Abstract
RNA-directed DNA methylation (RdDM) is an important de novo DNA methylation pathway in plants. RdDM mediates the transcriptional silencing of many endogenous genomic loci, most of which are transposon related. A forward genetics screen identified DTF1 (DNA-binding transcription factor 1) as a new component for RdDM in Arabidopsis. Loss-of-function mutations in DTF1 release the transcriptional silencing of RdDM target loci and reduce the accumulation of 24-nt small interfering RNAs (siRNAs) from some of the targets. Interestingly, in the dtf1 mutant plants, the release of transcriptional gene silencing at solo-LTR is accompanied by decreased siRNA accumulation but not by reduced DNA methylation. These results suggest that DTF1 is an atypical component of RdDM and has both DNA methylation-dependent and -independent roles in transcriptional gene silencing. We suggest that besides DNA methylation, siRNAs may cause some other uncharacterized epigenetic modifications that lead to transcriptional gene silencing.
Keywords: DTF1, siRNA, DNA methylation, transcriptional gene silencing, RdDM
Introduction
The RNA-directed DNA methylation (RdDM) and transcriptional gene-silencing pathway has been well characterized in plants 1, 2, 3, 4, 5. RdDM has been shown to be required for various epigenetic processes, which include transgene silencing at the transcriptional level, genomic imprinting and suppression of retrotransposition of transposable elements 6, 7, 8, 9, 10. 24-nt siRNAs and long noncoding RNAs are required for RdDM at target sequences 3, 6, 11. The plant-specific DNA-dependent RNA polymerase IV is presumably responsible for synthesizing the precursors of siRNAs 3, 12. The precursors are converted into double-stranded RNAs by the RNA-dependent RNA polymerase RDR2 1, 13. The double-stranded RNA is cleaved by Dicer-like 3 into 24-nt siRNAs, which are loaded into ARGONAUTE 4 (AGO4) 13, 14. Another DNA-dependent RNA polymerase, Pol V, is required for producing non-coding, presumably scaffold RNAs from RdDM targets 11. The chromatin-remodeling protein DRD1, structural-maintenance-of-chromosomes hinge domain-containing protein DMS3 and the novel plant-specific protein RDM1 form a tight protein complex (DDR), which is required for Pol V-dependent transcripts 15, 16, 17, 18. RDM1 is also associated with AGO4 and the DNA methytransferase DRM2 18. The largest subunit of Pol V, NRPE1 and the SPT5-like protein KTF1 have WG/GW repeats in their C-terminal regions, which bind AGO4 and the Pol V-dependent scaffold RNAs, and help to form RdDM effector complexes 19, 20. The yeast IWR1-like protein RDM4/DMS4 is a transcription factor that interacts with both Pol V and Pol II. The rdm4 mutation affects Pol V-dependent transcripts and plant development 21, 22. DRM2 and the recently characterized DRM3 are the de novo DNA methytransferases in the Arabidopsis RdDM pathway 23, 24, 25, 26.
siRNA-directed chromatin modification and TGS also occur in unicellular eukaryotes, such as the fission yeast 27. Several key components of plant RdDM appear to have counterparts in fission yeast, including Argonaute AGO1, Dicer DCR1 and RNA-dependent RNA polymerase RDP1 28. Moreover, like the Arabidopsis WG/GW motif-containing protein KTF1, fission yeast has a WG/GW-motif-containing protein TAS3, which is also required for interaction with Argonaute 29, 30. The fission yeast AGO1, TAS3 and the chromodomain protein CHP1 form an RNA-induced transcriptional silencing (RITS) complex, which is essential for heterochromatin formation at pericentromeric repeats 29. However, there is no DNA methylation in fission yeast, instead, H3K9 dimethylation catalyzed by the H3K9 methytransferase CLR4 is required for siRNA-directed repressive heterochromatin formation in fission yeast 27, 31. In plants, H3K9 dimethylation is catalyzed by histone H3K9 methytransferases such as KYP/SUVH4 32. KYP forms a self-enhancing loop with the DNA methytransferase CMT3, which catalyzes DNA methylation at CHG sites 33, 34.
At some RdDM-target loci, the transcriptional gene silencing depends on MOM1 35, 36. In mom1 mutants, although the silencing of transgenes and endogenous genomic target loci is released, neither DNA methylation nor histone modification is altered at these loci 35, 36, 37. MOM1 seems to act downstream of RdDM, although further studies are required to understand how MOM1 mediates transcriptional gene silencing independently of DNA methylation at RdDM-target loci. Interestingly, a recent study reported that MOM1 is required to prevent aberrant RNA transcriptional read-through 38. The minimal functional domain of MOM1, CMM2, forms a coiled-coil structure, which likely interacts with other proteins 39, 40.
ROS1 encodes a 5-methylcytosine DNA glycosylase, which functions as a DNA demethylase by initiating a base excision repair pathway for active DNA demethylation 41, 42, 43, 44. ros1 mutations cause transcriptional silencing of the transgenic RD29A promoter-driven luciferase reporter gene (RD29A-LUC) as well as the endogenous RD29A gene 41. By screening for second site suppressors of ros1 mutant plants, we have identified many RdDM components 45. Our results showed that the transcriptional silencing of the RD29A-LUC transgene in ros1 is dependent on RdDM.
In this study, we identified a novel component of RdDM, DTF1, by screening for suppressors of ros1. dtf1 mutations release the silencing of RD29A-LUC transgene as well as the endogenous RD29A in ros1. Moreover, DTF1 is required for silencing of endogenous transposable elements that are targeted by RdDM. The dtf1 mutations cause reduced siRNA accumulation and DNA methylation at some of the RdDM targets. Surprisingly, the role of DTF1 in transcriptional silencing at some RdDM targets is not dependent on DNA methylation. These results suggest that DTF1 is an atypical RdDM component, and functions in transcriptional silencing through both DNA methylation-dependent and -independent pathways.
Results
A putative DNA-binding transcription factor, DTF1, is required for transcriptional silencing of the RD29A-LUC transgene
Our previous results showed that the RD29A promoter-driven luciferase transgene is expressed in the wild-type C24 plants under stress conditions, whereas loss-of-function mutations in ROS1 result in transcriptional silencing of the RD29A-LUC transgene as well as the corresponding endogenous RD29A 41. RdDM is required for transcriptional silencing of transgenic RD29A-LUC and endogenous RD29A in ros1 45. Nearly all of the canonical RdDM components have been identified by screening for suppressors of ros1 in our previous 45 and current studies (unpublished data). These results suggest that ROS1-mediated DNA demethylation and RdDM function antagonistically in the control of DNA methylation and transcriptional gene silencing.
Our previous ros1 suppressor screens were carried out using an Agrobacteria-transformed population of ros1 45, 46. In order to identify new components of the RdDM pathway, we constructed an EMS-mutagenized library in the ros1 mutant background that has the silenced RD29A-LUC transgene. We found most of the known RdDM components by screening for ros1 suppressors from the library (unpublished data). In addition, we recovered a new mutant (referred to as dtf1) where the silencing of the RD29A-LUC transgene in the ros1 background is released.
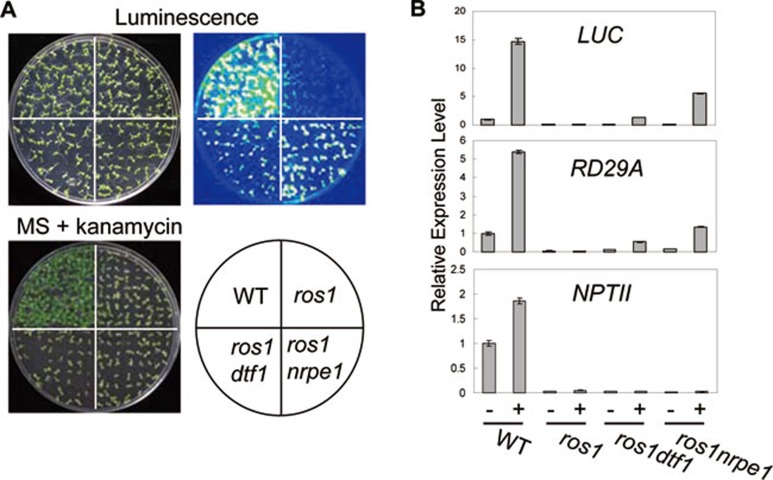
The expression of RD29A-LUC transgene and 35S promoter-driven NPTII (35S-NPTII) gene is indicated by bioluminescence intensity and kanamycin resistance, respectively 41, 46. In the wild-type C24, RD29A-LUC was highly expressed under stress conditions, and strong bioluminescence was emitted (Figure 1A). The 35S-NPTII transgene was normally expressed in the wild type, which is resistant to kanamycin (Figure 1A and 1B). ros1 mutations cause silencing of RD29A-LUC, corresponding endogenous RD29A and 35S-NPTII (Figure 1B) 41. In the ros1dtf1-1 double mutant, the silencing of RD29A-LUC and corresponding endogenous RD29A was partially released, whereas the silencing was released in ros1nrpe1 to a greater extent (Figure 1A and 1B). However, neither the dtf1-1 nor nrpe1 mutation suppressed silencing of the 35S-NPTII transgene, which is known to be independent of RdDM (Figure 1B) 45. The results suggest that DTF1 play an important role in transgene silencing in a manner similar to the canonical RdDM component NRPE1.
Figure 1.
The dtf1 mutation releases the silencing of RD29A-LUC and endogenous RD29A. (A) The dtf1 mutation releases the silencing of RD29A-LUC. The transgenic wild-type, ros1, ros1dtf1-1 and ros1nrpe1 were grown on MS medium for 2 weeks followed by treatment at 4 °C for 2 days. The treated plants were sprayed with luciferin for luminescence imaging. The same plants were grown on MS medium supplemented with 100 μg/ml kanamycin for 3 weeks and photographed. (B) Effect of dtf1 on expression of transgenic RD29A-LUC, endogenous RD29A and 35S-NPTII, as detected by real-time PCR. TUB4 was used as an internal control for normalization.
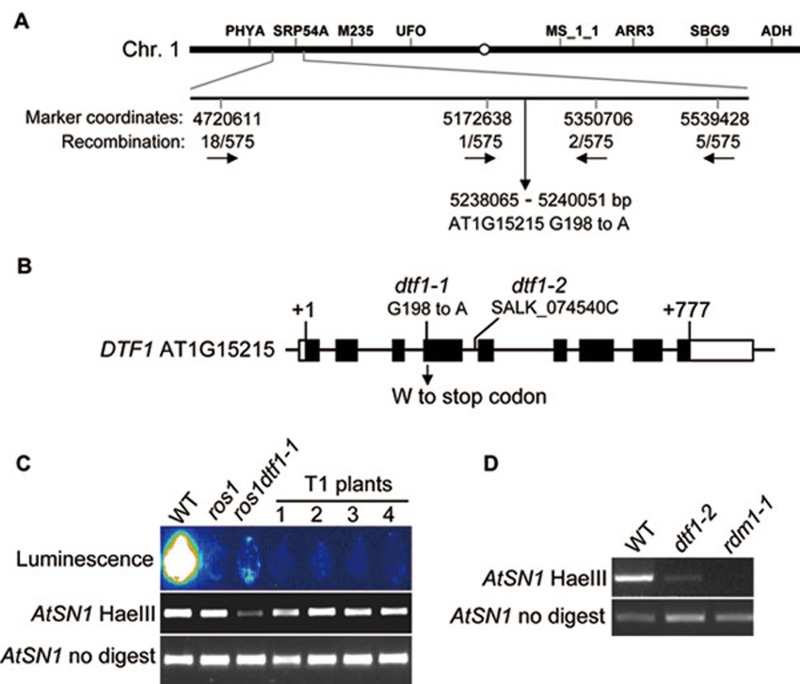
The ros1dtf1-1 mutant was crossed to a homozygous ros1 mutant (Salk_045303) in the Col-0 background, and the resulting F2 population was used for map-based cloning. The dtf1 mutation was mapped to a ∼180 kb region on chromosome 1 (Figure 2A). Next-generation high-throughput DNA sequencing was carried out to sequence the whole genome of the mutant. In the ∼180 kb mapping interval, we found a G to A mutation in the fourth exon of At1G15215, which creates a premature stop codon (Figure 2B). The mutation was confirmed by Sanger sequencing. The results suggest that At1g15215 might be the DTF1 gene. At1g15215 encodes a putative sequence-specific DNA-binding transcription factor (hence the name DTF1, for DNA-binding transcription factor 1).
Figure 2.
Map-based cloning and characterization of DTF1. (A) Map-based cloning of DTF1. The gene was located to the indicated ∼180 kb region of chromosome 1 by genetic mapping. A G-to-A mutation in the fourth exon of At1G15215 creates a premature stop codon. (B) Diagram of DTF1 and the dtf1-1 and dtf1-2 mutations. The diagram shows the DTF1 gene with the indicated positions of exons (boxes), introns (line) and open reading frame (solid boxes). The dtf1-1 and dtf1-2 mutations are labeled. (C) Complementation assay in ros1dtf1-1. The leaves from wild type, ros1, ros1dtf1-1, and four individual DTF1 transgenic T1 plants in ros1dtf1-1 were collected for luminescence imaging after treatment with 2% NaCl for 4 hours. The AtSN1 methylation was tested by chop-PCR. (D) The dtf1-2 mutation reduces AtSN1 DNA methylation. Genomic DNA from the indicated genotypes was digested with HaeIII, and used for amplification of AtSN1. The rdm1 mutant was used as a control.
To confirm that the mutation in At1g15215 is responsible for the silencing defect in ros1dtf1-1, the genomic fragment of At1g15215 was cloned from wild-type plants and transformed into ros1dtf1-1 for complementation test. Compared to ros1dtf1-1, the transgenic plants emitted much less luminescence (Figure 2C), suggesting that the silencing of RD29A-LUC transgene is restored by the wild-type transgene. In addition, we tested and found that AtSN1 methylation in ros1dtf1 was low, but the methylation in the transgenic plants was restored (Figure 2C). Furthermore, we obtained a homozygous T-DNA insertion mutant (Salk_074540C), and found that the T-DNA mutant also showed a reduction in DNA methylation at AtSN1 (Figure 2D). These results demonstrate that At1G15215 is DTF1, and is required for the silencing of RD29A-LUC transgene.
DTF1 encodes a putative DNA-binding transcription factor that contains a homeodomain in its N-terminal region. The homeodomain is known as the DNA-binding domain in some transcription factors. In addition to DTF1, there is a DTF1-like protein, At1G18380, encoded in the Arabidopsis genome. DTF1 and the DTF1-like protein are highly similar, except that the DTF1-like protein has a C-terminal extension (Supplementary information, Figure S1A). However, T-DNA insertion mutations in At1g18380 have no apparent effect on DNA methylation and transcriptional gene silencing (data not shown), suggesting that At1G18380 may have functions other than in transcriptional gene silencing.
DTF1 encodes a plant-specific gene, which is conserved in angiosperms, gymnosperms and bryophytes (Supplementary information, Figure S1B and S1C), but not in fungi and animals. DTF1 is conserved in the N-terminal homeodomain, which may function in DNA binding. In addition, the uncharacterized domain in the C-terminal half is also highly conserved.
DTF1 is required for the silencing of some endogenous RdDM targets and accumulation of 24-nt siRNAs
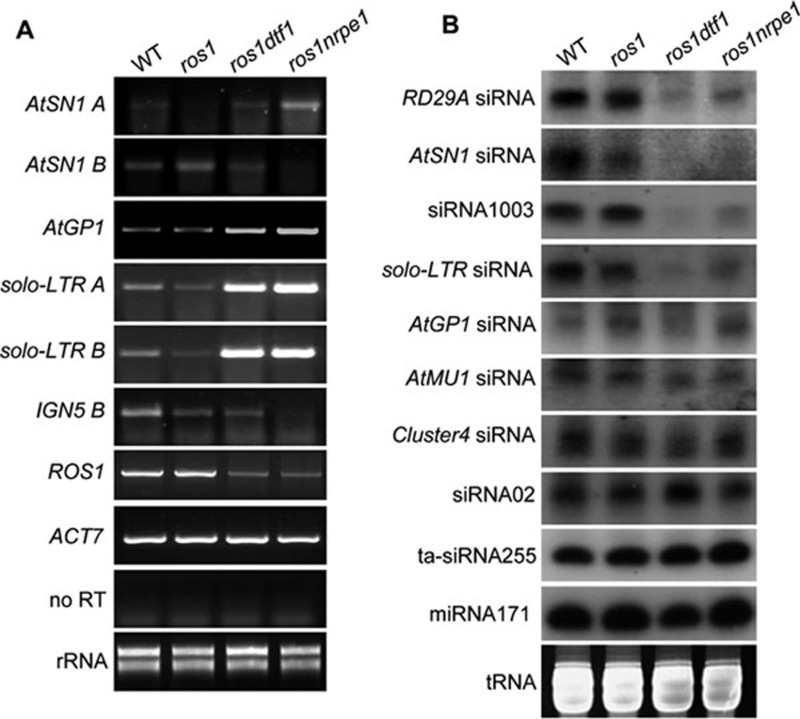
The above results showed that DTF1 is required for transgene silencing. Using semi-quantitative RT-PCR, we tested the effect of the dtf1-1 mutation on the silencing of endogenous RdDM targets. The results show that the dtf1-1 mutation released silencing of a subset of RdDM targets (Figure 3A). solo-LTR and AtGP1 were previously identified as RdDM targets, which show a low level of expression in wild-type and ros1 plants 45, 47. The dtf1-1 mutation released the silencing of solo-LTR and AtGP1, while it weakly released the silencing of AtSN1A (Figure 3A). As a control, the nrpe1 mutation caused more substantial expression of AtSN1 A. The results suggest that DTF1 is required for the silencing of a subset of RdDM targets.
Figure 3.
Effect of dtf1 mutation on transcriptional gene silencing and small RNA accumulation. (A) Effect of dtf1 on the transcript levels of endogenous RdDM targets was determined by semi-quantitative RT-PCR. ACT7 was used as an internal control. No RT amplification indicates that there was no DNA contamination in RNA samples. (B) Effect of dtf1 on accumulation of small RNAs. The levels of 24-nt siRNAs, 21-nt ta-siRNA255 and miRNA171 were tested in the indicated genotypes. The ethidium bromide-stained small RNA gel was used as a loading control.
Pol V-dependent noncoding RNA transcripts are required for RdDM and transcriptional gene silencing 11. We tested and found that the dtf1-1 mutation did not affect Pol V-dependent transcripts, while loss of the largest subunit of Pol V, NRPE1, blocked accumulation of the Pol V-dependent transcripts, including AtSN1 B and IGN5 B (Figure 3A). Previous studies showed that mutations in canonical RdDM components suppress ROS1 expression 21, 48, 49. We found that the dtf1-1 mutation also suppressed the expression of ROS1 (Figure 3A).
Accumulation of 24-nt siRNAs was detected in ros1dtf1-1 as well as in the wild type, ros1, and ros1nrpe1. 24-nt siRNAs generated from the transgenic RD29A promoter are required for RD29A-LUC silencing in ros1 45. RNA blot analysis showed that the level of RD29A promoter siRNAs was reduced in ros1dtf1-1 and ros1nrpe1 (Figure 3B). The result suggests that the role of DTF1 in RD29A-LUC trangene silencing is related to siRNA-directed DNA methylation and transcriptional silencing. The accumulation of 24-nt siRNAs was also reduced in ros1dtf1-1 at some endogenous RdDM targets, including AtSN1, solo-LTR and 5S rDNA, whereas the siRNAs from some other genomic loci (AtGP1, AtMU1, Cluster4, and siRNA02) were not affected by the dtf1 mutation (Figure 3B). The pattern of siRNA accumulation in ros1dtf1-1 was very similar to that in ros1nrpe1, although the dtf1 mutation affected siRNA accumulation to a greater extent than nrpe1 (Figure 3B). Neither trans-acting siRNA255 nor microRNA171 was affected in ros1dtf1-1 and ros1nrpe1 (Figure 3B), supporting that DTF1 specifically functions in 24-nt siRNA-directed transcriptional gene silencing.
DTF1 is required for DNA methylation at some endogenous RdDM targets
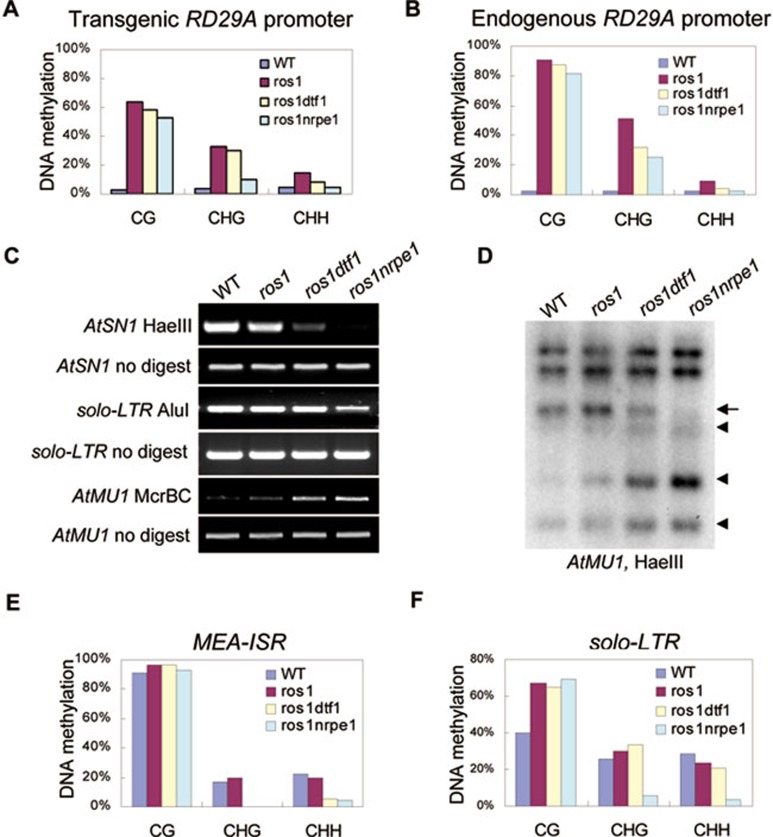
To test whether the suppression of transcriptional gene silencing in ros1dtf1-1 is correlated with a reduction in DNA methylation, we analyzed the DNA methylation status at the RD29A-LUC transgene, endogenous RD29A and several other endogenous RdDM targets. Consistent with our previous results 41, the DNA methylation level of both transgenic and endogenous RD29A promoters was very low in wild type, and the methylation in all cytosine contexts (CG, CHG, CHH) was substantially increased in ros1 (Figure 4A and 4B). The DNA methylation level was well correlated with the silencing of RD29A-LUC transgene and endogenous RD29A gene. We found that the RD29A promoter DNA methylation at non-CG sites was reduced in ros1dtf1-1, as in ros1nrpe1 (Figure 4A and 4B). The results further suggest that DTF1 functions in RdDM. Consistent with the weak reactivation of AtSN1 in dtf1 mutants (Figure 3A), the AtSN1 DNA methylation level was partially reduced by the dtf1 mutation (Figure 4C). The DNA methylation of the endogenous RdDM target AtMU1 was reduced in ros1dtf1-1 as well as in ros1nrpe1 (Figure 4C and 4D), although siRNA accumulation at the locus was not affected by dtf1 or nrpe1 (Figure 3B). These results suggest that like NRPE1, DTF1 functions downstream of siRNA generation to mediate DNA methylation and transcriptional gene silencing. As in ros1nrpe1, non-CG DNA methylation at MEA-ISR was reduced in ros1dtf1-1 (Figure 4E). Taken together, the results suggest that DTF1 is a new component of RdDM, and it functions in the DNA methylation and transcriptional silencing of a subset of RdDM targets, including AtMU1 and MEA-ISR.
Figure 4.
Effect of dtf1 on DNA methylation. (A, B) The effect of dtf1 on the promoter DNA methylation of RD29A-LUC (A) and endogenous RD29A (B) was determined by bisulfite sequencing. The percentage of methylated cytosine in different cytosine contexts is shown. (C) The DNA methylation level of AtSN1, solo-LTR and AtMU1 was determined by chop-PCR. The DNA methylation-sensitive restriction enzymes HaeIII and AluI, and DNA mehylation-insensitive enzyme McrBC were used for DNA methylation analysis as indicated. (D) The effect of dtf1 on AtMU1 DNA methylation was detected by Southern blot analysis. Genomic DNA from the indicated genotypes was digested by HaeIII followed by Southern hybridization. (E and F) The DNA methylation status of MEA-ISR and solo-LTR was determined by bisulfite sequencing in the indicated genotypes.
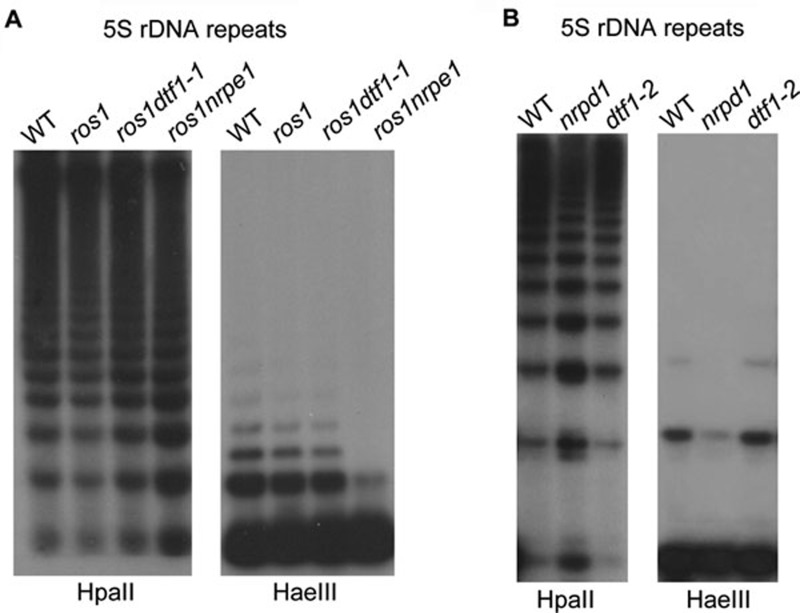
As the silencing of solo-LTR is substantially released in ros1dtf1-1, we tested whether the DNA methylation of solo-LTR is reduced in the mutant by bisulfite sequencing. The results show that the DNA methylation of solo-LTR was clearly reduced at CHG and CHH sites in ros1nrpe1, but not in ros1dtf1-1 (Figure 4F). This is in contrast to the silencing of solo-LTR, which was released to similar levels in ros1dtf1-1 and ros1nrpe1 (Figure 3A). Futhermore, the DNA methylation of 5S rDNA repeats was also not affected by the dtf1 mutations in ros1dtf1-1 as well as in dtf1-2, whereas the DNA methylation was significantly reduced in the canonical RdDM mutants nrpd1 and nrpe1 (Figure 5A and 5B). However, although the DNA methylation of both solo-LTR and 5S rDNA was not altered by dtf1, the accumulation of solo-LTR siRNAs and siRNA1003 (from 5S rDNA) was substantially reduced by dtf1. These results suggest that the function of DTF1 in siRNA-mediated transcriptional gene silencing at solo-LTR and 5S rDNA is independent of DNA methylation.
Figure 5.
Effect of dtf1 mutations on 5S rDNA methylation. Genomic DNA from the indicated genotypes was digested with HpaII (for CG and CHG methylation) and HaeIII (for CHH methylation), and used for Southern blot analysis. The effect of dtf1 mutations on the DNA methylation of 5S rDNA repeats was tested in ros1dtf1-1 (A) and dtf1-2 (B), respectively.
Discussion
In the canonical RdDM pathway, all previously characterized RdDM components are required for both the establishment of DNA methylation and for transcriptional gene silencing at target loci. solo-LTR is one of the RdDM targets, and was first discovered by the Matzke group 47. In nrpd1, nrpe1, rdr2 and drd1, the DNA methylation of solo-LTR is substantially reduced, resulting in the release of transcriptional gene silencing at solo-LTR in these mutants 11, 47, 50. However, different from the canonical RdDM components, we found that DTF1 is dispensable for DNA methylation at solo-LTR, although it is required for siRNA accumulation and transcriptional gene silencing at this locus. It is worth noticing that solo-LTR is a rather unusual RdDM target in that the repressive histone mark H3K27 monomethylation (H3K27me1) is substantially reduced at solo-LTR by mutations of the canonical RdDM components, whereas the mark is not changed in the mutants at the other tested RdDM targets 47. The dependence of H3K27me1 on RdDM at solo-LTR may reflect a special chromatin structure or DNA sequence specificity of the locus, which may be absent at the other tested RdDM target loci. The recently characterized Arabidopsis H3K27 monmethyltransferases, ATXR5 and ATXR6, are involved in transcriptional silencing of transposable elements and other repetitive DNA sequences 51. The silencing status of some RdDM targets such as solo-LTR may require both DNA methylaion and H3K27me1 catalyzed by ATXR5/ATXR6. In this study, we found that the dtf1 mutation suppresses silencing of solo-LTR without affecting DNA methylation, suggesting that at this locus DTF1 may function downstream of DNA methylation. As a homeodomain-containing transcription factor, DTF1 may recognize the special chromatin structure or DNA sequence of solo-LTR to help siRNAs guide H3K27 monomethylation and transcriptional silencing. At the other RdDM targets tested, DTF1 is required for both DNA methylation and silencing, just like the canonical RdDM components. One possibility is that DTF1 may be involved in siRNA-directed heterochromatic histone modifications, such as H3K9 methylation and H3K27 methylation, which may occur upstream (e.g., H3K9 methylation) or downstream (e.g., H3K27 methylation) of DNA methylation in the RdDM pathway. Regardless of the exact mechanism of function, our results suggest that DTF1 is an atypical RdDM component that mediates transcriptional gene silencing in both DNA methylation-dependent and -independent manners.
Our small RNA blot analysis showed that DTF1 is required for accumulation of 24-nt siRNAs at some RdDM targets but has no effect on the others. The siRNA pattern in ros1dtf1-1 appears the same as that in ros1nrpe1, suggesting that like Pol V, DTF1 may not be involved in the initial biogenesis of siRNAs, but may affect the accumulation of some siRNAs indirectly by influencing the heterochromatic marks of the loci. It is interesting that dtf1 affects siRNA accumulation to a greater extent than nrpe1, although the effect of dtf1 on DNA methylation is less or even undetectable. The disparaging effects of the two mutations (dtf1 and nrpe1) on siRNA accumulation and DNA methylation hint at unknown complexities and mechanisms of RdDM. During the preparation of this manuscript, Law et al. 52 reported that SHH1 is required for DNA methylation, and associates with Pol IV based on affinity purification of NRPD1. SHH1, which is the same protein as DTF1, was suggested to function in siRNA biogenesis as a transcription factor for Pol IV. In the Law et al. study, RDR2, CLSY1 and RDM4 were also shown to be associated with NRPD1. RDR2 is abundant in the affinity-purified NRPD1-containing complex, and likely functions together with Pol IV to produce siRNAs. The levels of CLSY1, RDM4 and SHH1 in the purified NRPD1-containing complex were very low 52. Moreover, RDM4 is known to be a Pol V-interacting protein required for the generation of Pol V-dependent transcripts 21. These observations suggest that CLSY1, RDM4 and SHH1 are unlikely to be core components of the NRPD1-containing complex. Our results show clearly that unlike Pol IV, DTF1 is only required for the accumulation of siRNAs at a subset of RdDM targets. Importantly, we discovered that dtf1 reduces siRNA accumulation and releases silencing at solo-LTR without affecting DNA methylation. In addition, we found that dtf1 reduces siRNA accumulation from 5S rDNA repeats but has no apparent effect on DNA methylation of the rDNA. These surprising observations suggest that besides causing de novo DNA methylation, siRNAs may also guide the establishment of other silencing epigenetic marks such as heterochromatic histone modifications. DTF1 may be involved in this later function of siRNAs in the RdDM pathway. Previous studies have found that some Pol IV-dependent 24-nt siRNAs do not cause DNA methylation 53. It would be of great interest to determine the DNA methylation-independent chromatin functions of the siRNAs and the role of DTF1 in these functions.
Materials and Methods
Plant materials, mutant screening and cloning
The wild-type C24 and ros1 mutant carry a homozygous RD29A promoter-driven luciferase transgene 54. The ros1 mutant with RD29-LUC transgene was mutagenized with ethyl methanesulfonate. Suppressors of ros1 were screened by luminescence imaging after the mutagenized M2 plants were cold-treated for 2 days at 4 °C. The selected plants were transplanted into soil, and the luminescence phenotype was confirmed in adult plants as previously described 45. Mutants of interest were crossed to the ros1 mutant in the Col-0 backcrossed for map-based cloning. High-throughput sequencing (Illumina) was carried out to detect the mutations in the mapped genomic interval. For complementation assay, the full-length DTF1 genomic sequence was amplified, and cloned into the binary vector pCAMBIA1300. The DTF1 construct was transformed into ros1dtf1-1 by the Agrobacterium strain GV3101. The transgenic T1 plants were tested for phenotypic complementation.
Analysis of RNA transcript level and small RNA accumulation
Total RNA was isolated from 2-week-old seedlings on MS plates as described previously 45. For RT-PCR, the sequences of primers are listed in Supplementary information, Table S1. After contaminating DNA was removed by DNase, total RNA was used for RT-PCR. For RT-PCR of protein-encoding genes, oligo-dT was used as the reverse primer to synthesize the first-stranded cDNA. RT-PCR of transposable elements and other DNA repeat sequences was carried out by a previously described one-step RT-PCR protocol 11. Briefly, the PCR condition is 95 °C for 5 min followed by 28-40 amplification cycles (95 °C for 30 s, 56 °C for 30 s, 72 °C for 1 min). The constitutively expressed ACT7 was used as an internal control. For small RNA analysis, small RNA was extracted as described previously 45, and was separated on a 15% polyacrylamide gel at 200 V for 3 h. The small RNA was electrotransfered to Hybond-N+ membranes (Amersham). The probes for small RNA hybridization are [γ-32P]ATP-labeled DNA oligonucleotides or [α-32P]dCTP-labeled amplified DNA. The small RNA hybridization was carried out in PerfectHyb buffer (Sigma) at 38 °C. The DNA primer and probe sequences are listed in Supplementary information, Table S1.
DNA methylation assay
The DNA methylation level was detected by chop-PCR, Southern blot analysis, or bisulfite sequencing. For chop-PCR, the genomic DNA was digested with DNA methylation-sensitive (HaeIII, AluI, and MspI) and -insensitive (McrBC) restriction enzymes, and used for amplification of target DNA sequences. For Southern blot analysis, 5 μg of genomic DNA was digested with indicated DNA methylation-sensitive enzymes, and resolved on 1.2% agarose gel at 40 V overnight, followed by a routine Southern blotting method. For bisulfite sequencing, 2 μg of genomic DNA was converted and purified with EpiTect Bisulfite Kit (Qiagen). The treated DNA was used for amplification and cloning. At least 10 individual clones were sequenced for each sample. The primers are described in Supplementary information, Table S1.
Acknowledgments
We thank the National Institute of Biological Sciences sequencing center for its excellent technical support on high-throughput DNA sequencing and analysis. This work is supported by a 973 program grant from the Chinese Ministry of Science and Technology (2011CB812600 to X-J H) and US National Institutes of Health grant (R01GM070795 to J-K Z).
Footnotes
(Supplementary information is linked to the online version of the paper on the Cell Research website.)
Supplementary Material
Sequence analysis of DTF1.
List of DNA oligos used in this study.
References
- Xie Z, Johansen L K, Gustafson A M, et al. Genetic and functional diversification of small RNA pathways in plants. PLoS Biol. 2004;2:E104. doi: 10.1371/journal.pbio.0020104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanno T, Huettel B, Mette MF, et al. Atypical RNA polymerase subunits required for RNA-directed DNA methylation. Nat Genet. 2005;37:761–765. doi: 10.1038/ng1580. [DOI] [PubMed] [Google Scholar]
- Herr AJ, Jensen MB, Dalmay T, Baulcombe DC. RNA polymerase IV directs silencing of endogenous DNA. Science. 2005;308:118–120. doi: 10.1126/science.1106910. [DOI] [PubMed] [Google Scholar]
- Law JA, Jacobsen SE. Establishing, maintaining and modifying DNA methylation patterns in plants and animals. Nature Rev Genet. 2010;11:204–220. doi: 10.1038/nrg2719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He XJ, Chen T, Zhu JK. Regulation of and function DNA methylation in plants and animals. Cell Res. 2011;21:442–465. doi: 10.1038/cr.2011.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matzke MA, Birchler JA. RNAi-mediated pathways in the nucleus. Nat Rev Genet. 2005;6:24–35. doi: 10.1038/nrg1500. [DOI] [PubMed] [Google Scholar]
- Mosher RA, Melnyk CW, Kelly KA, et al. Uniparental expression of PolIV-dependent siRNAs in developing endosperm of Arabidopsis. Nature. 2009;460:283–286. doi: 10.1038/nature08084. [DOI] [PubMed] [Google Scholar]
- Hsieh TF, Ibarra CA, Silva P, et al. Genome-wide demethylation of Arabidopsis endosperm. Science. 2009;324:1451–1454. doi: 10.1126/science.1172417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gehring M, Bubb KL, Henikoff S. Extensive demethylation of repetitive elements during seed development underlies gene imprinting. Science. 2009;324:1447–1451. doi: 10.1126/science.1171609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito H, Gaubert H, Bucher E, et al. An siRNA pathway prevents transgenerational retrotransposition in plants subjected to stress. Nature. 2011;472:115–119. doi: 10.1038/nature09861. [DOI] [PubMed] [Google Scholar]
- Wierzbicki AT, Haag JR, Pikaard CS. Non-coding transcription by RNA polymerase Pol IVb/Pol V mediates transcriptional silencing of overlapping and adjacent genes. Cell. 2008;135:635–648. doi: 10.1016/j.cell.2008.09.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pontier D, Yahubyan G, Vega D, et al. Reinforcement of silencing at transposons and highly repeated sequences requires the concerted action of two distinct RNA polymerases IV in Arabidopsis. Genes Dev. 2005;19:2030–2040. doi: 10.1101/gad.348405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pontes O, Li C F, Nunes PC, et al. The Arabidopsis chromatin-modifying nuclear siRNA pathway involves a nucleolar RNA processing center. Cell. 2006;126:79–92. doi: 10.1016/j.cell.2006.05.031. [DOI] [PubMed] [Google Scholar]
- Li CF, Pontes O, El-Shami M, et al. An ARGONAUTE4-containing nuclear processing center colocalized with Cajal bodies in Arabidopsis thaliana. Cell. 2006;126:93–106. doi: 10.1016/j.cell.2006.05.032. [DOI] [PubMed] [Google Scholar]
- Kanno T, Mette MF, Kreil DP, et al. Involvement of putative SNF2 chromatin remodeling protein DRD1 in RNA-directed DNA methylation. Curr Biol. 2004;14:801–805. doi: 10.1016/j.cub.2004.04.037. [DOI] [PubMed] [Google Scholar]
- Kanno T, Bucher E, Daxinger L, et al. A structural-maintenance-of chromosomes hinge domain-containing protein is required for RNA-directed DNA methylation. Nat Genet. 2008;40:670–675. doi: 10.1038/ng.119. [DOI] [PubMed] [Google Scholar]
- Law JA, Ausin I, Johnson LM, et al. A protein complex required for polymerase V transcripts and RNA-directed DNA methylation in Arabidopsis. Curr Biol. 2010;20:951–956. doi: 10.1016/j.cub.2010.03.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao Z, Liu HL, Daxinger L, et al. An RNA polymerase II- and AGO4-associated protein acts in RNA-directed DNA methylation. Nature. 2010;465:106–109. doi: 10.1038/nature09025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He XJ, Hsu YF, Zhu S, et al. An effector of RNA-directed DNA methylation in Arabidopsis is an ARGONAUTE 4- and RNA-binding protein. Cell. 2009;137:498–508. doi: 10.1016/j.cell.2009.04.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bies-Etheve N, Pontier D, Lahmy S, et al. RNA-directed DNA methylation requires an AGO4-interacting member of the SPT5 elongation factor family. EMBO Rep. 2009;10:649–654. doi: 10.1038/embor.2009.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He XJ, Hsu YF, Zhu S, et al. A conserved transcriptional regulator is required for RNA-directed DNA methylation and plant development. Genes Dev. 2009;23:2717–2722. doi: 10.1101/gad.1851809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanno T, Bucher E, Daxinger L, et al. RNA-directed DNA methylation and plant development require an IWR1-type transcription factor. EMBO Rep. 2010;11:65–71. doi: 10.1038/embor.2009.246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao X, Jacobsen SE. Role of the Arabidopsis DRM methyltransferases in de novo DNA methylation and gene silencing. Curr Biol. 2002;12:1138–1144. doi: 10.1016/s0960-9822(02)00925-9. [DOI] [PubMed] [Google Scholar]
- Naumann U, Daxinger L, Kanno T, et al. Genetic evidence that DNA methyltransferase DRM2 has a direct catalytic role in RNA-directed DNA methylation in Arabidopsis thaliana. Genetics. 2011;187:977–979. doi: 10.1534/genetics.110.125401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenberg MV, Ausin I, Chan SW, et al. Identification of genes required for de novo DNA methylation in Arabidopsis. Epigenetics. 2011;6:344–354. doi: 10.4161/epi.6.3.14242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henderson IR, Deleris A, Wong W, et al. The de novo cytosine methyltransferase DRM2 requires intact UBA domains and a catalytically mutated paralog DRM3 during RNA-directed DNA methylation in Arabidopsis thaliana. PLoS Genet. 2010;6:e1001182. doi: 10.1371/journal.pgen.1001182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moazed D. Small RNAs in transcriptional gene silencing and genome defence. Nature. 2009;457:413–420. doi: 10.1038/nature07756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Motamedi MR, Verdel A, Colmenares SU, et al. Two RNAi complexes, RITS and RDRC, physically interact and localize to noncoding centromeric RNAs. Cell. 2004;119:789–802. doi: 10.1016/j.cell.2004.11.034. [DOI] [PubMed] [Google Scholar]
- Verdel A, Jia S, Gerber S, et al. RNAi-mediated targeting of heterochromatin by the RITS complex. Science. 2004;303:672–676. doi: 10.1126/science.1093686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Debeauchamp JL, Moses A, Noffsinger VJ, et al. Chp1-Tas3 interaction is required to recruit RITS to fission yeast centromeres and for maintenance of centromeric heterochromatin. Mol Cell Biol. 2008;28:2154–2166. doi: 10.1128/MCB.01637-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakayama J, Rice JC, Strahl BD, Allis CD, Grewal SI. Role of histone H3 lysine 9 methylation in epigenetic control of heterochromatin assembly. Science. 2001;292:110–113. doi: 10.1126/science.1060118. [DOI] [PubMed] [Google Scholar]
- Cao X, Jacobsen SE. Control of CpNpG DNA methylation by the KRYPTONITE histone H3 methyltransferase. Nature. 2002;416:556–560. doi: 10.1038/nature731. [DOI] [PubMed] [Google Scholar]
- Lindroth AM, Shultis D, Jasencakova Z, et al. Dual histone H3 methylation marks at lysines 9 and 27 required for interaction with CHROMOMETHYLASE3. EMBO J. 2004;23:4286–4296. doi: 10.1038/sj.emboj.7600430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson LM, Bostick M, Zhang X, et al. The SRA methyl-cytosine-binding domain links DNA and histone methylation. Curr Biol. 2007;17:379–384. doi: 10.1016/j.cub.2007.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokthongwattana C, Bucher E, Caikovski M, et al. MOM1 and Pol-IV/V interactions regulate the intensity and specificity of transcriptional gene silencing. EMBO J. 2010;29:340–351. doi: 10.1038/emboj.2009.328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Numa H, Kim JM, Matsui A, et al. Transduction of RNA-directed DNA methylation signals to repressive histone marks in Arabidopsis thaliana. EMBO J. 2010;29:352–362. doi: 10.1038/emboj.2009.374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amedeo P, Habu Y, Afsar K, Mittelsten Scheid O, Paszkowski J. Disruption of the plant gene MOM releases transcriptional silencing of methylated genes. Nature. 2000;405:203–206. doi: 10.1038/35012108. [DOI] [PubMed] [Google Scholar]
- Zhou Y, Zhang J, Lin H, Guo G, Guo Y. MORPHEUS' MOLECULE1 is required to prevent aberrant RNA transcriptional read-through in Arabidopsis. Plant Physiol. 2010;154:1272–1280. doi: 10.1104/pp.110.162131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caikovski M, Yokthongwattana C, Habu Y, et al. Divergent evolution of CHD3 proteins resulted in MOM1 refining epigenetic control in vascular plants. PLoS Genet. 2008;4:e1000165. doi: 10.1371/journal.pgen.1000165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petty TJ, Nishimura T, Emamzadah S, et al. Expression, crystallization and preliminary X-ray diffraction analysis of the CMM2 region of the Arabidopsis thaliana Morpheus' molecule 1 protein. Acta Crystallogr Sect F Struct Biol Cryst Commun. 2010;66:916–918. doi: 10.1107/S1744309110021068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong Z, Morales-Ruiz T, Ariza RR, et al. ROS1, a repressor of transcriptional gene silencing in Arabidopsis, encodes a DNA glycosylase/lyase. Cell. 2002;111:803–814. doi: 10.1016/s0092-8674(02)01133-9. [DOI] [PubMed] [Google Scholar]
- Agius F, Kapoor A, Zhu JK. Role of the Arabidopsis DNA glycosylase/lyase ROS1 in active DNA demethylation. Proc Natl Acad Sci USA. 2006;103:11796–11801. doi: 10.1073/pnas.0603563103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu J, Kapoor A, Sridhar VV, et al. The DNA glycosylase/lyase ROS1 functions in pruning DNA methylation patterns in Arabidopsis. Curr Biol. 2007;17:54–59. doi: 10.1016/j.cub.2006.10.059. [DOI] [PubMed] [Google Scholar]
- Penterman J, Zilberman D, Huh JH, et al. DNA demethylation in the Arabidopsis genome. Proc Natl Acad Sci USA. 2007;104:6752–6757. doi: 10.1073/pnas.0701861104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He XJ, Hsu YF, Pontes O, et al. NRPD4, a protein related to the RPB4 subunit of RNA polymerase II, is a component of RNA polymerases IV and V and is required for RNA-directed DNA methylation. Genes Dev. 2009;23:318–330. doi: 10.1101/gad.1765209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapoor A, Agarwal M, Andreucci A, et al. Mutations in a conserved replication protein suppress transcriptional gene silencing in a DNA-methylation-independent manner in Arabidopsis. Curr Biol. 2005;15:1912–1918. doi: 10.1016/j.cub.2005.09.013. [DOI] [PubMed] [Google Scholar]
- Huettel B, Kanno T, Daxinger L, et al. Endogenous targets of RNA-directed DNA methylation and Pol IV in Arabidopsis. EMBO J. 2006;25:2828–2836. doi: 10.1038/sj.emboj.7601150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathieu O, Reinders J, Caikovski M, Smathajitt C, Paszkowski J. Transgenerational stability of the Arabidopsis epigenome is coordinated by CG methylation. Cell. 2007;130:851–862. doi: 10.1016/j.cell.2007.07.007. [DOI] [PubMed] [Google Scholar]
- Penterman J, Uzawa R, Fischer RL. Genetic interactions between DNA demethylation and methylation in Arabidopsis. Plant physiol. 2007;145:1549–1557. doi: 10.1104/pp.107.107730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng B, Wang Z, Li S, et al. Intergenic transcription by RNA polymerase II coordinates Pol IV and Pol V in siRNA-directed transcriptional gene silencing in Arabidopsis. Genes Dev. 2009;23:2850–2860. doi: 10.1101/gad.1868009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacob Y, Feng S, LeBlanc CA, et al. ATXR5 and ATXR6 are H3K27 monomethyltransferases required for chromatin structure and gene silencing. Nat Struct Mol Biol. 2009;16:763–768. doi: 10.1038/nsmb.1611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Law JA, Vashisht AA, Wohlschlegel JA, Jacobsen SE. SHH1, a homeodomain protein required for DNA methylation, as well as RDR2, RDM4, and chromatin remodeling factors, associate with RNA polymerase IV. PLoS Genet. 2011;7:e1002195. doi: 10.1371/journal.pgen.1002195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mosher RA, Schwach F, Studholme D, Baulcombe DC. PolIVb influences RNA-directed DNA methylation independently of its role in siRNA biogenesis. Proc Natl Acad Sci USA. 2008;105:3145–3150. doi: 10.1073/pnas.0709632105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishitani M, Xiong L, Stevenson B, Zhu JK. Genetic analysis of osmotic and cold stress signal transductionin Arabidopsis: Interactions and convergence of abscisic acid-dependent and abscisic acid-independent pathways. Plant Cell. 1997;9:1935–1949. doi: 10.1105/tpc.9.11.1935. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Sequence analysis of DTF1.
List of DNA oligos used in this study.