Summary
The histologic scoring of renal biopsies is still the gold standard for renal disease classification. The Banff classification scheme and the chronic allograft damage index are histopathologic scoring schemes widely used in renal transplantation. The determination of genome-wide gene expression profiles in human renal biopsies has the potential to serve as independent validation data sets and also provide a more precise evaluation of the functional status behind the visible morphologic alterations. It is expected that results from high-throughput -omics experiments will lead to improved classification schemes in the near future as also discussed at recent Banff meetings. In this review we give an overview on -omics studies, focusing on the association of molecular changes on the transcript as well as on the protein level and morphologic scoring schemes in renal disease and transplantation.
Keywords: Histopathologic classification, morphology, gene expression signatures, biomarkers, kidney function
The classification of renal disease currently is based on histologic scoring of kidney biopsies.1 Although this approach allows the evaluation of the morphology of all different compartments of the kidney such as the vasculature, the tubulointerstitium, and the glomeruli, the functional status and coregulation of genes cannot be uncovered by histology. In situ hybridization or immunohistochemical staining of few messenger RNAs and proteins are valuable tools in classification of disease states but usually are not helpful in prediction of the clinical course. The main reason is that many different pathophysiological entities yield a morphologically similar histopathology such as tubular atrophy or glomerular sclerosis.
Well-established classification schemes for native as well as for renal transplant biopsies were derived in the biannual Banff meetings.2 The Banff classification criteria have been updated sequentially to reflect the continuously increasing knowledge in the field.3 Besides the Banff criteria for the evaluation of renal allograft pathologies, the chronic allograft damage index has been developed and advocated by other researchers.4 Banff and the chronic allograft damage index have similar classification features and thus provide similar information for the clinical judgment of the described histologic features. Neither of those two scores, however, can provide the desired additive information on potential pathophysiological causes and consequences of morphologic changes.
Therefore, researchers started to investigate whether genome-wide gene expression studies of human renal biopsies may provide a more precise evaluation of the functional status and potential choreographed processes behind the visible morphologic alterations. Computational tools that integrate the experimentally uncovered regulation of molecular features were designed to derive useful information from the myriad of regulated single molecular features. The integration of information derived from molecular, functional, demographic, and clinical information is the aim of the booming field of systems biology.
Analysis that is -omics-based enabled the identification of predictors for renal injury such as the neutrophil gelatinase-associated lipocalin or the kidney injury molecule l.5,6 Although these individual markers are far from optimal in classification of renal cell injury from mild impairment to more severe damage to sublethal and lethal necrosis, combinations of these markers in a panel might better mirror the present state of renal cells and the kidney as a whole. The present article describes in detail how functional changes and morphologic consequences of tissue injury correlate with molecular features and how these features are interacting. Not surprisingly, many of the processes that were discovered on a genome-wide basis are precisely choreographed to direct functional and morphologic changes in renal cell states.
One clear application of such markers is the determination of the biological age of transplanted kidneys because the age of the deceased or living donor does not necessary reflect age-associated changes and thus transplant organ quality.7
BIOINFORMATICS ANALYSES SCHEMES FOR -OMICS DATA INTERPRETATION
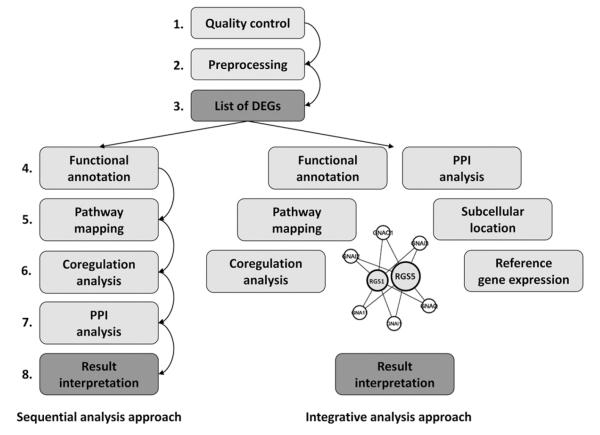
Since the advent of high-throughput -omics experiments a number of excellent tools for the analysis and interpretation of data have been developed as outlined in comprehensive reviews.8, 9 In short, the following analysis steps usually are conducted in the analysis chain of an -omics experiment. After initial quality checks and preprocessing routines a first list of differentially expressed genes is generated using statistical methods such as the standard t test or methods such as the Statistical Analysis of Microarrays.10 This list of differentially expressed genes is successively annotated on a functional level using gene ontology terms or the PANTHER Classification System and/or pathway databases as provided by KEGG.11-13 In silico analyses of the regulatory regions of deregulated genes can give hints on common regulatory mechanisms and certain master regulators on a transcriptional level.14,15 Protein-protein interaction databases can be interrogated to find links between differentially expressed genes.16,17 The prediction analysis for microarrays method calculates optimal gene sets for group classification and prediction based on expression data sets.18 A scheme of analysis approaches is given in Figure 1 with a detailed listing of -omics repositories and tools in Table 1.
Figure 1.
Bioinformatics analysis approaches. The sequential and integrative analysis approaches in data analysis. In the sequential approach the list of differentially genes is analyzed step by step to derive information. In the integrative approach data are combined and one large dependency network is generated for interpretation of differentially expressed genes. Color version available online.
Table 1. Tools and Resources for -Omics-Based Analysis.
SAM, Statistical Analysis of Microarrays; PAM, prediction analysis for microarrays.
Next to these single sequential analysis steps, data integration approaches have become more and more popular in recent years for the interpretation of -omics data.19-20 We have developed an analysis framework for linking gene/protein lists resulting from -omics experiments on the level of a protein-dependency network.21 Pairwise dependencies for all human protein-coding genes were calculated based on a gene expression data set in healthy human tissues, information on functional annotation based on the gene ontology as well as on assignment to molecular pathways, information on subcellular localization, reported protein-protein interaction data, as well as coregulation on the basis of joint transcription factor binding site profiles. Gene expression data as well as lists of differentially expressed features now can be analyzed with respect to their adjacent genes/proteins in the dependency network.
In a recent study by Rudnicki et al22 a network analysis approach resulted in the identification of deregulations on the transcript level in the hypoxia-inducible pathway and the connected vascular endothelial growth factor-receptor system in progressive chronic kidney disease.
In another study the network approach was used to analyze potential marker candidates for cardiovascular disease and bone metabolism disorders in chronic kidney disease patients.23
-OMICS AND HISTOMORPHOLOCY IN RENAL DISEASE AND TRANSPLANTATION
Delayed Allograft Function
A first approach of implementing the -omics technology in the field of renal transplantation was made by Hauser et al.24 The researchers studied the genome-wide gene expression in donor kidney biopsies obtained before transplantation at the end of cold ischemic time. The specific goal of this study was to elucidate the molecular signature that was associated with delayed allograft function (DGF), determined by the necessity of more than one posttransplant dialysis. DGF is highly associated with impaired long-term graft function and morphologic criteria of the donor organ cannot discriminate between the subsequent early graft function. For that purpose biopsies were obtained from 12 organs from deceased donors that subsequently showed DGF, 12 biopsies from deceased donors with primary allograft function, and 12 live donor kidneys, that almost always show immediate graft function. The investigators used generic complementary DNA microarrays holding 26,338 genes and 14,783 expressed sequence tags obtained from the Stanford University Functional Genomics core facility. Validation of array experiments was performed by real-time TaqMan polymerase chain reaction for five selected and differentially regulated genes from different gene ontology categories.
Unsupervised hierarchical clustering of expression profiles showed a clear separation of samples from deceased donors and DGF, primary function, and live donors. The linkage distance, calculated as 1 - Pearson correlation coefficient between paired kidneys, was less than 5%. A sensitivity analysis of the 48 differentially regulated genes was performed by Jack-knife procedures. These molecular features could be classified into the main gene ontology terms of cell-cycle regulation, cell growth/metabolism, and communication/signal transduction. Molecular network analysis suggested choreographed regulation of the identified features, that is, hierarchical activation of, for example, members of the complement cascade such as complement factor B, complement component 1, r + s subcomponents, complement component 2, and clusterin are well known to functionally interact in a top-down signal transduction. These findings suggest that choreographed inflammation in the donor kidneys occurring in deceased donors before organ retrieval is highly associated with DGF. Therefore, the authors suggested that a proof of causal inference should be performed by an interventional study that suppressed donor inflammation. This study was performed subsequently and showed that suppression of this inflammatory signature can be accomplished by pretreatment of the deceased donor with 1 g of methylprednisolone (Kainz, personal communication, 2010). This intervention, however, did not change the posttransplant course (ie, it did not reduce that rate of DGF).
Mas et al25 and Mueller et al26 repeated this gene expression study using different array platforms. Mas et al25 found 65 probe sets that were differentially regulated between DGF and immediate functioning kidneys that belong to the similar main functional categories as described by Hauser et al.24 Accordingly, Mueller et al26 showed in their data set of 87 biopsies that molecular features belonging to the inflammation cascade such as complement components and chemokines as well as acute phase proteins discriminated deceased from live donor biopsies but not primary function from DGF. When principal component analysis was used to compress information to a single eigenvector, deceased donors with DGF showed quantitative separation of molecular features. This prediction was not possible by clinical or histologic parameters.
From these studies it remained unclear which compartments in the kidney contribute to the inflammatory signature, therefore Kainz et al27 studied the molecular signature of micro-dissected donor kidney biopsies. For that purpose, the glomerular and tubulointerstitial compartments of five biopsies from deceased donor kidneys and five biopsies form live donor kidneys were isolated by manual microdissection and the different compartments were subjected to genomics analysis. Not surprisingly, the tubulointerstitial compartment showed a completely different gene expression profile than the glomeruli but, interestingly, the two compartments also showed a clear discrimination between live and deceased donors. The main biological function of these genes was counter-regulation of oxidative stress. Promoter analysis of these signatures showed coregulation of molecular features.
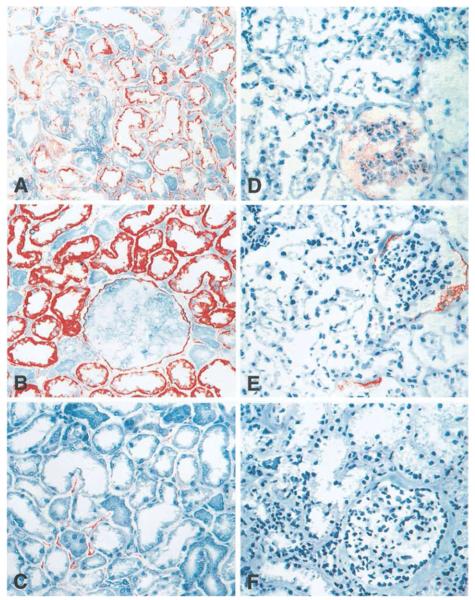
A clear limitation of these genomics data is the lack of information of compartment-specific regulation of transcripts on the protein level. A study supporting the findings of the transcriptomics experiments on the protein level was published by Schwarz et al,28 who performed immunohistochemical analysis of members of the inflammation cascade on implantation kidney biopsies from deceased and live donors. Schwarz et al28 showed that intercellular adhesion molecule 1 and vascular cell adhesion molecule 1 were significantly activated in the tubulointerstitial compartment of kidneys that developed subsequent DGF and between deceased and live donor kidneys. In addition, se-lectin E, which is expressed in the peritubular capillaries, was expressed mainly in kidneys with subsequent DGF and of deceased donor origin but not in allografts derived from live donors (Fig. 2).
Figure 2.
Immunohistochemistry of cell adhesion molecules. Immunohistochemical staining intensities in zero-hour renal transplant biopsies from (A-C) deceased and (D-F) living donors for the three cell adhesion molecules: (A and D) intercellular adhesion molecule 1, (B and E) vascular cell adhesion molecule 1, and (C and F) E-selectin. Reprinted with permission from Schwarz et al.28
Biopsy Confirmed Acute Rejection
Mueller et al29 used mouse transplants to annotate pathogenesis-based transcript sets (PBTs) involving cytotoxic T-cell activation, interferon effects, and parenchymal deterioration. The relationship between PBT expression and histopathologic lesions as well as clinical diagnoses was investigated in 143 consecutive indication human allograft biopsies. The transcripts found to be differentially regulated showed a stereotyped internal structure that led the investigators to conclude that transcriptome changes in rejection are features of a large-scale disturbance characteristic that are occurring at lower levels in many forms of injury from other causes.
In 67 human renal allografts with biopsy-conformed acute rejection, Sarwal et al30 showed that genome-wide gene expression could categorize morphologically indistinguishable biopsy-confirmed acute rejections in at least three different clusters. Specifically, a strong association of CD20-positive B-cell infiltrates was observed in samples of steroid-resistant acute rejection and also confirmed by subsequent immunohistochemical studies. These and the subsequent findings by Mengel et al (see later) suggest a prominent role of infiltrating B cells in biopsy-confirmed acute rejection with poor clinical prognosis.
Mengel et al31 assessed whether lymphocyte infiltrates in areas of scarring, a feature that is not classified by the Banff criteria, are associated with a distinct molecular signature and a poor clinical prognosis. The investigators studied 129 indication biopsies and validated findings in another 50 biopsy cases. The main finding was according to the results from Sarwal et al30 that the 172 identified signatures of B cells and immunoglobulins as well as mast cells in areas of scarring were associated with poor clinical outcomes.
Histogenomics
Mengel et al32 performed a balanced analysis of the Banff classification system. One of the main strengths of the Banff system is its self-organizing consensus and the ability to incorporate novel scientific findings as they emerge according to the authors. Weaknesses of the Banff classification on the other hand are the poor reproducibility and the lack of an independent validation.33 This provides an opportunity for novel high-throughput -omics technologies such as transcriptomics, proteomics, or metabolomics to function as external validation of existing classification systems but also to complement and improve existing scoring schemes. This issue already was addressed at the 2005 Banff meeting with a genomics-complemented Banff classification as the ultimate aim.34 To achieve this goal the participation of molecular biologists as well as bioinformaticians is probably necessary in future endeavors.
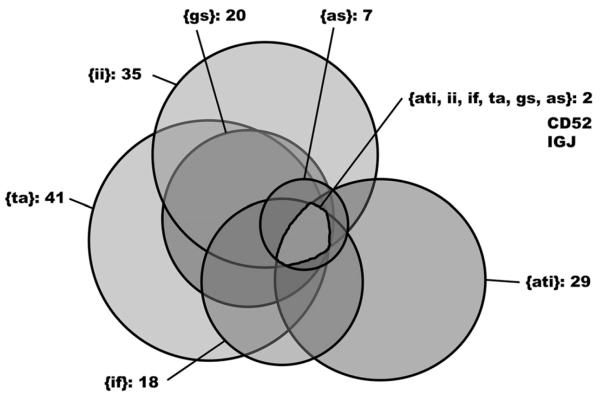
We recently determined the gene expression profiles in 82 zero-hour renal transplant biopsies that were scored histologically for six parameters as follows: degree of glomerulosclerosis, arteriolosclerosis, interstitial inflammation, interstitial fibrosis, tubular atrophy, and acute tubular injury.35 A positive correlation was found for the histologic parameters interstitial inflammation, interstitial fibrosis, arterioloselerosis, and degree of glomerulosclerosis. The degree of acute tubular damage was correlated weakly with tubular atrophy and not correlated with the other parameters that are indicative for chronic damage. Comparing samples with moderate to severe damage as compared with uninjured samples for each of the six parameters under study resulted in a list of 80 differentially expressed genes with two genes showing higher expression in damaged tissues no matter which of the six parameters was analyzed. The two genes are the CD52 molecule and the immunoglobulin J polypeptide (IGJ). Genes predominantly were up-regulated with members of the immune response being associated with all histologic damage categories. Up-regulated genes in tubular atrophy also were involved in cell structure, cell adhesion, and signal transduction. Large overlaps of differentially expressed genes could be found for the histomotphologic classifications tubular atrophy and interstitial inflammation, as well as for degree of glomerulosclerosis and interstitial inflammation (Fig. 3). In a successive analysis step we identified a marker combination consisting of the three molecules NLR family pyrin domain containing 2, regulator of G-protein signaling 5, and IGJ that was able to predict graft function better than a model consisting of clinical and histomotphologic data. The 1-year serum creatinine values after transplantation were used as graft function.
Figure 3.
Overlap of histologic parameters based on differentially expressed genes. Venn diagram of the six histopathologic scores based on shared identified differentially expressed genes between injured and healthy tissues. The two genes IGJ and CD52 showed up-regulated in damage tissues for all histopathologic scores under study. as, arteriolosclerosis; gs, degree of glomerulosclerosis; ii, interstitial inflammation; ta, tubular atrophy; if, interstitial fibrosis; ati, acute tubular injury. Color version available online. Reprinted with permission from Perco et al.35
Einecke et al36 used Asymetrix Gene Chips to study expression changes in cytotoxic T lymphocytes after renal transplantation in a mouse model. They focused on the association of gene expression patterns and the Banff lesions of interstitial infiltration and tubulitis. They determined a set of cytotoxic T lymphocyte-associated transcripts (GATs) that were expressed at higher levels in rejecting kidneys as compared with normal control organs. This increase in expression already could be observed at day three after transplantation with even higher levels at day five, whereas lesion for tubulitis developed much more slowly with a peak 14 days after transplantation.
Mueller et al29 analyzed PBT sets in a set of 143 human renal transplant biopsies. PBTs were derived from mouse transplant models and included CATs, interferon γ-dependent rejection-induced transcripts (GRITs), and two sets of transcripts highly expressed in normal mouse kidneys and with low expression in mouse inflammatory cells (KT1 and KT2). PBTs did show correlations with histopathologic scores for interstitial inflammation, tubulitis, and vasculitis. Also, a positive correlation between C4d staining and the GRIT1 score was observed. Expression in CATs and GRITs was increased markedly in samples with rejection episodes posttransplant, whether the rejection was T-cell-mediated or antibody-mediated. Classifiers were built based on these gene expression sets to predict the histopathologic diagnosis of rejection as well as clinically manifested rejection episodes. Sensitivity and specificity values were in the range between 66% and 79% and 68% and 88%, respectively.
People from the same group used an extended data set of 186 human renal transplant biopsies to build a classifier to distinguish between rejection and nonrejection using the prediction analysis of microarray tools.37 In 1,000 training and test runs the classification sets were found to consist of 24 genes on average. Genes mainly were induced by interferon γ or were associated with cytotoxic T cells. Classification was consistent in about 80% of samples between histologic scores and expression profiles. The transcriptomics classifier has the potential to yield additional information next to the histopathologic scoring, especially in the group of borderline T-cell-mediated rejection.
In an attempt to identify minimally invasive biomarkers for chronic allograft nephropathy (CAN), Kurian et al38 performed gene expression profiling and tandem mass spectroscopy proteomics in peripheral blood of renal transplant patients. CAN is characterized by interstitial fibrosis and tubular atrophy (IF/TA) and plays a dominant role in transplant rejection and diminished renal graft function. Kidney biopsies were classified into four CAN classes based on the Banff classification system: grade 0 depicts samples with no signs of CAN, mild CAN was designated as grade 1, moderate CAN was designated as grade 2, and severe CAN was designated as grade 3. Genes and proteins differentially expressed between CAN 0 and CAN 1 as well as between CAN 0 and CAN 2/3 were determined and analyzed further on a functional level. Differentially expressed features mainly belonged to the biological processes of immune response and inflammation, with the apoptosis pathway among the highest enriched canonical pathways. Top ranked genes were used for class prediction and yielded prediction accuracy values of 80% with sensitivity and specificity values of 85% and 77%, respectively, for mild CAN and accuracy values of 92% for the moderate/severe form. These results were confirmed in a second data set with slightly different distributions of clinical parameters such as the time of biopsy, immunosuppressive protocol, or the RNA extraction method used.
Similar patterns of deregulated genes previously were identified by Maluf et al39 when analyzing 47 renal biopsies with oligonucleotide arrays. Genes showing higher expression in the set of 17 allografts with IF/TA as compared with biopsies from normal kidneys and kidney without IF/TA mainly were involved in immune response, inflammation, and matrix deposition. On the level of molecular pathways the antigen presentation pathway, the leukocyte extravasation signaling cascade, and the natural killer cell signaling pathway were found to be enriched with differentially regulated genes.
Henger et al40 studied gene expression profiles in hydronephrotic kidneys with varying degrees of tubulointerstitial inflammation and fibrosis. Macroarrays holding complementary sequences for a number of inflammatory molecules as well as genes involved in cell-cell contact and matrix turnover were used to analyze samples that were well characterized with regard to their histomorphologic injury based on interstitial inflammation and chronic tubulointerstitial damage measured as tubular atrophy and interstitial fibrosis. A set of 31 genes was identified that could separate samples showing a high degree of inflammation and samples showing a high degree of fibrosis from control samples. A subset of nine genes was successively used to predict damage in an independent set of samples. These genes also showed markedly higher expression levels in the subset of samples with a progressive course of disease as compared with those patient samples with a stable course of disease.
FINAL COMMENTS AND OUTLOOK
Various groups have used gene expression profiling in recent years to perform the following: (1) to predict renal allograft function, (2) to assess the risk of or diagnose renal transplant rejection, and (3) to correlate expression patterns to histomorphological scoring schemes. Enriched biological processes that were involved in the different phenomena often were congruent along different studies from different groups, even though the identified differentially expressed genes were not always 100% identical. This is to a certain extent not surprising owing to different tissues and array platforms used in the experiments. To be used in clinical diagnostics the gene set(s) used for patient stratification, risk assessment, or early diagnosis need(s) to yield high sensitivity and specificity values and should be independent from the array platform used or from the laboratory where the experiment is conducted.
In the field of oncology various single morphoproteomic markers are used for guided therapeutic regimens such as the HER2/neu receptor in breast cancer or the absence or presence of the estrogen receptor α.41 The first microarray-based test system called Mamma-Print was cleared by the Food and Drug Administration in early 2007 for the prognosis of breast cancer patients and is making its way to the clinics.42,43 This test system has the potential to pave the way for microarray-based diagnostic or prognostic assays in the clinical setting, and also in the field of native renal disease and kidney transplantation.
Acknowledgments
Supported in part by the Austrian Science Fund (FWF project P-21436) and the BRIDGE program of the Austrian Research Promotion Agency (FFG project 814.289).
REFERENCES
- 1.Walker PD. The renal biopsy. Arch Pathol Lab Med. 2009;133:181–8. doi: 10.5858/133.2.181. [DOI] [PubMed] [Google Scholar]
- 2.Racusen LC, Solez K, Colvin RB, Bonsib SM, Castro MC, Cavallo T, et al. The Banff 97 working classification of renal allograft pathology. Kidney Int. 1999;55:713–23. doi: 10.1046/j.1523-1755.1999.00299.x. [DOI] [PubMed] [Google Scholar]
- 3.Solez K, Colvin RB, Racusen LC, Haas M, Sis B, Mengel M, et al. Banff 07 classification of renal allograft pathology: updates and future directions. Am J Transplant. 2008;8:753–60. doi: 10.1111/j.1600-6143.2008.02159.x. [DOI] [PubMed] [Google Scholar]
- 4.Yilmaz S, Tomlanovich S, Mathew T, Taskinen E, Paavonen T, Navarro M, et al. Protocol core needle biopsy and histologic Chronic Allograft Damage Index (CADI) as surrogate end point for long-term graft survival in multicenter studies. J Am Soc Nephrol. 2003;14:773–9. doi: 10.1097/01.asn.0000054496.68498.13. [DOI] [PubMed] [Google Scholar]
- 5.Mishra J, Ma Q, Prada A, Mitsnefes M, Zahedi K, Yang J, et al. Identification of neutrophil gelatinase-associated lipocalin as a novel early urinary biomarker for ischemic renal injury. J Am Soc Nephrol. 2003;14:2534–43. doi: 10.1097/01.asn.0000088027.54400.c6. [DOI] [PubMed] [Google Scholar]
- 6.Ichimura T, Bonventre JV, Bailly V, Wei H, Hession CA, Cate RL, et al. Kidney injury molecule-1 (KIM-1), a putative epithelial cell adhesion molecule containing a novel immunoglobulin domain, is up-regula-ted in renal cells after injury. J Biol Chem. 1998;273:4135–42. doi: 10.1074/jbc.273.7.4135. [DOI] [PubMed] [Google Scholar]
- 7.Koppelstaetter C, Schratzberger G, Perco P, Hofer J, Mark W, Ollinger R, et al. Markers of cellular senescence in zero hour biopsies predict outcome in renal transplantation. Aging Cell. 2008;7:491–7. doi: 10.1111/j.1474-9726.2008.00398.x. [DOI] [PubMed] [Google Scholar]
- 8.Perco P, Rapberger R, Siehs C, Lukas A, Oberbauer R, Mayer G, et al. Transforming omics data into context: bioinformatics on genomics and proteomics raw data. Electrophoresis. 2006;27:2659–75. doi: 10.1002/elps.200600064. [DOI] [PubMed] [Google Scholar]
- 9.Quackenbush J. Microarray analysis and tumor classification. N Engl J Med. 2006;354:2463–72. doi: 10.1056/NEJMra042342. [DOI] [PubMed] [Google Scholar]
- 10.Tusher VG, Tibshirani R, Chu G. Significance analysis of microarrays applied to the ionizing radiation response. Proc Natl Acad Sci USA. 2001;98:5116–21. doi: 10.1073/pnas.091062498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, et al. The Gene Ontology Consortium Gene ontology: tool for the unification of biology. Nat Genet. 2000;25:25–9. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kanehisa M, Goto S, Kawashima S, Nakaya A. The KEGG databases at GenomeNet. Nucleic Acids Res. 2002;30:42–6. doi: 10.1093/nar/30.1.42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mi H, Lazareva-Ulitsky B, Loo R, Kejariwal A, Vandergriff J, Rabkin S, et al. The PANTHER database of protein families, subfamilies, functions and pathways. Nucleic Acids Res. 2005;33:D284–8. doi: 10.1093/nar/gki078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ho Sui SJ, Fulton DL, Arenillas DJ, Kwon AT, Wasserman WW. oPOSSUM: integrated tools for analysis of regulatory motif over-representation. Nucleic Acids Res. 2007;35:W245–52. doi: 10.1093/nar/gkm427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cohen CD, Klingenhoff A, Boucherot A, Nitsche A, Henger A, Brunner B, et al. Comparative promoter analysis allows de novo identification of specialized cell junction-associated proteins. Proc Natl Acad Sci USA. 2006;103:5682–7. doi: 10.1073/pnas.0511257103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Brown KR, Jurisica I. Online predicted human interaction database. Bioinformatics. 2005;21:2076–82. doi: 10.1093/bioinformatics/bti273. [DOI] [PubMed] [Google Scholar]
- 17.Brown KR, Otasek D, Ali M, McGuffin M, Xie W, Devani B, et al. NAViGaTOR: Network Analysis, Visualization & Graphing Toronto. Bioinformatics. 2009;25:3327–9. doi: 10.1093/bioinformatics/btp595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tibshirani R, Hastie T, Narasimhan B, Chu G. Diagnosis of multiple cancer types by shrunken centroids of gene expression. Proc Natl Acad Sci USA. 2002;99:6567–72. doi: 10.1073/pnas.082099299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.von Mering C, Jensen LJ, Snel B, Hooper SD, Krupp M, Foglierini M, et al. STRING: known and predicted protein-protein associations, integrated and transferred across organisms. Nucleic Acids Res. 2005;33:D433–7. doi: 10.1093/nar/gki005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Alexeyenko A, Sonnhammer EL. Global networks of functional coupling in eukaryotes from comprehensive data integration. Genome Res. 2009;19:1107–16. doi: 10.1101/gr.087528.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bernthaler A, Muhlberger I, Fechete R, Perco P, Lukas A, Mayer B. A dependency graph approach for the analysis of differential gene expression profiles. Mol Biosyst. 2009;5:1720–31. doi: 10.1039/b903109j. [DOI] [PubMed] [Google Scholar]
- 22.Rudnicki M, Perco P, Enrich J, Eder S, Heininger D, Bernthaler A, et al. Hypoxia response and VEGF-A expression in human proximal tubular epithelial cells in stable and progressive renal disease. Lab Invest. 2009;89:337–46. doi: 10.1038/labinvest.2008.158. [DOI] [PubMed] [Google Scholar]
- 23.Perco P, Wilflingseder J, Bernthaler A, Wiesinger M, Rudnicki M, Wimmer B, et al. Biomarker candidates for cardiovascular disease and bone metabolism disorders in chronic kidney disease: a systems biology perspective. J Cell Mol Med. 2008;12:1177–87. doi: 10.1111/j.1582-4934.2008.00280.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hauser P, Schwarz C, Mitterbauer C, Regele HM, Muhlbacher F, Mayer G, et al. Genome-wide gene-expression patterns of donor kidney biopsies distinguish primary allograft function. Lab Invest. 2004;84:353–61. doi: 10.1038/labinvest.3700037. [DOI] [PubMed] [Google Scholar]
- 25.Mas VR, Archer KJ, Yanek K, Dumur CI, Capparuccini MI, Mangino MJ, et al. Gene expression patterns in deceased donor kidneys developing delayed graft function after kidney transplantation. Transplantation. 2008;85:626–35. doi: 10.1097/TP.0b013e318165491f. [DOI] [PubMed] [Google Scholar]
- 26.Mueller TF, Reeve J, Jhangri GS, Mengel M, Jacaj Z, Cairo L, et al. The transcriptome of the implant biopsy identifies donor kidneys at increased risk of delayed graft function. Am J Transplant. 2008;8:7885. doi: 10.1111/j.1600-6143.2007.02032.x. [DOI] [PubMed] [Google Scholar]
- 27.Kainz A, Mitterbauer C, Hauser P, Schwarz C, Regele HM, Berlakovich G, et al. Alterations in gene expression in cadaveric vs. live donor kidneys suggest impaired tubular counterbalance of oxidative stress at implantation. Am J Transplant. 2004;4:1595–604. doi: 10.1111/j.1600-6143.2004.00554.x. [DOI] [PubMed] [Google Scholar]
- 28.Schwarz C, Regele H, Steininger R, Hansmann C, Mayer G, Oberbauer R. The contribution of adhesion molecule expression in donor kidney biopsies to early allograft dysfunction. Transplantation. 2001;71:1666–70. doi: 10.1097/00007890-200106150-00028. [DOI] [PubMed] [Google Scholar]
- 29.Mueller TF, Einecke G, Reeve J, Sis B, Mengel M, Jhangri GS, et al. Microarray analysis of rejection in human kidney transplants using pathogenesis-based transcript sets. Am J Transplant. 2007;7:2712–22. doi: 10.1111/j.1600-6143.2007.02005.x. [DOI] [PubMed] [Google Scholar]
- 30.Sarwal M, Chua MS, Kambham N, Hsieh SC, Satterwhite T, Masek M, et al. Molecular heterogeneity in acute renal allograft rejection identified by DNA microarray profiling. N Engl J Med. 2003;349:125–38. doi: 10.1056/NEJMoa035588. [DOI] [PubMed] [Google Scholar]
- 31.Mengel M, Reeve J, Bunnag S, Einecke G, Sis B, Mueller T, et al. Molecular correlates of scarring in kidney transplants: the emergence of mast cell transcripts. Am J Transplant. 2009;9:169–78. doi: 10.1111/j.1600-6143.2008.02462.x. [DOI] [PubMed] [Google Scholar]
- 32.Mengel M, Sis B, Halloran PF. SWOT analysis of Banff: strengths, weaknesses, opportunities and threats of the international Banff consensus process and classification system for renal allograft pathology. Am J Transplant. 2007;7:2221–6. doi: 10.1111/j.1600-6143.2007.01924.x. [DOI] [PubMed] [Google Scholar]
- 33.Furness PN, Taub N. International variation in the interpretation of renal transplant biopsies: report of the CERTPAP Project. Kidney Int. 2001;60:1998–2012. doi: 10.1046/j.1523-1755.2001.00030.x. [DOI] [PubMed] [Google Scholar]
- 34.Solez K, Colvin RB, Racusen LC, Sis B, Halloran PF, Birk PE, et al. Banff ‘05 Meeting Report: differential diagnosis of chronic allograft injury and elimination of chronic allograft nephropathy (‘CAN’) Am J Transplant. 2007;7:518–26. doi: 10.1111/j.1600-6143.2006.01688.x. [DOI] [PubMed] [Google Scholar]
- 35.Perco P, Kainz A, Wilflingseder J, Soleiman A, Mayer B, Oberbauer R. Histogenomics: association of gene expression patterns with histological parameters in kidney biopsies. Transplantation. 2009;87:290–5. doi: 10.1097/TP.0b013e318191b4c0. [DOI] [PubMed] [Google Scholar]
- 36.Einecke G, Melk A, Ramassar V, Zhu LF, Bleackley RC, Famulski KS, et al. Expression of CTL associated transcripts precedes the development of tubulitis in T-cell mediated kidney graft rejection. Am J Transplant. 2005;5:1827–36. doi: 10.1111/j.1600-6143.2005.00974.x. [DOI] [PubMed] [Google Scholar]
- 37.Reeve J, Einecke G, Mengel M, Sis B, Kayser N, Kaplan B, et al. Diagnosing rejection in renal transplants: a comparison of molecular- and histopathology-based approaches. Am J Transplant. 2009;9:1802–10. doi: 10.1111/j.1600-6143.2009.02694.x. [DOI] [PubMed] [Google Scholar]
- 38.Kurian SM, Heilman R, Mondala TS, Nakorchevsky A, Hewel JA, Campbell D, et al. Biomarkers for early and late stage chronic allograft nephropathy by proteog-enomic profiling of peripheral blood. PLoS One. 2009;4:e6212. doi: 10.1371/journal.pone.0006212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Maluf DG, Mas VR, Archer KJ, Yanek K, Gibney EM, King AL, et al. Molecular pathways involved in loss of kidney graft function with tubular atrophy and interstitial fibrosis. Mol Med. 2008;14:276–85. doi: 10.2119/2007-00111.Maluf. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Henger A, Kretzler M, Doran P, Bonrouhi M, Schmid H, Kiss E, et al. Gene expression fingerprints in human tubulointerstitial inflammation and fibrosis as prognostic markers of disease progression. Kidney Int. 2004;65:904–17. doi: 10.1111/j.1523-1755.2004.00499.x. [DOI] [PubMed] [Google Scholar]
- 41.Brown RE. Morphogenomics and morphoproteom-ics: a role for anatomic pathology in personalized medicine. Arch Pathol Lab Med. 2009;133:568–79. doi: 10.5858/133.4.568. [DOI] [PubMed] [Google Scholar]
- 42.Slodkowska EA, Ross JS. MammaPrint 70-gene signature: another milestone in personalized medical care for breast cancer patients. Expert Rev Mol Diagn. 2009;9:417–22. doi: 10.1586/erm.09.32. [DOI] [PubMed] [Google Scholar]
- 43.Wittner BS, Sgroi DC, Ryan PD, Bruinsma TJ, Glas AM, Male A, et al. Analysis of the MammaPrint breast cancer assay in a predominantly postmenopausal cohort. Clin Cancer Res. 2008;14:2988–93. doi: 10.1158/1078-0432.CCR-07-4723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Martini S, Eichinger F, Nair V, Kretzler M. Defining human diabetic nephropathy on the molecular level: integration of transcriptomic profiles with biological knowledge. Rev Endocr Metab Disord. 2008;9:267–74. doi: 10.1007/s11154-008-9103-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Barrett T, Troup DB, Wilhite SE, Ledoux P, Rudnev D, Evangelista C, et al. NCBI GEO: archive for high-throughput functional genomic data. Nucleic Acids Res. 2009;37:D885–90. doi: 10.1093/nar/gkn764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Parkinson H, Kapushesky M, Shojatalab M, Abeygunawardena N, Coulson R, Fame A, et al. ArrayEx-press—a public database of microarray experiments and gene expression profiles. Nucleic Acids Res. 2007;35:D747–50. doi: 10.1093/nar/gkl995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Demeter J, Beauheim C, Gollub J, Hernandez-Bous-sard T, Jin H, Maier D, et al. The Stanford Microarray Database: implementation of new analysis tools and open source release of software. Nucleic Acids Res. 2007;35:D766–70. doi: 10.1093/nar/gkl1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gentleman RC, Carey VJ, Bates DM, Bolstad B, Dettling M, Dudoit S, et al. Bioconductor: open software development for computational biology and bioinformatics. Genome Biol. 2004;5:R80. doi: 10.1186/gb-2004-5-10-r80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Affymetrix . Statistical algorithms reference guide, technical report. Affymetrix; 2001. [Google Scholar]
- 50.Irizarry RA, Hobbs B, Collin F, Beazer-Barclay YD, Antonellis KJ, Scherf U, et al. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics. 2003;4:249–64. doi: 10.1093/biostatistics/4.2.249. [DOI] [PubMed] [Google Scholar]
- 51.Li C, Wong WH. Model-based analysis of oligonucleotide arrays: model validation, design issues and standard error application. Genome Biol. 2001;2 doi: 10.1186/gb-2001-2-8-research0032. RESEARCH0032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Saeed Al, Sharov V, White J, Li J, Liang W, Bhagabati N, et al. TM4: a free, open-source system for microarray data management and analysis. Biotechniques. 2003;34:374–8. doi: 10.2144/03342mt01. [DOI] [PubMed] [Google Scholar]
- 53.Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 54.Zeeberg BR, Feng W, Wang G, Wang MD, Fojo AT, Sunshine M, et al. GoMiner: a resource for biological interpretation of genomic and proteomic data. Genome Biol. 2003;4:R28. doi: 10.1186/gb-2003-4-4-r28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sherry ST, Ward MH, Kholodov M, Baker J, Phan L, Smigielski EM, et al. dbSNP: the NCBI database of genetic variation. Nucleic Acids Res. 2001;29:308–11. doi: 10.1093/nar/29.1.308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Thorisson GA, Smith AV, Krishnan L, Stein LD. The International HapMap Project web site. Genome Res. 2005;15:1592–3. doi: 10.1101/gr.4413105. [DOI] [PMC free article] [PubMed] [Google Scholar]