Abstract
Background
Hydrogen peroxide (H2O2) and hypochlorous acid (HOCl) are reactive oxygen species that are part of the oxidative burst encountered by Salmonella enterica serovar Typhimurium (S. Typhimurium) upon internalization by phagocytic cells. In order to survive, bacteria must sense these signals and modulate gene expression. Growing evidence indicates that the ArcAB two component system plays a role in the resistance to reactive oxygen species. We investigated the influx of H2O2 and HOCl through OmpW and the role of ArcAB in modulating its expression after exposure to both toxic compounds in S. Typhimurium.
Results
H2O2 and HOCl influx was determined both in vitro and in vivo. A S. Typhimurium ompW mutant strain (∆ompW) exposed to sub-lethal levels of H2O2 and HOCl showed a decreased influx of both compounds as compared to a wild type strain. Further evidence of H2O2 and HOCl diffusion through OmpW was obtained by using reconstituted proteoliposomes. We hypothesized that ompW expression should be negatively regulated upon exposure to H2O2 and HOCl to better exclude these compounds from the cell. As expected, qRT-PCR showed a negative regulation in a wild type strain treated with sub-lethal concentrations of these compounds. A bioinformatic analysis in search for potential negative regulators predicted the presence of three ArcA binding sites at the ompW promoter region. By electrophoretic mobility shift assay (EMSA) and using transcriptional fusions we demonstrated an interaction between ArcA and one site at the ompW promoter region. Moreover, qRT-PCR showed that the negative regulation observed in the wild type strain was lost in an arcA and in arcB mutant strains.
Conclusions
OmpW allows the influx of H2O2 and HOCl and is negatively regulated by ArcA by direct interaction with the ompW promoter region upon exposure to both toxic compounds.
Background
Hydrogen peroxide (H2O2) and hypochlorous acid (HOCl) are reactive oxygen species that are part of the oxidative burst encountered by S. Typhimurium upon internalization by phagocytic cells. Under acidic conditions, such as those found inside the phagosome, H2O2 is generated spontaneously by the reaction of two superoxide anion (O2−) molecules [1]. Moreover, S. Typhimurium encodes both periplasmic and cytoplasmic superoxide dismutases that catalyze O2− dismutation to generate H2O2 and molecular oxygen [2-4]. HOCl is produced by the action of myeloperoxidase (MPO) in a reaction that depends on H2O2, Cl−and acidic conditions [5,6]. Taken together, H2O2 and HOCl react with thiol and heme groups, copper and iron salts generating the reactive hydroxyl radical (OH.). As a consequence, they produce lipid peroxidation, chlorination of tyrosine residues, oxidation of iron centers, protein cross linking and DNA damage [5-8].
In order to enter Gram negative bacteria, H2O2 and HOCl must be able to cross the outer membrane (OM) and even though several biological membranes are permeable to H2O2, studies in E. coli and Saccharomyces cerevisiae showed that this compound cannot diffuse freely [9,10]. For HOCl, diffusion through the OM is reported to be limited [11]. One possibility for H2O2 and HOCl influx through the OM is diffusion through porins. In this context, we recently reported that OmpD, S. Typhimurium most abundant OM porin, allows H2O2 diffusion [12]. OM porins are organized as homo-trimers (classic porins) or monomers (small porins) forming aqueous channels that allow the influx of hydrophilic solutes with a molecular weight ≤ 600 Da [13]. Classic porins, including OmpC and OmpF, form β-barrels with 12–22 transmembrane segments while small porins (OmpW) are composed of 8–10 [14,15]. The crystal structure of OmpW from E. coli revealed that it forms an 8-stranded β-barrel and functions as an ion channel in lipid bilayers [16,17]. In Vibrio cholerae, OmpW was described as an immunogenic 22 KDa protein [18] and its expression is altered by factors such as temperature, salinity, nutrient availability and oxygen levels [19]. Additionally, several studies show that porins are regulated by ROS. Due its oxidant nature and diffusion through the OM, regulation of porin expression must be tightly regulated as a mechanism of controlling OM permeability. Accordingly, S. Typhimurium ompD and ompW expression is regulated in response to H2O2 and paraquat [12,20], respectively, and S. Enteritidis and Typhimurium exposure to HOCl results in lower levels of ompDompC and ompF transcripts [21].
The cellular response to oxidative stress is regulated at the transcriptional level by activating the SoxRS and OxyR regulons in response to O2− and H2O2, respectively [22,23], however, several studies have provided evidence for a role of the ArcAB two component system in the resistance to ROS induced damage [12,24-26]. ArcA is essential for S. Enteritidis, Typhimurium and E. coli resistance to ROS [24,26,27]. ArcB is a sensor member of the histidine kinase family that is anchored to the inner membrane [28]. In response to oxygen availability, ArcB autophosphorylates in an ATP dependant intramolecular reaction at position His-292 [29,30] and transfers the phosphate group to the cytoplasmic response regulator ArcA [31-33], which binds to promoter regions regulating gene expression [34,35]. ArcB activity is regulated in response to oxygen conditions by the redox state of both the ubiquinone and menaquinone pools [29,36-38]. However, recent studies in E. coli show that the system is regulated by the degree of aerobiosis but not by the redox state of the ubiquinone pool, challenging the idea that the system is inhibited by oxidized quinones [39].
In this work we provide further evidence of the role of the ArcAB two component system in the response to ROS under aerobic conditions and show that this system mediates regulation of ompW expression in response to a novel signal, HOCl. First we demonstrate, both in vivo and in vitro, that OmpW mediates diffusion of H2O2 and HOCl and that exposure of S. Typhimurium to these compounds results in a negative regulation of ompW. By EMSA and using transcriptional fusions, we demonstrate that the global regulator ArcA binds to the ompW promoter region. Furthermore, we show that ompW negative regulation observed in wild type cells treated with H2O2 and HOCl was not retained in an arcA or arcB mutant strain, indicating that the ArcAB two component system mediates ompW negative regulation in response to H2O2 and HOCl. These results further expand our knowledge in both the mechanisms of ROS resistance and the role of ArcAB in this process.
Results and discussion
The OmpW porin facilitates H2O2 and HOCl diffusion through the OM and reconstituted proteoliposomes
Hydrogen peroxide and hypochlorous acid are ROS generated by phagocytic cells and in order to enter Gram-negative bacteria they must be able to cross the OM. Even though several biological membranes are permeable to H2O2, studies in E. coli and S. cerevisiae demonstrate that this compound cannot diffuse freely [9,10]. Additionally, the dielectric properties of H2O2 are comparable to those of water and this compound has a slighter larger dipolar moment, further limiting its diffusion through the OM lipid bilayer. For HOCl, diffusion through the OM is also reported to be limited [11]. Therefore, H2O2 and HOCl must be channeled through the lipid bilayer and one possibility is the influx through porins. We recently demonstrated that the most abundant OM protein in S. Typhimurium, OmpD, allows H2O2 diffusion and is regulated by ArcAB [12]. Little is known about the diffusion of HOCl, but genetic evidence has suggested that in E. coli porins might be used as entry channels for hypothiocyanate ions (OSCN−), a molecule with a similar chemical structure generated by lactoperoxidase using thiocyanate and H2O2 as an oxidant [40]. In one study, ompC and ompF knockout mutants showed an increased resistance to OSCN−, however, a direct role of porins in mediating HOCl diffusion was not evaluated.
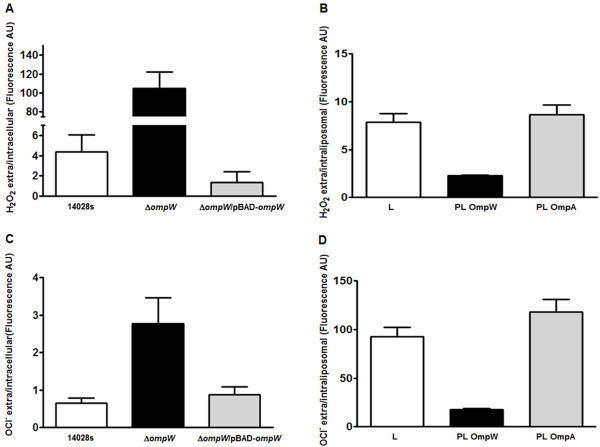
To assess whether OmpW allows the diffusion of H2O2 and HOCl, scopoletin and dihydrorhodamine (DHR)-123 probes, respectively, were used to measure uptake of both toxic compounds separately in a wild type, ∆ompW and a genetically complemented ∆ompW (pBAD-ompW) strain as described in methods. The ∆ompW strain showed an increase in extracellular fluorescence levels after exposure to H2O2 and HOCl resulting in higher extra/intracellular ratios (24 and 4-fold, respectively) as compared to the wild type strain, indicating that in the absence of OmpW the influx of both toxic compounds is decreased. Genetic complementation of ∆ompW resulted in nearly identical levels of both extra and intracellular fluorescence as those observed in the wild type strain, suggesting that OmpW is necessary for H2O2 and HOCl uptake (Figure 1A and C). Even though OmpW appears as a direct responsible for the influx of the compounds, a pleiotropic effect cannot be ruled out at this point because the absence of OmpW in the mutant strain could be producing a remodeling of the membrane organization.
Figure 1.
OmpW facilitates H2O2and HOCl diffusion through the outer membrane and reconstituted proteoliposomes. A and C. H2O2 and HOCl levels were measured indirectly by specific fluorescence assays in the wild type (14028s), mutant (∆ompW) and genetically complemented strains (∆ompW/pBAD-ompW + arabinose). Exponentially growing cells were exposed to H2O2(A) or NaOCl (C) for 5 min and fluorescence was determined in the extracellular (extra) and intracellular fractions. B and D. Free liposomes (L), proteoliposomes reconstituted with S. Typhimurium OmpW (PL OmpW) or OmpA (PL OmpA) proteins were incubated with H2O2(B) or NaOCl (D) for 5 min and fluorescence was determined in the extraliposomal (extra) and intraliposomal fractions. AU indicates arbitrary units. Values represent the average of four independent experiments ± SD.
To establish a direct contribution of OmpW in H2O2 and HOCl transport, we used reconstituted proteoliposomes. OmpW-proteoliposomes showed a decrease in H2O2 and HOCl extra/intraliposomal ratios (3.5 and 5-fold respectively) when compared to free liposomes (Figure 1B and D). Proteoliposomes with S. Typhimurium OmpA porin were used as a negative control as previously described [12]. As expected, OmpA-proteoliposomes showed similar levels to those of free liposomes, indicating that OmpW facilitates H2O2 and HOCl uptake.
Since OmpW channels both toxic compounds across the lipid bilayer, we hypothesized that a ∆ompW strain should be more resistant to both toxic compounds when compared to the wild type strain. As shown in Figure 2, exposure of ∆ompW to H2O2 4 mM or HOCl 5 mM resulted in an increase in the number of colony forming units (CFU) after 60 min of treatment. However, at longer periods the CFU count between strains 14028s and ∆ompW was similar. At 30 min post-treatment with either of the toxic compounds, strain ∆ompW showed an increase from 1×106 CFU/ml to approximately 6×107 CFU/ml. In contrast, the CFU/ml count for strain 14028s remained almost unaltered at 1×106, resulting in a 1.5-log10-fold increase in growth for ∆ompW. A similar result was observed after 60 min of treatment where the ompW mutant strain showed an increase from 6×107 to 1.5×109 CFU/ml while the wild type strain changed from 1×106 to 8×107 CFU/ml. Our results suggest that the absence of OmpW in the mutant strain represents an advantage at short time points due to a decreased permeability towards both H2O2 and HOCl. At longer periods, OM permeability should be reduced because exposure to both toxic compounds results in a negative regulation of S. Typhimurium porins including OmpD, OmpC and OmpF [12,21]. One important possibility that cannot be ruled out at this time is that in the ∆ompW strain, the expression of other porins or the OM lipid composition might be altered, therefore changing OM permeability. For example, a study conducted in E. coli showed that an ompC knockout mutant had increased levels of OmpA [40], however, changes in permeability were not evaluated. Furthermore, this has not been evaluated in a S. Typhimurium or E. coli ∆ompW strain.
Figure 2.
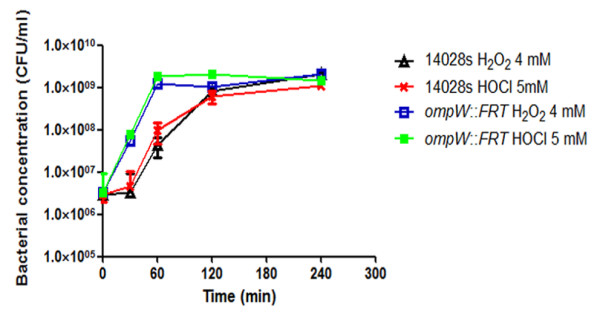
Bacterial concentration ofS. Typhimurium 14028s and ΔompWexposed to H2O2or NaOCl. Cultures of 14028s and ΔompW were grown to OD ~ 0.4 and treated with H2O2 4 mM or NaOCl 5 mM in LB medium. Time of treatment is indicated. Bacterial concentrations were determined by plating. The values are the concentrations of surviving bacteria after exposure to H2O2 or NaOCl. Experiments were performed in triplicate. Error bars indicate SD.
Our data supports the proposed model where OmpW allows the influx of small polar molecules, like H2O2 and HOCl. The crystal structure of OmpW from E. coli revealed that the cross-section of the barrel has approximate dimensions of 17 × 12 Å along the length of the barrel and although the interior of the channel has a hydrophobic character, the observed single channel activities shows that polar molecules traverse the barrel [17]. Taken together, these results provide biochemical and genetic evidence indicating that both toxic compounds are channeled through OmpW. From our knowledge, this is the first direct evidence of HOCl diffusion through porins. Furthermore, preliminary analyses indicate that H2O2 and HOCl channeling is common for S. Typhimurium OmpD, OmpC and OmpF porins (unpublished data).
Hydrogen peroxide and hypochlorous acid exposure results in ompW negative regulation
Since the OmpW porin channels H2O2 and HOCl through the OM and exposure to these molecules is detrimental to bacteria, we hypothesized that ompW should be negatively regulated when S. Typhimurium is exposed to H2O2 and HOCl. To study this effect, wild type S. Typhimurium cells were grown to mid-log phase, exposed to H2O2 or HOCl and ompW mRNA levels were measured by qRT-PCR. As seen in Figure 3, exposure to H2O2 and HOCl resulted in lower levels of ompW transcripts (0.27 ± 0.04 and 0.156 ± 0.079, respectively) relative to control untreated cells. In agreement with our results of ompW negative regulation, similar results were observed by Wang et al. (2010) who showed that S. Enteritidis and Typhimurium cells exposed to HOCl results in modulation of ompD, ompC, ompF (negatively) and ompA (positively) expression. Furthermore, Calderón et al. (2011) demonstrated that the S. Typhimurium ompD gene is negatively regulated in response to H2O2. Therefore, our and all the published data suggest that in the presence of OCl- or H2O2 there might be a general lowering in the concentration of porins in the outer membrane, in order to diminish the permeability. To assess the specificity of our assay, we evaluated ompD, ompC and arcB transcript levels as positive (ompD and ompC) and negative controls (arcB). The arcB gene was used as a negative control based on our microarray analysis which shows that it remains unaltered under these conditions and between strains 1408s and ΔarcA (unpublished data). Our results indicate that after exposure to both toxic compounds, arcB transcript levels remain unchanged while those of ompD and ompC are lowered as compared to untreated cells (Figure 3). Therefore, all the evidence indicates that OM permeability is tightly regulated in response to ROS and could represent a novel mechanism of resistance when bacteria are exposed to these toxic compounds.
Figure 3.
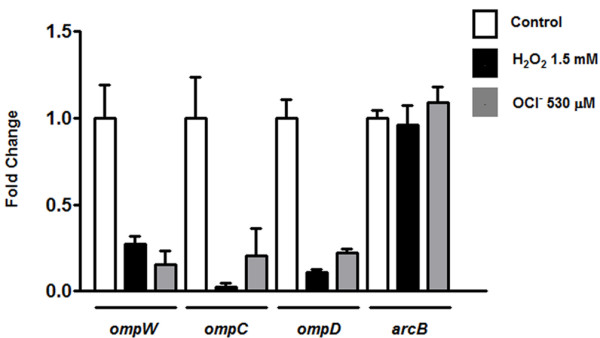
Effect of H2O2and HOCl onompWexpression. Wild type (14028s) exponentially growing cells were treated with H2O2 (1.5 mM) or NaOCl (530 μM) for 20 min and ompW, ompD, ompC and arcB mRNA levels were measured by qRT-PCR. Control cells received no treatment. 16S rRNA levels were used for normalization. Values represent the average of four independent experiments ± SD.
ArcA binds the ompW promoter region
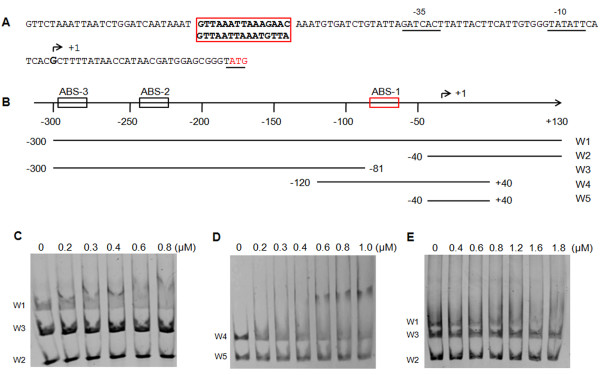
In addition to the soxRS and oxyR systems, several studies have provided evidence that the ArcAB two component system plays an important role in the resistance to ROS induced damage. For example, ArcA is essential for S. Enteritidis and Typhimurium resistance to ROS [24,27] and E. coli mutant strains of the sensor ArcB and the regulator ArcA, show an increased susceptibility to H2O2[26]. However, neither of these studies identified genes directly regulated by the system under oxidative stress. We recently demonstrated that ArcA negatively regulates the expression of S. Typhimurium ompD after H2O2 exposure by direct interaction with its promoter region [12]. To determine if ArcA mediates ompW down-regulation in response to H2O2 and HOCl, a search for putative ArcA binding sites at the ompW promoter region was performed using Virtual Footprint 3.0 [41]. The analysis predicted the presence of three ArcA binding sites (ABS) located at positions −61 to −70 (ABS-1, forward orientation), -230 to −239 (ABS-2) and −286 to −295 (ABS-3, both in reverse orientation) relative to the experimentally determined transcription start site [42]. Comparison with the extended core region 5′-GTTAATTAAATGTTA-3′ described by Evans et al. (2011) further revealed that only ABS-1 presented a high degree of identity (14 out of 15 nucleotides) with the consensus sequence. To confirm or rule out a direct interaction between ArcA and the predicted binding sites, deletions of the promoter region were generated by PCR (schematized in Figure 4B) and used to perform non-radioactive EMSAs with ArcA and phosphorylated ArcA (ArcA-P). The purity of the protein was assessed by PAGE and ArcA was the dominant product. Electrophoretic mobility shift with ArcA-P was only observed when incubated with fragments that included ABS-1 (Figure 4C and D, W1 and W4). No shifts were observed in fragments that include both ABS-2 and ABS-3 (W3, even at three-fold excess) or control fragments that did not include any ABS (W2 and W5). Non-phosphorylated ArcA only generated electrophoretic mobility shifts at higher concentrations (over 1200 nM) where the negative controls were also retarded as a result of non-specific binding (Figure 3E). Taken together our bioinformatic and EMSA analyses indicate that ArcA-P binds to the ompW promoter region at a site located between positions −80 and -41 and suggests that this site is ABS-1 which is located between positions −70 to −55.
Figure 4.
ArcA binding to theompWpromoter region. A.S. Typhimurium ompW promoter region. Black and red boxes indicate predicted ArcA binding sites. -10 and −35 boxes are underlined. The transcription start site is shown in bold and indicated as +1. The translation start site is underlined and in red. The consensus ArcA binding site is shown under the promoter sequence. B. Schematic representation of the ompW promoter region. Positions relative to the transcription start site are indicated. ArcA binding sites are indicated as in the text. PCR products used in EMSAs are shown and names of each fragment are indicated. C,D and E. EMSA of the ompW promoter region. A 3-fold excess (60 ng) of fragments W2 and W3 were incubated with W1 (C) and the fragment W4 was incubated with W5 (D) and increasing amounts of phosphorylated ArcA as indicated on the top of each gel. (E) W1, W2 and W3 were incubated with increasing amounts of non-phosphorylated ArcA
Evaluating ArcA binding site 1 (ABS-1) functionality
To further confirm that ABS-1 (Figure 4A) was the functional ArcA binding site mediating ompW negative regulation in response to ROS, we constructed transcriptional fusions of the ompW promoter region. We generated two different fusions which included the whole promoter from positions +1 to −600, with respect to the translation start site. One construction contained the native promoter (pompW-lacZ) while substitutions that mutated ABS-1 (shown in red and underlined, Figure 5A) were included in the second construction (pompW/ABS1-lacZ). The constructions were transformed into the wild type strain and β-galactosidase activity was measured in response to treatment with H2O2 and HOCl.
Figure 5.
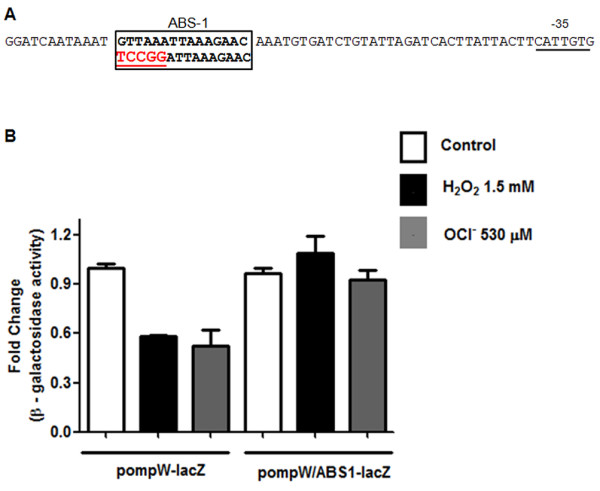
Evaluating ArcA binding site 1 (ABS-1) functionality at theompWpromoter. (A) Schematic representation of substitutions generated at the ompW promoter. Substituted bases are in red, underlined and shown below the core ArcA binding sequence. Black box indicates ABS-1. -35 is indicated. (B) Expression of the wild type and mutagenized regulatory region of ompW in S. Typhimurium. Strain 14028s was transformed with the reporter plasmids pompW-lacZ (wild type) or pompW/ABS1-lacZ (ABS-1 mutated). Cells were grown to OD ~ 0.4 and treated with H2O2 1.5 mM or NaOCl 530 μM for 20 min and β-galactosidase activity was measured. Values represent the average of three independent experiments ± SD.
The activity of the constructions was compared to the untreated 14028s strain with the wild type fusion. Treatment of this strain with H2O2 and HOCl resulted in lower activity levels (0.58 ± 0.008 and 0.53 ± 0.095, respectively), in agreement with qRT-PCR experiments. However, a 5 nucleotide substitution of the most conserved residues at ABS-1 site (pompW/ABS1-lacZ) resulted in no regulation after exposure to either of the toxic compounds (1,09 ± 0.104 and 0,93 ± 0.061), indicating that they are relevant for the transcriptional activity of ompW in response to H2O2 and HOCl (Figure 5B). Furthermore, these results are in agreement with EMSAs which indicate that ArcA only binds to fragments containing ABS-1.
The ArcAB two component system mediates ompW negative regulation
To establish a direct relationship between ompW negative regulation and ArcA-P binding to its promoter region, ompW expression was evaluated by qRT-PCR in a ∆arcA strain exposed to H2O2 and HOCl. The negative regulation observed in the wild type strain was not retained in an arcA mutant treated with either of the toxic compounds and ompW transcript levels were similar as those observed in untreated cells. Genetic complementation of ∆arcA restored the negative regulation observed in wild type cells exhibiting lower ompW mRNA levels (0.161 ± 0.068 and 0.488 ± 0.027, respectively) as compared to untreated cells (Figure 6A and C). Growth of the genetically complemented strain in the presence of glucose (non-induction) resulted in similar ompW mRNA levels between treated and untreated cells (data not shown). As controls, we measured ompDompC and arcB transcript levels after exposure to H2O2 and HOCl in a ∆arcA strain. Transcript levels of ompD were measured since its expression is regulated by ArcA under ROS conditions [12]. Our results indicate that neither ompD or arcB transcript levels were decreased after exposure to H2O2 or HOCl while those of ompC remained regulated in a ∆arcA strain treated with either of the toxic compounds (Figure 6A), confirming that ArcA mediates ompD regulation under ROS conditions and showing that the expression of ompC is ArcA independent and regulated by different mechanisms which remain unsolved to the date, and are under study in our laboratory. Furthermore, our bioinformatic analyses in search for ArcA motifs predicted binding sites in the promoter regions of ompW and ompD, but not for ompC ([12], data not shown).
Figure 6.
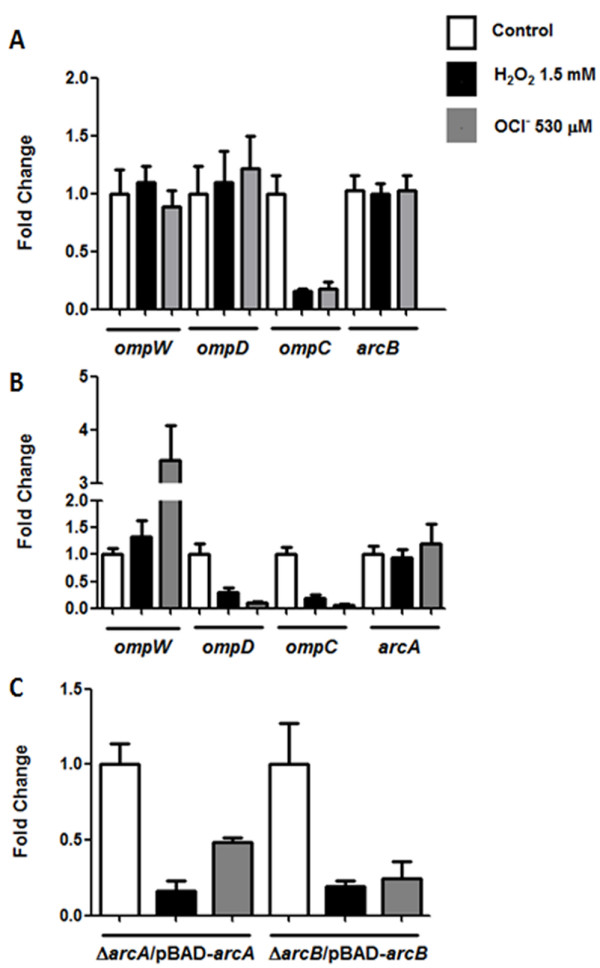
ArcAB-dependant expression ofompW.ompW, ompD, ompC, arcB and arcA mRNA levels were measured by qRT-PCR in a (A) ∆arcA, (B) ∆arcB and (C) ∆arcA/pBAD-arcA and ∆arcB/pBAD-arcB. arcB and arcA were used as negative controls in (A) and (B), respectively. Exponentially growing cells were treated with H2O2 1.5 mM or NaOCl 530 μM for 20 min and transcript levels were measured. Genetically complemented cells were grown in the presence of arabinose 1 mM. Control cells received no treatment. 16S rRNA levels were used for normalization. Values represent the average of three independent experiments ± SD.
To determine whether the negative regulation by ArcA was dependant on its cognate sensor ArcB, ompW mRNA levels were evaluated in a ∆arcB strain. In contrast to the negative regulation observed in wild type cells, ompW mRNA levels were further increased in a ∆arcB strain after exposure to HOCl (3.37 ± 1.09). Transcript levels after treatment with H2O2 were similar as those observed in untreated cells (Figure 6B). One possibility for this result is that in the absence of ArcA, ArcB might phosphorylate (i.e ArcB-OmpR, [43]) one or more response regulators, either unspecifically or due to cross-talk, which could bind to the promoter region and therefore prevent binding of positive regulators like SoxS, which has been demonstrated to regulate ompW and is up-regulated in response to HOCl [20,44]. This could result in constant ompW transcript levels as shown in Figure 6A. On the other hand, in the absence of ArcB no phosphorylation occurs and SoxS or other positive regulator(s) might have free accessibility to the ompW promoter and therefore increase its expression (Figure 6B), although this possibility has not been evaluated in this study. Genetic complementation of ∆arcB restored the negative regulation observed in wild type cells exposed to H2O2 and HOCl (0.19 ± 0.04 and 0.24 ± 0.11, respectively, Figure 6C). The ompD and ompC transcripts levels remained down-regulated after exposure to H2O2 and HOCl in the ∆arcB strain, while the negative control arcA remained unaltered (Figure 6B).
The ArcA regulon in anaerobically grown S. Typhimurium was recently determined [27]. Interestingly, neither ompD nor ompW expression was down-regulated in an ArcA dependant manner, suggesting that the ArcA regulon under anaerobic and aerobic ROS conditions could be different. Even in E. coliompW expression is suggested to be regulated by FNR in response to oxygen availability [39]. The difference between the ArcA regulons under aerobic and ROS conditions might be explained by studies suggesting that the mechanism of ArcA activation under aerobic conditions is different from those classically described. E. coli mutant strains in residue H-717 of ArcB are able to phosphorylate and activate ArcA through the transfer of the phosphate group from residue His-292 under aerobic conditions [45] and Loui et al. (2009) suggested that H2O2 resistance is independent of ArcA phosphorylation at residue Asp-54. To the date, the detailed molecular mechanism of ArcAB activation in response to ROS remains unsolved. Therefore, further experiments to unveil the molecular mechanism by which the S. Typhimurium ArcAB two component system is activated are needed and under way in our laboratory.
Conclusion
We provide both genetic and biochemical evidence indicating that the OM porin OmpW mediates the influx of H2O2 and HOCl. The results revealed that the S. Typhimurium ompW gene is negatively regulated upon exposure to both toxic compounds. Furthermore, we demonstrate that the response regulator ArcA mediates ompW negative regulation in response to H2O2 and HOCl via a direct interaction with the upstream region of ompW. Taken together, with our previous observation that OmpD mediates influx of H2O2 and is negatively regulated by ArcA in response to H2O2, these results further expand our knowledge regarding the coordinated regulatory mechanisms of ROS resistance and the role of ArcAB in this process.
Methods
Bacterial strains and growth conditions
Bacterial strains used in this work are listed in Table 1. Cells were grown aerobically with agitation in LB medium at 37°C. Solid media consisted of agar (20 g l−1) and plates were incubated at 37°C. Dilutions (1:100) of overnight cultures were used to initiate growth. When necessary, growth media was supplemented with the appropriate antibiotics (see below).
Table 1.
Bacterial strains used in this study
| Strain | Relevant characteristic(s) | Source |
|---|---|---|
|
S. Typhimurium |
|
|
| 14028s |
wild type strain |
G. Mora |
| 14028s/pompW-lacZ |
14028s transformed with a derivative of plasmid pLacZ-Basic carrying the ompW promoter (nt −600 to +1) |
This work |
| 14028s/pompW/ABS1-lacZ |
14028s transformed with a derivative of plasmid pLacZ-Basic carrying the ompW promoter (nt −600 to +1) with substitution GTTAA to TCCGG into position −70 to −66 |
This work |
| ΔompW |
ompW::kan |
C. Saavedra |
| ΔompW/pBAD-ompW |
ΔompW strain complemented with pBAD vector carrying the S. Typhimurium ompW gene |
C. Saavedra |
| ΔarcA |
arcA::cam |
[12] |
| ΔarcA/ pBAD-arcA |
ΔarcA strain complemented with pBAD vector carrying the S. Typhimurium arcA gene |
[12] |
| ΔarcB |
arcB::cam |
This work |
| ΔarcB/ pBAD-arcB |
ΔarcB strain complemented with pBAD vector carrying the S. Typhimurium arcB gene |
This work |
|
E. coli Top10 |
F- mcrA Δ(mrr-hsdRMS-mcrBC) Φ80lacZΔM15 ΔlacΧ74 recA1 araD139 Δ(ara-leu)7697 galU galK rpsL (StrR) endA1 nupG |
Invitrogen |
| Top10 pBAD-ompW |
Top10 transformed with the pBAD vector carrying the S. Typhimurium ompW gene |
C. Saavedra |
| Top10 pBAD-ompA |
Top10 transformed with the pBAD vector carrying the S. Typhimurium ompA gene |
C. Saavedra |
| Top10 pBAD-arcB |
Top10 transformed with the pBAD vector carrying the S. Typhimurium arcB gene |
This work |
| BL21 pET-TOPOArcA | BL21(DE3) transformed with the pET-TOPO101ArcA vector carrying the S. Typhimurium arcA gene | [12] |
Strain construction and genetic complementation
S. Typhimurium arcB gene was interrupted by gene disruption as previously described [46]. Strain 14028s (wild type) harboring plasmid pKD46 was grown in the presence of arabinose (10 mM) and ampicillin (100 μg ml−1) to OD600 ~ 0.4, made electrocompetent and transformed with a PCR product generated with plasmid pKD3 as template and primers 5′ ATTGGGTATTATGTGCGAAGTTGTGGTGAAGGAATCCTCTTGTAGGCTGGAGCTGCTTCG 3′ (WarcBF) and 5′ GGTGTTGGCGCAGTATTCGCGCACCCCGGTCAAACCGGGGCATATGAATATCCTCCTTAG 3′ (WarcBR). Transformants were selected on LB plates supplemented with chloramphenicol (20 μg ml−1) and confirmed by PCR using primers 5′ GCTACGCATATTTCGCACAA 3′ (arcBF) and 5′ GCGCCTTTGACATCATCATA 3′ (arcBR).
Genetic complementation of the ∆arcB strain was performed using plasmid pBAD-arcB. To generate this plasmid, S. Typhimurium arcB gene was amplified by PCR using primers 5′ ATGAAGCAAATTCGTATGCTG 3′ (pBADarcBF) and 5′ TCATTTTTTTTCCGCGTTTGCCACCC 3′ (pBADarcBR) and cloned into plasmid pBAD-TOPO TA® (Invitrogen) according to manufacturer’s instructions. Insertion was verified by DNA sequencing.
Bacterial survival after exposure to oxidative stress
Bacteria were cultured in 5 ml of LB medium at 37°C overnight with shaking. Antibiotics were added as appropriate. 1:1000 dilutions of the overnight cultures were grown in 25 ml to OD ~ 0.4 and H2O2 4 mM or NaOCl 5 mM (final concentration) were added. In all the assays the cultures were grown aerobically at 250 rpm. Aliquots of cultures were withdrawn at the different time points, diluted and plated in triplicate. Bacterial cultures were enumerated by counting the number of CFU after overnight incubation to determine the bacterial concentrations.
Construction of transcriptional fusions with reporter gene lacZ
The native ompW promoter region from positions +1 to −600 (with respect to the translation start) site was amplified by PCR with primers ompW_pLacZ_-600F_ATG 5′ CGGGGTACCCCCGATATCGAAAATTCGCG 3′ and ompW_pLacZ_-1R_ATG 5′ CCCAAGCTTACCCGCTCCATCGTTATGGT 3′ using genomic DNA from S. Typhimurium (strain 14028s). The restriction sites (KpnI and HindIII, respectively) at the ends of the DNA fragment were introduced by the PCR primers (underlined sequences) and digested with the corresponding enzymes. The digested PCR product was cloned into the multiple cloning site (MCS) of the β-galactosidase reporter vector pLacZ-Basic (GenBank accession no. U13184), Clontech, generating plasmid pompW-lacZ. To generate plasmid pompW/ABS1-lacZ, primers ompW_pLacZ_-600F_ATG with Mut_sit_arcAR 5′ TGTTCTTATAATGCGGAATTTATTGATCCAG 3′ and ompW_pLacZ_-1R_ATG with Mut_sit_arcAF 5′ CTGGATCAATAAATTCCGGAATTATAAGAACA 3′ were used to generate overlapping PCR products spanning the whole length of the ompW promoter. Mutation of ABS-1 was generated by incorporating substitutions in primers Mut_sit_arcAF and Mut_sit_arcAR (underlined sequences). The resulting PCR products were used as templates in a second reaction with primers ompW_pLacZ_-600F_ATG and ompW_pLacZ_-1R_ATG to generate the mutated ompW promoter, which was digested and cloned into the MCS of plasmid pLacZ-Basic. Constructions were confirmed by DNA sequencing. The generated constructs were transformed into wild type strain 14028s. To evaluate activity, cells at OD600 ~ 0.4 were grown for 20 min in the presence of H2O2 (1.5 mM) or NaOCl (530 μM). Control cells received no treatment. β-galactosidase activity was determined as previously described [20].
Protein purification
His-tagged ArcA used in EMSAs was purified as previously described [12]. Briefly E. coli BL21 cells harboring plasmid pET-TOPO-arcA were grown in 500 ml of LB medium supplemented with amplicillin (100 μg ml−1) to OD600 ~ 0.4 and protein overexpression was carried out by adding 1 mM IPTG and further growth for 6 h. Protein was purified by affinity chromatography as described by Georgellis et al., (1997).
Outer membrane proteins used in proteoliposomes were purified as described by Calderón et al. (2011). E. coli Top10 cells carrying pBAD-ompA or pBAD-ompW were grown in 500 ml to OD600 ~ 0.6 at 37°C and overexpression was performed for 5 h in the presence of 1 mM arabinose. His-tagged porins were purified by affinity chromatography using HisTrap HP columns (Amersham) according to the manufacturer’s instructions.
Plasmid pBAD-ompW was generated amplifying the coding region of S. Typhimurium ompW by PCR using primers 5′ ATGAAAAAATTTACAGTGGC 3′ (pBAD-ompWF) and 5′ GAAACGATAGCCTGCCGAG 3′ (pBAD-ompWR) and cloned into plasmid pBAD-TOPO TA® (Invitrogen) according to the manufacturer’s instructions. Insertion was verified by DNA sequencing.
RNA isolation and ompW mRNA detection
Overnight cultures were diluted (1:100) and cells were grown to OD600 ~ 0.4. Genetically complemented cells (∆arcA/pBAD-arcA and ∆arcB/pBAD-arcB) were grown in the presence of arabinose (1 mM) and ampicillin (100 μg ml-1). At this point, H2O2 (1.5 mM) or NaOCl (530 μM) was added and cells were grown for 20 min. Control cells received no treatment. After exposure to the toxic compounds, 4 ml were withdrawn from the culture and used to extract total RNA using GenElute Total RNA purification Kit® (Sigma). Total RNA treatment with DNase I and cDNA synthesis was performed as previously described [19].
Relative quantification of ompW mRNA was performed using Brilliant II SYBR Green QPCR Master Reagent Kit and the Mx3000P detection system (Stratagene). 16S rRNA was used for normalization. Specific primers were 5′ ATGAAAAAATTTACAGTGG 3′ (RTompWF) and 5′ GAAACGATAGCCTGCCGA 3′ (RTompWR) for the ompW gene; 5′ GTAGAATTCCAGGTGTAGCG 3′ (16SF) and 5′ TTATCACTGGCAGTCTCCTT 3′ (16SR) for 16S rRNA gene (16S). The reaction mixture was carried out in a final volume of 20 μl containing 1 μl of diluted cDNA (1:1000), 0.24 μl of each primer (120 nM), 10 μl of 10 x Master Mix, 0.14 μl of diluted ROX (1:200) and 8.38 μl of H2O. The reaction was performed under the following conditions: 10 min at 95°C followed by 40 cycles of 30 s at 95°C, 30 s at 53°C and 45 s at 72°C. Finally a melting cycle from 53 to 95°C was performed to check for amplification specificity. Amplification efficiency was calculated from a standard curve constructed by amplifying serial dilutions of RT-PCR products for each gene. These values were used to obtain the fold change in expression for the gene of interest normalized with 16S levels according to [47]. Experiments were performed in three biological and technical replicates.
DNA binding assays
Non-radioactive EMSAs were performed as described [48]. Briefly, increasing amounts of purified ArcA (phosphorylated and unphosphorylated) were incubated with 20 or 60 ng of PCR product(s) in binding buffer (100 mM Tris-Cl [pH 7.4], 100 mM KCl, 10 mM MgCl2, 10% glycerol, and 2 mM dithiothreitol) for 20 min at 30°C. Reaction mixtures were immediately loaded on prerun 4% native polyacrylamide gels. The DNA-protein complexes were visualized by ethidium bromide staining. PCR fragments used in EMSAs were generated by PCR using reverse primer 5′ ACCCGCTCCATCGTTATGGT 3′ (ompWR) in combination with 5′ GAGCAGACAAATATTTGCAT 3′ (300WF) or 5′ TATTAGATCACTTATTACTT 3′ (170WF) to generate fragments W1 and W2, respectively. Fragment W3 was generated using primers 300WF and 5′ GATCCAGATTAATTTAGAAC 3′. Fragments W4 and W5 were generated by using reverse primer 5′ AATTTTTTCATACCCGCTCC 3′ in combination with primers 5′ CCTATAACCAGGATTTTCAA 3′ and 170WF, respectively. ArcA phosphorylation was carried out as described by Linch and Lin (1996). Briefly purified ArcA was incubated with 50 mM disodium carbamoyl phosphate (Sigma) in a buffer containing 100 mM Tris-Cl (pH 7.4), 10 mM MgCl2, 125 mM KCl, for 1 h at 30°C and used immediately in EMSA assays.
In vivo and in vitro determination of hydrogen peroxide and hypochlorous acid diffusion
In vivo diffusion of H2O2 was assessed as previously described [12]. For HOCl detection, overnight cultures were diluted and cells were grown to OD600 ~ 0.5. Two ml of bacterial culture were centrifuged for 5 min at 4500 x g and resuspended in 1 ml of 100 mM phosphate buffer (pH 7.2). A 200 μl aliquot was incubated for 5 min with 530 μM NaOCl and constant agitation. Following, cells were vacuum filtered using polycarbonate filters of 0.025 μm (Millipore) and pass through was collected (extracellular fraction). Bacteria retained in the filter were recovered with 1 ml of 50 mM phosphate buffer (pH 7.2) and disrupted by sonication (intracellular fraction). Both fractions (190 μl) were incubated separately with dihydrorhodamine-123 to a final concentration of 5 μM as previously described [49]. The fluorescent product, rhodamine-123, was measured by fluorescence detection with excitation and emission wavelengths of 500 and 536 nm, respectively. HOCl and H2O2 uptake was determined as the extracellular/intracellular fluorescence ratio. The background fluorescence from a bacterial suspension not exposed to either of the toxic compounds was subtracted and results were normalized by protein concentration.
Proteoliposomes were prepared as described [50] with modifications [51]. For in vitro diffusion, proteoliposomes were incubated with 1.5 mM H2O2 or 530 μM NaOCl for 5 min, vacuum filtered and pass through was recovered (extraliposomal fraction). Proteoliposomes were recovered from the filters with 2 ml of 50 mM phosphate buffer (pH 7.2) and disrupted by sonication (intraliposomal fraction). Fluorescence was measured in both fractions as described above and H2O2 or HOCl uptake was determined as the extraliposomal/intraliposomal fluorescence ratio.
Misc
Eduardo H Morales and Iván L Calderón contributed equally to this work
Author’s contributions
EHM and CPS conceived the project. EHM, BC and ILC performed the experiments. FG and SPo conducted partial data analysis. EHM, ILC, MM and CPS wrote the paper. All authors read and approved the final manuscript.
Contributor Information
Eduardo H Morales, Email: ed.morales@uandresbello.edu.
Iván L Calderón, Email: lcalderon@unab.cl.
Bernardo Collao, Email: b.collao@uandresbello.edu.
Fernando Gil, Email: fernandogil@unab.cl.
Steffen Porwollik, Email: sporwollik@sdibr.org.
Michael McClelland, Email: mmcclelland@sdibr.org.
Claudia P Saavedra, Email: csaavedra@unab.cl.
Acknowledgments
This work was supported by grants from FONDECYT #1085131 and #1120384 (to CPS), Universidad Andres Bello DI-34-11/R (to CPS), POSTDOC FONDECYT 3095013 (to ILC) and Universidad Andres Bello DI-50-09/R (to ILC). EHM and BC received doctoral fellowships by CONICYT and MECESUP UAB0802 additionally to EHM. We would like to thank Nicolás Pacheco for his assistance in the UFC experiments. The authors have declared that no competing interests exist. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript. Publication fees were covered by FONDECYT grant # 1120384 and from Universidad Andres Bello DI-34-11/R (to CPS).
References
- Fridovich I. The biology of oxygen radicals. Science. 1978. pp. 875–880. [DOI] [PubMed]
- Hassett D, Cohen M. Bacterial adaptation to oxidative stress: implications for pathogenesis and interaction with phagocytic cells. FASEB J. 1989;3:2574–2582. doi: 10.1096/fasebj.3.14.2556311. [DOI] [PubMed] [Google Scholar]
- Canvin J, Langford PR, Wilks KE, Kroll JS. Identification of sodC encoding periplasmic [CuZn]-superoxide dismutase in Salmonella. FEMS Microbiol Lett. 1996;136:215–220. doi: 10.1111/j.1574-6968.1996.tb08052.x. [DOI] [PubMed] [Google Scholar]
- Storz G, Imlay JA. Oxidative stress. Curr Opin Microbiol. 1999;2:188–194. doi: 10.1016/S1369-5274(99)80033-2. [DOI] [PubMed] [Google Scholar]
- Thomas E. Myeloperoxidase: Hydrogen Peroxide, Chloride Antimicrobial System: Nitrogen-Chlorine Derivatives of Bacterial Components in Bactericidal Action Against Escherichia coli. Infect Immun. 1979;23:522–531. doi: 10.1128/iai.23.2.522-531.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosen H, Crowley J, Heinecke J. Human Neutrophils Use the Myeloperoxidase-Hydrogen Peroxide-Chloride System to Chlorinate but Not Nitrate Bacterial Proteins during Phagocytosis. J Biol Chem. 2002;277:30463–30468. doi: 10.1074/jbc.M202331200. [DOI] [PubMed] [Google Scholar]
- Hampton M, Kettle A, Winterbourn C. Inside the Neutrophil Phagosome: Oxidants, Myeloperoxidase and Bacterial Killing. Blood. 1998;92:3007–3017. [PubMed] [Google Scholar]
- Imlay J. Pathways of Oxidative Damage. Annu Rev Microbiol. 2003;57:395–418. doi: 10.1146/annurev.micro.57.030502.090938. [DOI] [PubMed] [Google Scholar]
- Seaver LC, Imlay JA. Hydrogen peroxide fluxes and compartmentalization inside growingEscherichia coli. J Bacteriol. 2001;183:7182–7189. doi: 10.1128/JB.183.24.7182-7189.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sousa-Lopes A, Antunes F, Cyrne L, Marinho HS. Decreased cellular permeability to H2O2protectsSaccharomyces cerevisiaecells in stationary phase against oxidative stress. FEBS Lett. 2004;578:152–156. doi: 10.1016/j.febslet.2004.10.090. [DOI] [PubMed] [Google Scholar]
- Leyer G, Johnson E. Acid Adaptation SensitizesSalmonellaTyphimurium to Hypochlorous Acid. Appl Environ Microbiol. 1997;63:461–467. doi: 10.1128/aem.63.2.461-467.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calderón IL, Morales E, Caro NJ, Chahuán CA, Collao B, Gil F, Villareal JM, Ipinza F, Mora GC, Saavedra CP. Response regulator ArcA ofSalmonella entericaserovar Typhimurium downregulates the expression of OmpD, a porin facilitating uptake of hydrogen peroxide. Res Microbiol. 2011;162:214–222. doi: 10.1016/j.resmic.2010.11.001. [DOI] [PubMed] [Google Scholar]
- Nikaido H. Multidrug efflux pumps of gram-negative bacteria. J Bacteriol. 1996;178:5853–5859. doi: 10.1128/jb.178.20.5853-5859.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shulz GE. β-barrel membrane proteins. Curr Opin Struct Biol. 2000;10:443–447. doi: 10.1016/S0959-440X(00)00120-2. [DOI] [PubMed] [Google Scholar]
- Klebba P. The Porinologist. 2005. pp. 8232–8236. [DOI] [PMC free article] [PubMed]
- Albrecht R, Zeth K, Soding J, Lupas A, Linke D. Expression, crystallization and preliminary X-ray crystallographic studies of the outer membrane protein OmpW fromEscherichia coli. Acta Cryst. 2006;62:415–418. doi: 10.1107/S1744309106010190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong H, Patel DR, Tamm LK, Van den Berg B. The outer membrane protein OmpW forms an eight-stranded beta-barrel with a hydrophobic channel. J Biol Chem. 2006;281:7568–7577. doi: 10.1074/jbc.M512365200. [DOI] [PubMed] [Google Scholar]
- Jalajakumari MB, Manning PA. Nucleotide sequence of the geneompW, encoding a 22kDa immunogenic outer membrane protein ofVibrio cholerae. Nucleic Acids Res. 1990;18:2180. doi: 10.1093/nar/18.8.2180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bisweswar N, Nandy RK, Sarkar A, Ghose AC. Structural features, properties and regulation of the outer-membrane protein W (OmpW) ofVibrio cholerae. Microbiology. 2005;151:2975–2986. doi: 10.1099/mic.0.27995-0. [DOI] [PubMed] [Google Scholar]
- Gil F, Ipinza P, Fuentes J, Fumeron R, Villareal JM, Aspée A, Mora GC, Vásquez CC, Saavedra C. The ompW (porin) gene mediates methyl viologen (paraquat) efflux in Salmonella enterica serovar Typhimurium. Res Microbiol. 2007;158:529–536. doi: 10.1016/j.resmic.2007.05.004. [DOI] [PubMed] [Google Scholar]
- Wang S, Phillippy A, Deng K, Rui X, Li Z, Tortorello ML, Zhang W. Transcriptomic Responses ofSalmonella entericSerovars Enteritidis and Typhimurium to Chlorine-Based Oxidative Stress. Appl Environ Microbiol. 2010;76:5013–5024. doi: 10.1128/AEM.00823-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christman MF, Morgan RW, Jacobson FS, Ames BN. Positive control of a regulon for defenses against oxidative stress and some heat-shock proteins inSalmonella typhimurium. Cell. 1985;41:753–762. doi: 10.1016/S0092-8674(85)80056-8. [DOI] [PubMed] [Google Scholar]
- Greenberg JT, Monach P, Chou JH, Josephy PD, Demple B. Positive control of a global antioxidant defense regulon activated by superoxide-generating agents inEscherichia coli. Proc Natl Acad Sci. 1990;87:6181–6185. doi: 10.1073/pnas.87.16.6181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu S, Killoran PB, Fang FC, Riley LW. The global regulator ArcA controls resistance to reactive nitrogen and oxygen intermediates inSalmonella entericaserovar Enteritidis. Infect Immun. 2002;70:451–461. doi: 10.1128/IAI.70.2.451-461.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong SM, Alugupalli KR, Ram S, Akerley BJ. The ArcA regulon and oxidative stress resistance inHaemophilus influenzae. Mol Microbiol. 2007;64:1375–1390. doi: 10.1111/j.1365-2958.2007.05747.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loui C, Chang AC, Lu S. Role of the ArcAB two-component system in the resistance ofEscherichia colito reactive oxygen stress. BMC Microbiol. 2009;9:183. doi: 10.1186/1471-2180-9-183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans MR, Fink RC, Vazquez-Torres A, Porwollik S, Jones-Carson J, McClelland M, Hassan HM. Analysis of the ArcA regulon in anaerobically grown Salmonella enterica sv. Typhimurium. BMC Microbiol. 2011;21:11–58. doi: 10.1186/1471-2180-11-58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iuchi S, Matsuda Z, Fujiwara T, Lin EC. ThearcBgene ofEscherichia coliencodes a sensor-regulator protein for anaerobic repression of the arc modulon. Mol Microbiol. 1990;4:715–727. doi: 10.1111/j.1365-2958.1990.tb00642.x. [DOI] [PubMed] [Google Scholar]
- Malpica R, Franco B, Rodriguez C, Kwon O, Georgellis D. Identification of a quinone-sensitive redox switch in the ArcB sensor kinase. Proc Natl Acad Sci. 2004;101:13318–13323. doi: 10.1073/pnas.0403064101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peña-Sandoval G, Georgellis D. The ArcB Sensor Kinase ofEscherichia coliAutophosphorylates by an Intramolecular Reaction. J Bacteriol. 2010;192:1735–1739. doi: 10.1128/JB.01401-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iuchi S, Lin EC. Purification and phosphorylation of the Arc regulatory components ofEscherichiacoli. J Bacteriol. 1992;174:5617–5623. doi: 10.1128/jb.174.17.5617-5623.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Georgellis D, Lynch AS, Lin EC. In vitro phosphorylation study of the arc two-component signal transduction system ofEscherichia coli. J Bacteriol. 1997;179:5429–5435. doi: 10.1128/jb.179.17.5429-5435.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon O, Georgellis D, Lin EC. Phosphorelay as the Sole Physiological Route of Signal Transmission by the Arc Two-Component system ofEscherichia coli. J Bacteriol. 2000;182:3858–3862. doi: 10.1128/JB.182.13.3858-3862.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch AS, Lin EC. Transcriptional control mediated by the ArcA two-component response regulator protein ofEscherichia coli: characterization of DNA binding at target promoters. J Bacteriol. 1996;178:6238–6249. doi: 10.1128/jb.178.21.6238-6249.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeon Y, Lee Y, Han J, Kim J, Hwang D. Multimerization of Phosphorylated and Non-phosphorylated ArcA is Necessary for the Response Regulator Function of the Arc Two-Component Signal Transduction System. J Biol Chem. 2001;276:40873–40879. doi: 10.1074/jbc.M104855200. [DOI] [PubMed] [Google Scholar]
- Georgellis D, Kwon O, Lin EC. Quinones as the Redox Signal for the Arc Two-Component System of Bacteria. Science. 2001;292:2314–2316. doi: 10.1126/science.1059361. [DOI] [PubMed] [Google Scholar]
- Alexeeva S, Hellingwerf K, Mattos JT. Requirement of ArcA for Redox Regulation inEscherichia coliunder Microaerobic but Not Anaerobic or Aerobic Conditions. J Bacteriol. 2003;185:204–209. doi: 10.1128/JB.185.1.204-209.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bekker M, Alexeeva S, Laan W, Sawers G, Mattos JT, Hellingwerf K. The ArcBA Two-Component System ofEscherichia coliIs Regulated by the Redox State of both the Ubiquinone and the Menaquinone Pool. J Bacteriol. 2010;191:746–754. doi: 10.1128/JB.01156-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rolfe MD, Ter Beek A, Graham AI, Trotter EW. Shahzad Asif HM, SysMO-SUMO, Sanguinetti G, Teixeira de Mattos J, Poole RK, Green J: Transcript Profiling and Inference ofEscherichia coliK-12 ArcA Activity across the Range of Physiologically Relevant Oxygen Concentrations. J Biol Chem. 2011;286:10147–10154. doi: 10.1074/jbc.M110.211144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Spiegeleer P, Sermon J, Vanoirbeek K, Aertsen A, Michiels CW. Role of porins in sensitivity of Escherichia coli to antibacterial activity of the lactoperoxidase enzyme system. Appl Environ Microbiol. 2005;71:3512–3518. doi: 10.1128/AEM.71.7.3512-3518.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Münch R, Hiller K, Grote A, Scheer M, Klein J, Schobert M, Jahn D. Virtual Footprint and PRODORIC: an integrative framework for regulon prediction in prokaryotes. Bioinformatics. 2005;21:4187–4189. doi: 10.1093/bioinformatics/bti635. [DOI] [PubMed] [Google Scholar]
- Gil F, Hernández-Lucas I, Polanco R, Pacheco N, Collao B, Villareal JM, Nardocci G, Calva E, Saavedra CP. SoxS regulates the expression of theSalmonella entericaserovar TyphimuriumompWgene. Microbiology. 2009;155:2490–2497. doi: 10.1099/mic.0.027433-0. [DOI] [PubMed] [Google Scholar]
- Matsubara M, Kitaoka SI, Takeda SI, Mizuno T. Tuning of the porin expression under anaerobic growth conditions by his-to-Asp cross-phosphorelay through both the EnvZ-osmosensor and ArcB-anaerosensor inEscherichia coli. Genes Cells. 2000;5:555–569. doi: 10.1046/j.1365-2443.2000.00347.x. [DOI] [PubMed] [Google Scholar]
- Dukan S, Dadon S, Smulski DR, Belkin S. Hypochlorous Acid Activates the Heat Shock and soxRS Systems ofEscherichia coli. Appl Environ Microbiol. 1996;62:4003–4008. doi: 10.1128/aem.62.11.4003-4008.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsushika A, Mizuno T. A dual-signaling mechanism mediated by the ArcB hybrid sensor kinase containing the histidine-containing phosphotransfer domain inEscherichia coli. J Bacteriol. 1998;180:3973–3977. doi: 10.1128/jb.180.15.3973-3977.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Datsenko KA, Wanner BL. One-step inactivation of chromosomal genes inEscherichia coliK-12 using PCR products. Proc Natl Acad Sci. 2000;97:6640–6645. doi: 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfaffl MW. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001;29:e45. doi: 10.1093/nar/29.9.e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mika F, Hengge R. A btwo-component phosphotransfer network involving ArcB, ArcA, and RssB coordinates synthesis and proteolysis of σS (RpoS) in E. coli. Genes Dev. 2005;19:2770–2781. doi: 10.1101/gad.353705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rezk BM, Haenen G, van der Vijgh W, Bast A. Lipoic Acid Protects Efficiently Only against a Specific Form of Peroxynitrite-induced Damage. J Biol Chem. 2004;279:9693–9697. doi: 10.1074/jbc.M312289200. [DOI] [PubMed] [Google Scholar]
- Nikaido H, Rosenberg EY. Porin channels in Escherichia coli: studies with liposomes reconstituted from purified proteins. J Bacteriol. 1983;153:241–252. doi: 10.1128/jb.153.1.241-252.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cubillos MA, Lissi EA, Abuin EB. Kinetics of peroxidation of linoleic acid incorporated into DPPC vesicles initiated by the thermal decomposition of 2,2'-azobis(2-amidinopropane) dihydrochloride. Chem Phys Lipids. 2001;112:41–46. doi: 10.1016/S0009-3084(01)00161-X. [DOI] [PubMed] [Google Scholar]