Abstract
In recent years, the biomedical community has witnessed a rapid scientific and technologic evolution after the development and refinement of high-throughput methodologies. Concurrently and consequentially, the scientific perspective has changed from the reductionist approach of meticulously analyzing the fine details of a single component of biology to the “holistic” approach of broadmindedly examining the globally interacting elements of biological systems. The emergence of this new way of thinking has brought about a scientific revolution in which genomics, proteomics, metabolomics, and other “omics” have become the predominant tools by which large amounts of data are amassed, analyzed, and applied to complex questions of biology that were previously unsolvable. This enormous transformation of basic science research and the ensuing plethora of promising data, especially in the realm of human health and disease, have unfortunately not been followed by a parallel increase in the clinical application of this information. On the contrary, the number of new potential drugs in development has been decreasing steadily, suggesting the existence of roadblocks that prevent the translation of promising research into medically relevant therapeutic or diagnostic application. In this article, we will review, in a noninclusive fashion, several recent scientific advancements in the field of translational research, with a specific focus on how they relate to infectious disease. We will also present a current picture of the limitations and challenges that exist for translational research, as well as ways that have been proposed by the National Institutes of Health to improve the state of this field.
Abbreviations: 2-DE, 2-dimensional electrophoresis; 2-D DIGE, 2-dimensional differential in-gel electrophoresis; CF, cystic fibrosis; CTSA, Clinical and Translational Science Awards program; EBV, Epstein-Barr virus; FDA, U.S. Food and Drug Administration; GWAS, genome-wide association studies; HCV, hepatitis C virus; HMP, Human Microbiome Project; HPLC, high-pressure liquid chromatography; LC, liquid chromatography; LSB, Laboratory of Systems Biology; mAb, monoclonal antibody; MRM/SRM, multiple reaction monitoring/selective reaction monitoring; MS, mass spectrometry; MS/MS, tandem mass spectrometry; NCATS, National Center for Advancing Translational Sciences; NCRR, National Center of Research Resources; NIAID, National Institute of Allergy and Infectious Disease; NIH, National Institutes of Health; NME, new molecular entity; NMR, nuclear magnetic resonance; PBMC, peripheral blood mononuclear cell; PCR, polymerase chain reaction; PRR, pathogen recognition receptor; QQQ, triple quadrupole mass spectrometry; SARS-CoV, coronavirus associated with severe acute respiratory syndrome; SNP, single nucleotide polymorphism; TB, tuberculosis; UTI, urinary tract infection; YFV, yellow fever virus
Translational research is defined precisely by the National Institutes of Health (NIH) as “the process of applying ideas, insights and discoveries generated through basic scientific inquiry to the treatment or prevention of human disease.”1 As the field of translational research has become increasingly popular in recent years, it has undergone numerous reiterations, such that the specific meaning of the term “translational research” has itself been redefined several times.1, 2 The recent evolution in next-generation sequencing techniques and the introduction of high-throughput methods has resulted in an explosive cascade of research applications, spanning from target identification to diagnostics and therapeutics. These technical advances have provided the impetus for some radical changes in the way research itself is conceived and performed. As a result, enhanced interactions and broader collaborations among researchers with different expertise will be required just to keep up with the rapidly changing state of science. For a multidisciplinary approach to be effective, better ways to collect and share data (eg, biorepositories) must be identified. In addition, a more rapid translation of information from basic science into useful clinical applications will require the removal of communication barriers and financial roadblocks that currently prevent basic science teams from working with each other and with clinical researchers. Finally, regulatory changes will be necessary to promote faster approval of new molecular entities (NMEs) resulting from such scientific collaborations.
A major turning point in the validation of the field of translational research came with the creation of the NIH’s Clinical and Translational Science Awards program (CTSA) in 2006, which supports a national consortium of 60 medical research institutions in 30 states.3 CTSA is the largest program of the National Center of Research Resources (NCRR), which was established initially in 1990 for the purpose of providing resources, tools, and networks to NIH researchers nationwide as well as support for other programs, such as biomedical technology and research infrastructure.4, 5 In a climate of rapid scientific development, these programs aim to provide infrastructure, resources, training, and opportunities for collaboration among scientists and clinicians from a wide variety of disciplines and across different institutions. The common goal of these programs is to facilitate the transition from basic science discoveries to clinical studies, community practice, and health care decision making.4, 5, 6, 7 Although the programs of the NCRR have promoted higher quality training, improved research management, and enhanced community participation in clinical research, many roadblocks remain for translational research.6 In March 2011, Francis Collins, director of the NIH, announced the creation of a new National Center for Advancing Translational Sciences (NCATS), which will assume the CTSA and dissolve the NCRR by redistributing its other programs throughout other institutes and centers. The aim of this controversial new program will be to catalyze the progression from basic scientific discovery to clinically useful therapeutic or diagnostic tools by identifying bottlenecks in translational research and exploring innovative methods to approach and remove them.8 Thus, the creation of the NCATS is an attempt to shift the current paradigm in which most basic science discoveries take more than 10 years before they are translated into a clinically relevant drug.
The long time periods and costs necessary for the development and approval of useful drugs have also affected pharmaceutical research and development negatively, particularly in the area of infectious disease, where drug companies are curtailing or ceasing antimicrobial development despite increasing antibiotic resistance and “bad bugs.”9, 10, 11 Similarly, despite the Orphan Drug Act, which was passed in 1983 to stimulate the development of drugs for rare diseases,12 the pipeline for new drugs targeting diseases not of interest to pharmaceutical companies is also dry. Ultimately, whereas the creation of programs like the NCATS is a step in the right direction, the changes proposed by this organization still leave several problems unresolved and generate a new series of concerns.
In this article, we will review, in a noninclusive fashion, several examples of basic science achievements that have been brought to the forefront of translational research. A complete review of the translational applications of all recent scientific discoveries transcends the purpose of this article, so we will limit our discussion to the field of infectious disease. Overall, we draw attention to the general challenges facing translational research and define some of the recently realized achievements and future prospects of this field.
The “Omics” Revolution of Translational Research
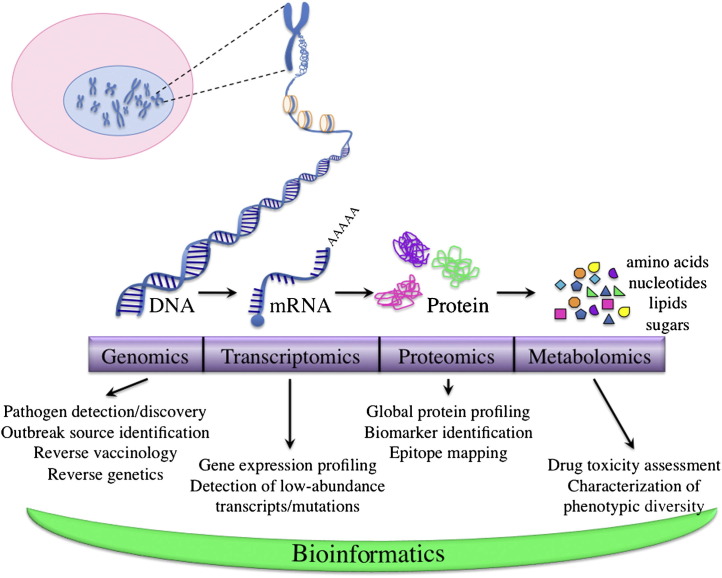
The Human Genome Project was conceived initially in 198513 and is considered to be one of the most important steps in the start of the genomic era. By definition, genomics characterizes the hereditary information of an organism. Research in proteomics, which defines the structure and function of proteins encoded by the genome of an organism, is the next logical step after genomics for the study of biological systems, because proteins are the major catalysts of intracellular processes.14 Metabolomics uses the systematic analysis of the complete set of chemical fingerprints left behind by certain cellular processes to determine key aspects of the function and regulation of those processes.15 Together, genomics, proteomics, and metabolomics comprise the 3 major systems biology approaches at the forefront of translational research (Fig 1 ).
Fig 1.
A “top-down” explanation of “omics.” Genomics is the study of the complete set of hereditary genetic information in an organism. With the advent of microarray technology and next-generation sequencing, numerous applications have arisen from the field of genomics, including pathogen discovery, epidemiologic advances, and a variety of molecular techniques that allow for precise manipulation of microbial genomes. As DNA begets RNA and protein, so do the fields of transcriptomics and proteomics follow logically from genomics. Transcriptomics, which is a subset of the field of genomics and concerns the collection of messenger RNA transcripts expressed within an organism, has emerged as a result of more sophisticated techniques that allow for the highly sensitive determination of low-abundance mutations, transcripts, and SNPs. Proteomics is the next major “omics” field after genomics, and it focuses on the complement of proteins, their modifications, and their interactions within an organism. With the aid of analytical techniques such as 2-DE, MS/MS, and shotgun proteomics, the field of proteomics has found useful application for the identification of biomarkers and mapping of epitopes that may provide targets for antimicrobial drug development. The field of metabolomics is a natural offshoot of proteomics as it uses many of the same MS techniques. A major difference, however, is that whereas genomics and proteomics provide insight into the potential of cellular processes, metabolomics gives an instantaneous snapshot of what is actually happening in a cell. Each one of these “omics” fields generates massive amounts of rich, detailed data that surpass the capabilities of manual data analysis. This therefore necessitates the incorporation of bioinformatics for the development of computer algorithms that are used to analyze and model data. (Color version of figure is available online.)
Genomics
Since it was first invented in the late 1970s, DNA sequencing has been one of the most influential innovations in biological research.16, 17, 18 This rapidly evolving technique has provided a feasible approach to a multitude of problems concerning biology, including infectious disease. Polymerase chain reaction (PCR) is the most common method for amplifying nucleotide sequences based on defined primer sets that recognize specific regions of the DNA of a sample. Physicians routinely refer to data from PCR and sequencing methods as an integral part of the standard of care. For example, HIV medications are chosen after analyzing the virus genotype for the presence of mutations that would confer resistance to some of the antiretroviral drugs on the market, and viral loads (viral levels measured by PCR) are used to help predict the responses to therapy.19 Similar approaches are also being used for other viral diseases such as hepatitis B virus,20, 21, 22 hepatitis C (HCV) virus,23, 24, 25, 26 and influenza.27, 28, 29 PCR and sequencing techniques also play a crucial role in pathogen discovery and outbreak investigation. In the recent years, the importance of this field has been highlighted by the growing number of emerging pathogens and their rapid spread as a consequence of globally mobile populations, climatic changes, improved surveillance and threats of bioterrorism. Both basic research and clinical microbiology laboratories use uniplex and multiplex PCR assays for the rapid identification and typing of many bacterial and viral pathogens. Conventional uniplex PCR methods use specifically targeted sets of primers (1 set per reaction) to amplify conserved genetic regions of a sample and identify close variants of known pathogens. Multiplex PCR assays, which first emerged in the late 1990s,30 are based on the same theory but can amplify multiple regions of DNA at once from a single sample, thus producing additional information that would otherwise require several times the amount of reagents to run using uniplex PCR.31 Although multiplex PCR techniques were a significant improvement compared with conventional PCR, both approaches are still limited in their ability to detect new pathogens and to screen large numbers of sequences in an efficient manner.
The introduction of microarray technology in the middle to late 1990s allowed for the development of microarray-based pathogen detection platforms and high-density, pan-microbial arrays, which truly made rapid screening for a large number of pathogens feasible.32 In this application of microarray technology, sample material suspected to contain pathogen genomes is hybridized to a 2-dimensional (2-D) array (or gene chip) of hundreds or thousands of miniaturized spots containing nucleic acid probes specific for various pathogen sequences. Microarray-based pathogen-detection platforms, such as the GreeneChip and the Virochip,33, 34, 35, 36 use 70-mer oligonucleotide probes of conserved genetic regions within each taxonomic group of viruses. Because of their length, these probes are more tolerant of sequence mismatches and therefore allow for the detection of unknown microbial targets.37 Furthermore, the probes are updated continuously when new viruses and sequences are added to their associated databases. To identify a pathogen from a clinical specimen, nucleic acids are amplified randomly and then hybridized to the chips. Given the large number of probes and possible hybridization successes, the results are analyzed with the help of computer programs that use algorithms and statistics to identify virus hybridization patterns. Although microarray-based pathogen-detection platforms have been instrumental in the detection of numerous pathogens, including novel ones like the coronavirus associated with severe acute respiratory syndrome (SARS-CoV),34, 38 they still rely on some prior knowledge of relevant genome sequences.39, 40, 41 The introduction of high-throughput sequencing has revolutionized the ability to detect novel infectious agents whose genomic sequences are completely unknown or are present in extremely low numbers. The high-throughput sequencing revolution started in 1987 with the introduction of the first automatic sequencer.42 The first of the next-generation sequencing technologies was developed only a few years later,43 but it took nearly 15 years of technologic improvements before it became commercially available.44, 45 Currently, the most frequently used platforms of this type include the 454 large-scale parallel pyrosequencing system (454 Life Sciences/Roche, Branford, Conn) and the Solexa (Illumina, Inc, San Diego, Calif) sequencing-by-synthesis system, but this field is evolving rapidly with new platform generations regularly being developed. These techniques allow for the “deep” sequencing of clinical samples. The term “deep” refers to the depth of coverage or the average number of times that a single nucleotide is sequenced, and it allows for high levels of accuracy in sequence determination.46, 47, 48 “Deep” sequencing methods can, therefore, achieve thousands to millions of simultaneous sequence reads per run, allowing for the precise determination of the entire genome of an organism. “Ultra-deep” sequencing, which is largely being enabled by platforms such as the 454 sequencing system, provides an even deeper sequence coverage and allows for several additional applications, including amplicon and transcript sequencing, which can detect extremely low-abundance genetic variations.49 Because pyrosequencing techniques are based on the detection of light produced whenever a nucleotide is incorporated and do not rely on physical separation of DNA bases, these platforms can be run in parallel and miniaturized to any reaction volume that generates detectable levels of light, thus driving down the overall cost of sequencing. Nevertheless, the shorter reads necessitated by the deeper sequencing capacity of next-generation sequencing creates additional challenges for sequence assembly and gene annotation.49 Although the cost of sequencing is decreasing, the analysis of the accumulated data is costly, time consuming, and computationally challenging. The analysis is complicated by the fact that computer systems will have to discriminate between the nucleic acids of the host and those of the newly discovered infectious agent. Despite these challenges, several new viruses have been identified using “deep” sequencing systems.50, 51, 52, 53 Additionally, these systems have made subsequent steps in pathogen discovery, including sequencing the entire genome to facilitate the development of diagnostic techniques and the identification of potential therapeutic targets, more time and cost effective.
Beyond pathogen discovery, sequencing-based methods have the ability to discriminate between closely related strains and can therefore be employed successfully to study viral variations and evolution. Next-generation sequencing techniques also provide an important tool that enables epidemiologists to follow person-to-person transmission more closely and eventually identify the source of an outbreak. This was the case in the recent cholera outbreak in Haiti, where the sequences of the Vibrio cholerae genome from the Haitian outbreak were compared with the sequences of previously isolated strains and found to be more closely related to strains isolated in Bangladesh in 2002 and 2008 than to the South American isolates.54 Similarly, high-throughput sequencing techniques have allowed for the fast sequencing of the 2009 H1N1 “swine” flu isolates and for the determination of the exact origin of each of its gene segments.55
New sequencing techniques, in combination with other advances in molecular medicine, have also improved the field of vaccine discovery drastically. In the past, vaccines were constituted by crude or partially purified, inactivated viral or bacterial preparations, and often carried the risk of serious allergic reactions or autoimmune diseases because of impurities in the formulation.56 Furthermore, administration of the vaccine itself could sometimes cause disease, as illustrated by vaccine-induced paralytic polio.57 Although some of these problems still exist with present-day vaccines, the field of genomics has paved the way for more powerful and focused techniques that allow for a better understanding of the nature of pathogen antigenicity and how our immune system can be tuned to respond optimally.
With the explosion of sequence data from the genomic era, the genomes of a large number of medically important microbes have been decoded and made readily available. As a result, techniques such as reverse vaccinology have emerged that allow for the design of more specifically targeted vaccines.58 The power of reverse vaccinology lies in its ability to identify novel vaccine targets based on computer-facilitated predictions of the immunogenicity and prevalence of all antigens in a given pathogen without ever having to cultivate the specific microorganism. Once potentially suitable vaccine targets are identified, each gene from the infectious agent that encodes one of these targets is cloned into an expression vector, and the resulting proteins are expressed in vitro and used to immunize an animal model from which antisera will be derived. Each corresponding antisera is then tested for its ability to neutralize or protect against the original infectious agent. Using this information, vaccines can be designed to elicit specific immune responses against the specified target and are combined with rationally designed adjuvants whose mechanisms of action could improve vaccine immunogenicity. One example of the successful use of this process is the recently developed vaccine for serogroup B Neisseria meningitides. 59, 60, 61 An array of other potential vaccines, including those targeting group B Streptococcus and Streptococcus pneumoniae species, are currently under study.62
One technique that has derived from the newly sequenced genomes of viral pathogens is that of reverse genetics, which has made it possible to generate viruses entirely by co-transfection of specifically engineered plasmid DNA in cell culture. This technique has very important implications for vaccine production because it means that very specific changes can be reverse engineered into the sequences of the plasmid DNA used for co-transfection, allowing extremely fine control over the pathogenicity and immunogenicity of the resulting virus.63 Reverse genetics has been used successfully in the development of promising candidates for vaccines targeting influenza viruses. Several groups have explored reverse genetics strategies to design live-attenuated H5N1 influenza virus vaccines,64 and a recent phase I clinical trial has demonstrated the safety and immunogenicity of a reverse genetics-derived NS1-truncated variant of an H1N1 influenza virus in male volunteers.65 Reverse genetics has also been used in pathogenetic studies. In this case, the insertion, mutagenesis, or deletion of specific sequences in the plasmids before transfection can clarify the effect of such modifications on viral infectivity and can help to elucidate the function of any modified protein in the context of the viral life cycle.66 Finally, the ability to exert such exquisite control over a viral sequence would permit the identification of specific sequence alterations that promote viral attenuation, which is information that can be used for the design and generation of new vaccines.67, 68, 69, 70, 71
Additional applications that have risen from the genomic revolution include the use of genetically altered viral vectors as an alternative approach to vaccination72, 73 or as a conduit for delivering antibodies as passive immunotherapy74, 75 and the use of sequence data in the design of specific inhibitors and antivirals. These approaches are currently under study and are likely come to the forefront in the near future.
The genomic revolution has improved significantly the way we detect, prevent, and treat infectious diseases. Future advancements in this field will continue to decrease the time necessary to identify newly emerging human pathogens and produce new vaccines for clinically relevant infections.
Proteomics
Proteomics is a predominantly mass spectrometry (MS)-based technique that deals with the large-scale evaluation of cellular function at the protein level. MS analysis is accomplished by ionizing a sample into its molecular components (eg, with an electron beam, laser, etc) and then passing them through an electromagnetic field that separates them by their ratio of molecular mass to charge (m/z).76 Proteomic analysis (as opposed to other MS-based techniques) seeks specifically to elucidate the entire complement of proteins and protein modifications and/or interactions in a specimen at a given time. Because proteins are the major effectors of cellular pathways, proteomics provides a way to interpret information encoded within the genome of an organism. Since the term proteomics was first coined in 1995,14 the field has expanded tremendously, largely because of the development of ionization and analytic methodologies, which have allowed for the more efficient separation and detection of almost all protein components within a sample of interest.77 Unlike genomics, no method or instrument currently exists that can identify and quantify the components of a complex protein sample in a single step.77, 78 Instead, a combination of steps with varied methodologies and instrumentation are used.
Proteomic platforms are characterized by 2 key steps: (1) protein separation, mostly obtained through either electrophoretic or chromatographic methods, and (2) protein analysis using primarily MS-based techniques. Two-dimensional electrophoresis (2-DE), which was developed in the 1970s, is a method that allows all proteins contained within a complex biological mixture to be separated by their isoelectric points and molecular weights, and then visualized by staining.79, 80 Today, 2-DE-based proteomic approaches remain the gold standard for the separation of complex protein mixtures despite several limitations, including the detection of only relatively abundant proteins in a complex mix, poor separation of insoluble and/or hydrophobic proteins, protein comigration, and the labor-intensive nature of identifying multiple protein spots on the resulting gels.79 To address the problem of comigration, a modification of 2-DE, known 2-D differential in-gel electrophoresis (2-D DIGE) was developed. In this technique, multiple protein samples are labeled separately with fluorescent dyes, mixed together, and then resolved on a single gel, eliminating the need for cross-gel comparisons and increasing the overall statistical power by the inclusion of a greater number of samples.77, 81, 82 Whereas the development of 2-D DIGE represented some improvement, the need for the increased sensitivity of detection and more efficient separation of proteins within a complex mixture remained and was the driving force behind the development of “gel-free” technologies.79
Gel-free proteomic approaches were made possible by coupling the physical separation capabilities of chromatographic techniques to the mass analysis capabilities of tandem mass spectrometry (MS/MS).79, 80 Chromatography is a technique that involves moving the components to be separated (mobile phase) across an immobile support (stationary phase) such that the migration rates of solutes (ie, proteins) vary depending on their chemical properties.83 Liquid chromatography (LC), in which the mobile phase is liquid, is one of the most widely used protein separation techniques in proteomics.83 With the use of smaller particles (less than 2 μm), monolithic columns (using porous channels rather than beads) and extremely high pressures (up to 19,000 p.s.i.), high-performance LC (HPLC) and ultrahigh-performance LC have improved the efficiency and resolution of protein separation drastically.84 In HPLC, complex protein mixtures are digested by trypsin into their peptide components, which are then moved in a liquid mobile phase at high pressures across a stationary phase that allows for their separation based on polarity. Next, the sorted peptide components are analyzed by MS/MS. The first mass spectrometer is programmed to detect peptides over a range of predetermined masses to distinguish targets of interest from the vast array of proteins typically present in complex biological fluids. The resulting MS1 data are used to pinpoint ions of interest, which are then fragmented, measured, and recorded by the second mass spectrometer (MS2 data). Finally, computer algorithms are used to aid researchers in identifying individual proteins from combined MS1 and MS2 data.77, 79, 81, 85 As an example, the SEQUEST algorithm, developed in 1994, uses the fragmentation spectra of peptides to infer amino acid sequences and match them to translated genomic sequences. This approach has an identification rate of greater than 100 proteins per run.86 The newest iteration of this software, MacroSEQUEST, features enhanced capabilities (eg, posttranslational modification scanning) and a dramatic improvement in processing speed compared with the original version.87
The combination of HPLC and MS/MS, which is now known as “shotgun proteomics,” has proven to be a fast and highly sensitive approach, especially when scaled down to microflow or nanoflow rates.88, 89 As such, shotgun proteomics has not only allowed for the identification of thousands of individual proteins within a given sample with essentially no prior knowledge of the component peptides but also has greatly facilitated the analysis of entire proteomes. Although shotgun proteomics represents a significant technological advancement in the analysis of complex biological mixtures, the concentrations of individual proteins within such samples can be low enough to require levels of sensitivity beyond what even standard HPLC conditions can achieve. By decreasing flow rates and column diameters, this level of sensitivity can be obtained, but only by adapting the system to the special requirements of nanoliter flow rates (20–1000 nL/min).88 Another shortcoming of shotgun proteomics is the inability of one-dimensional HPLC to provide sufficient separating power to manage the vast complexity of proteins inherent in most biological specimens. To address this issue, multidimensional separating approaches, such as multidimensional protein identification technology (MudPit), which combines strong cation exchange chromatography with HPLC, were developed.79, 85 Even with such improvements, these advanced shotgun proteomics platforms can still identify only approximately 40% to 70% of the proteins predicted by a microbial genome (less for proteomes of higher complexity). Furthermore, the actual proteome size may be smaller or larger than predicted by the genome because of the absence of transcription of some proteins and the alternative splicing and/or posttranslational modification of others.77, 81 Therefore, whereas shotgun proteomics does enable the identification and relative abundance of a large number of proteins in a sample, it is not optimal for absolute protein quantitation.
In addition to identifying proteins within complex biological mixture, the need to quantitate these components can provide information regarding their dynamics within the context of a biological system. This added focus has led to the emergence of a derivative field known as quantitative proteomics. Protein quantitation can be relative or absolute. Relative protein abundance may be estimated by several methods, including by measuring the total number of repeat observations of MS/MS spectra from all peptides in a protein (spectral counting), calculating the percentage of possible peptides per protein observed (peptide counting), or by comparing directly the extracted ion chromatograms (XIC, aligned using both mass and elution time) of one sample with that of another sample.90, 91 These label-free techniques have been facilitated greatly by the development of new software programs, such as MZmine, MapQuant, and SuperHirn.91, 92, 93 Alternatively, protein quantification may be accomplished by combining shotgun proteomic techniques with stable isotope labeling.90, 91 Using this approach, samples are labeled differentially with stable isotopes (heavy vs light), then pooled and analyzed together. Peptides common to both samples, but labeled with different isotopes, behave the same on analysis and are detected as peak pairs. By comparing the sizes of these peaks, the relative abundance of the peptides in each sample can be compared.79 The methods for absolute quantitation of protein abundance are more desirable in many cases (eg, for determining copies per cell). One technology that addresses this need and is now commonplace in clinical settings is called multiple reaction monitoring, also known as selective reaction monitoring (MRM/SRM). This technique exploits the unique capabilities triple quadrupole (QQQ) MS to analyze precursor and fragment ion pairs. QQQ MS uses 3 sets of 4 circular metal rods (quadrupoles) that run parallel to each other. The first quadrupole (Q1) acts specifically to select peptide ions with predefined m/z values. These peptides are then fragmented by the second quadrupole (Q2), which serves as a collision cell. The third quadrupole (Q3) can be set either to scan the entire m/z range to provide information about the structure of the original ion or to filter out fragments with specific m/z values that may be indicative of particular characteristics (eg, functional groups). MRM/SRM can provide targeted information (sensitive to the attomole level) about multiple peptides across a wide concentration scale within a single sample.90, 94, 95 The sensitivity of the MRM/SRM technique can also be improved by enriching the target from a complex background using antibody-mediated approaches such as stable isotype capture by antipeptide antibodies and immuno-matrix-assisted laser desorption/ionization.90 As with relative protein quantitation, absolute protein quantitation possesses its own arsenal of computer programs designed to assist in the analysis of MRM/SRM data, including Skyline MRM/SRM builder, MRMpilot (Applied Biosystems), Peptide Optimizer, and Dynamic MRM software (Agilent Technologies, Santa Clara, Calif), MaRiMba, MRMaid, and MRMer.90
To date, these analytical platforms have given rise to 4 major proteomic applications: (1) global protein profiling—analysis of all proteins within a given biological unit or sample, (2) analysis of protein modifications (phosphorylation, ubiquitination, nitrosylation, etc), (3) analysis of protein–protein interactions, and (4) analysis of protein–gene interactions.77, 78 With regard to the study of infectious diseases, global protein profiling using shotgun proteomics has been used recently to identify and quantitate protein expression in the malaria parasite, Plasmodium falciparum, according to the stage of its life cycle and its host compartment. These studies resulted in more than 200 candidate proteins for possible stage-specific drug or vaccine targets. In addition, they generated information on a large set of “orphan” proteins (proteins that could be mapped to the genome but were not found in the set of previously known proteins), protein interactions, and protein expression at each stage of the parasite’s life cycle.96, 97 An analysis of protein modifications has been used to identify phosphoprotein signaling pathways that are altered in Rift Valley fever virus-infected airway epithelial cells and to uncover potential targets of reactive nitrogen intermediates in Mycobacterium tuberculosis (M. tubercolosis), thus informing on potential targets of novel antiviral and antimycobacterial compounds.98, 99 Finally, analyses of protein-protein and protein-gene interactions have been used to study the relationship between Epstein-Barr virus (EBV) infection and nasopharyngeal carcinoma, as well as alterations in host cellular processes associated with HIV-1 viral replication.100, 101 Recent studies have aimed to establish the context in which these types of interactions occur by combining proteomic-based techniques with either cryo-electron tomography or “membrane shaving” with proteinase K digestion to examine specific cellular localization.81 Such approaches have been used successfully to map the spatial proteome of Leptospira interrogans 102 and the membrane proteome of Staphylococcus aureus (S. aureus). 103 Studies such as these ultimately might lead to an improved understanding of microbial pathogenesis and provide important clues for the identification of novel biomarkers and antivirals.
Because proteins carry out many functional biological activities, the proteomic analysis of an organism provides a layer of detail beyond that of genomics. Although it is still an emerging technology, proteomics has already had a profound impact on clinical and biological research by informing on the pathogenesis of disease, facilitating biomarker discovery and aiding in the identification of potential drug targets. New developments in MS platforms, MS-based tissue imaging techniques, and combined proteomic/genomic approaches will likely extend the impact of proteomics to drug design and testing as well as pathogen identification and discovery, which will continue to advance our understanding of complex biological processes.77, 78
Metabolomics
The newest field of systems biology, metabolomics, has been defined as “the comprehensive and simultaneous systematic determination of metabolite levels in whole organisms and their changes over time as a consequence of stimuli such as diet, lifestyle, environment, genetic effect and pharmaceutical intervention, both beneficial and adverse.”104 A more concise definition of metabolomics might be the global profiling of all small molecule metabolites contained within a sample of interest at a given time. Metabolites, which are the intermediates and products of all cellular processes, typically fall into one of 4 categories: amino acids, nucleotides, lipids, and sugars. Moreover, metabolites represent the level at which most pharmaceuticals exert their effects. Therefore, by studying the production and consumption of these chemical fingerprints, one can gain insight into the dynamic functions of any biological system.104, 105
The analytic techniques employed in metabolomic studies are largely similar to those used in proteomic studies and are based primarily on either nuclear magnetic resonance (NMR) spectroscopy or MS. The typical workflow involves mechanical extraction coupled to a rapid cooling step that halts metabolism in its native state, followed by detection (either via NMR or MS) and data analysis. MS-based metabolomics requires the additional separation of components by either gas chromatography or LC for enhanced molecular identification. For each molecule within a sample of interest, LC-MS-based metabolomics generates 3 pieces of data: (1) chromatographic retention time, which informs on the chemical structure; (2) a m/z ratio, which informs on the molecular mass; and (3) abundance. The retention time and m/z ratio may then be queried against a library of known standards to determine the identity of each metabolite.106 An NMR-based metabolomic analysis, by contrast, provides a nondestructive way to view changes in metabolite levels either in vitro or in vivo more broadly. Moreover, it provides more detailed structural information than MS. Because MS analysis provides better resolution and sensitivity, it remains the predominant methodology for analyzing metabolic data. All metabolomic analyses result in complex, multivariable data sets that require visualization software for spectral analysis as well as bioinformatics and statistical methodologies for interpretation.104, 107
Metabolomic analysis of data sets produced by NMR and MS yields 2 major applications, steady-state metabolite profiling and labeled flux analysis. Steady-state metabolite profiling may be global (profiling of all small molecule metabolites within a sample) or targeted (analysis of only prespecified metabolites of interest), and it has been used to discover new biomarkers, inform on dysregulated pathways associated with disease, and generate information about cellular functions. Labeled flux analysis, in which radiolabeled isotopes are used to analyze changes in the amount of a given metabolite that is used and produced in a particular metabolic pathway, is an approach that yields more detailed information about the dynamic nature of biological pathways and can be used to better identify potential therapeutic targets.107 Recently, Henderson et al108 used a steady-state LC-MS-based metabolomics approach to identify how bacterial strains associated with urinary tract infections (UTIs) differed from those that colonize the gastrointestinal tract. Their results indicated that urinary Escherichia coli (E. coli) strains preferentially express 2 small molecules, yersiniabactin and salmochelin, which are known to be important in iron scavenging and support bacterial survival and growth. These findings suggest that new antibiotics directed against yersiniabactin- or salmochelin-producing E. coli strains might be an improved and more targeted strategy to prevent recurrent UTIs. In another example, Munger et al109 used changes in metabolic flux between uninfected and human cytomegalovirus-infected cells to identify pathways that were upregulated by viral infection, thus providing important information regarding potential targets for novel antivirals aimed at blocking viral replication.
Lipidomics and glycomics are two growing fields related to metabolomics that merit special mention.110, 111 Lipidomics is the large-scale study of the networks and pathways employed by all cellular lipids, and it employs largely the same tools as those used for metabolomics, but with specific modifications that accommodate the unique characteristics of lipids.112 Lipids have key functions in signaling pathways, energy storage, and the structural integrity of cell membranes. They also play important roles in host–pathogen interactions and immunomodulation.111, 113 Glycomics is the comprehensive study of all sugars in an organism.114, 115 Because cellular sugars can be simple sugars, glycoproteins, or glycolipids, there is no common experimental approach for their analysis. Instead, glycomic studies use a combination of the techniques found in meta bolomics, proteomics, and lipidomics.110 Glycoconjugates participate in a variety of biological processes associated with cell adhesion and migration, bacterial and viral recognition, signaling pathways, and innate immunity.115 Therefore, the study of changes in the lipid and sugar components of a biological system can provide powerful clues about disease-related mechanisms and novel therapeutic targets. Indeed, these approaches have shown particular promise in the study of infectious disease. Lipidomics has been used to uncover the mechanisms involved in enhancing the drug susceptibility of Candida albicans 116 to gain insight regarding the intracellular processes and lipid composition of HIV,117, 118 CMV,119 and HCV,120, 121 and to identify lipidomic modifications that occur in the host as a result of bacterial or viral infection.122 The utility of glycomics has also been demonstrated recently in the characterization of specific interactions between viral proteins and various forms of sialic acid,123, 124 the detailed analysis of how lipopolysaccharide from Coxiella burnetii may contribute to pathogenesis,125 and the discovery that the glycan coats of HIV-1 virions match that of the T cell immunomodulatory microvesicles from which they derive, suggesting a mechanism by which the virus can evade the antiviral immune response.126
The aforementioned examples demonstrate the potential of metabolomics as a tool for enhancing development of novel diagnostics and therapeutics. Like proteomics, metabolomics has already been used as a vehicle for investigating disease pathogenesis, potential biomarkers, and therapeutic targets. Moving forward, improvements in automated metabolite identification will expand exponentially the potential uses of metabolomics-based approaches. Additionally, the integration of metabolomics data with other global profiling techniques, such as genomics and proteomics, will allow for a deeper understanding of biological pathways than possible to date, with unparalleled possibilities for clinical application.
The “Omics” of Infectious Disease
With the advent of genomics, proteomics, and metabolomics came an explosion of other “omics” sciences. This convention arose, in part, because the suffixes “ome” and “omics” succinctly describe a holistic way of looking at relationships that exist within a relatively complex scientific domain; however, this trend also reflects the growing involvement of bioinformatics and the internet as a way to integrate complex biological data.127 Concurrent to the significant advances in “omics” sciences that have allowed for a better understanding of the molecular and cellular processes that occur within humans, researchers also began to appreciate the number and complexity of organisms that resided on and in the human body. After the publication of the human genome sequence in 2001,128, 129 scientists began to call for a “second human genome project” that would catalog the complete inventory of microbial genomes at major sites of colonization on the human body.130, 131 Because microbiota are believed to play a fundamentally important role in human health and disease,132 a microbial inventory in combination with a comprehensive analysis of host gene expression would provide insights into the mechanisms of this interaction. A landmark study that was published in 2005 by Eckburg et al133 used the 16S ribosomal RNA sequencing technique to characterize the endogenous gastrointestinal microbial flora in 3 individuals. The authors concluded that the bacterial communities varied tremendously from one individual to the next. This and other studies that came after indicated that a larger scale investigation of the human microbial ecosystem was warranted. Thus, the NIH initiated a 5-year project called the Human Microbiome Project (HMP) in 2007.134
The HMP uses various “omics” approaches to characterize the complexity of microbial communities, determine the existence of a core microbiome at various colonization sites on and in the human body, and examine the relationship between changes in the microbiome and human health (Fig 2 ).135, 136 To date, the HMP has determined that substantial alterations in the human microbiome are important for a variety of disease states, including psoriasis,137 sexually transmitted infection of the male urogenital tract,138 Crohn disease,139 gastroesophageal reflux disease,140 and others. There is also a growing interest regarding the effects of the gut microbiome on the development of the immune system. A recent study showed that mice treated with antibiotics that depleted specific gut bacterial populations would have an impairment of virus-specific cell-mediated and humoral responses in the lung after infection with influenza virus.141 These data will require confirmation and more in-depth studies that address mechanism; however, they raise a series of new questions about the role of the bacterial microbiota in human health. Not only has the HMP made major advances in our understanding of the human microbiome, but also it has contributed significantly to the growing field of metagenomics by developing broadly applicable techniques for analyzing massive amounts of sequence data. For example, Langmead et al142 designed a cloud-computing software tool called Crossbow that can analyze large DNA sequence data sets more rapidly (38-fold coverage of the human genome in 3 h) to perform the alignment and detection of single-nucleotide polymorphisms (SNPs) without sacrificing accuracy. Another group developed a robust software program called QIIME (pronounced “chime” and stands for quantitative insights into microbial ecology) that can analyze pyrosequencing data from thousands of heterogeneous experimental data sets in order to acquire information rapidly about various microbial communities.143
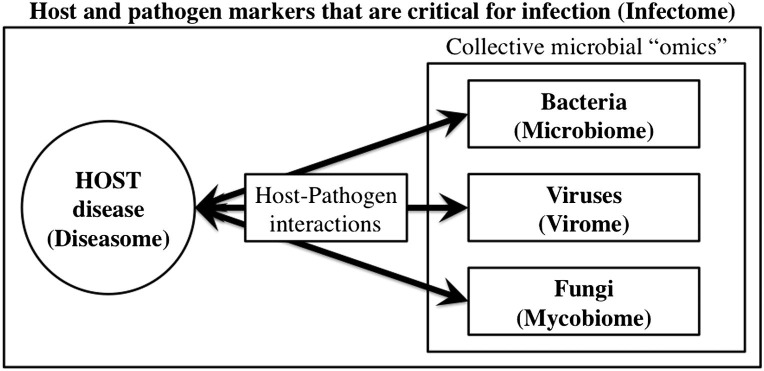
Fig 2.
The “omics” paradigm for host–microbe interactions. State-of-the-art “omics” technologies have paved the way for the comprehensive characterization of a variety of hosts and microbes. The microbiome, virome, and mycobiome are 3 microorganism-specific data sets that have arisen from the “omics” revolution. Although each subset comprises distinct information about its target (bacteria, viruses, fungi, etc), they do not exist in isolation. An increasing number of studies is focused on the intersection between the collective set of microbial “omics” and the profile of gene expression that is associated with human disease (diseasome). Thus, investigation into the complex dynamics that occur during infection (infectome) can provide significant insight regarding the etiology and progression of disease, as well as lead to the identification of targets at which to aim therapeutic intervention.
Although the HMP does fund at least 1 project aimed at characterizing the relationship between viruses and human illness,144 the primary focus of this project is currently with bacterial populations. Therefore, although not as well-defined as the HMP, at least 2 other infectious disease-related “omes” have emerged in parallel: the human virome and the human mycobiome, which comprise the myriad of viruses and fungi, respectively, that inhabit the human biosphere and play roles in health and disease (Fig 2). In 2003, Anderson et al36 proposed a system for the global screening of viruses in large human populations. Although this global screening is not limited to viruses that persist chronically in the human body as is the case for the commensal bacteria populations included in the HMP, such a system could lay the groundwork for the rapid detection of outbreaks and the systematic discovery of novel human viruses, as well as provide better clues about etiologies in human disease. For example, 1 study investigating the human virome uncovered differences in the metabolic states of DNA viromes in the respiratory tract of cystic fibrosis (CF) compared with non-CF patients.145 Another group demonstrated that bacteriophage populations became more similar in the guts of individuals who were on a diet of similar fat content, thus raising the possibility that virus populations could be engineered to combat obesity.146 In contrast, studies regarding the human mycobiome seem to still be in their infancy as the first study describing a baseline mycobiome in humans was published only recently.147 In this study, the authors used multitag pyrosequencing to characterize the fungi present in the oral cavity of healthy individuals and identified differences and similarities in white and Asian populations.
In light of major advancements in the genotypic characterization of bacterial, viral, and mycotic pathogens associated with humans, it is clear that an additional holistic approach will be required to understand the phenotype of this interaction more fully and how it may affect human health and disease. Thus, the emerging field of infectomics will aim to study the structural and functional genomics and proteomics of microbial infections more efficiently, accurately, and integratively (Fig 2). Whereas the microbiome, virome, and mycobiome will identify the comprehensive inventories of microbes that inhabit the human body, the infectome will examine the distinct signatures of each microbe as they vary depending on host or microbial gene expression, tissue type infected, and other dynamic characteristics of both host and pathogen.148 By networking the infectome with the human diseasome (Fig 2), which links certain diseases together based on common gene expression profiles,149 it will be possible to identify links between microbial infection and the etiology of human disease, which could inform the design of better vaccines and therapeutics.150
Taking it all Together: A Look at Systems Biology
The need to manage the rapidly accumulating number of sequences and massive amounts of data from high-throughput platforms has required the development of more sophisticated computer programs to analyze and integrate the data. Thus, systems biology has developed into a new field that aims to understand the complexity of pathogen–host interactions by using computational integration of high-throughput experimental data and by modeling molecular networks via bioinformatics. Intrinsic to this approach is the idea that biological systems display “emergent properties” which are complex patterns that arise from a multiplicity of relatively simple interactions. Therefore, a major objective of the systems biology approach with regard to infectious disease is to make predictions about the dynamic behavior of biochemical networks involved during infection with any pathogen.151 Thus, whereas small-scale studies look only at one side of the interaction, system biology gives a global perspective of the events.
Under the umbrella of systems biology, 4 main approaches are relevant to the study of infectious diseases: the systems biology of viral and bacterial pathogens, systems immunology, systems vaccinology, and high-throughput drug discovery. Common to each approach is the goal of integrating high-throughput multiomics data to construct predictive models of the networks and dynamic interactions between pathogens and their hosts to identify preventive and therapeutic targets for clinical development more quickly.
Systems biology of viral and bacterial pathogens
To recognize the importance of systems biology for the understanding, integration and practical application of the results of high-throughput screenings in the field of infectious disease and host-pathogen interaction, the National Institute of Allergy and Infectious Disease (NIAID) has created the Systems Biology for Infectious Diseases Research program. The research activities of this program were separated initially into 4 centers: The Tuberculosis (TB) Systems Biology Center, The Systems Virology Center, The Center for Systems Influenza, and The Center for Systems Biology for Enteropathogens. Each center will use both computational and experimental methodologies to analyze and model host-pathogen interactions comprehensively to determine the extent to which molecular and cellular networks are induced or altered during the course of infection. The overarching goal of this program is to generate and develop reagents, protocols, and statistical models that will be rapidly made available to the scientific community to promote swift advancements in scientific research and translation to clinical practice.152
The purpose of the TB Systems Biology Center is to model the molecular pathways of M. tuberculosis under conditions relevant to TB pathogenesis using a combination of experimental and computational techniques. For example, chromatin immunoprecipitation followed by next-generation DNA sequencing analysis (ChIP-seq) is a powerful technique that can be used to map global DNA binding sites precisely for any protein of interest so that the regulation of gene expression within the context of pathogenesis can be more fully understood.153 By combining this technique with a variety of “omics” data (transcriptomics, proteomics, glycomics, metabolomics, lipidomics, etc) during in vitro and in vivo growth of M. tuberculosis, the TB Systems Biology Center aims to measure the baseline metabolic and topologic state of the gene regulatory network of M. tuberculosis, delineate global alterations that occur in both the pathogen and host during infection of host macrophages, and integrate profiling data to develop predictive computational models of bacterial regulatory and metabolic networks. Using these models, predictions can be made regarding the state of these networks during infection with M. tuberculosis, leading to additional hypothesis-driven research.151, 154
The Systems Virology Center and the Center for Systems Influenza are similarly aimed at using systems biology to analyze and model alterations comprehensively that occur in molecular and cellular events during the course of virus infection and to provide deeper understanding into the processes that lead to the initiation and progression of infectious disease. The Systems Virology Center will focus on creating a unified view of the mechanisms of pathogenesis for highly pathogenic respiratory viruses including the 2009 pandemic H1N1 influenza virus, the H5N1 avian influenza virus, and SARS-CoV.151, 155 In contrast, the Center for Systems Influenza will focus more specifically on defining host innate immune responses to infection with influenza virus strains of varying pathogenicities, as well as characterizing regulatory networks associated with concurrent secondary infections like S. aureus. 151, 156 To date, several specific achievements have come out of these centers. Using a technique called weighted gene correlation network analysis, in which relationships between genes that are coexpressed are weighted based on their strengths or capacities,157 the transcriptional response of human bronchial epithelial cells infected with a highly pathogenic avian H5N1 influenza virus was profiled and compared with cells infected with a less virulent strain to identify potential novel mediators of pathogenesis.158 Another group performed a comprehensive proteomic analysis using LC-MS/MS to identify a large number of human host proteins associated with the polymerase complex of an H5N1 influenza virus. In addition to previously published interactions, this method identified novel interactions of the viral polymerase with host mitochondrial, apoptosis-inducing, innate antiviral, and RNA polymerase accessory proteins that could provide new insight into mechanisms by which the viral polymerase may contribute to host cell pathogenicity.159 Additionally, this center has used next-generation sequencing and analysis of the host transcriptome in response to influenza virus and SARS-CoV infections to investigate beyond the proteome and characterize the roles of non–protein-coding RNAs in regulating the host innate immune response.160
Finally, the Center for Systems Biology for Enteropathogens uses computational and experimental “omics” methodologies to analyze and model the interactions that occur between a host macrophage and the Salmonella enterica (S. enterica) and Yersinia pestis bacterial species to shed light on mechanisms behind the outcome of infection (either bacterial replication and host cell death or containment of the pathogen).151, 161 Researchers affiliated with this center have developed and used constraint-based reconstruction and analysis methods to simulate, analyze, and predict the way metabolic processes behave in various microorganisms.162, 163 In another study, novel virulence factors of S. enterica, including a new class of translocated effectors, were discovered and characterized through sample-matched multiomic measurements of various mutant strains.164 Other computational approaches, such as support vector machine-based identification and evaluation of virulence effectors, have also been used to facilitate the identification of secreted bacterial effector proteins from genomics sequence information.165
Systems Immunology
To understand fully the mechanisms by which drugs and vaccines combat disease, one must also grasp the way in which the human immune system responds to its infectome. Unfortunately, as science analyzes the fine details of immunity with higher and higher resolution, there is actually a decreasing ability to predict how the system behaves as a whole in the context of infection. To that end, researchers have begun to take systems biology approaches to immunology. One example of an effort to meet the challenge of amassing high-quality quantitative data using the available technical advances that “omics” sciences have provided while organizing and analyzing that data in ways that are meaningful to the study of immunology is the development of the NIAID Program in Systems Immunology and Infectious Disease Modeling in 2006, which became the Laboratory of Systems Biology (LSB) in 2011.166, 167 Rather than being a collection of independent laboratories, the LSB is an integrated group of scientists and staff who play a major role in fostering the growth of systems biology efforts both within the United States and abroad.167
At the forefront of this field is a rapidly increasing collection of systems biology techniques that have been applied to the unique set of problems encountered at the intersection between the human immune system and infectious disease. For example, computer simulations of the molecular dynamics of association, dissociation and posttranslational modifications of large peptide folding processes are used frequently in the field of systems immunology to characterize receptor–ligand interactions that occur between the host and pathogen. One study used this technique and the model system of EBV to demonstrate that some T cell receptor structures appear more frequently than others because of certain pattern recognition receptor (PRR)-like traits that occur within the T cell repertoire.168 Another group used constant pH molecular dynamics simulations to investigate the mechanism behind the ligand-binding activity of an innate immune system PRR that recognizes a pH-sensitive microbial protein to understand more completely how this type of interaction plays a role in immune surveillance and clearance of apoptotic cells.169 In addition to simulations that model the dynamics of molecular-level interactions, several programs have emerged that combine the techniques of systems biology with information provided by data-driven prediction methods. Because of advances in the predictive capacity of systems biology research, the newest version of the computer simulation software, C-ImmSim, can model the immune response in silico more accurately. Recently, the creators of C-ImmSim demonstrated a variety of new applications now accessible by their improved program, including the modeling of primary and secondary responses to prime/boost immunization, the way in which immunodominant peptides lead to affinity maturation and the relationship of diversity in MHC to “time to AIDS” in HIV-infected patients.170 Another simulation framework that has made its way into the public domain is the Multiscale Systems Immunology platform, which aims to model the early immune response to vaccination and natural infection by incorporating realistic biophysics and intracellular dynamics.171
Systems Vaccinology
The growing field of systems vaccinology combines modern analytic tools of systems biology with data on human immunological response patterns to vaccines in order to uncover molecular signatures of vaccine efficacy and to guide the design and evaluation of new vaccines. Most recently, systems vaccinology has been used to study the immune responses to vaccines against yellow fever virus (YFV).172, 173 Although the YFV vaccine has been used in humans since 1937, little was known about how the vaccine protected from infection. With the advent of systems vaccinology, scientists examined comprehensively the initial molecular signatures in individuals vaccinated against YFV not only to elucidate the mechanisms behind the vaccine’s efficacy but also to establish predictive correlates for the later development of protective responses.172, 173 Systems vaccinology was also used to establish the predictive correlates of a protective antibody response after administration of the trivalent inactivated influenza virus vaccine,174 and similar approaches have been applied to the study of immune responses to Brucella melitensis and fungal infections.175, 176, 177 Ideally, subsequent development of these models will define inherent and infection-mediated perturbations in the genetic regulatory networks of the host and will enable predictions about how the host will respond to vaccines. Based on these models, we can design vaccines that induce optimal immune responses without toxic effects, thus improving vaccine safety profiles.75, 178
Once the correlates of vaccine-induced protective immune responses against infection are well defined with the help of systems vaccinology, new methodologies must be identified that can apply this information to the design and development of new vaccines. The likeliness that commonalities exist among many different vaccines regarding the correlates of successful immune responses will drive faster and more accurate ways of screening vaccine candidates for their effectiveness. For example, Pulendran et al179 predicted the development of a vaccine chip microarray, similar to the MammaPrint prognostic chip that was developed for breast cancer, which will be able to predict the immunogenicity of any vaccine. Although this specific technology may so far still exist only in theory, several systems biology approaches are being used currently to aid with antigen discovery and vaccine development. Phage display is one such technique that enables the screening and identification of unique molecules with highly specific affinities for targets of interest.180 This approach works by introducing a foreign sequence at an appropriate site within a bacteriophage coat protein gene that results in the display of that protein on the phage surface. If random sequences are inserted, then it is possible to obtain a library containing a large number of distinct proteins, each displayed on a different phage. By immobilizing a DNA or protein target to a solid surface, libraries can be “panned” for phages that recognize the target. After washing, phages that do not bind to the target will be washed away, whereas specific phages can be eluted and used to produce more phage, thus enriching the population for relevant binding phages and enabling the isolation of specific proteins of interest for use in subsequent applications.181 Phage display is a powerful technique because of its many applications for vaccine and drug development. One application of phage display technology is the identification of the epitope repertoire of antibodies in postvaccination human sera. To this end, Khurana et al182 used H5N1 influenza virus whole genome-fragment phage display libraries to evaluate the quality of antibody responses in humans after immunization with a novel virus-like particle vaccine. This approach also enabled the authors to determine the most appropriate dosage of the vaccine to induce the desired antibody repertoire and will help them to evaluate the usefulness of incorporating an adjuvant into the vaccine.
High-Throughput Drug Discovery
In the war against infectious disease, the ability to develop nontoxic and fast-acting therapeutic and prophylactic drugs is just as important as being able to design safe and efficacious vaccines. Besides its usefulness in evaluating the efficacy of vaccine-induced immune responses, phage display can also be used to identify and synthesize monoclonal antibodies (mAbs) that can be used for therapeutic purposes in a variety of infections. An additional benefit of antibody identification through phage display is that because phage libraries can be generated from any set of sequences, it is possible to display human antibody fragments, thus avoiding the side effects inherent in humanized or chimeric antibodies.183 This approach has produced several mAbs with potential therapeutic applications against agents of infectious disease, including influenza A virus,184 Clostridium difficile, HIV, viral hepatitis, rabies, Pseudomonas aeruginosa, methicillin-resistant S. aureus, and Bacillus anthracis (B. anthracis). 185 Of these, the mAb targeting the protective antigen of B. anthracis, raxibacumab, is so far the only one that has met all criteria for approval by the U.S. Food and Drug Administration (FDA).186
With the growing emergence of antibiotic-resistant bacterial strains and the ever-present public health threat of pandemic viral infections, there has been an increasing pressure on the scientific community to discover, test, and produce novel antimicrobial drugs. High-throughput methods for synthesis and screening an increased number of targets resulting from the “omic” revolution have significantly advanced the search for new therapeutics. Combinatorial chemistry and diversity-oriented synthesis are 2 of the most important new methodologies in drug discovery because they have drastically reduced the time and costs associated with producing effective, marketable, and competitive new drugs. The power of these techniques comes from the ability to synthesize extremely large libraries of high-quality compounds with complex molecular diversity that increases their likelihood of interacting with biological macromolecules in a selective manner with high affinity.187 Phage display libraries have also been used widely in screening for novel drug candidates based on specific protein–protein interactions. Although they are frequently cheaper than most chemical synthesis methodologies, there are still several limitations regarding stop codon placement within displayed sequences, inefficient transformation methods, peptide length, and restricted use of naturally occurring amino acids. Nevertheless, several proof-of-principle studies have been performed recently to overcome these limitations, including 1 study that demonstrated that the incorporation of genetically encoded unusual amino acids might be feasible.188
After the synthesis of potential molecular targets, high-throughput screening is necessary to test the compounds in these large, natural, or synthetic libraries in in vitro or in silico assays and identify those that are highly specific and biologically active against a defined target of interest. Although many different assays are used in high-throughput screening, the general principle is that the specific molecular target or pathway of interest is combined systematically with each possible drug compound, then verified in manual or automated platforms for a positive result, or “hit.” The resulting “hits” would then be selected based on how strongly they interacted with the target, and additional confirmatory experiments would be performed to characterize the interaction.189 The incorporation of automation, computational methods, and nanotechnology has allowed for enormously increased efficiency in both the development of combinatorial libraries and the high-throughput screening of potentially useful drugs. To illustrate the rapidly accelerating success of this method, a recent paper has reported achieving ultrahigh-throughput screening speeds of 100 million individual enzyme reactions in 10 h using only small reagent volumes; this represents a 1000-fold increase in speed and a 1 million-fold reduction in cost compared with standard high-throughput techniques.190 A negative consequence of the massive amounts of data generated from high- and ultrahigh-throughput technologies is that bottlenecks will occur at the level of drug development and optimization. In the past decade, more than 3000 expressed proteins have been described as potential drug targets, yet only 20% of those have been exploited commercially.191 Therefore, despite the optimism engendered by high-throughput screening and other advances in drug discovery and validation, numerous obstacles remain for the translation of this vast amount of data into clinical application.
From the Side of the Host: Genetic Predisposition to Infectious Disease and Personalized Medicine
The human genome project brought about a conceptual shift in the focus of medicine, from treatment to the prediction of disease and prevention of side effects. The resulting principle is that if the manifestation of disease is influenced by interactions between the host and the infectious agent, then characterizing the host genetic correlates to disease would help to predict individual susceptibility to specific infection-related pathologies. The recent technological advances in sequencing and computational biology have facilitated the identification of host factors that predispose or affect the response to disease. Therefore, therapies can now be tailored according to the genetic makeup of the host and the characteristics of the microbe responsible for disease. Francis Collins suggested that genetics could be integrated with medicine either by using classic or “old” genetics to develop genetic tests for monogenic diseases and prenatal diagnostics or by using “new” genetics to predict patient responses and implement personalized medicine.192 The clinical application of personalized medicine principles may have seemed almost visionary a few years ago, but now this approach is used routinely in the treatment of some diseases. For example, genomic data are used to make accurate predictions about the susceptibility of a patient to disease or about how he/she will respond to treatment. Specifically, the application of personalized medicine is common in diseases such as cancer caused by some solid tumors or hematologic malignancies where patients are genetically tested to determine their likelihood of responding to therapy or having a serious adverse reaction to various drugs.193, 194
Personalized medicine has also been applied to the field of infectious disease to define host predisposition to some infections and to predict successful responses as well as adverse reactions to treatment. Evidence that certain infectious diseases have a genetic predisposition initially came from a study that showed an approximately 6-fold increase in the risk of mortality from infectious diseases in adopted children if the biological parents also died from an infection.195 Other groups later confirmed these results, and since then, the susceptibility to several infectious diseases, including S. aureus and M. tuberculosis, has been linked to rare defects in genes encoding effectors of the immune response.152, 196, 197, 198, 199, 200, 201, 202, 203 The completion of the Human Genome Project and the map of human genetic variation204 has promoted the evolution of highdensity SNP arrays that analyze the association of certain polymorphisms with susceptibility to infectious disease, as well as the linkage disequilibrium, or correlation between 2 SNPs. These increasingly common genome-wide association studies (GWAS) have identified a series of genes implicated in the predisposition of different bacterial and viral diseases.205, 206
Although GWAS was first applied to the fields of rheumatic and autoimmune diseases, recently this approach has identified novel genetic loci involved in susceptibility to infectious disease. In addition, the use of GWAS is beginning to be demonstrated in the identification of markers that indicate the quality of a patient’s response to treatment. For example, a recent GWAS showed that an SNP located upstream of the IL28B gene, which codes for type III interferon, is associated strongly with more than a 2-fold difference in response to anti-HCV drug treatment.207 Other studies showed a relationship between IL28B gene polymorphisms and the spontaneous clearance of acute HCV.208 Because HCV, which almost invariably leads to a chronic disease with liver failure in approximately 70% of individuals, is treated most effectively during the acute phase of the disease, this study was a significant step toward allowing physicians to predict spontaneous clearance accurately and to identify a subgroup of patients that will benefit from early treatment.208
Microarray technology has paved the way for the development of the field of transcriptomics, which is the large-scale profiling of all RNA transcripts in a cell to characterize gene expression (Fig 1). Transcriptomics has been applied widely to the study of the host response and identification of biomarkers after an infection or treatment. In this case, patterns of gene expression in human cells, most frequently peripheral blood mononuclear cells (PBMCs), are studied by hybridizing biological material to defined gene chips.209, 210, 211 With regard to infectious disease, microarray technology has made a huge contribution to our understanding of the changes evident in the transcriptome of humans during infection. Lempicki et al212 used microarray-derived gene expression patterns to demonstrate that the increased expression of interferon-stimulated genes in patient PBMCs could be one explanation for why anti-HCV therapy in patients coinfected with HCV and HIV has lower success rates. In another study, Jacobsen et al213 used microarray technology to identify the biomarkers expressed during infection with M. tuberculosis that would allow for more accurate predictions regarding the outcome of infection. Thus, microarray technology has been broadly applicable in studies aimed at characterizing the infection profiles of a variety of microorganisms, including SARS-COV,214 dengue,215 influenza,216 S. aureus, 217 and S. enterica. 218 The advent of next-generation deep and ultra-deep sequencing techniques has also improved the ability of researchers to identify splice variants and low abundance transcriptomic changes in response to infectious disease.49 Data such as these can be used to inform improvements in the diagnosis and prognosis of disease, the design of antiviral and antibiotic therapies, and the construction of vaccines that can target susceptible populations more specifically.
The application of new genomic techniques in predicting the response of a patient to drug therapy based on his/her genetic profile (pharmacogenomics) has found practical application in identifying populations that are susceptible to short- and long-term toxicities of specific HIV antiretroviral drugs. One example is the reaction to abacavir, which is a nucleoside analog reverse transcriptase inhibitor. The major toxicity manifests as a hypersensitivity reaction that occurs in approximately 5% to 8% of recipients within 6 weeks of commencing therapy.219 Mallal et al220 showed that patients carrying the HLA-B5701 allele are more likely to experience this hypersensitivity reaction to abacavir. This marker had a negative predictive value of 100%, indicating that genetic tests can provide unequivocal information that can help to predict and prevent otherwise unpredictable drug reactions. Another example of the use of pharmacogenomics to guide therapy is in the case of the antiretroviral drug maraviroc, which is a CCR5-specific HIV entry inhibitor. This drug is effective only in patients who are infected with strains of HIV that use the CCR5 coreceptor for entry, but not in patients who are infected with the CCR4-tropic viruses. Therefore, by screening for the specific tropism of the strain of HIV with which the patient is infected, clinicians can target drug therapy more specifically, for example, by using maraviroc only in patients who are infected with the susceptible strain of HIV.221 These results underline the success of GWAS and personalized medicine in predicting which populations should avoid treatment with specific drugs because they are more prone to side effects.
Although the clinical application of “omics” and systems biology holds great potential for medical discovery, the introduction of high-throughput technologies has also introduced new questions and challenges regarding the transformation of the large volumes of data produced by these approaches into useful information for the development of diagnostic, prognostic, and therapeutic devices. The “large n, small p” paradigm, in which the number of study subjects (n) is higher than the variables (p) generated from the study itself, currently serves as a fundamental analytic principle for data modeling. With a shift in this ratio toward smaller n values compared with the large number of variables amassed through high-throughput data acquisition, it is becoming clear that this paradigm will need to be revised to preserve statistical accuracy.222 To address this problem, some researchers have proposed changes in the mathematical principles used for data analysis, such as using continuous variables222, 223, 224 and nonlinear modeling.222, 225, 226 Some also recommend switching from deterministic to probability-based modeling strategies to account for the uncertainty that is typical of common clinical scenarios and to predict the relative probabilities of various outcomes given the same initial situation.222, 227 In addition to the necessary evolution of data modeling strategies, another critical factor in the successful translation of research data is the capability to access and analyze large sets of data that originated from different sources and contain diverse types of information. There is, therefore, a need to develop methods that enable the construction of hybrid data sets that contain both clinical and biological data, and are organized in ways that are accessible to all translational researchers and compatible among different systems. Translation bioinformatics is an emerging area that proposes to offer a holistic approach to this issue by providing integrative methods for incorporating multiple data modalities and enabling the development of predictive models for therapeutic responses.228 Although a detailed description of the methods used for data integration and modeling is beyond the scope of our article, other articles by Yan,228 Kanehisa et al,229 and Sarkar et al230 offer a more complete review of the topic. These developments are still the object of research and far from completion, but as the data accumulate, only the full development of new analytic strategies will enable their full diagnostic and clinical use.
Here, we mentioned some advances in translational research that are applicable to infectious disease studies. There have been several other major achievements in translational research with other diseases; some have been especially relevant in the fields of cancer and neuroscience, specifically with regard to epigenomics and stem cell research. Although a thorough discussion of these fields is beyond the scope of this article, their impact on translational research has also been profound. Regardless of the field of study or research technique, however, the promise of translational research remains hindered by a few key factors, which we will now discuss.
Bridging the Gaps: Turning Scientific Promise into Clinical Practice
Whereas the aforementioned studies illustrate the incredible promise of new technologies in translational research, their full potential remains largely untapped partly because of the high failure rates, long timelines, and costs of converting basic science research into clinically applicable methodologies. The recently proposed NCATS, a new NIH entity that would include the CTSA (and eliminate the NCRR), is an effort to circumvent these roadblocks. In the remainder of this review, we will discuss how time frames, costs, and poor success rates have persistently hindered the translation of basic research advances into clinically useful products, how the newly proposed NCATS aims to circumvent these roadblocks, and the additional challenges facing the field of translational medicine.8, 11
Failure rates
Over the past decade, the number of NMEs approved by the FDA and comparable regulatory bodies around the world has decreased progressively. In the second half of the 1990s, for example, the FDA approved more than 30 NMEs per year.231 By contrast, the number of NMEs approved since 2001 has averaged slightly less than 23 per year, with the exception of 36 approved in 2004, despite the increasing number of potential therapeutic compounds identified from high-throughput technologies.231, 232, 233 In addition to the decline in approvals, the number of NME applications submitted to the FDA over the past decade has decreased, with 2010 producing one of the lowest numbers in more than 15 years.233 Because of this decline in approvals and applications, as well as the increasing use of generic drugs, it is estimated that large cap pharmaceutical companies only can replace, on average, 26 cents to every dollar lost to patent expirations by new product revenues.234 According to current estimates, only 8% of NMEs will make it from candidate selection (in the preclinical stage) to a successful launch.232, 235 This decline in drug development threatens to take not only an economic toll but also a medical one, as approximately 40% of the 2-year increase in life expectancy from 1986 to 2000 was attributable to the introduction of new drugs.236 This lack of new drugs is especially worrisome in the field of infectious disease, where the emergence of multidrug resistant pathogens has led to a situation in which formerly curable infections are now untreatable. Paradoxically, some pharmaceutical companies have indicated that they are curtailing or curbing anti-infective research.10 A major reason for this is that anti-infective agents, which are only needed for a short time in smaller populations, make less economically attractive targets than drugs targeting chronic diseases that appear in the larger aging population (eg, antihypertensives and antidepressants).
At this time, approximately 85% of the failures in translating promising NMEs into clinical application occur during early clinical trials. According to a recent editorial,237 many of these NMEs fail because of outdated clinical trial designs. Drug testing requires the study of large sample populations to show statistically significant benefits compared with existing pharmaceuticals. The introduction of specific markers for personalized medicine makes it even more difficult to recruit patients who qualify for these tests. Therefore, many clinical trials take years to complete their enrollment, or they are cancelled because of the inability to recruit a sufficient number of subjects. These failures therefore contribute to the total costs and failure rates of translational research. As a result, many NMEs are never developed, and companies are removing more products from their pipelines than ever before.237
To circumvent this problem and increase the success rate of proposed NMEs, Francis Collins has proposed several priorities of the new NCATS. First, the NCATS will support studies that investigate broadly applicable, rather than disease-specific, target validation approaches (ie, targets that may be relevant to multiple diseases), as well as targets that are considered to be “too risky” for industry investment, such as those identified by GWAS. Second, the NCATS will encourage the use of “omics” and systems biology approaches in order to help design and validate new diagnostics and therapeutics for translation into clinical application. Third, the NCATS would also house the recently established NIH-FDA Regulatory Science Initiative, which was launched in 2010 with the goals of facilitating improvements in regulatory science and providing new insights to broadly benefit the field of translational research.8, 238, 239, 240 Finally, together with the NIH-EPA-FDA Tox21 Consortium, the NCATS would support the use of new cell-based approaches as a means of pursuing preclinical toxicology studies.241
Timelines
By current estimates, the average timeline for discovering and developing an NME is approximately 13.5 years. This estimate includes the regulatory review but not the time it takes to identify and validate the target pathway.232 Time spent in clinical trials, unnecessary bureaucratic procedures associated with research and design management, and inefficient regulatory review have all been identified as contributors to this problem.9, 232, 242, 243 To minimize these problems, the proposed NCATS would seek to encourage the “reengineering of the translational process, from initial target identification to first-in-human application of small molecules, biologics, diagnostics, and devices” to streamline this process and attract pharmaceutical involvement.8 Additionally, the center would aim to act as an honest broker, hastening Institutional Review Board (IRB) approval for multicenter trials, further decreasing the time that NMEs spend in clinical trials and facilitating the rescue and repurposing of abandoned compounds. Last, the center would support collaborative efforts for the testing of new entities to act as a “proof of concept” in the hopes that promising new drugs would be picked up by industry.8, 244
Costs
The cost of a translational endeavor is inexorably linked to both high failure rates and long timelines. Indeed, the average cost to bring an NME to market is now estimated to be $1.8 billion. This estimate seems to be the same regardless of whether the NME is produced by a big pharmaceutical company or small biotechnology firm.232 The NCATs aim to decrease these costs in a number of ways. First, it would encourage the use of early human trials, also called “phase zero” clinical trials, in which “microdoses” of a new drug are administered in a small number of patients.237 In these trials, the absorption, tissue distribution, and toxicity of the drug are assessed by highly sensitive methodologies, such as molecular imaging, metabolomics, or proteomics.8 The feasibility of such endeavors has been demonstrated already by the Consortium for Metabonomic Toxicology at the Imperial College in London, which showed it is possible to design a prediction model for clinical toxicity using metabolomic data.104 Another way in which the NCATS would decrease development costs is by encouraging innovative models, including human tissue biobanks, stem cell models, and tissue-engineered organoids. These models would potentially allow researchers to decrease or omit the use of animal models, which have been criticized as both costly and not accurately predictive of efficacy in humans.8, 237 Finally, the NCATS would encourage the development of technologies aimed at biomarker identification. These have been used already by researchers to design a new style of “adaptive” clinical trial, in which the study design changes as new data are collected, allowing patients who are more likely to respond to be assigned to treatment with a given compound.237 Beyond encouraging the development of new technologies aimed at decreasing costs, the NCATS would promote cross-collaborations between academic institutions, biotechnology firms, philanthropic organizations, and pharmaceutical companies to split research and development costs. Last, the proposed Cures Acceleration Network, which will be assessed for congressional funding in the next fiscal year, would provide the NIH with the ability to award up to $15 billion per year to academic and private consortia to direct the development of certain especially promising therapeutics.8
Are NCATS the Future of Translational Research?
Although the need to encourage the development of new drugs and therapeutics is clear, some have expressed concerns that the proposed NCATS represents a change in priority for the NIH away from basic, discovery-based science and toward application-focused research, thus neglecting the fact that basic science lays the foundation for translational research. Indeed, this has been a criticism since the development of the CTSA.245 Another criticism is that the support of GWAS and other high-throughput methodologies causes a dramatic shift from research that is hypothesis based to that which is data driven, placing an overemphasis on screening for drugs and biomarkers at the expense of discovery-based science.246 For example, studies based on high-throughput technologies are typically not the result of experiments designed to test a hypothesis. Rather, they aim to identify systematically all the molecular effectors that are involved in a specific system in the most comprehensive and efficient way possible. To address this critique, some authors have added an extra step of complexity to their high-throughput methods by determining a mechanistic function for the molecules they have identified, switching in this way from “fishing expedition” to discovery-based research. In the end, despite assertions that the establishment of the NCATS will reinforce, rather than detract from basic science, many investigators continue to have concerns, mostly related to money. In today’s climate of funding cuts, some wonder: Will it be possible to receive funding for more basic (ie, less obviously applicable) research? Will the emphasis on collaboration and multidisciplinary approaches shift most of the funding toward multicenter groups of established scientists, making it difficult for new investigators to survive? Will the current emphasis on clinical applicability result in a narrowing of ideas and perspectives that will result in most funding being allocated to well-established scientists pursuing traditional approaches? Beyond these concerns regarding a change in the focus of NIH, concerns regarding policy and infrastructure exist, such as whether funding NCATS programs will take money away from other programs that were previously housed within the NCRR. As the NCRR currently awards funds of approximately $1.25 billion annually, this is a substantial concern.247 Although none of the NCRR programs are currently slated to be discontinued, the perceived lack of transparency surrounding the establishment of the NCATS, the dissolution of the NCRR, and where programs formerly housed within the NCRR will now be located has resulted in persistent scrutiny from both NIH stakeholders and Congress.248, 249, 250
Perhaps the greatest issue of debate is where to draw the line between public and private industry. Many criticize the proposed NCATS because they believe it is designed to help companies develop new drugs by decreasing their financial risks, and it is believed that it will do so by using taxpayers’ money. Also, they argue that the NIH only has experience with basic science research and not with drug development. Indeed, some have argued that the decline in research and development productivity is not related to increasing costs, because public investment in the pharmaceutical industry has increased proportionally since 1970, nor is it to the result of time or attritional rates, which have been steady since the 1970s. Instead, they argue that it may be caused by poor prioritization and mismanagement within the pharmaceutical industry.9 Specifically, changes in management techniques in the pharmaceutical industry hierarchy have led to corporate policies that discourage innovation in favor of cost cutting and risk minimization.9, 246 This together with a shift of control from scientists and researchers to marketers, a focus on short-term profits and fast sales growth, and the discontinuation of compounds for nontechnical reasons, have led to the decreased translation of scientific discoveries to clinical applications.9 As such, some have argued that the development of the NCATS might make it even less likely that the pharmaceutical industry will try to solve these problems. Ultimately, although the NCATS might serve as a champion for key therapeutic compounds, the pharmaceutical industry itself will need to reform certain practices if the pace of drug development is to keep up with the pace of translational research.
Translational research has made incredible progress in recent years, both with the development of new techniques and with the expansion of more holistic ways of viewing the data that stem from those approaches. For the most part, the discovery of novel technologies, the development of new infrastructures, and the training of budding scientists have supported this evolution. The transition is not complete and roadblocks still exist on the path to scientific progress. It remains to be seen whether the newly proposed NCATS, which has raised as many objections as it has hopes, will be the answer to these problems. What is evident, however, is that translational research must be reprioritized and bolstered in a way that increases the speed at which promising drugs and technologies move through the pipeline into clinical application, while at the same time refraining from stifling the breadth of ideas and creativity of basic researchers, which is necessary for the progression of science itself.
Acknowledgments
We thank Ann Beeder, MD for useful discussion.
Footnotes
Supported by U.S. NIAID Grant 1R21AI073926-01A2 and by NIH Grant UL1RR024996 (for work in the MS laboratory); and by NIHKL2RR024997 (to E.A.).
References
- 1.Lost in clinical translation. Nat Med. 2004;10:879. doi: 10.1038/nm0904-879. [No authors listed] [DOI] [PubMed] [Google Scholar]
- 2.Woolf S.H. The meaning of translational research and why it matters. JAMA. 2008;299:211–213. doi: 10.1001/jama.2007.26. [DOI] [PubMed] [Google Scholar]
- 3.National Center for Research Resources. Clinical and Translational Science Awards. 2011.
- 4.National Institutes of Health . NIH; Bethesda, Md: 2011. National Center for Research Resources. [Google Scholar]
- 5.Steele S.J. Working with the CTSA Consortium: what we bring to the table. Sci Transl Med. 2010;2:63mr5. doi: 10.1126/scitranslmed.3001635. [DOI] [PubMed] [Google Scholar]
- 6.Pienta K.J., Spork A.L., Scheske J. The Clinical and Translational Science Awards (CTSAs) are transforming the way academic medical institutions approach translational research: the University of Michigan Experience. Clin Transl Sci. 2011;4:233–235. doi: 10.1111/j.1752-8062.2011.00319.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schubert C. Four years on, clinical partnerships program proves worth. Nat Med. 2010;16:612–613. doi: 10.1038/nm0610-612a. [DOI] [PubMed] [Google Scholar]
- 8.Collins F.S. Reengineering translational science: the time is right. Sci Transl Med. 2011;3:90cm17. doi: 10.1126/scitranslmed.3002747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cuatrecasas P. Drug discovery in jeopardy. J Clin Invest. 2006;116:2837–2842. doi: 10.1172/JCI29999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Spellberg B., Powers J.H., Brass E.P., Miller L.G., Edwards J.E., Jr. Trends in antimicrobial drug development: implications for the future. Clin Infect Dis. 2004;38:1279–1286. doi: 10.1086/420937. [DOI] [PubMed] [Google Scholar]
- 11.Harris G. Federal research center will help develop medicines. New York Times. January 23, 2011:A1. [Google Scholar]
- 12.Office of Inspector General . Department of Health and Human Services; Washington, DC: 1983. The orphan drug act: implementation and impact. 13. [Google Scholar]
- 13.Watson J.D., Cook-Deegan R.M. Origins of the human genome project. FASEB J. 1991;5:8–11. doi: 10.1096/fasebj.5.1.1991595. [DOI] [PubMed] [Google Scholar]
- 14.Springer; Berlin, Germany: 1997. Proteome Research: New Frontiers in Functional Genomics. [Google Scholar]
- 15.Oliver S.G., Winson M.K., Kell D.B., Baganz F. Systematic functional analysis of the yeast genome. Trends Biotechnol. 1998;16:373–378. doi: 10.1016/s0167-7799(98)01214-1. [DOI] [PubMed] [Google Scholar]
- 16.Hutchison C.A., 3rd DNA sequencing: bench to bedside and beyond. Nucleic Acids Res. 2007;35:6227–6237. doi: 10.1093/nar/gkm688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Maxam A.M., Gilbert W. A new method for sequencing DNA. Proc Natl Acad Sci U S A. 1977;74:560–564. doi: 10.1073/pnas.74.2.560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sanger F., Nicklen S., Coulson A.R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gulick R.M. Antiretroviral treatment 2010: progress and controversies. J Acquir Immune Defic Syndr. 2010;55:S43–S48. doi: 10.1097/QAI.0b013e3181f9c09e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Deng L., Tang H. Hepatitis B virus drug resistance to current nucleos(t)ide analogs: mechanisms and mutation sites. Hepatol Res. 2011;41:1017–1024. doi: 10.1111/j.1872-034X.2011.00873.x. [DOI] [PubMed] [Google Scholar]
- 21.Lin C.L., Kao J.H. The clinical implications of hepatitis B virus genotype: recent advances. J Gastroenterol Hepatol. 2011;26:123–130. doi: 10.1111/j.1440-1746.2010.06541.x. [DOI] [PubMed] [Google Scholar]
- 22.Vivekanandan P., Singh O.V. Molecular methods in the diagnosis and management of chronic hepatitis B. Expert Rev Mol Diagn. 2010;10:921–935. doi: 10.1586/erm.10.75. [DOI] [PubMed] [Google Scholar]
- 23.Hiraga N., Imamura M., Abe H. Rapid emergence of telaprevir resistant hepatitis C virus strain from wildtype clone in vivo. Hepatology. 2011;54:781–788. doi: 10.1002/hep.24460. [DOI] [PubMed] [Google Scholar]
- 24.Susser S., Vermehren J., Forestier N. Analysis of long-term persistence of resistance mutations within the hepatitis C virus NS3 protease after treatment with telaprevir or boceprevir. J Clin Virol. 2011;52:321–327. doi: 10.1016/j.jcv.2011.08.015. [DOI] [PubMed] [Google Scholar]
- 25.Kieffer T.L., Kwong A.D., Picchio G.R. Viral resistance to specifically targeted antiviral therapies for hepatitis C (STAT-Cs) J Antimicrob Chemother. 2010;65:202–212. doi: 10.1093/jac/dkp388. [DOI] [PubMed] [Google Scholar]
- 26.Rosen H.R. Clinical practice. Chronic hepatitis C infection. N Engl J Med. 2011;364:2429–2438. doi: 10.1056/NEJMcp1006613. [DOI] [PubMed] [Google Scholar]
- 27.Duan S., Boltz D.A., Li J. Novel genotyping and quantitative analysis of neuraminidase inhibitor resistance-associated mutations in influenza a viruses by single-nucleotide polymorphism analysis. Antimicrob Agents Chemother. 2011;55:4718–4727. doi: 10.1128/AAC.00316-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tong S.Y., Dakh F., Hurt A.C. Rapid detection of the H275Y oseltamivir resistance mutation in influenza A/H1N1 2009 by single base pair RT-PCR and high-resolution melting. PLoS One. 2011;6:e21446. doi: 10.1371/journal.pone.0021446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wijaya L., Chua Y.Y., Cui L., Chan K., Tan B.H. Intravenous zanamivir in critically ill patients due to pandemic 2009 (H1N1) influenza A virus. Singapore Med J. 2011;52:481–485. [PubMed] [Google Scholar]
- 30.Grondahl B., Puppe W., Hoppe A. Rapid identification of nine microorganisms causing acute respiratory tract infections by single-tube multiplex reverse transcription-PCR: feasibility study. J Clin Microbiol. 1999;37:1–7. doi: 10.1128/jcm.37.1.1-7.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Edwards M.C., Gibbs R.A. Multiplex PCR: advantages, development, and applications. PCR Methods Appl. 1994;3:S65–S75. doi: 10.1101/gr.3.4.s65. [DOI] [PubMed] [Google Scholar]
- 32.Uttamchandani M., Neo J.L., Ong B.N., Moochhala S. Applications of microarrays in pathogen detection and biodefence. Trends Biotechnol. 2009;27:53–61. doi: 10.1016/j.tibtech.2008.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang D., Coscoy L., Zylberberg M. Microarray-based detection and genotyping of viral pathogens. Proc Natl Acad Sci U S A. 2002;99:15687–15692. doi: 10.1073/pnas.242579699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang D., Urisman A., Liu Y.T. Viral discovery and sequence recovery using DNA microarrays. PLoS Biol. 2003;1:E2. doi: 10.1371/journal.pbio.0000002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Palacios G., Quan P.L., Jabado O.J. Panmicrobial oligonucleotide array for diagnosis of infectious diseases. Emerg Infect Dis. 2007;13:73–81. doi: 10.3201/eid1301.060837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Anderson N.G., Gerin J.L., Anderson N.L. Global screening for human viral pathogens. Emerg Infect Dis. 2003;9:768–774. doi: 10.3201/eid0907.030004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lipkin W.I. Microbe hunting. Microbiol Mol Biol Rev. 2010;74:363–377. doi: 10.1128/MMBR.00007-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rota P.A., Oberste M.S., Monroe S.S. Characterization of a novel coronavirus associated with severe acute respiratory syndrome. Science. 2003;300:1394–1399. doi: 10.1126/science.1085952. [DOI] [PubMed] [Google Scholar]
- 39.Mihindukulasuriya K.A., Wu G., St Leger J., Nordhausen R.W., Wang D. Identification of a novel coronavirus from a beluga whale by using a panviral microarray. J Virol. 2008;82:5084–5088. doi: 10.1128/JVI.02722-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kistler A., Avila P.C., Rouskin S. Pan-viral screening of respiratory tract infections in adults with and without asthma reveals unexpected human coronavirus and human rhinovirus diversity. J Infect Dis. 2007;196:817–825. doi: 10.1086/520816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chiu C.Y., Greninger A.L., Kanada K. Identification of cardioviruses related to Theiler’s murine encephalomyelitis virus in human infections. Proc Natl Acad Sci U S A. 2008;105:14124–14129. doi: 10.1073/pnas.0805968105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Connell C., Fung S., Heiner C. Automated DNA-sequence analysis. BioTechniques. 1987;5:342–348. [Google Scholar]
- 43.Brenner S., Johnson M., Bridgham J. Gene expression analysis by massively parallel signature sequencing (MPSS) on microbead arrays. Nat Biotechnol. 2000;18:630–634. doi: 10.1038/76469. [DOI] [PubMed] [Google Scholar]
- 44.Stein RA. Next-generation sequencing update. Genetic Engineering & Biotechnology News 2008;28.
- 45.Margulies M., Egholm M., Altman W.E. Genome sequencing in microfabricated high-density picolitre reactors. Nature. 2005;437:376–380. doi: 10.1038/nature03959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ansorge W.J. Next-generation DNA sequencing techniques. N Biotechnol. 2009;25:195–203. doi: 10.1016/j.nbt.2008.12.009. [DOI] [PubMed] [Google Scholar]
- 47.Nakamura S., Yang C.S., Sakon N. Direct metagenomic detection of viral pathogens in nasal and fecal specimens using an unbiased high-throughput sequencing approach. PLoS One. 2009;4:e4219. doi: 10.1371/journal.pone.0004219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sorber K., Chiu C., Webster D. The long march: a sample preparation technique that enhances contig length and coverage by high-throughput short-read sequencing. PLoS One. 2008;3:e3495. doi: 10.1371/journal.pone.0003495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rothberg J.M., Leamon J.H. The development and impact of 454 sequencing. Nat Biotechnol. 2008;26:1117–1124. doi: 10.1038/nbt1485. [DOI] [PubMed] [Google Scholar]
- 50.Palacios G., Druce J., Du L. A new arenavirus in a cluster of fatal transplant-associated diseases. N Engl J Med. 2008;358:991–998. doi: 10.1056/NEJMoa073785. [DOI] [PubMed] [Google Scholar]
- 51.Briese T., Paweska J.T., McMullan L.K. Genetic detection and characterization of Lujo virus, a new hemorrhagic fever-associated arenavirus from southern Africa. PLoS Pathog. 2009;5:e1000455. doi: 10.1371/journal.ppat.1000455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Greninger A.L., Runckel C., Chiu C.Y. The complete genome of klassevirus - a novel picornavirus in pediatric stool. Virol J. 2009;6:82. doi: 10.1186/1743-422X-6-82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Feng H., Shuda M., Chang Y., Moore P.S. Clonal integration of a polyomavirus in human Merkel cell carcinoma. Science. 2008;319:1096–1100. doi: 10.1126/science.1152586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Chin C.S., Sorenson J., Harris J.B. The origin of the Haitian cholera outbreak strain. N Engl J Med. 2011;364:33–42. doi: 10.1056/NEJMoa1012928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Garten R.J., Davis C.T., Russell C.A. Antigenic and genetic characteristics of swine-origin 2009 A(H1N1) influenza viruses circulating in humans. Science. 2009;325:197–201. doi: 10.1126/science.1176225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Bell J.F., Wright J.T., Habel K. Rabies vaccine freed of the factor causing allergic encephalitis. Proc Soc Exp Biol Med. 1949;70:457–461. doi: 10.3181/00379727-70-16960. [DOI] [PubMed] [Google Scholar]
- 57.Alexander L.N., Seward J.F., Santibanez T.A. Vaccine policy changes and epidemiology of poliomyelitis in the United States. JAMA. 2004;292:1696–1701. doi: 10.1001/jama.292.14.1696. [DOI] [PubMed] [Google Scholar]
- 58.Rappuoli R. Reverse vaccinology, a genome-based approach to vaccine development. Vaccine. 2001;19:2688–2691. doi: 10.1016/s0264-410x(00)00554-5. [DOI] [PubMed] [Google Scholar]
- 59.Danzig L. Reverse vaccinology–in search of a genome-derived meningococcal vaccine. Vaccine. 2006;24:11–12. doi: 10.1016/j.vaccine.2005.01.103. [DOI] [PubMed] [Google Scholar]
- 60.Yogev R., Tan T. Meningococcal disease: the advances and challenges of meningococcal disease prevention. Hum Vaccin. 2011;7:828–837. doi: 10.4161/hv.7.8.16270. [DOI] [PubMed] [Google Scholar]
- 61.Pizza M., Scarlato V., Masignani V. Identification of vaccine candidates against serogroup B meningococcus by whole-genome sequencing. Science. 2000;287:1816–1820. doi: 10.1126/science.287.5459.1816. [DOI] [PubMed] [Google Scholar]
- 62.Tettelin H., Medini D., Donati C., Masignani V. Towards a universal group B Streptococcus vaccine using multistrain genome analysis. Expert Rev Vaccines. 2006;5:687–694. doi: 10.1586/14760584.5.5.687. [DOI] [PubMed] [Google Scholar]
- 63.Subbarao K., Katz J.M. Influenza vaccines generated by reverse genetics. Curr Top Microbiol Immunol. 2004;283:313–342. doi: 10.1007/978-3-662-06099-5_9. [DOI] [PubMed] [Google Scholar]
- 64.Steel J. New strategies for the development of H5N1 subtype influenza vaccines: progress and challenges. BioDrugs. 2011;25:285–298. doi: 10.1007/BF03256169. [DOI] [PubMed] [Google Scholar]
- 65.Wacheck V., Egorov A., Groiss F. A novel type of influenza vaccine: safety and immunogenicity of replication-deficient influenza virus created by deletion of the interferon antagonist NS1. J Infect Dis. 2010;201:354–362. doi: 10.1086/649428. [DOI] [PubMed] [Google Scholar]
- 66.Emonet S.E., Urata S., de la Torre J.C. Arenavirus reverse genetics: new approaches for the investigation of arenavirus biology and development of antiviral strategies. Virology. 2011;411:416–425. doi: 10.1016/j.virol.2011.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.McGettigan J.P. Experimental rabies vaccines for humans. Expert Rev Vaccines. 2010;9:1177–1186. doi: 10.1586/erv.10.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Horimoto T., Kawaoka Y. Designing vaccines for pandemic influenza. Curr Top Microbiol Immunol. 2009;333:165–176. doi: 10.1007/978-3-540-92165-3_8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Richt J.A., Garcia-Sastre A. Attenuated influenza virus vaccines with modified NS1 proteins. Curr Top Microbiol Immunol. 2009;333:177–195. doi: 10.1007/978-3-540-92165-3_9. [DOI] [PubMed] [Google Scholar]
- 70.Bouloy M., Flick R. Reverse genetics technology for Rift Valley fever virus: current and future applications for the development of therapeutics and vaccines. Antiviral Res. 2009;84:101–118. doi: 10.1016/j.antiviral.2009.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Billeter M.A., Naim H.Y., Udem S.A. Reverse genetics of measles virus and resulting multivalent recombinant vaccines: applications of recombinant measles viruses. Curr Top Microbiol Immunol. 2009;329:129–162. doi: 10.1007/978-3-540-70523-9_7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Draper S.J., Heeney J.L. Viruses as vaccine vectors for infectious diseases and cancer. Nat Rev Microbiol. 2010;8:62–73. doi: 10.1038/nrmicro2240. [DOI] [PubMed] [Google Scholar]
- 73.Rollier C.S., Reyes-Sandoval A., Cottingham M.G., Ewer K., Hill A.V. Viral vectors as vaccine platforms: deployment in sight. Curr Opin Immunol. 2011;23:377–382. doi: 10.1016/j.coi.2011.03.006. [DOI] [PubMed] [Google Scholar]
- 74.Sofer-Podesta C., Ang J., Hackett N.R. Adenovirus-mediated delivery of an anti-V antigen monoclonal antibody protects mice against a lethal Yersinia pestis challenge. Infect Immun. 2009;77:1561–1568. doi: 10.1128/IAI.00856-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Kasuya K., Boyer J.L., Tan Y. Passive immunotherapy for anthrax toxin mediated by an adenovirus expressing an anti-protective antigen single-chain antibody. Mol Ther. 2005;11:237–244. doi: 10.1016/j.ymthe.2004.10.018. [DOI] [PubMed] [Google Scholar]
- 76.Sparkman D. 2nd ed. Global View Publishing; Pittsburgh, Pa: 2006. Mass spectrometry desk reference. [Google Scholar]
- 77.Aebersold R., Mann M. Mass spectrometry-based proteomics. Nature. 2003;422:198–207. doi: 10.1038/nature01511. [DOI] [PubMed] [Google Scholar]
- 78.Blackstock W.P., Weir M.P. Proteomics: quantitative and physical mapping of cellular proteins. Trends Biotechnol. 1999;17:121–127. doi: 10.1016/s0167-7799(98)01245-1. [DOI] [PubMed] [Google Scholar]
- 79.Roe M.R., Griffin T.J. Gel-free mass spectrometry-based high throughput proteomics: tools for studying biological response of proteins and proteomes. Proteomics. 2006;6:4678–4687. doi: 10.1002/pmic.200500876. [DOI] [PubMed] [Google Scholar]
- 80.Brewis I.A., Brennan P. Proteomics technologies for the global identification and quantification of proteins. Adv Protein Chem Struct Biol. 2010;80:1–44. doi: 10.1016/B978-0-12-381264-3.00001-1. [DOI] [PubMed] [Google Scholar]
- 81.Malmstrom L., Malmstrom J., Aebersold R. Quantitative proteomics of microbes: principles and applications to virulence. Proteomics. 2011;11:2947–2956. doi: 10.1002/pmic.201100088. [DOI] [PubMed] [Google Scholar]
- 82.Alban A., David S.O., Bjorkesten L. A novel experimental design for comparative two-dimensional gel analysis: two-dimensional difference gel electrophoresis incorporating a pooled internal standard. Proteomics. 2003;3:36–44. doi: 10.1002/pmic.200390006. [DOI] [PubMed] [Google Scholar]
- 83.Ly L., Wasinger V.C. Protein and peptide fractionation, enrichment and depletion: tools for the complex proteome. Proteomics. 2011;11:513–534. doi: 10.1002/pmic.201000394. [DOI] [PubMed] [Google Scholar]
- 84.Carr P.W., Stoll D.R., Wang X. Perspectives on recent advances in the speed of high-performance liquid chromatography. Anal Chem. 2011;83:1890–1900. doi: 10.1021/ac102570t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Link A.J., Eng J., Schieltz D.M. Direct analysis of protein complexes using mass spectrometry. Nat Biotechnol. 1999;17:676–682. doi: 10.1038/10890. [DOI] [PubMed] [Google Scholar]
- 86.Eng J.K.M., McCormack A.L., Yates J.R. An approach to correlate tandem mass spectral data of peptides with amino acid sequences is a protein database. J Am Soc Mass Spectrom. 1994;5:976–989. doi: 10.1016/1044-0305(94)80016-2. [DOI] [PubMed] [Google Scholar]
- 87.Faherty B.K., Gerber S.A. MacroSEQUEST: efficient candidate-centric searching and high-resolution correlation analysis for large-scale proteomics data sets. Anal Chem. 2010;82:6821–6829. doi: 10.1021/ac100783x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Frohlich T., Arnold G.J. Proteome research based on modern liquid chromatography–tandem mass spectrometry: separation, identification and quantification. J Neural Transm. 2006;113:973–994. doi: 10.1007/s00702-006-0509-3. [DOI] [PubMed] [Google Scholar]
- 89.Chiou S.H., Wu C.Y. Clinical proteomics: current status, challenges, and future perspectives. Kaohsiung J Med Sci. 2011;27:1–14. doi: 10.1016/j.kjms.2010.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Elliott M.H., Smith D.S., Parker C.E., Borchers C. Current trends in quantitative proteomics. J Mass Spectrom. 2009;44:1637–1660. doi: 10.1002/jms.1692. [DOI] [PubMed] [Google Scholar]
- 91.Vogel C., Marcotte E.M. Calculating absolute and relative protein abundance from mass spectrometry-based protein expression data. Nat Protoc. 2008;3:1444–1451. doi: 10.1038/nport.2008.132. [DOI] [PubMed] [Google Scholar]
- 92.Matthiesen R., Carvalho A.S. Methods and algorithms for relative quantitative proteomics by mass spectrometry. Methods Mol Biol. 2010;593:187–204. doi: 10.1007/978-1-60327-194-3_10. [DOI] [PubMed] [Google Scholar]
- 93.Mueller L.N., Rinner O., Schmidt A. SuperHirn - a novel tool for high resolution LC-MS-based peptide/protein profiling. Proteomics. 2007;7:3470–3480. doi: 10.1002/pmic.200700057. [DOI] [PubMed] [Google Scholar]
- 94.Lange V., Picotti P., Domon B., Aebersold R. Selected reaction monitoring for quantitative proteomics: a tutorial. Mol Syst Biol. 2008;4:222. doi: 10.1038/msb.2008.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Kuzyk M.A., Smith D., Yang J. Multiple reaction monitoring-based, multiplexed, absolute quantitation of 45 proteins in human plasma. Mol Cell Proteomics. 2009;8:1860–1877. doi: 10.1074/mcp.M800540-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Lasonder E., Ishihama Y., Andersen J.S. Analysis of the Plasmodium falciparum proteome by high-accuracy mass spectrometry. Nature. 2002;419:537–542. doi: 10.1038/nature01111. [DOI] [PubMed] [Google Scholar]
- 97.Florens L., Washburn M.P., Raine J.D. A proteomic view of the Plasmodium falciparum life cycle. Nature. 2002;419:520–526. doi: 10.1038/nature01107. [DOI] [PubMed] [Google Scholar]
- 98.Rhee K.Y., Erdjument-Bromage H., Tempst P., Nathan C.F. S-nitroso proteome of Mycobacterium tuberculosis: enzymes of intermediary metabolism and antioxidant defense. Proc Natl Acad Sci U S A. 2005;102:467–472. doi: 10.1073/pnas.0406133102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Popova T.G., Turell M.J., Espina V. Reverse-phase phosphoproteome analysis of signaling pathways induced by Rift valley fever virus in human small airway epithelial cells. PLoS One. 2010;5:e13805. doi: 10.1371/journal.pone.0013805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Chan E.Y., Qian W.J., Diamond D.L. Quantitative analysis of human immunodeficiency virus type 1-infected CD4+ cell proteome: dysregulated cell cycle progression and nuclear transport coincide with robust virus production. J Virol. 2007;81:7571–7583. doi: 10.1128/JVI.00288-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Feng X., Zhang J., Chen W.N., Ching C.B. Proteome profiling of Epstein-Barr virus infected nasopharyngeal carcinoma cell line: identification of potential biomarkers by comparative iTRAQ-coupled 2D LC/MS-MS analysis. J Proteomics. 2011;74:567–576. doi: 10.1016/j.jprot.2011.01.017. [DOI] [PubMed] [Google Scholar]
- 102.Malmstrom J., Beck M., Schmidt A. Proteome-wide cellular protein concentrations of the human pathogen Leptospira interrogans. Nature. 2009;460:762–765. doi: 10.1038/nature08184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Francois P., Scherl A., Hochstrasser D., Schrenzel J. Proteomic approaches to study Staphylococcus aureus pathogenesis. J Proteomics. 2010;73:701–708. doi: 10.1016/j.jprot.2009.10.007. [DOI] [PubMed] [Google Scholar]
- 104.Lindon J.C., Holmes E., Nicholson J.K. Metabonomics in pharmaceutical R&D. Febs J. 2007;274:1140–1151. doi: 10.1111/j.1742-4658.2007.05673.x. [DOI] [PubMed] [Google Scholar]
- 105.Nicholson J.K., Wilson I.D., Lindon J.C. Pharmacometabonomics as an effector for personalized medicine. Pharmacogenomics. 2011;12:103–111. doi: 10.2217/pgs.10.157. [DOI] [PubMed] [Google Scholar]
- 106.van der Werf M.J., Jellema R.H., Hankemeier T. Microbial metabolomics: replacing trial-and-error by the unbiased selection and ranking of targets. J Ind Microbiol Biotechnol. 2005;32:234–252. doi: 10.1007/s10295-005-0231-4. [DOI] [PubMed] [Google Scholar]
- 107.Vinayavekhin N., Homan E.A., Saghatelian A. Exploring disease through metabolomics. ACS Chem Biol. 2010;5:91–103. doi: 10.1021/cb900271r. [DOI] [PubMed] [Google Scholar]
- 108.Henderson J.P., Crowley J.R., Pinkner J.S. Quantitative metabolomics reveals an epigenetic blueprint for iron acquisition in uropathogenic Escherichia coli. PLoS Pathog. 2009;5:e1000305. doi: 10.1371/journal.ppat.1000305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Munger J., Bennett B.D., Parikh A. Systems-level metabolic flux profiling identifies fatty acid synthesis as a target for antiviral therapy. Nat Biotechnol. 2008;26:1179–1186. doi: 10.1038/nbt.1500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Aoki-Kinoshita K.F. An introduction to bioinformatics for glycomics research. PLoS Comput Biol. 2008;4:e1000075. doi: 10.1371/journal.pcbi.1000075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Wenk M.R. The emerging field of lipidomics. Nat Rev Drug Discov. 2005;4:594–610. doi: 10.1038/nrd1776. [DOI] [PubMed] [Google Scholar]
- 112.Wenk M.R. Lipidomics: new tools and applications. Cell. 2010;143:888–895. doi: 10.1016/j.cell.2010.11.033. [DOI] [PubMed] [Google Scholar]
- 113.Wenk M.R. Lipidomics of host-pathogen interactions. FEBS Lett. 2006;580:5541–5551. doi: 10.1016/j.febslet.2006.07.007. [DOI] [PubMed] [Google Scholar]
- 114.Srivastava S. Move over proteomics, here comes glycomics. J Proteome Res. 2008;7:1799. doi: 10.1021/pr083696k. [DOI] [PubMed] [Google Scholar]
- 115.Hart G.W., Copeland R.J. Glycomics hits the big time. Cell. 2010;143:672–676. doi: 10.1016/j.cell.2010.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Hameed S., Dhamgaye S., Singh A., Goswami S.K., Prasad R. Calcineurin signaling and membrane lipid homeostasis regulates iron mediated multidrug resistance mechanisms in Candida albicans. PLoS One. 2011;6:e18684. doi: 10.1371/journal.pone.0018684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Brugger B., Glass B., Haberkant P. The HIV lipidome: a raft with an unusual composition. Proc Natl Acad Sci U S A. 2006;103:2641–2646. doi: 10.1073/pnas.0511136103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Brugger B., Krautkramer E., Tibroni N. Human immunodeficiency virus type 1 Nef protein modulates the lipid composition of virions and host cell membrane microdomains. Retrovirology. 2007;4:70. doi: 10.1186/1742-4690-4-70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Liu S.T., Sharon-Friling R., Ivanova P. Synaptic vesicle-like lipidome of human cytomegalovirus virions reveals a role for SNARE machinery in virion egress. Proc Natl Acad Sci U S A. 2011;108:12869–12874. doi: 10.1073/pnas.1109796108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Diamond D.L., Syder A.J., Jacobs J.M. Temporal proteome and lipidome profiles reveal hepatitis C virus-associated reprogramming of hepatocellular metabolism and bioenergetics. PLoS Pathog. 2010;6:e1000719. doi: 10.1371/journal.ppat.1000719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Merz A., Long G., Hiet M.S. Biochemical and morphological properties of hepatitis C virus particles and determination of their lipidome. J Biol Chem. 2011;286:3018–3032. doi: 10.1074/jbc.M110.175018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.van der Meer-Janssen Y.P., van Galen J., Batenburg J.J., Helms J.B. Lipids in host-pathogen interactions: pathogens exploit the complexity of the host cell lipidome. Prog Lipid Res. 2010;49:1–26. doi: 10.1016/j.plipres.2009.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Song X., Yu H., Chen X. A sialylated glycan microarray reveals novel interactions of modified sialic acids with proteins and viruses. J Biol Chem. 2011;286:31610–31622. doi: 10.1074/jbc.M111.274217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Yu X., Coulson B.S., Fleming F.E. Novel structural insights into rotavirus recognition of ganglioside glycan receptors. J Mol Biol. 2011;413:929–939. doi: 10.1016/j.jmb.2011.09.005. [DOI] [PubMed] [Google Scholar]
- 125.Toman R., Skultety L., Ihnatko R. Coxiella burnetii glycomics and proteomics–tools for linking structure to function. Ann N Y Acad Sci. 2009;1166:67–78. doi: 10.1111/j.1749-6632.2009.04512.x. [DOI] [PubMed] [Google Scholar]
- 126.Krishnamoorthy L., Bess J.W., Jr., Preston A.B., Nagashima K., Mahal L.K. HIV-1 and microvesicles from T cells share a common glycome, arguing for a common origin. Nat Chem Biol. 2009;5:244–250. doi: 10.1038/nchembio.151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Bhak J. History of omics. Available from: http://omics.org. Accessed September 28, 2011.
- 128.Lander E.S., Linton L.M., Birren B. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- 129.Venter J.C., Adams M.D., Myers E.W. The sequence of the human genome. Science. 2001;291:1304–1351. doi: 10.1126/science.1058040. [DOI] [PubMed] [Google Scholar]
- 130.Relman D.A., Falkow S. The meaning and impact of the human genome sequence for microbiology. Trends Microbiol. 2001;9:206–208. doi: 10.1016/s0966-842x(01)02041-8. [DOI] [PubMed] [Google Scholar]
- 131.Relman D.A. New technologies, human-microbe interactions, and the search for previously unrecognized pathogens. J Infect Dis. 2002;186:S254–S258. doi: 10.1086/344935. [DOI] [PubMed] [Google Scholar]
- 132.Hooper L.V., Gordon J.I. Commensal host-bacterial relationships in the gut. Science. 2001;292:1115–1118. doi: 10.1126/science.1058709. [DOI] [PubMed] [Google Scholar]
- 133.Eckburg P.B., Bik E.M., Bernstein C.N. Diversity of the human intestinal microbial flora. Science. 2005;308:1635–1638. doi: 10.1126/science.1110591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.National Institutes of Health . National Institutes of Health; Bethesda, MD: 2007. NIH launches human microbiome project. [Google Scholar]
- 135.National Institutes of Health . National Institutes of Health; Bethesda, MD: 2007. Human microbiome project. [Google Scholar]
- 136.Peterson J., Garges S., Giovanni M. The NIH human microbiome project. Genome Res. 2009;19:2317–2323. doi: 10.1101/gr.096651.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Gao Z., Tseng C.H., Strober B.E., Pei Z., Blaser M.J. Substantial alterations of the cutaneous bacterial biota in psoriatic lesions. PLoS One. 2008;3:e2719. doi: 10.1371/journal.pone.0002719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Nelson D.E., Van Der Pol B., Dong Q. Characteristic male urine microbiomes associate with asymptomatic sexually transmitted infection. PLoS One. 2010;5:e14116. doi: 10.1371/journal.pone.0014116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Frank D.N., Robertson C.E., Hamm C.M. Disease phenotype and genotype are associated with shifts in intestinal-associated microbiota in inflammatory bowel diseases. Inflamm Bowel Dis. 2011;17:179–184. doi: 10.1002/ibd.21339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Yang L., Lu X., Nossa C.W. Inflammation and intestinal metaplasia of the distal esophagus are associated with alterations in the microbiome. Gastroenterology. 2009;137:588–597. doi: 10.1053/j.gastro.2009.04.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Ichinohe T., Pang I.K., Kumamoto Y. Microbiota regulates immune defense against respiratory tract influenza A virus infection. Proc Natl Acad Sci U S A. 2011;108:5354–5359. doi: 10.1073/pnas.1019378108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Langmead B., Schatz M.C., Lin J., Pop M., Salzberg S.L. Searching for SNPs with cloud computing. Genome Biol. 2009;10:R134. doi: 10.1186/gb-2009-10-11-r134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Caporaso J.G., Kuczynski J., Stombaugh J. QIIME allows analysis of high-throughput community sequencing data. Nat Methods. 2010;7:335–336. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Storch G., Weinstock G., Sodergren E. The human virome in children and its relationship to febrile illness. Human Microbiome Project. 2010 [Google Scholar]
- 145.Willner D., Furlan M., Haynes M. Metagenomic analysis of respiratory tract DNA viral communities in cystic fibrosis and non-cystic fibrosis individuals. PLoS One. 2009;4:e7370. doi: 10.1371/journal.pone.0007370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Minot S., Sinha R., Chen J., Li H. The human gut virome: inter-individual variation and dynamic response to diet. Genome Res. 2011;21:1616–1625. doi: 10.1101/gr.122705.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Ghannoum M.A., Jurevic R.J., Mukherjee P.K. Characterization of the oral fungal microbiome (mycobiome) in healthy individuals. PLoS Pathog. 2010;6:e1000713. doi: 10.1371/journal.ppat.1000713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Huang S.H., Triche T., Jong A.Y. Infectomics: genomics and proteomics of microbial infections. Funct Integr Genomics. 2002;1:331–344. doi: 10.1007/s10142-002-0048-4. [DOI] [PubMed] [Google Scholar]
- 149.Goh K.I., Cusick M.E., Valle D. The human disease network. Proc Natl Acad Sci U S A. 2007;104:8685–8690. doi: 10.1073/pnas.0701361104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Navratil V., de Chassey B., Combe C.R., Lotteau V. When the human viral infectome and diseasome networks collide: towards a systems biology platform for the aetiology of human diseases. BMC Syst Biol. 2011;5:13. doi: 10.1186/1752-0509-5-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Aderem A., Adkins J.N., Ansong C. A systems biology approach to infectious disease research: innovating the pathogen-host research paradigm. MBio. 2011;2 doi: 10.1128/mBio.00325-10. e00325–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Haverkamp M.H., van Dissel J.T., Holland S.M. Human host genetic factors in nontuberculous mycobacterial infection: lessons from single gene disorders affecting innate and adaptive immunity and lessons from molecular defects in interferon-gamma-dependent signaling. Microbes Infect. 2006;8:1157–1166. doi: 10.1016/j.micinf.2005.10.029. [DOI] [PubMed] [Google Scholar]
- 153.Kidder B.L., Hu G., Zhao K. ChIP-Seq: technical considerations for obtaining high-quality data. Nat Immunol. 2011;12:918–922. doi: 10.1038/ni.2117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.National Institute of Allergy and Infectious Disease . NIAID; Bethesda, Md: 2011. TB systems biology program. [Google Scholar]
- 155.National Institute of Allergy and Infectious Disease . NIAID; Bethesda, Md: 2011. Systems virology. [Google Scholar]
- 156.National Institute of Allergy and Infectious Disease . NIAID; Bethesda, Md: 2011. Systems influenza. [Google Scholar]
- 157.Zhang B., Horvath S. A general framework for weighted gene co-expression network analysis. Stat Appl Genet Mol Biol. 2005;4:17. doi: 10.2202/1544-6115.1128. [DOI] [PubMed] [Google Scholar]
- 158.Li C., Bankhead A., 3rd, Eisfeld A.J. Host regulatory network response to infection with highly pathogenic H5N1 avian influenza virus. J Virol. 2011;85:10955–10967. doi: 10.1128/JVI.05792-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Bradel-Tretheway B.G., Kelley Z., Chakraborty-Sett S. The human H5N1 influenza A virus polymerase complex is active in vitro over a broad range of temperatures, in contrast to the WSN complex, and this property can be attributed to the PB2 subunit. J Gen Virol. 2008;89:2923–2932. doi: 10.1099/vir.0.2008/006254-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Peng X., Gralinski L., Armour C.D. Unique signatures of long noncoding RNA expression in response to virus infection and altered innate immune signaling. MBio. 2010;1:e00206–e00210. doi: 10.1128/mBio.00206-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.National Institute of Allergy and Infectious Disease . NIAID; Bethesda, Md: 2011. Center for Systems Biology of EnteroPathogens.http://www.sysbep.org Available from: Accessed September 28, 2011. [Google Scholar]
- 162.Schellenberger J., Que R., Fleming R.M. Quantitative prediction of cellular metabolism with constraint-based models: the COBRA Toolbox v2.0. Nat Protoc. 2011;6:1290–1307. doi: 10.1038/nprot.2011.308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Becker S.A., Feist A.M., Mo M.L. Quantitative prediction of cellular metabolism with constraint-based models: the COBRA Toolbox. Nat Protoc. 2007;2:727–738. doi: 10.1038/nprot.2007.99. [DOI] [PubMed] [Google Scholar]
- 164.Yoon H., Ansong C., McDermott J.E. Systems analysis of multiple regulator perturbations allows discovery of virulence factors in Salmonella. BMC Syst Biol. 2011;5:100. doi: 10.1186/1752-0509-5-100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Buchko G.W., Niemann G., Baker E.S. A multi-pronged search for a common structural motif in the secretion signal of Salmonella enterica serovar Typhimurium type III effector proteins. Mol Biosyst. 2010;6:2448–2458. doi: 10.1039/c0mb00097c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Germain R. Ron Germain: towards a grand unified theory. Interview by Amy Maxmen. J Exp Med. 2010;207:266–267. doi: 10.1084/jem.2072pi. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.National Institute of Allergy and Infectious Disease . NIAID; Bethesda, Md: 2011. Laboratory of Systems Biology (LSB)http://www.niaid.nih.gov/labsandresources/labs/aboutlabs/lsb/pages/default.aspx?wt.ac=bcLsb Available from: Accessed September 30, 2011. [Google Scholar]
- 168.Miles J.J., Bulek A.M., Cole D.K. Genetic and structural basis for selection of a ubiquitous T cell receptor deployed in Epstein-Barr virus infection. PLoS pathogens. 2010;6:e1001198. doi: 10.1371/journal.ppat.1001198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Yang L., Zhang J., Ho B., Ding J.L. Histidine-mediated pH-sensitive regulation of M-ficolin:GlcNAc binding activity in innate immunity examined by molecular dynamics simulations. PLoS One. 2011;6:e19647. doi: 10.1371/journal.pone.0019647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Rapin N., Lund O., Bernaschi M., Castiglione F. Computational immunology meets bioinformatics: the use of prediction tools for molecular binding in the simulation of the immune system. PLoS One. 2010;5:e9862. doi: 10.1371/journal.pone.0009862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Mitha F., Lucas T.A., Feng F., Kepler T.B., Chan C. The multiscale systems immunology project: software for cell-based immunological simulation. Source Code Biol Med. 2008;3:6. doi: 10.1186/1751-0473-3-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Pulendran B. Learning immunology from the yellow fever vaccine: innate immunity to systems vaccinology. Nat Rev Immunol. 2009;9:741–747. doi: 10.1038/nri2629. [DOI] [PubMed] [Google Scholar]
- 173.Querec T.D., Akondy R.S., Lee E.K. Systems biology approach predicts immunogenicity of the yellow fever vaccine in humans. Nat Immunol. 2009;10:116–125. doi: 10.1038/ni.1688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Nakaya H.I., Wrammert J., Lee E.K. Systems biology of vaccination for seasonal influenza in humans. Nat Immunol. 2011;12:786–795. doi: 10.1038/ni.2067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Santamaria R., Rizzetto L., Bromley M. Systems biology of infectious diseases: a focus on fungal infections. Immunobiology. 2011;216:1212–1227. doi: 10.1016/j.imbio.2011.08.004. [DOI] [PubMed] [Google Scholar]
- 176.Liang L., Tan X., Juarez S. Systems biology approach predicts antibody signature associated with Brucella melitensis infection in humans. J Proteome Res. 2011;10:4813–4824. doi: 10.1021/pr200619r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Adams L.G., Khare S., Lawhon S.D. Enhancing the role of veterinary vaccines reducing zoonotic diseases of humans: linking systems biology with vaccine development. Vaccine. 2011;29:7197–7206. doi: 10.1016/j.vaccine.2011.05.080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Rappuoli R., Black S., Lambert P.H. Vaccine discovery and translation of new vaccine technology. Lancet. 2011;378:360–368. doi: 10.1016/S0140-6736(11)60440-6. [DOI] [PubMed] [Google Scholar]
- 179.Pulendran B., Li S., Nakaya H.I. Systems vaccinology. Immunity. 2010;33:516–529. doi: 10.1016/j.immuni.2010.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Smith G.P. Filamentous fusion phage: novel expression vectors that display cloned antigens on the virion surface. Science. 1985;228:1315–1317. doi: 10.1126/science.4001944. [DOI] [PubMed] [Google Scholar]
- 181.Wang L.F., Yu M. Epitope identification and discovery using phage display libraries: applications in vaccine development and diagnostics. Curr Drug Targets. 2004;5:1–15. doi: 10.2174/1389450043490668. [DOI] [PubMed] [Google Scholar]
- 182.Khurana S., Wu J., Verma N. H5N1 virus-like particle vaccine elicits cross-reactive neutralizing antibodies that preferentially bind to the oligomeric form of influenza virus hemagglutinin in humans. J Virol. 2011;85:10945–10954. doi: 10.1128/JVI.05406-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Barbas C.F., 3rd, Kang A.S., Lerner R.A., Benkovic S.J. Assembly of combinatorial antibody libraries on phage surfaces: the gene III site. Proc Natl Acad Sci U S A. 1991;88:7978–7982. doi: 10.1073/pnas.88.18.7978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Sun L., Lu X., Li C. Generation, characterization and epitope mapping of two neutralizing and protective human recombinant antibodies against influenza A H5N1 viruses. PLoS One. 2009;4:e5476. doi: 10.1371/journal.pone.0005476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Nelson A.L., Dhimolea E., Reichert J.M. Development trends for human monoclonal antibody therapeutics. Nat Rev Drug Discov. 2010;9:767–774. doi: 10.1038/nrd3229. [DOI] [PubMed] [Google Scholar]
- 186.Migone T.S., Subramanian G.M., Zhong J. Raxibacumab for the treatment of inhalational anthrax. N Engl J Med. 2009;361:135–144. doi: 10.1056/NEJMoa0810603. [DOI] [PubMed] [Google Scholar]
- 187.Galloway W.R., Bender A., Welch M., Spring D.R. The discovery of antibacterial agents using diversity-oriented synthesis. Chem Commun (Camb) 2009:2446–2462. doi: 10.1039/b816852k. [DOI] [PubMed] [Google Scholar]
- 188.Bratkovic T. Progress in phage display: evolution of the technique and its applications. Cell Mol Life Sci. 2010;67:749–767. doi: 10.1007/s00018-009-0192-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Pereira D.A., Williams J.A. Origin and evolution of high throughput screening. Br J Pharmacol. 2007;152:53–61. doi: 10.1038/sj.bjp.0707373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Agresti J.J., Antipov E., Abate A.R. Ultrahigh-throughput screening in drop-based microfluidics for directed evolution. Proc Natl Acad Sci U S A. 2010;107:4004–4009. doi: 10.1073/pnas.0910781107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Perrior T. Overcoming bottlenecks in drug discovery. Drug Discov World. 2010:29–33. [Google Scholar]
- 192.Collins F.S. The Genome Project and human health. FASEB J. 1991;5:77. doi: 10.1096/fasebj.5.1.1991592. [DOI] [PubMed] [Google Scholar]
- 193.Schilsky R.L. Personalized medicine in oncology: the future is now. Nat Rev Drug Discov. 2010;9:363–366. doi: 10.1038/nrd3181. [DOI] [PubMed] [Google Scholar]
- 194.Cronin M., Ross J.S. Comprehensive next-generation cancer genome sequencing in the era of targeted therapy and personalized oncology. Biomark Med. 2011;5:293–305. doi: 10.2217/bmm.11.37. [DOI] [PubMed] [Google Scholar]
- 195.Sorensen T.I., Nielsen G.G., Andersen P.K., Teasdale T.W. Genetic and environmental influences on premature death in adult adoptees. N Engl J Med. 1988;318:727–732. doi: 10.1056/NEJM198803243181202. [DOI] [PubMed] [Google Scholar]
- 196.Casanova J.L., Abel L., Quintana-Murci L. Human TLRs and IL-1Rs in host defense: natural insights from evolutionary, epidemiological, and clinical genetics. Annu Rev Immunol. 2011;29:447–491. doi: 10.1146/annurev-immunol-030409-101335. [DOI] [PubMed] [Google Scholar]
- 197.Guo Y., Audry M., Ciancanelli M. Herpes simplex virus encephalitis in a patient with complete TLR3 deficiency: TLR3 is otherwise redundant in protective immunity. J Exp Med. 2011;208:2083–2098. doi: 10.1084/jem.20101568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Picard C., Casanova J.L., Puel A. Infectious diseases in patients with IRAK-4, MyD88, NEMO, or IkappaBalpha deficiency. Clin Microbiol Rev. 2011;24:490–497. doi: 10.1128/CMR.00001-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Tabarsi P., Marjani M., Mansouri N. Lethal tuberculosis in a previously healthy adult with IL-12 receptor deficiency. J Clin Immunol. 2011;31:537–539. doi: 10.1007/s10875-011-9523-9. [DOI] [PubMed] [Google Scholar]
- 200.Burbelo P.D., Browne S.K., Sampaio E.P. Anti-cytokine autoantibodies are associated with opportunistic infection in patients with thymic neoplasia. Blood. 2010;116:4848–4858. doi: 10.1182/blood-2010-05-286161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Holland S.M. Interferon gamma, IL-12, IL-12R and STAT-1 immunodeficiency diseases: disorders of the interface of innate and adaptive immunity. Immunol Res. 2007;38:342–346. doi: 10.1007/s12026-007-0045-8. [DOI] [PubMed] [Google Scholar]
- 202.Vinh D.C., Schwartz B., Hsu A.P. Interleukin-12 receptor beta1 deficiency predisposing to disseminated Coccidioidomycosis. Clinical infectious diseases: an official publication of the Infectious Diseases Society of America. 2011;52:e99–e102. doi: 10.1093/cid/ciq215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Holland S.M., DeLeo F.R., Elloumi H.Z. STAT3 mutations in the hyper-IgE syndrome. N Engl J Med. 2007;357:1608–1619. doi: 10.1056/NEJMoa073687. [DOI] [PubMed] [Google Scholar]
- 204.International HapMap Consortium A haplotype map of the human genome. Nature. 2005;437:1299–1320. doi: 10.1038/nature04226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Vannberg F.O., Chapman S.J., Hill A.V. Human genetic susceptibility to intracellular pathogens. Immunol Rev. 2011;240:105–116. doi: 10.1111/j.1600-065X.2010.00996.x. [DOI] [PubMed] [Google Scholar]
- 206.Oki N.O., Motsinger-Reif A.A., Antas P.R. Novel human genetic variants associated with extrapulmonary tuberculosis: a pilot genome wide association study. BMC Res Notes. 2011;4:28. doi: 10.1186/1756-0500-4-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Rauch A., Kutalik Z., Descombes P. Genetic variation in IL28B is associated with chronic hepatitis C and treatment failure: a genome-wide association study. Gastroenterology. 2010;138:1338–1345. doi: 10.1053/j.gastro.2009.12.056. 45 e1-7. [DOI] [PubMed] [Google Scholar]
- 208.Thomas D.L., Thio C.L., Martin M.P. Genetic variation in IL28B and spontaneous clearance of hepatitis C virus. Nature. 2009;461:798–801. doi: 10.1038/nature08463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Poustka A., Pohl T., Barlow D.P. Molecular approaches to mammalian genetics. Cold Spring Harb Symp Quant Biol. 1986;51:131–139. doi: 10.1101/sqb.1986.051.01.016. [DOI] [PubMed] [Google Scholar]
- 210.Cantor C.R., Mirzabekov A., Southern E. Report on the sequencing by hybridization workshop. Genomics. 1992;13:1378–1383. doi: 10.1016/0888-7543(92)90079-8. [DOI] [PubMed] [Google Scholar]
- 211.Schena M., Shalon D., Davis R.W., Brown P.O. Quantitative monitoring of gene expression patterns with a complementary DNA microarray. Science. 1995;270:467–470. doi: 10.1126/science.270.5235.467. [DOI] [PubMed] [Google Scholar]
- 212.Lempicki R.A., Polis M.A., Yang J. Gene expression profiles in hepatitis C virus (HCV) and HIV coinfection: class prediction analyses before treatment predict the outcome of anti-HCV therapy among HIV-coinfected persons. J Infect Dis. 2006;193:1172–1177. doi: 10.1086/501365. [DOI] [PubMed] [Google Scholar]
- 213.Jacobsen M., Repsilber D., Gutschmidt A. Candidate biomarkers for discrimination between infection and disease caused by Mycobacterium tuberculosis. J Mol Med (Berl) 2007;85:613–621. doi: 10.1007/s00109-007-0157-6. [DOI] [PubMed] [Google Scholar]
- 214.Reghunathan R., Jayapal M., Hsu L.Y. Expression profile of immune response genes in patients with severe acute respiratory syndrome. BMC Immunol. 2005;6:2. doi: 10.1186/1471-2172-6-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Simmons C.P., Popper S., Dolocek C. Patterns of host genome-wide gene transcript abundance in the peripheral blood of patients with acute dengue hemorrhagic fever. J Infect Dis. 2007;195:1097–1107. doi: 10.1086/512162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Zaas A.K., Chen M., Varkey J. Gene expression signatures diagnose influenza and other symptomatic respiratory viral infections in humans. Cell Host Microbe. 2009;6:207–217. doi: 10.1016/j.chom.2009.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Ardura M.I., Banchereau R., Mejias A. Enhanced monocyte response and decreased central memory T cells in children with invasive Staphylococcus aureus infections. PLoS One. 2009;4:e5446. doi: 10.1371/journal.pone.0005446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Thompson L.J., Dunstan S.J., Dolecek C. Transcriptional response in the peripheral blood of patients infected with Salmonella enterica serovar Typhi. Proc Natl Acad Sci U S A. 2009;106:22433–22438. doi: 10.1073/pnas.0912386106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.Hetherington S., McGuirk S., Powell G. Hypersensitivity reactions during therapy with the nucleoside reverse transcriptase inhibitor abacavir. Clin Ther. 2001;23:1603–1614. doi: 10.1016/s0149-2918(01)80132-6. [DOI] [PubMed] [Google Scholar]
- 220.Mallal S., Phillips E., Carosi G. HLA-B∗5701 screening for hypersensitivity to abacavir. N Engl J Med. 2008;358:568–579. doi: 10.1056/NEJMoa0706135. [DOI] [PubMed] [Google Scholar]
- 221.Genebat M., Ruiz-Mateos E., Leon J.A. Correlation between the Trofile test and virological response to a short-term maraviroc exposure in HIV-infected patients. J Antimicrob Chemother. 2009;64:845–849. doi: 10.1093/jac/dkp293. [DOI] [PubMed] [Google Scholar]
- 222.Marko N.F., Weil R.J. Mathematical modeling of molecular data in translational medicine: theoretical considerations. Sci Transl Med. 2010;2:56rv4. doi: 10.1126/scitranslmed.3001207. [DOI] [PubMed] [Google Scholar]
- 223.Royston P., Altman D.G., Sauerbrei W. Dichotomizing continuous predictors in multiple regression: a bad idea. Stat Med. 2006;25:127–141. doi: 10.1002/sim.2331. [DOI] [PubMed] [Google Scholar]
- 224.MacCallum R.C., Zhang S., Preacher K.J., Rucker D.D. On the practice of dichotomization of quantitative variables. Psychol Methods. 2002;7:19–40. doi: 10.1037/1082-989x.7.1.19. [DOI] [PubMed] [Google Scholar]
- 225.Zhao X., Cheung L.W. Kernel-imbedded Gaussian processes for disease classification using microarray gene expression data. BMC Bioinformatics. 2007;8:67. doi: 10.1186/1471-2105-8-67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226.Kong W., Vanderburg C.R., Gunshin H., Rogers J.T., Huang X. A review of independent component analysis application to microarray gene expression data. BioTechniques. 2008;45:501–520. doi: 10.2144/000112950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227.Lawler G.F. 2nd ed. CRC; Boca Raton, FL: 2006. Introduction to stochastic processes. [Google Scholar]
- 228.Yan Q. Translational bioinformatics and systems biology approaches for personalized medicine. Methods Mol Biol. 2010;662:167–178. doi: 10.1007/978-1-60761-800-3_8. [DOI] [PubMed] [Google Scholar]
- 229.Kanehisa M., Goto S., Sato Y., Furumichi M., Tanabe M. KEGG for integration and interpretation of large-scale molecular data sets. Nucleic Acids Res. 2011;40:D109–D114. doi: 10.1093/nar/gkr988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 230.Sarkar I.N., Butte A.J., Lussier Y.A., Tarczy-Hornoch P., Ohno-Machado L. Translational bioinformatics: linking knowledge across biological and clinical realms. J Am Med Inform Assoc. 2011;18:354–357. doi: 10.1136/amiajnl-2011-000245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 231.Raising the game. Nat Biotechnol. 2008;26:137. doi: 10.1038/nbt0208-137. [No authors listed] [DOI] [PubMed] [Google Scholar]
- 232.Paul S.M., Mytelka D.S., Dunwiddie C.T. How to improve R&D productivity: the pharmaceutical industry’s grand challenge. Nat Rev Drug Discov. 2010;9:203–214. doi: 10.1038/nrd3078. [DOI] [PubMed] [Google Scholar]
- 233.US Food and Drug Administration . FDA; Silver Spring, Md: 2011. NME drug and new biologic approvals. [Google Scholar]
- 234.Goodman M. Market watch: pharma industry strategic performance: 2007-2012E. Nat Rev Drug Discov. 2008;7:967. doi: 10.1038/nrd2768. [DOI] [PubMed] [Google Scholar]
- 235.Munos B. Lessons from 60 years of pharmaceutical innovation. Nat Rev Drug Discov. 2009;8:959–968. doi: 10.1038/nrd2961. [DOI] [PubMed] [Google Scholar]
- 236.Lichtenberg F.R. The impact of new drug launches on longevity: evidence from longitudinal, disease-level data from 52 countries, 1982-2001. Int J Health Care Finance Econ. 2005;5:47–73. doi: 10.1007/s10754-005-6601-7. [DOI] [PubMed] [Google Scholar]
- 237.Ledford H. Translational research: 4 ways to fix the clinical trial. Nature. 2011;477:526–528. doi: 10.1038/477526a. [DOI] [PubMed] [Google Scholar]
- 238.National Institutes of Health . NIH; Bethesda, Md: 2011. Regulatory science. [Google Scholar]
- 239.US Food and Drug Administration . FDA; Silver Spring, Md: 2011. FDA-NIH Joint Leadership Council Charter. [Google Scholar]
- 240.US Food and Drug Administration NIoH . FDA; Silver Spring, Md: 2010. FDA-NIH News Release: NIH and FDA announce collaborative initiative to fast-track innovations to the public. [Google Scholar]
- 241.Collins F.S., Gray G.M., Bucher J.R. Toxicology. Transforming environmental health protection. Science. 2008;319:906–907. doi: 10.1126/science.1154619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 242.Kennedy D. FDA centennial. Science. 2006;312:19. doi: 10.1126/science.1127486. [DOI] [PubMed] [Google Scholar]
- 243.Wood A.J. A proposal for radical changes in the drug-approval process. N Engl J Med. 2006;355:618–623. doi: 10.1056/NEJMsb055203. [DOI] [PubMed] [Google Scholar]
- 244.Collins F.S. Mining for therapeutic gold. Nat Rev Drug Discov. 2011;10:397. doi: 10.1038/nrd3461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 245.Fang F.C., Casadevall A. Lost in translation–basic science in the era of translational research. Infect Immun. 2010;78:563–566. doi: 10.1128/IAI.01318-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 246.Petsko G.A. Herding CATS. Sci Transl Med. 2011;3:97cm24. doi: 10.1126/scitranslmed.3002837. [DOI] [PubMed] [Google Scholar]
- 247.Redhead C.S., Smith P.W. Congressional Research Service; Washington, DC: 2011. Public Health Service (PHS) Agencies: Overview and Funding, FY2010–FY2012. [Google Scholar]
- 248.Smrb Hears From The Public About Ncats. Available from: http://feedback.nih.gov/index.php/ncats/smrb-hears-from-the-public-about-ncats/. Accessed March 1, 2011.
- 249.Kaiser J. NIH reorganization draws scrutiny from Congress. Science Insider. January 27, 2011 [Google Scholar]
- 250.FitzGerald G.A. NCATS purrs: emerging signs of form and function. Sci Transl Med. 2011;3:83ed2. doi: 10.1126/scitranslmed.3002232. [DOI] [PubMed] [Google Scholar]