Abstract
Layer-by-layer polyelectrolyte adsorption is a simple, convenient method for introducing ion-exchange sites in porous membranes. This study demonstrates that adsorption of poly(acrylic acid) (PAA)-containing films at pH 3 rather than pH 5 increases the protein-binding capacity of such polyelectrolyte-modified membranes 3- to 6-fold. The low adsorption pH generates a high density of –COOH groups that function as either ion-exchange sites or points for covalent immobilization of metal-ion complexes that selectively bind tagged proteins. When functionalized with nitrilotriacetate (NTA)-Ni2+ complexes, membranes containing PAA/polyethyleneimine (PEI)/PAA films bind 93 mg of histidine6-tagged (His-tagged) ubiquitin per cm3 of membrane. Additionally these membranes isolate His-tagged COP9 signalosome complex subunit 8 from cell extracts and show >90% recovery of His-tagged ubiquitin. Although modification with polyelectrolyte films occurs by simply passing polyelectrolyte solutions through the membrane for as little as 5 min, with low-pH deposition the protein binding capacities of such membranes are as high as for membranes modified with polymer brushes and 2–3 fold higher than for commercially available IMAC resins. Moreover, the buffer permeabilities of polyelectrolyte-modified membranes that bind His-tagged protein are ~30% of the corresponding permeabilities of unmodified membranes, so protein capture can occur rapidly with low pressure drops. Even at a solution linear velocity of 570 cm/h, membranes modified with PAA/PEI/PAA exhibit a lysozyme dynamic binding capacity (capacity at 10% breakthrough) of ~ 40 mg/cm3. Preliminary studies suggest that these membranes are stable under depyrogenation conditions (1 M NaOH).
Introduction
Affinity adsorption of tagged recombinant proteins is a vital step in their purification.1,2 Remarkably, specific binding of the tagged protein to ligands immobilized in packed columns often leads to eluted protein purities >90%. However, slow diffusion of large macromolecules into the affinity resin sometimes results in long separation times that are particularly deleterious for purification of sensitive proteins or their complexes.2–5 In large-scale affinity adsorption, column packing is also challenging, and high pressure drops may occur.
Porous membranes modified with affinity ligands offer a potential solution to some of the challenges in column-based affinity separations.4,6–13 Convection through the membrane pores and short radial diffusion distances provide rapid protein transport to binding sites, and increasing the membrane surface area is a relatively straightforward strategy to scale up membrane processes. Unfortunately, membrane adsorbers suffer from low binding capacities relative to traditional columns. A number of research groups successfully modified membranes with polymer brushes to increase the number of binding sites and enhance binding capacity,13–23 but brush growth is a relatively cumbersome process, frequently requiring both deposition of initiator molecules and polymerization under anaerobic conditions.15,17,18
We recently examined whether layer-by-layer (LbL) adsorption of polyelectrolyte multilayers in nylon with 5 μm pores could effectively create ion-exchange membranes.24 Modification of membranes using LbL adsorption, which simply involves passing a few aqueous solutions through the membrane, is extremely convenient,25 but the lysozyme binding capacities of those membranes were at most 16 mg/cm3.24 Commercial ion-exchange Mustang S membranes already show lysozyme binding capacities of 45–50 mg/cm3.26
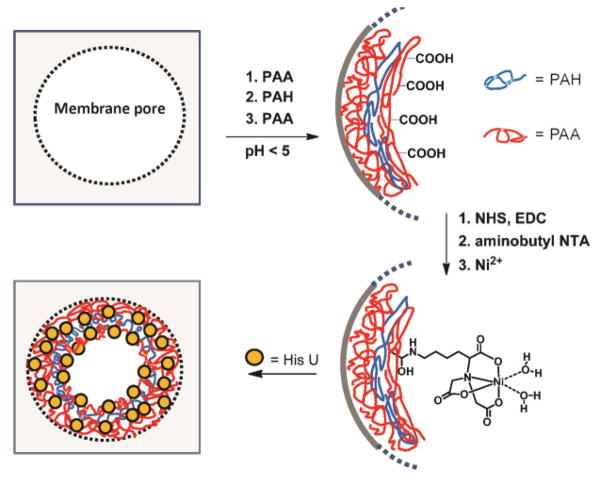
This study demonstrates that control of the pH employed during deposition of weak polyelectrolytes can greatly increase the protein-binding capacities of membranes modified with polyelectrolyte multilayers. A number of papers report that changes in the deposition pH of poly(acrylic acid) (PAA)/protonated poly(allyl amine) (PAH) multilayer coatings greatly alter film properties including thickness, swelling, metal adsorption capacity, permeability, and biocompatibility.27–34 Additionally, our recent work shows that deposition of (PAH/PAA)n films at pH 3 rather than pH 5 leads to a ~6-fold increase in lysozyme adsorption.35 Thus we thought that in membranes, PAH/PAA adsorption at low pH would give a high density of free –COOH groups that bind cationic proteins through ion-exchange interactions. (By free, we mean that the –COOH groups are not deprotonated and ion-paired with neighboring ammonium groups of PAH during deposition.) Moreover, derivatization of the free –COOH groups by reaction with aminobutyl nitrilotriacetate (NTA) should yield metal-ion complexes that selectively bind tagged proteins (Figure 1). Remarkably, membranes modified with PAA/polycation/PAA films deposited at pH 3 bind as much as 120 mg lysozyme per cm3 of membrane, which is comparable to the capacities of the best membranes modified with polymer brushes.18 Additionally, after derivatization with NTA-Ni2+ complexes, these membranes can capture His-tagged proteins from cell extracts and facilitate 95% protein recovery at high purity. The simplicity of LbL adsorption and the high performance of these membranes make them very attractive for protein purification.
Figure 1.
Schematic showing polyelectrolyte immobilization within membrane pores, derivatization of the surface layer of PAA with NTA-Ni2+ complexes, and protein binding to the modified membrane. Abbreviations: PAA- poly(acrylic acid), PAH- protonated poly(allylamine), NHS- N-hydroxysuccinimide, EDC- N-(3-Dimethylaminopropyl)-N′-ethylcarbodiimide hydrochloride NTA- nitrilotriacetate, HisU- His-tagged ubiquitin.
EXPERIMENTAL
Materials
Hydroxylated nylon (LoProdyne® LP, Pall, 1.2 μm pore size, 110 μm thick), nylon (GE, non-hydroxylated, 1.2 μm pore size, average thickness 95 μm), and polyethersulfone (GE, 1.2 μm pore size, average thickness 130 μm) membranes were cut into 25 mm-diameter discs prior to use. Unless specified, all proteins and chemicals were obtained from Sigma-Aldrich. Coomassie protein assay reagent (Thermo Scientific), Histidine6-tagged Ubiquitin (HisU, human recombinant, Enzo Life Sciences), conconavalin A (Con A) from Canavaliaensiformis (Jack bean), albumin from chicken egg white; lysozyme from chicken egg white; bovine serum albumin (BSA), and β-Lactoglobulin B from Bovine milk were used as received. His-tagged COP9 signalosome complex subunit 8 (CSN 8) was overexpressed in BL21DE3 cells as described below. Buffers were prepared using analytical grade chemicals and deionized water (Milli-Q, 18.2 MΩ cm). Poly(sodium 4-styrenesulfonate) (Mw = 70,000), poly(allylamine hydrochloride) (Mw=120,000–210,000, Alfa-Aesar), polyethyleneimine (branched, Mw = 25,000), poly(acrylic acid) (Mw = 90,000, 25% aqueous solution, Polysciences), TWEEN-20 surfactant, N-(3-dimethylaminopropyl)-N′-ethylcarbodiimide hydrochloride (EDC), N-hydroxysuccinimide, and Nα, Nα-bis(carboxymethyl)-L-lysine hydrate (aminobutyl NTA) were used without further purification.
Membrane modification
Membrane discs were cleaned for 10 min with UV/ozone and placed in a homemade Teflon holder (similar to an Amicon cell) that exposed 3.1 cm2 of external membrane surface area. Subsequently, a 20 mL solution containing 0.02 M PSS and 0.5 M NaCl was circulated through the membrane for 40 min at a flow rate of 1 mL/min using a peristaltic pump. Additional polycation (PAH or PEI) and polyanion (PAA) layers were deposited similarly using solutions containing 0.01 M PAA and 0.5 M NaCl or 0.02 M PAH or 2 mg/mL PEI (no NaCl was added to polycation solutions). After deposition of each polyelectrolyte, 20 mL of water was passed through the membrane at the same flow rate. The pH of the PSS solution was 4.7, and PAA, PAH, and PEI deposition solutions were adjusted to different values with 1 M NaOH or 1M HCl. Membrane hydraulic permeabilities were determined as described previously.36
To derivatize PAA side chains in adsorbed films, 10 mL of 0.1 M NHS, 0.1 M EDC in water was circulated through the membrane for 1 h prior to rinsing with 20 mL of deionized water and 10 mL of ethanol. Subsequently, 10 mL of aqueous aminobutyl NTA (0.1 M, pH 10.2) was circulated through the NHS-modified substrate for 1 h followed by rinsing with 20 mL of water. Finally, the NTA-Cu2+ (or Ni2+) complex was formed by circulating 10 mL of aqueous 0.1 M CuSO4 (or NiSO4) through the membrane for 2 h followed by rinsing with water. The substrate was dried with N2 prior to protein binding.
Protein Binding
A solution of lysozyme (0.30 or 0.45 mg/mL) in 20 mM phosphate buffer (pH 7.4) was pumped through the modified membrane at a flow rate of 1 or 30 mL/min, and the permeate was collected for analysis at specific time intervals. Subsequently, the membrane was rinsed with 20 mL of washing buffer A (20 mM phosphate buffer with 0.1 % Tween 20, pH 7.4) followed by 20 mL of phosphate buffer. The protein was then eluted using 5–10 mL of 20 mM phosphate buffer (pH 7.4) containing 1 M KSCN. Unless specified, flow rates were 1 mL/min.
Con A and HisU solutions (0.3 mg/mL) were prepared in 20 mM phosphate buffer at pH 6 and 7.4, respectively. For Con A, washing buffer B (20 mM phosphate buffer containing 0.1% Tween-20 surfactant and 0.15 M NaCl) and elution buffer (20 mM phosphate buffer containing 50 mM EDTA) were also adjusted to pH 6. In the case of HisU, the washing buffer B and elution buffer (0.5 M NaCl, 0.5 M imidazole in 20 mM phosphate buffer) were maintained at pH 7.4. For both Con A and HisU, membranes were loaded with buffered protein solution, rinsed with 20 mL washing buffer B followed by 20 mL phosphate buffer at pH 6 or 7.4, and eluted with 8–9 mL of elution buffer. The concentrations of protein in loading, rinsing, and eluate solutions were determined using a Bradford assay.36 Each Con A binding capacity was determined with two membranes, and the ± values represent the difference between the average and the data points.
Purification of His-U from a Model Protein Mixture
To test protein binding specificity and recovery, we prepared a solution containing 0.05 mg/mL (each) His-U, Con A, BSA, Ovalbumin, and β-Lactoglobulin B in 20 mM phosphate buffer (pH 7.4). Ten mL of this protein solution was passed through a PAA/PEI/PAA-NTA-Ni2+-modified membrane at 1.5 mL/min after the membrane was equilibrated with 20 mL phosphate buffer. Subsequently, the membrane was washed with 20 mL washing buffer C (20 mM phosphate buffer containing 0.1% Tween-20 surfactant and 150 mM NaCl, pH 8.0) and 20 mL phosphate buffer. The bound protein was eluted with 10 mL (2 mL for each fraction) elution buffer (20 mM phosphate buffer with 500 mM imidazole and 500 mM NaCl, pH 8.0). The purity of eluted protein was examined by SDS-PAGE (4–20% gradient gel from Bio-Rad with standard Coomassie blue staining protocols).
Cell Culture
The CSN8 orf was obtained through the Drosophila Genomics Resource Center and sub-cloned into the his-SUMO modified37 pet28b vector (Novagen). The plasmid was transformed into BL21DE3 codon plus (Stratagene) competent cells. Colonies were grown in LB broth (with Kanamycin and Chloramphenicol) at 37 °C until an O. D. of 0.8 was reached. The growth was induced with 0.4 mM isopropyl-thio-2-D-galactopyranoside (IPTG) for 16 hours at 16 °C. The growth was pelleted by centrifugation. The pellet was resuspended in denaturing buffer containing 6 M urea, 10 mM Tris-HCl, and 100 mM NaH2PO4, at a pH of 8.0. The lysate was sonicated and then centrifuged to pellet the debris. The resulting supernatant was diluted 4:1 with 20 mM pH 8 phosphate buffer containing 10 mM imidazole, 300 mM NaCl, and 10% glycerol and stored in a −80 °C freezer until use.
Protein Isolation from a Cell Extract
At a flow rate of 1.5 mL/min, the diluted lysate supernatant described above was passed through a PAA/PEI/PAA-NTA-Ni2+-modified membrane that was equilibrated with lysate buffer (20 mM pH 8 phosphate buffer containing 10 mM imidazole and 300 mM NaCl). After washing with 20 mL washing buffer B, 20 mL washing buffer C (20 mM phosphate buffer containing 45 mM imidazole and 150 mM NaCl), and 20 mL phosphate buffer at a flow rate of 5 mL/min, protein elution and gel electrophoresis followed the same procedure for HisU purification.
RESULTS AND DISCUSSION
Adsorption of Polyelectrolytes in Hydroxylated Nylon Membranes
In previous work we showed that PSS serves as a robust adhesion layer for deposition of polyelectrolyte multilayers in a variety of membranes.24 Multiple hydrophobic interactions likely lead to strong PSS adsorption. Thus we initially deposited a PAH/PAA bilayer on PSS adsorbed in a 1.2 μm nylon membrane. This procedure requires no organic solvents or anaerobic conditions and is much simpler than modification of membranes with polymer brushes. Moreover, because the fraction of ionized –COOH groups on an adsorbed PAA monolayer varies from <10 % to > 60 % on going from pH 2 to pH 5,31 variation of deposition pH provides an important variable for modifying film properties.29 Adsorption of PAA at low pH leads to films that contain free –COOH groups,38 and subsequent derivatization or deprotonation of these groups should lead to a high density of protein-binding sites. Deposition pH also affects the degree of protonation of PAH, but at pH values ≤ 5, this polycation will be >90% protonated in the film.29,39,40
Monitoring polyelectrolyte adsorption in nylon membranes is challenging. SEM images suggest a decrease in porosity after deposition of polyelectrolytes (Figure S-1), but such images are only qualitative and do not reflect film swelling. The water permeability of nylon membranes decreases significantly after polyelectrolyte adsorption (Tables S-1, S-3, and S-5), and the decrease is most significant for deposition of polyelectrolytes at low pH. Moreover, with film deposition at low pH, membrane permeability increases after derivatization with NTA-Cu2+ complexes, presumably because of a decrease in swelling (Tables S-2, S-4, and S-6). Nevertheless, in control experiments, even bare (unmodified) nylon membranes showed a 30–50% decline in hydraulic pure water permeability after exposure to 20 mM phosphate buffer (pH 7.4). Thus, although water permeabilities suggest greater polyelectrolyte adsorption at low pH, they do not provide a quantitative measure of polyelectrolyte adsorption. Previous studies show that PAA/PAH multilayer films on flat surface are softer and swell more when deposited at low pH,31,33 but such results may not quantitatively describe very thin coatings in membranes.
We can, however, reliably determine the amount of Cu2+ that binds to membranes modified with PSS/PAH/PAA-NTA-Cu2+. Table 1 (column 5) shows that the quantity of Cu2+ captured in these membranes increases with a decrease in the pH of PAH and PAA adsorption. This increase likely reflects enhancements in both the film thickness and availability of free –COOH or –COO− groups for activation and reaction with aminobutyl NTA.29,31,38,41,42 Considering the membrane modified with PSS/PAH/PAA-NTA using a deposition pH of 2, the Cu2+ binding capacity of 15 mg/cm3 suggests that there is ~75 mg/cm3 of polymer in the membrane. This estimation assumes complete derivatization to give a repeat unit molecular weight of 316 for PAA-NTA and neglects the amount of PSS and PAH in the membranes as well as PAA repeat units that interact with PAH and cannot be derivatized. The total amount of polymer in the membrane could easily be twice the calculated value.
Table 1.
Lysozyme and Cu2+ binding capacities of nylon membranes modified with different films.a
| Polyelectrolyte films in nylon membrane | pH of PAH and/or PAA deposition solutions | Lysozyme binding from breakthrough curves (mg/cm3) | Lysozyme binding From elution (mg/cm3) | Cu2+ binding (mg/cm3)c |
|---|---|---|---|---|
| PSS/PAH/PAA | 2 | 90±1 | 87±1 | 15±1 |
| 3 | 106±2 | 106±6 | 11±2 | |
| 4 | 49±2 | 49±1 | 6±1 | |
| 5 | 37±4 | 33±3 | 3±2 | |
| PAA | 2 | 78±1 | 77±1 | 12+2 |
| 3 | 89±4 | 89±2 | 13±2 | |
| 4 | 22±9 | 22±6 | 4±2 | |
| 5 | 14±6 | 14±2 | 2±1 | |
| PAA/PAH/PAA | 3 | 107±2 | 115±2 | 14±1 |
| PAA/PEI/PAA | 3 | 120±6 | 130±4 | 18±2 |
| PAA/PEI/PAAb | 3 | 101±5 | 112±4 | - |
Each experiment was performed with two different membranes, and the ± values represent the difference between the average and the data points.
Lysozyme flow rate of 30 mL/min. In all other cases the lysozyme flow rate was 1 mL/min.
Binding capacity after derivatization with aminobutyl NTA.
Although we initially thought that a PSS adhesion layer is important for forming stable polyelectrolyte films in nylon membranes, adsorption of PAA directly in nylon pores also provides a remarkably simple way to introduce a high density of functional groups in these systems. In principle PAA might adsorb to nylon membranes through hydrophobic interactions or hydrogen bonds. Similar to membranes modified with PSS/PAH/PAA, the Cu2+-binding capacities of PAA films deposited at pH 2 and pH 3 and subsequently modified with aminobutyl NTA are 4- to 6-fold higher than the capacities of corresponding films deposited at pH 4 and 5 (Table 1, column 5). Thus the total amount of adsorbed PAA may be ~5-fold higher when comparing films deposited at pH 2 or 3 with films deposited at pH 5. Notably, for films deposited at low pH, the Cu2+ binding is similar for membranes modified with PAA and PSS/PAH/PAA films.
Adsorption of a PAH/PAA or PEI/PAA bilayer on a PAA base layer can in principle increase the number of free –COOH groups in a membrane. However, membranes modified with PAA-NTA, PAA/PAH/PAA-NTA, and PAA/PEI/PAA-NTA bind only 13±2, 14±1, and 18±2 mg Cu2+/cm3, respectively (these films were deposited at pH 3, as we were concerned about film stability at pH 2). Only the PAA/PEI/PAA-NTA coating shows significantly more Cu2+ sorption than simple PAA-NTA films. The relatively small increase in bound Cu2+ with the addition of the polycation/PAA bilayer reflects the formation of ion pairs between the polycations and underlying PAA and perhaps less extension of the outer PAA layer when adsorption occurs on the polycation rather than directly on a membrane. Notably, adsorption of PAA on the branched PEI apparently leads to more derivatizable –COOH groups than adsorption on linear PAH.
In addition to NTA, Cu2+ may bind to underivatized –COOH groups and complicate the interpretation of Cu2+ binding data. Thus, we compared Cu2+ binding to PAA/PEI/PAA and PAA/PEI/PAA-NTA membranes. The binding was ~30% higher for the NTA-derivatized membrane, but significant binding does occur to the PAA/PEI/PAA film. This is not surprising given the large number of free –COOH groups in the film. Even with the NTA-derivatized coating some fraction of the Cu2+ binding likely occurs to underivatized –COOH groups. We tried unsuccessfully to selectively elute the Cu2+ bound to underivatized –COOH groups.
Plugging of membrane pores is always a potential problem when modifying membranes by adsorption. The permeability of membranes to pH 7.4 phosphate buffer (20 mM) decreases from around 70 to 20 mL/(cm2 min atm) when comparing a bare membrane and a membrane containing a PAA/PEI/PAA film. Although this is a significant decline in permeability, rapid flow through the membrane can still occur using a simple peristaltic pump, even after derivatization with NTA. In contrast, the permeability of PAA/PEI/PAA-modified membranes to deionized water after treatment with buffer is <1 mL/(cm2 min atm). Extension of deprotonated PAA at low ionic strength evidently blocks pores, so filtration should occur with at least small amounts of salt. Rinsing membranes with 2.7 mM HCl protonates –COOH groups, and the resulting collapse of polymers restores water permeability to around 100 mL/(cm2 min atm). Subsequent exposure to pH 7.4 buffer again decreases permeability. These results are consistent with prior studies of pH-responsive membranes.43–45
Lysozyme Binding to Hydroxylated Nylon Membranes Modified with Polyelectrolyte Films
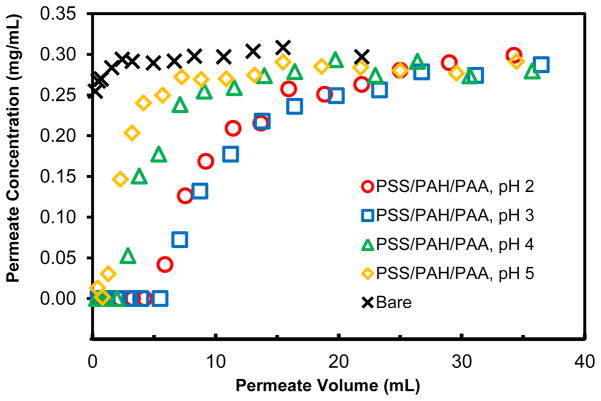
Deposition of polyanion-terminated films in membrane pores creates cation-exchange sites18 that bind positively charged proteins such as lysozyme (molecular weight 14.3 kDa), which at pH 7.4 has a charge of +8.46 Figure 2 presents breakthrough curves for passage of 0.3 mg/mL lysozyme (in pH 7.4 buffer) through nylon membranes modified with PSS/PAH/PAA films deposited at several pH values. When protein begins to saturate the binding sites, the lysozyme breaks through the membrane, and its effluent concentration eventually reaches that of the feed solution. The later breakthrough in the case of films deposited at pH 2 and 3 demonstrates the higher binding capacities in these systems. Integration of the differences between the feed concentration and the effluent concentration gives the membrane binding capacity, and Table 1 (column 3) shows that the lysozyme binding capacity for PSS/PAH/PAA films deposited at pH 3 is 3 times that for films adsorbed at pH 5. Thicker films and higher concentrations of free –COOH groups, as indicated by Cu2+ binding capacities (Table 1, column 5), presumably lead to more binding sites for membranes modified by polyelectrolyte adsorption at low pH. The highest binding at pH 3 rather than pH 2 might relate to film conformation. Binding capacities determined from elution of the lysozyme with 20 mM phosphate buffer (pH 7.4) containing 1 M KSCN agree well with those determined from the breakthrough curves (compare columns 3 and 4 of Table 1).
Figure 2.
Breakthrough curves for the passage of 0.3 mg/mL lysozyme through a bare nylon membrane and nylon membranes modified with PSS/PAH/PAA films deposited at different pH values. The protein-solution flow rate was 1 mL/min, which corresponds to a linear velocity of 19 cm/h above the membrane.
Table 1 also shows the lysozyme binding capacities of membranes modified by adsorption of PAA, PAA/PAH/PAA, and PAA/PEI/PAA films. Figures S-2 and S-3 show representative breakthrough curves. Maximum binding using a single PAA layer occurs for films deposited at pH 3, which is consistent with the high Cu2+ binding for this membrane (see Table 1, column 5). PAA/PAH/PAA or PAA/PEI/PAA multilayers provide 20–30% higher binding capacities than single PAA layers, with the PAA/PEI/PAA film showing especially high capacities. This high lysozyme adsorption with PAA/PEI/PAA agrees well with data for Cu2+ binding.
To further simplify film formation, we reduced the adsorption time from 40 min to 5 min for deposition of each polyelectrolyte. We also decreased the rinsing time from 20 min to 5 min. The binding capacity of PAA/PEI/PAA-containing membranes modified using the short deposition times is 108±1 mg/cm3, so reducing the total deposition time 6-fold decreased the binding capacity only 10%. By reducing the adsorption time, complete deposition of a PAA/PEI/PAA film requires only 30 min. We should note that these binding capacities are more than 6-fold greater than those in our prior study in part due to the low-pH deposition but also because the membranes have smaller pores (1.2 μm versus 5 μm) that lead to higher surface areas.24
Protein Binding as a Function of Flow Rate
Compared to column-based methods, membrane adsorbers are particularly attractive for rapid protein capture because radial diffusion distances are short, and convection brings proteins to binding sites. Moreover, rapid flow rates are possible because of modest pressure drops.11 If we define the dynamic capacity as the amount of protein bound when breakthrough reaches 10%, typical dynamic capacities for the protein binding studies described above are about 1/3 of the equilibrium binding capacities. However, these experiments all employed flow rates of 1 mL/min. To better examine the dynamics of protein binding, we compared the breakthrough curves for lysozyme binding to PAA/PEI/PAA-modified membranes at solution flow rates of 1 and 30 mL/min. These flow rates correspond to linear velocities of 19 cm/h and 570 cm/h, and residence times of ~1000 msec and 35 msec, respectively. (Note that these residence times assume a membrane porosity of 50%, whereas the linear velocity is that above the membrane.) As Figure 3 shows, the breakthrough curves are not very different at the two flow rates and dynamic capacities are similar (within about 25%).
Figure 3.
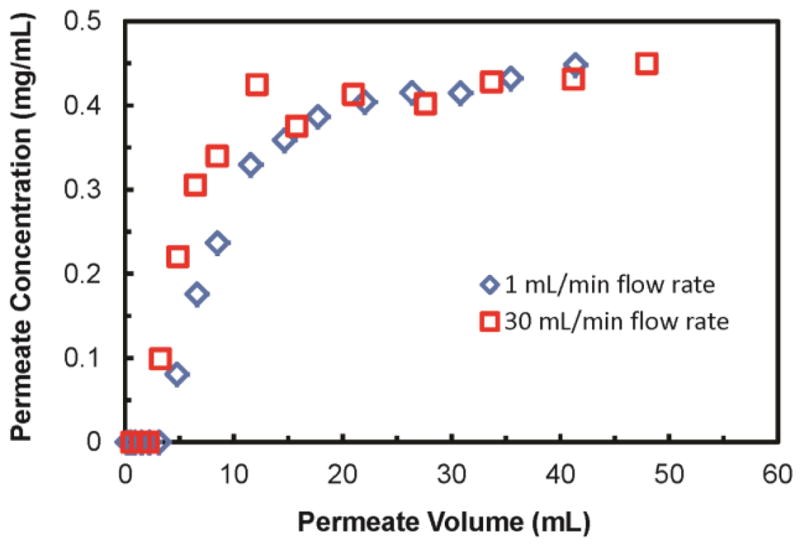
Breakthrough curves for the passage of 0.45 mg/mL lysozyme through a PAA/PEI/PAA-modified (deposition pH of 3) nylon membrane at flow rates of 1 mL/min (blue diamonds) and 30 mL/min (red squares).
Polyelectrolyte Films in Other Membrane Materials
As a test of the versatility of layer-by-layer adsorption for membrane modification, we immobilized polyelectrolyte layers in PES and non-hydroxylated nylon membranes and studied lysozyme binding to these systems. For non-hydroxylated membranes, we deposited PSS/PAH/PAA and PAA/PEI/PAA films using a deposition pH of 3 for all polyelectrolytes except PSS. The lysozyme binding capacities for the PSS/PAH/PAA- and PAA/PEI/PAA-modified membranes were 68±3 mg/cm3 and 72±5 mg/cm3, respectively, or about 60% of the binding capacities for corresponding hydroxylated nylon membranes. The drop in binding capacity could stem either from less adsorption to the non-hydroxylated membrane or a difference in the surface areas of the two substrates. After treating the non-hydroxylated membranes with phosphoric acid in formalin to introduce hydroxyl groups,47 PAA/PEI/PAA adsorption leads to a lysozyme binding capacity of 103±2 (105±7 from elution) mg/cm3. Assuming that the formaldehyde treatment does not increase surface area, this result suggests that hydroxylation increases the quantity of polyelectrolyte adsorption. Unfortunately, PES membranes plugged during deposition of PSS/PAH/PAA and PAA/PEI/PAA films. The membrane geometry is obviously a crucial factor in determining whether polyelectrolyte adsorption can occur without plugging the membrane. When a monolayer of PAA (deposition pH of 3) was immobilized in the PES membrane, the lysozyme binding capacity was only 21±7 mg/cm3, or about 25% of that for a similar hydroxylated nylon membrane. Selection of the appropriate membrane substrate is thus vital to optimizing membranes modified with polyelectrolytes.
Con A Binding to Membranes with Films Containing NTA-Cu2+
To increase the specificity of protein binding, we derivatize PAA films with metal-ion complexes that bind proteins containing accessible histidine groups. We initially examined capture of a readily available protein, Con A, through interaction with NTA-Cu2+ complexes. Figures S-4, S-5, and S-6 show representative breakthrough curves for passage of Con A solutions through different modified nylon membranes, and Table 2 presents the protein-binding capacities determined both from breakthrough curves and protein elution. For modification with either PSS/PAH/PAA-NTA-Cu2+ or PAA-NTA-Cu2+, the Con A binding capacities decrease with an increase in deposition pH, following the trend in the amount of Cu2+ bound in the different membranes (Table 1, column 5). Membranes modified with PAA/PEI/PAA-NTA-Cu2+ and PAA/PAH/PAA-NTA-Cu2+ show the highest Con A-binding capacities (Table 2) of the membranes tested. Deposition of even more polyelectrolyte bilayers might increase capacity, but it would also lead to plugging of membrane pores or large decreases in permeability. Overall, Con A binding capacities are ~35% lower than for lysozyme, presumably because the large size of Con A (108 kDa)48 prevents access to some binding sites. Con A also interacts with different species (metal-ion complexes) in the film.
Table 2.
Con A binding capacities of hydroxylated nylon membranes modified by different polyelectrolyte films.a
| Polyelectrolyte films | pH of PAH and/or PAA deposition solution | Con A binding from breakthrough curve (mg/cm3) | Con A binding from elution (mg/cm3) |
|---|---|---|---|
| PSS/PAH/PAA-NTA-Cu2+ | 2 | 65±1 | 69±2 |
| 3 | 53±1 | 52±2 | |
| 4 | 36±4 | 33±2 | |
| 5 | 19±4 | 21±4 | |
| PAA-NTA-Cu2+ | 2 | 60±1 | 59±2 |
| 3 | 52±2 | 52±6 | |
| 4 | 37±4 | 37±5 | |
| 5 | 12±2 | 11±3 | |
| PAA/PAH/PAA-NTA-Cu2+ | 3 | 69±1 | 71±2 |
| PAA/PEI/PAA-NTA-Cu2+ | 3 | 73±4 | 71±7 |
Each experiment was repeated with two different membranes, and the ± values represent the difference between the average and the data points.
HisU Binding to PAA/PEI/PAA-NTA-Ni2+-modified Membranes
Interactions with histidine residues are weaker for NTA-Ni2+ than NTA-Cu2+, so the incorporation of NTA-Ni2+complexes in columns and membranes allows highly selective binding of proteins that contain polyhistidine tags. In fact, polyhistidine is the most common tag for recombinant protein purification. We employed HisU as a model His-tagged protein to determine the binding capacity of modified nylon membranes. Unfortunately, even though HisU is the least expensive His-tagged protein of which we are aware, the high cost of this protein prohibits determining binding capacities on multiple membranes. We selected a PAA/PEI/PAA-NTA-Ni2+-modified membrane to determine HisU binding capacity because the related membranes modified with PAA/PEI/PAA-NTA-Cu2+ exhibit the most extensive binding of Con A. The breakthrough curve for HisU binding to a PAA/PEI/PAA-NTA-Ni2+-modified nylon membrane (Figure 4) reveals a HisU binding capacity of 93 mg/cm3. The corresponding capacity determined from HisU elution, 97 mg/cm3, is about twice the value of typical binding capacities of commercial IMAC resins49 and similar to the capacities (88 ± 4 mg/cm3) that we obtained by modifying nylon membranes with polymer brushes.36 Additionally, the brush-containing membranes are more difficult to prepare and less permeable.
Figure 4.
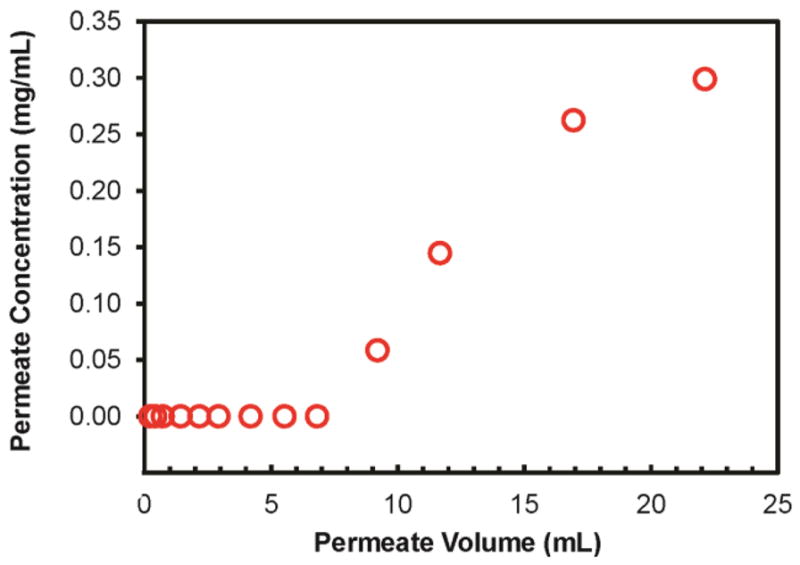
Breakthrough curve for passage of a 0.3 mg/mL HisU solution through a hydroxylated nylon membrane modified with PAA/PEI/PAA-NTA-Ni2+. The feed solution contained 0.3 mg protein/mL, and the solution flow rate was 1.0 mL/min.
Purification of His-Tagged CSN8 and HisU from Cell Extracts and Protein Mixtures
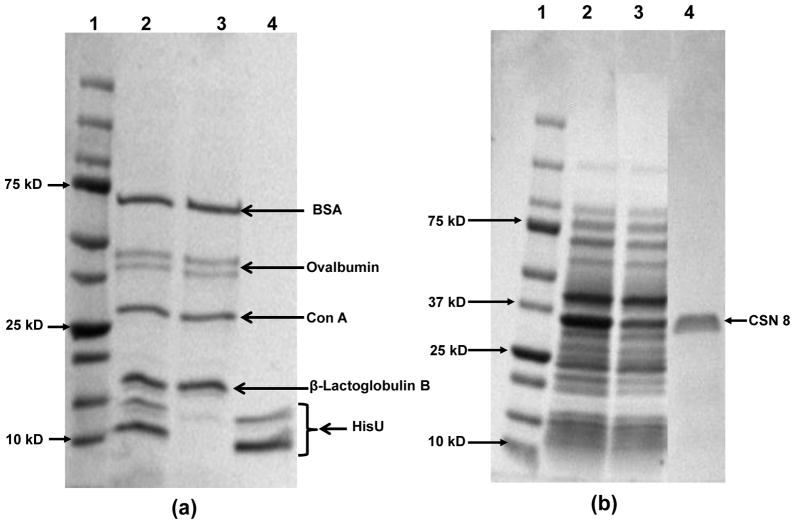
To demonstrate the selectivity of PAA/PEI/PAA-Ni2+-modified membranes for capture of His-tagged proteins, we first separated HisU from a mixture of HisU, Con A, BSA, Ovalbumin, and β-Lactoglobulin B. These model proteins, except HisU, do not have His-tags and serve as contaminating proteins in this experiment. SDS-PAGE analysis of the mixed-protein solution exiting the membrane suggests successful removal of HisU (Figure 5(a) lane 3), while the eluate shows only bands due to HisU (Figure 5(a) lane 4, note that even the as received HisU shows two bands). Thus, the membranes are highly selective for capturing HisU.
Figure 5.
SDS-PAGE analysis (Coomassie blue staining) of (a) purification of a mixture of BSA, Ovalbumin, Con A, β-Lactoglobulin B, and HisU: lane 1- a protein ladder; lane2- the protein solution; lane 3- the protein solution that passed through the membrane; and lane 4-the eluate from the membrane. (b) purification of CSN 8 from a cell extract: lane 1- a protein ladder; lane 2- a cell extract form BL21DE3 cells with overexpressed His-tagged CSN8 protein; lane 3- the cell extract after passing through from the membrane; and lane 4- the eluate from the membrane. Figure S-8 shows the original gel for Figure 5(b).
In addition to selectivity, high recovery is important in most protein purifications. Because we do not know the concentration of CSN8 in the cell extract, we determined recovery for capture of HisU from the protein mixture. The high purity of the eluted HisU (as demonstrated by gel electrophoresis) allowed us to use a Bradford assay to demonstrate that recovery was 95 ± 3%, even with the loading, washing, and elution steps.
Purification of His-tagged CSN8 from whole cell extracts further demonstrates the high selectivity and potential applications of membranes modified with PAA/PEI/PAA-Ni2+. Figure 5(b) shows the SDS-PAGE analysis of the cell extract (lane 2) and the eluate from a membrane that was loaded with the cell extract and washed with buffers (lane 4). Remarkably, the eluate contains only one strong band, suggesting that the purity of the captured CSN8 is above 95%. Moreover, the complete membrane purification process requires less than 20 minutes, including loading cell lysate on the membrane, washing with 3 different buffers, and eluting.
Stability of Membranes Modified with PAA/PEI/PAA
In studies of the stability of membranes modified with PAA/PEI/PAA, we employed two membranes for 6 repetitions of lysozyme binding and elution. The binding capacity ranged from 125 to 141 mg/cm3 over the 6 replicates (see Figure S-7). Thus the membranes are stable, but we observed gradually declining flow rates during the 6th experiment for both modified membranes. Note that this stability occurs even when using 1 M KSCN for protein elution.
We also tested the stability of modified membranes under depyrogenation conditions. In this case we first bound lysozyme in nylon membranes modified with PAA/PEI/PAA films deposited at pH 3. After lysozyme elution with 1 M KSCN, we circulated 10 mL 1 N NaOH through the modified membranes for 1 h and repeated the binding experiment. The binding capacities before and after treatment with 1 M NaOH were 131±1 mg/cm3 and 129±3 mg/cm3 respectively. Thus, treatment of the membranes with NaOH to remove or disable toxins prior to reuse might be feasible, but more research is needed in this area.
Conclusions
At pH 3, adsorption of as little as one layer of PAA in a porous membrane creates a high density of –COOH groups that function as either ion-exchange sites or points for attachment of metal-ion complexes that selectively bind proteins. Increasing the adsorption pH leads to much less protein binding, whereas adsorption of a PEI/PAA bilayer on the initial PAA layer increases lysozyme binding from 89 to 120 mg/cm3 of membrane. Polyelectrolyte adsorption at low pH is much simpler than growth of polymer brushes in membranes, and the binding capacities that result from the two modification methods are similar. Derivatization of PAA/PEI/PAA-modified membranes with NTA-Ni2+ complexes yields materials that selectively capture His-tagged protein with >90% recovery.
Supplementary Material
Acknowledgments
We are grateful to the U.S. National Institutes of Health (GM080511) for funding this work.
Footnotes
Supporting Information Available
Tables S1–S6- studies of flux through bare and modified membranes, Figures S1- SEM images of bare and modified membranes, Figures S2–S6- breakthrough curves, Figure S7- membrane reuse, and Figure S8- an SDS PAGE gel. This material is available free of charge via the Internet at http:/pubs.acs.org.
References
- 1.Clarke W, Hage DS. Clinical Applications of Affinity Chromatography. Sep Pur Rev. 2003;32:19–60. [Google Scholar]
- 2.Kawai T, Saito K, Lee W. Protein Binding to Polymer Brush, Based on Ion-Exchange, Hydrophobic, and Affinity Interactions. J Chromatogr B. 2003;790:131–142. doi: 10.1016/s1570-0232(03)00090-4. [DOI] [PubMed] [Google Scholar]
- 3.Bhut BV, Husson SM. Dramatic Performance Improvement of Weak Anion-Exchange Membranes for Chromatographic Bioseparations. J Membr Sci. 2009;337:215–223. [Google Scholar]
- 4.Ghosh R. Protein Separation Using Membrane Chromatography: Opportunities and Challenges. J Chromatogr A. 2002;952:13–27. doi: 10.1016/s0021-9673(02)00057-2. [DOI] [PubMed] [Google Scholar]
- 5.Datta S, Bhattacharyya D, Ray PD, Nath A, Toborek M. Effect of Pre-Filtration on Selective Isolation of Tat Protein by Affinity Membrane Separation: Analysis of Flux, Separation Efficiency, and Processing Time. Sep Sci Technol. 2007;42:2451–2471. [Google Scholar]
- 6.Brandt S, Goffe RA, Kessler SB, O’Connor JL, Zale SE. Membrane-Based Affinity Technology for Commercial Scale Purifications. Nat Biotechnol. 1988;6:779– 782. [Google Scholar]
- 7.Thömmes J, Etzel M. Alternatives to Chromatographic Separations. Biotechnol Prog. 2007;23:42–45. doi: 10.1021/bp0603661. [DOI] [PubMed] [Google Scholar]
- 8.Roper DK, Lightfoot EN. Separation of Biomolecules Using Adsorptive Membranes. J Chromatogr A. 1995;702:3–26. [Google Scholar]
- 9.Lightfoot EN, Moscariello JS. Bioseparations. Biotechnol Bioeng. 2004;87:259–273. doi: 10.1002/bit.20111. [DOI] [PubMed] [Google Scholar]
- 10.Thömmes J, Kula MR. Membrane Chromatography-an Integrative Concept in the Downstream Processing of Proteins. Biotechnol Prog. 1995;11:357–367. [Google Scholar]
- 11.Saxena A, Tripathi BP, Kumar M, Shahi VK. Membrane-Based Techniques for the Separation and Purification of Proteins: An Overview. Adv Colloid Interface Sci. 2009;145:1–22. doi: 10.1016/j.cis.2008.07.004. [DOI] [PubMed] [Google Scholar]
- 12.Zeng XF, Ruckenstein E. Membrane Chromatography: Preparation and Applications to Protein Separation. Biotechnol Prog. 1999;15:1003–1019. doi: 10.1021/bp990120e. [DOI] [PubMed] [Google Scholar]
- 13.Sun L, Dai J, Baker GL, Bruening ML. High-Capacity, Protein-Binding Membranes Based on Polymer Brushes Grown in Porous Substrates. Chem Mater. 2006;18:4033–4039. [Google Scholar]
- 14.Jain P, Baker GL, Bruening ML. Applications of Polymer Brushes in Protein Analysis and Purification. Ann Rev Anal Chem. 2009;2:387–408. doi: 10.1146/annurev-anchem-060908-155153. [DOI] [PubMed] [Google Scholar]
- 15.Jain P, Dai J, Baker GL, Bruening ML. Rapid Synthesis of Functional Polymer Brushes by Surface-Initiated Atom Transfer Radical Polymerization of an Acidic Monomer. Macromolecules. 2008;41:8413–8417. [Google Scholar]
- 16.Bruening ML, Dotzauer DM, Jain P, Lu O, Baker GL. Creation of Functional Membranes Using Polyelectrolyte Multilayers and Polymer Brushes. Langmuir. 2008;24:7663–7673. doi: 10.1021/la800179z. [DOI] [PubMed] [Google Scholar]
- 17.Jain P, Sun L, Dai J, Baker GL, Bruening ML. High-Capacity Purification of His-Tagged Proteins by Affinity Membranes Containing Functionalized Polymer Brushes. Biomacromolecules. 2007;8:3102–3107. doi: 10.1021/bm700515m. [DOI] [PubMed] [Google Scholar]
- 18.Jain P, Vyas MK, Geiger JH, Baker GL, Bruening ML. Protein Purification with Polymeric Affinity Membranes Containing Functionalized Poly(Acid) Brushes. Biomacromolecules. 2010;11:1019–1026. doi: 10.1021/bm9014792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bhut BV, Christensen KA, Husson SM. Membrane Chromatography Protein Purification from E. Coli Lysate Using Newly Designed and Commercial Anion-Exchange Stationary Phases. J Chromatogr, A. 2010;1217:12. doi: 10.1016/j.chroma.2010.05.049. [DOI] [PubMed] [Google Scholar]
- 20.Ulbricht M, Yang H. Porous Polypropylene Membranes with Different Carboxyl Polymer Brush Layers for Reversible Protein Binding Via Surface-Initiated Graft Copolymerization. Chem Mater. 2005;17:2622–2631. [Google Scholar]
- 21.Bhut BV, Wickramasinghe SR, Husson SM. Preparation of High-Capacity, Weak Anion-Exchange Membranes for Protein Separations Using Surface-Initiated Atom Transfer Radical Polymerization. J Membr Sci. 2008;325:176–183. [Google Scholar]
- 22.He DM, Ulbricht M. Preparation and Characterization of Porous Anion-Exchange Membrane Adsorbers with High Protein-Binding Capacity. J Membr Sci. 2008;315:155–163. [Google Scholar]
- 23.Dai J, Bao Z, Sun L, Hong SU, Baker GL, Bruening ML. High-Capacity Binding of Proteins by Poly(Acrylic Acid) Brushes and Their Derivatives. Langmuir. 2006;22:4274–4281. doi: 10.1021/la0600550. [DOI] [PubMed] [Google Scholar]
- 24.Liu G, Dotzauer DM, Bruening ML. Ion-Exchange Membranes Prepared Using Layer-by-Layer Polyelectrolyte Deposition. J Membr Sci. 2010;354:198–205. doi: 10.1016/j.memsci.2010.02.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Datta S, Cecil C, Bhattacharyya D. Functionalized Membranes by Layer-by-Layer Assembly of Polyelectrolytes and in Situ Polymerization of Acrylic Acid for Applications in Enzymatic Catalysis. Ind Eng Chem Res. 2008;47:4586–4597. doi: 10.1021/ie800142d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. [Accessed April 2: 2012];Mustang® Membrane Chromatography Starter Kits. http://www.pall.com/main/Biopharmaceuticals/Product.page?id=33053 (Mustang S membranes have a protein binding capacity of 45–50 mg/mL.
- 27.Secrist KE, Nolte AJ. Humidity Swelling/Deswelling Hysteresis in a Polyelectrolyte Multilayer Film. Macromolecules. 2011;44:2859–2865. [Google Scholar]
- 28.Quinn A, Such GK, Quinn JF, Caruso F. Polyelectrolyte Blend Multilayers: A Versatile Route to Engineering Interfaces and Films. Adv Funct Mater. 2008;18:17–26. [Google Scholar]
- 29.Choi J, Rubner MF. Influence of the Degree of Ionization on Weak Polyelectrolyte Multilayer Assembly. Macromolecules. 2005;38:116–124. [Google Scholar]
- 30.Dubas ST, Schlenoff JB. Polyelectrolyte Multilayers Containing a Weak Polyacid: Construction and Deconstruction. Macromolecules. 2001;34:3736–3740. [Google Scholar]
- 31.Bieker P, Schönhoff M. Linear and Exponential Growth Regimes of Multilayers of Weak Polyelectrolytes in Dependence on pH. Macromolecules. 2010;43:5052–5059. [Google Scholar]
- 32.Harris JJ, Bruening ML. Electrochemical and in Situ Ellipsometric Investigation of the Permeability and Stability of Layered Polyelectrolyte Films. Langmuir. 2000;16:2006–2013. [Google Scholar]
- 33.Mendelsohn JD, Yang SY, Hiller JA, Hochbaum AI, Rubner MF. Rational Design of Cytophilic and Cytophobic Polyelectrolyte Multilayer Thin Films. Biomacromolecules. 2003;4:96–106. doi: 10.1021/bm0256101. [DOI] [PubMed] [Google Scholar]
- 34.Salloum DS, Schlenoff JB. Protein Adsorption Modalities on Polyelectrolyte Multilayers. Biomacromolecules. 2004;5:1089–1096. doi: 10.1021/bm034522t. [DOI] [PubMed] [Google Scholar]
- 35.Ma Y, Bhattacharjee S, Wijeratne S, Bruening ML, Baker GL. to be submitted. [Google Scholar]
- 36.Anuraj N, Bhattacharjee S, Geiger JH, Baker GL, Bruening ML. An All-Aqueous Route to Polymer Brush-Modified Membranes with Remarkable Permeabilites and Protein Capture Rates. J Membr Sci. 2012;389:117–125. doi: 10.1016/j.memsci.2011.10.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mossessova E, Lima CD. Ulp1-SUMO Crystal Structure and Genetic Analysis Reveal Conserved Interactions and a Regulatory Element Essential for Cell Growth in Yeast. Mol Cell. 2000;5:865–876. doi: 10.1016/s1097-2765(00)80326-3. [DOI] [PubMed] [Google Scholar]
- 38.Wang TC, Rubner MF, Cohen RE. Polyelectrolyte Multilayer Nanoreactors for Preparing Silver Nanoparticle Composites: Controlling Metal Concentration and Nanoparticle Size. Langmuir. 2002;18:3370–3375. [Google Scholar]
- 39.Burke SE, Barrett CJ. Acid-Base Equilibria of Weak Polyelectrolytes in Multilayer Thin Films. Langmuir. 2003;19:3297–3303. [Google Scholar]
- 40.Petrov AI, Antipov AA, Sukhorukov GB. Base-Acid Equilibria in Polyelectrolyte Systems: From Weak Polyelectrolytes to Interpolyelectrolyte Complexes and Multilayered Polyelectrolyte Shells. Macromolecules. 2003;36:10079–10086. [Google Scholar]
- 41.Yoo D, Shiratori SS, Rubner MF. Controlling Bilayer Composition and Surface Wettability of Sequentially Adsorbed Multilayers of Weak Polyelectrolytes. Macromolecules. 1998;31:4309–4318. [Google Scholar]
- 42.Shiratori SS, Rubner MF. pH-Dependent Thickness Behavior of Sequentially Adsorbed Layers of Weak Polyelectrolytes. Macromolecules. 2000;33:4213–4219. [Google Scholar]
- 43.Lee D, Nolte AJ, Kunz AL, Rubner MF, Cohen RE. pH-Induced Hysteretic Gating of Track-Etched Polycarbonate Membranes: Swelling/Deswelling Behavior of Polyelectrolyte Multilayers in Confined Geometry. J Am Chem Soc. 2006;128:8521–8529. doi: 10.1021/ja0608803. [DOI] [PubMed] [Google Scholar]
- 44.Ito Y, Park YS, Imanishi Y. Visualization of Critical pH-Controlled Gating of a Porous Membrane Grafted with Polyelectrolyte Brushes. J Am Chem Soc. 1997;119:2739–2740. [Google Scholar]
- 45.Ito Y, Inaba M, Chung DJ, Imanishi Y. Control of Water Permeation by pH and Ionic Strength through a Porous Membrane Having Poly(Carboxylic Acid) Surface-Grafted. Macromolecules. 1992;25:7313–7316. [Google Scholar]
- 46.Bergers JJ, Vingerhoeds MH, van Bloois L, Herron JN, Janssen LHM, Fischer MJE, Crommelin DJA. The Role of Protein Charge in Protein-Lipid Interactions. pH-Dependent Changes of the Electrophoretic Mobility of Liposomes through Adsorption of Water-Soluble, Globular Proteins. Biochemistry. 1993;32:4641–4649. doi: 10.1021/bi00068a023. [DOI] [PubMed] [Google Scholar]
- 47.Xu FJ, Zhao JP, Kang ET, Neoh KG, Li J. Functionalization of Nylon Membranes Via Surface-Initiated Atom-Transfer Radical Polymerization. Langmuir. 2007;23:8585–8592. doi: 10.1021/la7011342. [DOI] [PubMed] [Google Scholar]
- 48.Hardman KD, Wood MK, Schiffer M, Edmundson AB, Ainsworth CF. Structure of Concanavalin A at 4.25-Ångström Resolution. Proc Natl Acad Sci U S A. 1971;68:1393–1397. doi: 10.1073/pnas.68.7.1393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. [accessed April 2: 2012];Qiagen Ni-NTA — Binding Capacity of up to 50 mg/ml. http://www1.qiagen.com/products/protein/purification/ni-ntabinding.aspx.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.