Abstract
Dietary interventions are effective ways to extend or shorten lifespan. By examining midlife hepatic gene expressions in mice under different dietary conditions, which resulted in different lifespans and aging-related phenotypes, we were able to identify genes and pathways that modulate the aging process. We found that pathways transcriptionally correlated with diet-modulated lifespan and physiological changes were enriched for lifespan-modifying genes. Intriguingly, mitochondrial gene expression correlated with lifespan and anticorrelated with aging-related pathological changes, whereas peroxisomal gene expression showed an opposite trend. Both organelles produce reactive oxygen species, a proposed causative factor of aging. This finding implicates a contribution of peroxisome to aging. Consistent with this hypothesis, lowering the expression levels of peroxisome proliferation genes decreased the cellular peroxide levels and extended the lifespan of Drosophila melanogaster and Caenorhabditis elegans. These findings show that transcriptional changes resulting from dietary interventions can effectively reflect causal factors in aging and identify previously unknown or under-appreciated longevity pathways, such as the peroxisome pathway.
Keywords: systems biology, diet-induced obesity
Aging-related gene expressions have been examined by microarray analysis for various human and mouse tissues (1–5), fruit flies (6), and worms (7, 8). These studies have revealed hundreds to thousands of genes and numerous biological functions that change as an organism ages. Some of the changes are similar across different species. Expressions of genes involved in stress response and inflammation consistently increase in animals, and those expressions involved in tissue-specific functions gradually decrease, reflecting functional decline of tissues or organs (9). However, most of these changes reflect the consequence of aging (some can serve as biomarkers of aging) rather than the cause or regulatory factors of aging. For instance, the key aging regulatory genes identified by genetic approaches are rarely identifiable merely by the expression level changes during aging (10). However, interventions of the aging process by genetic, dietary, or reproductive measures can effectively modulate lifespan and aging (11, 12).
Caloric restriction (CR) is the best-studied intervention for modulating aging and has been reported to prolong both mean and maximal lifespans in most organisms examined (11, 12). In contrast, feeding mice a high-fat, high-calorie diet results in age-related obesity, cardiovascular diseases, and other metabolic disorders, and it shortens lifespan (13–15). Exercise, however, can increase energy expenditure, reduce body weight, and prevent some age-related functional declines (16). It is, therefore, not surprising that nutrient- and energy-sensing pathways have been identified by genetic approaches to be key regulators of lifespan and aging (12, 17).
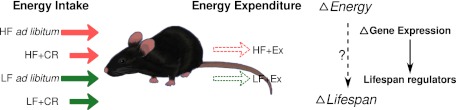
Similar to the powerful genetic manipulations, manipulations of energy input or expenditure also provide a means to modify the phenotypes and course of aging. Analogous to the concept of quantitative trait loci (QTL), the incremental changes in lifespan and aging-related phenotypes under various dietary/environmental conditions from low- to high-energy input could be explained by gene expression changes of lifespan/aging regulators that are responsive to energy input changes. Unlike genetic manipulations that only examine one (or at most, a few) loci at a time, the incremental changes of gene expression that are correlated with lifespan and other aging-related phenotypes under different energy regimens may capture many regulatory changes at the same time. Because of the heterogeneous genetic background of QTL analysis, this method often requires hundreds of samples to infer putative causal variants from naturally occurring variants. We expect that the sample size required for environmental manipulations is much smaller than the sample size required for QTL because of the homogeneous genetic background of the animals, the standardization of treatments, and the averaging effect within group to minimize individual variations. Under this assumption, we subjected mice to six different diet/energy regimens (30 mice per group) that led to the following order of increasing lifespan: high-fat diet (HF) fed ad libitum high-fat diet combined with voluntary exercise (HF+Ex), low-fat diet (LF) fed ad libitum low-fat diet with voluntary Ex (LF+Ex), high-fat diet combined with 70% CR (HF+CR), and low-fat diet with 70% CR (LF+CR). In addition to lifespan, health span parameters were assessed for each cohort, including liver and metabolic functions, and gene expression profiles were obtained at midlife before the increase in mortality.
In this context, we first asked if the six different dietary groups give rise to different lifespans according to their energy input and output levels; if so, we asked whether we can predict the lifespan differences across these groups from the midlife liver phenotypes and hepatic gene expressions and finally, whether the genes or pathways that predict the lifespan differences are regulators of lifespan. The fact that all of the intervention experiments were carried out in parallel rather than in different laboratories with variable and/or noncomparable conditions enabled us to conduct an integrative analysis that was free of system variations in the data; we searched for common target genes of different dietary interventions that contribute to the consequent lifespan differences through changes in their gene expression levels. Our results indicate that midlife liver gene expressions showing positive or negative correlation with mean lifespan across the six groups indeed identified not only many genes previously implicated in aging but at least one previously unknown or under-appreciated aging-related pathway that implicates peroxisomal biogenesis as a key determinant of longevity.
Results
Lifespan and Metabolic Phenotypes of Mice Under Different Intervention Regimens.
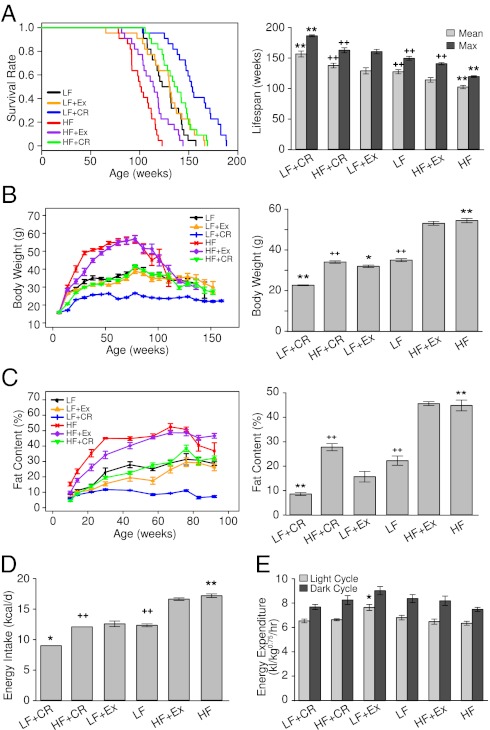
To arrive at six groups with different energy-input and -expenditure states, we maintained male C57BL/6J mice (n = 30 per group) on either a regular LF diet or a HF diet; the food was fed ad libitum or calorically restricted to 70% of the amount of food offered daily to the ad libitum group (CR) or the group subjected to voluntary wheel-running Ex. Kaplan–Meier survival curves (Fig. 1A, Left) showed that lifelong feeding of HF diet markedly reduced the lifespan of mice (by ∼20% for both the mean and maximal lifespan) (Fig. 1A, Right), and CR exerted more striking effects than Ex (its effects were monitored and recorded as shown in SI Appendix, Fig. S1A) on extending the lifespan of animals fed either LF or HF diets. Overall, the order of the mean and maximal lifespan (weeks) under the six regimens was as follows: HF (mean = 101, 118.8 ± 1.5 maximum) < HF+Ex (mean = 114, 139.7 ± 1.9 maximum) < LF (mean = 127, 148.7 ± 3.1 maximum) < LF+Ex (mean = 131, 159.6 ± 3.7 maximum) < HF+CR (mean = 137, 161.9 ± 1.5 maximum) < LF+CR (153 mean, 185.5 ± 1.6 maximum) (Fig. 1A). While CR under LF diet feeding (LF+CR) increased the mean and maximum lifespan by ∼20% and ∼25%, respectively, relative to the LF group, restricted HF intake (HF+CR) extended both the mean and maximum lifespan by ∼36% compared with the HF group.
Fig. 1.
Effects of diet and Ex on lifespan and energy state. (A, Left) Kaplan–Meier survival curves (n = 22 per group) showing significant changes in lifespan between intervention pairs of HF/LF (P = 0.9E-6), HF+CR/HF (P = 0.87E-9), HF+Ex/HF (P = 0.006), and LF+CR/LF (P = 0.42E-5) by log-rank test. (A, Right) Mean and maximum lifespan of mice in the six intervention groups as derived from the Kaplan–Meier survival curves. Maximum lifespan was calculated from the average of the oldest 20% of mice in each group. (B) Body weight was monitored throughout the (Left) indicated ages and (Right) midlife (62-wk age) until the number of mice alive in each group was less than six. (C) Total body fat content was measured by NMR (n = 8–10 per group) at (Left) each indicated age and (Right) midlife (62-wk age) until 92 wk of age. (D) Energy intake was calculated based on the daily food consumption of mice during the intervention before 62 wk of age (n = 8 per group). (E) Oxygen consumption was determined at midlife when mice were 57–58 wk of age (n = 5 per group). Data in B–D are presented as means ± SEM. *P < 0.05, **P < 0.01 vs. LF, ++P < 0.01 vs. HF by ANOVAs.
Physiological analyses revealed the expected improving effects of CR and Ex on animals’ body weight and whole-body fat content throughout their lifetime (Fig. 1 B and C). Notably, glucose tolerance tests indicated a close association of their adiposity levels with glucose intolerance when examined at 21 and 30 wk of age across the six intervention groups; however, only animals in the LF+CR group exhibited significant improvement of glucose tolerance after 60 wk of age (SI Appendix, Fig. S1B). When assessed at the midlife ages of 60–62 wk, CR not only markedly prevented HF diet-induced obesity (Fig. 1 B and C) but also protected against obesity-associated hyperleptinemia, hyperinsulinemia, and dyslipidemia in the HF+CR mice, which contrasted the moderate effects of Ex (SI Appendix, Table S1).
The effects of dietary interventions on the lifespan and metabolic syndromes also agreed with the aging phenotypes of animals under these conditions, which were shown by aging-associated biomarkers including hair regrowth, rotarod, and tightrope assays performed in mice at the ages of 85–90 wk (SI Appendix, Fig. S2 A–C). Thus, dietary modifications, particularly CR, most dramatically affected aging rate and lifespan in this model.
To assess the contribution of energy balance to lifespan, we quantified the daily energy intake of each group of mice before 62 wk of age, which displayed the following order: HF (17.14 kcal) > HF+Ex (16.58 kcal) > LF+Ex (12.53 kcal) > LF (12.29 kcal) > HF+CR (12.03 kcal) > LF+CR (8.95 kcal) (Fig. 1D). Using the comprehensive laboratory animal monitoring system, we also measured the energy expenditure for mice at 57–58 wk of age. Because of the different body size of mice in each group, we compared the metabolic rate by normalizing to the metabolic size as reflected by body weight0.75 (18) of each animal. CR did not significantly affect oxygen consumption in mice fed either LF or HF (Fig. 1E), whereas voluntary Ex significantly increased it in LF-fed mice compared with their corresponding feeding group. Additionally, CR led to an increase in physical activity during the light cycle in the LF-fed group (SI Appendix, Fig. S2D), and it resulted in significantly decreased body temperature in both LF- and HF-fed groups at 90 wk of age (SI Appendix, Fig. S2E).
Liver Health at Midlife in Mice Under Different Intervention Regimens.
Reasoning that the liver is central to metabolic homeostasis and may serve as a functional organ indicator of whole-body health, we decided to examine whether liver pathology at midlife under different intervention regimens correlates with the lifespan phenotype. At around midlife age of 62 wk, mice of the HF group had enlarged liver, with a twofold increase in liver weight relative to the LF group, whereas CR more potently antagonized HF-induced hepatomegaly than Ex (SI Appendix, Table S1). Next, we evaluated the hepatic function by measuring serum levels of alanine aminotransferase (ALT) and aspartate aminotransferase (AST), the well-established markers of liver injury or damage. In the HF group of mice, serum ALT or AST levels were markedly elevated, and in contrast, both were significantly lower in the HF+CR and HF+Ex groups (SI Appendix, Fig. S3A), suggesting reductions of HF-associated hepatocellular damages by CR and Ex. Again, CR had a more pronounced effect of reversing this consequence of HF feeding than Ex (SI Appendix, Fig. S3A).
Consistently, CR completely prevented HF-induced hepatic overload of triglycerides (TG; i.e., hepatosteatosis) and considerably reduced liver cholesterol content in the HF+CR group, while also decreasing liver TG accumulation in the LF-CR group (SI Appendix, Table S1 and Fig. S3B). Because chronic liver damage induced by nonalcoholic fatty liver can lead to liver fibrosis, we wondered whether CR or Ex could exert an effect on this process. Assessment by Sirius Red staining of fibrotic collagen deposition showed that ∼12% of the livers from the HF group were Sirius Red-positive, representing an almost 10-fold increase relative to the LF group that exhibited very low levels of fibrosis (SI Appendix, Fig. S3C). Both CR and Ex could markedly reduce HF-induced liver fibrosis from ∼12% to ∼2%, which was comparable with the level of the LF group of mice (SI Appendix, Fig. S3C).
Perturbations of mitochondrial function are known to contribute to metabolic dysfunctions and may play a causal role in the aging process. In accordance with this notion, mice from the HF group showed an ∼50% reduction in the mitochondrial density in hepatocytes relative to the LF group but without apparent differences in their size (SI Appendix, Fig. S3D). CR increased the mitochondrial density in livers from HF-fed but not LF-fed animals, whereas Ex displayed no detectable effect regardless of diets (SI Appendix, Fig. S3D). Notably, both CR and Ex increased the mitochondrial size on either diet, with CR eliciting a larger effect. Therefore, CR could elevate both the density and size of mitochondria in livers of HF-fed mice (SI Appendix, Fig. S3D).
Correlation of Midlife Metabolic Phenotypes with Lifespan.
Taken together, the resultant physical and physiological phenotypes at both the organismal and liver organ levels were highly coordinated across the six intervention regimens, which could be clustered into three major groups (Fig. 2). The largest group reflected the midlife pathological states of the liver and other associated serum and whole-body parameters such as serum cholesterol, body weight, and fat content, which positively correlated with energy intake (Fig. 2). The second group, one-half of the size of the aforementioned group, consisted of mitochondria size and density, activity during the light cycle, and other performance parameters in the aged animals (i.e., tightrope, rotarod, and hair regrowth), which positively correlated with lifespan (Fig. 2). The average z score values in these two groups were highly negatively correlated [average Pearson correlation coefficient (PCC) = −0.97]. Finally, loosely clustered with the second group, the third and smallest cluster included energy expenditure and activity during the dark cycle, which did not show a strong correlation with lifespan (Fig. 2).
Fig. 2.
Midlife physiological parameters correlated with lifespan. Hierarchical clustering of the diet intervention groups and the indicated physiological parameters including body weight, fat content, midlife liver and serum measurements, energy intake and expenditure, and aging-sensitive markers based on the group mean values as well as mean lifespans.
Midlife Liver Gene Expression Profiles Under Different Intervention Regimens.
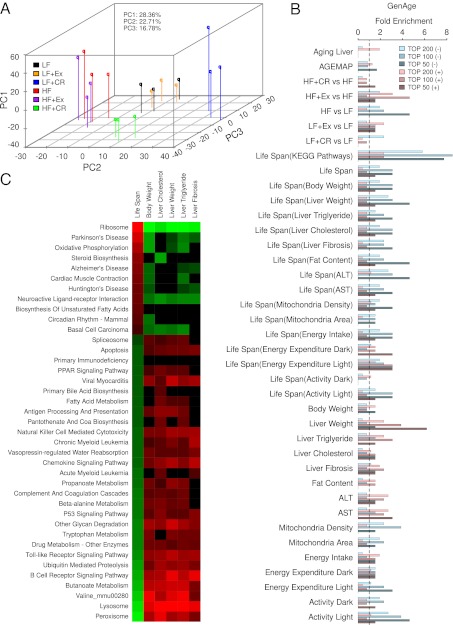
To understand the molecular basis of the physiology and lifespan differences under the six intervention regimens, we measured the liver transcriptome of three mice from each group at 62 wk of age using whole-genome microarrays (Materials and Methods). We clustered the profiles of genes differentially expressed between any two groups (Materials and Methods and Dataset S1) in the 18 samples using principle component analysis (PCA). Strikingly, the 18 samples were separated into four clusters that reflected intervention regimens (Fig. 3A), which was consistent with hierarchical clustering (SI Appendix, Fig. S4). The first three principle components (PCs) accounted for 68% of the gene expression variations. Generally, PC1 mainly reflected the individual variation among samples, whereas PC2 accounted for 22.7% of the variations and was most correlated with the average lifespans of the six intervention groups (PCC = 0.98). PC3 mostly distinguished the HF+CR group from LF-fed groups. In particular, LF+CR samples were the farthest from other samples, indicating that they had a distinctive common expression pattern (Fig. 3A). Moreover, the HF+CR group was separated from HF-fed groups on PC1 and PC2, and it was more similar to the LF group than the HF group (Fig. 3A). This finding suggests that CR might counteract the effect of HF feeding and shift the transcriptomic profile of HF mice to the profile of LF mice. Consistent with the relatively weak effect of physical Ex on liver physiology and lifespan, Ex seemed to exhibit subtle effects on the transcriptome, especially for LF-fed mice (Fig. 3A). Therefore, the distances between the overall expression profiles of the six groups did reflect the physiology and lifespan differences among these intervention groups.
Fig. 3.
Midlife liver gene expression correlations to lifespan are predictive of aging regulators. (A) PCA analysis of the profiles of the differentially expressed genes (Materials and Methods and Dataset S1) for the indicated 18 samples. (B) Fold enrichment over genome-wide background for aging regulatory genes from GenAge database among the top 50, 100, or 200 genes with expressions that are most positively (+) or negatively (−) correlated with lifespan or liver physiological parameters. Enrichments are shown for genes in pathways significantly positively/negatively correlated with lifespan with (as indicated by the labels inside parentheses) or without prefiltering for significant correlation with other phenotypes. Significance of correlations was determined by PCCs (P < 0.05). (C) The KEGG pathways with gene expressions that significantly correlated or anticorrelated with the physiological parameters identified by the GSEA analysis.
Differences in Midlife Liver Gene Expressions Are Predictive of Diet-Induced Lifespan Differences.
The high correlation between lifespan and PC2 (22.7% total variations) indicates that a large portion of the observed transcriptional differences is associated with lifespan/aging differences introduced by the dietary interventions. We, therefore, asked whether incremental changes of gene expressions in the midlife liver under the six dietary regimens might not only reflect the pace of aging but could also identify potential modulators of aging and longevity, which are not readily identifiable through routine age-dependency analysis.
We tested this idea first by determining whether known lifespan-modifying genes were enriched among those genes with expression changes that are positively or negatively correlated with lifespan and other phenotypic differences among the six dietary regimens. Previously identified lifespan-modifying genes were obtained from the GenAge database (19), which we refer to as GenAge regulators, and are used as a gold-standard positive gene set. We used PCC to rank the genes’ expression (log2-transformed) correlation with phenotypes or the mean lifespan in each of the six dietary intervention groups. As shown in Fig. 3B, genes with expression that showed the highest negative correlation with lifespan (measured by PCC between a gene’s expression level and the mean lifespan across the six groups; Dataset S2) were two- to threefold more enriched for the genetically identified lifespan-modifying genes compared with the background genome-wide average (approximately twofold in the top 200 and approximately threefold in the top 100 and top 50). These enrichment levels (approximately threefold over the background) are much higher than the levels for differentially expressed genes between any single pair of dietary regimens (approximately twofold on average) as determined and ranked by RankProd test percentage of false-positive values (20) (Fig. 3B). This finding indicates that expression correlation or anticorrelation with lifespan is a better predictor for aging regulators. Because many of the phenotypes that we measured strongly correlated with the mean lifespan in the six groups (Fig. 2), it is not surprising that genes with expression that correlated positively/negatively with liver weight, liver TG, ALT, AST, mitochondria density, activity, and energy expenditure in the light cycle also had higher than background enrichment for aging regulators (Fig. 3B). Consistently, the combination of correlation to lifespan and other phenotypes in general did not further increase the enrichment for GenAge lifespan regulators, except for lifespan correlation together with liver weight or fat content (Fig. 3B). Lifespan-modifying genes from the mouse genome informatics (MGI) database were also overrepresented among genes that either negatively or positively correlated with lifespan (SI Appendix, Fig. S5). One drawback of the MGI-curated gene set is that it includes genes for which mutation shortens lifespan. Lifespan-shortening mutations may not affect aging per se, because nonspecific defects in development or overall health of the animals can also reduce lifespan.
To determine how our dietary intervention-based transcriptomic approach for predicting aging regulatory genes compares with a more traditional approach of using age-dependent transcriptional changes, we examined the hepatic gene expression changes in LF-fed mice during aging at 4, 8, 13, and 21 mo. At each age point, we pooled liver RNAs from six mice of the same age to obtain an average microarray profile (Materials and Methods). Using the Short Time Series Expression Miner software (21), we identified five expression patterns that were significantly age-dependent (P < 0.05) (SI Appendix, Fig. S6A). Genes that displayed these significant age-related expression changes were not significantly enriched for lifespan-modifying genes (Fig. 3B and SI Appendix, Fig. S6A) (P = 0.306, P = 0.438, P = 0.149, P = 0.184, and P = 0.048, respectively). Similar results were obtained using another more comprehensive liver gene expression dataset: the AGEMAP dataset (4) (SI Appendix, Fig. S6B). These results are consistent with our previous findings on the human brain and the fruit fly aging gene expression profiles (10), suggesting that most of these changes reflect the consequences of aging rather than causal or regulatory mechanisms of aging.
Potential Pathways Contributing to Lifespan Alterations Under Different Dietary Regimens.
We next used Gene Set Enrichment Analysis (GSEA) (22) to identify pathways with gene expressions that are positively or negatively correlated with lifespan of the six intervention groups (Materials and Methods). Unexpectedly, genes in these pathways were as high as eightfold enriched for GenAge lifespan regulators relative to the genomic background (Fig. 3 B and C), suggesting that the statistical power at the pathway level brings more enrichment for lifespan regulators compared with the single-gene level.
Our approach of directly measuring lifespan as well as measuring dietary and other metabolic conditions, although extremely tedious and costly (180 mice over a time span of 3 y), allowed us to search for pathways that may directly explain lifespan changes instead of other phenotypic changes. For instance, although body weight is highly correlated with lifespan, we were able to identify pathways, such as steroid biosynthesis, biosynthesis of unsaturated fatty acids, primary immunodeficiency, primary bile acid biosynthesis, and acute myeloid leukemia pathways, etc., that are correlated with lifespan but independent of body weight (Fig. 3C). Furthermore, we also observed some lifespan-associated pathways that are independent of most other lifespan-associated phenotypes. For example, the primary immunodeficiency pathway is not associated with any phenotypes examined other than lifespan, whereas the steroid synthesis pathway is only negatively correlated with liver cholesterol level; also, the circadian rhythm pathway is only negatively correlated with body weight (Fig. 3C). Steroid hormones, such as pregnenolone, can act independently or synergistically with insulin/IGF1 signaling to extend lifespan (23). Thus, increased expression of steroid synthesis genes in response to CR under both HF and LF diet feeding conditions and the significant correlation with lifespan under the six different treatments (Fig. 3C) may be other important components of the dietary effects on longevity. The relationship between circadian rhythm and aging has also been under intense investigation (24).
Pathways with gene expressions that were highly positively correlated with lifespan across the six groups included age-associated neurodegenerative disorders such as Huntington disease, Alzheimer’s disease, and Parkinson disease. Also positively correlated with lifespan were the known aging-related oxidative phosphorylation (25) and ribosome pathways (26). In fact, the ribosome pathway showed the strongest positive correlation with lifespan and strongest negative correlation with many physiological parameters of liver damage (Fig. 3C).
Interestingly, the number of pathways negatively correlated with lifespan was three times the number of pathways positively correlated with lifespan, although the number of genes positively and negatively correlated with lifespan was similar [586 genes were positively correlated with the mean lifespan (PCC > 0.866 or P < 0.05), whereas 429 genes were negatively correlated (PCC < −0.888 or P < 0.05)]. This finding suggests that genes negatively correlating with lifespan are more functionally diverse (Fig. 3C and Dataset S1). In addition, several pathways that negatively correlated with lifespan were related to inflammation and apoptosis, which have been previously implicated in aging and age-related diseases. Moreover, we also identified peroxisome proliferator-activated receptor (PPAR) signaling and various metabolic pathways, particularly those pathways involved in fat metabolism, as negatively correlated with lifespan. Genes inside these pathways frequently displayed coordinated changes that correlated with lifespan and other phenotypes [for example, within the peroxisome pathway (SI Appendix, Fig. S7), which is the pathway that most negatively correlated with lifespan].
Validation of Lifespan Regulation by Peroxisome Biogenesis Genes.
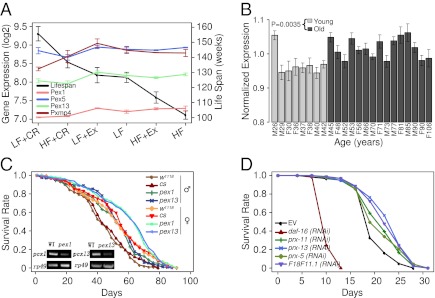
Fatty acids are activators for the nuclear receptor PPARα, which promotes peroxisome proliferation. During PPARα activation, peroxide-generating enzymes for fatty acid β-oxidation are induced 10- to 30-fold; the peroxide-catabolizing enzyme catalase is induced only one- to twofold. Therefore, peroxisome proliferation may increase cellular levels of peroxide (27). Among the peroxisome genes, those genes involved in peroxisome biogenesis were most homogeneously anticorrelated with lifespan (Fig. 4A and SI Appendix, Figs. S7 and S8 A and B), suggesting that peroxisome biogenesis may negatively influence longevity. Consistent with this idea, we observed an overall increase in peroxisomal gene expression in the aged brain, especially for genes that participate in peroxisome biosynthesis and proliferation (the PEX genes), based on previously published microarray gene profiles of 30 human brain samples (2) with (Fig. 4B) or without outliers (SI Appendix, Fig. S9) as described previously (28).
Fig. 4.
Peroxisomal pathway’s gene expression correlation with diet-induced lifespan changes is predictive of its regulation of lifespan. (A) The mean lifespan and expression levels of Pex1, Pex5, Pex13, and Pxmp4 among the six groups. (B) Expression level changes of the proxisomal proliferation genes (PEX) during human brain aging. Average and SD of the log2 expression levels of the 11 PEX genes in each of the human brain samples, excluding seven outliers. The outliers were identified as described previously (28). Analysis including the outliers can be found in SI Appendix, Fig. S8B. (C) Survival curves for male and female WT (red-eyed cs and white-eyed w1118 strains) and pex1S4868 and pex13KG04339 mutant flies (log-rank test: P = 0.0006 for male and P = 0.0075 for female pex1 vs. cs; P = 7.59E-5 for male and P = 0.015 for female pex13 vs. cs; P = 1.41E-6 for male and P = 0.0021 for female pex1 vs. w1118; and P = 5.22E-9 for male and P = 0.0053 for female pex13 vs. w1118). Insets indicate that mutations in pex1 and pex13 promoters reduced their expression levels to 20% of the expression level in WT. (D) Survival curves of C. elegans fed with bacteria containing vector control (EV) or dsRNA constructs directed against daf-16 (the negative control) or genes encoding the indicated peroxisomal biogenesis proteins. RNAi knockdown of prx-13, F18F11.1 (PXMP4 homolog), prx-11, and prx-5 significantly extended worm’s lifespan (log rank test: P = 2.3E-7, P = 3.5E-6, P = 0.002, and P = 0.009, respectively). All RNAi constructs were confirmed by sequencing.
To test experimentally whether peroxisome biogenesis negatively affects lifespan, we used two Drosophila melanogaster mutants pex1S4868 and pex13KG04339 harboring mutations in the promoter regions of pex1 and pex13 that resulted in lowered expression levels of these genes (Fig. 4C, Inset). Compared with the WT strains, both mutants had lifespan that increased by 16% in male flies and 13% in female flies (Fig. 4C).
We also identified the Caenorhabditis elegans orthologs of peroxisomal proliferation genes, with expression levels (as measured by our microarray analysis) that showed negative correlation with lifespan, and tested the effect of RNAi knockdown of their expression on lifespan. Indeed, knockdown of four of eight genes resulted in significantly increased lifespan relative to the empty vector control. In particular, knockdown of prx-13 or F18F11.1, which correspond to the mouse genes showing the strongest negative correlation with the mean lifespan (Pex13 and Pxmp4), resulted in significant lifespan extensions (17% and 15% longer mean lifespan relative to vector control, respectively) (Fig. 4D). These results are also supported by a previous genome-wide screen in the enhanced RNAi strain eri-1(mg366) showing that postdevelopmental knockdown of prx-5, a C. elegans homolog for mouse Pex5, increased the worm’s lifespan (29). Consistent with this report, we observed an 8% extension of the mean lifespan of N2 animals after prx-5 knockdown initiated on the first day of adulthood (Fig. 4D). Interestingly, knockdown of these genes from hatching had no effect on lifespan or slightly shortened lifespan (SI Appendix, Fig. S10A), indicating that these genes are important during development but may exert postdevelopmental detrimental effects on longevity.
Knockdown of Peroxisomal Genes Leads to Decreased Cellular Peroxide Level and Increased Tolerance of Oxidative Stress.
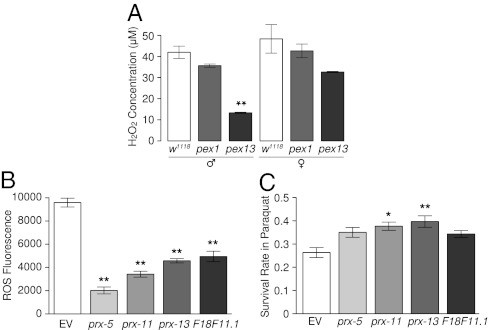
We next asked whether RNAi knockdown of PEX genes led to reduction of peroxide in the animals or increased the tolerance of the animals to peroxide. Compared with the WT parental flies, both the homozygous pex1 and pex13 mutants (pex1S4868 and pex13KG04339) had reduced levels of hydrogen peroxide (Fig. 5A), which is the major metabolic end product from peroxisomes. The reduction was stronger in males than females. The pex13 mutants showed more substantial reduction in peroxide than pex1 mutants for either sex but particularly for males (Fig. 5A). This finding is consistent with the relatively longer lifespan extension in pex13 males (Fig. 4C). In C. elegans, compared with the vector control, RNAi knockdown of prx-5, prx-11, prx-13, or F18F11.1 decreased the endogenous levels of reactive oxidative species (ROS) (Fig. 5B) and increased the stress tolerance to paraquat (Fig. 5C). In contrast, knockdown of PEX genes did not consistently increase the resistance to heat shock (SI Appendix, Fig. S10B).
Fig. 5.
RNAi knockdown of peroxisome biogenesis genes decreases endogenous peroxide level and increases oxidative stress tolerance. (A) Reduced H2O2 levels in pex1 and pex13 mutant flies. **Significant (P < 0.01) changes compared with the corresponding WT flies (w1118). (B and C) Decreased endogenous ROS levels (B) as measured by DCF-DA (Materials and Methods) and enhanced resistance to paraquat (C) in worms with RNAi knockdown of prx-5, prx-11, prx-13, or F18F11.1. Asterisks indicate significance of difference compared with the EV control worms (*P < 0.05, **P < 0.01).
Taken together, these findings show that reduced expression of peroxisomal biogenesis genes during adulthood could decrease cellular ROS levels, enhance resistance to ROS, and increase lifespan in both D. melanogaster and C. elegans. This finding suggests that the negative correlation between the expression of peroxisomal biogenesis genes and lifespan in mice may reflect an important, evolutionarily conserved role for peroxisome biogenesis in longevity control.
Discussion
In this study, we found that dietary interventions led to concordant changes in aging-related physical and physiological phenotypes. These changes were reflected by the midlife gene expression differences under six different dietary regimens. The correlation between incremental changes of hepatic gene expression and changes of the mean lifespans under these conditions was predictive of lifespan-modifying/regulating genes. We verified this conclusion in two ways. First, we showed that known longevity-modifying genes were enriched among genes and pathways with expression that was correlated or anticorrelated with lifespan. Second, we predicted that high expression of peroximal biogenesis genes might negatively influence lifespan, and we verified this prediction experimentally in D. melanogaster and C. elegans, two different model organisms that are commonly used in aging studies. Thus, our dietary perturbation-based method of gene expression analysis has successfully identified peroxisomal biogenesis factors as an important class of conserved longevity-modifying genes, providing insight into the molecular mechanisms linking diet, disease, and aging.
Mitochondria and peroxisomes are two major organelles generating ROS. Intriguingly, contrary to peroxisomal genes, mitochondrial genes were up-regulated by CR and down-regulated by HF feeding, which is consistent with the changes in the total area of liver mitochondria under these conditions (Fig. 2 and SI Appendix, Fig. S3D). Curiously, although SIRT1 and PGC1α (encoded by the Ppargc1a gene) are proposed to mediate the response to Ex and CR through regulating mitochondrial biogenesis (30, 31), we did not observe changes in the hepatic expression of Sirt1 at midlife across the six different dietary groups as detected by quantitative RT-PCR analysis (SI Appendix, Fig. S11); moreover, whereas both CR and Ex led to significantly increased liver expression of Ppargc1a in LF- but not HF-fed mice (SI Appendix, Fig. S11), these changes did not correlate with the lifespan differences. However, CR has been shown to reduce ROS generation in mitochondria, perhaps by decreasing proton leak (32). Equally important, reducing ROS from peroxisomes by suppressing peroxisome proliferation might contribute to lifespan extension by CR, whereas increased peroxisome proliferation might accelerate aging in response to HF feeding. Although speculative, this idea is supported by both the experimental data in worms and flies and the highly negative correlation of peroxisome gene expression with lifespan under different dietary regimens in mice. This finding is also supported by the suppression of many aging-related metabolic phenotypes, including insulin resistance, in PPARα KO mice when fed a HF diet (33). Intuitively, one might think that the negative correlation of PEX genes expression with lifespan would result from induction of these genes in response to a HF diet, because fatty acids are activators for PPARα; however, in reality, it could be mostly attributed to the strong suppression of these genes under CR (both LF+CR and HF+CR) (Fig. 4A).
Mitochondria and peroxisomes are not only linked in metabolic reactions, such as fatty acid metabolism, but also share essential proteins for biogenesis (34). A number of recent studies have revealed the crucial role of mitochondrial biogenesis in CR- or diet-responsive regulation of longevity (13–15, 35–37), ROS generation, and aging-associated metabolic dysfunction (38–40). Therefore, it is interesting to note that, in our mouse model, the strong anticorrelation of PEX genes expression with lifespan is coupled with the strong positive correlation of mitochondrial gene expression, mitochondrial number, and size with lifespan. This finding suggests that the biogenesis of these two organelles may be competitive and that this competitive relationship may be coupled to modulate lifespan in response to dietary cues.
Now that large-scale genome-wide experiments can be routinely conducted, how to design a systems biology experiment to effectively capture critical regulatory pathways for aging and lifespan is a pressing issue. In this study, we have shown that, consistent with the fact that dietary intervention is a powerful approach to modulating aging and lifespan in nearly all organisms studied, analyzing gene expression changes in response to dietary interventions is an efficient way to identify aging and lifespan modulators. Using such a design paradigm, additional experiments with more dietary conditions and/or measurements at the level of individuals may help elucidate the complex networks of aging and lifespan control under various nutritional conditions. Such an approach also offers the potential to define mechanisms that underlie individual variations in response to environmental/dietary conditions, and it may provide an avenue to developing personalized dietary regimens to optimize health span.
Materials and Methods
Some of the experimental procedures and materials for animal studies and bioinformatic analyses are described in SI Appendix.
Dietary Interventions in Mice.
Male C57BL/6J mice at 4 wk of age were purchased from Shanghai Animal Co, Ltd. Mice were maintained under a 12-h dark/light cycle (lights on at 6:30 AM) at a temperature of 22 °C ± 3 °C in accredited animal facilities. All experimental procedures and protocols were approved by the Institutional Animal Care and Use Committee of the Institute for Nutritional Sciences, Chinese Academy of Sciences. Before the start of the experiment, mice were maintained on an LF diet (Research Diets) for 1 wk. At the age of 5 wk, animals were randomly assigned to one of the six intervention groups (n = 30 for each group): feeding of an LF diet (10% fat, D12450B; Research Diets) ad libitum or with 30% CR (LF+CR) or voluntary running Ex (LF+Ex) or feeding of a HF diet (60% fat, D12492l; Research Diets) ad libitum or with 30% CR (HF+CR) or voluntary running Ex (HF+Ex). All mice were housed individually during the study. The daily consumption of food in LF and HF groups was recorded over 1 wk and averaged to determine the amount of food for the following week for the LF+CR and HF+CR groups, respectively. After 1 wk acclimation in cage with the locked running wheels, mice in the LF+Ex and HF+Ex groups were allowed free access to a running wheel, and the running distance and time were recorded automatically by the equipment.
Determination of Lifespan in Mice.
After eight mice were randomly selected from each group for analyses of midlife metabolic phenotypes at 62 wk of age, survival was assessed from the remaining animals (n = 22 per group), which were under close surveillance daily for death recording. Survival curves were plotted using the Kaplan–Meier method, and lifespan differences between groups were evaluated by the log-rank test. Maximum lifespan was calculated as the mean age of the oldest 20% of mice within each group.
Statistical Analysis.
All of the physiological data for animal intervention studies were presented as means ± SEM and analyzed by two-way ANOVA. Differences were considered statistically significant when P < 0.05.
Microarray Analysis.
The hepatic transcriptional level for three mice from each intervention group at 62 wk of age was analyzed using Affymetrix Mouse Genome 430 2.0 Arrays. The raw data were log2-transformed and normalized using the affy package in bioconductor. Differentially expressed genes between different groups were determined by RankProd (41) with proportion of false positive (pfp) < 0.1. PCA and hierarchical clustering based on the differentially expressed genes were performed to evaluate the relationships of those samples with different interventions.
Aging Regulatory Genes.
We compiled a gold-standard set of aging regulatory genes from three sources: 264 human and mouse lifespan or aging-related genes in GenAge, 319 genes annotated with lifespan or aging (MP:0005372) in MGI, and another 7 genes by literature survey as described (10).
Enrichment of KEGG Pathways.
KEGG pathways were downloaded from the KEGG database on October 9, 2009. The significance of enrichment was calculated using the GSEA program (22).
Peroxisome Functional Categories.
Peroxisome functional genes and categories were downloaded from http://www.peroxisomedb.org on January 7, 2010.
Drosophila Stocks and Lifespan Analysis.
pex1S4868 and pex13KG04339 mutants (42) were obtained from the Bloomington Stock Center and backcrossed six times to remove background differences. Flies were reared on standard corn meal at 25 °C. Both males and females were collected within 24 h of eclosion and randomly allocated to glass vials at a density of 20 flies per vial and 10 vials per genotype (n = 200). Flies were transferred to fresh vials every 3 d, and the number of dead was recorded.
Measurement of H2O2 Level.
pex1S4868, pex13KG04339, and w1118 flies (42) were cultured on standard corn meal at 25 °C. Adult males and females were separated at 1 d after eclosion. At 7–8 d after eclosion, male and female flies of each genotype were divided into four groups with 10 flies in each group. Flies in each group were then homogenized together, and H2O2 level was determined using a detection kit from Beyotime (S0038) following the manufacturer’s instructions.
C. elegans Strains and RNAi.
C. elegans strains were maintained at 20 °C as described (43) unless otherwise noted. The worm strains N2, daf-2(e1370), and daf-16(mu86) were obtained from the Caenorhabditis Genetics Center. RNAi was performed essentially as described previously (10, 44) with Ahringer’s RNAi feeding library (45).
C. elegans Lifespan Analysis.
The lifespan of C. elegans strains was determined as described previously (10) with minor modifications. Briefly, worms were cultured at 20 °C for two or more generations in the standard nematode growth medium (NGM) plates with E. coli OP50 before the assays. Adult worms at the beginning of egg laying (day 0) were transferred to the standard RNAi NGM plates with RNAi bacteria. At least 100 gravid worms per treatment in three plates were moved to fresh plates every 2–3 d. Worms that bagged, exploded, or crawled off the plate were excluded from the analysis. The significance of survival curves were calculated by log-rank test using the Survival package in R (http://www.r-project.org/).
Measurement of Endogenous ROS Level.
The endogenous ROS level in C. elegans was measured using 2′,7′-dichlorofluorescin diacetate (DCF-DA, D6883; Sigma) as described previously (46) with modifications. About 1,000 adult worms (day 4) were collected in M9 buffer and washed more than three times to eliminate bacteria. Then, the worms were washed one time in PBS, transferred to a 1.5-mL tube, and immediately frozen in liquid nitrogen. After thawing at room temperature, the worms were broken up by sonication (Misonix). Supernatants were collected after centrifugation (Eppendorf F-45-24-11 rotor; 12,000 rpm at 4 °C for 5 min) and transferred to new tubes. The supernatant containing 5 μg protein was incubated with 10 μL 100 μM DCF-DA in PBS at room temperature for 10 min before centrifugation. Fluorescence intensity of the supernatant of the mixture was measured using an Agilent Stratagene MX3000P at the standard detection range for SYBR Green. Six measurements were performed for each sample at 5-min intervals to examine the DCF-DA fluorescence signal linearity as previously described (46). The ROS level was determined by the fluorescence intensity at the last time point after subtracting the background fluorescence detected without the supernatant.
Oxidative Stress Resistance Assays.
About 200 adult worms (day 4) per group fed RNAi bacteria were collected in 300 μL M9 buffer and washed more than three times. About 50 worms were transferred to the 24-well plates with 500 μL 0.4 M paraquat (Sigma) in M9 per well. Survival was determined after 8 h of treatment.
Supplementary Material
Acknowledgments
We thank Prof. Hong Zhang from the National Institute for Biological Sciences and Dangsheng Li from Cell Research for invaluable suggestions. We also thank Christopher B. Newgard (Duke University) for initial design of the intervention study in mice. This work was supported by Chinese Ministry of Science and Technology Grants 2011CB910900 (to Y.L.), 2011CB504206 (to J.-D.J.H.), and 973 Program 2012CB524900; National Natural Science Foundation of China Grants 81021002 (to Y.L.), 30988002 (to Y.L.), 30830033 (to Y.L.), 30890033 (to J.-D.J.H.), and 91019019 (to J.-D.J.H.); and Chinese Academy of Sciences Grants KSCX2-EW-R-09 (to Y.L.), KSCX2-EW-R-02 (to J.-D.J.H.), KSCX2-EW-J-15 (to J.-D.J.H.), and XDA01010303 (to J.-D.J.H.). M.K. is an Ellison Medical Foundation New Scholar in Aging.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission. V.D.L. is a guest editor invited by the Editorial Board.
Data deposition: The microarray data reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, www.ncbi.nlm.nih.gov/geo (accession nos. GSE36836 and GSE36838).
See Author Summary on page 7154 (volume 109, number 19).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1119304109/-/DCSupplemental.
References
- 1.Khaitovich P, et al. Regional patterns of gene expression in human and chimpanzee brains. Genome Res. 2004;14:1462–1473. doi: 10.1101/gr.2538704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lu T, et al. Gene regulation and DNA damage in the ageing human brain. Nature. 2004;429:883–891. doi: 10.1038/nature02661. [DOI] [PubMed] [Google Scholar]
- 3.Somel M, et al. Transcriptional neoteny in the human brain. Proc Natl Acad Sci USA. 2009;106:5743–5748. doi: 10.1073/pnas.0900544106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zahn JM, et al. AGEMAP: A gene expression database for aging in mice. PLoS Genet. 2007;3:e201. doi: 10.1371/journal.pgen.0030201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zahn JM, et al. Transcriptional profiling of aging in human muscle reveals a common aging signature. PLoS Genet. 2006;2:e115. doi: 10.1371/journal.pgen.0020115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pletcher SD, et al. Genome-wide transcript profiles in aging and calorically restricted Drosophila melanogaster. Curr Biol. 2002;12:712–723. doi: 10.1016/s0960-9822(02)00808-4. [DOI] [PubMed] [Google Scholar]
- 7.Lund J, et al. Transcriptional profile of aging in C. elegans. Curr Biol. 2002;12:1566–1573. doi: 10.1016/s0960-9822(02)01146-6. [DOI] [PubMed] [Google Scholar]
- 8.McCarroll SA, et al. Comparing genomic expression patterns across species identifies shared transcriptional profile in aging. Nat Genet. 2004;36:197–204. doi: 10.1038/ng1291. [DOI] [PubMed] [Google Scholar]
- 9.Kim SK. Common aging pathways in worms, flies, mice and humans. J Exp Biol. 2007;210:1607–1612. doi: 10.1242/jeb.004887. [DOI] [PubMed] [Google Scholar]
- 10.Xue H, et al. A modular network model of aging. Mol Syst Biol. 2007;3:147. doi: 10.1038/msb4100189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fontana L, Partridge L, Longo VD. Extending healthy life span—from yeast to humans. Science. 2010;328:321–326. doi: 10.1126/science.1172539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kenyon C. The plasticity of aging: Insights from long-lived mutants. Cell. 2005;120:449–460. doi: 10.1016/j.cell.2005.02.002. [DOI] [PubMed] [Google Scholar]
- 13.Baur JA, et al. Resveratrol improves health and survival of mice on a high-calorie diet. Nature. 2006;444:337–342. doi: 10.1038/nature05354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Milne JC, et al. Small molecule activators of SIRT1 as therapeutics for the treatment of type 2 diabetes. Nature. 2007;450:712–716. doi: 10.1038/nature06261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pearson KJ, et al. Resveratrol delays age-related deterioration and mimics transcriptional aspects of dietary restriction without extending life span. Cell Metab. 2008;8:157–168. doi: 10.1016/j.cmet.2008.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Goodrick CL, Ingram DK, Reynolds MA, Freeman JR, Cider NL. Differential effects of intermittent feeding and voluntary exercise on body weight and lifespan in adult rats. J Gerontol. 1983;38:36–45. doi: 10.1093/geronj/38.1.36. [DOI] [PubMed] [Google Scholar]
- 17.Hietakangas V, Cohen SM. Regulation of tissue growth through nutrient sensing. Annu Rev Genet. 2009;43:389–410. doi: 10.1146/annurev-genet-102108-134815. [DOI] [PubMed] [Google Scholar]
- 18.Kleiber M. Body size and metabolic rate. Physiol Rev. 1947;27:511–541. doi: 10.1152/physrev.1947.27.4.511. [DOI] [PubMed] [Google Scholar]
- 19.de Magalhães JP, Costa J, Toussaint O. HAGR: The Human Ageing Genomic Resources. Nucleic Acids Res. 2005;33:D537–D543. doi: 10.1093/nar/gki017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Breitling R, Armengaud P, Amtmann A, Herzyk P. Rank products: A simple, yet powerful, new method to detect differentially regulated genes in replicated microarray experiments. FEBS Lett. 2004;573:83–92. doi: 10.1016/j.febslet.2004.07.055. [DOI] [PubMed] [Google Scholar]
- 21.Ernst J, Bar-Joseph Z. STEM: A tool for the analysis of short time series gene expression data. BMC Bioinformatics. 2006;7:191. doi: 10.1186/1471-2105-7-191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Subramanian A, et al. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Russell SJ, Kahn CR. Endocrine regulation of ageing. Nat Rev Mol Cell Biol. 2007;8:681–691. doi: 10.1038/nrm2234. [DOI] [PubMed] [Google Scholar]
- 24.Schibler U. The daily rhythms of genes, cells and organs. Biological clocks and circadian timing in cells. EMBO Rep. 2005;6:S9–S13. doi: 10.1038/sj.embor.7400424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lee SS, et al. A systematic RNAi screen identifies a critical role for mitochondria in C. elegans longevity. Nat Genet. 2003;33:40–48. doi: 10.1038/ng1056. [DOI] [PubMed] [Google Scholar]
- 26.Steffen KK, et al. Yeast life span extension by depletion of 60s ribosomal subunits is mediated by Gcn4. Cell. 2008;133:292–302. doi: 10.1016/j.cell.2008.02.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schrader M, Fahimi HD. Mammalian peroxisomes and reactive oxygen species. Histochem Cell Biol. 2004;122:383–393. doi: 10.1007/s00418-004-0673-1. [DOI] [PubMed] [Google Scholar]
- 28.Jin C, et al. Histone demethylase UTX-1 regulates C. elegans life span by targeting the insulin/IGF-1 signaling pathway. Cell Metab. 2011;14:161–172. doi: 10.1016/j.cmet.2011.07.001. [DOI] [PubMed] [Google Scholar]
- 29.Curran SP, Ruvkun G. Lifespan regulation by evolutionarily conserved genes essential for viability. PLoS Genet. 2007;3:e56. doi: 10.1371/journal.pgen.0030056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cantó C, Auwerx J. Caloric restriction, SIRT1 and longevity. Trends Endocrinol Metab. 2009;20:325–331. doi: 10.1016/j.tem.2009.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gurd BJ. Deacetylation of PGC-1α by SIRT1: Importance for skeletal muscle function and exercise-induced mitochondrial biogenesis. Appl Physiol Nutr Metab. 2011;36:589–597. doi: 10.1139/h11-070. [DOI] [PubMed] [Google Scholar]
- 32.Guarente LP, Partridge L, Wallace DC. Molecular Biology of Aging. Plainview, NY: Cold Spring Harbor Laboratory Press; 2007. pp. 409–425. [Google Scholar]
- 33.Cha DR, et al. Peroxisome proliferator-activated receptor-alpha deficiency protects aged mice from insulin resistance induced by high-fat diet. Am J Nephrol. 2007;27:479–482. doi: 10.1159/000106485. [DOI] [PubMed] [Google Scholar]
- 34.Delille HK, Alves R, Schrader M. Biogenesis of peroxisomes and mitochondria: Linked by division. Histochem Cell Biol. 2009;131:441–446. doi: 10.1007/s00418-009-0561-9. [DOI] [PubMed] [Google Scholar]
- 35.D’Antona G, et al. Branched-chain amino acid supplementation promotes survival and supports cardiac and skeletal muscle mitochondrial biogenesis in middle-aged mice. Cell Metab. 2010;12:362–372. doi: 10.1016/j.cmet.2010.08.016. [DOI] [PubMed] [Google Scholar]
- 36.Nisoli E, et al. Calorie restriction promotes mitochondrial biogenesis by inducing the expression of eNOS. Science. 2005;310:314–317. doi: 10.1126/science.1117728. [DOI] [PubMed] [Google Scholar]
- 37.López-Lluch G, et al. Calorie restriction induces mitochondrial biogenesis and bioenergetic efficiency. Proc Natl Acad Sci USA. 2006;103:1768–1773. doi: 10.1073/pnas.0510452103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hagopian K, et al. Caloric restriction influences hydrogen peroxide generation in mitochondrial sub-populations from mouse liver. J Bioenerg Biomembr. 2011;43:227–236. doi: 10.1007/s10863-011-9353-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hagopian K, et al. Long-term calorie restriction reduces proton leak and hydrogen peroxide production in liver mitochondria. Am J Physiol Endocrinol Metab. 2005;288:E674–E684. doi: 10.1152/ajpendo.00382.2004. [DOI] [PubMed] [Google Scholar]
- 40.Civitarese AE, et al. Calorie restriction increases muscle mitochondrial biogenesis in healthy humans. PLoS Med. 2007;4:e76. doi: 10.1371/journal.pmed.0040076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hong F, et al. RankProd: A bioconductor package for detecting differentially expressed genes in meta-analysis. Bioinformatics. 2006;22:2825–2827. doi: 10.1093/bioinformatics/btl476. [DOI] [PubMed] [Google Scholar]
- 42.Chen H, Liu Z, Huang X. Drosophila models of peroxisomal biogenesis disorder: Peroxins are required for spermatogenesis and very-long-chain fatty acid metabolism. Hum Mol Genet. 2010;19:494–505. doi: 10.1093/hmg/ddp518. [DOI] [PubMed] [Google Scholar]
- 43.Brenner S. The genetics of Caenorhabditis elegans. Genetics. 1974;77:71–94. doi: 10.1093/genetics/77.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kamath RS, Martinez-Campos M, Zipperlen P, Fraser AG, Ahringer J. Effectiveness of specific RNA-mediated interference through ingested double-stranded RNA in Caenorhabditis elegans. Genome Biol. 2001;2:RESEARCH0002. doi: 10.1186/gb-2000-2-1-research0002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kamath RS, et al. Systematic functional analysis of the Caenorhabditis elegans genome using RNAi. Nature. 2003;421:231–237. doi: 10.1038/nature01278. [DOI] [PubMed] [Google Scholar]
- 46.Lee SJ, Hwang AB, Kenyon C. Inhibition of respiration extends C. elegans life span via reactive oxygen species that increase HIF-1 activity. Curr Biol. 2010;20:2131–2136. doi: 10.1016/j.cub.2010.10.057. [DOI] [PMC free article] [PubMed] [Google Scholar]