Abstract
The bacterial tmRNA quality control system monitors protein synthesis and recycles stalled translation complexes in a process termed “ribosome rescue”. During rescue, tmRNA acts first as a transfer RNA to bind stalled ribosomes, then as a messenger RNA to add the ssrA peptide tag to the C-terminus of the nascent polypeptide chain. The ssrA peptide targets tagged peptides for proteolysis, ensuring rapid degradation of potentially deleterious truncated polypeptides. Ribosome rescue also facilitates turnover of the damaged messages responsible for translational arrest. Thus, tmRNA increases the fidelity of gene expression by promoting the synthesis of full-length proteins. In addition to serving as a global quality control system, tmRNA also plays important roles in bacterial development, pathogenesis and environmental stress responses. This review focuses on the mechanism of tmRNA-mediated ribosome rescue and the role of tmRNA in bacterial physiology.
Keywords: mRNA turnover, proteolysis, ribonuclease, ribosome rescue, ssrA peptide, SmpB, tmRNA, trans-translation
I. Introduction
Quality control during protein synthesis is important to maintain both the speed and fidelity of gene expression. It has long been recognized that translation rates vary and that ribosomes pause at specific sites along messenger RNAs. In some instances, ribosome pausing is exploited to regulate protein synthesis through programmed frameshifting and other recoding events. However, paused ribosomes are often the manifestation of defective protein synthesis. Such a problem occurs during the translation of non-stop mRNAs, which are truncated transcripts lacking in-frame stop codons. Non-stop messages are produced by premature transcription termination, ribonuclease activity and chemical/physical damage due to environmental stresses. Ribosomes translate to the 3′-end of non-stop mRNA and then arrest with an incomplete codon in the aminoacyl-tRNA acceptor site (A site). Because release factor (RF) mediated translation termination depends upon an intact A-site stop codon, specialized systems are required to recycle these stalled ribosomes. Additionally, cells must also dispose of the truncated polypeptide chains produced from non-stop translation. All eubacteria deploy tmRNA, a specialized RNA that functions as both a tRNA and an mRNA, to recycle stalled ribosomes and tag incomplete nascent chains for degradation. Although tmRNA is only found in the eubacteria and some plastids, non-stop mRNA is a universal problem and all organisms have systems dedicated to the recycling of stalled ribosomes (Doma and Parker, 2007; Shoemaker et al., 2010; Pisareva et al., 2011).
tmRNA was first identified as 10S RNA by David Apirion and colleagues, who used a radiolabeling approach to discover stable RNAs in Escherichia coli (Ray and Apirion, 1979). Subsequently, it was found that 10S RNA is comprised of two species: 10Sa RNA representing tmRNA, and 10Sb RNA, which is the catalytic subunit of RNase P (Jain et al., 1982; Ray and Apirion, 1979). Apirion’s group also identified the gene encoding 10Sa RNA and named it ssrA for small stable RNA (Chauhan and Apirion, 1989; Oh et al., 1990). Slow growth and carbon starvation phenotypes were observed in E. coli ssrA disruption mutants (Oh and Apirion, 1991), but the function of 10Sa RNA remained unknown for several years. Two key papers published in 1994 and 1995 by Hachiro Inokuchi and Richard Simpson (respectively) led to the discovery of tmRNA function. Inokuchi and colleagues recognized that 10Sa RNA had the potential to form a tRNA-like structure and demonstrated that the RNA could be aminoacylated by alanine tRNA synthetase (Komine et al., 1994). Simpson and colleagues discovered that 10Sa RNA mediated a novel process in which truncated proteins were tagged at their C-termini with the AANDENYALAA peptide sequence (Tu et al., 1995). The last ten residues of this peptide tag are encoded by the ssrA gene, and Tu et al. demonstrated that tagging was abolished in an ssrA mutant (Tu et al., 1995). Although this latter paper clearly showed the ssrA gene encodes a peptide, the authors did not propose a messenger RNA function for 10Sa RNA because prior work had concluded that 10Sa RNA did not associate with ribosomes (Ray and Apirion, 1979).
Soon after the discovery of 10Sa RNA mediated tagging, Ken Keiler postulated that the ssrA peptide targets tagged proteins for degradation. This hypothesis was partly based on the hydrophobic nature of the ssrA tag, which is similar to the C-terminal WVAAA peptide motif recognized by the E. coli Tsp protease (Keiler and Sauer, 1996; Silber et al., 1992). Synthesizing the findings of Inokuchi and Simpson, Keiler et al. proposed that 10Sa RNA acts as a hybrid tRNA-mRNA to recycle stalled ribosomes from truncated mRNA and tag the incomplete nascent chains for degradation (Keiler et al., 1996). This paper outlined the biological rationale for ribosome rescue, and also proposed a mechanistic model for tagging. According to the transfer-messenger RNA model (Fig. 1), tmRNA enters the A site of stalled ribosomes by virtue of its alanine-charged tRNA-like domain and accepts the nascent chain through ribosomal peptidyltransferase activity. After the nascent chain transfer to tmRNA, the truncated transcript is released from the ribosome and translation resumes using the small open reading frame (ORF) within tmRNA. The tmRNA ORF is terminated by a stop codon, which allows translation termination and recycling of the 70S ribosome into 50S and 30S subunits. In this manner, tmRNA provides the stop codon for truncated messages in trans; and therefore this activity is often referred to as trans-translation in the literature (Dulebohn et al., 2007; Keiler, 2008; Moore and Sauer, 2007).
Figure 1. The trans-translation model of tmRNA activity.
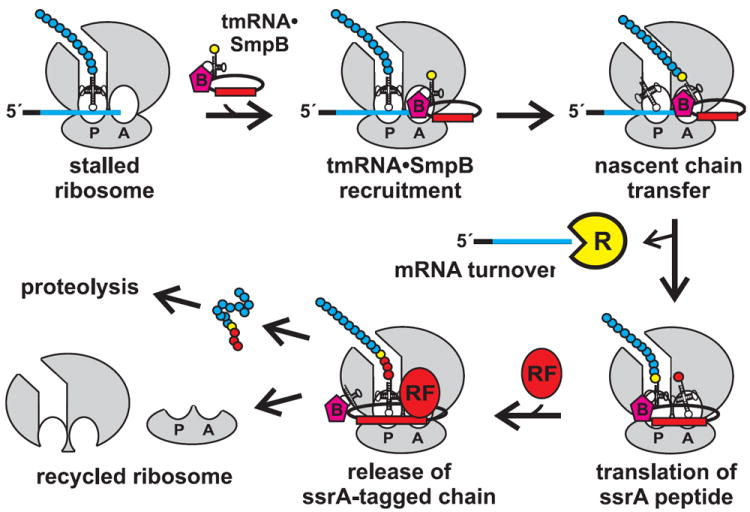
Translation of non-stop mRNA leads to ribosome arrest. tmRNA•SmpB binds the A site of stalled ribosomes and accepts the nascent chain. The non-stop mRNA is released and preferentially degraded by RNase R. Translation then resumes using the open reading frame found within tmRNA. After synthesis of the ssrA peptide, release factors (RF) terminate translation and the ribosome is recycled into large and small subunits. The ssrA-tagged chain is degraded by a number of proteases.
II. tmRNA•SmpB and the mechanism of trans-translation
1. tmRNA
The ssrA gene has been found in all eubacterial genomes sequenced to date, and is also retained in some plastid chromosomes (Gueneau de Novoa and Williams, 2004; Oudot-Le Secq et al., 2007). Most tmRNA molecules are between 325 and 400 nucleotides in length, although reductive evolution has resulted in significantly shorter (~250 nucleotides) species in plastids and cyanobacteria (Gueneau de Novoa and Williams, 2004). Other structural variations occur in α-proteobacteria and some cyanobacteria, whose functional tmRNAs are comprised of two non-covalently linked RNA chains (Keiler et al., 2000; Sharkady and Williams, 2004). tmRNA is synthesized initially as a precursor RNA, which is rapidly processed into the mature form by a variety of RNases. Like canonical tRNAs, the 5′-end of mature tmRNA is generated by RNase P cleavage (Gimple and Schon, 2001; Komine et al., 1994). Processing of the 3′-end has only been examined in E. coli, where RNase E and 3′-to-5′ exoribonuclease activity are required for tmRNA maturation (Li et al., 1998; Lin-Chao et al., 1999).
The tmRNA from E. coli has been studied most extensively and serves as a structural and biochemical model. E. coli tmRNA is composed of a tRNA-like domain (TLD), a short ORF encoding the ssrA peptide, and four pseudoknot structures (Fig. 2). The TLD is comprised of an aminoacyl acceptor stem, a T-loop and an open D-loop, but lacks an anticodon stem-loop. The acceptor stem contains a G3•U357 (E. coli numbering) wobble base pair, which is the recognition determinant for alanine tRNA synthetase (Komine et al., 1994; Nameki et al., 1999b). Additionally, the T-loop contains the thymine and pseudouridine nucleotides found in most tRNAs (Felden et al., 1998). Pseudoknot-1 is located upstream of the ORF and is important for tmRNA function (Nameki et al., 1999a; Wower et al., 2009), though other secondary structure elements in this position also support trans-translation (Tanner et al., 2006). The remaining pseudoknots are arrayed downstream of the ORF and each can be individually replaced with single-stranded RNA without abrogating tmRNA function (Nameki et al., 2000). The ORF itself can also be modified without affecting trans-translation. Several groups have developed tmRNA variants that encode various epitope tags that are resistant to proteolysis (Fig. 2 and Section III) (Barends et al., 2010; Fujihara et al., 2002; Gottesman et al., 1998; Roche and Sauer, 2001; Thibonnier et al., 2008; Yang and Glover, 2009). These tmRNA variants allow tagging activity to be assessed by immunoblot and have facilitated the identification of several ssrA-tagged proteins in a variety of bacteria (Barends et al., 2010; Collier et al., 2002; Fujihara et al., 2002; Roche and Sauer, 2001).
Figure 2. Structure of tmRNA and SmpB.
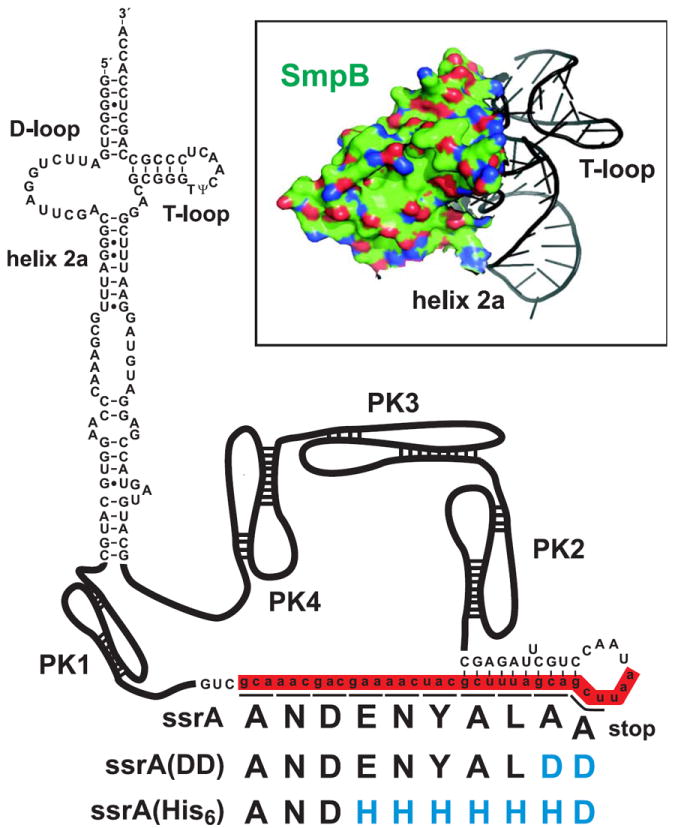
The secondary structure of E. coli tmRNA is shown, highlighting the tRNA-like domain (TLD) and the ssrA open reading frame (Nameki et al., 1999a). Pseudoknot (PK) structures are shown schematically to simplify the image. The wild-type ssrA peptide is shown in one-letter code, as are the modified ssrA(DD) and ssrA(His6) peptide tags. The inset shows the crystal structure of SmpB bound to the tmRNA TLD (1P6V) (Gutmann et al., 2003).
2. SmpB
In 1999, Wali Karzai, Mimi Susskind and Bob Sauer reported that tmRNA-mediated ribosome rescue requires a small protein called SmpB (Karzai et al., 1999). SmpB is a tmRNA-binding protein and is encoded immediately upstream of ssrA in E. coli (Moore and Sauer, 2005). SmpB stabilizes tmRNA, facilitates its charging by alanine tRNA synthetase, and is required for tmRNA binding to the ribosome (Barends et al., 2001; Hanawa-Suetsugu et al., 2002; Karzai et al., 1999). High-resolution structural studies show that SmpB adopts a β-barrel oligonucleotide-binding (OB) fold and has a flexible C-terminal tail (Dong et al., 2002; Someya et al., 2003). Co-crystal structures from Nenad Ban and colleagues show that SmpB binds to the D-loop of the tmRNA TLD and holds it an open and extended conformation (Fig. 2) (Gutmann et al., 2003). This binding site in the TLD elbow region allows simultaneous binding of EF-Tu, which is also required for delivery of tmRNA to the ribosome (Barends et al., 2001). SmpB binds tmRNA at other sites outside of the TLD, and these contacts may be important for establishing the correct reading frame during ssrA peptide translation (Konno et al., 2007; Metzinger et al., 2005, 2008; Watts et al., 2009; Wower et al., 2002). SmpB can also bind both ribosomal subunits in a tmRNA-independent manner, leading Brice Felden and colleagues to propose SmpB binds stalled ribosomes prior to tmRNA recruitment (Hallier et al., 2006; Hallier et al., 2004). However, this model is controversial; and work from Sundermeier & Karzai indicates that SmpB-ribosome interactions require tmRNA (Sundermeier and Karzai, 2007). Finally, the flexible C-terminal tail of SmpB is critical for trans-translation. SmpB variants lacking the C-terminal tail are still able to bind and deliver tmRNA to the ribosome, but transfer of the nascent chain to tmRNA is blocked (Jacob et al., 2005; Kurita et al., 2010; Sundermeier et al., 2005). These observations indicate that the C-terminal tail is required for accommodation of tmRNA into the ribosome, perhaps by promoting the GTPase activity of EF-Tu (Shimizu and Ueda, 2006; Sundermeier et al., 2005). Accordingly, Brice Felden and colleagues have shown that SmpB interacts with residues in the decoding center and have suggested that the C-terminal tail plays a role in mimicking the missing A-site codon during trans-translation (Nonin-Lecomte et al., 2009).
3. Mechanism of trans-translation
The majority of protein chains are tagged by tmRNA when synthesized from non-stop mRNA in vivo, suggesting that ribosome stalling upon truncated mRNA is required for tmRNA•SmpB recruitment. Måns Ehrenberg and colleagues have tested this model in vitro by measuring the rate of nascent peptide transfer to tmRNA as a function of transcript length (Ivanova et al., 2004). As expected, trans-translation occurs very rapidly when there is no codon, or an incomplete codon, present in the ribosome A-site. However, transfer still occurs when the transcript extends up to six nucleotides downstream of the A-site codon. This latter activity appears to be significant in vivo, because ssrA-tagging is often associated with mRNA cleavage at the 3′-leading edge of paused ribosomes (Garza-Sánchez et al., 2008; Garza-Sánchez et al., 2006; Garza-Sánchez et al., 2009; Li et al., 2006; Sunohara et al., 2004a). Longer stretches of downstream mRNA do not support trans-translation in vitro, demonstrating that tmRNA•SmpB is not recruited if ribosomes are paused on full-length mRNA (Asano et al., 2005; Ivanova et al., 2004). Thus, truncated mRNA greatly stimulates trans-translation, but the 3′-end of transcript need not be positioned in the ribosome A-site for tmRNA•SmpB recruitment.
The transfer-messenger RNA model has been tested and verified independently by several research groups. Although the model is fundamentally correct, the structural and mechanistic details of trans-translation continue to be the most important problems in the field. The first structural model of tmRNA•SmpB bound to the ribosome was obtained in a collaborative effort between Venki Ramakrishnan and Joachim Frank (Valle et al., 2003). Their pre-accommodation model shows that the TLD and EF-Tu adopt conformations similar to those found in canonical ternary complexes, and that pseudoknots 2, 3 and 4 are draped over the beak of the 30S subunit in an arc (Valle et al., 2003). This model places SmpB in close proximity to helices H69, H71 and H89 of the large ribosome subunit. Subsequent structural and biochemical data suggest that two SmpB molecules are present during the pre-accommodation step: one interacting with the large subunit GTPase activating center and the other close to the 30S A-site (Cheng et al., 2010; Ivanova et al., 2005; Kaur et al., 2006). After accommodation of the TLD into the A-site, it appears that the SmpB associated with the GTPase activating center of the 50S subunit is released (Bugaeva et al., 2008; Cheng et al., 2010; Ivanova et al., 2005; Kaur et al., 2006), whereas the other SmpB remains bound to the D-loop throughout trans-translation (Bugaeva et al., 2008; Ivanova et al., 2007). Cryo-EM analyses of the post-accommodation and resume stages of trans-translation have been challenging due to ribosome heterogeneity and low tmRNA•SmpB occupancy, but the resulting models show that SmpB continues mimic an anticodon loop throughout the process (Cheng et al., 2010; Weis et al., 2010a; Weis et al., 2010b). These models also show that the loop formed by pseudoknots 2, 3 and 4 remains draped over the head of the 30S subunit after translocation of the TLD into the P site (Cheng et al., 2010; Fu et al., 2010; Weis et al., 2010b). It is unclear how the pseudoknots traverse the ribosome, and it has been suggested that these elements unfold during trans-translation (Wower et al., 2005). However, chemical protection and footprinting analyses indicate that tmRNA secondary structure remains largely intact as the complex moves through the ribosome (Ivanova et al., 2007; Shpanchenko et al., 2005).
4. Ribosomal mutations influence tmRNA activity
A small number of mutations have been shown to modulate trans-translation. Holberger & Hayes examined the effects of streptomycin-resistant rpsL mutations on tmRNA activity (Holberger and Hayes, 2009). The rpsL gene encodes ribosomal protein S12, which is located near the decoding center and has long been known to influence translational fidelity (Kurland et al., 1996). Most mutations that alter S12 residues Lys42 and Pro90 result in decreased tmRNA activity, but the Lys42Arg and Pro90Arg mutations actually increase the efficiency of trans-translation relative to wild-type S12 (Holberger and Hayes, 2009). Additionally, tmRNA activity in a subset of the S12 Pro90 mutants is restored to wild-type levels when the cells are treated with streptomycin. Classical rpsL mutants usually exhibit the error-restrictive phenotype, characterized by decreased ribosome affinity for aminoacyl-tRNA and hyper-accurate decoding (Kurland et al., 1996). Error-restriction could explain the observed defects in trans-translation if the rpsL mutations significantly reduce A-site affinity for tmRNA•SmpB. However, there is no correlation between error-restriction and tmRNA activity in the examined mutants (Holberger and Hayes, 2009). Perhaps the unique interaction between SmpB and the decoding center plays a role in these phenomena, such that the S12 variants modulate tmRNA•SmpB binding to the ribosome in manner that is distinct from the effects on tRNA binding.
Allen Buskirk and colleagues have also identified ribosome residues that are important for tmRNA function. Using a clever genetic selection that links trans-translation to cell death, they screened ribosomal RNA libraries for mutations that inhibit tmRNA activity (Crandall et al., 2010). Three mutations were identified in 16S rRNA (A1150G, ΔA1150, and ΔU1123), all of which interfere with tmRNA activity. The affected residues map to helix 39 and are located in the 30S subunit head, close to where the tmRNA pseudoknots are positioned during the early stages of trans-translation. Two additional mutations (U846C and C889U) were isolated in the 23S rRNA. These residues are within the A-site finger, which makes contact with A-site tRNA and also forms part of intersubunit bridge B1a (Yusupov et al., 2001). The position of the A-site finger and its conformational dynamics during translocation are both suggestive of a role in ribosome pausing. Intriguingly, the C889U mutation only affects tmRNA activity when ribosomes pause on full-length mRNA (Crandall et al., 2010). This latter finding suggests that the C889U mutation interferes with the mRNA processing events required for tmRNA recruitment to the paused ribosome.
III. Degradation of ssrA-tagged proteins and truncated mRNA
In addition to facilitating ribosome recycling, tmRNA also plays an important role in the degradation of incomplete protein chains. Simpson and colleagues were able to isolate and characterize ssrA-tagged proteins because the chains in their study formed protease-resistant inclusion bodies (Tu et al., 1995). Keiler et al. first demonstrated that ssrA-tagged proteins are rapidly degraded, and discovered that Tsp (tail specific protease) is responsible for the turnover of tagged proteins in the periplasm (Keiler et al., 1996). Tsp recognizes hydrophobic residues at the C-termini of its substrates, and replacement of the C-terminal alanine residues in ssrA with aspartate prevents Tsp-dependent degradation (Keiler and Sauer, 1996; Silber et al., 1992). This ssrA(DD) peptide tag is also resistant to degradation in the cytoplasm (Keiler et al., 1996), and several studies have shown that the hydrophobic C-terminus of ssrA is critical for degradation by cytoplasmic proteases (Flynn et al., 2001; Gottesman et al., 1998; Herman et al., 1998). This recognition mode is highly conserved throughout eubacteria and tmRNA variants encoding charged C-terminal residues have been used to stabilize tagged proteins in a variety of species (Fujihara et al., 2002; Hong et al., 2007; Keiler and Shapiro, 2003b; Thibonnier et al., 2008; Wiegert and Schumann, 2001; Yang and Glover, 2009).
Tagged proteins in the cytoplasm are degraded by a number of proteases belonging to the AAA+ ATPase family. In general, these enzymes use the free energy of ATP hydrolysis to unfold and translocate substrates into an enclosed proteolytic compartment (Sauer and Baker, 2010; Sauer et al., 2004). The ClpXP protease degrades most ssrA-tagged proteins in the cytoplasm of E. coli and B. subtilis (Gottesman et al., 1998; Wiegert and Schumann, 2001). ClpXP is a compound protease built from ClpX ATPase and ClpP protease subunits (Wojtkowiak et al., 1993). ClpX binds the ssrA peptide and translocates tagged proteins into the lumen of the ClpP protease (Sauer and Baker, 2010). ClpP can also degrade ssrA-tagged proteins in conjunction with the ClpA ATPase, but does so less efficiently than ClpXP in vivo (Gottesman et al., 1998). ClpX and ClpA recognize partially overlapping determinants within the ssrA peptide; ClpX binds the C-terminal residues (AANDENYALAA), whereas ClpA also makes additional contacts with the N-terminal portion of the ssrA peptide (AANDENYALAA) (Flynn et al., 2001). Degradation is also influenced by the adaptor protein SspB, which delivers ssrA-tagged proteins to ClpXP (Levchenko et al., 2000; Wah et al., 2002). SspB binds to the N-terminal portion of the ssrA peptide (AANDENYALAA) and thus blocks recognition of tagged proteins by ClpAP (Flynn et al., 2001).
Although ClpXP is critical for the degradation of ssrA-tagged proteins in many species, it is not found some bacteria. Two other AAA+ proteases, FtsH (HflB) and Lon, are more highly conserved in eubacteria and are known to degrade ssrA-tagged proteins in E. coli (Choy et al., 2007; Herman et al., 1998). FtsH is an integral membrane protein found in all eubacteria and is probably responsible for degradation of all ssrA-tagged membrane proteins. Mycoplasma species have lost the Clp proteases through reductive evolution and instead rely upon Lon for degradation of ssrA-tagged proteins (Karzai et al., 2000). Accordingly, the ssrA tags in Mycoplasma species have acquired distinct features that facilitate recognition by Lon protease (Ge and Karzai, 2009; Gur and Sauer, 2008). The evolution of Mycoplasma ssrA tags highlights the selective advantage of coupling proteolysis to ribosome rescue.
Non-stop mRNA is also degraded rapidly as a consequence of ribosome rescue by tmRNA•SmpB (Fig. 1). Yamamoto et al. showed that truncated mRNA is stabilized in ΔssrA cells, presumably because stalled ribosomes protect the 3′-ends of these messages from 3′-to-5′ exoribonuclease activity (Yamamoto et al., 2003). Subsequent work from Wali Karzai’s group has shown that RNase R specifically mediates turnover of non-stop mRNA (Richards et al., 2006). RNase R is transiently associated with ribosomes and is able to rapidly bind and degrade non-stop messages released by tmRNA•SmpB during ribosome rescue (Ge et al., 2010). Thus, tmRNA activity serves to rid the cell of damaged messages that cause translational arrest.
IV. tmRNA-independent ribosome rescue pathways
The tmRNA system is essential for the viability of several bacterial species including Haemophilus influenzae, Neisseria gonorrhoeae, Mycoplasma genitalium, Mycoplasma pulmonis, and Helicobacter pylori (Akerley et al., 2002; French et al., 2008; Glass et al., 2006; Huang et al., 2000; Thibonnier et al., 2008). In contrast, the ssrA and/or smpB genes can be deleted from E. coli, Bacillus subtilis and Caulobacter crescentus (Keiler and Shapiro, 2003b; Oh and Apirion, 1991; Wiegert and Schumann, 2001). These observations suggest that some bacteria possess a tmRNA-independent ribosome rescue system(s), whereas others rely solely upon the tmRNA pathway. Several reports have shown that proteins can be overproduced to very high levels when synthesized from non-stop mRNA in E. coli ΔssrA mutants (Janssen and Hayes, 2009; Karzai et al., 1999; Keiler et al., 1996; Roche and Sauer, 1999; Yamamoto et al., 2003), indicating that E. coli contains a tmRNA-independent system capable of releasing nascent chains from stalled ribosomes. A number of known translation factors have been proposed to mediate tmRNA-independent recycling (Heurgue-Hamard et al., 1998; Karimi et al., 1998; Singh and Varshney, 2004), but it has been difficult to define specific roles in ribosome rescue because these proteins are also required for normal recycling.
Tatsuhiko Abo and colleagues recently identified a novel factor that appears dedicated to tmRNA-independent ribosome rescue (Chadani et al., 2010). Reasoning that the tmRNA-independent ribosome rescue factor would be essential in cells lacking tmRNA, Abo and colleagues identified the yhdL gene in a genetic screen for mutations that are synthetically lethal in ΔssrA mutants. The yhdL gene has subsequently been renamed arfA for alternative ribosome-rescue factor A. ArfA competes with tmRNA•SmpB for the release of nascent chains, and deletion of arfA increases the efficiency of ssrA tagging. Although ArfA is required for tmRNA-independent rescue, it is not sufficient to release nascent chains in purified translation reactions (Chadani et al., 2011). These latter results indicate that ArfA activity requires another unidentified factor. Genetic screens have failed to identify this factor, and it has been proposed that class I release factors or peptidyl-tRNA hydrolase collaborate with ArfA to release nascent chains (Chadani et al., 2011; Garza-Sánchez et al., 2011).
Intriguingly, ArfA is synthesized from a non-stop mRNA and is therefore regulated by tmRNA-mediated ssrA tagging and proteolysis (Fig. 3) (Garza-Sánchez et al., 2011). The arfA message contains a hairpin structure that is efficiently cleaved by RNase III to generate non-stop mRNA. In ssrA+ cells, virtually all ArfA is ssrA-tagged and degraded, whereas C-terminal truncated forms of ArfA are produced in ΔssrA cells. Truncated ArfA is fully functional in nascent chain release and supports the viability of ΔssrA mutants. These results indicate that ArfA is only deployed under conditions in which the tmRNA system is overwhelmed or otherwise incapacitated, suggesting that ArfA functions as a fail-safe ribosome rescue system. The arfA genes in other γ-proteobacteria also encode hairpin structures. The arfA transcripts from Salmonella typhimurium LT2, Proteus mirabilis, Yersinia pestis, Mannheimia haemolytica, Haemophilus influenzae, and Dickeya dadantii 3937 are all cleaved by RNase III in vitro (F. Garza-Sánchez, R. Schaub and C.S.H., unpublished results) (Garza-Sánchez et al., 2011). In contrast, the hairpin found within the Neisseria gonorrhoeae arfA transcript is not cleaved by RNase III and instead functions as an intrinsic transcription terminator to generate non-stop mRNA (R. Schaub and C.S.H., unpublished results). Taken together, these observations strongly suggest that ArfA synthesis is regulated by tmRNA activity in many different bacteria.
Figure 3. tmRNA regulates the synthesis of ArfA.
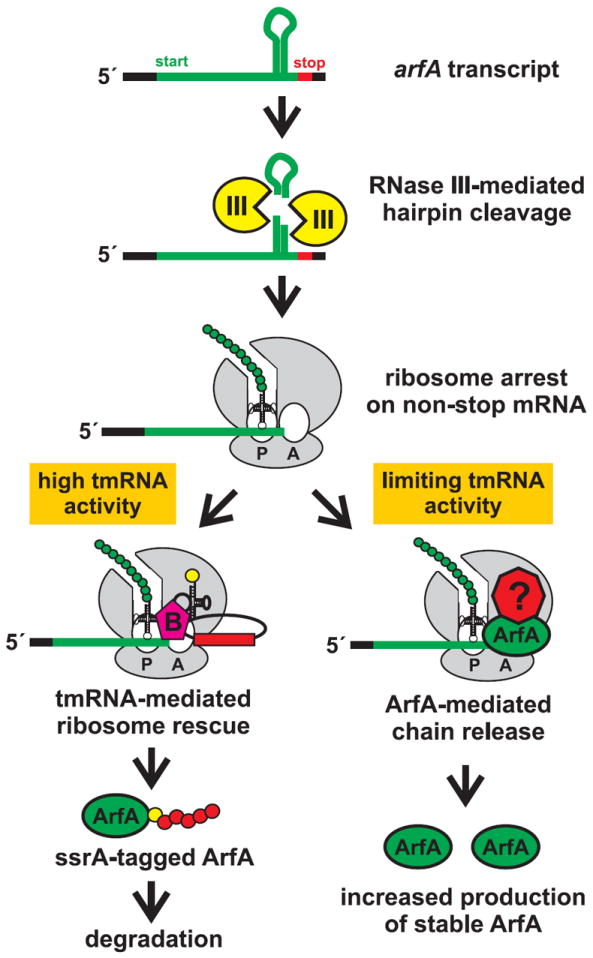
The arfA transcript coding sequence contains a hairpin structure, which is cleaved by RNase III to generate non-stop mRNA. Ribosomes stalled at the 3′-end of the truncated arfA message have at least two fates. tmRNA-mediated rescue results in tagging of the ArfA nascent chain, thereby maintaining ArfA at low levels in the cell. When tmRNA activity is limiting or absent, the incomplete ArfA nascent chain is released from the ribosome. ArfA chain release may require pre-existing cytosolic ArfA in collaboration with an unknown factor.
The identification of ArfA has resolved the mystery of tmRNA-independent ribosome rescue in E. coli. However, recognizable homologues of arfA are only found within a subset of γ- and β-proteobacteria. If ribosome rescue is indeed an essential function, then other species in which ssrA is dispensable (e.g. B. subtilis and C. crescentus) must possess tmRNA-independent systems that are distinct from ArfA. A candidate for this activity has been characterized independently by Nobukazu Nameki and Tatsuhiko Abo (Chadani et al., 2011; Handa et al., 2011). These research groups showed that the E. coli YaeJ protein is able to directly release nascent chains from stalled ribosomes. YaeJ contains the catalytic domain of canonical protein release factors, but lacks the domains required for stop codon recognition (Baranov et al., 2006; Hayes and Keiler, 2010). YaeJ is widely distributed throughout the proteobacteria (α, β, γ and δ), bacteroidetes, actinobacteria, and cyanobacteria, but is largely absent from the firmicutes. Remarkably, YaeJ homologs are also found in most eukaryotes, where they are thought to function in mitochondrial protein synthesis. ICT1, the human homolog of YaeJ, is an essential protein and an integral component of the mitochondrial ribosome (Richter et al., 2010). Both YaeJ and ICT1 retain stop-codon independent peptidyl-tRNA hydrolase activity and presumably function to release nascent chains from stalled ribosome complexes (Chadani et al., 2011; Handa et al., 2011; Richter et al., 2010). E. coli YaeJ is not essential in either ssrA+ or ΔssrA genetic backgrounds (Handa et al., 2011). In principle, YaeJ function could be redundant with that of PrfH, which is another putative stop-codon independent release factor in E. coli (Baranov et al., 2006). However, the E. coli ΔssrA ΔyaeJ ΔprfH triple mutant is viable and exhibits no growth phenotype compared to ΔssrA cells under standard laboratory conditions (B.D.J. and C.S.H., unpublished results). These observations suggest that YaeJ responds to translational problems that are distinct from those resolved by tmRNA and ArfA. Alternatively, YaeJ (and PrfH) mediated ribosome rescue may be critical under certain growth conditions or in response to environmental stresses such as heat shock. Translating ribosomes can dissociate during heat shock, producing isolated 50S subunits that carry peptidyl-tRNA. Jiang et al. have proposed that YaeJ/PrfH collaborates with Hsp15 to release nascent chains from these dissociated subunits (Jiang et al., 2009). Although the role of YaeJ in ribosome rescue remains to be clarified, the wide distribution of this alternative translation factor suggests that it mediates an important function.
V. Origins of non-stop mRNA in bacteria
1. Natural non-stop messages and stop codon readthrough
There are a number of pathways that produce truncated mRNAs in bacteria. In the original study that defined tmRNA activity, Keiler et al. fused an intrinsic transcription terminator downstream of an ORF to generate an artificial transcript lacking in-frame stop codons (Keiler et al., 1996). A similar type of non-stop mRNA occurs naturally in an undomesticated strain of Bacillus subtilis. The kinA gene in Bacillus subtilis ATCC 6051 has acquired a mutation that changes the ancestral stop codon to a Tyr codon (Kobayashi et al., 2008). An intrinsic transcription terminator is encoded immediately downstream of the kinA ORF, and therefore the missense mutation leads to non-stop mRNA production. KinA is a histidine kinase that plays a role in entry into sporulation, and therefore the ssrA-tagging and degradation of KinA partially accounts for the sporulation defect in this strain. Decoding errors during translation termination can also convert a normal transcript into non-stop mRNA if there is no in-frame stop codon in the 3′-untranslated region. This can occur under physiological conditions if cells express suppressor tRNAs, which decode stop codons as sense codons. Hiroji Aiba and colleagues have shown that suppressor tRNAs allow some ribosomes to translate to the 3′-ends of transcripts and consequently induce tmRNA activity (Abo et al., 2002). The same phenomenon occurs in cells treated with aminoglycoside antibiotics, which induce stop codon readthrough by promoting decoding errors (Abo et al., 2002; Holberger and Hayes, 2009). Not surprisingly, ΔssrA mutants are more sensitive than ssrA+ cells to aminoglycosides and other antibiotics that target translation (Abo et al., 2002; de la Cruz and Vioque, 2001).
2. Premature transcription termination and road-blocks to RNA polymerase
In bacteria, the synthesis of mRNA and protein are coupled. Therefore, ribosomes initiate translation on transcripts that are still in the process of being synthesized. Although this strategy allows for rapid changes in gene expression, it also increases the burden of non-stop mRNA through pre-mature transcription termination (Fig. 4). Hiroji Aiba’s group first reported that pre-mature transcription termination leads to ssrA-tagging of the lac repressor (LacI) (Abo et al., 2000). LacI represses transcription of the lac operon by binding to two operator sites that overlap the lacZYA promoter. However, an additional low-affinity operator site is found upstream of the lac promoter at a position that overlaps with the 3′-end of the lacI coding sequence. At high levels, LacI binds to this third operator site and promotes the production of non-stop lacI messages by acting as a physical barrier, or “road-block”, to transcription. Thus, LacI negatively regulates its own expression by inducing pre-mature transcription termination, which leads to the ssrA-tagging and degradation of LacI nascent chains. This feedback loop appears to maintain LacI at optimal levels, because ssrA+ cells are able to respond to lac operon inducers more rapidly than ΔssrA mutants (Abo et al., 2000).
Figure 4. Premature transcription termination and tmRNA activity.
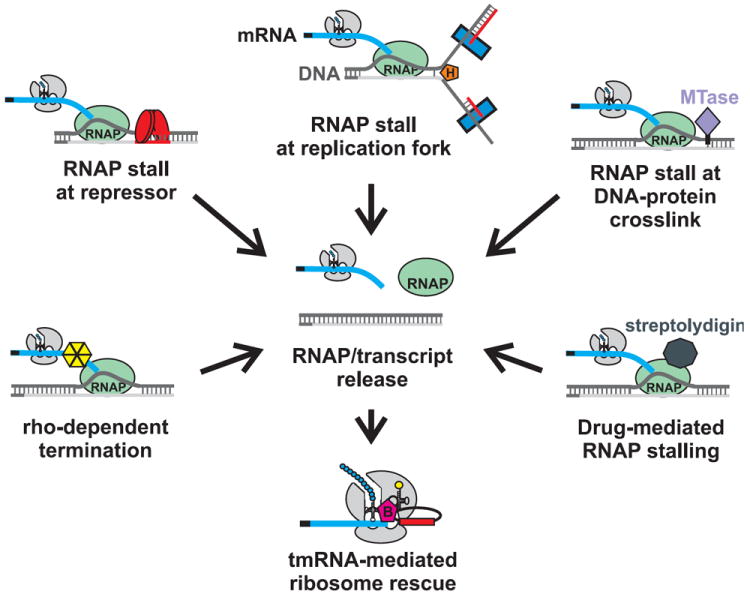
Potential mechanisms of premature transcription termination in bacteria. Collisions between RNA polymerase (RNAP) and DNA replication forks, DNA-bound transcription factors, or covalently cross-linked proteins are predicted to generate stalled transcription complexes. Antibiotics (e.g. streptolydigin) and adventitious Rho activity can also promote premature transcription termination. Termination within coding sequences generates non-stop mRNA, which ultimately leads to tmRNA-mediated ribosome rescue.
Transcription road-blocks have also been reported to induce tmRNA tagging activity in B. subtilis. The catabolite control protein, CcpA, is a transcription factor that regulates several carbon-utilization genes in Gram-positive bacteria. CcpA binds to cre (catabolite-responsive element) operator sites, which not only overlap regulated promoters but are also found within the coding region of several genes. Akira Muto and colleagues have shown that CcpA induces pre-mature transcription termination immediately upstream of a cre site within the trehalose permease gene (treP) (Ujiie et al., 2009). CcpA is likely responsible for the tagging of other B. subtilis proteins, such as YtoQ and FolA, whose DNA coding sequences also contain cre sites (Fujihara et al., 2002). It is unclear whether the tmRNA activity associated with CcpA road-blocks serves a regulatory function analogous to that of LacI in E. coli. Alternatively, non-stop mRNA may be a deleterious consequence of CcpA transcriptional control, and tmRNA is serving a quality control function to remove truncated transcripts and polypeptides.
An intriguing twist on the transcription road-block theme was recently reported by Ken Kreuzer, Ashok Bhagwat and their colleagues. They discovered that E. coli ssrA mutants are significantly more sensitive to 5-azacytidine than are ssrA+ cells (Kuo et al., 2010). 5-azacytidine is incorporated into the chromosome during replication and forms specific covalent cross-links with the cytosine DNA methyltransferase. These cross-links are required for the hypersensitivity phenotype, because an ssrA mutant lacking cytosine DNA methyltransferase was no more sensitive to 5-azacytidine than an isogenic ssrA+ strain. Kreuzer and colleagues propose that the DNA-methyltransferase cross-links block transcription elongation and tmRNA plays a role in disassembling the stalled RNA polymerase/polysome complex (Kuo et al., 2010). In support of this model, they also find that ssrA mutants have increased sensitivity to streptolydigin, an antibiotic that arrests RNA polymerase during elongation. Because nascent transcripts remain bound to streptolydigin inhibited polymerases, it is not clear whether tmRNA•SmpB can directly release ribosomes from the stalled complexes. The Rho transcription termination factor and the Mfd helicase are known to dissociate nascent transcripts from stalled polymerases, but neither of these factors affects the sensitivity of ssrA cells to 5-azacytidine (Kuo et al., 2010). Although it is possible that tmRNA•SmpB acts directly to release ribosomes associated with stalled transcription complexes, another unknown factor(s) could also play a role. Recent work from Jade Wang and Christophe Herman shows that DksA facilitates disassembly of transcriptional complexes that have collided with replication forks (Tehranchi et al., 2010). DksA is a transcription factor that, like the Gre transcription elongation factors, directly interacts with the polymerase active site via the secondary channel (Paul et al., 2004; Perederina et al., 2004). Therefore, DksA could conceivably release nascent transcripts from stalled polymerase complexes, and thereby facilitate subsequent tmRNA-mediated ribosome rescue.
3. RNase activity and non-stop mRNA
In principle, non-stop transcripts can also be generated during normal mRNA turnover. The degradation pathway for any given message is unique and depends upon its secondary structure content as well as the presence and location of specific endoribonuclease recognition sequences (Belasco, 2010; Regnier and Arraiano, 2000). It is generally accepted that mRNA degradation in E. coli is rate-limited by RNase E, which initiates turnover through endonucleolytic cleavage near the 5′-end of transcripts. RNase E then scans the downstream cleavage fragment in search of additional recognition sites (Carpousis et al., 2009). Thus, RNase E activity produces a series of mRNA fragments, which are subsequently degraded by a variety of 3′-to-5′ exonucleases (Andrade et al., 2009). Because RNase E mediated degradation occurs in the 5′-to-3′ direction, this pathway minimizes non-stop mRNA production. E. coli has long been used as a model for mRNA turnover, but it is increasingly clear that degradation pathways can be dramatically different in other bacteria. For example, B. subtilis has a unique complement of RNases, which mediate turnover pathways distinct from those in E. coli (Bechhofer, 2009; Condon, 2007, 2010). Although turnover pathways are diverse, bacterial messages are typically degraded rapidly without detectable intermediates. However, perturbations to turnover can generate significant levels of non-stop mRNA. David Bechhofer and colleagues have shown that B. subtilis mutants lacking polynucleotide phosphorylase (PNPase) accumulate mRNA degradation intermediates, and the level of ssrA-tagged proteins increases concomitantly (Oussenko et al., 2005). These results also underscore the differences in mRNA turnover between B. subtilis and E. coli, where PNPase is quantitatively less important for mRNA turnover (Deutscher and Reuven, 1991). E. coli cells lacking PNPase show no obvious increase in turnover intermediates and ssrA-tagging is unaffected in these mutant cells (Donovan and Kushner, 1986; Garza-Sánchez et al., 2009).
4. Toxin-antitoxin (TA) modules and mRNA interferase activity
Although normal mRNA turnover does not typically generate translatable degradation intermediates, small RNases encoded by toxin-antitoxin (TA) modules can produce non-stop mRNA. TA modules are widely distributed throughout the eubacteria and archaea and appear to play a role in controlling cell growth, but their precise physiological function(s) remains controversial (Engelberg-Kulka et al., 2005; Gerdes et al., 2005; Magnuson, 2007; Van Melderen and Saavedra De Bast, 2009). Antitoxins bind to their cognate toxins and neutralize toxin-mediated growth inhibition. Toxins are typically activated under stress conditions (e.g. starvation and antibiotic treatment) through ClpAP- and Lon-dependent degradation of antitoxins (Aizenman et al., 1996; Christensen and Gerdes, 2004; Christensen et al., 2001; Christensen et al., 2003; Sat et al., 2001; Sat et al., 2003). The majority of toxins characterized thus far are small RNases that preferentially cleave mRNA and therefore these enzymes are often referred to as “mRNA interferases” in the literature (Yamaguchi and Inouye, 2009). Overproduced mRNA interferases are cytotoxic and a series of papers from Hannah Engelberg-Kulka and colleagues suggest that some toxins mediate programmed cell death in bacteria (Aizenman et al., 1996; Engelberg-Kulka et al., 2005; Sat et al., 2001; Sat et al., 2003). This appears to be the case during Myxococcus xanthus fruiting body formation and sporulation, in which approximately 80% of cells activate the MazF toxin and undergo cell death (Nariya and Inouye, 2008). Another model proposed by Kenn Gerdes postulates that TA modules are response regulators that facilitate changes in gene expression in response to environmental stress (Christensen et al., 2003; Christensen-Dalsgaard et al., 2008; Gerdes, 2000; Gerdes et al., 2005; Pandey and Gerdes, 2005). This latter model is supported by data showing that cells can recover from toxin activation through a transcriptional feedback loop that increases antitoxin production (Christensen et al., 2003; Gerdes et al., 2005; Overgaard et al., 2009; Pedersen et al., 2002). Regardless of their physiological function, mRNA interferases produce non-stop mRNA and tmRNA-mediated ribosome rescue assists in the recovery from toxin activation (Christensen and Gerdes, 2003; Christensen et al., 2003).
The mRNA interferases cleave transcripts in either a ribosome-independent or ribosome-dependent manner. Ribosome-independent mRNA interferases are typified by E. coli MazF, which Masayori Inouye’s group showed is a site-specific RNase that cleaves at the 5′-side of ACA sequences in single-stranded RNA (Zhang et al., 2003). Although ribosomal RNAs and other stable RNAs contain ACA sequences, these motifs are usually found within secondary structures and therefore mRNAs are the primary MazF substrates in vivo (Suzuki et al., 2005). Intriguingly, there is a bias against ACA triplets in ssrA from the Enterobacteriaceae, suggesting the selection for tmRNAs that are resistant to MazF cleavage (Baik et al., 2009). Ribosome-dependent mRNA interferases use the ribosome as a scaffold to identify and cleave specific codons. This activity was first demonstrated for the E. coli RelE toxin in an elegant study by Måns Ehrenberg, Kenn Gerdes and their colleagues (Pedersen et al., 2003). They found that purified RelE cleaves A-site codons between the 2nd and 3rd nucleotides. RelE cleaves several different codons, but A-site CAG and UAG codons are cleaved most efficiently (Pedersen et al., 2003). The E. coli YoeB toxin appears to be related to RelE and also has A-site nuclease activity (Christensen-Dalsgaard and Gerdes, 2008; Zhang and Inouye, 2009). Recently, Inouye and colleagues have identified another class of ribosome-dependent mRNA interferase activity. The E. coli YafQ toxin cleaves mRNA at the 3′-border of the translating ribosome (Prysak et al., 2009). YafQ induced cleavages tend to occur during the initiation phase of protein synthesis, suggesting the enzyme may recognize initiation complexes (Prysak et al., 2009). Although the cleavage mechanisms are varied, all mRNA interferases appear to inhibit cell growth by producing non-stop mRNA.
VI. mRNA cleavage resulting from translational arrest
The original model of ribosome rescue postulated that tmRNA acts on ribosomes stalled at the ends of truncated mRNAs. Shortly thereafter, it became clear that translational pausing on full-length messages could also elicit ribosome rescue activity. Because truncated mRNA is required for tmRNA activity, the message must be cleaved (or degraded) subsequent to ribosome arrest. As described below, mRNA cleavage occurs at a variety of positions with respect to the paused ribosome, and in some instances these cleavages induce tmRNA-mediated ribosome rescue.
1. Inefficient translation termination and A-site mRNA cleavage
Leif Isaksson and colleagues first discovered that the last two residues of the nascent chain influence translation termination (Bjornsson et al., 1996; Mottagui-Tabar et al., 1994). In general, C-terminal Pro, Asp and Gly residues interfere with release factor activity and promote stop codon readthrough. These nascent peptide sequences also induce ssrA tagging of full-length proteins in E. coli, with C-terminal Asp-Pro and Pro-Pro sequences leading to particularly high levels of tagging (Hayes et al., 2002a; Roche and Sauer, 2001). Ribosome bound peptidyl prolyl-tRNA reacts with puromycin and aminoacyl-tRNA more slowly than other peptidyl-tRNA species (Muto and Ito, 2008; Wohlgemuth et al., 2008), perhaps accounting for the alternative reactions that occur during termination in vivo. Nascent peptide induced ribosome pausing is associated with cleavage of the A-site stop codon (Hayes and Sauer, 2003; Sunohara et al., 2004b). This RNase activity requires the ribosome and only occurs in response to translational pausing (Hayes and Sauer, 2003; Janssen and Hayes, 2009). A-site cleavage products accumulate to high levels in ΔssrA cells, but are difficult to detect in ssrA+ cells. Presumably, tmRNA activity facilitates the turnover of A-site truncated mRNA by releasing stalled ribosomes from the 3′-ends of these transcripts (Yamamoto et al., 2003). Together, these observations suggest that A-site mRNA cleavage provides a mechanism for tmRNA recruitment when ribosomes pause on full-length transcripts. A-site mRNA cleavage activity is very similar to that of ribosome-dependent mRNA interferases like RelE and YoeB. However, A-site cleavage still occurs in cells lacking the RelE, MazF, ChpBK, YoeB, YafQ, and YhaV toxins (Garza-Sánchez et al., 2008; Hayes and Sauer, 2003). Because the known mRNA interferases are not involved, we and Hiroji Aiba’s group proposed that the ribosome itself may catalyze this A-site cleavage reaction (Hayes and Sauer, 2003; Sunohara et al., 2004a).
Subsequent work has shown that A-site mRNA cleavage is a more complicated process than originally appreciated. A-site cleavage does not occur in Δrnb mutants, which lack RNase II (Garza-Sánchez et al., 2009). RNase II is the major 3′-to-5′ exoribonuclease responsible for mRNA turnover in E. coli (Deutscher and Reuven, 1991). In Δrnb cells, prolonged translational pausing produces transcripts that are truncated +12 nucleotides downstream of the A-site codon. This position corresponds to the ribosome leading edge or “toeprint” (Yusupova et al., 2001), and suggests that downstream mRNA is degraded to the 3′-border of the stalled ribosome. The effect of the Δrnb mutation on A-site mRNA cleavage is indirect because RNase II itself is unable to degrade mRNA into the ribosomal A-site. Purified RNase II only degrades mRNA to the +18 position with respect to the A-site codon within the stalled ribosome (Garza-Sánchez et al., 2009). There are at least two models to explain these findings. Firstly, deletion of rnb could alter the expression of other RNases such that +12 cleavage is favored over A-site mRNA cleavage. This model is consistent with microarray data showing that global transcription is significantly altered in E. coli Δrnb mutants (Mohanty and Kushner, 2003). Alternatively, RNase II mediated degradation of downstream mRNA could be a precondition for further degradation into the A-site codon, perhaps by facilitating the activity of another RNase.
2. Rare codons and amino acid starvation
Ribosomes also pause and recruit tmRNA during the translation of rare codon clusters (Collier et al., 2002; Hayes et al., 2002b; Roche and Sauer, 1999). Rare codons are under-represented in the genome and are generally decoded by low abundance tRNA species (Dong et al., 1996). However, ribosome pausing at a given codon not necessarily correlated to the absolute concentration of cognate tRNA in the cell. For example, although tRNAHis and tRNA3Arg are processed from the same transcript and present at approximately the same levels in E. coli tRNA (Dong et al., 1996); ribosomes rapidly synthesize hexa-histidine (His6) sequences, but pause during the translation of tandem CGG arginine codons (Garza-Sánchez et al., 2008). Acid-urea gel analysis reveals that most cellular tRNA3Arg is deacylated under these conditions, whereas about 70% of tRNAHis is charged. Deacylated tRNA3Arg is probably charged less efficiently because it must compete with the more abundant tRNA2Arg species for aminoacylation. In contrast, there is only one tRNAHis species and therefore no competition for charging. These and other observations suggest that the kinetics of tRNA recycling and aminoacylation influence translational pausing at specific codons (Elf et al., 2003).
Several groups have shown that tmRNA acts during the translation of rare arginine codon clusters, including AGG, AGA, CGA and CGG (Garza-Sánchez et al., 2008; Li et al., 2006; Roche and Sauer, 1999). In each instance, translational pausing results in mRNA truncation, either within the A-site codon or at downstream positions corresponding to the leading edge of the ribosome (Garza-Sánchez et al., 2008; Li et al., 2006). Truncations at each position appear to be sufficient for tmRNA•SmpB recruitment and nascent chain tagging. Translational pausing at sense codons also occurs under conditions that promote deacylation of cellular tRNA. Aminoacyl-tRNA synthetase inhibitors (e.g. serine hydroxamate) and acute amino acid starvation both induce site-specific translational arrest and ssrA-peptide tagging in E. coli (Garza-Sánchez et al., 2008; Li et al., 2008). Thus, the tmRNA system may play an important role in the physiological response to starvation. Tagging ensures that the incomplete nascent chains will be degraded, thereby replenishing amino acid pools to support synthesis of protein involved in de novo amino acid biosynthesis.
3. tmRNA activity during programmed ribosome pauses
Translational pausing is commonly used to regulate gene expression in bacteria. The classical example of this type of regulation is transcriptional attenuation, in which paused ribosomes control transcriptional elongation by influencing a switch between terminator and anti-terminator structures within the nascent transcript (Henkin and Yanofsky, 2002). Ribosome pausing during the synthesis of leader peptides can also regulate translation initiation of downstream cistrons in a process termed translational attenuation (Ramu et al., 2009). In principle, the ribosome rescue activity of tmRNA could interfere with regulatory pathways that rely on ribosome pausing. However, ribosomes undergoing regulatory arrest appear to be refractory to tmRNA activity. The E. coli SecM nascent chain mediates a conditional ribosome pause that controls translation of the downstream secA ORF through an attenuation mechanism (Nakatogawa and Ito, 2002; Nakatogawa et al., 2004). The regulation of SecA synthesis is identical in ssrA+ and ΔssrA backgrounds, strongly suggesting that the duration of the SecM-mediated translational arrest is unaffected by tmRNA (Garza-Sánchez et al., 2006; Nakatogawa and Ito, 2001, 2002). Indeed, tmRNA•SmpB is unable to bind to SecM-arrested ribosomes because the A site is filled with prolyl-tRNAPro (Garza-Sánchez et al., 2006; Muto et al., 2006). Similarly, the E. coli TnaC nascent peptide induces a regulated ribosome arrest during translation termination, but tmRNA activity is blocked by release factor-2 bound in the ribosome A-site (Gong et al., 2007). Taken together, these observations suggest that regulated ribosome pauses may be generally refractory to tmRNA-mediated ribosome rescue.
Other programmed translational arrests lead to mRNA cleavage at the 5′-edge of the ribosome. The E. coli daa operon encodes proteins required for biogenesis of F1845 fimbriae. Remarkably, proper expression of the fimbrial adhesin (encoded by the daaE ORF) requires endonucleolytic processing of the polycistronic daa transcript (Bilge et al., 1993). Steve Moseley and colleagues have shown this processing requires a programmed ribosome arrest during translation of the short daaP ORF found upstream of daaE (Loomis et al., 2001; Loomis and Moseley, 1998). A critical Gly-Pro-Pro motif in the DaaP nascent peptide induces ribosome arrest, and the transcript is subsequently cleaved at a position 12 to 15 nucleotides upstream of the A-site codon (Loomis et al., 2001). This cleavage site corresponds to the 5′-edge of the stalled ribosome. Because E. coli lacks a 5′-to-3′ exoribonuclease capable of degrading mRNA to the 5′-border of stalled ribosomes, this activity is presumably is mediated by an unknown endonuclease (Bilge et al., 1993). Although it has been suggested that tmRNA plays a role in daa mRNA processing (Koo et al., 2004; Loomis et al., 2001), the DaaP-arrested ribosome should not recruit tmRNA•SmpB because the downstream transcript encoding daaE is intact. A similar mRNA processing event occurs in B. subtilis when ribosomes pause during translation of the ermC leader sequence. In the presence of sub-inhibitory concentrations of erythromycin, the ErmC nascent peptide mediates a site-specific ribosome arrest that facilitates translation of the downstream ermA rRNA methyltransferase through an attenuation mechanism (Dubnau, 1985; Hahn et al., 1982). Similar to the daa operon, pausing during ermC translation results in mRNA cleavage upstream of the paused ribosome and stabilization of the downstream sequence (Bechhofer and Zen, 1989; Drider et al., 2002). David Bechhofer and colleagues have discovered that this mRNA processing is mediated by RNase J1 (Yao et al., 2009), an essential enzyme with both endonuclease and 5′-to-3′ exonuclease activities (Mathy et al., 2007). Although tmRNA activity has not been examined during ErmC-mediated ribosome pausing, like the DaaP-arrested ribosome, the intact downstream message should preclude ribosome rescue.
VII. Other determinants of tmRNA activity
Recent work suggests that co-translational protein folding may also influence tmRNA activity. The E. coli GalE protein (UDP-galactose 4-epimerse) is ssrA-tagged at dozens of sites in distinct clusters throughout the C-terminal domain (Ruhe and Hayes, 2010). Codon usage and mRNA secondary structure do not account for ribosome pausing in this system because synonymous recoding of the galE gene has no effect on ssrA tagging. GalE tagging is unaffected by both the overproduction and deletion of RNases responsible for mRNA turnover, suggesting that tagging is not due to the translation of mRNA degradation intermediates. Genetic fusions revealed that the N-terminal domain of GalE is sufficient to induce ssrA tagging of a heterologous thioredoxin (TrxA) domain fused at the C-terminus (Ruhe and Hayes, 2010). Moreover, the C-terminal domain of GalE is no longer tagged when fused to the C-terminus of TrxA. Together, these findings demonstrate that the 3′-coding sequence of galE is not sufficient for tagging, suggesting that this tmRNA activity depends on the larger genetic or molecular context. One explanation for these results is that co-translational misfolding of the GalE N-terminal domain induces ribosome pausing during synthesis of the C-terminal domain (Hayes and Keiler, 2010; Ruhe and Hayes, 2010). This model implies that the ribosome is able to monitor co-translational events and modulate its activity accordingly. Intriguingly, multidomain proteins are tagged at significantly higher levels in cells lacking DnaK, which has co-translational chaperone activity (Z. Ruhe and C.S.H, unpublished results). Although this latter observation is broadly consistent with the co-translational misfolding model, DnaK could also influence tmRNA activity by modulating mRNA turnover pathways or ribosome assembly (Maki et al., 2002).
Another unique ssrA-tagging determinant has been discovered in C. crescentus by Ken Keiler and colleagues. Hong et al. used a proteomic approach to identify ssrA(His6) tagging sites in 73 different C. crescentus proteins (Hong et al., 2007). A handful of these tagging sites correspond to tmRNA activity during translation of rare and stop codons, but the mechanisms underlying tagging of the remaining proteins were not readily apparent. Sequence analysis revealed a common motif (CGACAAGATCGTCGTG) present within many of the genes that encode tagged proteins (Hong et al., 2007). This DNA sequence could represent a protein-binding site, but a transcription road-block cannot be involved because the motif is always found upstream of the corresponding tagging sites. Similarly, the motif is unlikely to act as an RNase recognition determinant due to its variable position (9 to 180 nucleotides) upstream of the observed tagging sites. It is possible that the motif is a loading site for Rho or another transcription termination factor, thereby promoting termination at the downstream sites (Hong et al., 2007). Because many of the tagged proteins are involved in DNA replication and repair, perhaps these phenomena represent a concerted regulatory strategy acting at the level of transcription termination.
VIII. Role of tmRNA in physiology and the regulation of gene expression
In E. coli, approximately 0.4% of all protein chains are tagged by the tmRNA system (Moore and Sauer, 2005), indicating that translational problems arise fairly frequently. Roche and Sauer have shown that several hundred protein species are ssrA-tagged in E. coli during logarithmic growth conditions (Roche and Sauer, 2001). Because tmRNA activity can impinge on the synthesis of any protein in the cell, the system is important for gene expression. Although somewhat counterintuitive given its role in proteolysis, tmRNA activity promotes higher yields of full-length protein from problematic messages (Ranquet et al., 2001). As a consequence, the deletion of ssrA leads to synthetic phenotypes with a variety of temperature sensitive mutations in essential genes (Ando et al., 1996; Nakano et al., 2001; Singh and Varshney, 2004). tmRNA•SmpB also plays important roles during stress response. In B. subtilis, tmRNA activity is important for cell growth at both low and high temperatures, and exposure to ethanol and CdCl2 increases the transcription of tmRNA (Muto et al., 2000; Shin and Price, 2007). trans-Translation has also been implicated in the stress responses of Streptomyces coelicolor (Barends et al., 2010; Paget et al., 2001) and Helicobacter pylori (Thibonnier et al., 2008). Presumably, environmental stress promotes the generation of non-stop mRNA or otherwise interferes with translation, thereby increasing the demand for tmRNA-mediated ribosome rescue.
The tmRNA system also plays critical roles in cell cycle control and bacterial development. tmRNA activity is required for the correct timing of DNA replication during the C. crescentus cell cycle (Keiler and Shapiro, 2003b). In ΔssrA mutants, there is a profound delay in dnaA transcription, which is required to initiate replication (Cheng and Keiler, 2009). This temporal control is dependent upon a cis-acting element in the dnaA promoter, and also requires degradation of ssrA-tagged proteins (Cheng and Keiler, 2009; Keiler and Shapiro, 2003b). Together, these observations suggest that tmRNA activity regulates a repressor of dnaA transcription, perhaps by promoting its degradation. tmRNA undergoes cycles of synthesis and degradation during the C. crescentus cell cycle, and presumably these oscillations are directly related to transcriptional control of dnaA and DNA replication (Keiler and Shapiro, 2001, 2003a). Although tmRNA is a global molecular quality control system, these findings illustrate how trans-translation can also regulate gene expression and thereby influence complex cellular processes.
The tmRNA system is required for developmental pathways in B. subtilis and Streptomyces coelicolor. Hyouta Himeno and colleagues have discovered that B. subtilis ΔssrA mutants sporulate at a greatly reduced efficiency compared to ssrA+ cells (Abe et al., 2008). This defect is due to reduced production of SpoIVCA, the recombinase responsible for excision of the chromosomal skin element during sporulation. The skin element interrupts the sigK gene and must be removed to produce functional σK (Abe et al., 2008), which is a transcription factor required for gene expression during later stages of sporulation. These observations suggest that ribosomes stall during the synthesis of SpoIVCA or one (or more) of its regulators. Additionally, B. subtilis ΔssrA and ΔsmpB mutants are not competent to take up foreign DNA, though the mechanism(s) underlying this phenotype is still unclear (C.S.H, unpublished results). Sporulation in Streptomyces coelicolor is also regulated by tmRNA activity. The S. coelicolor ΔssrA mutant does not sporulate efficiently; and this phenotype may be related to ssrA-tagging of the SsgR, SsgA and SsgF proteins, which regulate sporulation (Barends et al., 2010).
Because tmRNA•SmpB functions in global quality control and stress responses, it is not surprising that the system is critical for bacterial pathogenesis. Before the function of tmRNA or SmpB was appreciated, the smpB gene in Salmonella typhimurium was found to be important for bacterial survival within macrophages and for resistance to oxidative stress (Baumler et al., 1994; Fields et al., 1986). These findings were subsequently confirmed by Mike Mahan, whose group demonstrated that the Salmonella enterica ΔssrA mutant is ~200-fold less virulent in a mouse model (Julio et al., 2000). Wali Karzai and colleagues have found that ΔsmpB-ssrA mutants of Yersinia pestis and Yersinia pseudotuberculosis are also dramatically less virulent than wild-type bacteria (Okan et al., 2006; Okan et al., 2010). These virulence phenotypes are due to defects in flagella assembly and reduced secretion of and expression of Type III effector proteins (Yops) (Okan et al., 2006; Okan et al., 2010). The Y. pestis ΔsmpB-ssrA mutant elicits a vigorous antibody response in a mouse model, and challenged animals are well protected upon subsequent inoculation with virulent Y. pestis (Okan et al., 2010). Thus, the Y. pestis ΔsmpB-ssrA mutant appears to be an effective attenuated live vaccine against plague. Although tmRNA activity has been examined in only a handful of pathogens thus far, ribosome rescue is likely to play an important and general role in bacterial pathogenesis.
IX. Concluding remarks
The emerging view is that ribosome rescue is an essential function for bacteria and probably all cells. In bacteria, the tmRNA quality control system is the primary mediator of ribosome rescue and thus is critical for several aspects of bacterial fitness, including development, stress response and pathogenesis. Much has been learned about the function and biological roles of tmRNA, however several outstanding questions remain. Although excellent progress has been made developing structural models of tmRNA function, we still lack high-resolution structures of tmRNA•SmpB at various stages of trans-translation. Such structures would illuminate the various functions ascribed to SmpB and demonstrate precisely how tmRNA moves through the ribosome. The existence and identities of tmRNA-independent ribosome rescue systems outside of E. coli and related γ-proteobacteria remain to be discovered. Finally, the bases for many ssrA phenotypes are incompletely understood. In principle, tmRNA can control the levels of specific proteins through ssrA-tagging and subsequent proteolysis. In some instances, the identification of ssrA-tagged proteins has provided some insight in ssrA phenotypes. However, it is not known how some phenotypes are achieved when only a small fraction of some protein chains is tagged and degraded. Clearly, much remains to be learned before we have a complete understanding of the biochemical mechanism and physiological importance of trans-translation.
Acknowledgments
Research in the authors’ laboratory is supported by grants GM078634 and U54 AI065359 from the NIH. The content of this work is the sole responsibility of the authors and does not necessarily represent the official views of the National Institute of Allergy and Infectious Diseases or the National Institutes of Health.
References
- Abe T, Sakaki K, Fujihara A, Ujiie H, Ushida C, Himeno H, Sato T, Muto A. tmRNA-dependent trans-translation is required for sporulation in Bacillus subtilis. Molecular Microbiology. 2008;69:1491–1498. doi: 10.1111/j.1365-2958.2008.06381.x. [DOI] [PubMed] [Google Scholar]
- Abo T, Inada T, Ogawa K, Aiba H. SsrA-mediated tagging and proteolysis of LacI and its role in the regulation of lac operon. EMBO Journal. 2000;19:3762–3769. doi: 10.1093/emboj/19.14.3762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abo T, Ueda K, Sunohara T, Ogawa K, Aiba H. SsrA-mediated protein tagging in the presence of miscoding drugs and its physiological role in Escherichia coli. Genes to Cells. 2002;7:629–638. doi: 10.1046/j.1365-2443.2002.00549.x. [DOI] [PubMed] [Google Scholar]
- Aizenman E, Engelberg-Kulka H, Glaser G. An Escherichia coli chromosomal “addiction module” regulated by guanosine [corrected] 3’,5’-bispyrophosphate: a model for programmed bacterial cell death. Proceedings of the National Academy of Sciences U S A. 1996;93:6059–6063. doi: 10.1073/pnas.93.12.6059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akerley BJ, Rubin EJ, Novick VL, Amaya K, Judson N, Mekalanos JJ. A genome-scale analysis for identification of genes required for growth or survival of Haemophilus influenzae. Proceedings of the National Academy of Sciences U S A. 2002;99:966–971. doi: 10.1073/pnas.012602299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ando H, Kitabatake M, Inokuchi H. 10Sa RNA complements the temperature-sensitive phenotype caused by a mutation in the phosphoribosyl pyrophosphate synthetase (prs) gene in Escherichia coli. Genes & Genetic Systems. 1996;71:47–50. doi: 10.1266/ggs.71.47. [DOI] [PubMed] [Google Scholar]
- Andrade JM, Pobre V, Silva IJ, Domingues S, Arraiano CM. The role of 3’-5’ exoribonucleases in RNA degradation. Progress in Molecular Biology and Translational Science. 2009;85:187–229. doi: 10.1016/S0079-6603(08)00805-2. [DOI] [PubMed] [Google Scholar]
- Asano K, Kurita D, Takada K, Konno T, Muto A, Himeno H. Competition between trans-translation and termination or elongation of translation. Nucleic Acids Research. 2005;33:5544–5552. doi: 10.1093/nar/gki871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baik S, Inoue K, Ouyang M, Inouye M. Significant bias against the ACA triplet in the tmRNA sequence of Escherichia coli K-12. Journal of Bacteriology. 2009;191:6157–6166. doi: 10.1128/JB.00699-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baranov PV, Vestergaard B, Hamelryck T, Gesteland RF, Nyborg J, Atkins JF. Diverse bacterial genomes encode an operon of two genes, one of which is an unusual class-I release factor that potentially recognizes atypical mRNA signals other than normal stop codons. Biology Direct. 2006;1:28. doi: 10.1186/1745-6150-1-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barends S, Karzai AW, Sauer RT, Wower J, Kraal B. Simultaneous and functional binding of SmpB and EF-Tu-TP to the alanyl acceptor arm of tmRNA. Journal of Molecular Biology. 2001;314:9–21. doi: 10.1006/jmbi.2001.5114. [DOI] [PubMed] [Google Scholar]
- Barends S, Zehl M, Bialek S, de Waal E, Traag BA, Willemse J, Jensen ON, Vijgenboom E, van Wezel GP. Transfer-messenger RNA controls the translation of cell-cycle and stress proteins in Streptomyces. EMBO Reports. 2010;11:119–125. doi: 10.1038/embor.2009.255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baumler AJ, Kusters JG, Stojiljkovic I, Heffron F. Salmonella typhimurium loci involved in survival within macrophages. Infection and Immunity. 1994;62:1623–1630. doi: 10.1128/iai.62.5.1623-1630.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bechhofer DH. Messenger RNA decay and maturation in Bacillus subtilis. Progress in Molecular Biology and Translational Science. 2009;85:231–273. doi: 10.1016/S0079-6603(08)00806-4. [DOI] [PubMed] [Google Scholar]
- Bechhofer DH, Zen KH. Mechanism of erythromycin-induced ermC mRNA stability in Bacillus subtilis. Journal of Bacteriology. 1989;171:5803–5811. doi: 10.1128/jb.171.11.5803-5811.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belasco JG. All things must pass: contrasts and commonalities in eukaryotic and bacterial mRNA decay. Nature Reviews Molecular Cell Biology. 2010;11:467–478. doi: 10.1038/nrm2917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bilge SS, Apostol JM, Jr, Aldape MA, Moseley SL. mRNA processing independent of RNase III and RNase E in the expression of the F1845 fimbrial adhesin of Escherichia coli. Proceedings of the National Academy of Sciences U S A. 1993;90:1455–1459. doi: 10.1073/pnas.90.4.1455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bjornsson A, Mottagui-Tabar S, Isaksson LA. Structure of the C-terminal end of the nascent peptide influences translation termination. EMBO Journal. 1996;15:1696–1704. [PMC free article] [PubMed] [Google Scholar]
- Bugaeva EY, Shpanchenko OV, Felden B, Isaksson LA, Dontsova OA. One SmpB molecule accompanies tmRNA during its passage through the ribosomes. FEBS Letters. 2008;582:1532–1536. doi: 10.1016/j.febslet.2008.03.049. [DOI] [PubMed] [Google Scholar]
- Carpousis AJ, Luisi BF, McDowall KJ. Endonucleolytic initiation of mRNA decay in Escherichia coli. Progress in Molecular Biology and Translational Science. 2009;85:91–135. doi: 10.1016/S0079-6603(08)00803-9. [DOI] [PubMed] [Google Scholar]
- Chadani Y, Ono K, Kutsukake K, Abo T. Escherichia coli YaeJ protein mediates a novel ribosome-rescue pathway distinct from SsrA- and ArfA-mediated pathways. Mol Microbiol. 2011;80:772–785. doi: 10.1111/j.1365-2958.2011.07607.x. [DOI] [PubMed] [Google Scholar]
- Chadani Y, Ono K, Ozawa S, Takahashi Y, Takai K, Nanamiya H, Tozawa Y, Kutsukake K, Abo T. Ribosome rescue by Escherichia coli ArfA (YhdL) in the absence of trans-translation system. Molecular Microbiology. 2010;78:796–808. doi: 10.1111/j.1365-2958.2010.07375.x. [DOI] [PubMed] [Google Scholar]
- Chauhan AK, Apirion D. The gene for a small stable RNA (10Sa RNA) of Escherichia coli. Molecular Microbiology. 1989;3:1481–1485. doi: 10.1111/j.1365-2958.1989.tb00133.x. [DOI] [PubMed] [Google Scholar]
- Cheng K, Ivanova N, Scheres SH, Pavlov MY, Carazo JM, Hebert H, Ehrenberg M, Lindahl M. tmRNA•SmpB complex mimics native aminoacyl-tRNAs in the A site of stalled ribosomes. Journal of Structural Biology. 2010;169:342–348. doi: 10.1016/j.jsb.2009.10.015. [DOI] [PubMed] [Google Scholar]
- Cheng L, Keiler KC. Correct timing of dnaA transcription and initiation of DNA replication requires trans translation. Journal of Bacteriology. 2009;191:4268–4275. doi: 10.1128/JB.00362-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choy JS, Aung LL, Karzai AW. Lon protease degrades transfer-messenger RNA-tagged proteins. Journal of Bacteriology. 2007;189:6564–6571. doi: 10.1128/JB.00860-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christensen SK, Gerdes K. RelE toxins from bacteria and Archaea cleave mRNAs on translating ribosomes, which are rescued by tmRNA. Molecular Microbiology. 2003;48:1389–1400. doi: 10.1046/j.1365-2958.2003.03512.x. [DOI] [PubMed] [Google Scholar]
- Christensen SK, Gerdes K. Delayed-relaxed response explained by hyperactivation of RelE. Molecular Microbiology. 2004;53:587–597. doi: 10.1111/j.1365-2958.2004.04127.x. [DOI] [PubMed] [Google Scholar]
- Christensen SK, Mikkelsen M, Pedersen K, Gerdes K. RelE, a global inhibitor of translation, is activated during nutritional stress. Proceedings of the National Academy of Sciences U S A. 2001;98:14328–14333. doi: 10.1073/pnas.251327898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christensen SK, Pedersen K, Hansen FG, Gerdes K. Toxin-antitoxin loci as stress-response-elements: ChpAK/MazF and ChpBK cleave translated RNAs and are counteracted by tmRNA. Journal of Molecular Biology. 2003;332:809–819. doi: 10.1016/s0022-2836(03)00922-7. [DOI] [PubMed] [Google Scholar]
- Christensen-Dalsgaard M, Gerdes K. Translation affects YoeB and MazF messenger RNA interferase activities by different mechanisms. Nucleic Acids Research. 2008;36:6472–6481. doi: 10.1093/nar/gkn667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christensen-Dalsgaard M, Overgaard M, Winther KS, Gerdes K. RNA decay by messenger RNA interferases. Methods Enzymol. 2008;447:521–535. doi: 10.1016/S0076-6879(08)02225-8. [DOI] [PubMed] [Google Scholar]
- Collier J, Binet E, Bouloc P. Competition between SsrA tagging and translational termination at weak stop codons in Escherichia coli. Molecular Microbiology. 2002;45:745–754. doi: 10.1046/j.1365-2958.2002.03045.x. [DOI] [PubMed] [Google Scholar]
- Condon C. Maturation and degradation of RNA in bacteria. Current Opinion in Microbiology. 2007;10:271–278. doi: 10.1016/j.mib.2007.05.008. [DOI] [PubMed] [Google Scholar]
- Condon C. What is the role of RNase J in mRNA turnover? RNA Biology. 2010;7:316–321. doi: 10.4161/rna.7.3.11913. [DOI] [PubMed] [Google Scholar]
- Crandall J, Rodriguez-Lopez M, Pfeiffer M, Mortensen B, Buskirk A. rRNA mutations that inhibit transfer-messenger RNA activity on stalled ribosomes. Journal of Bacteriology. 2010;192:553–559. doi: 10.1128/JB.01178-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de la Cruz J, Vioque A. Increased sensitivity to protein synthesis inhibitors in cells lacking tmRNA. RNA. 2001;7:1708–1716. [PMC free article] [PubMed] [Google Scholar]
- Deutscher MP, Reuven NB. Enzymatic basis for hydrolytic versus phosphorolytic mRNA degradation in Escherichia coli and Bacillus subtilis. Proceedings of the National Academy of Sciences U S A. 1991;88:3277–3280. doi: 10.1073/pnas.88.8.3277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doma MK, Parker R. RNA quality control in eukaryotes. Cell. 2007;131:660–668. doi: 10.1016/j.cell.2007.10.041. [DOI] [PubMed] [Google Scholar]
- Dong G, Nowakowski J, Hoffman DW. Structure of small protein B: the protein component of the tmRNA-SmpB system for ribosome rescue. EMBO Journal. 2002;21:1845–1854. doi: 10.1093/emboj/21.7.1845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong H, Nilsson L, Kurland CG. Co-variation of tRNA abundance and codon usage in Escherichia coli at different growth rates. Journal of Molecular Biology. 1996;260:649–663. doi: 10.1006/jmbi.1996.0428. [DOI] [PubMed] [Google Scholar]
- Donovan WP, Kushner SR. Polynucleotide phosphorylase and ribonuclease II are required for cell viability and mRNA turnover in Escherichia coli K-12. Proceedings of the National Academy of Sciences U S A. 1986;83:120–124. doi: 10.1073/pnas.83.1.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drider D, DiChiara JM, Wei J, Sharp JS, Bechhofer DH. Endonuclease cleavage of messenger RNA in Bacillus subtilis. Molecular Microbiology. 2002;43:1319–1329. doi: 10.1046/j.1365-2958.2002.02830.x. [DOI] [PubMed] [Google Scholar]
- Dubnau D. Induction of ermC requires translation of the leader peptide. EMBO J. 1985;4:533–537. doi: 10.1002/j.1460-2075.1985.tb03661.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dulebohn D, Choy J, Sundermeier T, Okan N, Karzai AW. Trans-translation: the tmRNA-mediated surveillance mechanism for ribosome rescue, directed protein degradation, and nonstop mRNA decay. Biochemistry. 2007;46:4681–4693. doi: 10.1021/bi6026055. [DOI] [PubMed] [Google Scholar]
- Elf J, Nilsson D, Tenson T, Ehrenberg M. Selective charging of tRNA isoacceptors explains patterns of codon usage. Science. 2003;300:1718–1722. doi: 10.1126/science.1083811. [DOI] [PubMed] [Google Scholar]
- Engelberg-Kulka H, Hazan R, Amitai S. mazEF: a chromosomal toxin-antitoxin module that triggers programmed cell death in bacteria. J Cell Sci. 2005;118:4327–4332. doi: 10.1242/jcs.02619. [DOI] [PubMed] [Google Scholar]
- Felden B, Hanawa K, Atkins JF, Himeno H, Muto A, Gesteland RF, McCloskey JA, Crain PF. Presence and location of modified nucleotides in Escherichia coli tmRNA: structural mimicry with tRNA acceptor branches. EMBO Journal. 1998;17:3188–3196. doi: 10.1093/emboj/17.11.3188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fields PI, Swanson RV, Haidaris CG, Heffron F. Mutants of Salmonella typhimurium that cannot survive within the macrophage are avirulent. Proceedings of the National Academy of Sciences U S A. 1986;83:5189–5193. doi: 10.1073/pnas.83.14.5189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flynn JM, Levchenko I, Seidel M, Wickner SH, Sauer RT, Baker TA. Overlapping recognition determinants within the ssrA degradation tag allow modulation of proteolysis. Proceedings of the National Academy of Sciences U S A. 2001;98:10584–10589. doi: 10.1073/pnas.191375298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- French CT, Lao P, Loraine AE, Matthews BT, Yu H, Dybvig K. Large-scale transposon mutagenesis of Mycoplasma pulmonis. Molecular Microbiology. 2008;69:67–76. doi: 10.1111/j.1365-2958.2008.06262.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu J, Hashem Y, Wower I, Lei J, Liao HY, Zwieb C, Wower J, Frank J. Visualizing the transfer-messenger RNA as the ribosome resumes translation. EMBO Journal. 2010;29:3819–3825. doi: 10.1038/emboj.2010.255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujihara A, Tomatsu H, Inagaki S, Tadaki T, Ushida C, Himeno H, Muto A. Detection of tmRNA-mediated trans-translation products in Bacillus subtilis. Genes to Cells. 2002;7:343–350. doi: 10.1046/j.1365-2443.2002.00523.x. [DOI] [PubMed] [Google Scholar]
- Garza-Sánchez F, Gin JG, Hayes CS. Amino acid starvation and colicin D treatment induce A-site mRNA cleavage in Escherichia coli. Journal of Molecular Biology. 2008;378:505–519. doi: 10.1016/j.jmb.2008.02.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garza-Sánchez F, Janssen BD, Hayes CS. Prolyl-tRNAPro in the A-site of SecM-arrested ribosomes inhibits the recruitment of transfer-messenger RNA. Journal of Biological Chemistry. 2006;281:34258–34268. doi: 10.1074/jbc.M608052200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garza-Sánchez F, Schaub RE, Janssen BD, Hayes CS. tmRNA regulates synthesis of the ArfA ribosome rescue factor. Molecular Microbiology. 2011 doi: 10.1111/j.1365-2958.2011.07638.x. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garza-Sánchez F, Shoji S, Fredrick K, Hayes CS. RNase II is important for A-site mRNA cleavage during ribosome pausing. Molecular Microbiology. 2009;73:882–897. doi: 10.1111/j.1365-2958.2009.06813.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge Z, Karzai AW. Co-evolution of multipartite interactions between an extended tmRNA tag and a robust Lon protease in Mycoplasma. Molecular Microbiology. 2009;74:1083–1099. doi: 10.1111/j.1365-2958.2009.06923.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge Z, Mehta P, Richards J, Karzai AW. Non-stop mRNA decay initiates at the ribosome. Molecular Microbiology. 2010;78:1159–1170. doi: 10.1111/j.1365-2958.2010.07396.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerdes K. Toxin-antitoxin modules may regulate synthesis of macromolecules during nutritional stress. J Bacteriol. 2000;182:561–572. doi: 10.1128/jb.182.3.561-572.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerdes K, Christensen SK, Lobner-Olesen A. Prokaryotic toxin-antitoxin stress response loci. Nature Reviews Microbiology. 2005;3:371–382. doi: 10.1038/nrmicro1147. [DOI] [PubMed] [Google Scholar]
- Gimple O, Schon A. In vitro and in vivo processing of cyanelle tmRNA by RNase P. Biol Chem. 2001;382:1421–1429. doi: 10.1515/BC.2001.175. [DOI] [PubMed] [Google Scholar]
- Glass JI, Assad-Garcia N, Alperovich N, Yooseph S, Lewis MR, Maruf M, Hutchison CA, 3rd, Smith HO, Venter JC. Essential genes of a minimal bacterium. Proceedings of the National Academy of Sciences U S A. 2006;103:425–430. doi: 10.1073/pnas.0510013103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong M, Cruz-Vera LR, Yanofsky C. Ribosome recycling factor and release factor 3 action promotes TnaC-peptidyl-tRNA Dropoff and relieves ribosome stalling during tryptophan induction of tna operon expression in Escherichia coli. Journal of Bacteriology. 2007;189:3147–3155. doi: 10.1128/JB.01868-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gottesman S, Roche E, Zhou Y, Sauer RT. The ClpXP and ClpAP proteases degrade proteins with carboxy-terminal peptide tails added by the SsrA-tagging system. Genes and Development. 1998;12:1338–1347. doi: 10.1101/gad.12.9.1338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gueneau de Novoa P, Williams KP. The tmRNA website: reductive evolution of tmRNA in plastids and other endosymbionts. Nucleic Acids Research. 2004;32:D104–108. doi: 10.1093/nar/gkh102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gur E, Sauer RT. Evolution of the ssrA degradation tag in Mycoplasma: specificity switch to a different protease. Proceedings of the National Academy of Sciences U S A. 2008;105:16113–16118. doi: 10.1073/pnas.0808802105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutmann S, Haebel PW, Metzinger L, Sutter M, Felden B, Ban N. Crystal structure of the transfer-RNA domain of transfer-messenger RNA in complex with SmpB. Nature. 2003;424:699–703. doi: 10.1038/nature01831. [DOI] [PubMed] [Google Scholar]
- Hahn J, Grandi G, Gryczan TJ, Dubnau D. Translational attenuation of ermC: a deletion analysis. Molecular and General Genetics. 1982;186:204–216. doi: 10.1007/BF00331851. [DOI] [PubMed] [Google Scholar]
- Hallier M, Desreac J, Felden B. Small protein B interacts with the large and the small subunits of a stalled ribosome during trans-translation. Nucleic Acids Research. 2006;34:1935–1943. doi: 10.1093/nar/gkl097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hallier M, Ivanova N, Rametti A, Pavlov M, Ehrenberg M, Felden B. Pre-binding of small protein B to a stalled ribosome triggers trans-translation. Journal of Biological Chemistry. 2004;279:25978–25985. doi: 10.1074/jbc.M314086200. [DOI] [PubMed] [Google Scholar]
- Hanawa-Suetsugu K, Takagi M, Inokuchi H, Himeno H, Muto A. SmpB functions in various steps of trans-translation. Nucleic Acids Research. 2002;30:1620–1629. doi: 10.1093/nar/30.7.1620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Handa Y, Inaho N, Nameki N. YaeJ is a novel ribosome-associated protein in Escherichia coli that can hydrolyze peptidyl-tRNA on stalled ribosomes. Nucleic Acids Research. 2011;39:1739–1748. doi: 10.1093/nar/gkq1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayes CS, Bose B, Sauer RT. Proline residues at the C terminus of nascent chains induce SsrA tagging during translation termination. Journal of Biological Chemistry. 2002a;277:33825–33832. doi: 10.1074/jbc.M205405200. [DOI] [PubMed] [Google Scholar]
- Hayes CS, Bose B, Sauer RT. Stop codons preceded by rare arginine codons are efficient determinants of SsrA tagging in Escherichia coli. Proceedings of the National Academy of Sciences U S A. 2002b;99:3440–3445. doi: 10.1073/pnas.052707199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayes CS, Keiler KC. Beyond ribosome rescue: tmRNA and co-translational processes. FEBS Letters. 2010;584:413–419. doi: 10.1016/j.febslet.2009.11.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayes CS, Sauer RT. Cleavage of the A site mRNA codon during ribosome pausing provides a mechanism for translational quality control. Molecular Cell. 2003;12:903–911. doi: 10.1016/s1097-2765(03)00385-x. [DOI] [PubMed] [Google Scholar]
- Henkin TM, Yanofsky C. Regulation by transcription attenuation in bacteria: how RNA provides instructions for transcription termination/antitermination decisions. Bioessays. 2002;24:700–707. doi: 10.1002/bies.10125. [DOI] [PubMed] [Google Scholar]
- Herman C, Thevenet D, Bouloc P, Walker GC, D’Ari R. Degradation of carboxy-terminal-tagged cytoplasmic proteins by the Escherichia coli protease HflB (FtsH) Genes and Development. 1998;12:1348–1355. doi: 10.1101/gad.12.9.1348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heurgue-Hamard V, Karimi R, Mora L, MacDougall J, Leboeuf C, Grentzmann G, Ehrenberg M, Buckingham RH. Ribosome release factor RF4 and termination factor RF3 are involved in dissociation of peptidyl-tRNA from the ribosome. EMBO Journal. 1998;17:808–816. doi: 10.1093/emboj/17.3.808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holberger LE, Hayes CS. Ribosomal protein S12 and aminoglycoside antibiotics modulate A-site mRNA cleavage and transfer-messenger RNA activity in Escherichia coli. Journal of Biological Chemistry. 2009;284:32188–32200. doi: 10.1074/jbc.M109.062745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong SJ, Lessner FH, Mahen EM, Keiler KC. Proteomic identification of tmRNA substrates. Proceedings of the National Academy of Sciences U S A. 2007;104:17128–17133. doi: 10.1073/pnas.0707671104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang C, Wolfgang MC, Withey J, Koomey M, Friedman DI. Charged tmRNA but not tmRNA-mediated proteolysis is essential for Neisseria gonorrhoeae viability. EMBO Journal. 2000;19:1098–1107. doi: 10.1093/emboj/19.5.1098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanova N, Lindell M, Pavlov M, Holmberg Schiavone L, Wagner EG, Ehrenberg M. Structure probing of tmRNA in distinct stages of trans-translation. RNA. 2007;13:713–722. doi: 10.1261/rna.451507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanova N, Pavlov MY, Bouakaz E, Ehrenberg M, Schiavone LH. Mapping the interaction of SmpB with ribosomes by footprinting of ribosomal RNA. Nucleic Acids Research. 2005;33:3529–3539. doi: 10.1093/nar/gki666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanova N, Pavlov MY, Felden B, Ehrenberg M. Ribosome rescue by tmRNA requires truncated mRNAs. Journal of Molecular Biology. 2004;338:33–41. doi: 10.1016/j.jmb.2004.02.043. [DOI] [PubMed] [Google Scholar]
- Jacob Y, Sharkady SM, Bhardwaj K, Sanda A, Williams KP. Function of the SmpB tail in transfer-messenger RNA translation revealed by a nucleus-encoded form. Journal of Biological Chemistry. 2005;280:5503–5509. doi: 10.1074/jbc.M409277200. [DOI] [PubMed] [Google Scholar]
- Jain SK, Gurevitz M, Apirion D. A small RNA that complements mutants in the RNA processing enzyme ribonuclease P. Journal of Molecular Biology. 1982;162:515–533. doi: 10.1016/0022-2836(82)90386-2. [DOI] [PubMed] [Google Scholar]
- Janssen BD, Hayes CS. Kinetics of paused ribosome recycling in Escherichia coli. Journal of Molecular Biology. 2009;394:251–267. doi: 10.1016/j.jmb.2009.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang L, Schaffitzel C, Bingel-Erlenmeyer R, Ban N, Korber P, Koning RI, de Geus DC, Plaisier JR, Abrahams JP. Recycling of aborted ribosomal 50S subunit-nascent chain-tRNA complexes by the heat shock protein Hsp15. Journal of Molecular Biology. 2009;386:1357–1367. doi: 10.1016/j.jmb.2008.10.079. [DOI] [PubMed] [Google Scholar]
- Julio SM, Heithoff DM, Mahan MJ. ssrA (tmRNA) plays a role in Salmonella enterica serovar Typhimurium pathogenesis. Journal of Bacteriology. 2000;182:1558–1563. doi: 10.1128/jb.182.6.1558-1563.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karimi R, Pavlov MY, Heurgue-Hamard V, Buckingham RH, Ehrenberg M. Initiation factors IF1 and IF2 synergistically remove peptidyl-tRNAs with short polypeptides from the P-site of translating Escherichia coli ribosomes. Journal of Molecular Biology. 1998;281:241–252. doi: 10.1006/jmbi.1998.1953. [DOI] [PubMed] [Google Scholar]
- Karzai AW, Roche ED, Sauer RT. The SsrA-SmpB system for protein tagging, directed degradation and ribosome rescue. Nature Structural Biology. 2000;7:449–455. doi: 10.1038/75843. [DOI] [PubMed] [Google Scholar]
- Karzai AW, Susskind MM, Sauer RT. SmpB, a unique RNA-binding protein essential for the peptide-tagging activity of SsrA (tmRNA) EMBO Journal. 1999;18:3793–3799. doi: 10.1093/emboj/18.13.3793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaur S, Gillet R, Li W, Gursky R, Frank J. Cryo-EM visualization of transfer messenger RNA with two SmpBs in a stalled ribosome. Proceedings of the National Academy of Sciences U S A. 2006;103:16484–16489. doi: 10.1073/pnas.0607438103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keiler KC. Biology of trans-translation. Annual Review of Microbiology. 2008;62:133–151. doi: 10.1146/annurev.micro.62.081307.162948. [DOI] [PubMed] [Google Scholar]
- Keiler KC, Sauer RT. Sequence determinants of C-terminal substrate recognition by the Tsp protease. Journal of Biological Chemistry. 1996;271:2589–2593. doi: 10.1074/jbc.271.5.2589. [DOI] [PubMed] [Google Scholar]
- Keiler KC, Shapiro L. Conserved promoter motif is required for cell cycle timing of dnaX transcription in Caulobacter. Journal of Bacteriology. 2001;183:4860–4865. doi: 10.1128/JB.183.16.4860-4865.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keiler KC, Shapiro L. tmRNA in Caulobacter crescentus is cell cycle regulated by temporally controlled transcription and RNA degradation. Journal of Bacteriology. 2003a;185:1825–1830. doi: 10.1128/JB.185.6.1825-1830.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keiler KC, Shapiro L. tmRNA is required for correct timing of DNA replication in Caulobacter crescentus. Journal of Bacteriology. 2003b;185:573–580. doi: 10.1128/JB.185.2.573-580.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keiler KC, Shapiro L, Williams KP. tmRNAs that encode proteolysis-inducing tags are found in all known bacterial genomes: A two-piece tmRNA functions in Caulobacter. Proceedings of the National Academy of Sciences U S A. 2000;97:7778–7783. doi: 10.1073/pnas.97.14.7778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keiler KC, Waller PR, Sauer RT. Role of a peptide tagging system in degradation of proteins synthesized from damaged messenger RNA. Science. 1996;271:990–993. doi: 10.1126/science.271.5251.990. [DOI] [PubMed] [Google Scholar]
- Kobayashi K, Kuwana R, Takamatsu H. kinA mRNA is missing a stop codon in the undomesticated Bacillus subtilis strain ATCC 6051. Microbiology. 2008;154:54–63. doi: 10.1099/mic.0.2007/011783-0. [DOI] [PubMed] [Google Scholar]
- Komine Y, Kitabatake M, Yokogawa T, Nishikawa K, Inokuchi H. A tRNA-like structure is present in 10Sa RNA, a small stable RNA from Escherichia coli. Proceedings of the National Academy of Sciences U S A. 1994;91:9223–9227. doi: 10.1073/pnas.91.20.9223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konno T, Kurita D, Takada K, Muto A, Himeno H. A functional interaction of SmpB with tmRNA for determination of the resuming point of trans-translation. RNA. 2007;13:1723–1731. doi: 10.1261/rna.604907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koo JT, Choe J, Moseley SL. HrpA, a DEAH-box RNA helicase, is involved in mRNA processing of a fimbrial operon in Escherichia coli. Molecular Microbiology. 2004;52:1813–1826. doi: 10.1111/j.1365-2958.2004.04099.x. [DOI] [PubMed] [Google Scholar]
- Kuo HK, Krasich R, Bhagwat AS, Kreuzer KN. Importance of the tmRNA system for cell survival when transcription is blocked by DNA-protein cross-links. Molecular Microbiology. 2010;78:686–700. doi: 10.1111/j.1365-2958.2010.07355.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurita D, Muto A, Himeno H. Role of the C-terminal tail of SmpB in the early stage of trans-translation. RNA. 2010;16:980–990. doi: 10.1261/rna.1916610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurland CG, Hughes D, Ehrenberg M. Limitations of translational accuracy. In: Neidhardt FC, Curtiss R, editors. Escherichia coli and Salmonella: Cellular and Molecular Biology. ASM Press; Washington, D. C.: 1996. pp. 979–1004. [Google Scholar]
- Levchenko I, Seidel M, Sauer RT, Baker TA. A specificity-enhancing factor for the ClpXP degradation machine. Science. 2000;289:2354–2356. doi: 10.1126/science.289.5488.2354. [DOI] [PubMed] [Google Scholar]
- Li X, Hirano R, Tagami H, Aiba H. Protein tagging at rare codons is caused by tmRNA action at the 3’ end of nonstop mRNA generated in response to ribosome stalling. RNA. 2006;12:248–255. doi: 10.1261/rna.2212606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Yagi M, Morita T, Aiba H. Cleavage of mRNAs and role of tmRNA system under amino acid starvation in Escherichia coli. Molecular Microbiology. 2008;68:462–473. doi: 10.1111/j.1365-2958.2008.06167.x. [DOI] [PubMed] [Google Scholar]
- Li Z, Pandit S, Deutscher MP. 3’ exoribonucleolytic trimming is a common feature of the maturation of small, stable RNAs in Escherichia coli. Proceedings of the National Academy of Sciences U S A. 1998;95:2856–2861. doi: 10.1073/pnas.95.6.2856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin-Chao S, Wei CL, Lin YT. RNase E is required for the maturation of ssrA RNA and normal ssrA RNA peptide-tagging activity. Proceedings of the National Academy of Sciences U S A. 1999;96:12406–12411. doi: 10.1073/pnas.96.22.12406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loomis WP, Koo JT, Cheung TP, Moseley SL. A tripeptide sequence within the nascent DaaP protein is required for mRNA processing of a fimbrial operon in Escherichia coli. Molecular Microbiology. 2001;39:693–707. doi: 10.1046/j.1365-2958.2001.02241.x. [DOI] [PubMed] [Google Scholar]
- Loomis WP, Moseley SL. Translational control of mRNA processing in the F1845 fimbrial operon of Escherichia coli. Molecular Microbiology. 1998;30:843–853. doi: 10.1046/j.1365-2958.1998.01117.x. [DOI] [PubMed] [Google Scholar]
- Magnuson RD. Hypothetical functions of toxin-antitoxin systems. Journal of Bacteriology. 2007;189:6089–6092. doi: 10.1128/JB.00958-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maki JA, Schnobrich DJ, Culver GM. The DnaK chaperone system facilitates 30S ribosomal subunit assembly. Molecular Cell. 2002;10:129–138. doi: 10.1016/s1097-2765(02)00562-2. [DOI] [PubMed] [Google Scholar]
- Mathy N, Benard L, Pellegrini O, Daou R, Wen T, Condon C. 5’-to-3’ exoribonuclease activity in bacteria: role of RNase J1 in rRNA maturation and 5’ stability of mRNA. Cell. 2007;129:681–692. doi: 10.1016/j.cell.2007.02.051. [DOI] [PubMed] [Google Scholar]
- Metzinger L, Hallier M, Felden B. Independent binding sites of small protein B onto transfer-messenger RNA during trans-translation. Nucleic Acids Research. 2005;33:2384–2394. doi: 10.1093/nar/gki534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metzinger L, Hallier M, Felden B. The highest affinity binding site of small protein B on transfer messenger RNA is outside the tRNA domain. RNA. 2008;14:1761–1772. doi: 10.1261/rna.1185808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohanty BK, Kushner SR. Genomic analysis in Escherichia coli demonstrates differential roles for polynucleotide phosphorylase and RNase II in mRNA abundance and decay. Molecular Microbiology. 2003;50:645–658. doi: 10.1046/j.1365-2958.2003.03724.x. [DOI] [PubMed] [Google Scholar]
- Moore SD, Sauer RT. Ribosome rescue: tmRNA tagging activity and capacity in Escherichia coli. Molecular Microbiology. 2005;58:456–466. doi: 10.1111/j.1365-2958.2005.04832.x. [DOI] [PubMed] [Google Scholar]
- Moore SD, Sauer RT. The tmRNA system for translational surveillance and ribosome rescue. Annual Review of Biochemistry. 2007;76:101–124. doi: 10.1146/annurev.biochem.75.103004.142733. [DOI] [PubMed] [Google Scholar]
- Mottagui-Tabar S, Bjornsson A, Isaksson LA. The second to last amino acid in the nascent peptide as a codon context determinant. EMBO Journal. 1994;13:249–257. doi: 10.1002/j.1460-2075.1994.tb06255.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muto A, Fujihara A, Ito KI, Matsuno J, Ushida C, Himeno H. Requirement of transfer-messenger RNA for the growth of Bacillus subtilis under stresses. Genes to Cells. 2000;5:627–635. doi: 10.1046/j.1365-2443.2000.00356.x. [DOI] [PubMed] [Google Scholar]
- Muto H, Ito K. Peptidyl-prolyl-tRNA at the ribosomal P-site reacts poorly with puromycin. Biochemical and Biophysical Research Communications. 2008;366:1043–1047. doi: 10.1016/j.bbrc.2007.12.072. [DOI] [PubMed] [Google Scholar]
- Muto H, Nakatogawa H, Ito K. Genetically encoded but nonpolypeptide prolyl-tRNA functions in the A site for SecM-mediated ribosomal stall. Molecular Cell. 2006;22:545–552. doi: 10.1016/j.molcel.2006.03.033. [DOI] [PubMed] [Google Scholar]
- Nakano H, Goto S, Nakayashiki T, Inokuchi H. Temperature-sensitive mutations in various genes of Escherichia coli K12 can be suppressed by the ssrA gene for 10Sa RNA (tmRNA) Molecular Genetics and Genomics. 2001;265:615–621. doi: 10.1007/s004380100453. [DOI] [PubMed] [Google Scholar]
- Nakatogawa H, Ito K. Secretion monitor, SecM, undergoes self-translation arrest in the cytosol. Molecular Cell. 2001;7:185–192. doi: 10.1016/s1097-2765(01)00166-6. [DOI] [PubMed] [Google Scholar]
- Nakatogawa H, Ito K. The ribosomal exit tunnel functions as a discriminating gate. Cell. 2002;108:629–636. doi: 10.1016/s0092-8674(02)00649-9. [DOI] [PubMed] [Google Scholar]
- Nakatogawa H, Murakami A, Ito K. Control of SecA and SecM translation by protein secretion. Current Opinion in Microbiology. 2004;7:145–150. doi: 10.1016/j.mib.2004.01.001. [DOI] [PubMed] [Google Scholar]
- Nameki N, Felden B, Atkins JF, Gesteland RF, Himeno H, Muto A. Functional and structural analysis of a pseudoknot upstream of the tag-encoded sequence in E. coli tmRNA. Journal of Molecular Biology. 1999a;286:733–744. doi: 10.1006/jmbi.1998.2487. [DOI] [PubMed] [Google Scholar]
- Nameki N, Tadaki T, Himeno H, Muto A. Three of four pseudoknots in tmRNA are interchangeable and are substitutable with single-stranded RNAs. FEBS Letters. 2000;470:345–349. doi: 10.1016/s0014-5793(00)01349-1. [DOI] [PubMed] [Google Scholar]
- Nameki N, Tadaki T, Muto A, Himeno H. Amino acid acceptor identity switch of Escherichia coli tmRNA from alanine to histidine in vitro. Journal of Molecular Biology. 1999b;289:1–7. doi: 10.1006/jmbi.1999.2754. [DOI] [PubMed] [Google Scholar]
- Nariya H, Inouye M. MazF, an mRNA interferase, mediates programmed cell death during multicellular Myxococcus development. Cell. 2008;132:55–66. doi: 10.1016/j.cell.2007.11.044. [DOI] [PubMed] [Google Scholar]
- Nonin-Lecomte S, Germain-Amiot N, Gillet R, Hallier M, Ponchon L, Dardel F, Felden B. Ribosome hijacking: a role for small protein B during trans-translation. EMBO Reports. 2009;10:160–165. doi: 10.1038/embor.2008.243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh BK, Apirion D. 10Sa RNA, a small stable RNA of Escherichia coli, is functional. Molecular and General Genetics. 1991;229:52–56. doi: 10.1007/BF00264212. [DOI] [PubMed] [Google Scholar]
- Oh BK, Chauhan AK, Isono K, Apirion D. Location of a gene (ssrA) for a small, stable RNA (10Sa RNA) in the Escherichia coli chromosome. Journal of Bacteriology. 1990;172:4708–4709. doi: 10.1128/jb.172.8.4708-4709.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okan NA, Bliska JB, Karzai AW. A role for the SmpB-SsrA system in Yersinia pseudotuberculosis pathogenesis. PLoS Pathogens. 2006;2:e6. doi: 10.1371/journal.ppat.0020006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okan NA, Mena P, Benach JL, Bliska JB, Karzai AW. The smpB-ssrA mutant of Yersinia pestis functions as a live attenuated vaccine to protect mice against pulmonary plague infection. Infection and Immunity. 2010;78:1284–1293. doi: 10.1128/IAI.00976-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oudot-Le Secq MP, Grimwood J, Shapiro H, Armbrust EV, Bowler C, Green BR. Chloroplast genomes of the diatoms Phaeodactylum tricornutum and Thalassiosira pseudonana: comparison with other plastid genomes of the red lineage. Molecular Genetics and Genomics. 2007;277:427–439. doi: 10.1007/s00438-006-0199-4. [DOI] [PubMed] [Google Scholar]
- Oussenko IA, Abe T, Ujiie H, Muto A, Bechhofer DH. Participation of 3’-to-5’ exoribonucleases in the turnover of Bacillus subtilis mRNA. Journal of Bacteriology. 2005;187:2758–2767. doi: 10.1128/JB.187.8.2758-2767.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Overgaard M, Borch J, Gerdes K. RelB and RelE of Escherichia coli form a tight complex that represses transcription via the ribbon-helix-helix motif in RelB. Journal of Molecular Biology. 2009;394:183–196. doi: 10.1016/j.jmb.2009.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paget MS, Molle V, Cohen G, Aharonowitz Y, Buttner MJ. Defining the disulphide stress response in Streptomyces coelicolor A3(2): identification of the sigmaR regulon. Molecular Microbiology. 2001;42:1007–1020. doi: 10.1046/j.1365-2958.2001.02675.x. [DOI] [PubMed] [Google Scholar]
- Pandey DP, Gerdes K. Toxin-antitoxin loci are highly abundant in free-living but lost from host-associated prokaryotes. Nucleic Acids Res. 2005;33:966–976. doi: 10.1093/nar/gki201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paul BJ, Barker MM, Ross W, Schneider DA, Webb C, Foster JW, Gourse RL. DksA: a critical component of the transcription initiation machinery that potentiates the regulation of rRNA promoters by ppGpp and the initiating NTP. Cell. 2004;118:311–322. doi: 10.1016/j.cell.2004.07.009. [DOI] [PubMed] [Google Scholar]
- Pedersen K, Christensen SK, Gerdes K. Rapid induction and reversal of a bacteriostatic condition by controlled expression of toxins and antitoxins. Molecular Microbiology. 2002;45:501–510. doi: 10.1046/j.1365-2958.2002.03027.x. [DOI] [PubMed] [Google Scholar]
- Pedersen K, Zavialov AV, Pavlov MY, Elf J, Gerdes K, Ehrenberg M. The bacterial toxin RelE displays codon-specific cleavage of mRNAs in the ribosomal A site. Cell. 2003;112:131–140. doi: 10.1016/s0092-8674(02)01248-5. [DOI] [PubMed] [Google Scholar]
- Perederina A, Svetlov V, Vassylyeva MN, Tahirov TH, Yokoyama S, Artsimovitch I, Vassylyev DG. Regulation through the secondary channel--structural framework for ppGpp-DksA synergism during transcription. Cell. 2004;118:297–309. doi: 10.1016/j.cell.2004.06.030. [DOI] [PubMed] [Google Scholar]
- Pisareva VP, Skabkin MA, Hellen CU, Pestova TV, Pisarev AV. Dissociation by Pelota, Hbs1 and ABCE1 of mammalian vacant 80S ribosomes and stalled elongation complexes. EMBO Journal. 2011;30:1804–1817. doi: 10.1038/emboj.2011.93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prysak MH, Mozdzierz CJ, Cook AM, Zhu L, Zhang Y, Inouye M, Woychik NA. Bacterial toxin YafQ is an endoribonuclease that associates with the ribosome and blocks translation elongation through sequence-specific and frame-dependent mRNA cleavage. Molecular Microbiology. 2009;71:1071–1087. doi: 10.1111/j.1365-2958.2008.06572.x. [DOI] [PubMed] [Google Scholar]
- Ramu H, Mankin A, Vazquez-Laslop N. Programmed drug-dependent ribosome stalling. Molecular Microbiology. 2009;71:811–824. doi: 10.1111/j.1365-2958.2008.06576.x. [DOI] [PubMed] [Google Scholar]
- Ranquet C, Geiselmann J, Toussaint A. The tRNA function of SsrA contributes to controlling repression of bacteriophage Mu prophage. Proceedings of the National Academy of Sciences U S A. 2001;98:10220–10225. doi: 10.1073/pnas.171620598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ray BK, Apirion D. Characterization of 10S RNA: a new stable RNA molecule from Escherichia coli. Molecular and General Genetics. 1979;174:25–32. doi: 10.1007/BF00433301. [DOI] [PubMed] [Google Scholar]
- Regnier P, Arraiano CM. Degradation of mRNA in bacteria: emergence of ubiquitous features. Bioessays. 2000;22:235–244. doi: 10.1002/(SICI)1521-1878(200003)22:3<235::AID-BIES5>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- Richards J, Mehta P, Karzai AW. RNase R degrades non-stop mRNAs selectively in an SmpB-tmRNA-dependent manner. Molecular Microbiology. 2006;62:1700–1712. doi: 10.1111/j.1365-2958.2006.05472.x. [DOI] [PubMed] [Google Scholar]
- Richter R, Rorbach J, Pajak A, Smith PM, Wessels HJ, Huynen MA, Smeitink JA, Lightowlers RN, Chrzanowska-Lightowlers ZM. A functional peptidyl-tRNA hydrolase, ICT1, has been recruited into the human mitochondrial ribosome. EMBO Journal. 2010;29:1116–1125. doi: 10.1038/emboj.2010.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roche ED, Sauer RT. SsrA-mediated peptide tagging caused by rare codons and tRNA scarcity. EMBO Journal. 1999;18:4579–4589. doi: 10.1093/emboj/18.16.4579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roche ED, Sauer RT. Identification of endogenous SsrA-tagged proteins reveals tagging at positions corresponding to stop codons. Journal of Biological Chemistry. 2001;276:28509–28515. doi: 10.1074/jbc.M103864200. [DOI] [PubMed] [Google Scholar]
- Ruhe ZC, Hayes CS. The N-terminus of GalE induces tmRNA activity in Escherichia coli. PLoS ONE. 2010;5:e15207. doi: 10.1371/journal.pone.0015207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sat B, Hazan R, Fisher T, Khaner H, Glaser G, Engelberg-Kulka H. Programmed cell death in Escherichia coli: some antibiotics can trigger mazEF lethality. Journal of Bacteriology. 2001;183:2041–2045. doi: 10.1128/JB.183.6.2041-2045.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sat B, Reches M, Engelberg-Kulka H. The Escherichia coli mazEF suicide module mediates thymineless death. Journal of Bacteriology. 2003;185:1803–1807. doi: 10.1128/JB.185.6.1803-1807.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauer RT, Baker TA. AAA+ Proteases: ATP-Fueled Machines of Protein Destruction. Annual Review of Biochemistry. 2010 doi: 10.1146/annurev-biochem-060408-172623. [DOI] [PubMed] [Google Scholar]
- Sauer RT, Bolon DN, Burton BM, Burton RE, Flynn JM, Grant RA, Hersch GL, Joshi SA, Kenniston JA, Levchenko I, Neher SB, Oakes ES, Siddiqui SM, Wah DA, Baker TA. Sculpting the proteome with AAA(+) proteases and disassembly machines. Cell. 2004;119:9–18. doi: 10.1016/j.cell.2004.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharkady SM, Williams KP. A third lineage with two-piece tmRNA. Nucleic Acids Res. 2004;32:4531–4538. doi: 10.1093/nar/gkh795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimizu Y, Ueda T. SmpB triggers GTP hydrolysis of elongation factor Tu on ribosomes by compensating for the lack of codon-anticodon interaction during trans-translation initiation. Journal of Biological Chemistry. 2006;281:15987–15996. doi: 10.1074/jbc.M512165200. [DOI] [PubMed] [Google Scholar]
- Shin JH, Price CW. The SsrA-SmpB ribosome rescue system is important for growth of Bacillus subtilis at low and high temperatures. Journal of Bacteriology. 2007;189:3729–3737. doi: 10.1128/JB.00062-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shoemaker CJ, Eyler DE, Green R. Dom34:Hbs1 promotes subunit dissociation and peptidyl-tRNA drop-off to initiate no-go decay. Science. 2010;330:369–372. doi: 10.1126/science.1192430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shpanchenko OV, Zvereva MI, Ivanov PV, Bugaeva EY, Rozov AS, Bogdanov AA, Kalkum M, Isaksson LA, Nierhaus KH, Dontsova OA. Stepping transfer messenger RNA through the ribosome. Journal of Biological Chemistry. 2005;280:18368–18374. doi: 10.1074/jbc.M409094200. [DOI] [PubMed] [Google Scholar]
- Silber KR, Keiler KC, Sauer RT. Tsp: a tail-specific protease that selectively degrades proteins with nonpolar C termini. Proceedings of the National Academy of Sciences U S A. 1992;89:295–299. doi: 10.1073/pnas.89.1.295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh NS, Varshney U. A physiological connection between tmRNA and peptidyl-tRNA hydrolase functions in Escherichia coli. Nucleic Acids Research. 2004;32:6028–6037. doi: 10.1093/nar/gkh924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Someya T, Nameki N, Hosoi H, Suzuki S, Hatanaka H, Fujii M, Terada T, Shirouzu M, Inoue Y, Shibata T, Kuramitsu S, Yokoyama S, Kawai G. Solution structure of a tmRNA-binding protein, SmpB, from Thermus thermophilus. FEBS Letters. 2003;535:94–100. doi: 10.1016/s0014-5793(02)03880-2. [DOI] [PubMed] [Google Scholar]
- Sundermeier TR, Dulebohn DP, Cho HJ, Karzai AW. A previously uncharacterized role for small protein B (SmpB) in transfer messenger RNA-mediated trans-translation. Proceedings of the National Academy of Sciences U S A. 2005;102:2316–2321. doi: 10.1073/pnas.0409694102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sundermeier TR, Karzai AW. Functional SmpB-ribosome interactions require tmRNA. Journal of Biological Chemistry. 2007;282:34779–34786. doi: 10.1074/jbc.M707256200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sunohara T, Jojima K, Tagami H, Inada T, Aiba H. Ribosome stalling during translation elongation induces cleavage of mRNA being translated in Escherichia coli. Journal of Biological Chemistry. 2004a;279:15368–15375. doi: 10.1074/jbc.M312805200. [DOI] [PubMed] [Google Scholar]
- Sunohara T, Jojima K, Yamamoto Y, Inada T, Aiba H. Nascent-peptide-mediated ribosome stalling at a stop codon induces mRNA cleavage resulting in nonstop mRNA that is recognized by tmRNA. RNA. 2004b;10:378–386. doi: 10.1261/rna.5169404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki M, Zhang J, Liu M, Woychik NA, Inouye M. Single protein production in living cells facilitated by an mRNA interferase. Molecular Cell. 2005;18:253–261. doi: 10.1016/j.molcel.2005.03.011. [DOI] [PubMed] [Google Scholar]
- Tanner DR, Dewey JD, Miller MR, Buskirk AR. Genetic analysis of the structure and function of transfer messenger RNA pseudoknot 1. Journal of Biological Chemistry. 2006;281:10561–10566. doi: 10.1074/jbc.M600167200. [DOI] [PubMed] [Google Scholar]
- Tehranchi AK, Blankschien MD, Zhang Y, Halliday JA, Srivatsan A, Peng J, Herman C, Wang JD. The transcription factor DksA prevents conflicts between DNA replication and transcription machinery. Cell. 2010;141:595–605. doi: 10.1016/j.cell.2010.03.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thibonnier M, Thiberge JM, De Reuse H. Trans-translation in Helicobacter pylori: essentiality of ribosome rescue and requirement of protein tagging for stress resistance and competence. PLoS ONE. 2008;3:e3810. doi: 10.1371/journal.pone.0003810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tu GF, Reid GE, Zhang JG, Moritz RL, Simpson RJ. C-terminal extension of truncated recombinant proteins in Escherichia coli with a 10Sa RNA decapeptide. Journal of Biological Chemistry. 1995;270:9322–9326. doi: 10.1074/jbc.270.16.9322. [DOI] [PubMed] [Google Scholar]
- Ujiie H, Matsutani T, Tomatsu H, Fujihara A, Ushida C, Miwa Y, Fujita Y, Himeno H, Muto A. Trans-translation is involved in the CcpA-dependent tagging and degradation of TreP in Bacillus subtilis. Journal of Biochemistry. 2009;145:59–66. doi: 10.1093/jb/mvn143. [DOI] [PubMed] [Google Scholar]
- Valle M, Gillet R, Kaur S, Henne A, Ramakrishnan V, Frank J. Visualizing tmRNA entry into a stalled ribosome. Science. 2003;300:127–130. doi: 10.1126/science.1081798. [DOI] [PubMed] [Google Scholar]
- Van Melderen L, Saavedra De Bast M. Bacterial toxin-antitoxin systems: more than selfish entities? PLoS Genetics. 2009;5:e1000437. doi: 10.1371/journal.pgen.1000437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wah DA, Levchenko I, Baker TA, Sauer RT. Characterization of a specificity factor for an AAA+ ATPase: assembly of SspB dimers with ssrA-tagged proteins and the ClpX hexamer. Chemical Biology. 2002;9:1237–1245. doi: 10.1016/s1074-5521(02)00268-5. [DOI] [PubMed] [Google Scholar]
- Watts T, Cazier D, Healey D, Buskirk A. SmpB contributes to reading frame selection in the translation of transfer-messenger RNA. Journal of Molecular Biology. 2009;391:275–281. doi: 10.1016/j.jmb.2009.06.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weis F, Bron P, Giudice E, Rolland JP, Thomas D, Felden B, Gillet R. tmRNA-SmpB: a journey to the centre of the bacterial ribosome. EMBO Journal. 2010a;29:3810–3818. doi: 10.1038/emboj.2010.252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weis F, Bron P, Rolland JP, Thomas D, Felden B, Gillet R. Accommodation of tmRNA-SmpB into stalled ribosomes: a cryo-EM study. RNA. 2010b;16:299–306. doi: 10.1261/rna.1757410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiegert T, Schumann W. SsrA-mediated tagging in Bacillus subtilis. Journal of Bacteriology. 2001;183:3885–3889. doi: 10.1128/JB.183.13.3885-3889.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wohlgemuth I, Brenner S, Beringer M, Rodnina MV. Modulation of the rate of peptidyl transfer on the ribosome by the nature of substrates. Journal of Biological Chemistry. 2008;283:32229–32235. doi: 10.1074/jbc.M805316200. [DOI] [PubMed] [Google Scholar]
- Wojtkowiak D, Georgopoulos C, Zylicz M. Isolation and characterization of ClpX, a new ATP-dependent specificity component of the Clp protease of Escherichia coli. Journal of Biological Chemistry. 1993;268:22609–22617. [PubMed] [Google Scholar]
- Wower IK, Zwieb C, Wower J. Transfer-messenger RNA unfolds as it transits the ribosome. RNA. 2005;11:668–673. doi: 10.1261/rna.7269305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wower IK, Zwieb C, Wower J. Escherichia coli tmRNA lacking pseudoknot 1 tags truncated proteins in vivo and in vitro. RNA. 2009;15:128–137. doi: 10.1261/rna.1192409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wower J, Zwieb CW, Hoffman DW, Wower IK. SmpB: a protein that binds to double-stranded segments in tmRNA and tRNA. Biochemistry. 2002;41:8826–8836. doi: 10.1021/bi0201365. [DOI] [PubMed] [Google Scholar]
- Yamaguchi Y, Inouye M. mRNA interferases, sequence-specific endoribonucleases from the toxin-antitoxin systems. Progress in Molecular Biology and Translational Science. 2009;85:467–500. doi: 10.1016/S0079-6603(08)00812-X. [DOI] [PubMed] [Google Scholar]
- Yamamoto Y, Sunohara T, Jojima K, Inada T, Aiba H. SsrA-mediated trans-translation plays a role in mRNA quality control by facilitating degradation of truncated mRNAs. RNA. 2003;9:408–418. doi: 10.1261/rna.2174803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang C, Glover JR. The SmpB-tmRNA tagging system plays important roles in Streptomyces coelicolor growth and development. PLoS ONE. 2009;4:e4459. doi: 10.1371/journal.pone.0004459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao S, Sharp JS, Bechhofer DH. Bacillus subtilis RNase J1 endonuclease and 5’ exonuclease activities in the turnover of ΔermC mRNA. RNA. 2009;15:2331–2339. doi: 10.1261/rna.1749109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yusupov MM, Yusupova GZ, Baucom A, Lieberman K, Earnest TN, Cate JH, Noller HF. Crystal structure of the ribosome at 5.5 A resolution. Science. 2001;292:883–896. doi: 10.1126/science.1060089. [DOI] [PubMed] [Google Scholar]
- Yusupova GZ, Yusupov MM, Cate JH, Noller HF. The path of messenger RNA through the ribosome. Cell. 2001;106:233–241. doi: 10.1016/s0092-8674(01)00435-4. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Inouye M. The inhibitory mechanism of protein synthesis by YoeB, an Escherichia coli toxin. Journal of Biological Chemistry. 2009;284:6627–6638. doi: 10.1074/jbc.M808779200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Zhang J, Hoeflich KP, Ikura M, Qing G, Inouye M. MazF cleaves cellular mRNAs specifically at ACA to block protein synthesis in Escherichia coli. Molecular Cell. 2003;12:913–923. doi: 10.1016/s1097-2765(03)00402-7. [DOI] [PubMed] [Google Scholar]


