Abstract
Applied forces and the biophysical nature of the cellular microenvironment play a central role in determining cellular behavior. Specifically, forces due to cell contraction are transmitted into structural ECM proteins and these forces are presumed to activate integrin “switches.” The mechanism of such switches is thought to be the partial unfolding of integrin-binding domains within fibronectin (Fn). However, integrin switches remain largely hypothetical due to a dearth of evidence for their existence, and relevance, in vivo. By using phage display in combination with the controlled deposition and extension of Fn fibers, we report the discovery of peptide-based molecular probes capable of selectively discriminating Fn fibers under different strain states. Importantly, we show that the probes are functional in both in vitro and ex vivo tissue contexts. The development of such tools represents a critical step in establishing the relevance of theoretical mechanotransduction events within the cellular microenvironment.
Interactions between cells and their ECM are crucial for regulating cell phenotypes and determining cell and tissue fate. In addition to its scaffolding role, it is now appreciated that the ECM actively signals cells through transmembrane receptors and that the ECM itself is dynamically regulated in both structure and composition (1). Through these dynamic changes, the ECM is thought to play a vital role in maintenance of the normal tissue microenvironment and its misregulation leads to pathological conditions such as cancer and fibrosis (2).
Biophysical dynamics of the ECM are controlled in large part by macromolecular cell adhesive structures, termed focal adhesions, that couple ECM proteins such as fibronectin (Fn) (3) to the cellular cytoskeleton through transmembrane receptors, termed integrins. These cell–ECM interactions are inherently physical/mechanical and, as such, directly link the state of a cell to its microenvironmental ECM and vice versa. Recent work suggests that under mechanical load, presumably due to forces transmitted from the ECM, the conformation of specific intracellular focal adhesion proteins, e.g., vinculin (4), are altered and may result in the maturation or growth of focal adhesion structures. Because of the reciprocity of force transmission across the focal adhesion, ECM proteins within structural fibers may also serve similar roles as “force sensors” (5). Indeed, recent work of our own and others indicate that engineered hypersensitization or stabilization of the force-sensitive integrin-binding domain of Fn regulates integrin specificity and concomitant cell phenotype. These findings implicate the Fn integrin-binding domain as one possible force sensor capable of regulating cell phenotype (6, 7).
Recent evidence indicates that Fn molecules within intact fibers experience cell-derived forces and in response exhibit molecular strain (8). Methods developed by Discher and coworkers demonstrated that cysteine accessibility may be used as a probe for screening the conformational states of a large number of proteins in cells, including Fn (9, 10). Importantly, molecular strain in Fn fibers may also be an important regulator of specific protein binding to Fn, as recent work (11) has suggested that the binding sites of two bacterial adhesin-derived peptides are destroyed with high fiber strain. This strain can also presumably cause unfolding of the same force-sensitive domains that comprise the integrin-binding domain, namely Fn type III domains. Furthermore, cells displaying altered physiological states are capable of unfolding Fn type III domains to a greater or lesser extent depending on the activation state of their contractile machinery (12). In addition to the likely disruption of receptor binding motifs, unfolding events within Fn domains are also known to unmask cryptic sites crucial for Fn–Fn interactions during fiber assembly (13–16). Despite the prevailing view in the literature of the importance of force-mediated Fn molecular alterations, the presence of such events and their relevance in vivo has yet to be demonstrated.
There is currently only one approach, intramolecular FRET, enabling the detection of molecular strain events in Fn fibers (17, 18). By using chemically modified Fn, the force-induced separation of donor and acceptor fluorophores located on the same Fn molecule is directly related to changes in FRET intensity (8, 19, 20). A limitation of this FRET approach is that Fn molecular strain events are only detectable in vitro, and only truly feasible with abundantly available and chemically modified plasma-derived Fn or genetically manipulated and recombinant Fn. Furthermore, there are significant technical challenges in using FRET to investigate Fn molecular strain events in complex environments such as ex vivo tissue samples. Because of the above concerns, there is significant debate on whether Fn strain and partial unfolding events observed in silico (13) and in vitro (8) are physiologically relevant in native tissues (i.e., in vivo) during morphogenesis, wound repair, and pathological progression.
Radically different approaches are therefore needed to develop probes that can detect molecular signatures representative of Fn fiber strain in intact ex vivo tissues and intravital staining of living tissues. Here, we report such a demonstration using random phage display in combination with the controlled deposition and extension of aligned Fn fibers. The resulting phage peptide-based molecular probes exhibit strain-selective binding to manually extruded Fn fibers, cell-derived Fn ECM, and ex vivo living lung slices. In principle, these probes can be used to map molecular strain events in unmodified native ECM microenvironments, as well as for molecular targeting of Fn (ECM) in altered structural states associated with disease.
Results
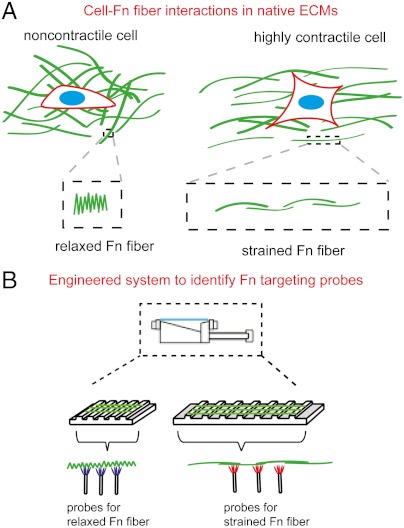
The development of molecular probes for the detection of strain events within Fn fibers will ultimately enable the analysis of native Fn ECM in vitro and in ex vivo tissue. Fn molecules within fibers of a native ECM are thought to adopt a range of conformations induced by applied force (Fig. 1A). In order to realize the goal of identifying strain-selective probes to Fn fibers, methods are needed for reproducible and controlled mechanical straining of pure Fn fibers. We manually extruded Fn fibers onto flexible polydimethylsiloxane (PDMS) substrates and aligned perpendicular to the long axis of micropatterned plateau/trough structures as previously described with minor modifications (21–23). Subsequent cross-linking of Fn fibers to the PDMS allowed for suspension and controlled uniaxial straining (Fig. 1B). The second element of our strategy consisted of a random phage display library to interrogate the Fn fiber surface (Fig. 1B). Strained or relaxed fibers were placed in a holding chamber to enable phage incubation under controlled conditions. The phage enrichment process followed a typical workflow starting with a randomized library (1011 clones of 6.4 × 107 diversity).
Fig. 1.
Integrated system for identifying conformation sensitive molecular probes for Fn fibers. (A) Schematic for single Fn fibers in native ECM under different mechanical environments. (Left) A relaxed Fn fiber. (Right) A strained Fn fiber under loading of the ECM in the longitudinal direction. (B) Fn fibers were deposited onto chemically treated, patterned PDMS surfaces. PDMS with Fn fibers were then strained using a homemade PDMS straining device capable of applying strain of up to 300%. Schematic of freely suspended Fn fibers on relaxed (Left) and strained (Right) patterned PDMS substrates on which phage panning were performed to identify strain-specific probes for Fn fibers.
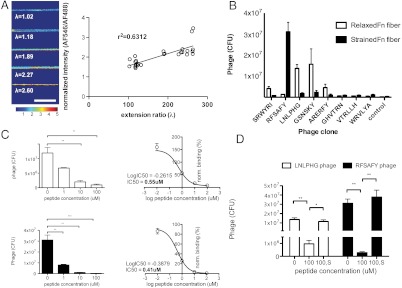
Using the straining device, we were able to apply uniaxial tension to the PDMS substrate and achieve extension of the deposited Fn fibers exceeding 2.6 times original length without significant breaking of Fn fibers (i.e., 50% breakage). To verify the Fn fiber strain-dependent increase in Fn type III domain unfolding, we followed the exposure of cryptic cysteine residues in 7th and 15th type III domains using a modified cysteine shotgun method (24) (Fig. 2A). Normalized heat map images of the reporter dye showed increasing intensity with increasing strain of the PDMS (top to bottom). Because the Fn fibers are cross-linked to the underlying PDMS, they experience the same strain. Analysis of at least 25 Fn fibers displayed a robust and linear (r2 = 0.63) relationship between Fn fiber strain and exposure of cryptic cysteines. We note that it is likely for other domains of Fn to also be partially unfolded as a result of the applied macroscopic strain.
Fig. 2.
Isolation of specific phage clones targeting Fn fibers under varying strain. (A) Increased strain in Fn fibers correlates with unfolding of Fn type III domains. A thiol-reactive compound (AlexaFluor 546-maleimide) was used to detect the unmasking of the buried free cysteine residues in FnIII7 and FnIII15. Amount of free cysteine detected (unfolding of FnIII7 and/or FnIII15) correlated with Fn fiber strain. Images were acquired at 63×, shown is normalized intensity of AF546 channel divided by AF488 channel. (Scale bar: 20 μm.) (B) Binding of individual clones to Fn fibers under varying strain. Eight clones were individually assayed for their binding to Fn fibers. Each experiment used 1 × 1011 phage. Control phage was phage population after round three of panning. (C) Competitive inhibition of phage clones with soluble peptides at increasing concentrations. Two phage clones that show the greatest dynamic range in difference between binding relaxed (Upper, LNLPHG phage) and strained (Lower, RFSAFY phage) Fn fibers were characterized for their binding specificity. Each phage clone was coincubated with its corresponding displayed peptide at increasing concentrations. Data were fitted using a nonlinear log (inhibitor) vs. normalized response fit and IC50 values were calculated to 0.55 μM for LNLPHG phage, and 0.41 μM for RFSAFY phage. N > 3 for all samples, error bars are SEM. Statistics were performed using a one-way ANOVA with Bonferroni posttest correction. (**, p < 0.01) (D) Scrambled soluble peptides do not inhibit binding of phage clone to Fn fibers. Phage clones displaying either the LNLPHG or RFSAFY peptide were incubated with its corresponding peptide or a scrambled peptide (HLNPGL or AYSRFF) at 100 μM in the presence of relaxed (λ = 0.93) or strained (λ = 2.64) Fn fibers, respectively. Incubation with 100 μM peptide that matched the phage-displayed peptide showed competitive inhibition (labeled “100”), whereas incubation with scrambled peptides did not (labeled “100, S”). N > 3 for all samples, error bars are SEM. Statistics were performed using a one-way ANOVA with Bonferroni posttest correction (*, p < 0.05, ** p < 0.001).
We then performed phage display screens to identify unique peptides that discriminate Fn fibers under varying strain, specifically a “relaxed” and a “strained” state. The fuse5 6-mer phage peptide library was chosen because the random peptides are fused to phage pIII coat proteins located at the tip of the phage, and are therefore thought to be sterically favorable to probing unfolded domains. Furthermore, the relatively low number of copies of pIII per phage makes multivalent binding less likely and as a result yields higher affinity interactions. Our strategy is an initial negative selection step to remove phage that bound to other targets besides Fn fibers, specifically gelatin and serum albumin used to passivate the surface. Supernatant phages (i.e., unbound phage) from this negative selection were then amplified and used for round one of positive selection. Selection was performed on both relaxed (extension ratio = 1.0; λ = ℓ/L) and strained (λ = 2.6) Fn fibers. After each round, all phages were collected and accounted for by phage titers, ensuring enrichment of a population of phage that bound strongly to the Fn fibers under each condition (Fig. S1). After three rounds of selection, each with increasing stringency, phage clones were isolated from individual Tg1 Escherichia coli colonies and phage DNA was sequenced (Table S1). Translated sequences revealed multiple identical sequences recovered from different colonies, suggesting enrichment of selective sequences from the initial repertoire of random peptides. No clear homology was identified comparing sequences within each population (derived from relaxed or strained fibers). However, considering the short length of the randomized peptide sequence and the structural complexity of Fn fibers, this result was not unexpected.
Eight of the identified phage clones were individually produced and purified, and their binding to Fn fibers characterized by titer analysis (Fig. 2B). Five clones of the eight tested displayed significant (> 107 cfu) and strain-selective binding to Fn fibers. Of particular interest were clones displaying peptide sequences LNLPHG and RFSAFY. These two phage clones showed the greatest binding to Fn fibers and the greatest efficiency in discriminating between relaxed versus strained Fn fibers. The clone RFSAFY preferentially bound to strained fibers over relaxed fibers (3.1 × 107 compared to 1.4 × 106 cfu, or a selectivity of 22.1) whereas clone LNLPHG preferentially bound relaxed fibers over strained fibers (1.4 × 107 compared to 1.8 × 106 cfu, or a selectivity of 7.8). Clones displaying SRWYRI and ARERFY peptides showed good discrimination between relaxed and strained fibers (both binding relaxed), but their overall binding efficiency to Fn fibers was significantly lower. The only other clone that displayed significant binding to Fn fibers was GSNSKY, which bound preferentially to relaxed fibers (1.5 × 107 cfu) but had greater variability, as evidenced by the standard error of repeated titers. Based on these observations, clones LNLPHG and RFSAFY were chosen for competitive binding assays with their corresponding soluble peptides (Fig. 2C). Binding of each phage clone to its antigen [LNLPHG to relaxed Fn fibers (Upper), RFSAFY to strained Fn fibers (Lower)] was found to be inhibited in a dose-dependent manner when coincubated with its corresponding soluble peptide, indicative of specific peptide-mediated binding. Calculation of the IC50 from the competitive binding data indicates nanomolar IC50 for both LNLPHG- and RFSAFY-antigen interactions (550 and 410 nM, respectively). Furthermore, inhibition was not observed when phage clones were incubated with scrambled versions of the peptides at 100 μM concentration (Fig. 2D), demonstrating specificity of the interactions. Binding of the LNLPHG phage clone to relaxed Fn fibers, and of the RFSAFY phage clone to strained Fn fibers were found to be reversible (Fig. S2), whereby strain/relaxation of Fn fibers following phage binding was found to displace the bound phage.
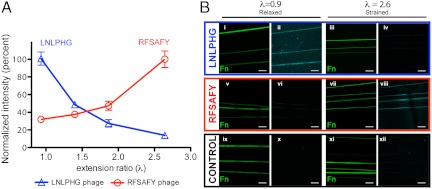
To directly evaluate phage targeting to Fn fibers under varying strain, we used a semiquantitative approach using fluorescently labeled phage. Phage binding to Fn fibers deposited on micropatterned PDMS substrates was assessed on increasingly strained Fn fibers (from λ = 0.9 to 2.6; Fig. 3A). Fluorescence intensity of the Fn fiber at 629–672 nm (AF633-labeled phage) was normalized to the amount of Fn in the fiber (5% AF488-labeled tracer Fn). The Fn fibers labeled with the LNLPHG clone showed a reproducible nonlinear decrease in fluorescence intensity as strain increased (Fig. 3B, i–iv). Conversely, as expected, Fn fibers incubated with the RFSAFY clone showed an increased signal with fiber strain (Fig. 3B, v–viii). The intensity of Fn fibers labeled with a control (random) phage was negligible (nondetectable) at all strains tested (Fig. 3B, ix–xii). Targeting selectivity for the LNLPHG clone, defined as [signal at λ = 0.9]/[signal at λ = 2.6], was determined to be 7.29. Targeting selectivity for the RFSAFY clone, defined as [signal at λ = 2.6]/[signal at λ = 0.9] was determined to be 3.13.
Fig. 3.
Staining of specific phage clones correlates with Fn fiber strain. Phage clones displaying the LNLPHG and RFSAFY peptides were labeled with AlexaFluor 633 SE and incubated on Fn fibers (1 × 1011 phage per sample). (A) Staining of labeled LNLPHG phage decreases with increasing fiber strain, and staining of RFSAFY phage increases with increasing fiber strain. (B, i–xii) Fluorescence images of labeled phage clones on Fn fibers under varying strain. LNLPHG phage binding to relaxed (λ = 0.93) (i, ii) and strained (λ = 2.64) Fn fibers (iii, iv). RFSAFY phage binding to relaxed (λ = 0.93) (v, vi) and strained (λ = 2.64) Fn fibers (vii, viii). Labeled control phage show minimal binding (ix–xii). Error bars are SD. All images acquired with 63× oil immersion objective. (Scale bar: 20 μm.)
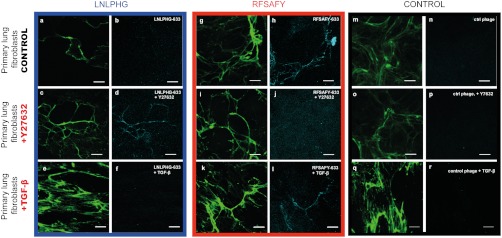
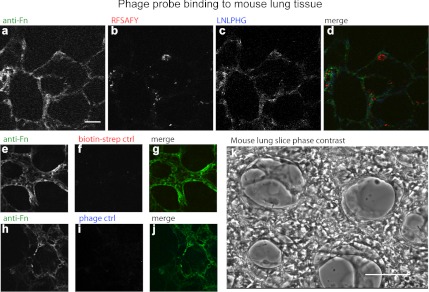
As a result of combining phage display screens with control over Fn fiber strain, we discovered simple probes that are capable of detecting varying states of strain in Fn fibers. Importantly, our approach does not require Fn to be chemically labeled (a requirement and limitation of the FRET method). Such probes enable the interrogation of native Fn ECM, allowing us to address a critical gap in the determination of the relevance of force-mediated Fn structural modifications. As a first demonstration of this principle, we used our phage probes to discriminate Fn fibers within native cell-assembled ECM assembled by contractile primary lung fibroblasts cultured in the presence of tracer Fn (AF488-labeled; Fig. 4). Labeled (AF633) phage clones were detectable primarily within larger diameter fibers and their binding was correlated with cell-mediated tensional strain of the Fn ECM as demonstrated by the addition of the contractility inhibitor Y-27632 (inhibitor of Rho-kinase). Specifically, staining of native, cell-derived Fn fibers with the LNLPHG phage was not detectable at steady state and increased upon addition of Y-27632, which results in a more relaxed (lower strain) Fn ECM (Fig. 4 A–D), and decreased upon stimulation of cell contraction with TGF-β (Fig. 4 E and F). Concomitantly, staining of native, cell-derived Fn fibers with the RFSAFY phage clone demonstrated robust staining at steady state and significantly decreased upon Y-27632 addition (Fig. 4 G–J), but was not changed with TGF-β stimulation (Fig. 4 K and L). Staining of Fn fibers with a labeled control phage was nondetectable with or without Y-27632 or TGF-β treatment (Fig. 4 M–R). As a further proof of concept demonstration, mouse lung slices were prepared (Fig. 5), and staining of our phage clones were evaluated in concert with a commercially available anti-Fn antibody. Qualitatively, the phage probes were able to delineate Fn matrix morphology, similar to the staining patterns of the Fn antibody (Fig. 5 A–D). Interestingly, the staining of RFSAFY phage (Fig. 5B) appeared more punctuate and more spatially heterogeneous when compared to the staining of Fn antibody (Fig. 5A). Additionally, labeled control phages displayed no detectable binding regardless of labeling species (Fig. 5 E–J). Coincubation with an excess of a polyclonal anti-Fn antibody were able to inhibit binding of both phage probes demonstrating the specificity of the probes to Fn (Fig. S3). Similar results were obtained using a molecular probe approach (Fig. S4). Specifically, quantum dot-peptide conjugated probes displaying either LNLPHG or RFSAFY peptides similarly costained with anti-Fn antibody (Fig. S4 A–F). In addition, no staining was observed for scrambled versions of both peptide probes (Fig. S4 G–J). Importantly, no colocalization of the two phage-based probes or peptide-based probes were observed, suggesting their ability to discriminate regions within Fn fibers in native in vivo tissue. Collectively, these data strongly suggest that Fn fibers under strain display markedly different biochemical signatures that can be used for the molecular-level detection of Fn fiber strain. Because the molecular imaging probes developed here do not rely on chemical labeling of Fn molecules (which also requires partial denaturation of the protein) it is conceivable that native tissue (ex vivo) can be interrogated for mechanochemical alterations and their potential role in physiological and pathological progression established.
Fig. 4.
Staining of specific phage clones on cell-assembled ECM. Phage clones displaying the LNLPHG and RFSAFY peptide motifs were labeled with AlexaFluor 633 SE and incubated on ECM assembled by primary lung fibroblasts (1 × 1011 phage per sample). Experiments performed in the presence of the ROCK inhibitor Y27632 (Middle) to relax the matrix, and in the presence of TGF-β to contract the matrix (Bottom). Staining of LNLPHG phage observed primarily on large diameter fibers, and increased after matrix relaxation (C and D). Staining of RFSAFY phage observed primarily on large diameter fibers. Staining is minimal after matrix relaxation by Y27632 (I and J). Staining of a labeled control phage (M–R). All images acquired with 63× oil immersion objective. (Scale bar: 20 μm.)
Fig. 5.
Staining of specific phage clones on prepared living lung slices. (A–D) Costaining of multiple Fn targeting probes on a representative mouse lung slice. (A) Staining of polyclonal anti-mouse Fn antibody, (B) staining of labeled RFSAFY phage, detected with AF 546, (C) staining of labeled LNLPHG phage, detected with AF633. (E–G) Staining of labeled random phage clone detected with AF546. H–J) Staining of labeled random phage clone detected with AF633. (K) Representative phase micrograph of a prepared living lung slice. (Scale bar: 100 μm.) Images in A–J acquired with 63× oil immersion objective. (Scale bars: 20 μm.)
Discussion
The role of mechanical forces in mediating cell–ECM interactions is becoming increasingly important in understanding how the ECM directs cell behavior and cell fate. In particular, forces emanating from contractile cells such as pathogenic myofibroblasts have been hypothesized to partially unfold ECM proteins like Fn, thus engaging/disengaging theorized integrin switches (25). Despite considerable in silico and in vitro evidence for the extensibility of Fn within fibers and Fn type III domain unfolding (26–28), there is still no direct evidence that such molecular events occur in vivo, a fact that perpetuates the debate regarding the validity of such observations. To fill this void, we combined controlled Fn fiber straining with random peptide phage display to isolate peptide-based molecular probes capable of discriminating Fn fibers under relaxed and strained conditions. We discovered two probes (LNLPHG and RFSAFY) that displayed highly specific binding to Fn fibers in a strain-selective manner. These two probes display highly reproducible binding characteristics to Fn fibers; with increasing (decreasing) strain, LNLPHG binding is reduced (increased) and RFSAFY binding is increased (reduced). Therefore these two probes can be used in concert to achieve exceptionally high resolution of the strain state of Fn fibers through cross comparison of their binding to the same fiber (e.g., Fig. 5D), assuming no steric hindrance between the two probes. Most importantly, these peptide-based probes are capable of discriminating native Fn fibers, enabling the detection of Fn fiber strain events in vivo.
Even with the selectivity of each identified probe in detecting and discriminating Fn fibers of variable strain, the specific epitopes on Fn fibers to which these probes are targeting cannot be determined with the present system. Indeed, this uncertainty is not a trivial matter because the structure and molecular packing of Fn fibers themselves have not been elucidated. Fn fibrillogenesis is known to involve self-association of Fn molecules through an integrin-mediated, mechanically active process (29) yet recent evidence suggests as many as eight different Fn–Fn interactions can occur, further complicating a basic understanding of Fn fiber arrangement (30). Thus, under force, the extensibility of Fn fibers may be because of a combination of rearrangement of Fn-Fn interactions within the fiber, rearrangement of the heterotypic type III domain interactions within a single Fn molecule (28), or unfolding of Fn type III domains (8). Any of these events could potentially create new epitopes that can be recognized by the phage probes. Additionally, recent evidence suggests that Fn fibers become fouling–i.e., exhibit significant nonspecific and presumably hydrophobic protein–protein interactions—upon straining (24). However, our data suggest that this nonspecific fouling effect does not contribute significantly to the observed specific binding of our probes because we observed a saturable binding of our probes with increasing concentration (Fig. S5 A and C). Furthermore, at the saturation point, incubations with increasing amounts of BSA (Fig. S5 B and D) did not significantly affect either probe signal, suggesting that it was not capable of competing for specific binding to fibers with our probes. Costaining of both phage probes with the fibrillar adhesion marker integrin β-1 and the focal adhesion marker vinculin did not appear to show significant differential phage accumulation at sites of adhesion (Fig. S6), suggesting that the epitope targeted by the phage probes is likely along the entire length of the Fn fiber and not specifically localized or excluded at sites of Fn-adhesion linkages. Furthermore, the phage probes display minimal binding to collagen, gelatin, and the negative control BSA (Fig. S7).
In conclusion, we present here the identification and characterization of two molecular probes capable of discriminating Fn fibers in a relaxed versus a strained state. Looking forward, using these and other future mechanosensitive probes it is conceivable to “map” dynamic Fn molecular strain events. With such tools in hand, one can correlate these events with specific cellular phenotypic alterations associated with tissue pathology ex vivo, yielding insights into the roles of mechanotransduction in the progression of disease. Furthermore, although the intent of these efforts was to address the fundamental gaps in Fn mechanotransduction in vivo, it is conceivable that such Fn strain-specific probes can be used to target therapeutics to Fn fibers based on their mechanical signature or “fingerprint.” Although this area obviously needs work, such capabilities could transform how diseases are managed.
Materials and Methods
Materials.
All chemicals were obtained from VWR International and Fisher unless otherwise noted. Dyes for fluorescent conjugation were from Invitrogen. Peptides were synthesized by Genscript Corportation. The fuse5 phage 6-mer peptide library was a generous gift from G. Smith (University of Missouri, Columbia, MO).
Fn Fiber Deposition.
Substrates for Fn fiber deposition were prepared on PDMS by soft lithography with features of 10 × 100 μm and 50 μm spacing. Masks were fabricated by standard photolithography. Fn was purified from frozen human plasma by gelatin-sepharose affinity chromatography. Fn fibers were deposited on PDMS from 1 mg/mL solutions as previously described (22, 23).
Fn III Domain Unfolding in Fn Fibers.
Fn was fluorescently labeled with AlexaFluor 488 (AF488) tetrafluorophenyl-ester and dialyzed against PBS buffer. Labeled Fn was then diluted in a 5∶95 ratio with unlabeled Fn. AF488-labeled, extruded Fn fibers were cross-linked to micropatterned PDMS substrates and strained to defined amounts. Fn fibers were incubated with AlexaFluor 546 (AF546) C5-maleimde (100 μM) for 15 min at room temperature. Excess dye was removed by extensive washes with PBS and samples were mounted in Prolong gold mounting medium (Invitrogen). Image analysis was performed by confocal microscopy (Zeiss; LSM 510 NLO). Images were acquired using a 1.4 N.A. 63× oil immersion objective. Emitted light was detected using photomultiplier tubes (PMTs) at 500–530 nm (AF488) and 554–597 nm (AF546). Images were generally acquired at 1,024 × 1,024 pixels for a field of view of 142 × 142 μm. Pinhole diameter was 178 μm. Quantitative analysis was performed by normalizing AF546 intensity to AF488 intensity. Image acquisition settings were consistent for all samples.
Phage Display.
A negative screen was first performed with BSA and gelatin-blocked PDMS substrate. The fuse5 6-mer library (1 × 1011 cfu) was incubated for 1 h on the substrate. Supernatant phage were collected and amplified by infection into Tg1. Escherichia coli and grown overnight in LB broth, 15 μg/mL tetracycline, and 1 mM IPTG. Phage were precipitated from overnight cultures by standard PEG/NaCl precipitation (31). Phage physical concentration was determined using UV/visible spectrometry (cfu/mL = (A269-A320 nm)/9,225 × 6 × 1017) and 1 × 1011 cfu was used as the input for first round of positive selection. Phage were allowed to incubate on deposited Fn fibers for 1 h, followed by incubation with wash buffer (PBS, 0.05% Tween-20) for 10 min, and eluted with 0.2 M glycine, pH 2.1, and neutralized with 1 M Tris•HCl, pH 9. Eluted phage were propagated into Tg1 cells, and purified as above. Three rounds of selection were performed in parallel on both relaxed and strained Fn fibers. Stringency of selection was controlled by subsequently increasing the number of wash steps prior to phage elution for each successive screen.
Phage Clone Binding and Phage Peptide Competition.
After three rounds of selection, 40 clones were randomly picked and sequenced. Primers used for PCR were 5′-aagctgataaaccgatacaatt-3′ and 5′-ccgtaacactgagtttcgtc-3′. Sequenced clones were produced and binding specificity for to Fn fibers determined by phage titering for individual clones under relaxed (λ = 1.0) and strained (λ = 2.6) conditions. Soluble peptides corresponding to the phage-displayed sequence were produced by solid phase synthesis (Genscript Corp.) and incubated with their corresponding phage in competition experiments at 1, 10, and 100 μM. Competitive exclusion of phage binding by peptides was determined by phage titers. Corresponding scrambled peptide sequences were used as controls for nonspecific peptide inhibition.
Phage Clone Staining on in Vitro Generated Fibers.
For imaging purposes, phage probes were labeled with AlexaFluor 633 (AF633) SE (Invitrogen) for 1 h per manufacturer’s recommendations, and purified by PEG/NaCl precipitation. AF488-labeled Fn fibers were extruded onto micropatterned PDMS and strained to appropriate amounts. Phage clones were incubated with Fn fibers for 1 h (1 × 1012 cfu per sample), washed 3× with PBS + 0.1% Tween-20, and mounted in Prolong gold mounting medium (Invitrogen). Imaging was performed by confocal microscopy, emitted light was detected using PMTs at 500–530 (AF488), 565–587 (AF546), and 629–672 nm (AF633). Semiquantitative analysis was performed by normalizing the AF546 and AF633 intensity to AF488 intensity. Image acquisition settings were consistent for all samples.
Phage Clone Staining on Cell-Assembled ECM.
Chambered coverslides (BD Biosciences) used for cell culture were incubated with 500 μL of unlabeled Fn at 0.03 g/L overnight to allow Fn to adsorb to surface. Mouse primary lung fibroblasts were seeded at a density of 20,000 cells per cm2. After 30 min, medium was exchanged with Fn-depleted medium, supplemented with 1 μg/mL of AF488-labeled Fn. Cells were allowed to assemble ECM for 48 h, washed with PBS, and blocked with 1 mg/mL BSA for 30 min on ice. Labeled phage were incubated at 1 × 1012 cfu per well for 30 min, and washed 3× with PBS + 0.1% Tween-20. All cells were fixed with 4% formaldehyde for 20 min and mounted in Prolong gold mounting medium (Invitrogen) prior to imaging.
Phage Clone Staining on Prepared Mouse Lung Slices.
Lungs were inflated using 2% ultra-low-melting temperature agarose (SeaPrep; Lonza) warmed to 37 °C and subsequently allowed to solidify on ice. The left lobe was dissected into approximately 1-cm3 blocks, and 100-μm thick slices were generated using a VT100S vibratome (Leica). Vital lung slices were placed in warm growth medium (DMEM, 10% FBS, 1% penicillin/streptomycin) to dissolve the agarose. Immediately prior to staining, lung slices were fixed with 4% formaldehyde for 20 min and washed with PBS. Staining was performed using labeled phage clones (1 × 1012 cfu/sample), and a commercially available polyclonal anti-Fn antibody (rabbit anti-rat, AB2040, lot number LV1580997; Millipore).
Supplementary Material
Acknowledgments.
We thank Drs. Treniece Terry and Michael Smith for enlightening discussions and helpful suggestions. This work was supported by National Institutes of Health (NIH) T32-GM008433 and NIH T32-EB006343 training grants to L.C., an Armstrong Fund for Science award to H.B., and a Georgia Tech Integrative BioSystems Institute seed grant to T.H.B.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1118088109/-/DCSupplemental.
References
- 1.Frantz C, Stewart KM, Weaver VM. The extracellular matrix at a glance. J Cell Sci. 2010;123:4195–4200. doi: 10.1242/jcs.023820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Raghow R. The role of extracellular-matrix in postinflammatory wound-healing and fibrosis. FASEB J. 1994;8:823–831. doi: 10.1096/fasebj.8.11.8070631. [DOI] [PubMed] [Google Scholar]
- 3.Pankov R, Yamada KM. Fibronectin at a glance. J Cell Sci. 2002;115:3861–3863. doi: 10.1242/jcs.00059. [DOI] [PubMed] [Google Scholar]
- 4.Grashoff C, et al. Measuring mechanical tension across vinculin reveals regulation of focal adhesion dynamics. Nature. 2010;466:263–266. doi: 10.1038/nature09198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Vogel V. Mechanotransduction involving multimodular proteins: Converting force into biochemical signals. Annu Rev Biophys Biomol Struct. 2006;35:459–488. doi: 10.1146/annurev.biophys.35.040405.102013. [DOI] [PubMed] [Google Scholar]
- 6.Martino MM, et al. Controlling integrin specificity and stem cell differentiation in 2D and 3D environments through regulation of fibronectin domain stability. Biomaterials. 2009;30:1089–1097. doi: 10.1016/j.biomaterials.2008.10.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brown AC, Rowe JA, Barker TH. Guiding epithelial cell phenotypes with engineered integrin-specific recombinant fibronectin fragments. Tissue Eng Part A. 2010;17:139–150. doi: 10.1089/ten.tea.2010.0199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Smith ML, et al. Force-induced unfolding of fibronectin in the extracellular matrix of living cells. PLoS Biol. 2007;5:2243–2254. doi: 10.1371/journal.pbio.0050268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Johnson CP, Tang HY, Carag C, Speicher DW, Discher DE. Forced unfolding of proteins within cells. Science. 2007;317:663–666. doi: 10.1126/science.1139857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Brown AEX, Discher DE. Conformational changes and signaling in cell and matrix physics. Curr Biol. 2009;19:R781–R789. doi: 10.1016/j.cub.2009.06.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chabria M, Hertig S, Smith ML, Vogel V. Stretching fibronectin fibres disrupts binding of bacterial adhesins by physically destroying an epitope. Nat Commun. 2010;1:135. doi: 10.1038/ncomms1135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Barker TH, et al. SPARC regulates extracellular matrix organization through its modulation of integrin-linked kinase activity. J Biol Chem. 2005;280:36483–36493. doi: 10.1074/jbc.M504663200. [DOI] [PubMed] [Google Scholar]
- 13.Craig D, Gao M, Schulten K, Vogel V. Tuning the mechanical stability of fibronectin type III modules through sequence variations. Structure. 2004;12:21–30. doi: 10.1016/j.str.2003.11.024. [DOI] [PubMed] [Google Scholar]
- 14.Vakonakis I, Staunton D, Rooney LM, Campbell ID. Interdomain association in fibronectin: Insight into cryptic sites and fibrillogenesis. EMBO J. 2007;26:2575–2583. doi: 10.1038/sj.emboj.7601694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ingham KC, Brew SA, Huff S, Litvinovich SV. Cryptic self-association sites in type III modules of fibronectin. J Biol Chem. 1997;272:1718–1724. doi: 10.1074/jbc.272.3.1718. [DOI] [PubMed] [Google Scholar]
- 16.Zhong CL, et al. Rho-mediated contractility exposes a cryptic site in fibronectin and induces fibronectin matrix assembly. J Cell Biol. 1998;141:539–551. doi: 10.1083/jcb.141.2.539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Baneyx G, Baugh L, Vogel V. Fibronectin extension and unfolding within cell matrix fibrils controlled by cytoskeletal tension. Proc Natl Acad Sci USA. 2002;99:5139–5143. doi: 10.1073/pnas.072650799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Baneyx G, Vogel V. Self-assembly of fibronectin into fibrillar networks underneath dipalmitoyl phosphatidylcholine monolayers: Role of lipid matrix and tensile forces. Proc Natl Acad Sci USA. 1999;96:12518–12523. doi: 10.1073/pnas.96.22.12518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Little WC, Smith ML, Ebneter U, Vogel V. Assay to mechanically tune and optically probe fibrillar fibronectin conformations from fully relaxed to breakage. Matrix Biol. 2008;27:451–461. doi: 10.1016/j.matbio.2008.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Baneyx G, Baugh L, Vogel V. Coexisting conformations of fibronectin in cell culture imaged using fluorescence resonance energy transfer. Proc Natl Acad Sci USA. 2001;98:14464–14468. doi: 10.1073/pnas.251422998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ulmer J, Geiger B, Spatz JP. Force-induced fibronectin fibrillogenesis in vitro. Soft Matter. 2008;4:1998–2007. [Google Scholar]
- 22.Ahmed Z, Brown RA. Adhesion, alignment, and migration of cultured Schwann cells on ultrathin fibronectin fibres. Cell Motil Cytoskeleton. 1999;42:331–343. doi: 10.1002/(SICI)1097-0169(1999)42:4<331::AID-CM6>3.0.CO;2-7. [DOI] [PubMed] [Google Scholar]
- 23.Ejim OS, Blunn GW, Brown RA. Production of artificial-oriented mats and strands from plasma fibronectin—a morphological study. Biomaterials. 1993;14:743–748. doi: 10.1016/0142-9612(93)90038-4. [DOI] [PubMed] [Google Scholar]
- 24.Little WC, Schwartlander R, Smith ML, Gourdon D, Vogel V. Stretched extracellular matrix proteins turn fouling and are functionally rescued by the chaperones albumin and casein. Nano Lett. 2009;9:4158–4167. doi: 10.1021/nl902365z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Klotzsch E, et al. Fibronectin forms the most extensible biological fibers displaying switchable force-exposed cryptic binding sites. Proc Natl Acad Sci USA. 2009;106:18267–18272. doi: 10.1073/pnas.0907518106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Krammer A, Lu H, Isralewitz B, Schulten K, Vogel V. Forced unfolding of the fibronectin type III module reveals a tensile molecular recognition switch. Proc Natl Acad Sci USA. 1999;96:1351–1356. doi: 10.1073/pnas.96.4.1351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Craig D, Krammer A, Schulten K, Vogel V. Comparison of the early stages of forced unfolding for fibronectin type III modules. Proc Natl Acad Sci USA. 2001;98:5590–5595. doi: 10.1073/pnas.101582198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Erickson HP. Stretching fibronectin. J Muscle Res Cell Motil. 2002;23:575–580. doi: 10.1023/a:1023427026818. [DOI] [PubMed] [Google Scholar]
- 29.Mao Y, Schwarzbauer JE. Fibronectin fibrillogenesis, a cell-mediated matrix assembly process. Matrix Biol. 2005;24:389–399. doi: 10.1016/j.matbio.2005.06.008. [DOI] [PubMed] [Google Scholar]
- 30.Singh P, Carraher C, Schwarzbauer JE. Assembly of fibronectin extracellular matrix. Annu Rev Cell Dev Biol. 2010;26:397–419. doi: 10.1146/annurev-cellbio-100109-104020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Brissette R, Goldstein NI. The Use of Phage Display Peptide Libraries for Basic and Translational Research. Methods Mol Biol. 2007;383:203–213. doi: 10.1007/978-1-59745-335-6_13. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.