Abstract
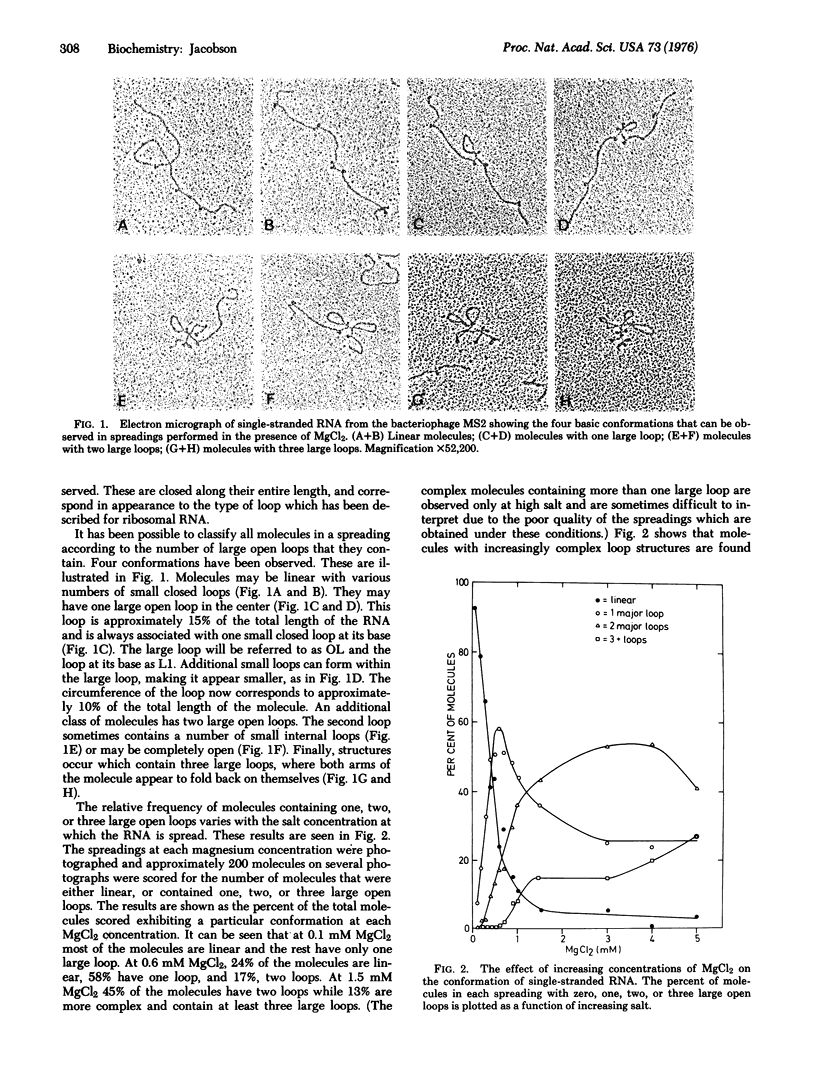
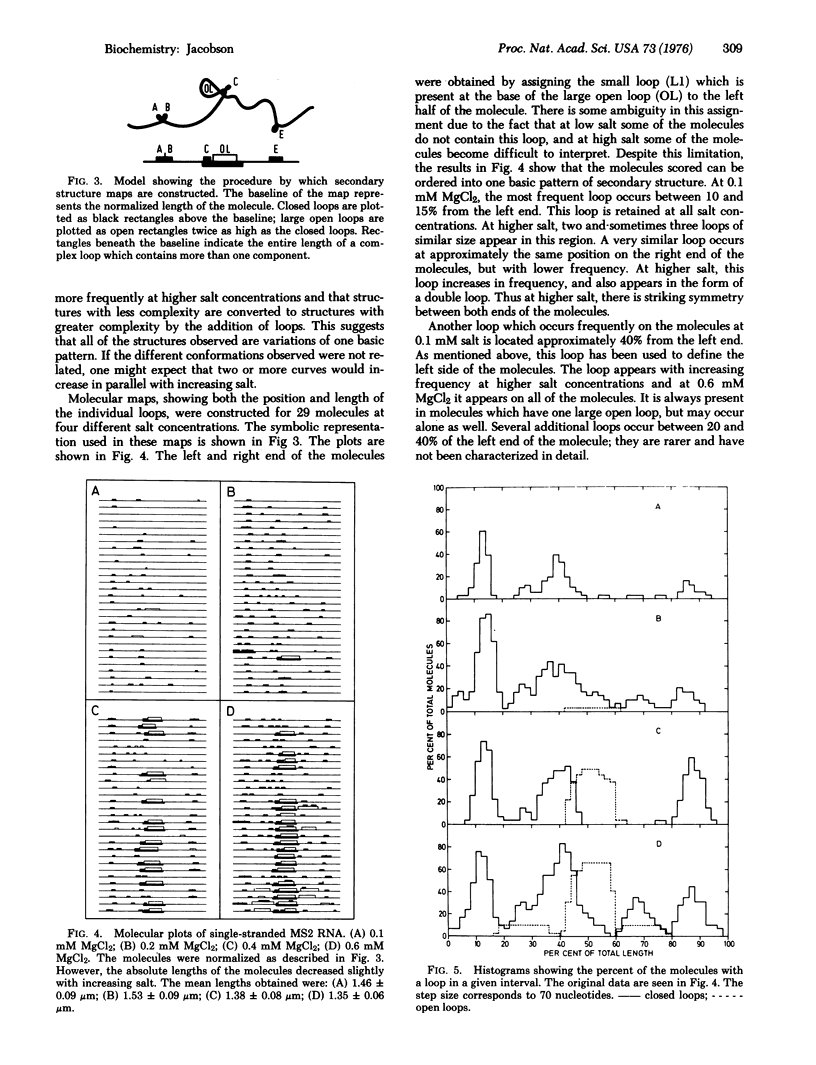
A method allowing the demonstration and study by electron microscopy of secondary structure of viral RNA has been developed. Single-stranded RNA from the bacteriophage MS2 has been analyzed in the electron microscope in the presence of various concentrations of MgCl2. Depending on the salt concentration, the molecules display one to three large open loops which range in size from 10 to 20% of the total RNA length, and smaller closed loops which are approximately 3-5% of the total RNA length. Within one spreading, the conformation of the molecules is variable. However, the average complexity of the molecules increases with increasing salt, and individual loops which are infrequent at low salt increase in frequency with increasing salt. By analyzing the manner in which the individual loop appeared, it was possible to show that all molecules could be described by one basic pattern of secondary structure formation.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Cole P. E., Yang S. K., Crothers D. M. Conformational changes of transfer ribonucleic acid. Equilibrium phase diagrams. Biochemistry. 1972 Nov 7;11(23):4358–4368. doi: 10.1021/bi00773a024. [DOI] [PubMed] [Google Scholar]
- Delius H., Westphal H., Axelrod N. Length measurements of RNA synthesized in vitro by Escherichia coli RNA polymerase. J Mol Biol. 1973 Mar 15;74(4):677–687. doi: 10.1016/0022-2836(73)90056-9. [DOI] [PubMed] [Google Scholar]
- Fresco J. R., Adams A., Ascione R., Henley D., Lindahl T. Tertiary structure in transfer ribonucleic acids. Cold Spring Harb Symp Quant Biol. 1966;31:527–537. doi: 10.1101/sqb.1966.031.01.068. [DOI] [PubMed] [Google Scholar]
- GESTELAND R. F., BOEDTKER H. SOME PHYSICAL PROPERTIES OF BACTERIOPHAGE R17 AND ITS RIBONUCLEIC ACID. J Mol Biol. 1964 Apr;8:496–507. doi: 10.1016/s0022-2836(64)80007-3. [DOI] [PubMed] [Google Scholar]
- Gralla J., Steitz J. A., Crothers D. M. Direct physical evidence for secondary structure in an isolated fragment of R17 bacteriophage mRNA. Nature. 1974 Mar 15;248(445):204–208. doi: 10.1038/248204a0. [DOI] [PubMed] [Google Scholar]
- Hsu M. T., Kung H. J., Davidson N. An electron microscope study of Sindbis virus RNA. Cold Spring Harb Symp Quant Biol. 1974;38:943–950. doi: 10.1101/sqb.1974.038.01.096. [DOI] [PubMed] [Google Scholar]
- Isenberg H., Cotter R. I., Gratzer W. B. Secondary structure and interaction of RNA and protein in a bacteriophage. Biochim Biophys Acta. 1971 Feb 25;232(1):184–191. doi: 10.1016/0005-2787(71)90502-8. [DOI] [PubMed] [Google Scholar]
- Jeppesen P. G., Steitz J. A., Gesteland R. F., Spahr P. F. Gene order in the bacteriophage R17 RNA: 5'-a protein-coat protein-synthetase-3'. Nature. 1970 Apr 18;226(5242):230–237. doi: 10.1038/226230a0. [DOI] [PubMed] [Google Scholar]
- Kolakofsky D., Weissmann C. Possible mechanism for transition of viral RNA from polysome to replication complex. Nat New Biol. 1971 May 12;231(19):42–46. doi: 10.1038/newbio231042a0. [DOI] [PubMed] [Google Scholar]
- Kozak M., Nathans D. Translation of the genome of a ribonucleic acid bacteriophage. Bacteriol Rev. 1972 Mar;36(1):109–134. doi: 10.1128/br.36.1.109-134.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kung H. J., Bailey J. M., Davidson N., Nicolson M. O., McAllister R. M. Structure, subunit composition, and molecular weight of RD-114 RNA. J Virol. 1975 Aug;16(2):397–411. doi: 10.1128/jvi.16.2.397-411.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Min Jou W., Haegeman G., Ysebaert M., Fiers W. Nucleotide sequence of the gene coding for the bacteriophage MS2 coat protein. Nature. 1972 May 12;237(5350):82–88. doi: 10.1038/237082a0. [DOI] [PubMed] [Google Scholar]
- Mitra S., Enger M. D., Kaesberg P. PHYSICAL AND CHEMICAL PROPERTIES OF RNA FROM THE BACTERIAL VIRUS R17. Proc Natl Acad Sci U S A. 1963 Jul;50(1):68–75. doi: 10.1073/pnas.50.1.68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- STRAUSS J. H., Jr, SINSHEIMER R. L. Purification and properties of bacteriophage MS2 and of its ribonucleic acid. J Mol Biol. 1963 Jul;7:43–54. doi: 10.1016/s0022-2836(63)80017-0. [DOI] [PubMed] [Google Scholar]
- Vandenberghe A., Min Jou W., Fiers W. 3'-Terminal nucletide sequence (n equals 361) of bacteriophage MS2 RNA. Proc Natl Acad Sci U S A. 1975 Jul;72(7):2559–2562. doi: 10.1073/pnas.72.7.2559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vollenweider H. J., Sogo J. M., Koller T. A routine method for protein-free spreading of double- and single-stranded nucleic acid molecules. Proc Natl Acad Sci U S A. 1975 Jan;72(1):83–87. doi: 10.1073/pnas.72.1.83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weissmann C., Billeter M. A., Goodman H. M., Hindley J., Weber H. Structure and function of phage RNA. Annu Rev Biochem. 1973;42:303–328. doi: 10.1146/annurev.bi.42.070173.001511. [DOI] [PubMed] [Google Scholar]
- Wellauer P. K., Dawid I. B., Kelley D. E., Perry R. P. Secondary structure maps of ribosomal RNA. II. Processing of mouse L-cell ribosomal RNA and variations in the processing pathway. J Mol Biol. 1974 Oct 25;89(2):397–407. doi: 10.1016/0022-2836(74)90527-0. [DOI] [PubMed] [Google Scholar]
- Wellauer P. K., Dawid I. B. Secondary structure maps of ribosomal RNA and DNA. I. Processing of Xenopus laevis ribosomal RNA and structure of single-stranded ribosomal DNA. J Mol Biol. 1974 Oct 25;89(2):379–395. doi: 10.1016/0022-2836(74)90526-9. [DOI] [PubMed] [Google Scholar]





