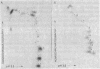
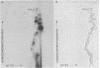
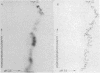
Abstract
A procedure is described by which a discrete high-molecular-weight RNA transcription product can be used as a primer by DNA polymerase (DNA nucleotidyltransferase; EC 2.7.7.7; deoxynucleoside triphosphate: DNA deoxynucleotidyltransferase) for determining nucleic acid sequence in the template DNA beyond the 3'-terminus of the transcript. This procedure is applied to two lambda phage transcripts, the 4S "oop" RNA [Short l-strand RNA transcript from the region of origin of replication (ori) and the 6S RNA. Sequences of 35 and 19 nucleotides, respectively, following the sites at which these two transcripts terminate, are determined. Little structural homology is apparent in the template DNA beyond the 3'-ends of these two transcripts. The lack of homology suggests that this region might not be important to the termination process.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bertrand K., Korn L., Lee F., Platt T., Squires C. L., Squires C., Yanofsky C. New features of the regulation of the tryptophan operon. Science. 1975 Jul 4;189(4196):22–26. doi: 10.1126/science.1094538. [DOI] [PubMed] [Google Scholar]
- Brownlee G. G., Sanger F. Chromatography of 32P-labelled oligonucleotides on thin layers of DEAE-cellulose. Eur J Biochem. 1969 Dec;11(2):395–399. doi: 10.1111/j.1432-1033.1969.tb00786.x. [DOI] [PubMed] [Google Scholar]
- Dingman C. W. A convenient program for the rapid calculation of sedimentation coefficients in linear salt or sucrose gradients. Anal Biochem. 1972 Sep;49(1):124–133. doi: 10.1016/0003-2697(72)90249-7. [DOI] [PubMed] [Google Scholar]
- Galibert F., Sedat J., Ziff E. Direct determination of DNA nucleotide sequences: structure of a fragment of bacteriophage phiX172 DNA. J Mol Biol. 1974 Aug 15;87(3):377–407. doi: 10.1016/0022-2836(74)90093-x. [DOI] [PubMed] [Google Scholar]
- Hayes S., Szybalski W. Control of short leftward transcripts from the immunity and ori regions in induced coliphage lambda. Mol Gen Genet. 1973 Nov 22;126(4):275–290. doi: 10.1007/BF00269438. [DOI] [PubMed] [Google Scholar]
- Ikemura T., Dahlberg J. E. Small ribonucleic acids of Escherichia coli. I. Characterization by polyacrylamide gel electrophoresis and fingerprint analysis. J Biol Chem. 1973 Jul 25;248(14):5024–5032. [PubMed] [Google Scholar]
- Kleid D., Humayun Z., Jeffrey A., Ptashne M. Novel properties of a restriction endonuclease isolated from Haemophilus parahaemolyticus. Proc Natl Acad Sci U S A. 1976 Feb;73(2):293–297. doi: 10.1073/pnas.73.2.293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kramer R. A., Rosenberg M., Steitz J. A. Nucleotide sequences of the 5' and 3' termini of bacteriophage T7 early messenger RNAs synthesized in vivo: evidence for sequence specificity in RNA processing. J Mol Biol. 1974 Nov 15;89(4):767–776. doi: 10.1016/0022-2836(74)90051-5. [DOI] [PubMed] [Google Scholar]
- Lebowitz P., Weissman S. M., Radding C. M. Nucleotide sequence of a ribonucleic acid transcribed in vitro from lambda phage deoxyribonucleic acid. J Biol Chem. 1971 Aug 25;246(16):5120–5139. [PubMed] [Google Scholar]
- Loewen P. C., Khorana H. G. Studies on polynucleotides. CXXII. The dodecanucleotide sequence adjoining the C-C-A end of the tyrosine transfer ribonucleic acid gene. J Biol Chem. 1973 May 25;248(10):3489–3499. [PubMed] [Google Scholar]
- Pieczenik G., Barrell B. G., Gefter M. L. Bacteriophage phi 80-induced low molecular weight RNA. Arch Biochem Biophys. 1972 Sep;152(1):152–165. doi: 10.1016/0003-9861(72)90203-2. [DOI] [PubMed] [Google Scholar]
- Roberts J. W. Termination factor for RNA synthesis. Nature. 1969 Dec 20;224(5225):1168–1174. doi: 10.1038/2241168a0. [DOI] [PubMed] [Google Scholar]
- Roberts J. W. Transcription termination and late control in phage lambda. Proc Natl Acad Sci U S A. 1975 Sep;72(9):3300–3304. doi: 10.1073/pnas.72.9.3300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenberg M., Weissman S., deCrombrugghe B. Termination of transcription in bacteriophage lambda. Heterogeneous, 3'-terminal oligo-adenylate additions and the effects of rho factor. J Biol Chem. 1975 Jun 25;250(12):4755–4764. [PubMed] [Google Scholar]
- Salser W., Fry K., Brunk C., Poon R. Nucleotide sequencing of DNA: preliminary characterization of the products of specific cleavages at guanine, cytosine, or adenine residues (bacteriophage M13-ribosubstitution-DNA polymerase I-electrophoresis-two-dimensional fingerprinting). Proc Natl Acad Sci U S A. 1972 Jan;69(1):238–242. doi: 10.1073/pnas.69.1.238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanger F., Donelson J. E., Coulson A. R., Kössel H., Fischer D. Determination of a nucleotide sequence in bacteriophage f1 DNA by primed synthesis with DNA polymerase. J Mol Biol. 1974 Dec 5;90(2):315–333. doi: 10.1016/0022-2836(74)90376-3. [DOI] [PubMed] [Google Scholar]
- Sklar J., Yot P., Weissman S. M. Determination of genes, restriction sites, and DNA sequences surrounding the 6S RNA template of bacteriophage lambda. Proc Natl Acad Sci U S A. 1975 May;72(5):1817–1821. doi: 10.1073/pnas.72.5.1817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith G. R., Hedgpeth J. Oligo(A) not coded by DNA generating 3'-terminal heterogeneity in a lambda phage RNA. J Biol Chem. 1975 Jun 25;250(12):4818–4821. [PubMed] [Google Scholar]