Abstract
Congenital fibrosis of the extraocular muscles type 1 (CFEOM1) is a rare inherited strabismus syndrome characterized by non-progressive ophthalmoplegia. We previously identified that CFEOM1 results from heterozygous missense mutations in KIF21A, which encodes a kinesin motor protein. Here we evaluate the expression pattern of KIF21A in human brain and muscles of control and CFEOM1 patients, and during human and mouse embryonic development. KIF21A is expressed in the cell bodies, axons, and dendrites of many neuronal populations including those in the hippocampus, cerebral cortex, cerebellum, striatum, and motor neurons of the oculomotor, trochlear, and abducens nuclei from early development into maturity, and its spatial distribution is not altered in the CFEOM1 tissues available for study. Multiple splice isoforms of KIF21A are identified in human fetal brain, but none of the reported CFEOM1 mutations are located in or near the alternatively spliced exons. KIF21A immunoreactivity is also observed in extraocular and skeletal muscle biopsies of control and CFEOM1 patients, where it co-localizes with triadin, a marker of the excitation-contractile coupling system. The diffuse and widespread expression of KIF21A in the developing human and mouse central and peripheral nervous system as well as in extraocular muscle does not account for the restricted ocular phenotype observed in CFEOM1, nor does it permit the formal exclusion of a myogenic etiology based on expression patterns alone.
Section 1. Introduction
Congenital fibrosis of the extraocular muscles type 1 (CFEOM1) is a rare strabismus syndrome characterized by congenital non-progressive ophthalmoplegia, with both active limitation and passive restriction of globe movement. Individuals are born with bilateral ptosis and their eyes fixed downward, with the inability to elevate either eye above the midline and often with aberrant residual eye movements. Some children affected by CFEOM1 have mild hypotonia and gross motor delays, but no other associations have been found to consistently co-segregate with the eye disorder.
Traditionally, CFEOM1 was thought to be a myopathic disorder characterized by extraocular muscle (EOM) replacement with fibrous tissue (Apt and Axelrod, 1978; Brown, 1950; Crawford, 1970; Harley et al., 1978; Laughlin, 1956). A neurogenic etiology for CFEOM1 is, however, suggested by clinical signs of misinnervation (Yamada et al., 2005) and by anatomical studies of human pathological tissues from individuals affected by CFEOM1 (Engle et al., 1997). Postmortem findings in an affected adult from a large family with CFEOM1 revealed selective absence of the levator palpebrae superioris and superior rectus subnuclei of the oculomotor nucleus, and loss of the superior division of the oculomotor nerve, which is comprised of axons from these subnuclei only. Consistent with the absence of the superior division of the oculomotor nerve, there was absence of the levator palpebrae superioris muscle and reduced mass of the superior rectus muscle, which contained fat, connective tissue, and some residual myofibers. The remaining EOMs were not fibrotic. Light microscopic findings from EOM surgical biopsies of two affected members of the same large family revealed fairly normal appearing inferior and lateral recti with increased central nuclei and select fibers with central mitochondrial clumping. Together, these data suggested but did not prove that CFEOM1 is neurogenic, resulting from failure of the normal development of the superior division of the oculomotor nerve and its corresponding oculomotor subnuclei.
CFEOM1 is inherited as a dominant trait, and results from heterozygous missense mutations in KIF21A (Yamada et al., 2003). KIF21A encodes an anterograde kinesin motor protein (Marszalek et al., 1999), and the mutations underlying CFEOM1 are remarkable in that they cluster primarily in the 3rd coiled-coil domain of the KIF21A stalk, with two additional mutations reported in the motor domain (Chan et al., 2007; Lu et al., 2008; Yamada et al., 2003). Moreover, many of the mutations recur in unrelated individuals, and over 70% of probands with CFEOM1 harbor the heterozygous 2860C>T mutation resulting in a R954W amino acid substitution in the 3rd coiled-coil domain (Chan et al., 2007; Yamada et al., 2003), including those individuals whose postmortem and biopsy tissues are described above (Engle et al., 1997).
Kif21a was identified and cloned from a PCR based screen of a mouse retinal cDNA library (Marszalek et al., 1999), and its initial expression pattern was defined using an antibody directed against a mouse fusion protein that recognized a 6.2 kb transcript and a 180 kDa protein. By western blot analysis of adult mouse tissue lysates, protein expression was high in brain, intermediate in kidney and testes, low in liver, lung, and heart, and absent in spleen. By immunohistochemistry, Kif21a was found in neuronal cell bodies and processes extending from adult retinal ganglion cells, mitral cells, pyramidal cells, motor neurons, and absent from glial cells (Marszalek et al., 1999). Analysis of Kif21a mouse and KIF21A human mRNA by northern-blot and RP-PCR (Yamada et al., 2003) revealed that expression in mouse whole embryos began at ~E10.5 and then slowly decreased, while expression in mouse brain started embryonically and gradually increased. In human fetal and adult tissue, KIF21A mRNA was most abundant in brain, and within adult human brain, mRNA expression was high in cerebral cortex, cerebellum, putamen, lower in the medulla, and absent from spinal cord. Lower levels of expression were noted in liver, kidney, and heart. Notably, expression was also present in skeletal muscle.
The CFEOM1 phenotype is restricted spatially and temporally to developing ocular cranial nerves and extraocular muscles. The identification of KIF21A transcripts in most regions of the adult human brain and in skeletal muscle led us to ask about its developmental expression pattern in mouse and human, in order to determine whether this may provide insight into a myogenic versus neurogenic etiology, and help to explain the restricted CFEOM1 phenotype. Moreover, we wondered if we could detect a difference in the expression of KIF21A protein in CFEOM1 tissues compared to control tissues. Thus, we have further defined the spatial and temporal expression pattern of KIF21A/Kif21 protein in both the developing and mature nervous and skeletal system of mouse and human, and in postmortem and biopsy tissues of individuals with CFEOM1.
Section 2. Results
2.1 Immunoreactivity of Kif21a/KIF21A in mouse and human neuronal and muscle tissues
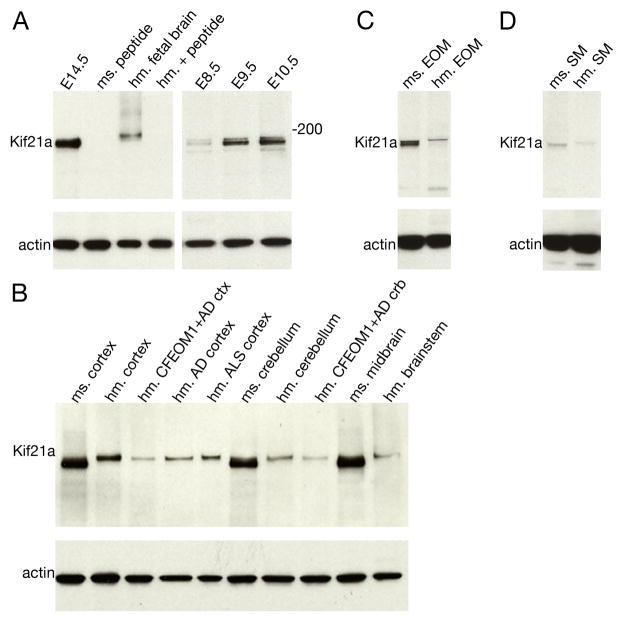
Using a polyclonal antibody generated against the C-terminus of mouse Kif21a with one amino acid mismatch against human KIF21A (Tischfield et al., 2010), we detect a ~180 kDa mouse protein with somewhat greater affinity than a ~185 kDa human protein (Fig. 1A). Analysis of protein lysates obtained from several human and mouse neuronal and non-neuronal and embryonic tissues reveal Kif21a protein expression is first detected at high levels at E9.5 in the mouse embryo (Fig. 1A) and continues to be expressed in adulthood, where it is detected at high levels in the human cerebral cortex, cerebellum, and spinal cord in the adult human and mouse brain (Fig 1B). We then examined KIF21A expression in patient tissue from an individual with CFEOM1 and Alzheimer’s disease whose autopsy was previously reported (Engle et al., 1997), and who harbored the R954W amino acid substitution in KIF21A (Yamada et al., 2003). KIF21A is reduced in lysates from the CFEOM1 cerebral cortical tissue as well as in cortical tissue from Alzheimer and ALS patients without CFEOM1 (Fig 1B). KIF21A/Kif21a is also detected in both extraocular (Fig. 1C) and, to a lesser degree, skeletal muscle (Fig. 1D) in mouse and human.
Figure 1. Western blot analysis of Kif21A antibody in mouse and human tissues.
(A) Western blot analysis using polyclonal Kif21a/KIF21A antibody (Tischfield et al., 2010) on mouse and human tissue lysates. Specific bands at 180 kDa and 185 kDa are detected from E14.5 mouse and human fetal brain tissue lysates, respectively, and both the mouse and human bands are absent following antigen competition using a 50 fold molar excess of the peptide, indicating that the antibody recognizes endogenous KIF21A/Kif21a. (B) Immunoblot with anit-KIF21A antibody in tissue lysates obtained from control mouse brain, and postmortem human brain from a control individual, and individuals with CFEOM1 and Alzheimer’s disease, Alzheimer’s disease without CFEOM1, and ALS without CFEOM1. (C and D) Western blot analysis of mouse and human extraocular muscle (C) and skeletal muscle (D) tissue lysates. KIF21A/Kif21a is detected in both muscle types, with higher signal in mouse than human, and in EOM than skeletal muscle. Actin was used a loading control. Abbreviations used are as follows: E = embryonic age; ms = mouse; hm = human, EOM = extraocular muscle; SM = skeletal muscle; AD = Alzheimer’s disease; ALS = amyotrophic lateral sclerosis; ctx = cortex; crb = cerebellum.
2.2 KIF21A splice variants in the fetal brain
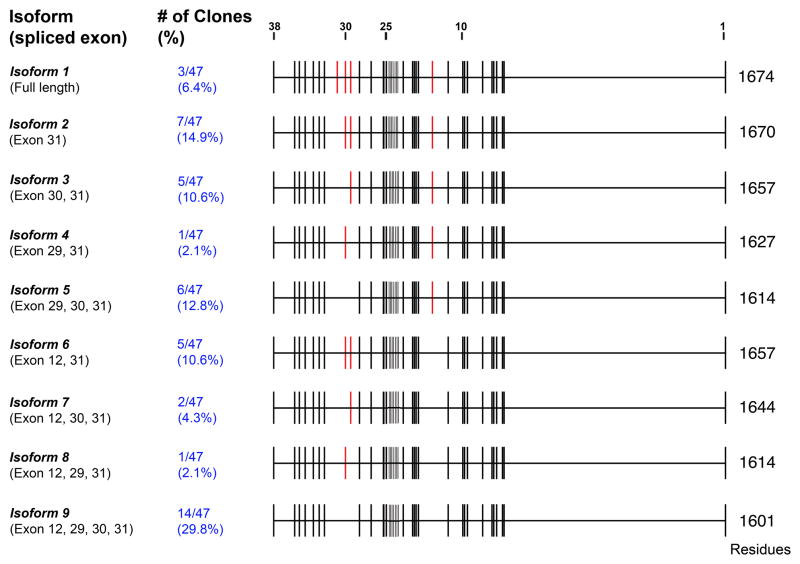
We primarily detect a single band in our western blot analysis, however occasionally a doublet is observed likely reflecting alternative splicing and/or post-transcriptional modification of KIF21A/Kif21A. In order to investigate whether KIF21A is alternatively spliced and to identify spliced isoforms, we amplified and performed sequence analysis on the ~5kb full-length KIF21A open reading frame from a human fetal brain cDNA library (Fig. 2). Sequence analysis of 47 isolated clones identifies a complex pattern of KIF21A splicing, with at least 9 isoforms resulting from alternative splicing of exon 12 and exons 29–31. The alternatively spliced exons encode regions between the first and second coil-coil domain and in the linker region between the last coil-coil and the start of the WD40 repeat domain of KIF21A. We did not find alternative splicing of any of the exons altered by CFEOM1 mutations (exons 2, 8, 20, and 21), and none of the CFEOM1 mutations identified to date are located in or near the alternatively spliced exons.
Figure 2. KIF21A splice variants by in-frame alternative splicing.
Nine splice isoforms of KIF21A were identified from sequencing of 47 cDNA clones from a 16-week human fetal brain library. Each isoform with its corresponding spliced exons is listed at the left, while the number of each clone identified and the corresponding percentage of all clones are indicated in blue text. The genomic structure of KIF21A is provided in the schematic, with each of the 38 exons denoted by a black vertical line from beginning with exon 1 on the right. Red vertical lines denote the four alternatively spliced exons 12, 29, 30, and 31. Three clones were not abled to be classified.
2.3 Kif21a/KIF21A expression in developing mouse and human brain
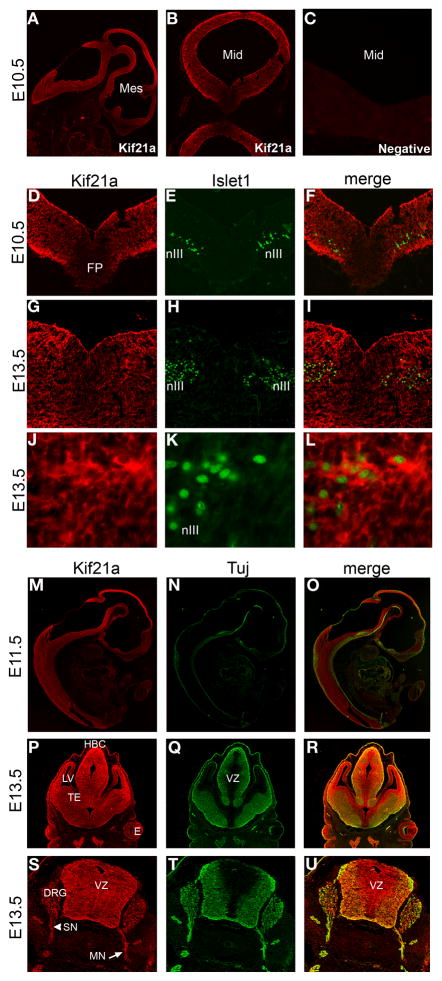
By immunohistochemistry of mouse embryonic tissues, Kif21a expression is observed in both the mitotically active ventricular zone and in post-mitotic neurons throughout the mouse central nervous system (Fig. 3). Expression is observed in a diffuse staining pattern, as shown at E10.5 (Fig. 3A,B,D–F), E11.5 (Fig. 3M–O), and E13.5 (Fig. 3G–L and P–U), and corresponding control sections incubated with secondary antibody only were negative for signal (Fig. 3C). Kif21a expression in the midbrain includes expression in the cytoplasm of oculomotor nuclei (co-labeled with Islet1), shown at E10.5 and E13.5 (Fig. 3D–I). At E13.5, expression can be seen in peripheral cranial and spinal motor and sensory nerves (Fig. 3S–U).
Figure 3. Spatial distribution of Kif21a in the developing mouse embryo.
(A–C) Sagittal and coronal sections of E10.5 mouse embryo immunostained with Kif21a (A,B) and 40X image of negative control without primary antibody (C). (D–L) Coronal sections through the midbrain of E10.5 (D–F) and E13.5 (G–L) mouse embryos co-immunostained with Kif21a and Islet1. Islet1 marks the nuclei of oculomotor neurons (nIII) seen at low power images in (D–I) and high power images 60X magnification in (J–L). (M–U) Cryosections of mouse embryos co-immunostained with Kif21a and Tuj1 antibodies. (M–O) Mid-sagittal sections through an E11.5 embryo. Kif21a protein is observed in a diffuse staining pattern throughout the developing CNS. (P–R) Coronal sections of E13.5 developing forebrain. Kif21A immunoreactivity is detected in both the ventricular and post mitotic mantle zones with only a few areas of negative staining. Kif21a is detected at higher levels in the regions of the habenular commissure and lateral ventricle. (S–U) Transverse sections through E13.5 spinal cord, highlighting expression in spinal motor and sensory axonal projections and dorsal root ganglia. All images are 10X magnification unless otherwise indicated. Abbreviations used are as follows: Mes = mesencephalon; Mid = midbrain; SC = spinal cord; FP = floor plate; nIII = oculomotor nucleus; HBC = habenular commissure; LV = lateral ventricle; TE = thalamic eminence; E = eye; VZ = ventricular zone; DRG = dorsal root ganglion; SN = DRG sensory nerve; MN = spinal motor nerve.
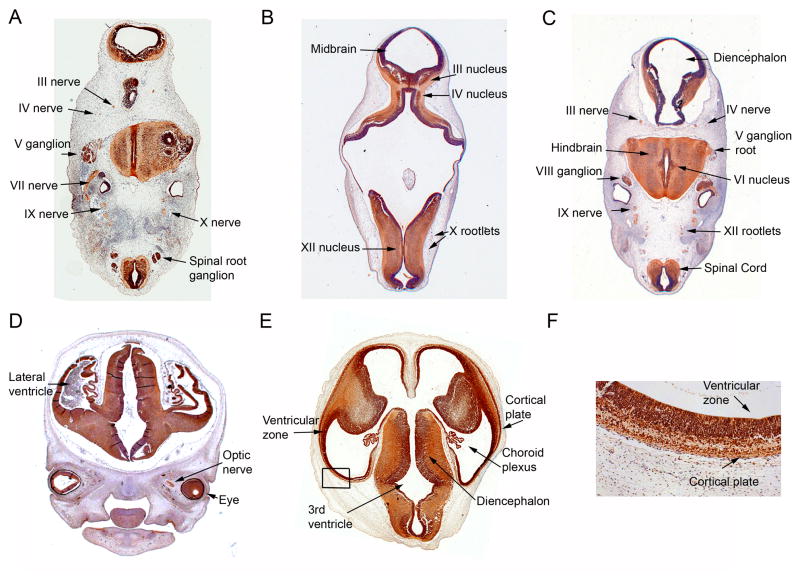
Immunohistochemistry analysis of KIF21A in CS16, CS17, and CS22 human embryo tissue indicates the expression pattern of KIF21A is similarly widespread in the developing central and peripheral nervous system of humans (Fig. 4A–F). In CS16 and CS17 human embryonic sections, KIF21A expression is not restricted to any one specific region or neuronal cell type; we observe KIF21A reactivity in both mitotic and postmitotic neurons throughout the developing CNS, and in both neuronal cell bodies and their processes (Fig. 4A–C). KIF21A expression is detected in all cranial and spinal motor (e.g. oculomotor, trochlear and abducens [Fig. 4A,C]) and sensory (e.g. vestibular [Fig. 4C]) nuclei and nerves (e.g. oculomotor and trochlear nerves [Fig. 4A–C] and optic nerve [Fig. 3D]). This expression pattern of KIF21A is maintained in later stages of human embryonic development examined (Fig. 4D–F), including in the developing cortex at CS22, where expression is seen in both the ventricular zone and in the cortical plate (Fig. 4F), layers that contain mitotic and post-mitotic cells, respectively.
Figure 4. Spatial distribution of KIF21A in the developing human nervous system.
Transverse sections of CS16 (A), CS17 (B,C), and CS22 (D–F), human embryos immunostained with anti-KIF21A/Kif21a antibody. Sections were counterstained with haematoxylin or 1% toluidine blue. KIF21A expression is detected in the structures labeled, including cranial and peripheral nerves and in mitotic and postmitotic neurons throughout the developing CNS at CS16 (A), and CS17 (B, C). (D, E) Representative sections through CS22 human fetus demonstrating KIF21A expression in the developing cortex, eye, and optic nerve. (F) Higher magnification of boxed region of (E) shows expression of KIF21A in neurons of the developing cortical plate and ventricular zone. Abbreviations used are as follows: III =oculomotor; IV = trochlear; V = trigeminal; VI = abducens; VII = facial; VIII = vestibular; IX = glossopharangeal; X = vagus; XII = hypoglossal.
2.4 KIF21A expression in the mature human nervous system
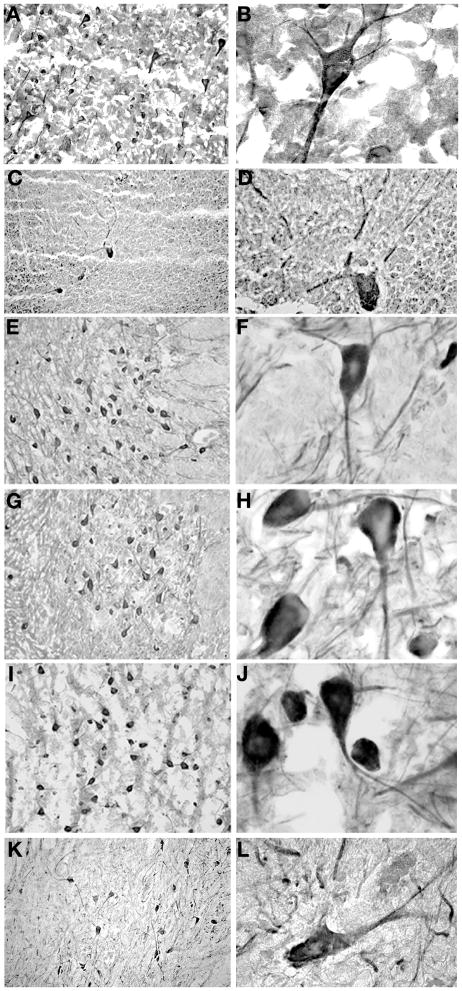
By Western blot analysis, Kif21a/KIF21A is detected at high levels in the adult mouse and human cerebral cortex, cerebellum, and spinal cord (Fig. 1B). To further assess the spatial distribution of KIF21A in the mature human brain and compare this to the published expression in mouse (Marszalek et al., 1999), we surveyed postmortem brain tissue from a 3.5 year old and a 65 year old, both of who died of non-neurological causes and did not have CFEOM. Similar to expression in mouse, we find human KIF21A to be expressed widely in neuronal cell bodies and their processes within both the central and peripheral nervous system, including all layers of the frontal and occipital cortex (Fig. 5A,B), hippocampus, striatum, and in the Purkinje cells of the cerebellum (Fig. 5C,D). Most neurons in the brainstem, including oculomotor, trochlear, and abducens motor neurons, express KIF21A (Fig. 5E–J). Throughout all levels of the spinal cord, alpha-motor neurons and their processes were immunoreactive for KIF21A (Fig 5K–L). Cerebellar granule cells (Fig. 5C,D) and neurons in the superior and inferior colliculi were among the few cell populations found to be KIF21A negative in the human brain, providing an internal control for this broadly expressed protein. No glial expression was observed and corresponding control sections incubated with secondary antibody only were negative for signal. Table 1 summarizes KIF21A expression in the mature human brainstem.
Figure 5. Spatial distribution of KIF21A in the mature human nervous system.
Immunohistochemistry using anti-KIF21A antibody of control human postmortem brain sections from cortex, cerebellum, brainstem, and spinal cord. (A–B) KIF21A immunoreactivity in neuronal cell bodies, axons, and dendrites of the occipital cortex. (C–D) Immunoreactivity in Purkinje cells and their processes in the cerebellum. Cerebellar granular cells are negative for KIF21A. (E–L) Immunoreactivity in motor neurons and their processes of the oculomotor (E, F), trochlear (G, H) and abducens (I, J) nuclei in the brainstem, and anterior horn cells (K–L) of the spinal cord. (A, C, E, G, I, and K are 20X magnification and B, D, F, H, J, and L are 100X magnification).
Table 1.
| Structure | KIF21A expression | Signal |
|---|---|---|
|
Midbrain Level of superior colliculus |
• Superior Colliculus | (negative) |
| • Round Cells | + | |
| • Oculomotor nucleus | ++ | |
| • Mesencephalic trigeminal nucleus | ++ | |
| • Edinger-Westphal | + | |
| • Substantia Nigra | + | |
| • Red Nucleus | ++ | |
| Level of inferior colliculus | • Inferior colliculus | (negative) |
| • Dorsal nucleus of raphe | + | |
| • Trochlear nucleus | ++ | |
| • Interpeducular nucleus | + | |
| • Mesencephalic trigeminal (sensory) | + | |
| • Substantia Nigra | + | |
| Cerebellum | • Granule Cells | (negative) |
| • Purkinje Cells | + | |
|
Pons Level of cranial nerve VI and VII |
• Abducens nucleus | ++ |
| • Facial nucleus | ++ | |
| • Pontine nucleus | + | |
| • Reticular formation | + | |
| • Raphe pontine | + | |
| • Ventral gray matter | + | |
|
Medulla Level of cranial nerve X and XII |
• Hypoglossal nucleus | + |
| • Dorsal efferent nucleus of Vagus | + | |
| • Solitary tract | + | |
| • Nucleus ambiguous | + | |
| • Cuneate nucleus | + | |
| • Inferior olivary nucleus | + | |
2.5 KIF21A/Kif21a expression in extraocular and skeletal muscles
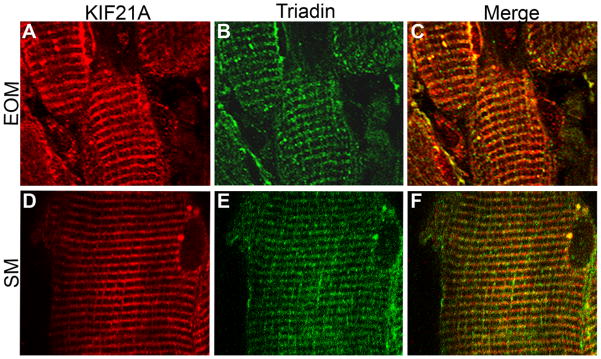
Extraocular muscle (EOM) is fundamentally different from other skeletal muscles (Porter and Baker, 1996; Porter et al., 1995; Porter et al., 2001b; Spencer and McNeer, 1980) with a unique gene expression profile (Porter et al., 2001a). Thus, we examined KIF21A expression in human skeletal and extraocular muscle biopsies and postmortem tissues to determine if differences may account for the selective CFEOM1 phenotype. By western blot analysis, Kif21a/KIF21A is expressed in both extraocular and skeletal muscles of both mouse and human (Fig. 1C,D). By immunofluorescence, Kif21a has a sarcomeric expression pattern in both muscle types (Fig. 6A–F), and its expression does not differ among different EOMs. Co-staining with antibodies to components of the sarcomere, including myosin, tropomyosin, and actinin, and to a component of the calcium-coupling system, triadin, we find that Kif21a/KIF21A co-localizes most precisely with triadin in both muscle isotypes (Fig. 6A–F).
Figure 6. Spatial distribution of KIF21A in human extraocular and skeletal muscle tissues.
Longitudinal cryostat sections of extraocular (EOM) and skeletal (SK) muscles obtained via biopsy of individuals without CFEOM are immunofluorescently labeled with antibodies to KIF21A (A, D) and triadin (B, E) and photographed at 60X magnification. KIF21A and triadin co-localize to the membrane system between sarcomeres in EOM (C) and SK (F) muscles.
2.6 Expression of KIF21A in CFEOM1 tissue
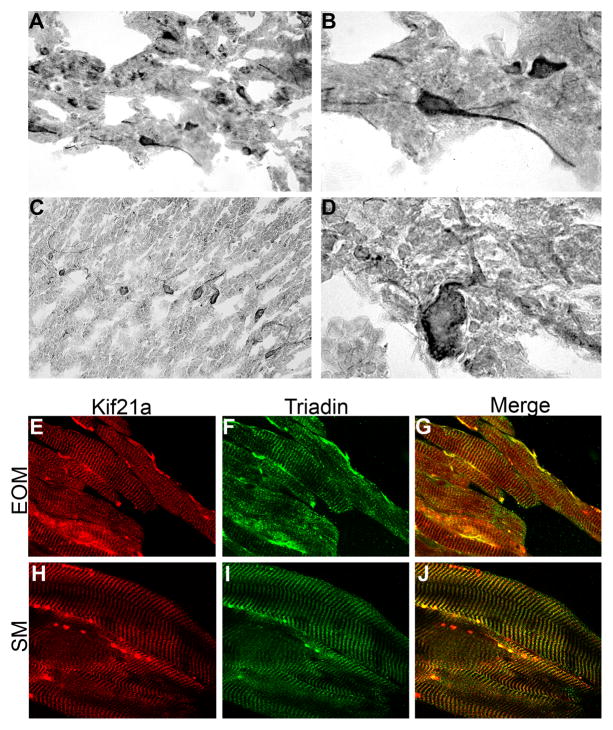
To determine if we can detect differences in KIF21A expression in individuals with CFEOM1 who harbor KIF21A missense mutations compared to controls, we examined KIF21A expression in postmortem brain tissue, and in extraocular and skeletal muscle biopsies obtained from patients who harbored the KIF21A R954W amino acid substitution (Engle et al., 1997; Yamada et al., 2003), and compared these to the expression patterns described above. First, we found that the size of the KIF21A protein is unchanged by Western blot analysis (Fig 1B). Next, we examined the spatial pattern of KIF21A immunoreactivity in the cerebral cortex, hippocampus, striatum, and cerebellum from postmortem CFEOM1 tissue (Engle et al., 1997; Yamada et al., 2003) and found that expression did not appear to differ between CFEOM1 and control tissue (Fig 7A–D). CFEOM1 postmortem brainstem and spinal cord were not available for study. Finally, we examined the EOM and skeletal muscles from CFEOM1 biopsy and postmortem tissues. KIF21A co-localizes with triadin, and no differences were found in its distribution in CFEOM1 EOM and skeletal muscles compared with controls (Fig 7E–J).
Figure 7. Spatial distribution of mutant KIF21A in human CFEOM1-diseased brain and muscles.
Post-mortem (A–D) and biopsy (E–J) tissues from individuals with CFEOM1 who harbor the KIF21A exon 21 2860 C>T (R954W) mutation. Similar to control tissues, immunostaining of KIF21A is present in neuronal cell bodies and processes in the cerebral cortex (A,B) and Purkinje cells (C,D) of the cerebellum at 20X and 100X magnification. Longitudinal cryostat sections of EOM (E–G) and SK (H–J) muscle biopsies immunostained with KIF21A and triadin reveal similar spatial distribution to control muscles at 60X magnification.
Section 3. Discussion
We find Kif21a/KIF21A to be widely expressed in neurons of both the central and peripheral nervous system during development and in maturity, and identify no spatial restriction of human expression compared to mouse that might account for the human CFEOM1 phenotype. Moreover, we do not detect a difference in size or distribution of KIF21A in tissues from individuals who harbor the CFEOM1 R954W amino acid substitution, nor could we detect differences in the localization of KIF21A by immunohistochemistry between normal and CFEOM1 diseased brain and muscle tissues. These data indicate CFEOM1 is not likely to result from gross changes in the spatial or temporal distribution of KIF21A. Although we did observe decreases in KIF21A protein levels in CFEOM1 patients compared to healthy controls, the unknowns of harvesting, handling, and processing of postmortem tissue and the co-affliction of Alzheimer’s in this patient confound quantitative analysis of these data. We cannot, however, formally rule out a specific reduction in KIF21A protein level as a result of the KIF21A R954W substitution.
Kif21a has been demonstrated to move in an anterograde direction in axons (Marszalek et al., 1999), and to interact with Kank1 and Big1 in vitro (Kakinuma and Kiyama, 2009; Li et al., 2011; Shen et al., 2008). Interestingly, we have also found KIF21A/kif21a to co-localize with triadin in both extraocular and skeletal muscles. Triadin is located at the triad formed by sarcoplasmic reticulum to both sides of a T-tubule, and serves as a component of the muscle’s excitation-contraction coupling mechanism. The only other kinesin with reported subcellular localization in skeletal muscle is Kif3b (Ginkel and Wordeman, 2000); it too co-localizes with triadin and has been proposed to assemble and maintain these membrane structures in myogenic cells by participating in the transport and/or recycling of specialized membrane components. Thus, our data support an additional role for KIF21A in the calcium coupling system and muscle physiology.
In conclusion, the role of KIF21A in the development and maintenance of neurons and muscle and how these roles may relate to the pathogenesis of CFEOM1 remain unknown. The expression pattern of KIF21A/Kif21a in both neurons and muscles of human and mouse does account for the restricted ocular phenotype observed in patients with CFEOM1 who harbor mutation in the KIF21A gene, and does not permit the formal exclusion of a myogenic etiology for this rare disorder. Because Kif21a/KIF21A expression does not appear to vary between mouse and human, generation of appropriate Kif21a mouse models should provide insight into specific features of developing ocular motor neurons and extraocular muscles and help to elucidate the etiology of CFEOM1 and the role of KIF21A in health and disease.
Experimental Procedures
Antibody Production
We previously generated a polyclonal rabbit anti-Kif21a affinity purified antibody by producing a synthetic peptide corresponding to the N terminal mouse Kif21a amino acids (aa) 1655–1674 (NH2-{C}NLQDGQLSDTGDLGEDIASN-COOH)/human KIF21A aa 1653–1672 with an L/I mismatch at aa 1654 (Tischfield et al., 2010). The peptide was purified by HPLC, conjugated to KLH and injected to immunize rabbits. The antiserum was affinity purified and ELISA testing by Bethyl Laboratories (www.bethyl.com) and the affinity purified antibodies were filtered with a final concentration of 1 mg/ml.
Human Tissue
The collection and processing of postmortem brain and muscle tissues from a 71-year old with CFEOM1 and Alzheimer’s Disease, and extraocular and skeletal muscle biopsy tissue from individuals with CFEOM1, all of whom harbored the R954W amino acid substitution, as well as control autopsy tissue from individuals who died from unrelated disorders were previously reported (Engle et al., 1997). The study was conducted with IRB approval from Children’s Hospital Boston and appropriate informed consent was obtained.
Human embryos ranging in age from Carnegie Stage (CS) 16 to CS23 (approximately 37–56 days post-conception, respectively) were obtained from the MRC/Wellcome-Trust funded Human Developmental Biology Resource at Newcastle University (HBDR, http://www.hdbr.org), with appropriate maternal written consent and approval from the Newcastle and North Tyneside NHS Health Authority Joint Ethics Committee. HDBR is regulated by the UK Human Tissue Authority (HTA; www.hta.gov.uk) and operates in accordance with the relevant HTA Codes of Practice.
Western Blot Analysis
Under a Children’s Hospital Boston IACUC approved protocol, mouse tissue/embryos of varying ages were harvested, protein was isolated and protein concentration was determined using the Protein Assay Kit (Sigma) according to the manufacture’s recommendations. Immunoblotting was performed by loading equal amount of protein in each lane, separating protein samples by 3–8% Tris-actetate gradient gels (Invitrogen), and transferring to a nitrocellulose membrane. Blots were blocked then incubated overnight with either 1:5000 or 1:10,000 dilutions of anti-KIF21A antibody followed by 1:5000 dilution of HRP-conjugated secondary antibody and visualized using Chemiluminescence Reagent Plus (Perkin-Elmer).
Alternative Splicing of KIF21A
The various alternative isoforms of KIF21A were amplified from 16-week human fetal brain cDNA library (Clontech) with LA Taq (Takara). RT-PCR was performed in a 12-μl reaction using Advantage2 Polymerase Mix (Clontech). The ~5kb full-length KIF21A ORF was amplified using primers to either side of the open reading frame (KIF5′UTR2F, 5′-TCAGCTCTGCGGTGCCGAGG -3′;KIF5′UTR2R, 5′-TCTGGGTACTGGCAGTTGTCCGGGAT -3′), and isolated clones were sequenced by ABI 377 DNA sequencer (PE-Applied Biosystems).
Immunohistochemistry
Mouse Embryos
Mouse Embryos were fixed in 4% paraformaldehyde and embedded in OCT for cryo-sectioning. Frozen sections at 8–10um were cut on a Leica 3050S cryostat, and thaw-mounted on charged superfrost slides. Sections were blocked with 1–5% normal goat/donkey serum in 0.1% Triton/PBS at room temperature for 1 hr and incubated overnight with primary antibody (anti-Kif21a 1:1500, Anti-Tuj 1/1500, and Islet-1 1:300) in blocking buffer at 4°C. After removal of primary antibody, 484 and 594 conjugated anti-rabbit and anti mouse (Molecular Probes) secondary antibodies were added at a 1/2000 dilution at room temperature for 1 hr. Sections were mounted with Vectashield (Vector Labs) for microscopy.
Human Embryos
Samples were fixed overnight at 4°C in 0.1M phosphate buffered saline (PBS) containing 4% paraformaldehyde (PFA; Sigma Aldrich, Poole, UK) before short-term storage at 4°C in 70% ethanol. Placental tissue for karyotype analysis was sampled prior to fixation of the tissue. The stage of development was assessed on the basis of external features according to the Carnegie staging protocol (O’Rahilly and Muller, 1987) modified for use with individual embryonic samples (Bullen and Wilson, 1997). Positive and negative controls from quality checked embryos were included in each experiment. Control anti-Ki67 antibody was used on a known quality checked embryo as a positive control. Briefly, paraffin sections were dewaxed and were rinsed in two changes in absolute ethanol. Endogenous peroxidase activity was blocked with methanol peroxide solution for 10 min at RT. Sections were then rinsed in TBS and incubated with 10% normal serum in TBS for 10 min at RT. Primary anti-KIF21A antibody was added in normal serum blocking solution in TBS at a 1:1500 dilution and incubated for 1–2 hr at RT, or overnight at 4°C. Sections were washed and incubated with Goat or Horse anti- rabbit secondary antibody in normal serum in TBS at 1/500 dilution for 30 min at RT. Sections were then incubated with tertiary complex utilizing standard VECTASTAIN Elite ABC kit PK6100 in TBS for 30 min at RT and developed with Sigma DAB kit (D-4293) for 10 min at RT. Sections were then counterstained with haematoxylin or 1% toluidine blue (2mins) then washed, dehydrated, cleared, and mounted for light microscopy.
Human Adult Brain Tissue
Frozen sections from postmortem and biopsy human tissue were cut on a Leica 3050S cryostat at 10um to 20um thickness, thaw-mounted on SuperFrost*/Plus slides and then left to dry at RT (slides were stored at −80 °C). All steps were then carried out at RT in a humidified chamber. Frozen sections were post fixed with 4% paraformaldehyde for 30 min. Sections were hydrated with three 15 min washes in PBS and endogenous peroxidase activity was quenched with 0.5% hydrogen peroxide solution in methanol for 30 min. After three additional washes in PBS sections were blocked with 10% goat or fetal calf serum in PBS with 0.1% Trition X 100 (Sigma) for 1 hr. Primary anti-KIF21A antibody at a 1:400 dilution in blocking buffer was applied to sections overnight at 4°C. The sections were washed three times in PBS for 15 min and then incubated for 1 hr with biotin or horseradish peroxidase conjugated secondary antibodies (Jackson Immuno) at a dilution of 1:1000. Sections were then drained and washed with PBST (PBS, 0.05% Tween-20) × 3 and signal visualization was observed by DAB Fast (Sigma) or Vectastain ABC reagent (Vector Labs) according to manufacturer’s recommendations.
Human Muscle Immunocytochemistry
Surgical and postmortem extraocular muscle biopsies were restricted to the distal segments and did not include the orbital layer. Skeletal muscle biopsies were taken from the quadriceps and postmortem muscle from the psoas or quadriceps. Longitudinal sections 10–20um in thickness were cut on a Leica 3050S cryostat. Sections were washed briefly with PBS, post-fixed with 4% PFA and washed 3 times in PBS. Sections were subsequently blocked for 1 hr with 20% HI-FCS (heat inactivated fetal calf serum) in PBST (PBS, 0.05% Tween-20). Primary antibody (1:400 anti-KIF21A and 1:1000 anti-Triadin) was diluted in PBST- 5% normal blocking serum and sections were incubated overnight at 4 C in primary antibody. The following day, slides were washed with PBST-20% HIFCS and PBS and incubated with secondary antibody fluorescently labeled with FITC or Cy3 at 1:400 dilution in PBS 5% FCS at RT. Slides were drained and washed was with PBST and mounted with Vectashield mounting media.
Highlights.
KIF21A, the protein product of the KIF21A gene mutated in Congenital Fibrosis of the
Extraocular Muscles type 1 (CFEOM1), is widely expressed in developing and mature neurons and muscle in both human and mouse.
KIF21A co-localizes with triadin in skeletal and extraocular muscle.
The spatial pattern of KIF21A expression in available brain and muscle tissues from individuals with CFEOM1 does not vary from control tissues.
The expression pattern of KIF21A does not account for the restricted CFEOM1 phenotype nor does it clarify if the primary etiology of CFEOM1 is neurogenic or myogenic.
Acknowledgments
We thank Dr. Hannah C. Kinney for critical review and assistance in interpretation of brainstem expression patterns. Supported by R01 EY013583 (ECE) and the Children’s Hospital Boston Intellectual and Developmental Disabilities Research Center (2P30HD018655). The human embryonic and fetal material was provided by the Joint MRC (grant # G0700089)/Wellcome Trust (grant # GR082557) Human Developmental Biology Resource (http://hdbr.org). ECE is an investigator of the Howard Hughes Medical Institute.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Contributor Information
Jigar Desai, Email: jdesai@enders.tch.harvard.edu.
Marie Pia Rogines Velo, Email: mroginesvelo@tuftsmedicalcenter.org.
Koki Yamada, Email: kyamada-ngs@umin.ac.jp.
Lynne M Overman, Email: lynne.overman@newcastle.ac.uk.
References
- Apt L, Axelrod RN. Generalized fibrosis of the extraocular muscles. Am J Ophthalmol. 1978;85:822–829. doi: 10.1016/s0002-9394(14)78112-7. [DOI] [PubMed] [Google Scholar]
- Brown HW. Congenital structural muscle anomalies. In: Allen JH, editor. Strabismus Ophthalmic Symposium. C. V. Mosby Co; St. Louis: 1950. pp. 205–236. [Google Scholar]
- Bullen P, Wilson D. The carnegie staging of human embryos: a practical guide. In: Strachan T, Lindsay S, Wilson DI, Strachan T, Lindsay S, Wilson DI, editors. Molecular Genetics of Early Human Development. Bios Scientific Publishers Ltd; 1997. [Google Scholar]
- Chan WM, Andrews C, Dragan L, Fredrick D, Armstrong L, Lyons C, Geraghty MT, Hunter DG, Yazdani A, Traboulsi EI, Pott JW, Gutowski NJ, Ellard S, Young E, Hanisch F, Koc F, Schnall B, Engle EC. Three novel mutations in KIF21A highlight the importance of the third coiled-coil stalk domain in the etiology of CFEOM1. BMC genetics. 2007;8:26. doi: 10.1186/1471-2156-8-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crawford JS. Congenital fibrosis syndrome. Can J Ophthalmol. 1970;5:331–336. [PubMed] [Google Scholar]
- Engle EC, Goumnerov BC, McKeown CA, Schatz M, Johns DR, Porter JD, Beggs AH. Oculomotor nerve and muscle abnormalities in congenital fibrosis of the extraocular muscles. Ann Neurol. 1997;41:314–325. doi: 10.1002/ana.410410306. [DOI] [PubMed] [Google Scholar]
- Ginkel LM, Wordeman L. Expression and partial characterization of kinesin-related proteins in differentiating and adult skeletal muscle. Mol Biol Cell. 2000;11:4143–4158. doi: 10.1091/mbc.11.12.4143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harley RD, Rodrigues MM, Crawford JS. Congenital fibrosis of the extraocular muscles. J Pediatr Ophthalmol Strabismus. 1978;15:346–358. doi: 10.3928/0191-3913-19781101-04. [DOI] [PubMed] [Google Scholar]
- Kakinuma N, Kiyama R. A major mutation of KIF21A associated with congenital fibrosis of the extraocular muscles type 1 (CFEOM1) enhances translocation of Kank1 to the membrane. Biochem Biophys Res Commun. 2009 doi: 10.1016/j.bbrc.2009.06.109. [DOI] [PubMed] [Google Scholar]
- Laughlin RC. Congenital fibrosis of the extraocular muscles; a report of six cases. Amer J Ophthalmol. 1956;41:432–438. doi: 10.1016/0002-9394(56)91259-4. [DOI] [PubMed] [Google Scholar]
- Li CC, Kuo JC, Waterman CM, Kiyama R, Moss J, Vaughan M. Effects of brefeldin A-inhibited guanine nucleotide-exchange (BIG) 1 and KANK1 proteins on cell polarity and directed migration during wound healing. Proc Natl Acad Sci U S A. 2011 doi: 10.1073/pnas.1117011108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu S, Zhao C, Zhao K, Li N, Larsson C. Novel and recurrent KIF21A mutations in congenital fibrosis of the extraocular muscles type 1 and 3. Arch Ophthalmol. 2008;126:388–394. doi: 10.1001/archopht.126.3.388. [DOI] [PubMed] [Google Scholar]
- Marszalek JR, Weiner JA, Farlow SJ, Chun J, Goldstein LS. Novel dendritic kinesin sorting identified by different process targeting of two related kinesins: KIF21A and KIF21B. J Cell Biol. 1999;145:469–479. doi: 10.1083/jcb.145.3.469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Rahilly R, Muller F. Developmental stages in human embryos. Carnegie Institute Publication no 637 1987 [Google Scholar]
- Porter J, Baker R. Muscles of a different ‘color’: The unusual properties of the extraocular muscles may predispose or protect them in neurogenic and myogenic disease. Neurology. 1996;46:30–37. doi: 10.1212/wnl.46.1.30. [DOI] [PubMed] [Google Scholar]
- Porter J, Baker R, Ragusa R, Brueckner J. Extraocular muscles: basic and clinical aspects of structure and function. Surv Ophthalmol. 1995;39:451–484. doi: 10.1016/s0039-6257(05)80055-4. [DOI] [PubMed] [Google Scholar]
- Porter J, Khanna S, Kaminski H, et al. Extraocular muscle is defined by a fundamentally distinct gene expression profile. Proc Natl Acad Sci USA. 2001a doi: 10.1073/pnas.211257298. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porter JD, Khanna S, Kaminski HJ, Rao JS, Merriam AP, Richmonds CR, Leahy P, Li J, Andrade FH. Extraocular muscle is defined by a fundamentally distinct gene expression profile. Proc Natl Acad Sci U S A. 2001b;98:12062–12067. doi: 10.1073/pnas.211257298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen X, Meza-Carmen V, Puxeddu E, Wang G, Moss J, Vaughan M. Interaction of brefeldin A-inhibited guanine nucleotide-exchange protein (BIG) 1 and kinesin motor protein KIF21A. Proc Natl Acad Sci U S A. 2008;105:18788–18793. doi: 10.1073/pnas.0810104105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spencer R, McNeer K. Structural alterations in overacting inferior oblique muscles. Arch Ophthalmol. 1980;98:128–133. doi: 10.1001/archopht.1980.01020030130015. [DOI] [PubMed] [Google Scholar]
- Tischfield MA, Baris HN, Wu C, Rudolph G, Van Maldergem L, He W, Chan WM, Andrews C, Demer JL, Robertson RL, Mackey DA, Ruddle JB, Bird TD, Gottlob I, Pieh C, Traboulsi EI, Pomeroy SL, Hunter DG, Soul JS, Newlin A, Sabol LJ, Doherty EJ, de Uzcategui CE, de Uzcategui N, Collins ML, Sener EC, Wabbels B, Hellebrand H, Meitinger T, de Berardinis T, Magli A, Schiavi C, Pastore-Trossello M, Koc F, Wong AM, Levin AV, Geraghty MT, Descartes M, Flaherty M, Jamieson RV, Moller HU, Meuthen I, Callen DF, Kerwin J, Lindsay S, Meindl A, Gupta ML, Jr, Pellman D, Engle EC. Human TUBB3 mutations perturb microtubule dynamics, kinesin interactions, and axon guidance. Cell. 2010;140:74–87. doi: 10.1016/j.cell.2009.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada K, Andrews C, Chan WM, McKeown CA, Magli A, De Berardinis T, Loewenstein A, Lazar M, O’Keefe M, Letson R, London A, Ruttum M, Matsumoto N, Saito N, Morris L, Monte MD, Johnson RH, Uyama E, Houtman WA, De Vries B, Carlow TJ, Hart BL, Krawiecki N, Shoffner J, Vogel MC, Katowitz J, Goldstein SM, Levin AV, Sener EC, Ozturk BT, Akarsu AN, Brodsky MC, Hanisch F, Cruse RP, Zubcov AA, Robb RM, Roggenkaemper P, Gottlob I, Kowal L, Battu R, Traboulsi EI, Franceschini P, Newlin A, Demer JL, Engle EC. Heterozygous mutations of the kinesin KIF21A in congenital fibrosis of the extraocular muscles type 1 (CFEOM1) Nat Genet. 2003;35:318–321. doi: 10.1038/ng1261. [DOI] [PubMed] [Google Scholar]
- Yamada K, Hunter DG, Andrews C, Engle EC. A novel KIF21A mutation in a patient with congenital fibrosis of the extraocular muscles and Marcus Gunn jaw-winking phenomenon. Arch Ophthalmol. 2005;123:1254–1259. doi: 10.1001/archopht.123.9.1254. [DOI] [PubMed] [Google Scholar]