Abstract
DNA methylation mediates imprinted gene expression by passing an epigenomic state across generations and differentially marking specific regulatory regions on maternal and paternal alleles. Imprinting has been tied to the evolution of the placenta in mammals and defects of imprinting have been associated with human diseases. Although recent advances in genome sequencing have revolutionized the study of DNA methylation, existing methylome data remain largely untapped in the study of imprinting. We present a statistical model to describe allele-specific methylation (ASM) in data from high-throughput short-read bisulfite sequencing. Simulation results indicate technical specifications of existing methylome data, such as read length and coverage, are sufficient for full-genome ASM profiling based on our model. We used our model to analyze methylomes for a diverse set of human cell types, including cultured and uncultured differentiated cells, embryonic stem cells and induced pluripotent stem cells. Regions of ASM identified most consistently across methylomes are tightly connected with known imprinted genes and precisely delineate the boundaries of several known imprinting control regions. Predicted regions of ASM common to multiple cell types frequently mark noncoding RNA promoters and represent promising starting points for targeted validation. More generally, our model provides the analytical complement to cutting-edge experimental technologies for surveying ASM in specific cell types and across species.
Keywords: epigenetics, genomic imprinting, computational prediction
Genomic imprinting refers to genes that are preferentially expressed from either the maternal or paternal allele without genotype dependence (1). In mammals, such parent-of-origin gene expression is believed to have evolved along with the placenta, serving to mediate resource distribution between a mother and her offspring (2, 3), though other theories have been proposed (4–6).
The connection between imprinting and DNA methylation was uncovered shortly after the first identification of imprinted genes in mammals (7). Imprinted gene expression, in all known cases, is regulated by allele-specific methylation (ASM) of some cis-acting regulatory regions. We use the term allelically methylated region (AMR) in reference to any genomic interval of ASM, whether or not it is associated with imprinted regulation. Typically, an entire imprinted locus is organized as a cluster and regulated by an imprinting control region (ICR) and several other AMRs. The allelic methylation patterns of ICRs are set during gametogenesis and stably maintained throughout somatic development in the offspring (8), irrespective of gene expression levels. The remaining AMRs may be established after fertilization (9), possibly under the control of nearby ICRs or other epigenetic signals.
The identification of imprinted genes and a detailed understanding of their regulation has become increasingly important, along with the realization that aberrant genomic imprinting contributes to several complex diseases (10). Much effort has been directed toward locating imprinted genes using expression screen-based approaches (11, 12). One limitation of such approaches is that many imprinted genes may only show allele-specific expressions in particular tissues at appropriate developmental stages (13). ASM screen-based approaches might overcome the effect of temporal and spatial expression patterns because the ICRs are expected to exist through developmental stages preceding the context in which they become active. Such methods have been successfully applied to identify unique imprinted genes (14–17).
Advances in DNA sequencing technology have been leveraged for high-throughput identification of imprinted genes. The “BS-seq” technology couples bisulfite treatment with high-throughput short-read sequencing, and has enabled genome-wide profiling of DNA methylation in mammalian genomes at single-CpG (cytosine guanine dinucleotide) resolution (18). Li et al. (19) produced a methylome from peripheral blood of a single individual and recognized the potential of using such data to profile ASM. They employed a method based on associating heterozygous SNPs with differential methylation, and identified hundreds of ASM regions. Methods such as this, however, must be applied to data from a single individual and for which matching genotypic data are available. There are two shortcomings of approaches that depend on genotype. First, they can be confounded by ASM that is associated with genotype, but which may not have any regulatory effect. The amount of ASM typically associated with genotype is not well understood, but recent reports suggest it is significant (20). More importantly, because imprinted methylation is not necessarily associated with genotypic variation, these methods will be inherently blind to some portion of ASM.
We present a probabilistic model to describe ASM based on data from BS-seq experiments. Our model is independent of genotype, and therefore has broad applicability to identify ASM in the context of imprinting. In essence, our model describes the degree to which methylation states in reads appear to reflect two distinct patterns, each pattern representing roughly half the data. We validated our method using semisimulated data in which methylation states were simulated within actual reads from BS-seq experiments. Our results indicate that technical characteristics of existing public methylomes (i.e., read length and coverage) are sufficient to accurately identify AMRs. By applying our model to 22 human methylomes, emphasizing those from uncultured cells, we identified a set of candidate AMRs involved in imprinted gene regulation. Candidates consistently identified across methylomes display remarkable concordance with known imprinted genes and allow boundaries of known AMRs to be precisely defined. Many candidates not associated with known imprinted genes mark the promoters of long noncoding RNAs (lncRNAs) and are also supported by similar analyses at orthologous regions in chimp; these provide a starting point for identifying additional imprinted genes, ICRs, and possibly imprinted clusters. Our model, therefore, is an essential analytical complement to recently emerged experimental methods for understanding the role of DNA methylation in genomic imprinting.
Modeling Allele-Specific Methylation in BS-Seq Data
We begin this section with a verbal description of our question and the main issues that are addressed by our model. We assume any read has been sequenced after bisulfite treatment and mapped uniquely to the reference genome. Because we are interested in mammalian methylation, we restrict our attention to CpG sites both in the genome and in the reads. Reads not mapping over a CpG are ignored. Our goal is to identify intervals of the genome where it appears that the two alleles have different methylation patterns—typically, in such a case, one allele will be highly methylated and the other not. There are two kinds of important information our model must capture: (i) The set of reads mapping into the interval should appear to represent two distinct methylation patterns, and (ii) the subsets of reads corresponding to those two patterns should be in roughly equal proportions because the alleles themselves are present in equal proportions. One can consider a methylation pattern as analogous to a haplotype, but with a strong stochastic component. Therefore, reads that contain only a single CpG will provide us with relatively little information, and we would like reads to cover as many CpGs as possible. We can then ask whether neighboring CpG sites on the same read tend to share methylation states, and whether other reads cover the same CpG sites but with the alternative shared methylation state. Our approach is to apply a single-allele model to the data, then apply an allele-specific model, and to compare the fit for these models to determine if the data support ASM.
Modeling Site-Specific DNA Methylation in a Single Allele.
We associate each CpG with a single parameter indicating the probability that the CpG is methylated in the cells of interest. For a genomic interval containing n CpGs, the single-allele model is Θ = (θ1,…,θn). Given a set of reads R, the likelihood in the single-allele model within the interval is
 |
[1] |
where m(R,i) and u(R,i) give the numbers of methylated and unmethylated observations from reads mapping over the ith CpG. Estimates for each θi are obtained assuming a binomial distribution for methylation states m(R,i).
Modeling Regions of Allele-Specific Methylation.
Within regions of ASM, we use a two-allele model that associates two distinct methylation probabilities with each CpG. Assuming there are n CpGs in the genomic interval, the two-allele model has the structure Θ = {(θ11,θ12),…,(θn1,θn2)}, with θi1 and θi2 representing the methylation probabilities at the ith CpG on allele one and allele two, respectively. Under this model, reads mapping over the same genomic CpG may have different probabilities of methylation for their CpGs depending on the allele from which they originate. The allele of origin for any read is missing data, and for a given set R of reads we express this missing data as the partition γ = {γ1,γ2} defined by R = γ1∪γ2, where |R| = |γ1| + |γ2|. For any r∈R, if r∈γj, we say that r originates from allele j. Because we are modeling alleles in the context of data from a diploid cell population, the probability that any read originates from a given allele is 0.5. Thus the likelihood is
| [2] |
because the partition γ is independent of Θ. The probability Pr(γ) is effectively a prior on the read partition assuming
because each allele is present with equal frequency. Therefore,
 |
[3] |
where the m and u are as defined for Eq. 1. Because the allele of origin for each read is missing data, we fit the two-allele model using expectation maximization (21), obtaining expectations on membership in γ1 and γ2. Details are provided in SI Text.
Identifying Intervals of Allele-Specific Methylation.
We use Bayesian information criterion (BIC) (22) as a model selection criterion in determining whether a fixed interval is best described using a single-allele [Eq. 1] or two-allele [Eq. 2] model. A single-allele model has one parameter for each of the n CpGs, and the number of observations is equal to |R|:
| [4] |
For the two-allele model, there are two parameters for each CpG:
| [5] |
An interval is identified as having allele-specific methylation if and only if BIC(pair) < BIC(single).
We identify regions of ASM genome-wide by using a fixed-width sliding window (i.e., fixed number of CpG sites) and determining for each whether the single- or two-allele model better describes the data. Results we present are based on a sliding window of 10 CpGs, and issues related to selecting a window size are discussed in the SI Text. Intervals in close proximity are merged, and we also excluded intervals overlapping large subunit ribosomal RNA (LSU rRNA) genes from our final analyses because we suspected problems with their assembly in the reference genome (see SI Text).
Semisimulated Allele-Specific Methylation Data
We conducted simulations to evaluate how the performance of our model relates to several critical parameters of the underlying dataset. To reflect performance characteristics on real datasets, we used a strategy called “semisimulated” data. The locations of mapped reads were taken from real data, as were the locations of CpGs within reads and the underlying reference genome. The methylation states inside those reads were determined according to randomly generated allele-specific or single-allele methylation profiles. Briefly, within a region designated as an AMR, we randomly generated two methylation profiles by sampling individual CpG methylation levels as βeta variants skewed toward 0 or 1. Then we assigned each read with equal probability to one of the two alleles, and the methylation states of the CpGs within the read were sampled according to probabilities given by the methylation profile corresponding to that allele. A full description of this procedure is provided in the SI Text.
With current methylomes from BS-seq, we expected the variation in coverage along chromosomes to be a critical factor for the performance of our model. In addition, the variation in inter-CpG distance may prevent our method from capturing ASM in regions of low CpG density for a fixed read length. We examined how well our method could identify ASM in a given genomic interval by manipulating three independent variables:
Mean coverages were {5×, 10×, 15×}, corresponding to current methylomes from BS-seq.
Read lengths were {50, 100, 150} bases corresponding roughly with current short-read sequencing technologies.
CpG density distributions took three different settings: CpG islands (CGIs) defined as in ref. 23, non-CGI promoters defined as 1 kb upstream of transcription start site (TSS) in National Center for Biotechnology Information reference sequences but not CGIs, and randomly sampled genomic background with CpG density (observed/expected) between 0.2 and 0.4.
Details concerning the number of simulated datasets for each parameter combination can be found in the SI Text.
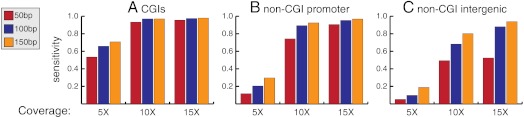
Specificity was generally very high (approximately 99%) for all simulation parameter combinations, reflecting our conservative model selection criterion (Eqs. 4 and 5). In contrast, sensitivity showed greater dependence on properties of the datasets. Sensitivity was higher for regions of higher CpG density, as expected because our model depends on the relationships between CpG states inside a read. As shown in Fig. 1, inside CGIs sensitivity reached above 95% for all read lengths when the mean coverage was above 10×. Sensitivity reached approximately 70% for intergenic regions but required both 10× coverage and read length 100, which compensates for the decrease in CpG density. As expected, greater coverage and read length improved accuracy, and the effect of read length is equivalent to that of CpG density. These results indicate that methylomes with read lengths around 100 bp and mean coverage above 10× appear sufficient for our model to accurately identify ASM. These criteria are met by most existing methylomes from BS-seq experiments.
Fig. 1.
Sensitivity of AMR identification based on semisimulated data. Coverages of 5, 10, and 15×, and read lengths of 50, 100, and 150 bp were used. CpG densities were controlled by simulating within (A) CGIs, (B) non-CGI promoter regions, and (C) non-CGI intergenic regions.
Properties of the Methylomes Analyzed
We analyzed 22 publicly available methylomes, including five uncultured primary cell types, eight cultured differentiated cell lines, four embryonic stem cells (ESCs), and five induced pluripotent stem cells (iPSCs) from following studies. Additional details about each of the methylomes can be found in Dataset S1.
Hodges et al. (24) produced four uncultured methylomes from blood cells: hematopoietic stem and progenitor cells (HSPC), B cells, neutrophils, and CD133+ cord blood cells. Because the first three samples were pooled from six unrelated individuals, ASM caused by genetic variants should not be apparent due to the effect of pooling. The last sample was generated from one individual. Li et al. (19) produced the other uncultured methylome from peripheral blood mononuclear cells (PBMC) of one individual.
The study of Laurent et al. (25) produced three methylomes: foreskin fibroblasts, H9 ESCs, and fibroblasts derived from H9 ESCs. Lister et al. (18) produced methylomes for IMR90 cells and H1 ESCs, two replicates each which we treat as distinct methylomes. In a separate study, Lister et al. (26) produced methylomes for 10 cell types. Included among these were H9 ESCs, adipose-derived stem cells (ADS), adipocytes derived from ADS cells (ADS Adipose) and foreskin fibroblasts (FF). Induced pluripotent stem cells derived from ADS, IMR90, and FF cells were also profiled, with FF iPSCs taken at three different times, the last of which were also profiled after being differentiated in the presence of bone morphogenic protein 4 (BMP4).
Allele-Specific Methylation on the X Chromosome
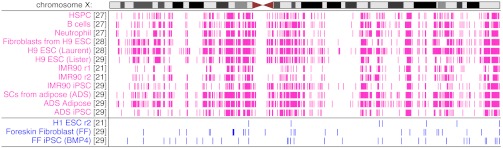
Though not a form of imprinting, dosage compensation is associated with ASM differentially marking one chromosome X (chrX) in female somatic cells (27). In contrast, only a single allele from chrX is represented in male methylome data. Comparing the results of our analyses between male and female X chromosomes therefore provides a measure of specificity: AMRs identified on chrX in males are likely false-positives. In total, 12 of the analyzed methylomes are female. Although coverage on chrX in males is reduced by half, three male methylomes approached 10× coverage on chrX (H1 ESC rep 2, FF, and FF iPSC BMP4; refs. 18 and 26).
The locations of identified AMRs on chrX are presented in Fig. 2. The fraction of AMRs from chrX in female methylomes ranges from 15% to 36% with a mean of 24%. For the three male methylomes tested, the fraction is in the range of 1% to 2%. These results further support our conclusion from simulations that specificity is high in our AMR prediction. X chromosome inactivation is regulated via the X-inactive specific transcript (XIST) gene, a lncRNA with random allele-specific expression in female somatic cells. Our analyses identified an AMR at the XIST promoter in each female differentiated methylome (Fig. S1), but not in any of the ESCs, iPSCs, or male methylomes as expected (28).
Fig. 2.
Locations of AMRs identified on chrX. All female data (pink) was included. Only male methylomes (blue) with sufficient coverage on chrX are shown because these have coverage reduced by 50% compared with autosomes. Numbers in brackets indicate references for data sources. See text and Dataset S1 for information about methylomes.
Genome-Wide AMR Identification Predicts Imprinted Genes
The full set of identified AMRs for each of the 22 methylomes is presented in Dataset S1. We emphasized the uncultured blood methylomes in compiling sets of high-confidence AMRs, and generally use the remaining methylomes to provide additional supporting evidence. We found 579 autosomal AMRs that are common to at least three of the five uncultured methylomes (HSPCs, B cells, neutrophils, CD133+ cord blood, and PBMCs), 247 common to at least four out of five, and 81 shared across all five. Table 1 presents the 39 AMRs common to all five uncultured methylomes and that are proximal to promoters (± 4 kb of a University of California Santa Cruz KnownGene TSS). Among these, 18 overlap a known imprinted gene and 20 mark a lncRNA promoter. The high concordance between our prediction and known imprinted genes further validates our model and provides strong support for the remaining predictions as candidate imprinted genes. The regulatory activity of lncRNAs has been observed for most imprinted clusters (29), and the frequent overlap of identified AMRs and lncRNA promoters suggests these might have similar activity (30). We also identified AMRs in low-coverage chimp blood cell data from the study of Hodges et al. (24), adding additional evidence to several of our predictions.
Table 1.
AMRs common to all uncultured cells that overlap gene promoters
| Gene symbols | NC | CGI | Sp | ESC | iPSC | Tot | Chimp |
| * GNAS,… | Y | Y | 4 | 5 | 22 | 3 | |
| * GNAS-AS1,… | Y | Y | 4 | 5 | 22 | 3 | |
| * MESTIT1/MEST,… | Y | Y | 4 | 5 | 22 | 3 | |
| * SGCE,PEG10 | Y | 4 | 5 | 22 | 3 | ||
| NHP2L1 | Y | 4 | 5 | 22 | 3 | ||
| * ZNF597,NAA60 | Y | 4 | 5 | 22 | 3 | ||
| * SNRPN,SNURF | Y | 4 | 5 | 22 | 2 | ||
| * AMPD3,… | Y | 4 | 5 | 22 | 0 | ||
| PMF1-BGLAP | Y | 4 | 5 | 22 | 0 | ||
| LOC554226,… | Y | Y | 4 | 5 | 22 | 0 | |
| UNC45B | 4 | 5 | 22 | 0 | |||
| LINC00273 | Y | Y | 4 | 5 | 22 | 0 | |
| * NAP1L5 | 4 | 5 | 21 | 3 | |||
| KCNQ1OT1 | Y | Y | 3 | 5 | 21 | 0 | |
| LOC284801,… | Y | Y | 4 | 4 | 21 | 0 | |
| * PSIMCT1 | Y | Y | 2 | 5 | 20 | 0 | |
| * H19,… | Y | Y | Y | 2 | 5 | 20 | 0 |
| TRAPPC9 | Y | 3 | 4 | 20 | 0 | ||
| CR590796 | Y | 4 | 3 | 20 | 0 | ||
| * DIRAS3 | Y | 2 | 5 | 19 | 2 | ||
| AX748049 | 2 | 5 | 19 | 2 | |||
| BC028329 | Y | 3 | 4 | 19 | 2 | ||
| ZNF718,ZNF595 | Y | 3 | 5 | 19 | 0 | ||
| BC023516 | Y | Y | 3 | 5 | 18 | 3 | |
| LOC100130522 | Y | Y | 2 | 4 | 18 | 3 | |
| * FANK1 | Y | 2 | 5 | 18 | 0 | ||
| * GNAS | Y | Y | 2 | 3 | 18 | 0 | |
| VTRNA2-1 | Y | Y | 3 | 4 | 17 | 3 | |
| MTRNR2L3 | Y | 4 | 1 | 15 | 0 | ||
| * BLCAP,NNAT | Y | 2 | 2 | 14 | 2 | ||
| LOC728024 | Y | 0 | 3 | 14 | 2 | ||
| RPS2P32 | Y | Y | 0 | 3 | 14 | 2 | |
| * ZIM2,PEG3,… | Y | Y | 1 | 0 | 12 | 0 | |
| * MEG3 | Y | 0 | 0 | 11 | 3 | ||
| LOC100132167 | Y | 1 | 0 | 11 | 0 | ||
| * HOXA6,HOXA5,… | Y | Y | 0 | 0 | 8 | 2 | |
| KIAA0934,DIP2C | Y | Y | 0 | 0 | 8 | 2 | |
| LOC440570,… | Y | 0 | 0 | 5 | 3 | ||
| LOC100335030 | Y | 0 | 0 | 5 | 1 |
Columns indicate whether the gene is noncoding (NC), the AMR overlaps a CGI promoter (CGI), or is hypermethylated in sperm (Sp). Counts indicate the number of ESC, iPSC, total human methylomes, and chimp methylomes in which the AMR is found. Ellipsis indicate additional gene names can be found in Dataset S1. Asterisk * indicates known imprinted gene.
We computed the methylation level in sperm at each of the identified AMRs using data from a previous study (31). Among the 579 autosomal AMRs common to three out of five uncultured methylomes, 146 were methylated (> 50%) in sperm. As indicated in Table 1, among the 39 predicted AMRs common across all uncultured cell types, only three are methylated in sperm. Among these is the H19 ICR, which is well known to be methylated on the paternal allele (32). If we use the methylation level in sperm as an indicator of methylation on the paternal allele, these results point to an asymmetry in the paternal and maternal mechanisms of imprinting DNA methylation.
One of the central questions related to the use of iPSCs, in research or therapeutically, is the degree to which they resemble true ESCs. The landmark study of Lister et al. demonstrated significant reprogramming variability between iPSCs (26). Evidence from cloning studies suggests that imprinting might be especially difficult to reprogram (33). We assembled the union of all identified AMRs in all methylomes. For each of these AMRs, we computed average methylation in each methylome. We then clustered the methylomes hierarchically according to correlation of methylation levels through these intervals (Fig. S2). The iPSCs correlated better with ESCs than with the somatic cells from which they are derived, suggesting that ASM has in general been successfully reprogrammed in these iPSCs.
However, we found several examples where the iPSCs appear to diverge from the ESCs in terms of ASM (Fig. S3). An AMR was identified at the GNAS-1 promoter in 18 of the methylomes, but this interval was hypomethylated in the ADS iPSCs. Similarly, for the AMR identified at the 3′ promoter of ZNF331 gene, the ADS iPSCs are methylated at 50% and resemble differentiated cells more closely than ESCs or other iPSCs, suggesting failed reprogramming of ADS iPSCs at these locations. It has been proposed that a single imprinted cluster might be sufficient to diagnose iPSC reprogramming in mouse (34). The diversity of ASM we observe between iPSCs and even between ESC lines suggests such diagnosis will be more complex in human.
Analysis of Known Imprinting Control Regions
There are approximately 65 human genes currently validated as imprinted and these reside in 32 imprinted clusters (see Dataset S1). We asked for what proportion of these clusters do we identify an AMR shared between cells, and do these shared AMRs coincide with experimentally validated AMRs? As can be seen from Table 2, 24 of the clusters contain validated AMRs, and in 21 of those cases we correctly identify a known AMR common to four out of five uncultured cells. For the IGF2R and INPP5F clusters, we only identified AMRs shared between two and three of the uncultured cells, respectively. The AMPD3 gene has no validated AMR to our knowledge. Our algorithm finds an AMR shared across all 22 methylomes, indicating a likely candidate for validation. To our knowledge, no AMRs have yet been identified for the remaining clusters, and our algorithm fails to predict any AMRs that are shared between methylomes.
Table 2.
Imprinted clusters and associated AMRs
| Cluster | Known | ICR | Unc | ESC | iPSC | Tot | Chimp |
| GNAS | Y | Y | 5 | 4 | 5 | 22 | 3 |
| SGCE/PEG10 | Y | Y | 5 | 4 | 5 | 22 | 3 |
| MESTIT1/MEST | Y | Y | 5 | 4 | 5 | 22 | 3 |
| ZNF597/NAA60 | Y | Y | 5 | 4 | 5 | 22 | 2 |
| SNRPN/SNURF | Y | Y | 5 | 4 | 5 | 22 | 1 |
| AMPD3 | 5 | 4 | 5 | 22 | 0 | ||
| NAP1L5 | Y | Y | 5 | 4 | 5 | 21 | 3 |
| KCNQ1OT1 | Y | Y | 5 | 3 | 5 | 21 | 0 |
| PSIMCT-1/HM13 | Y | Y | 5 | 2 | 5 | 20 | 3 |
| KCNK9 | Y | Y | 5 | 3 | 4 | 20 | 3 |
| INS-IGF2-H19 | Y | Y | 5 | 2 | 5 | 20 | 0 |
| DIRAS3 | Y | Y | 5 | 2 | 5 | 19 | 3 |
| ZDBF2 | Y | Y | 5 | 3 | 4 | 19 | 2 |
| FANK1 | Y | 5 | 2 | 5 | 18 | 0 | |
| BLCAP/NNAT | Y | Y | 5 | 2 | 2 | 14 | 2 |
| ZIM2/PEG3 | Y | Y | 5 | 1 | 0 | 12 | 0 |
| DLK1/MEG3 | Y | 5 | 0 | 0 | 11 | 3 | |
| RB1 | Y | Y | 5 | 0 | 0 | 7 | 3 |
| L3MBTL | Y | Y | 4 | 3 | 5 | 19 | 3 |
| DDC/GRB10 | Y | Y | 4 | 3 | 4 | 19 | 3 |
| PLAGL1/HYMAI | Y | Y | 4 | 2 | 4 | 18 | 3 |
| FAM50B | Y | Y | 4 | 0 | 1 | 12 | 3 |
| TCEB3C | Y | Y | 4 | 1 | 1 | 11 | 0 |
| INPP5F | Y | Y | 3 | 3 | 5 | 18 | 2 |
| IGF2R | Y | 2 | 0 | 4 | 13 | 1 | |
| DXLGAP2 | 1 | 1 | 0 | 2 | 0 | ||
| TP73 | 1 | 0 | 0 | 1 | 0 | ||
| ANKRD11 | 1 | 0 | 0 | 1 | 0 | ||
| DLX5 | 0 | 0 | 0 | 2 | 0 | ||
| ABCA1 | 0 | 0 | 0 | 1 | 0 | ||
| WT1 | 0 | 0 | 0 | 0 | 0 | ||
| RBP5 | 0 | 0 | 0 | 0 | 0 |
Columns indicate whether the AMR was previously known, an ICR, and the number of uncultured (Unc), ESCs, iPSCs, total (Tot), and chimp methylomes in which each AMR was found. Genomic locations of AMRs can be found in Dataset S1
Knowledge of the location of true AMRs around several of these imprinted genes allowed us to apply a more intensive analysis to examine them with greater sensitivity and precision. We designed a dynamic programming algorithm to optimize the locations of AMR boundaries by evaluating each possible AMR size rather than joining overlapping sliding windows. This algorithm uses a scoring function based on the likelihoods of Eqs. 1 and 2 but remains too computationally demanding for genome-wide application (details are provided in the SI Text). We refer to AMRs identified with this algorithm as “refined” AMRs.
The imprinted cluster on chr14 consists of seven genes controlled by the maternally expressed lncRNA MEG3. The region harbors an AMR at the MEG3 promoter, and another intergenic AMR approximately 15 kb upstream of the MEG3 TSS, both paternally methylated with the upstream AMR shown to act as an ICR (35). Our genome-wide scan found the MEG3 promoter AMR in 11/13 differentiated cells. The boundaries of refined AMRs were identified in each uncultured methylome at nearly the exact same location, covering an interval that is hypomethylated in sperm (Fig. S4). A refined AMR was identified in each uncultured methylome precisely at the known ICR location, which is methylated in sperm. Interestingly, each of the ESC/iPSC methylomes shows full methylation through the ICR, suggesting possible imprinting defects in these cells.
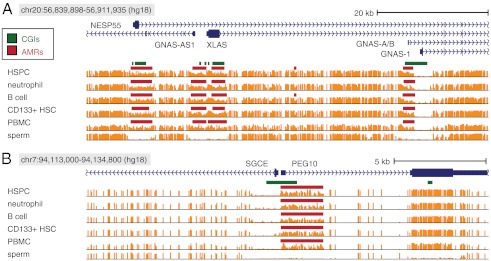
Imprinted expression in the GNAS locus is highly complex, with maternally, paternally, and biallelically expressed transcripts sharing sets of exons (36). This locus includes four AMRs at alternative promoters (NESP55, GNAS-AS1, XLαs, Exon A/B) (37). We identified refined AMRs at these locations in all uncultured methylomes (Fig. 3). Between different methylomes, boundaries of refined AMRs fluctuated by fewer than 10 CpGs and frequently were identified at identical locations. In each case, two separate refined AMRs were identified at the GNAS-AS1 and XLαs promoters. The consistent location of the refined AMR boundary between the GNAS-A/B and GNAS-1 TSS, which coincides with the center of a CGI, suggests a strict partition of regulatory sequence between these two transcripts.
Fig. 3.
Regions of allele-specific methylation through (A) the GNAS and (B) SGCE/PEG10 loci in five uncultured blood cells (HSPC, neutrophils, B cells, and CD 133+ cord blood cells). Vertical orange bars indicate methylation levels of CpGs. In both examples refined AMRs show highly consistent boundaries across methylomes, and each includes an AMR with a precise boundary inside a CGI, distinguishing the regulatory regions of distinct TSS.
The LTR-derived PEG10 and the adjacent SGCE are part of an imprinted gene cluster on chr7 sharing complete synteny with imprinted orthologs in mouse (38). PEG10 and SGCE are separated by less than 100 bp, are divergently transcribed, and have a single CGI overlapping both TSS. Our analyses revealed an AMR at their shared promoter in all 22 methylomes, with a positional bias in the direction of PEG10. As can be seen from Fig. 3, the refined AMRs in uncultured methylomes have identical boundaries precisely between the PEG10 and SGCE TSS at the center of a CGI similar to the GNAS-A/B case described above. Each refined AMR is fully contained inside the body of PEG10, consistent with the LTR origin of PEG10, which implies that PEG10 carries internal regulatory elements. This internal PEG10 promoter appears responsible for imprinted regulation of both genes, despite the hypomethylation reaching into SGCE in all methylomes. One plausible scenario is that regulatory elements within the AMR interact with those nearby in the hypomethylated portion of the CGI to regulate SGCE.
Discussion
We presented a computational strategy for identifying ASM in methylomes produced by BS-seq technology. Our method does not depend on the existence of genotypic variation and is therefore able to identify ASM associated solely with parent-of-origin. Results on simulated data indicate that our method has generally high specificity, and that sensitivity increases with read length as well as mean coverage throughout the genome. Our results also show that this model is accurate even for current read lengths and depths of coverage, both of which are critical technical parameters in connecting methylation states of individual molecules. We applied our method to 22 publicly available human methylomes and validated its accuracy on real data by comparing ASM identified on female and male X chromosomes. Our most consistent predictions across methylomes showed high concordance with known AMRs associated with imprinted genes. The remaining predictions represent likely candidate ICRs for imprinted loci, with several overlapping lncRNA promoters and supported by similar analysis at orthologous regions based on low-coverage methylomes from chimp.
Our top predicted autosomal AMRs show remarkable concordance with known AMRs controlling imprinted gene expression. Among the 39 common to all uncultured methylomes and proximal to annotated promoters, 18 are marking known imprinted genes. It appears as though the AMRs that are already known are also those identified most consistently across methylomes. This finding can be interpreted in several ways. One possibility is that a significant portion of the imprinted genes or clusters, possibly more than half, have already been identified. Estimates of the total number of imprinted genes in human hover around 100–200 (39, 40), and many parent-of-origin disease phenotypes have already been explained through known imprinted genes. Among our remaining predictions (several hundred putative AMRs) many could represent weaker ASM signals possibly without functional relevance, and these are identified with less consistency across datasets because of a lack of sensitivity in our method. Another possibility is that many genes are imprinted with cell-type specificity, and that the known AMRs are biased toward those that can be identified in a greater variety of cell types. Our top predictions were based on consistency across the available methylomes from uncultured cells, but these all happened to be from blood.
Another important finding to emerge from our analyses is the precision with which AMRs are defined across cell types. The GNAS and SGCE/PEG10 examples illustrate this strong consistency of AMR boundaries: In both of these examples, there are pairs of TSS in very close proximity, sharing CGIs, but for which one has ASM methylation and the other does not. Methods like ours that can delineate the boundaries of AMRs will assist future efforts to precisely map the elements inside these regulatory regions.
Among the 22 methylomes analyzed, the total number of identified AMRs varied substantially across methylomes. Much of this variation is likely due to variation in coverage and number of CpGs/read, and relates to sensitivity as illustrated in our simulations. A substantial part may also be due to epigenotypic variation between the cells: Those from culture will exhibit the associated epigenomic effects. It has been known for some time that permanent cell lines have altered DNA methylation (41, 42), and more recently it was shown that aberrant methylation is correlated with passage number (43). We also observed some examples which might indicate an effect of culture conditions, such as the MEG3 AMRs (Fig. S4) where cells differentiated in culture show different properties from other somatic cells. Moreover, any methylome datasets derived from single individuals might exhibit the effects of genotype (44).
Combining experimental approaches with analytic methods such as ours on appropriate biological samples will reveal the breadth of cell-type specific ASM in humans and further clarify the mechanisms underlying imprinted gene expression in human diseases. Moreover, conducting similar studies in appropriately selected mammalian species will help to elucidate the enigmatic role of sexual conflict influencing mammalian evolution.
Supplementary Material
Acknowledgments.
The authors thank Mike Waterman, Simon Tavaré, Fengzhu Sun, and members of the Smith, Hannon, and Dean labs for helpful discussions. The work was supported in part by grants from the National Institutes of Health (R01HG005238 and 5P01CA013106-45) and by a kind gift from Kathryn W. Davis.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1201310109/-/DCSupplemental.
References
- 1.Barlow DP, Stöger R, Herrmann BG, Saito K, Schweifer N. The mouse insulin-like growth factor type-2 receptor is imprinted and closely linked to the Tme locus. Nature. 1991;349:84–87. doi: 10.1038/349084a0. [DOI] [PubMed] [Google Scholar]
- 2.Haig D, Westoby M. Parent-specific gene expression and the triploid endosperm. Am Nat. 1989;134:147–155. [Google Scholar]
- 3.Moore T, Haig D. Genomic imprinting in mammalian development: a parental tug-of-war. Trends Genet. 1991;7:45–49. doi: 10.1016/0168-9525(91)90230-N. [DOI] [PubMed] [Google Scholar]
- 4.Barlow DP. Methylation and imprinting: From host defense to gene regulation? Science. 1993;260:309–310. doi: 10.1126/science.8469984. [DOI] [PubMed] [Google Scholar]
- 5.Varmuza S, Mann M. Genomic imprinting-defusing the ovarian time bomb. Trends Genet. 1994;10:118–123. doi: 10.1016/0168-9525(94)90212-7. [DOI] [PubMed] [Google Scholar]
- 6.Pardo-Manuel de Villena F, de la Casa-Esperón E, Sapienza C. Natural selection and the function of genome imprinting: Beyond the silenced minority. Trends Genet. 2000;16:573–579. doi: 10.1016/s0168-9525(00)02134-x. [DOI] [PubMed] [Google Scholar]
- 7.Zhang Y, et al. Imprinting of human H19: Allele-specific CpG methylation, loss of the active allele in Wilms tumor, and potential for somatic allele switching. Am J Hum Genet. 1993;53:113–124. [PMC free article] [PubMed] [Google Scholar]
- 8.Davis TL, Yang GJ, McCarrey JR, Bartolomei MS. The H19 methylation imprint is erased and re-established differentially on the parental alleles during male germ cell development. Hum Mol Genet. 2000;9:2885–2894. doi: 10.1093/hmg/9.19.2885. [DOI] [PubMed] [Google Scholar]
- 9.El-Maarri O, et al. Maternal methylation imprints on human chromosome 15 are established during or after fertilization. Nat Genet. 2001;27:341–344. doi: 10.1038/85927. [DOI] [PubMed] [Google Scholar]
- 10.Monk D. Deciphering the cancer imprintome. Brief Funct Genomics. 2010;9:329–339. doi: 10.1093/bfgp/elq013. [DOI] [PubMed] [Google Scholar]
- 11.Nikaido I, et al. Discovery of imprinted transcripts in the mouse transcriptome using large-scale expression profiling. Genome Res. 2003;13:1402–1409. doi: 10.1101/gr.1055303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pollard KS, et al. A genome-wide approach to identifying novel-imprinted genes. Hum Genet. 2008;122:625–634. doi: 10.1007/s00439-007-0440-1. [DOI] [PubMed] [Google Scholar]
- 13.Deltour L, Montagutelli X, Guenet JL, Jami J, Páldi A. Tissue-and developmental stage-specific imprinting of the mouse proinsulin gene, Ins2. Dev Biol. 1995;168:686–688. doi: 10.1006/dbio.1995.1114. [DOI] [PubMed] [Google Scholar]
- 14.Peters J, et al. A cluster of oppositely imprinted transcripts at the Gnas locus in the distal imprinting region of mouse chromosome 2. Proc Natl Acad Sci USA. 1999;96:3830–3835. doi: 10.1073/pnas.96.7.3830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Smith RJ, Dean W, Konfortova G, Kelsey G. Identification of novel imprinted genes in a genome-wide screen for maternal methylation. Genome Res. 2003;13:558–569. doi: 10.1101/gr.781503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shoemaker R, Deng J, Wang W, Zhang K. Allele-specific methylation is prevalent and is contributed by CpG-SNPs in the human genome. Genome Res. 2010;20:883–889. doi: 10.1101/gr.104695.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Choufani S, et al. A novel approach identifies new differentially methylated regions (DMRs) associated with imprinted genes. Genome Res. 2011;21:465–476. doi: 10.1101/gr.111922.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lister R, et al. Human DNA methylomes at base resolution show widespread epigenomic differences. Nature. 2009;462:315–322. doi: 10.1038/nature08514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li Y, et al. The DNA methylome of human peripheral blood mononuclear cells. PLoS Biol. 2010;8:e1000533. doi: 10.1371/journal.pbio.1000533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kerkel K, et al. Genomic surveys by methylation-sensitive SNP analysis identify sequence-dependent allele-specific DNA methylation. Nat Genet. 2008;40:904–908. doi: 10.1038/ng.174. [DOI] [PubMed] [Google Scholar]
- 21.Dempster AP, Laird NM, Rubin DB. Maximum likelihood from incomplete data via the EM algorithm. J R Stat Soc Series B Stat Methodol. 1977;39:1–38. [Google Scholar]
- 22.Schwarz G. Estimating the dimension of a model. Ann Stat. 1978;6:461–464. [Google Scholar]
- 23.Gardiner-Garden M, Frommer M. CpG islands in vertebrate genomes. J Mol Biol. 1987;196:261–282. doi: 10.1016/0022-2836(87)90689-9. [DOI] [PubMed] [Google Scholar]
- 24.Hodges E, et al. Directional DNA methylation changes and complex intermediate states accompany lineage specificity in the adult hematopoietic compartment. Mol Cell. 2011;44:17–28. doi: 10.1016/j.molcel.2011.08.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Laurent L, et al. Dynamic changes in the human methylome during differentiation. Genome Res. 2010;20:320–331. doi: 10.1101/gr.101907.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lister R, et al. Hotspots of aberrant epigenomic reprogramming in human induced pluripotent stem cells. Nature. 2011;471:68–73. doi: 10.1038/nature09798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Migeon BR. Insights into X chromosome inactivation from studies of species variation, DNA methylation and replication, and vice versa. Genet Res. 1990;56:91–98. doi: 10.1017/s0016672300035151. [DOI] [PubMed] [Google Scholar]
- 28.Wutz A. Gene silencing in X-chromosome inactivation: Advances in understanding facultative heterochromatin formation. Nat Rev Genet. 2011;12:542–553. doi: 10.1038/nrg3035. [DOI] [PubMed] [Google Scholar]
- 29.O’Neill MJ. The influence of non-coding RNAs on allele-specific gene expression in mammals. Hum Mol Genet. 2005;14(Suppl 1):R113–120. doi: 10.1093/hmg/ddi108. [DOI] [PubMed] [Google Scholar]
- 30.Koerner MV, Pauler FM, Huang R, Barlow DP. The function of non-coding RNAs in genomic imprinting. Development. 2009;136:1771–1783. doi: 10.1242/dev.030403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Molaro A, et al. Sperm methylation profiles reveal features of epigenetic inheritance and evolution in primates. Cell. 2011;146:1029–1041. doi: 10.1016/j.cell.2011.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tremblay KD, Saam JR, Ingram RS, Tilghman SM, Bartolomei MS. A paternal-specific methylation imprint marks the alleles of the mouse H19 gene. Nat Genet. 1995;9:407–413. doi: 10.1038/ng0495-407. [DOI] [PubMed] [Google Scholar]
- 33.Rideout WM, Eggan K, Jaenisch R. Nuclear cloning and epigenetic reprogramming of the genome. Science. 2001;293:1093–1098. doi: 10.1126/science.1063206. [DOI] [PubMed] [Google Scholar]
- 34.Stadtfeld M, et al. Aberrant silencing of imprinted genes on chromosome 12qF1 in mouse induced pluripotent stem cells. Nature. 2010;465:175–181. doi: 10.1038/nature09017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kagami M, et al. Deletions and epimutations affecting the human 14q322 imprinted region in individuals with paternal and maternal upd(14)-like phenotypes. Nat Genet. 2008;40:237–242. doi: 10.1038/ng.2007.56. [DOI] [PubMed] [Google Scholar]
- 36.Williamson CM, et al. Identification of an imprinting control region affecting the expression of all transcripts in the Gnas cluster. Nat Genet. 2006;38:350–355. doi: 10.1038/ng1731. [DOI] [PubMed] [Google Scholar]
- 37.Fröhlich LF, et al. Targeted deletion of the Nesp55 DMR defines another Gnas imprinting control region and provides a mouse model of autosomal dominant PHP-Ib. Proc Natl Acad Sci USA. 2010;107:9275–9280. doi: 10.1073/pnas.0910224107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ono R, et al. A retrotransposon-derived gene, PEG10, is a novel imprinted gene located on human chromosome 7q21. Genomics. 2001;73:232–237. doi: 10.1006/geno.2001.6494. [DOI] [PubMed] [Google Scholar]
- 39.Barlow DP. Gametic imprinting in mammals. Science. 1995;270:1610–1613. doi: 10.1126/science.270.5242.1610. [DOI] [PubMed] [Google Scholar]
- 40.Luedi PP, et al. Computational and experimental identification of novel human imprinted genes. Genome Res. 2007;17:1723–1730. doi: 10.1101/gr.6584707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Doherty AS, Mann MRW, Tremblay KD, Bartolomei MS, Schultz RM. Differential effects of culture on imprinted H19 expression in the preimplantation mouse embryo. Biol Reprod. 2000;62:1526–1535. doi: 10.1095/biolreprod62.6.1526. [DOI] [PubMed] [Google Scholar]
- 42.Fernández-Gonzalez R, et al. Long-term effect of in vitro culture of mouse embryos with serum on mRNA expression of imprinting genes, development, and behavior. Proc Natl Acad Sci USA. 2004;101:5880–5885. doi: 10.1073/pnas.0308560101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Meissner A, et al. Genome-scale DNA methylation maps of pluripotent and differentiated cells. Nature. 2008;454:766–770. doi: 10.1038/nature07107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gertz J, et al. Analysis of DNA methylation in a three-generation family reveals widespread genetic influence on epigenetic regulation. PLoS Genet. 2011;7:e1002228. doi: 10.1371/journal.pgen.1002228. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.