR. Introduction of foreign DNA into mycobacteria using a shuttle phasmid. Nature. 1987;327:532–5. doi: 10.1038/327532a0. [DOI] [PubMed] [Google Scholar]
- 66.Bardarov S, Kriakov J, Carriere C, Yu S, Vaamonde C, McAdam RA, et al. Conditionally replicating mycobacteriophages: a system for transposon delivery to Mycobacterium tuberculosis. Proc Natl Acad Sci USA. 1997;94:10961–6. doi: 10.1073/pnas.94.20.10961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Banaiee N, Bobadilla-del-Valle M, Riska PF, Bardarov S, Jr., Small PM, Ponce-de-Leon A, et al. Rapid identification and susceptibility testing of Mycobacterium tuberculosis from MGIT cultures with luciferase reporter mycobacteriophages. J Med Microbiol. 2003;52:557–61. doi: 10.1099/jmm.0.05149-0. [DOI] [PubMed] [Google Scholar]
- 68.Jacobs WR, Jr., Barletta RG, Udani R, Chan J, Kalkut G, Sosne G, et al. Rapid assessment of drug susceptibilities of Mycobacterium tuberculosis by means of luciferase reporter phages. Science. 1993;260:819–22. doi: 10.1126/science.8484123. [DOI] [PubMed] [Google Scholar]
- 69.Piuri M, Jacobs WR, Jr., Hatfull GF. Fluoromycobacteriophages for rapid, specific, and sensitive antibiotic susceptibility testing of Mycobacterium tuberculosis. PLoS ONE. 2009;4:e4870. doi: 10.1371/journal.pone.0004870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Bardarov S, Bardarov Jr S, Jr., Pavelka Jr MS, Jr., Sambandamurthy V, Larsen M, Tufariello J, et al. Specialized transduction: an efficient method for generating marked and unmarked targeted gene disruptions in Mycobacterium tuberculosis, M. bovis BCG and M. smegmatis. Microbiology. 2002;148:3007–17. doi: 10.1099/00221287-148-10-3007. [DOI] [PubMed] [Google Scholar]
- 71.Kim AI, Ghosh P, Aaron MA, Bibb LA, Jain S, Hatfull GF. Mycobacteriophage Bxb1 integrates into the Mycobacterium smegmatis groEL1 gene. Mol Microbiol. 2003;50:463–73. doi: 10.1046/j.1365-2958.2003.03723.x. [DOI] [PubMed] [Google Scholar]
- 72.Lee MH, Pascopella L, Jacobs WR, Jr., Hatfull GF. Site-specific integration of mycobacteriophage L5: integration- proficient vectors for Mycobacterium smegmatis, Mycobacterium tuberculosis, and bacille Calmette-Guerin. Proc Natl Acad Sci USA. 1991;88:3111–5. doi: 10.1073/pnas.88.8.3111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Sampson T, Broussard GW, Marinelli LJ, Jacobs-Sera D, Ray M, Ko CC, et al. Mycobacteriophages BPs, Angel and Halo: comparative genomics reveals a novel class of ultra-small mobile genetic elements. Microbiology. 2009;155:2962–77. doi: 10.1099/mic.0.030486-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Jacobs WR, Jr., Kalpana GV, Cirillo JD, Pascopella L, Snapper SB, Udani RA, et al. Genetic systems for mycobacteria. Methods Enzymol. 1991;204:537–55. doi: 10.1016/0076-6879(91)04027-L. [DOI] [PubMed] [Google Scholar]
- 75.Kalpana GV, Bloom BR, Jacobs WR., Jr Insertional mutagenesis and illegitimate recombination in mycobacteria. Proc Natl Acad Sci USA. 1991;88:5433–7. doi: 10.1073/pnas.88.12.5433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Payne K, Sun Q, Sacchettini J, Hatfull GF. Mycobacteriophage Lysin B is a novel mycolylarabinogalactan esterase. Mol Microbiol. 2009;73:367–81. doi: 10.1111/j.1365-2958.2009.06775.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Catalão MJ, Gil F, Moniz-Pereira J, Pimentel M. The mycobacteriophage Ms6 encodes a chaperone-like protein involved in the endolysin delivery to the peptidoglycan. Mol Microbiol. 2010;77:672–86. doi: 10.1111/j.1365-2958.2010.07239.x. [DOI] [PubMed] [Google Scholar]
- 78.Catalão MJ, Milho C, Gil F, Moniz-Pereira J, Pimentel M. A second endolysin gene is fully embedded in-frame with the lysA gene of mycobacteriophage Ms6. PLoS ONE. 2011;6:e20515. doi: 10.1371/journal.pone.0020515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Ghosh P, Wasil LR, Hatfull GF. Control of Phage Bxb1 Excision by a Novel Recombination Directionality Factor. PLoS Biol. 2006;4:e186. doi: 10.1371/journal.pbio.0040186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Hatfull GF. Mycobacteriophages: genes and genomes. Annu Rev Microbiol. 2010;64:331–56. doi: 10.1146/annurev.micro.112408.134233. [DOI] [PubMed] [Google Scholar]