The first entry in the λ-Int family album was an insightful sketch by Allan Campbell depicting a mechanism for the integration and excision of lambdoid phage chromosomes into and out of the chromosome of the host cell (1). Since that time the λ-Int family has grown to more than 100 site-specific recombinases whose biological roles also include the segregation of viral, plasmid, and cellular chromosomes, the control of gene expression, and the creation of novel gene combinations. From a distant perspective this family of pathways involves the alignment of two DNA target sites (e.g., att sites for λ and lox sites for the phage P1-encoded Cre targets), which then are cleaved, exchanged, and resealed as new DNA junctions. Depending on the arrangement of the recombination target sites the global result will be either the insertion of one DNA sequence into another or the inversion or deletion of DNA between two sites on the same chromosome. The products of recombination always reconstitute the recombination target sequences, thus setting the stage for recombination in the reverse direction (2).
In contrast to its more ubiquitous and less conservative relatives, homologous recombination and transposition, site-specific recombination uses specific DNA sequences on both of its DNA targets and requires no exogenous energy source. The λ-Int family recombinases use tyrosine as the DNA-cleaving nucleophile and exchange DNA strands sequentially, first forming and then resolving a four-way junction, or Holliday recombination intermediate (3–5). Understanding the metabolism of such junctions is a key element in any serious description of this pathway and is the main subject of the crystallographic studies extended in this issue of the Proceedings by Greg Van Duyne and his collaborators (6–8).
As shown by the work of R. Hoess, K. Abremski, and N. Sternberg (9, 10), the Cre-lox pathway of bacteriophage P1 comprises the simplest example of the λ-Int family reaction. Cre recombinase binds to each DNA lox site at a pair of inverted repeat binding sites that are separated by a 6-bp “overlap” or “crossover” region. After synapsis and the formation of a tetrameric bihelical complex, one strand of each DNA target is cleaved at the 5′ ends of the crossover region (forming a covalent 3′ phosphotyrosine intermediate) and the freed 5′ ends are exchanged and ligated to form the Holliday junction (HJ) intermediate. The HJ is resolved when the second pair of Cre monomers at the other end of the crossover region carries out an identical reaction on the unexchanged strands, thereby generating a pair of product helical junctions that are chemically heteroduplex within the 6-bp crossover region. Other λ-Int family members have different lengths of crossover regions (6–8 bp) and additional layers of complexity that are not essential to an appreciation of the structures discussed here.
1997 was a good year for the λ-Int family album as it saw the introduction of its first portraits: the x-ray crystal structures of the catalytic domains of λ Int (11) and HP1 integrase (12), the XerD recombinase (13), and the Cre-lox cocrystal (6). These structures, in conjunction with earlier genetic and biochemical studies and the structures of two closely related domains from eukaryotic topoisomerases (14, 15), provided a good picture of the catalytic pocket. Although this pocket as well as the overall fold of the catalytic domain appear to be quite well conserved among the λ-Int family recombinases (despite extensive divergence in their amino acid sequences), the carboxyl-terminal structural element that delivers the tyrosine nucleophile shows more variability between different recombinases. Indeed, this variability may reflect a structural flexibility that is important in the mechanism of recombination (see also below). It also may explain why some Ints evolved a monomeric active site (e.g., cis cleavage by λ Int and Cre) and others a composite active site (e.g., trans cleavage by Flp) (6, 16, 17).
Appropriately, the most mechanistically informative 1997 photo was of the Cre-lox complex, because it represents the fundamental exemplar of the λ-Int family reactions. To obtain cocrystals, Guo et al. (6) took advantage of nicked suicide substrates developed in the λ system to trap covalent complexes (18) (Fig. 1A). The tetrameric structure formed from these covalent Cre-lox complexes reveals that two of the Cre monomers have cleaved the DNA, and the resulting 5′ termini are seen invading their new helix partners. The four Cre monomers bend the antiparallel DNA helices into an incipient pseudo-square planar HJ and delimit a cavity where the released single strands are free of protein contacts. The 5′ OHs of these exchanging single strands are unable to attack the phosphotyrosine bonds of the partner Cre because they are one base too short (i.e., the suicide feature). Each Cre monomer clamps around its respective lox half-site such that the amino- and carboxyl-terminal domains cluster on opposite faces of the saucer-like HJ. The two most illuminating features of the cocrystal structure concerned the global disposition of the helical arms and the differences between the cleaving and noncleaving pairs of Cre monomers. The approximate 4-fold symmetry of the active sites in the cleaved covalent complex suggests that the strand exchange can be executed without large charges in quaternary structure. It would readily accommodate a model that involved swapping three bases at a time centered around very limited branch migration (19).
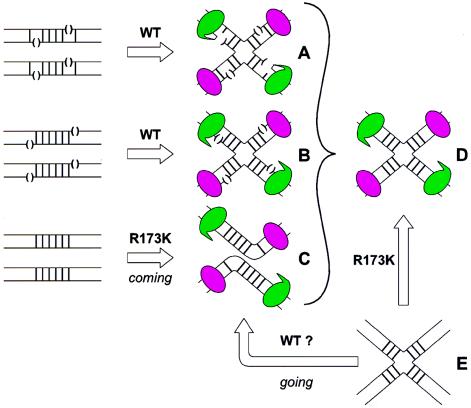
Figure 1.
Schematic comparison of the different Cre-lox complexes for which x-ray crystal structures have been obtained. DNA strands (thin lines) in the vicinity of the 6-bp crossover region (heavy vertical bars) of the lox site contain nicks where indicated by open parentheses. The Cre monomers are shown as being either in the noncleaving mode (magenta ellipses) or the cleaving mode (green ellipses with a beak). Covalent 3′ phosphotyrosine linkages are denoted by a beak that makes contact with a DNA strand (A). The covalent Cre-lox complexes (A) were formed by incubating suicide lox site DNAs with wild-type (WT) Cre (6). Nicked Cre-HJ complexes (B) were made by incubating WT Cre with lox sites containing a nick at the scissle phosphate while intact Cre-HJ complexes (D) were made by incubating HJ lox DNA (E) with a cleavage-incompetent Cre mutant (R173K) (7). Cre-lox synaptic complex (C) was made by incubating helical lox sites with the R173K Cre mutant (8). The question is raised as to whether the same synaptic complex (C) also might be generated by incubating HJ lox DNA (E) with WT Cre (see text).
The mechanism that enables Cre to sequentially swap top and then bottom strands can be nicely explained by subtle differences between the carboxyl termini of the cleaving versus the noncleaving pairs of Cre. The four Cre monomers are associated in a cyclic manner by a short carboxyl-terminal helix that emanates from the active site vicinity of one monomer and buries itself in a hydrophobic pocket near the active site of an adjacent monomer. These interactions mediate a dialog that nicely explains why double-strand cleavage does not occur (“I am cleaving, therefore you cannot”). The potential for intermolecular communication involving the active site tyrosine nucleophiles and carboxyl-terminal segments also was reflected in different ways in each of the other λ-Int family structures. Before leaving this image it is worth noting that the cleaving and noncleaving monomers are sufficiently different that they can be identified in subsequent pictures.
Additions to the family album from the Van Duyne laboratory also were made in 1998 (7) and now in 1999 (8). Each of the three papers has used a different device to capitalize on Cre’s strong propensity to form HJ-like structures and each has resulted in a unique and informative view of the recombination reaction. As discussed above, the first paper exploited a suicide substrate to trap Cre in a covalent, incipient HJ complex (Fig. 1A). The second paper used both an HJ that could not be processed further by Cre because its scissile phosphates had been replaced by nicks (Fig. 1B) and an intact HJ that could not be processed further because it was paired with a cleavage-incompetent Cre mutant (Fig. 1D). The third paper describes helical lox sites that are incubated with a cleavage-incompetent Cre mutant and are shown to form a tetrameric synaptic complex with a global structure that resembles the HJ complexes (Fig. 1C).
It is the second paper of the Van Duyne collection that shows a bona fide HJ complex (7). What we see is a 2-fold symmetric four-way DNA junction that is distinct both from the continuous duplexes that are fully stacked at the junction branch point in physiological magnesium and from the 4-fold symmetric square conformation seen in low salt and EDTA (20). It is interesting, but not surprising in light of the initial structures of covalent complexes, that Cre forms very similar structures with both the unnicked and nicked HJs. The latter is the result of Cre promoting synapsis and strand exchange to generate an HJ structure even in the absence of ligation.
The most interesting mechanistic insights from these structures concerned the subtle but well-delineated stereochemical link between reconfiguration of the complex and cleavage specificity. Although an HJ isomerization between pairs of strand exchanges had been predicted from solution studies (21, 22), the nature of the isomerization was a surprise. Especially gratifying was the ability to correlate the asymmetry in quaternary structure with differences in the conformation of the linker peptides that communicate between cleaving and noncleaving partners. Indeed, these differences could be viewed as rendering pairs of partners either competent or incompetent for DNA cleavage in a mutually exclusive toggle switch.
The new picture on view in this issue is a close-up (2.2 Å) and has more than its share of surprises and puzzles (8). This structure is truly a synaptic complex for there is no inclusion of, or avenue for, strand exchange or DNA cleavage. By now, the aspiring recombination maven will pretend not to be surprised that the synaptic structure looks so much like the HJs seen previously. Indeed, from the perspective of the overall quaternary structure, all of the structures in Fig. 1 (A–D) are virtually indistinguishable. This similarity is informative (and useful for the crystallographers) because it says that all of the deformation necessary to break substrate and remake product DNA cylinders is carried out by the Cre-tetramer before and subsequent to the execution of any chemistry. The bending of DNA helices by the related Flp recombinases was predicted by biochemical experiments (23) but the location and type of bend seen in this cocrystal structure was quite a surprise. The DNA helices are both sharply bent at a single base pair step in the crossover region; however, the bend is neither centrally located nor is it proximal to the scissile phosphates, but rather it is on the opposite end of the crossover region. Although this bend features an unpairing of DNA strands, it is not situated where strand swapping is programmed to occur. Also, this bend, whose location and direction govern which DNA strand is cleaved, is atypical of protein-induced DNA bends. It is interesting to note that the only way to distinguish which Cre monomers are programmed as cleaving versus noncleaving is by the characteristic structural signatures identified in the earlier covalent and nicked complexes. (We might also applaud the authors for their timing.)
While discussing this picture I have avoided using the future tense when referring to DNA cleavage because one cannot know whether this Cre-DNA complex most closely represents a reaction that is coming (i.e., about to make an HJ), or one that is going (i.e., has just resolved an HJ; see Fig. 1C). The authors have most reasonably adopted the former view, because it reflects the experimental assembly of the complex and suggests an attractive model for how the stereochemical energy stored in DNA bending and phosphate repulsion is used to promote the strand exchange step after DNA cleavage. Might this same complex also have been assembled by incubating a cleavage-competent Cre with HJ DNA? More significantly, is the recombination completely symmetrical? If so, there should be no distinction between a structure that is coming versus going (bottom row in Fig. 1). One also might ask whether there is a correlation between which strands in helical substrates are cleaved first and the propensity of different DNA sequences to form the characteristic kink at one or the other end of the crossover region.
The global similarity of the synaptic structure to that of the HJ complexes is (as noted above) most seductive. However, one must keep an open eye to the fact that all of these structures have crystallized in the same space group and consequently some, or much, of the observed global similarity might be exaggerated by the conditions and forces of crystallization. It sounds a bit greedy to suggest that one structure in a different space group would be nice. A more modest wish list would include a structure of Cre not bound to DNA, wild-type Cre on a full lox site (perhaps modeling the product of HJ resolution), or a synaptic complex with an asymmetric crossover region (to explore the sequence dependence of strand cleavage). The collection of structures from the Van Duyne laboratory have overlapping and reinforcing messages, but each one has provided important new insights into the central mechanisms used by this family of recombinases. Send more pictures.
Acknowledgments
I thank Marco Azaro for helpful comments and assistance with the manuscript.
Footnotes
A commentary on this article begins on page 7143.
References
- 1.Campbell A M. In: Advances in Genetics. Caspari W W, Thoday J M, editors. New York: Academic; 1962. pp. 101–137. [Google Scholar]
- 2.Nash H A. In: Escherichia coli and Salmonella. Neidhardt F C, editor. Washington, DC: Am. Soc. Microbiol.; 1996. pp. 2363–2376. [Google Scholar]
- 3.Hsu P-L, Landy A. Nature (London) 1984;311:721–726. doi: 10.1038/311721a0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kitts P A, Nash H A. Nature (London) 1987;329:346–348. doi: 10.1038/329346a0. [DOI] [PubMed] [Google Scholar]
- 5.Nunes-Düby S E, Matsumoto L, Landy A. Cell. 1987;50:779–788. doi: 10.1016/0092-8674(87)90336-9. [DOI] [PubMed] [Google Scholar]
- 6.Guo F, Gopaul D N, Van Duyne G D. Nature (London) 1997;389:40–46. doi: 10.1038/37925. [DOI] [PubMed] [Google Scholar]
- 7.Gopaul D N, Guo F, Van Duyne G D. EMBO J. 1998;17:4175–4187. doi: 10.1093/emboj/17.14.4175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Guo F, Gopaul D N, Van Duyne G D. Proc Natl Acad Sci USA. 1999;96:7143–7148. doi: 10.1073/pnas.96.13.7143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hoess R, Abremski K, Sternberg N. Cold Spring Harbor Symp Quant Biol. 1984;49:761–768. doi: 10.1101/sqb.1984.049.01.086. [DOI] [PubMed] [Google Scholar]
- 10.Abremski K, Hoess R. J Biol Chem. 1984;259:1509–1514. [PubMed] [Google Scholar]
- 11.Kwon H J, Tirumalai R S, Landy A, Ellenberger T. Science. 1997;276:126–131. doi: 10.1126/science.276.5309.126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hickman A B, Waninger S, Scocca J J, Dyda F. Cell. 1997;89:227–237. doi: 10.1016/s0092-8674(00)80202-0. [DOI] [PubMed] [Google Scholar]
- 13.Subramanya H S, Arciszewska L K, Baker R A, Bird L E, Sherratt D J, Wigley D B. EMBO J. 1997;16:5178–5187. doi: 10.1093/emboj/16.17.5178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Redinbo M R, Stewart L, Kuhn P, Champoux J J, Hol W G J. Science. 1998;279:1504–1513. doi: 10.1126/science.279.5356.1504. [DOI] [PubMed] [Google Scholar]
- 15.Cheng C, Kussie P, Pavletich N, Shuman S. Cell. 1998;92:841–850. doi: 10.1016/s0092-8674(00)81411-7. [DOI] [PubMed] [Google Scholar]
- 16.Nunes-Düby S E, Tirumalai R S, Dorgai L, Yagil R, Weisberg R, Landy A. EMBO J. 1994;13:4421–4430. doi: 10.1002/j.1460-2075.1994.tb06762.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chen J-W, Lee J, Jayaram M. Cell. 1992;69:647–658. doi: 10.1016/0092-8674(92)90228-5. [DOI] [PubMed] [Google Scholar]
- 18.Pargellis C A, Nunes-Düby S E, Moitoso de Vargas L, Landy A. J Biol Chem. 1988;263:7678–7685. [PubMed] [Google Scholar]
- 19.Nunes-Düby S, Azaro M, Landy A. Curr Biol. 1995;5:139–148. doi: 10.1016/s0960-9822(95)00035-2. [DOI] [PubMed] [Google Scholar]
- 20.Duckett D R, Murchie A I H, Lilley D M J. EMBO J. 1990;9:583–590. doi: 10.1002/j.1460-2075.1990.tb08146.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Azaro M A, Landy A. EMBO J. 1997;16:3744–3755. doi: 10.1093/emboj/16.12.3744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Arciszewska L K, Grainge I, Sherratt D J. EMBO J. 1997;16:3731–3743. doi: 10.1093/emboj/16.12.3731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Luetke K H, Sadowski P D. J Mol Biol. 1995;251:493–506. doi: 10.1006/jmbi.1995.0451. [DOI] [PubMed] [Google Scholar]