Abstract
B lymphocytes, like all mammalian cells, are equipped with the unfolded protein response (UPR), a complex signaling system allowing for both pro- and mal-adaptive responses to increased demands on the endoplasmic reticulum (ER). The UPR is comprised of three signaling pathways initiated by the ER transmembrane stress sensors, IRE1α/β, PERK and ATF6α/β. Activation of IRE1 yields XBP1(S), a transcription factor that directs expansion of the ER and enhances protein biosynthetic and secretory machinery. XBP1(S) is essential for the differentiation of B lymphocytes into antibody-secreting cells. In contrast, the PERK pathway, a regulator of translation and transcription, is dispensable for the generation of antibody-secreting cells. Functioning as a transcription factor, ATF6α can augment ER quality control processes and drive ER expansion, but the potential role of this UPR pathway in activated B cells has not been investigated. Here, we report studies of ATF6α-deficient B cells demonstrating that ATF6α is not required for the development of antibody-secreting cells. Thus, when B cells are stimulated to secrete antibody, a specialized UPR relies exclusively on the IRE1-XBP1 pathway to remodel the ER and expand cellular secretory capacity.
Keywords: B lymphocytes, plasma cells, endoplasmic reticulum, unfolded protein response, ATF6α
1. Introduction
The differentiation of B lymphocytes into antibody-secreting plasma cells is fundamental to the generation of humoral immunity (Calame et al., 2003). This developmental process is marked by substantial increases in rough endoplasmic reticulum (ER) and in production of immunoglobulin (Ig) heavy and light chains (Tagliavacca et al., 2003; Wiest et al., 1990), the building blocks of functional antibodies. The ER is a multi-functional organelle, serving as a platform for protein and lipid biosynthesis and as an oxidative, chaperone-rich protein folding compartment (Lykidis and Jackowski, 2001; van Anken and Braakman, 2005). As such, the ER is the site for Ig synthesis, folding and assembly. Thus, when B cells are stimulated to secrete antibody, the escalation in Ig production increases demands on the protein biosynthetic and folding capacity of the ER. The mechanisms by which differentiating B cells cope with these challenges and how such mechanisms influence antibody output, the efficiency of humoral responses and the lifespan of antibody-secreting cells are not fully understood.
All eukaryotic cell types are equipped with the unfolded protein response (UPR) signaling system that can assist in balancing client protein load with ER capacity (Walter and Ron, 2011). The UPR consists of three distinct pathways triggered by the ubiquitously expressed ER transmembrane proteins inositol-requiring enzyme 1 (IRE1α/β) (Tirasophon et al., 1998; Wang et al., 1998), protein kinase RNA activated (PKR)-like ER kinase (PERK) (Harding et al., 1999; Shi et al., 1998) and activating transcription factor 6 (ATF6α/β) (Haze et al., 2001; Haze et al., 1999). IRE1α is ubiquitously expressed (Tirasophon et al., 1998), whereas IRE1β is restricted to the gut epithelium (Wang et al., 1998). When IRE1 is activated, its cytoplasmic endoribonuclease domain initiates a novel mRNA splicing mechanism that modifies X-box binding protein 1 (XBP1) transcripts to encode XBP1(S) (Calfon et al., 2002; Shen et al., 2001; Yoshida et al., 2001). As a transcriptional activator, XBP1(S) up-regulates expression of many secretory pathway proteins including factors involved in entry of nascent polypeptides into the ER, protein folding, ER-associated degradation (ERAD) and vesicular transport (Shaffer et al., 2004; Sriburi et al., 2007). Also, XBP1(S) regulates lipid biosynthesis and plays a key role in directing expansion of rough ER (Lee et al., 2005; Shaffer et al., 2004; Sriburi et al., 2007; Sriburi et al., 2004). Therefore, the IRE1-XBP1 pathway can enhance cellular biosynthetic and secretory capacity. Upon PERK activation, its cytoplasmic serine/threonine kinase domain phosphorylates the α subunit of eukaryotic initiation factor 2 (eIF2α). This modification effectively attenuates total protein synthesis, thereby reducing the flow of new client polypeptides into the ER (Harding et al., 1999; Shi et al., 1998). The ATF6α and ATF6β proteins are activated upon transport to the Golgi where they undergo intramembrane cleavage by the site 1 and site 2 proteases (Ye et al., 2000). This event liberates the ATF6 cytosolic domain from the membrane, allowing it to traffic into the nucleus and function as a basic leucine zipper transcription factor. Both ATF6α and ATF6β are capable of regulating expression of certain ER stress-responsive genes (Haze et al., 2001; Haze et al., 1999). However, studies of gene knockout mouse embryo fibroblasts indicate that ATF6α, but not ATF6β, is required for induction of various ER resident molecular chaperones, folding assistants and ERAD components under conditions of ER stress (Wu et al., 2007; Yamamoto et al., 2007). In addition to enhancing ER quality control mechanisms (Adachi et al., 2008), ATF6α has the capacity to promote lipid biosynthesis and ER biogenesis (Bommiasamy et al., 2009; Maiuolo et al., 2011).
Signaling through Toll-like receptor 4 (Peng, 2005), bacterial lipopolysaccharide (LPS) activates mouse B cells to proliferate and to differentiate into antibody-secreting cells, a process involving increased expression of Ig heavy and light chains, elevated synthesis of membrane lipids and expansion of the rough ER (Fagone et al., 2009; Fagone et al., 2007; Lewis et al., 1985; Rush et al., 1991; van Anken et al., 2003; Wiest et al., 1990). Thus, LPS-stimulated differentiation provides an appropriate in vitro system in which to investigate the role of the UPR in the generation of antibody-secreting B cells. During this developmental process, the IRE1-XBP1 pathway is activated (Calfon et al., 2002; Gass et al., 2002) and XBP1(S) plays essential roles in driving ER expansion, Ig synthesis and antibody secretion (Hu et al., 2009; Iwakoshi et al., 2003; McGehee et al., 2009; Shaffer et al., 2004; Tirosh et al., 2005; Todd et al., 2009). In contrast, there is very little PERK activation in LPS-stimulated B cells (Gass et al., 2008; Ma et al., 2010) and the PERK pathway is not required for antibody secretion (Gass et al., 2008). ATF6α is activated in LPS-stimulated B cells (Brunsing et al., 2008; Gass et al., 2002; Gass et al., 2008), suggesting that this UPR transcription factor might participate in the expansion of the secretory machinery when B cells transition into antibody secretion. To investigate this hypothesis, we assessed the differentiation of ATF6α-deficient B cells in vitro and evaluated humoral immune responses in ATF6α-deficient mice.
2. Materials and methods
2.1. Animals
ATF6α knockout mice, as previously described (Yamamoto et al., 2007), are of a C57BL/6 background and are deleted for exons 8 and 9 which encode the entire basic leucine zipper domain and the majority of the ATF6α transmembrane domain. Adult mice (6 to 12 weeks of age) were used for experiments. Maintenance of breeding colonies and all procedures involving mice were perfomed according to protocols approved by the University of South Alabama Institutional Animal Care and Use Committee.
2.2. Cell culture
Splenic B cells were isolated from mice using erythrocyte lysis and positive selection with the MACS B cell isolation kit (Miltenyi Biotec, Auburn, CA). Cells were cultured in RPMI-1640 supplemented as described (Gass et al., 2002) at 1×106c/ml and stimulated with 10μg/ml LPS (E. coli 055:B5, Sigma, St. Louis, MO) for various intervals. Cells were counted using trypan blue dye exclusion to determine viability.
2.3. Flow cytometry and assessment of lymphocyte populations
Spleen preparations were subjected to erythrocyte lysis, and single cell suspensions of splenic mononuclear cells were subsequently stained with combinations of the following antibody conjugates: FITC anti-CD21/CD35 (clone 7G6), FITC anti-CD8α (53-6.7), PE anti-CD93 (AA4.1), PE anti-CD138 (281–2), PE-Cy7 anti-CD23 (B3B4), PE-Cy7 streptavidin, APC anti-IgM (Nov-41), Alexa 647 anti-GL7 (GL7), Pacific Blue anti-CD45R (RA3-6B2), biotin anti-IgD (11–26), biotin anti-CD45.2 (104), APC-Cy7 anti CD4 (GK1.5) and streptavidin (eBioscience, San Diego, CA and BD Biosciences, San Jose, CA). Peritoneal lavage cells were stained with FITC anti-CD11b (M1/70), PE anti-CD5 (53-7.3), in addition to anti-B220, anti-CD23, anti-IgD, and anti-IgM antibodies. Sample acquisition was performed using a FACSCanto II (BD Biosciences), using visible light scatter properties to distinguish lymphocytes (R1 = lymphocyte gate). Data were analyzed using FlowJo software (Tree Star Ashland, OR).
2.4. Analysis of IgM secreted in vitro
At various intervals of LPS stimulation, B cells were counted, washed twice in warm media and then replated at defined cell densities for various intervals. Culture supernatants were then harvested and assessed for IgM by ELISA as described (Gass et al., 2002).
2.5. RNA isolation, quantitative real-time RT-PCR and analysis of Xbp1 mRNA splicing
Total RNA was extracted from cells using the RNeasy® Plus Mini Kit (Qiagen, Valencia, CA). Equivalents amounts of RNA were reverse transcribed into cDNA using the Improm-II™ Reverse Transcription System (Promega, Madison, WI). Real-time PCR was performed using a C1000™ Thermocycler with a CFX96 Optic Module Real-Time Detection system (Bio-Rad, Hercules, CA). Reactions were done in triplicate using the IQ™ SYBR® Green Supermix (Bio-Rad) and the following mouse-specific forward and reverse primers: 5′-TAGAAAGAAAGCCCGGATGAGCGA-3′ and 5′-GTGTCCATTCCCAAGCGTGTTCTT-3′ (total Xbp1) 5′–CCCGCCTCACATTGAAATCC–3′ and 5′– GCGTATGTATCAGTCTCAGTGG–3′ (β2M); 5′-AGACTGCTGAGGCGTATTTGGGAA-3′ and 5′-CAGCATCTTTGGCTTGTCGCT-3′ (Hspa5); 5′-CACTCAAATCGAACACGGCTTGCT-3′ and 5′-AGAAGATTCCGCCTCCTTTCTGCT-3′ (Hsp90b1); 5′-TTGGCTTCATGTTTGGAGGAACCC-3′ and 5′-CTGGCCACAGGCTTGTTTCTAACT-3′ (Dnajb11); 5′-AAGTACTCGCAAGCAGCAAACAGC-3′ and 5′-TATCTCGCCCAGTCAATGAACGCT-3′ (Ero1lb). Transcript levels were normalized to β2- microglobulin mRNA levels (ΔCT) and the normalized data were used to determine changes in gene expression (2−ΔΔCT). Analysis of UPR-mediated splicing of Xbp1 mRNA splicing was performed as described (Gunn et al., 2004) using RT-PCR and primers that flank the 26 nt intron, yielding products of 237 and 211 nts from unspliced and spliced Xbp1 mRNA, respectively.
2.6 Preparation of cell extracts and immunoblotting
Cell lysates were prepared using lysis buffer containing 0.5% NP-40, 0.5% DOC, 150mM NaCl, 50mM Tris pH 7.5, sodium azide, 10 mM β-glycerol phosphate, and 1μl/ml protease inhibitor cocktail (Sigma). Clarified lysates from equivalent number of cells/sample were combined with an equal volume of 2x sample buffer (125mM Tris-HCl, pH 6.8, 10% 2-mercaptoethanol, 20% glycerol, 4% SDS, and 0.02% bromophenol blue) and separated by electrophoresis in 10% SDS-polyacrylamide gels. Proteins were electrophoretically transferred to Immobilon-P membranes (Millipore, Billerica, MA) using a CAPS-buffered system and placed in blocking buffer [PBS pH 7.4, 5% non-fat milk, 0.1% Tween-20 (PBS-T)]. Immunoblotting was performed using rabbit antibodies specific for BiP, GRP94, TRAPα, Ig μ heavy chain, Ig κ light chain (Gass et al., 2002; Gunn and Brewer, 2006), a rabbit anti-ATF6α antibody (kindly provided by Dr. Laurie H. Glimcher, Weill Cornell Medical College, New York, NY) and a mouse anti-β-actin antibody (Sigma) as primary reagents and horseradish peroxidase-conjugated donkey antibodies specific for either rabbit IgG or mouse IgG as secondary reagents (Jackson Immuno Research, West Grove, PA). Chemiluminescence signals were detected using Super Signal West Dura (Thermo Scientific, Rockford, IL) and captured using a Fuji-LAS-1000 imaging system (Fujifilm, Tokyo, Japan).
2.7. Analysis of ER abundance
On day 3 of LPS stimulation, splenic B cells were counted, washed in Hanks Balanced Salt Solution (HBSS) and then cultured at 1×106 c/ml in HBSS containing 1μM ER-Tracker™ Green (glibenclamide BODIPY® FL) (Molecular Probes™, Invitrogen, Eugene, OR) for 25 min. Stained cells were washed with PBS, resuspended in PBS and analyzed by flow cytometry using a Canto II (BD Biosciences, San Jose, CA).
2.8. Quantification of phosphatidylcholine
Cells (2 × 107) were pelleted by centrifugation, flash frozen and stored at −80°C until analysis. Lipids were isolated and then detected and quantified by flame ionization as described (Fagone et al., 2007).
2.9. Analysis of hapten-specific antibody responses
For analysis of T cell-dependent antibody responses, mice were immunized intraperitoneally with 50 μg of the hapten conjugate NP5-KLH (4-hydroxy-3-nitrophenylacetic-keyhole limpet hemocyanin) precipitated in alum and received a booster immunization (50μg NP5-KLH in alum) 3 weeks later. Sera samples were obtained by tail bleeds at day 0, 3 weeks post-immunization and at 3 and 24 weeks postboost. For analysis of T cell-independent antibody responses, mice were immunized intraperitoneally with 50 μg of the hapten conjugate NP-Ficoll (NP-aminoethylcarboxymethyl-FICOLL) (Biosearch Technologies, Novato, CA) in PBS. Sera samples were obtained by tail bleeds at day 0 and 3 weeks postimmunization. High-affinity NP-specific antibodies were assessed by ELISA using NP5-BSA as a capture reagent, biotinylated goat anti-mouse IgM, biotinylated goat anti-mouse IgG, streptavidin-horseradish peroxidase (HRP) and HRP-conjugated goat anti-mouse IgG3 antibodies (Southern Biotechnology Associates, Birmingham, AL). Serial dilutions of sera were assayed and absorbance signals ≥ 2 times background (pre-immune sera) were used to determine titers of anti-NP antibodies.
2.10. Statistical analysis
Statistical differences between groups were assessed using the Student’s t-test. A 95% confidence interval was considered statistically significant, and for all data sets P > 0.05. In figures, error bars represent mean ± standard deviation (S.D.).
3. Results and Discussion
3.1. Expression of ATF6α in B cells
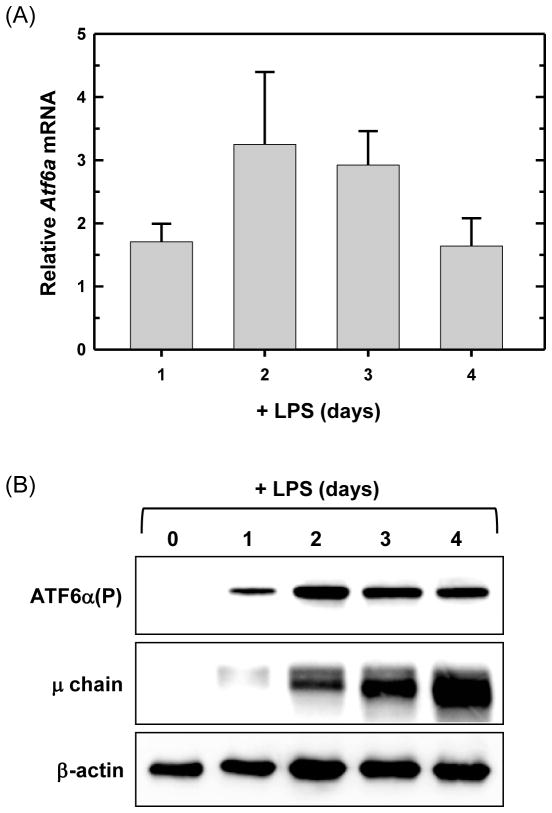
The active form of ATF6α which traffics into the nucleus (ATF6α(N)) is detectable in LPS-stimulated splenic B cells (Brunsing et al., 2008; Gass et al., 2002; Gass et al., 2008). Building on these data, we assessed the expression of ATF6α in B cells. We found that Atf6α mRNA is present in freshly isolated, resting splenic B cells and is up-regulated upon LPS stimulation (Fig. 1A). Interestingly, by our immunoblot assay, the full-length, precursor form of ATF6α (ATF6α(P)) is extremely difficult to detect in resting B cells, but increases substantially upon LPS stimulation (Fig. 1B). This is in sharp contrast to the IRE1α and PERK proteins, which are both readily detectable in resting B cells and remain at relatively constant levels over the course of LPS stimulation (Gass et al., 2008; Zhang et al., 2005). These data, coupled with previous evidence for ATF6α activation in activated B cells, suggested that the ATF6α branch of the UPR might play a vital role in the development of antibody-secreting B cells.
Fig. 1.
Expression of ATF6α in resting and LPS-stimulated splenic B cells. Splenic B cells were isolated from wild-type mice and cultured in the presence of LPS for the indicated intervals. (A) Atf6a expression was assessed by quantitative real-time RT-PCR (qRT-PCR). Data are plotted as the level of Atf6a mRNA in LPS-stimulated cells relative to that in freshly isolated, resting cells (set at 1) (mean ± S.D., n = 3). (B) Immunoblot analysis of ATF6α(P) (full-length, precursor form), Ig μ heavy chain as a positive control for differentiation and β-actin as a control for loading and sample integrity. Data are from a representative experiment out of three repeats.
3.2. Lymphocyte development in ATF6α-deficient mice
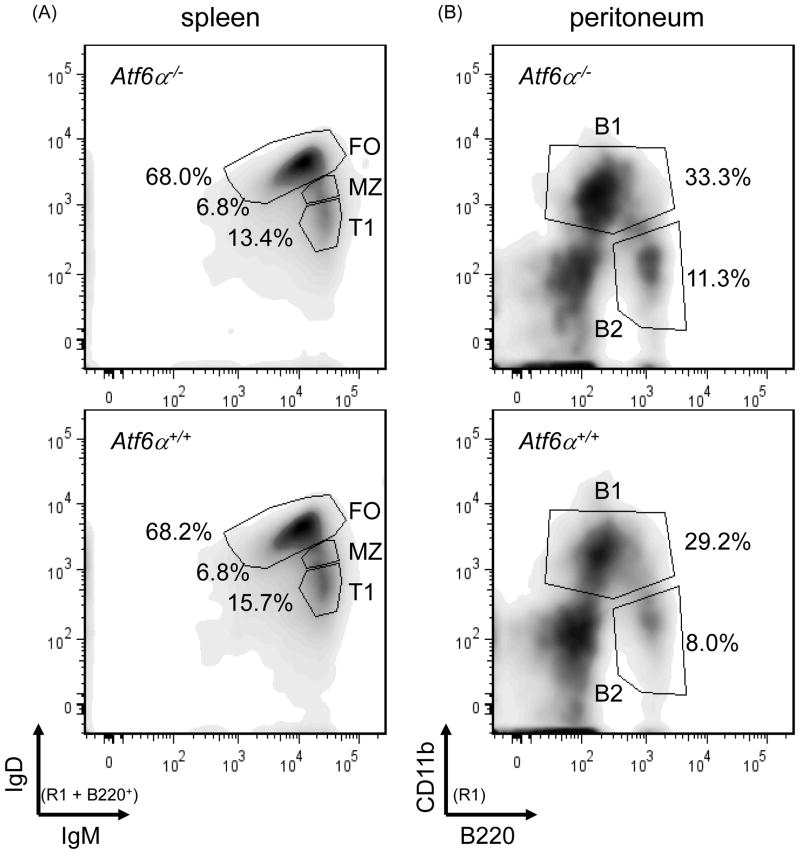
To further investigate the potential roles of ATF6α in B cells and humoral immunity, we utilized ATF6α knockout mice in which the entire basic leucine zipper domain and the majority of the transmembrane domain of ATF6α were removed by gene-targeted deletion (Yamamoto et al., 2007). As previously published (Wu et al., 2007; Yamamoto et al., 2007), ATF6α-deficient mice are generally healthy and do not exhibit obvious phenotypes under normal conditions. Flow cytometry analysis of splenocytes revealed that the frequencies of immature transitional (T1) B cells (IgMhighIgD−/low), mature follicular (FO) B cells (IgMlowIgDhigh) and marginal zone (MZ) B cells (IgMhighIgDlow) in the spleen are comparable in wild-type and ATF6α-deficient mice (Fig. 2, left panels and Table 1). Likewise, the frequencies of B1 (CD11b+) and B2 (Cd11b−) cells in the peritoneal cavity (Fig. 2, right panels and Table 1) and of mature CD4+ and CD8+ T cells in the spleen are normal in ATF6α-deficient mice (Table 1). These data demonstrate that ATF6α is not required for lymphocyte development.
Fig. 2.
Flow cytometry analysis of lymphocyte populations in wild-type and ATF6α-deficient mice. Splenocytes and peritoneal cells were isolated from age- and gender-matched Atf6a+/+ and Atf6a−/− mice, stained with fluorochrome-labeled antibodies specific for cell surface markers that discriminate lymphocyte populations and analyzed by flow cytometry (R1 = lymphocyte gate as determined by visible light scatter). (A) B220+ splenocytes were assessed for levels of IgM and IgD to identify follicular (FO), marginal zone (MZ) and transitional T1 subpopulations. These subsets were further analyzed for levels of CD21, CD23, and CD93 to validate their identity and/or to identify additional subsets (Table 1 and data not shown). (B) Peritoneal cells were stained with B220 and CD11b to reveal B1 and B2 B cells. Five mice for each genotype were analyzed and representative density plots are shown.
Table 1.
Lymphocyte populations in wild-type and ATF6α-deficient mice. Frequencies (percent of total lymphocytes) of major B and T cell populations in the spleen and peritoneal cavity. Results represent mean ± standard error. No significant differences were observed between ATF6α-sufficient and -deficient mice.
| Genotype | Spleen
|
Peritoneum
|
||||||
|---|---|---|---|---|---|---|---|---|
| % B220 | B220-positive
|
%CD4 | %CD8 | %B1 | %B2 | |||
| %FO | %MZ | %Transitional | ||||||
| Atf6α+/+ (n = 5) | 57.2 ± 7.2 | 68.8 ± 1.2 | 7.7 ± 0.9 | 8.5 ± 2.4 | 15.8 ± 5.3 | 7.5 ± 4.3 | 31.8 ± 5.3 | 12.4 ± 4.6 |
| Atf6 α−/− (n = 5) | 59.5 ± 6.1 | 67.6 ± 4.9 | 7.9 ± 0.8 | 8.7 ± 2.3 | 14.8 ± 5.1 | 7.7 ± 4.1 | 31.1 ± 8.6 | 12.9 ± 3.4 |
3.3. In vitro differentiation of LPS-stimulated ATF6α-deficient B cells
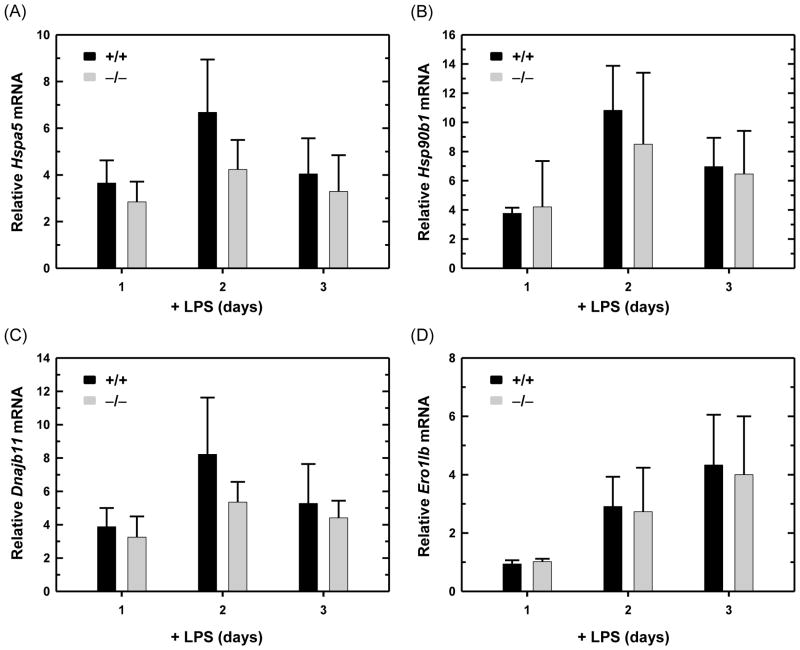
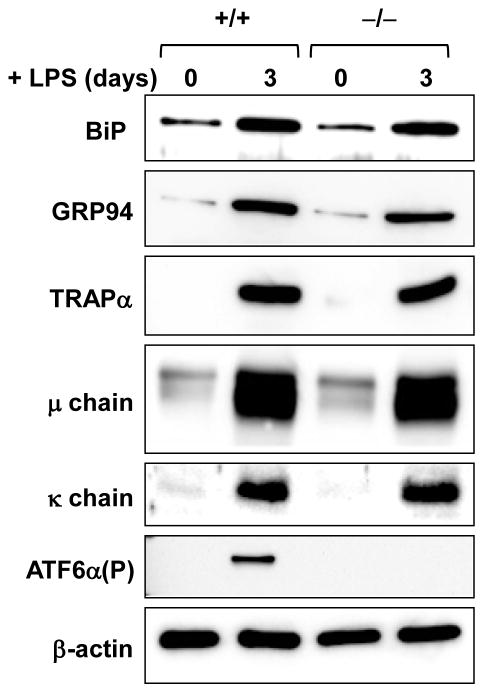
Normal lymphocyte development in ATF6α knockout mice made it possible for us to explore the functionality of ATF6α-deficient B cells. We began by utilizing the in vitro system of LPS-induced differentiation of antibody-secreting B cells. First, we assessed the expression of several genes encoding ER proteins that are up-regulated in the differentiation process. We found that transcripts for Hspa5 (encodes the ER chaperone BiP/GRP78), Hsp90b1 (encodes the ER chaperone GRP94), Dnajb11 (encodes the ER chaperone co-factor ERdj3) and Ero1lb (encodes the ER folding enzyme ERO1-like β) increase similarly in wild-type and ATF6α-deficient B cells cultured in the presence of LPS (Fig. 3). This correlates with comparable increases in the abundance of ER proteins in LPS-stimulated wild-type and ATF6α-deficient B cells as revealed by immunoblot analysis of BiP/GRP78, GRP94 and TRAPα, a transmembrane component of the ER translocon (Fig. 4). These data demonstrate that ATF6α is not essential for enhancement of ER protein folding machinery during the differentiation of antibody-secreting B cells.
Fig. 3.
Expression of ER protein folding machinery in LPS-stimulated wild-type and ATF6α-deficient B cells. Splenic B cells were isolated from Atf6a+/+ and Atf6a−/− mice and cultured in the presence of LPS for the indicated intervals. Expression of (A) Hspa5, encodes ER chaperone BiP/GRP78, (B) Hsp90b1, encodes ER chaperone GRP94, (C) Dnajb11, encodes ER chaperone co-factor ERdj3 and (D) Ero1lb, encodes ER folding enzyme ERO1-like β was assessed by qRT-PCR. Data are plotted as the level of each target mRNA in LPS-stimulated cells relative to that in freshly isolated, resting cells (set at 1) (mean ± S.D., n = 4).
Fig. 4.
Expression of ER proteins and Ig chains in LPS-stimulated wild-type and ATF6α-deficient B cells. Splenic B cells were isolated from Atf6a+/+ and Atf6a−/− mice and cultured in the presence of LPS for the indicated intervals. Immunoblotting was performed for the ER proteins BiP/GRP78 and GRP94, the ER translocon component TRAPα, Ig μ heavy and κ light chains, ATF6α(P) as an internal control for genotyping and β-actin as a control for loading and sample integrity. Data are from a representative experiment out of three repeats.
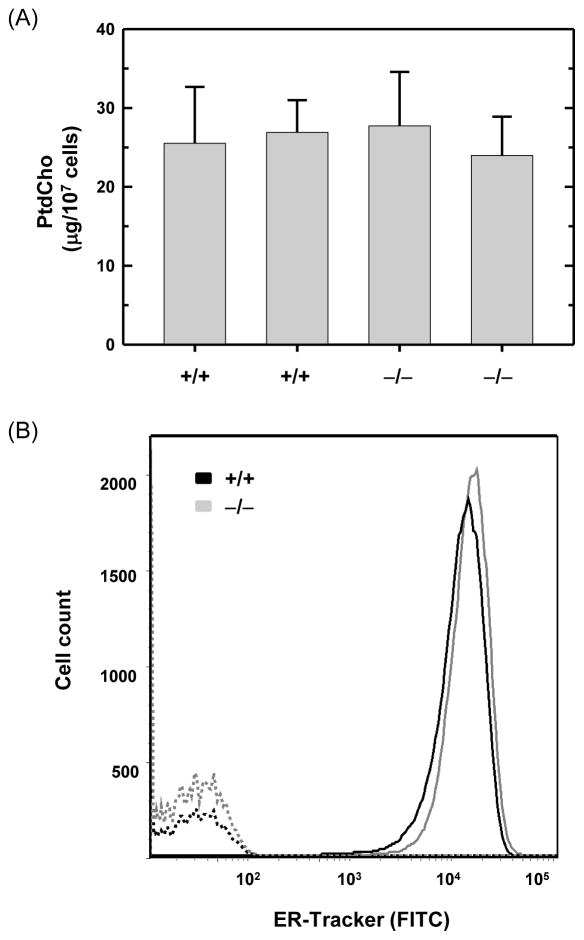
When B cells are stimulated to secrete antibody, proliferation of the rough ER requires an increased supply of membrane lipids as well ER resident proteins. Indeed, the synthesis and abundance of phosphatidylcholine (PtdCho), the major phospholipid in cellular membranes including ER membranes, increases markedly in LPS-stimulated B cells (Fagone et al., 2009; Fagone et al., 2007; Rush et al., 1991). We found similar levels of PtdCho in wild-type and ATF6α-deficient B cells that had been stimulated with LPS for three days (Fig. 5A). This correlated with comparable levels of ER as revealed by staining with ER-Tracker™, an ER-specific fluorescent dye (Fig. 5B). Therefore, while ATF6α has the ability to enhance PtdCho synthesis and ER biogenesis (Bommiasamy et al., 2009; Maiuolo et al., 2011), this UPR transcription factor is not required for either of these critical processes during the differentiation of antibody-secreting B cells.
Fig. 5.
Phosphatidylcholine levels and ER abundance in LPS-stimulated wild-type and ATF6α-deficient B cells. Splenic B cells were isolated from Atf6a+/+ and Atf6a−/− mice and cultured in the presence of LPS for 3 days. (A) Lipids were extracted from equivalent numbers of cells and the amount of phosphatidylcholine (PtdCho) was determined. The experiment was performed in duplicate using B cells from two mice of each genotype; data are plotted as mean ± S.D. of triplicate determinations. (B) Cells were left unstained (hashed lines) or stained with ER-Tracker™ (solid lines) and analyzed by flow cytometry for ER abundance. Data are from a representative experiment out of two repeats.
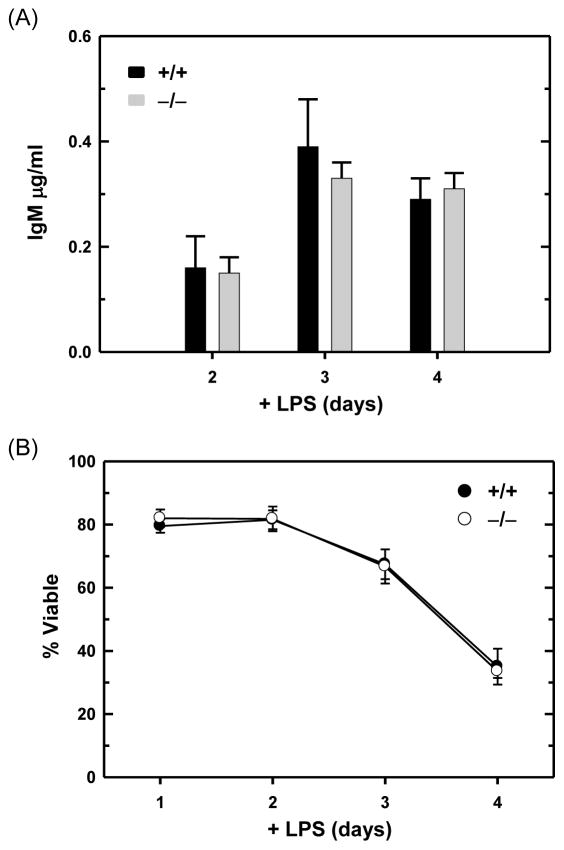
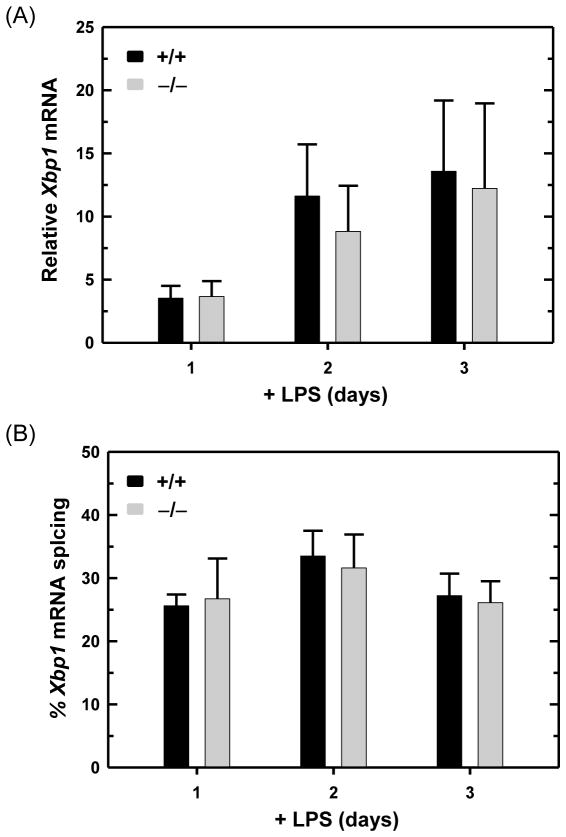
LPS-stimulated mouse B cells strongly up-regulate Ig expression, primarily synthesizing Ig μ heavy chains and κ light chains and secreting IgM antibodies. Consistent with the analysis of ER protein folding machinery and ER abundance (Figs. 3, 4 and 5), we found that wild-type and ATF6α-deficient B cells express similar levels of Ig μ and κ chains (Fig. 4) and secrete comparable amounts of IgM (Fig. 6A) in response to LPS activation. In addition, the viability of ATF6α-deficient B cells over four days of LPS stimulation was indistinguishable from that of wild-type B cells (Fig. 6B). These data establish that ATF6α is not essential for either development or survival of antibody-secreting B cells in vitro. Finally, the expression of Xbp1 mRNA (Fig. 7A) and its IRE1-mediated splicing (Fig. 7B) were up-regulated similarly in LPS-stimulated wild-type and ATF6α-deficient B cells. These findings argue that the normal differentiation and survival of ATF6α-deficient antibody B cells in vitro (Figs. 3, 4, 5, 6 and 7) is not due to compensatory effects of increased XBP1(S).
Fig. 6.
Antibody secretion and viability of LPS-stimulated wild-type and ATF6α-deficient B cells. Splenic B cells were isolated from Atf6a+/+ and Atf6a−/− mice and cultured in the presence of LPS. (A) On the indicated days, cells were harvested, washed and replated at equivalent densities for either 2 or 3 h. Culture supernatants were then collected and assessed for IgM content by ELISA; data are plotted as the amount of IgM secreted by 5×105 cells/ml/h (mean ± S.D., n = 5). (B) At the indicated intervals, cell viability was assessed by trypan blue dye exclusion (mean ± S.D., n ≥ 5 for each interval).
Fig. 7.
Expression and UPR-mediated splicing of Xbp1 mRNA in LPS-stimulated wild-type and ATF6α-deficient B cells. Splenic B cells were isolated from Atf6a+/+ and Atf6a−/− mice and cultured in the presence of LPS for the indicated intervals. (A) The level of total Xbp1 mRNA was assessed by qRTPCR. Data are plotted as the level of Xbp1 mRNA in LPS-stimulated cells relative to that in freshly isolated, resting cells (set at 1) (mean ± S.D., n = 3). (B) The percentage of Xbp1 transcripts modified by UPR-mediated splicing (mean ± S.D., n = 3) was determined by resolving RT-PCR products amplified from unspliced and spliced Xbp1 mRNA (237 and 211 nts, respectively) by gel electrophoresis followed by imaging and quantification.
3.4. In vivo antibody responses in ATF6α-deficient mice
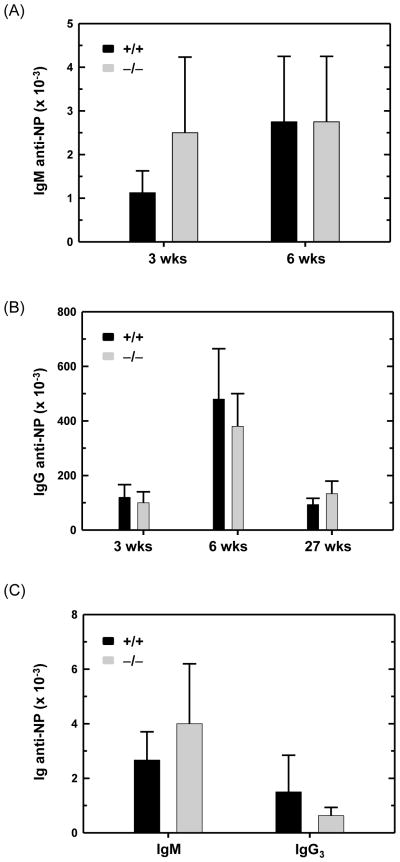
We then investigated the capacity of ATF6α-deficient B cells to differentiate into antibody-secreting plasma cells in vivo. Antibody responses to protein antigens are primarily mediated by follicular B cells and require the assistance of CD4+ T helper cells which promote Ig isotype switching and memory B cell development (Allman and Pillai, 2008; Jacob et al., 1991). The marginal zone and B1 subsets of B lymphocytes are responsible for responding to T cell-independent antigens, such as bacterial polysaccharides, and produce IgM and, in smaller amounts, IgG3 (Martin and Kearney, 2001; Martin and Kearney, 2002; Martin et al., 2001). Thus, we immunized mice with haptenated T cell-dependent and - independent antigens and measured specific antibody responses by hapten-specific ELISA. In response to the T cell-dependent antigen NP-KLH, wild-type and ATF6α-deficient mice generated similar primary responses (3 weeks post-immunization) including IgM anti-NP (Fig. 8A) and IgG anti-NP (Fig. 8B) antibodies. Upon re-challenge with NP-KLH, both genotypes of mice exhibited robust secondary responses (6 weeks post-immunization), yielding comparable titers of IgM anti-NP (Fig. 8A) and IgG anti-NP (Fig. 8B) antibodies. These data indicate that ATF6α is not essential for antibody secretion by antigen-activated follicular B cells in vivo. Moreover, these findings reveal that T cell help, development of memory B cells and isotype switching proceed normally in ATF6α-deficient mice. Importantly, the titer of IgG anti-NP antibodies several months (27 weeks) post-immunization with NP-KLH was analogous in wild-type and ATF6α-deficient mice (Fig. 8B), suggesting that ATF6α is not required for either plasma cell longevity or the maintenance of humoral immunity in vivo. In response to the T cell-independent antigen NP-Ficoll, wild-type and ATF6α-deficient mice mounted comparable IgM and IgG3 anti-NP antibody responses (Fig. 8C). These results indicate that ATF6α is not required for antibody secretion by antigen-activated marginal zone B cells and B1 cells in vivo. In agreement with these studies, we observed similar concentrations of total IgM and IgG in the serum of non-immunized wildtype and ATF6α-deficient mice (Supplemental Fig. S1).
Fig. 8.
Antigen-specific antibody responses in wild-type and ATF6α-deficient mice. Age- and gender-matched Atf6a+/+ and Atf6a−/− mice were immunized intraperitoneally with haptenated antigens. Blood samples were obtained at various intervals, and serial dilutions of sera were assayed by hapten-specific ELISA. Absorbance signals ≥ 2 times background (pre-immune sera) were used to determine titers of anti-NP antibodies. (A and B) For the T cell-dependent antigen NP5-KLH, mice received a primary immunization and a booster immunization 3 weeks later. Serum samples were collected at 3, 6 and 27 weeks (wks) post-primary immunization and analyzed for titers of (A) IgM and (B) IgG anti-NP antibodies (mean ± S.D., n = 4). (C) For the T cell-independent antigen NP-Ficoll, serum samples were harvested 3 weeks post-immunization and analyzed for titers of IgM and IgG3 anti-NP antibodies (mean ± S.D., n = 6).
4. Conclusions
The UPR transcription factor ATF6α has been implicated in the physiology of several cell types including hepatocytes (Cinaroglu et al., 2011; Yamamoto et al., 2010; Wang et al., 2009; Wu et al., 2007), dopaminergic neurons (Egawa et al., 2011), skeletal muscle cells (Wu et al., 2011), pancreatice β-cells (Teodoro et al., 2011) and dormant tumor cells (Schewe and Aguirre-Ghiso, 2008). The ability of ATF6α to up-regulate many genes associated with ER quality control processes (Adachi et al., 2008), promote lipid synthesis and drive ER biogenesis (Bommiasamy et al., 2009; Maiuolo et al., 2011), coupled with the strong induction of ATF6α expression in LPS-stimulated B cells (Fig. 1), suggested that this UPR pathway might also play a critical role when B cells are stimulated to secrete antibody. However, our studies of ATF6α-deficient B cells demonstrate that ATF6α, like the PERK pathway, is dispensable for B cell development and the differentiation of antibody-secreting B cells.
It is important to note that questions remain regarding ATF6α and its possible role in B cells. First, our analysis of gene expression in ATF6α-deficient B cells was restricted to a subset of UPR-regulated targets. A more comprehensive assessment might reveal that ATF6α does contribute to the regulation of certain genes, such as additional ERAD components or factors involved in lipid metabolism, in activated B cells. Next, while our data indicate that Ig production is not compromised in ATF6α-deficient B cells, it should not be assumed that the synthesis and maturation of all secretory pathway proteins proceeds normally under these conditions. Further analysis of ER protein folding, including the level of chaperone-associated proteins, in ATF6α-deficient B cells that have been stimulated to secrete antibody and/or treated with pharmacologic agents that disrupt the ER environment is warranted. Finally, we cannot exclude the possibility that ATF6β might play a compensatory role in ATF6α-deficient B cells. ATF6α-and ATF6β-single knockout mice develop normally, whereas ATF6α/β-double knockout animals exhibit embryonic lethality (Yamamoto et al., 2007). Thus, ATF6α and ATF6β can compensate for each other, at least during certain stages of embryonic development. Whether such compensatory activities occur in cell types such as B lymphocytes remains to be investigated. To this end, we have found that Atf6β mRNA is expressed at similar levels in freshly isolated, resting wild-type and ATF6α-deficient B cells and is comparably up-regulated in response to LPS in both B cell genotypes (data not shown). However, it would certainly be interesting to assess the level of ATF6β(N) protein in activated wild-type and ATF6α-deficient B cells and, potentially, to employ a conditional deletion approach to generate B cells lacking both ATF6α and ATF6β.
In summary, our findings further support the idea that a specialized, limited UPR is utilized when B cells are stimulated to secrete antibody (Gass et al., 2002; Gass et al., 2008; Ma et al., 2010; Masciarelli et al., 2010). In this normal, developmental process, a physiologic UPR featuring the IRE1-XBP1 pathway effectively copes with the escalation of Ig synthesis and antibody assembly necessary for high-rate antibody output. The possibility that the entire UPR or distinct UPR pathways might play significant roles in dysfunctional B cells involved in pathophysiologic processes such as autoimmunity or malignancy awaits further study.
Supplementary Material
Fig. S1. Levels of IgM and IgG in serum of non-immunized ATF6α-deficient mice. Serum samples were collected from non-immunized Atf6a+/+ and Atf6a−/− mice (n = 11 for each genotype). The concentrations of IgM and IgG were determined by ELISA using purified mouse IgM and IgG as standards. Data are plotted as the level of serum IgM and IgG in Atf6a−/− mice relative to that of Atf6a+/+ mice, expressed as percentage of wild-type control (mean ± S.D.).
Highlights.
The UPR factor ATF6α is up-regulated and activated in LPS-stimulated B cells.
ATF6α is not required for lymphocyte development.
ATF6α is not required for development of antibody-secreting B cells in vitro.
ATF6α is not essential for humoral immune responses in vivo.
The specialized UPR of differentiating B cells is not dependent on ATF6α.
Acknowledgments
The authors thank LeeTerry Moore for technical assistance, Cynthia Van Hook (University of South Alabama, Department of Comparative Medicine) for assistance with animal husbandry, Bryant Hanks (University of South Alabama, Flow Cytometry Core Facility) for assistance with flow cytometry and Karen Miller (St. Jude Children’s Research Hospital) for technical assistance with lipid analysis. The work was supported by funds provided by grants from the US National Institutes of Health to S. J. (GM062896) and J. W. B. (GM061970).
Abbreviations
- ATF6
activating transcription factor 6
- ER
endoplasmic reticulum
- ERAD
ER-associated degradation
- IRE1
inositol-requiring enzyme 1
- KLH
keyhole limpet hemocyanin
- MEFs
mouse embryo fibroblasts
- NP
nitrophenyl
- nt
nucleotide
- PERK
PKR-like ER kinase
- PKR
protein kinase RNA-activated
- PtdCho
phosphatidylcholine
- qRT-PCR
quantitative RT-PCR
- UPR
unfolded protein response
- XBP1
X-box binding protein 1
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Adachi Y, Yamamoto K, Okada T, Yoshida H, Harada A, Mori K. ATF6 is a transcription factor specializing in the regulation of quality control proteins in the endoplasmic reticulum. Cell Struct Funct. 2008;33:75–89. doi: 10.1247/csf.07044. [DOI] [PubMed] [Google Scholar]
- Allman D, Pillai S. Peripheral B cell subsets. Curr Opin Immunol. 2008;20:149–57. doi: 10.1016/j.coi.2008.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bommiasamy H, Back SH, Fagone P, Lee K, Meshinchi S, Vink E, Sriburi R, Frank M, Jackowski S, Kaufman RJ, Brewer JW. ATF6α induces XBP1-independent expansion of the endoplasmic reticulum. J Cell Sci. 2009;122:1626–1636. doi: 10.1242/jcs.045625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunsing R, Omori SA, Weber F, Bicknell A, Friend L, Rickert R, Niwa M. B- and T-cell development both involve activity of the unfolded protein response pathway. J Biol Chem. 2008;283:17954–17961. doi: 10.1074/jbc.M801395200. [DOI] [PubMed] [Google Scholar]
- Calame KL, Lin KI, Tunyaplin C. Regulatory mechanisms that determine the development and function of plasma cells. Annu Rev Immunol. 2003;21:205–230. doi: 10.1146/annurev.immunol.21.120601.141138. [DOI] [PubMed] [Google Scholar]
- Calfon M, Zeng H, Urano F, Till JH, Hubbard SR, Harding HP, Clark SG, Ron D. IRE1 couples endoplasmic reticulum load to secretory capacity by processing the XBP-1 mRNA. Nature. 2002;415:92–96. doi: 10.1038/415092a. [DOI] [PubMed] [Google Scholar]
- Cinaroglu A, Gao C, Imrie D, Sadler KC. Activating transcription factor 6 plays protective and pathological roles in steatosis due to endoplasmic reticulum stress in zebrafish. Hepatol. 2011;54:495–508. doi: 10.1002/hep.24396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egawa N, Yamamoto K, Inoue H, Hikawa R, Nishi K, Mori K, Takahashi R. The endoplasmic reticulum stress sensor, ATF6α, protects against neurotoxin-induced dopaminergic neuronal death. J Biol Chem. 2011;286:7947–7957. doi: 10.1074/jbc.M110.156430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fagone P, Gunter C, Sage CR, Gunn KE, Brewer JW, Jackowski S. CTP:Phosphocholine cytidylyltransferase α is required for B-cell proliferation and class switch recombination. J Biol Chem. 2009;284:6847–6854. doi: 10.1074/jbc.M807338200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fagone P, Sriburi R, Ward-Chapman C, Frank M, Wang J, Gunter C, Brewer JW, Jackowski S. Phospholipid biosynthesis program underlying membrane expansion during B-lymphocyte differentiation. J Biol Chem. 2007;282:7591–7605. doi: 10.1074/jbc.M608175200. [DOI] [PubMed] [Google Scholar]
- Gass JN, Gifford NM, Brewer JW. Activation of an unfolded protein response during differentiation of antibody-secreting B cells. J Biol Chem. 2002;277:49047–49054. doi: 10.1074/jbc.M205011200. [DOI] [PubMed] [Google Scholar]
- Gass JN, Jiang HY, Wek RC, Brewer JW. The unfolded protein response of Blymphocytes: PERK-independent development of antibody-secreting cells. Mol Immunol. 2008;45:1035–1043. doi: 10.1016/j.molimm.2007.07.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gunn KE, Brewer JW. Evidence that marginal zone B cells possess an enhanced secretory apparatus and exhibit superior secretory activity. J Immunol. 2006;177:3791–3798. doi: 10.4049/jimmunol.177.6.3791. [DOI] [PubMed] [Google Scholar]
- Gunn KE, Gifford NM, Mori K, Brewer JW. A role for the unfolded protein response in optimizing antibody secretion. Mol Immunol. 2004;41:919–927. doi: 10.1016/j.molimm.2004.04.023. [DOI] [PubMed] [Google Scholar]
- Harding HP, Zhang Y, Ron D. Protein translation and folding are coupled by an endoplasmic-reticulum- resident kinase. Nature. 1999;397:271–274. doi: 10.1038/16729. [DOI] [PubMed] [Google Scholar]
- Haze K, Okada T, Yoshida H, Yanagi H, Yura T, Negishi M, Mori K. Identification of the G13 (cAMP-response-element-binding protein-related protein) gene product related to activating transcription factor 6 as a transcriptional activator of the mammalian unfolded protein response. Biochem J. 2001;355:19–28. doi: 10.1042/0264-6021:3550019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haze K, Yoshida H, Yanagi H, Yura T, Mori K. Mammalian transcription factor ATF6 is synthesized as a transmembrane protein and activated by proteolysis in response to endoplasmic reticulum stress. Mol Biol Cell. 1999;10:3787–3799. doi: 10.1091/mbc.10.11.3787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu CCA, Dougan SK, McGehee AM, Love JC, Ploegh HL. XBP-1 regulates signal transduction, transcription factors and bone marrow colonization in B cells. EMBO J. 2009;28:1624– 1636. doi: 10.1038/emboj.2009.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwakoshi NN, Lee AH, Vallabhajosyula P, Otipoby KL, Rajewsky K, Glimcher LH. Plasma cell differentiation and the unfolded protein response intersect at the transcription factor XBP-1. Nat Immunol. 2003;4:321–329. doi: 10.1038/ni907. [DOI] [PubMed] [Google Scholar]
- Jacob J, Kassir R, Kelsoe G. In situ studies of the primary immune response to (4-hydroxy-3- nitrophenyl)acetyl. I The architecture and dynamics of responding cell populations. J Exp Med. 1991;173:1165–1175. doi: 10.1084/jem.173.5.1165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee AH, Chu GC, Iwakoshi NN, Glimcher LH. XBP-1 is required for biogenesis of cellular secretory machinery of exocrine glands. EMBO J. 2005;24:4368–4380. doi: 10.1038/sj.emboj.7600903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis MJ, Mazzarella RA, Green M. Structure and assembly of the endoplasmic reticulum. The synthesis of three major endoplasmic reticulum proteins during lipopolysaccharide-induced differentiation of murine lymphocytes. J Biol Chem. 1985;260:3050–3057. [PubMed] [Google Scholar]
- Lykidis A, Jackowski S. Regulation of mammalian cell membrane biosynthesis. Prog Nucleic Acid Res Mol Biol. 2001;65:361–393. doi: 10.1016/s0079-6603(00)65010-9. [DOI] [PubMed] [Google Scholar]
- Ma Y, Shimizu Y, Mann MJ, Jin Y, Hendershot LM. Plasma cell differentiation initiates a limited ER stress response by specifically suppressing the PERK-dependent branch of the unfolded protein response. Cell Stress Chaperones. 2010;15:281–293. doi: 10.1007/s12192-009-0142-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maiuolo J, Bulotta S, Verderio C, Benfante R, Borgese N. Selective activation of the transcription factor ATF6 mediates endoplasmic reticulum proliferation triggered by a membrane protein. Proc Natl Acad Sci U S A. 2011;108:7832–7837. doi: 10.1073/pnas.1101379108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin F, Kearney JF. B1 cells: similarities and differences with other B cell subsets. Curr Opin Immunol. 2001a;13:195–201. doi: 10.1016/s0952-7915(00)00204-1. [DOI] [PubMed] [Google Scholar]
- Martin F, Kearney JF. Marginal-zone B cells. Nat Rev Immunol. 2002;2:323–335. doi: 10.1038/nri799. [DOI] [PubMed] [Google Scholar]
- Martin F, Oliver AM, Kearney JF. Marginal zone and B1 B cells unite in the early response against T-independent blood-borne particulate antigens. Immunity. 2001b;14:617–629. doi: 10.1016/s1074-7613(01)00129-7. [DOI] [PubMed] [Google Scholar]
- Masciarelli S, Fra AM, Pengo N, Bertolotti M, Cenci S, Fagioli C, Ron D, Hendershot LM, Sitia R. CHOP-independent apoptosis and pathway-selective induction of the UPR in developing plasma cells. Mol Immunol. 2010;47:1356–1365. doi: 10.1016/j.molimm.2009.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGehee AM, Dougan SK, Klemm EJ, Shui G, Park B, Kim YM, Watson N, Wenk MR, Ploegh HL, Hu CCA. XBP-1-deficient plasmablasts show normal protein folding but altered glycosylation and lipid synthesis. J Immunol. 2009;183:3690–3699. doi: 10.4049/jimmunol.0900953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng SL. Signaling in B cells via Toll-like receptors. Curr Opin Immunol. 2005;17:230–236. doi: 10.1016/j.coi.2005.03.003. [DOI] [PubMed] [Google Scholar]
- Rush JS, Sweitzer T, Kent C, Decker GL, Waechter CJ. Biogenesis of the endoplasmic reticulum in activated B lymphocytes: temporal relationships between the induction of protein Nglycosylation activity and the biosynthesis of membrane protein and phospholipid. Arch Biochem Biophys. 1991;284:63–70. doi: 10.1016/0003-9861(91)90264-j. [DOI] [PubMed] [Google Scholar]
- Schewe DM, Aguirre-Ghiso JA. ATF6α-Rheb-mTOR signaling promotes survival of dormant tumor cells in vivo. Proc Natl Acad Sci U S A. 2008;105:10519–10524. doi: 10.1073/pnas.0800939105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaffer AL, Shapiro-Shelef M, Iwakoshi NN, Lee AH, Qian SB, Zhao H, Yu X, Yang L, Tan BK, Rosenwald A, Hurt EM, Petroulakis E, Sonenberg N, Yewdell JW, Calame K, Glimcher LH, Staudt LM. XBP1, downstream of Blimp-1, expands the secretory apparatus and other organelles, and increases protein synthesis in plasma cell differentiation. Immunity. 2004;21:81–93. doi: 10.1016/j.immuni.2004.06.010. [DOI] [PubMed] [Google Scholar]
- Shen X, Ellis R, Lee K, Liu CY, Yang K, Solomon A, Yoshida H, Morimoto R, Kurnit DM, Mori K, Kaufman RJ. Complementary signaling pathways regulate the unfolded protein response and are required for C. elegans development. Cell. 2001;107:893–903. doi: 10.1016/s0092-8674(01)00612-2. [DOI] [PubMed] [Google Scholar]
- Shi Y, Vattem KM, Sood R, An J, Liang J, Stramm L, Wek RC. Identification and characterization of pancreatic eukaryotic initiation factor 2 alpha-subunit kinase, PEK, involved in translational control. Mol Cell Biol. 1998;18:7499–7509. doi: 10.1128/mcb.18.12.7499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sriburi R, Bommiasamy H, Buldak GL, Robbins GR, Frank M, Jackowski S, Brewer JW. Coordinate regulation of phospholipid biosynthesis and secretory pathway gene expression in XBP-1(S)-induced endoplasmic reticulum biogenesis. J Biol Chem. 2007;282:7024–7034. doi: 10.1074/jbc.M609490200. [DOI] [PubMed] [Google Scholar]
- Sriburi R, Jackowski S, Mori K, Brewer JW. XBP1: a link between the unfolded protein response, lipid biosynthesis, and biogenesis of the endoplasmic reticulum. J Cell Biol. 2004;167:35– 41. doi: 10.1083/jcb.200406136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tagliavacca L, Anelli T, Fagioli C, Mezghrani A, Ruffato E, Sitia R. The making of a professional secretory cell: architectural and functional changes in the ER during B lymphocyte plasma cell differentiation. Biol Chem. 2003;384:1273–1277. doi: 10.1515/BC.2003.141. [DOI] [PubMed] [Google Scholar]
- Teodoro T, Odisho T, Sidorova E, Volchuk A. Pancreatic β-cells depend on basal expression of active ATF6α-p50 for cell survival even under non-stress conditions. Am J Physiol Cell Physiol. 2011 doi: 10.1152/ajpcell.00160.2011. in press. Epub ahead of print. [DOI] [PubMed] [Google Scholar]
- Tirasophon W, Welihinda AA, Kaufman RJ. A stress response pathway from the endoplasmic reticulum to the nucleus requires a novel bifunctional protein kinase/endoribonuclease (Ire1p) in mammalian cells. Genes Dev. 1998;12:1812–1824. doi: 10.1101/gad.12.12.1812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tirosh B, Iwakoshi NN, Glimcher LH, Ploegh HL. XBP-1 specifically promotes IgM synthesis and secretion, but is dispensable for degradation of glycoproteins in primary B cells. J Exp Med. 2005;202:505–516. doi: 10.1084/jem.20050575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Todd DJ, McHeyzer-Williams LJ, Kowal C, Lee AH, Volpe BT, Diamond B, McHeyzer- Williams MG, Glimcher LH. XBP1 governs late events in plasma cell differentiation and is not required for antigen-specific memory B cell development. J Exp Med. 2009;206:2151– 2159. doi: 10.1084/jem.20090738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Anken E, Braakman I. Versatility of the endoplasmic reticulum protein folding factory. Crit Rev Biochem Mol Biol. 2005;40:191–228. doi: 10.1080/10409230591008161. [DOI] [PubMed] [Google Scholar]
- van Anken E, Romijn EP, Maggioni C, Mezghrani A, Sitia R, Braakman I, Heck AJ. Sequential waves of functionally related proteins are expressed when B cells prepare for antibody secretion. Immunity. 2003;18:243–253. doi: 10.1016/s1074-7613(03)00024-4. [DOI] [PubMed] [Google Scholar]
- Walter P, Ron D. The unfolded protein response: from stress pathway to homeostatic regulation. Science. 2011;334:1081–1086. doi: 10.1126/science.1209038. [DOI] [PubMed] [Google Scholar]
- Wang XZ, Harding HP, Zhang Y, Jolicoeur EM, Kuroda M, Ron D. Cloning of mammalian Ire1 reveals diversity in the ER stress responses. EMBO J. 1998;17:5708–5717. doi: 10.1093/emboj/17.19.5708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Vera L, Fischer WH, Montminy M. The CREB coactivator CRTC2 links hepatic ER stress and fasting gluconeogenesis. Nature. 2009;460:534–537. doi: 10.1038/nature08111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiest DL, Burkhardt JK, Hester S, Hortsch M, Meyer DI, Argon Y. Membrane biogenesis during B cell differentiation: most endoplasmic reticulum proteins are expressed coordinately. J Cell Biol. 1990;110:1501–1511. doi: 10.1083/jcb.110.5.1501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J, Ruas JL, Estall JL, Rasbach KA, Choi JH, Ye L, Bostrom P, Tyra HM, Crawford RW, Campbell KP, Rutkowski DT, Kaufman RJ, Spiegelman BM. The unfolded protein response mediates adaptation to exercise in skeletal muscle through a PGC- 1α/ATF6α complex. Cell Metab. 2011;13:160–169. doi: 10.1016/j.cmet.2011.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J, Rutkowski DT, Dubois M, Swathirajan J, Saunders T, Wang J, Song B, Yau GD, Kaufman RJ. ATF6α optimizes long-term endoplasmic reticulum function to protect cells from chronic stress. Dev Cell. 2007;13:351–364. doi: 10.1016/j.devcel.2007.07.005. [DOI] [PubMed] [Google Scholar]
- Yamamoto K, Sato T, Matsui T, Sato M, Okada T, Yoshida H, Harada A, Mori K. Transcriptional induction of mammalian ER quality control proteins is mediated by single or combined action of ATF6α and XBP1. Dev Cell. 2007;13:365–376. doi: 10.1016/j.devcel.2007.07.018. [DOI] [PubMed] [Google Scholar]
- Yamamoto K, Takahara K, Oyadomari S, Okada T, Sato T, Harada A, Mori K. Induction of liver steatosis and lipid droplet formation in ATF6α-knockout mice burdened with pharmacological endoplasmic reticulum stress. Mol Biol Cell. 2010;21:2975–2986. doi: 10.1091/mbc.E09-02-0133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye J, Rawson RB, Komuro R, Chen X, Dave UP, Prywes R, Brown MS, Goldstein JL. ER stress induces cleavage of membrane-bound ATF6 by the same proteases that process SREBPs. Mol Cell. 2000;6:1355–1364. doi: 10.1016/s1097-2765(00)00133-7. [DOI] [PubMed] [Google Scholar]
- Yoshida H, Matsui T, Yamamoto A, Okada T, Mori K. XBP1 mRNA is induced by ATF6 and spliced by IRE1 in response to ER stress to produce a highly active transcription factor. Cell. 2001;107:881–891. doi: 10.1016/s0092-8674(01)00611-0. [DOI] [PubMed] [Google Scholar]
- Zhang K, Wong HN, Song B, Miller CN, Scheuner D, Kaufman RJ. The unfolded protein response sensor IRE1α is required at 2 distinct steps in B cell lymphopoiesis. J Clin Invest. 2005;115:268–281. doi: 10.1172/JCI21848. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1. Levels of IgM and IgG in serum of non-immunized ATF6α-deficient mice. Serum samples were collected from non-immunized Atf6a+/+ and Atf6a−/− mice (n = 11 for each genotype). The concentrations of IgM and IgG were determined by ELISA using purified mouse IgM and IgG as standards. Data are plotted as the level of serum IgM and IgG in Atf6a−/− mice relative to that of Atf6a+/+ mice, expressed as percentage of wild-type control (mean ± S.D.).