Abstract
The divalent metal transporter 1 (DMT1) is essential for cellular uptake of iron, mediating iron absorption across the duodenal brush border membrane. We have previously shown that with iron feeding DMT1 in the brush border membrane undergoes endocytosis into the subapical compartment of enterocytes. To understand the mechanisms of iron-induced endocytosis of DMT1, we used the yeast two-hybrid system to find proteins that interact with DMT1 and isolated from a rat duodenal cDNA library a protein that interacts specifically with the IRE containing isoform of DMT1 {DMT1 [iron-responsive element (IRE)]}. The protein (Genbank AY336075) is 97.5% identical with peripheral benzodiazepine receptor-associated protein 7 (PAP7), a protein that interacts with the peripheral benzodiazepine receptor. PAP7 is ubiquitously expressed in the rat and in multiple cell lines with consensus sequences including a nuclear localization signal and a Golgi dynamic domain. PAP7, expressed on the brush border of rat duodenum, copurified with DMT1 in brush border membrane vesicles, and following iron feeding, was internalized in parallel with the internalization of DMT1. To determine if PAP7 plays a role in cellular iron metabolism, we downregulated PAP7 expression in K562 cells with small interfering RNA. Following the decrease in PAP7 protein, DMT1 (IRE) protein but not mRNA was significantly downregulated but without effect on DMT1 (non-IRE), transferin (Tf)R1, or ferritin expression. Lowered levels of PAP7 resulted also in decreased cell proliferation and G1 cell cycle arrest. These data are consistent with PAP7 interacting with DMT1 (IRE) and regulating DMT1 (IRE) expression in K562 cells by modulating expression of DMT1 (IRE) protein.
Keywords: divalent metal transporter 1, acyl-coenzyme A binding domain containing 3, iron transport, small interfering ribonucleic acid
iron is essential for cell growth and survival with various ferriproteins and heme-containing proteins being involved in a myriad of cell functions including DNA synthesis, oxygen transport, and cell respiration. As a consequence, a deficiency of iron has pleiotropic detrimental effects on the organism while an excess of iron is also deleterious and through oxidative stress may injure many tissues including, for example, the production of hepatic cirrhosis, diabetes mellitus, and cardiomyopathies. The human organism uses a number of strategies to maintain iron homeostasis. Principally, however, iron stores are maintained by regulating iron absorption from the duodenum, which is the only site in which regulation of body iron stores can occur. The transport of iron across the brush border membrane (BBM) of the duodenum is via the divalent metal transporter 1 (DMT1; SLC11A2; Ref. 10). DMT1 is a cotransporter of protons and divalent metals with a pH optimum of about pH 5.5 for iron transport (9, 19). The exact mechanisms by which iron is transported by DMT1 are obscure. We, and others (22, 30), have observed that in intestinal epithelial cells with iron feeding DMT1 is internalized from the BBM into vesicles within the subapical cytosolic space. It is not known whether the internalization of DMT1 is required for iron transport and/or is part of the iron uptake regulatory mechanism, although transcytosis of vesicles derived from the apical BBM and vesicles derived from the basolateral surface has been described. In order for DMT1 to exhibit such movement, it is likely that DMT1 interacts with other cellular proteins. These interactions would enable DMT1 in response to a signal generated by the flux of iron into the cell to undergo translocation to a new cellular localization. To begin to understand the interactions of DMT1 with other intestinal cellular proteins, we have used the yeast two-hybrid system as a method for detecting proteins that can possibly interact with DMT1. DMT1 is a complex protein with 12 putative transmembrane regions. Both the N- and COOH-terminal portions of the protein are cytoplasmic. There are multiple DMT1 mRNA isoforms formed by alternative splice mechanisms (13). Isoform I contains an iron-responsive element (IRE) in the 3′-untranslated region and has an 18 amino acid COOH terminus that is uniquely different from the 21 amino acid COOH terminus of isoform II (non-IRE). In intestinal epithelium, isoform I is primarily expressed while isoform II predominates in erythroid cells (3, 5). In the current studies, we elected to use the COOH terminus of isoform I as the bait in the yeast two-hybrid system with prey from a rat intestinal cDNA library to determine other proteins that interact with DMT1. In these studies, cDNA clones were obtained representing three different proteins, one of which we designated as DMT1-associated protein (DAP; AY336075). DAP is 97.5% identical to the previously described mouse peripheral-type benzodiazepine receptor (PBR)-associated protein 7 (PAP7). DAP was ubiquitously expressed in various organs. Because of the very high homology between DAP and PAP7 (17) and the observation that PAP7 interacts with DMT1 in the neuronal PC12 cell line affecting iron uptake in neuronal cells (6), we now refer to DAP as PAP7. In addition, PAP7 has also been designated as acyl-coenzyme A binding domain containing 3 (ACBD3), golgi complex-associated protein (GOCAP), and golgi resident protein 60 (GCP60; Refs. 8, 14, 25). In the current studies, we demonstrate by immunohistochemistry that in both rat intestine and in Caco2 cells, a cell line used as a model for intestinal iron uptake, that iron feeding affects both the expression and cellular location of PAP7. Further, we show that in BBM vesicles (BBMV) isolated from rat duodenum PAP7 and DMT1 can be copurified by immunoabsorption. To determine if PAP7 is involved in cellular iron metabolism, we employed small interferin (si)RNAs to specifically decrease PAP7 expression in K562 cells, a cell line that can be induced to undergo erythroid differentiation. In these cells, decreased levels of PAP7 decreased the expression of DMT1 (IRE) protein but not DMT1 (IRE) mRNA. Decreased levels of PAP7 had no effect on expression of DMT1 (non-IRE), transferrin receptor 1, or ferritin, suggesting that the effects of PAP7 on DMT1 (IRE) expression were specific. Transfection with the PAP7 siRNA resulted also in the more protean effects of decreased cell proliferation and arrest of cells in G1 of the cell cycle. These data are consistent with PAP7 regulating DMT1 expression in K562 cells by modulating turnover of DMT1 (IRE) protein and of PAP7 also having more global effects on cellular metabolism.
MATERIALS AND METHODS
Materials.
Restriction enzymes were from New England Biolabs, (Beverly, MA). Electrophoresis reagents and materials were obtained from Bio-Rad Laboratories, (Hercules, CA). Nitrocellulose (0.45 μm) was from Osmonics (Westborough, MA). S·Tag thrombin purification kits, recombinant enterokinase kits, pET30a(+) bacterial expression vector, and Single Tube Protein System 3, T7 kits were obtained from Novagen (San Diego, CA). Seize X protein A immunoprecipitation kit and SuperSignal West Pico chemiluminescent substrate were from Pierce Chemical (Rockford, IL). The various oligonucleotides were obtained from IDT (Coralville, IA). The nucleofector and cell line nucleofector Kit V were from Amaxa (Gaithersburg, MD). The Matchmaker yeast two-hybrid system, multiple tissue expression array (MTE), RNA I-Ready pSIREN-Retro-Q-ZsGreen vector, and enhanced green fluorescent protein (EGFP)-C2 vector were from Clontech (Mountain View, CA). Cells were enumerated using a ISOTON II and a cell counter a from Beckman Coulter (Fullerton, CA). TRIzol, Superscript III first-strand synthesis system for RT-PCR, SYBR Green ER qPCR SuperMix for icylcer instrument, Mito-tracker, TOPRO-3, ProLong antifade kit, and RNase A were from Invitrogen (Carlsbad, CA). FBS was from Atlanta Biologicals (Lawrenceville, GA). RPMI 1640 was from Mediatech (Herndon, VA), and paraformaldehyde was from Fisher Scientific (Pittsburgh, PA). BSA, propidium iodide, (holo-Tf), insulin, selenium dioxide (SeO2), protease inhibitor cocktails, and all other chemicals were obtained from Sigma-Aldrich (St. Louis, MO).
Antibodies to DMT1 (total), DMT1 (non-IRE), and DMT1 (IRE) were described previously (30). Other antibodies included anti-transferrin receptor 1 (TfR1), anti-Golgi97, Alexa 488 labeled anti-rabbit IgG, and Alexa 594 labeled anti-mouse IgG from Invitrogen (Carlsbad, CA), anti-actin from Sigma-Aldrich, (St. Louis, MO), anti-ferritin heavy chain from Abcam (Cambridge, MA), and anti-GFP, horseradish peroxidase labeled anti-rabbit, and mouse IgG from Santa Cruz Biotechnology (Santa Cruz, CA).
Animals.
Phenotypically normal heterozygous (+/b) and anemic homozygous (b/b) Belgrade rats were bred and maintained in the animal quarters of Louisiana State University Health Sciences Center. The animal protocol for the experiments was approved by the Committee of Animal Care and Use at Louisiana State University Health Sciences Center. Both the heterozygous and homozygous rats were obtained by mating homozygous (b/b) males with heterozygous (+/b) females. The homozygous (b/b) pups were identified by high reticulocyte counts and low hematocrits. The rats used for the study were weaned onto a standard Harlan Tekled 22/5 rodent diet for 2 mo. The iron loading of +/+ rats was achieved with a rat diet supplemented with carbonyl iron (3.0 g iron per kg diet) plus two weekly intraperitoneal injections of 0.1 mg/g Imferon.
Tissue samples were taken between 10:00 AM and 1:00 PM. The duodenum and colon were removed and rinsed with ice-cold saline and the mucosa scraped and frozen at −70°C. Other tissues such as brain, spleen, kidney, liver, and lung were also removed and immediately frozen.
Cell culture.
Caco2 (HTB 37) and K562 (CCL-243) cell lines from the American Type Culture Collection (Rockville, MD) were grown and maintained in DMEM or RPMI 1640 supplemented with 10% FBS (Atlanta Biologicals, Lawrenceville, GA) and antibiotics (100 U/ml penicillin-G and 100 U/ml streptomycin).
Yeast two-hybrid screen.
The MATCHMAKER yeast two-hybrid system (Clontech Laboratories) was utilized in this study with screening performed according to the manufacturer's instructions. The COOH-terminal cytoplasmic domain (corresponding to amino acids 512–561) of rat DMT1-IRE was cloned into pGBKT7 as the bait construct (BD-CT512). The prey was constructed from the insertion of a rat duodenal cDNA library into pGADT7 to form the AD-library.
The cDNA library was made from high quality poly(A)+ RNA isolated from total RNA from freshly collected rat duodenum and purified twice on a oligo(dT)-cellulose column. The poly(A)+ RNA in effluent fractions was verified to have homogeneously distributed RNA from large (>6 kb) to small RNA, and a cDNA library was synthesized from the poly(A)+ mRNA with a TimeSaver cDNA synthesis kit according to the manufacturer's instructions (Amersham Pharmacia Biotech, NJ). To ensure that the cDNA-library had both 3′- and 5′-cDNAs the library was then mixed with oligo(dT)12–18 (0.5 μg) and random hexamers (0.037 μg) were used as primers to synthesize the first-strand cDNA from 5 μg poly(A)+ RNA. After the second strand synthesis, cDNAs >400 bp were ligated with EcoRI/NotI adapters, inserted into dephosphorylated EcoRI-digested pGADT7 plasmids, and amplified once with ElectroMax DH10B cells (Life Technologies, MD). The AD-library was transformed into the AH109 host previously transformed with BD-CT512 and grown under high stringency conditions. With interaction of bait and prey, BD and AD are brought together activating the downstream β-galactosidase reporter gene and generating a blue color for selection. cDNA clones were obtained representing three different proteins, Snapin, PAP7, and rat fatty acid binding protein. The interaction of these cDNAs with BD-CT512 non-IRE, which represents the COOH terminus of DMT1 isoform II was analyzed by the two-hybrid system and failed to demonstrate any interaction. The clones of PAP7 encoded amino acids 171–536 (accession no. AY336075).
PAP7 cDNA constructs and preparation of anti-PAP7 antiserum.
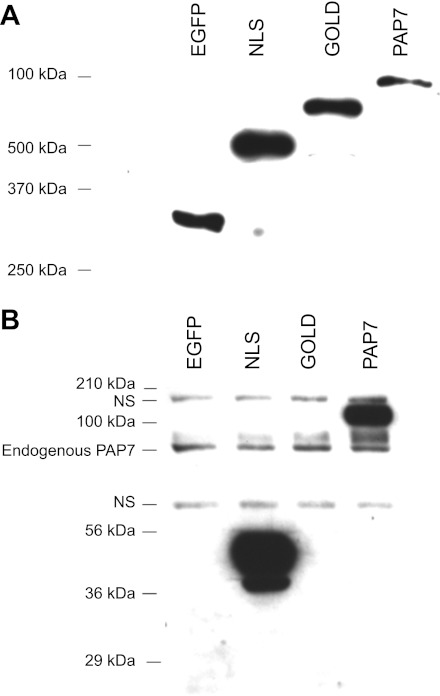
The PAP7 partial cDNA from the original clone (including the COOH-terminal amino acid sequence from amino acids 171 to 526) was fused into the EcoRI/EcoRI site of pET-30a(+) bacterial expression vector (Novagen, San Diego, CA) to form pET-30a(+)-PAP7–31 with an N-terminal His·tag and S·tag. The fusion site was verified by sequencing. The pET30a(+)-PAP7–31 was transformed into Escherichia coli (BL21) for expression of a His·tagged and S·tagged partial PAP7 (amino acids 171–526) fusion protein (SHisPAP7–31). The purified fusion protein was confirmed by matrix-assisted laser desorption ionization time-of-flight analysis. The His and S·tags was removed by recombinant enterokinase, the purified protein emulsified with Freund's complete adjuvant and then inoculated intradermally into a New Zealand white rabbit. The rabbit was boosted on days 30 and 60 after the first injection using a rabbit whose preimmune serum gave no reactivity with Caco2 cell and rat duodenal lysates. In preliminary experiments to confirm the specificity of the antisera with Western blot analysis of K562 cell lysates separated by SDS-PAGE demonstrated a single band of approximate molecular mass of 75 kDa, which was abolished by preincubation of the protein used for immunization. Using Prosite software and the National Center for Biotechnology Information search engine, we identified functional domains in PAP7 including a nuclear localization signal (NLS) and a Golgi dynamics domain (GOLD), which are described in more detail in the results. Fusion proteins with EGFP representing these domains were constructed consisting of amino acids 171–268 [EGFP-C2-PAP7(NLS)] and amino acids 269–526 [EGFP-C2- PAP7(GOLD)] as well as a full-length cDNA (EGFP-C2-PAP7). To verify that the resulting antiserum was specific against PAP7 the GFP fusion proteins (EGFP-NLS, EGFP-GOLD, and EGFP-PAP7) were separated by SDS-PAGE and Western blot analysis performed with anti-GFP antibodies and the anti-PAP7 serum (see Fig. 3, A and B, respectively).
Fig. 3.
Confirmation of the specificity of the anti-PAP7 antiserum. K562 cells were transfected with the various enhanced green fluorescent protein (EGFP)-PAP7 constructs EGFP vector alone, EGFP-nuclear localization signal (NLS), EGFP-Golgi dynamics domain (GOLD), and EGFP-PAP7 as described in the materials and methods. At day 1 after transfection cell lysates were prepared, proteins separated by SDS-PAGE and Western blots performed with anti-GFP antibody (A) or anti-PAP7 antiserum (B). NS refers to nonspecific bands that were always present with the anti-PAP7 antiserum. Molecular weight (MW) markers are also shown for A and B as is the endogenous PAP7 present in the K562 cells.
K562 cells were transfected by Nucleofector using 3 × 106 cells and 5 μg plasmid DNA consisting of pEGFP-C2 fused to PAP7 or to the PAP7 NLS or GOLD domains in 100 μl nucleofector kit V solution. The cells were then cultured in RPMI 1640 with 10% FBS and antibiotics at 37°C with 5% CO2 and harvested at different times as indicated for analysis. Transfection efficiency was checked by fluorescent microscopy daily after transfection. For subcellular localization study, nonfixed cells were examined by confocal microscopy 24 h after transfection.
Immunohistochemical staining.
Frozen sections of rat duodenum and Caco2 cells grown on coverslips were fixed with 2% paraformaldehyde for 10 min at room temperature (RT). Frozen sections of Caco2 cells grown on the semiporous membrane of bicameral chambers, the model system used for studying iron transport across the intestine, were fixed with acetone:methanol (9:1) for 20 min at −20°C (18). The sections and cells on coverslips were blocked with 5% BSA and 1% goat serum in PBS for 30 min at RT and subsequently incubated with anti-PAP7 serum (1:500) for 2 h at RT. After being extensively washed with PBS, the samples were incubated with Alexa488-labeled goat anti-rabbit IgG (1:500) and with TOPRO-3 (1:1,000) for 1 h at RT and mounted with ProLong antifade kit after being extensively washed with PBS, and fluorescence images were obtained by digital photomicroscopy on an Olympus BX-60 epifluorescence microscope. Golgi apparatus was similarly detected using mouse anti-Golgi97 antibody, and mitochondria were detected with Mito-Tracker probe (Invitrogen).
siRNA constructs and transfections.
Desalted DNA oligonucleotides were obtained from IDT. The targeted region of siRNA was the coding sequence of PAP7 cDNA (human ACBD 3; NM_022735) avoiding the start codon and regions in 5′- and-3′-UTRs, which may be rich in regulatory protein binding sites. The following sense and antisense primers were designed to form a small hairpin (sh)RNA and include Eco RI and Bam HI ligation sites: siPAP7–3 sense: 5′-GATCCGGAAACATGTCTAAAGATTTCAAGAGAATCCTCTTTAGACATGTTTCCTTTTTTACGCGTG-3′; siPAP7–3 antisense: 5′-AATTCACGCGTAAAAAAGGAAACATGTCTAAAGAGGATTCTCTTGAAATCCTCTTTAGACATGTTTCCG-3′; siPAP7–8 sense: 5′GATCCGCCTACTCTTTGTGGCGGTCAATTCAAGAGATTGACCGCCACAAAGAGTAGGTTTTTTACGCGTG-3′; siPAP7–8 antisense: 5′-AATTCACGCGTAAAAAACCTACTCTTTGTGGCGGTCAATCTCTTGAATTGACCGCCACAAAGAGTAGGCG-3′; siPAP7–10 sense: 5′-GATCCGTTGTATCAAGTCCAGCTTGCTTCAAGAGAGCAAGCTGGACTTGATACAACTTTTTTACGCGTG-3′; and siPAP7–10 antisense: 5′-AATTCACGCGTAAAAAAGTTGTATCAAGTCCAGCTTGCTCTCTTGAAGCAAGCTGGACTTGATACAACG-3′;
Western blot analysis.
K562 cells were lysed in RIPA buffer [10 mM phosphate buffer pH 7.2, 2 mM EDTA, 150 mM NaCl, 0.1% SDS (wt/vol), 50 mM sodium fluoride, 1% sodium deoxycholate (wt/vol), 1% Triton X-100 (vol/vol) with protease inhibitor cocktail (1% vol/vol)], and protein concentrations were measured with the BCA assay kit. The lysates were separated by SDS-PAGE, transferred to nitrocellulose membranes, and the membranes were incubated with 5% skimmed milk overnight. The membranes were then incubated with the primary antibodies for TfR1 (1:20,000), ferritin (1:5,000), DMT1 (total) (1:3,000), DMT1 (non-IRE) (1:3,000), and DMT1 (IRE) (1:5,000) in 5% skimmed milk overnight and for PAP7 (1:5,000), GFP (1:1,000), and actin (1:5,000) in 5% skimmed milk for 3 h. After being washed with PBS three times, membranes were incubated with the appropriate secondary antibodies in 5% skimmed milk. The signals were visualized by a chemiluminescent method. The intensities of specific bands were analyzed by ImageJ software downloaded from http://rsb.info.nih.gov/ij/.
Immunoabsorption of DMT1 by anti-PAP7 antiserum.
BBMV from +/+ rat duodenum were isolated as previously described (11). The isolated vesicles were solubilized in 0.1% Triton X-100 in PBS and incubated with anti-PAP7 antisera previously cross-lined to protein A using the Seize X protein A immunoprecipitation kit according to the manufacturer's directions. The agarose beads were washed extensively in microcentrifuge tubes, and bound protein was eluted with the supplied elution buffer. After purification, proteins in the BBMV were separated by SDS-PAGE and Western blot analysis was performed with anti-DMT1 (IRE) antisera. After visualization of signals, the first and second antibodies were removed from the membranes by incubation with stripping buffer [62.5 mM Tris·HCl (pH 6.8), 100 mM 2-mercaptoethanol and 2% SDS (wt/vol)] for 10 min, and the membranes were washed with PBS, blocked with 5% skimmed milk and Western blot analysis then repeated using anti-PAP7 antisera to confirm the purification of PAP7-containing BBMV.
Quantitative real-time PCR.
RNA was extracted from K562 by TRIzol, according to manufacturer's protocol. After extraction, cDNA were synthesized from 5 μg of total RNA by Superscript III first-strand system for RT-PCR. The expression levels of mRNA were examined by quantitative real-time PCR using the following primers: PAP7: forward primer 5-GTC AGT GAG TCC AGC GAT GA 3-, reverse primer 5-ATT TTG ACC GCC ACA AAG AG 3-; DMT1 (IRE): forward primer 5-TGG CTT ATC TGG GCT TTG TG 3-, reverse primer 5-CAC ACT GGC TCT GAT GGC TA 3-; DMT1 (non-IRE): forward primer 5-GTG GCA TTA TAT GTG GTG GC 3-, reverse primer 5-CAG CGT CCA TGG TGT TCA GA 3-; TfR 1: forward primer 5-CCC TTC CTT CAA TCA CAC TCA G 3-, reverse primer 5-CAG TGA GCT TCA CAT TCT TGC T 3-; ferritin heavy chain: forward primer 5-CTG GAG CTC TAC GCC TCC TA 3-, reverse primer 5-TGG TTC TGC AGC TTC ATC AG 3-; and GAPDH: forward primer 5-GAA GGT GAA GGT CGG AGT C 3-, reverse primer 5-GAC AAG CTT CCC GTT CTC AG 3-.
Cell cycle analysis and cell proliferation after siRNA transfection.
After transfection with siPAP7 vectors, cells were cultured at a density of 1 × 106 cells/ml. Following a 24-h recovery period, the cells were seeded at a 1:10 dilution and cell numbers determined daily with a Coulter counter. To analyze the effect of decreased PAP7 expression on cell cycle, aliquots of transfected cells were synchronized by withdrawal of FBS from the medium at day 1. Cells were then seeded at a 1:10 dilution (1 × 105 /ml) with serum-free medium (RPMI 1640 with 4 mg/ml BSA, 50 μg/ml holo-Tf, insulin 5 μg/ml, 30 nM SeO2, and antibiotics) for 24 h. At day 2, 48 h after transfection, culture medium was changed to RPMI 1640–10% FCS, and at day 3, cells were harvested and fixed with 1% paraformaldehyde on ice for 10 min. The fixed cells were pelleted, resuspended in 70% ethanol on ice for 30 min, washed with PBS and resuspended in 1 ml of RNase A (0.5 mg/ml), and incubated at 37°C for 60 min. The cells were again washed, resuspended in 400 μl of propidium iodide solution (50 μg/ml), and analyzed by flow cytometry with cell cycle analysis by ModFitLT v3.1 for Mac.
RESULTS
Isolation and identification of proteins that interact with the COOH terminus of DMT1.
The IRE-containing isoforms of DMT1 have a unique sequence of 18 amino acids in the COOH terminus compared with 25 amino acids in the non-IRE isoforms and are the predominant isoforms expressed in the intestinal epithelium. To identify proteins that interact with the unique amino acid sequence in the IRE isoforms and that might therefore be important in intestinal iron transport, we employed the yeast two-hybrid system. The bait, designated as BD-CT512, was formed by cloning into pGBKT7 a cDNA fragment coding the COOH-terminal cytoplasmic domain of DMT1 (IRE) (amino acids 512–561). BD-CT512 was used to screen a rat duodenal cDNA library constructed into the pGADT7 vector. From a screening of ∼3 × 106 transformants, 18 β-galactosidase-positive cDNA clones were obtained. The plasmids that activated both His+ and LacZ genes were tested for specificity by pairing them with a plasmid encoding a hybrid of the Gal4 DNA-binding domain with COOH-terminal of DMT1 (non-IRE) (BD-CT512-non-IRE). None of the positive clones encoded proteins that interacted with COOH-terminal of DMT1 (non-IRE). Sequence analysis revealed that the cDNAs encoded three different proteins: Snapin (accession no. AF086837), rat intestinal fatty acid binding protein (accession no. M35992), and a third protein that had been identified previously both as human GCP60 (accession no. AB043587) and mouse PBR PAP7 (accession no. AF022770). Because the protein putatively interacted with DMT1, we initially designated the protein as DMT1 associated protein or DAP. The 88.1 or 97.5% amino acid identity between human PAP7 (GCP60 and ACBD3) and rat DAP or rat PAP7 and rat DAP, respectively, suggested that DAP and PAP7 were the same protein. Therefore, because of the high degree of homology and to avoid the confusion of another name for the same protein, we subsequently refer to DAP as PAP7. The function of this protein has previously been described as involved in maintenance of Golgi structure and endoplasmic reticulum-Golgi transport by its interaction with the integral Golgi protein giantin (1, 12, 21). In addition, the protein has been strongly implicated as involved in hormone-induced steroid biosynthesis (23).
Both of the PAP7 clones isolated contained a 2,892-bp insert. The only translation start site of the insert in-frame with the GAL4 activating domain encoded a 349-amino acid protein with the stop codon located 1,813 nucleotides upstream from the poly(A) tail. After 5′-rapid amplification of cDNA ends was performed, a full-length PAP7 cDNA was assembled from sequencing of both strands of the original clones. The complete PAP7 cDNA is 3,461 bp and contains an open reading frame encoding a predicted protein of 526 amino acids with a calculated molecular mass of 60.5 kDa and an acid isoelectric point of pI = 4.99. The amino acid sequence analysis showed that PAP7 has high percentages of glutamic acid (14.3%), glutamine (7.8%), and arginine (7.4%) residues. A Kyte-Doolittle hydropathy plot analysis of the amino acid sequence revealed that PAP7 was a hydrophilic protein without any obvious transmembrane domain. Several characteristic protein motifs were identified either with Prosite software or the NCBI search engine including an acyl-CoA binding protein/diazepam binding inhibitor motif (ACBP/DBI; amino acids 101–166), a bipartite nuclear targeting sequence (amino acids 212–228), an arginine-rich region (amino acids 186–235), a glutamine-rich region (amino acids 238–305), a glutamic acid-rich region (amino acids 179–237), and a GOLD (Golgi dynamic) domain (amino acids 381–524). In addition, several sites for potential posttranslational modification were present including two N-glycosylation sites, four protein kinase C phosphorylation sites, eight casein kinase II phosphorylation sites, two tyrosine kinase phosphorylation sites, and three N-myristoylation sites.
Expression of PAP7 in various rat tissues.
The cDNA for PAP7 was isolated from a rat duodenal cDNA library. As DMT1 is important in the intestinal uptake of iron and as proteins that interact with DMT1 might be involved in iron uptake, we examined by Western blot analysis the levels of PAP7 protein in the duodenum compared with other tissues (Fig. 1A). PAP7 protein was present in all tissues examined with the polyclonal anti-PAP7 antiserum raised against the truncated rat PAP7 protein demonstrating a single band (Fig. 1A) in rat tissues including the duodenum, colon, and brain. The ∼75-kDa molecular mass protein detected by the anti-PAP7 antiserum was slightly higher than calculated molecular mass of the PAP7 protein. No isoforms were detected in any tissue.
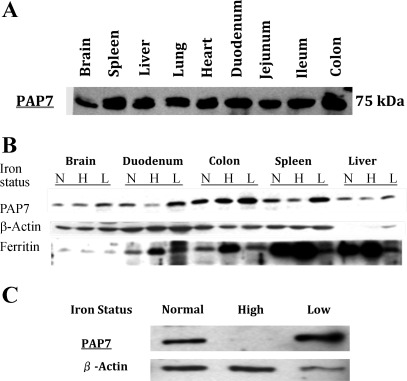
Fig. 1.
Distribution of peripheral benzodiazepine receptor associated protein 7 (PAP7) protein in rat tissues and cell lines. A: Western blot analysis of PAP7 in various rat tissues. Protein from the lysates of the indicated rat tissues were separated by SDS-PAGE, transferred to nitrocellulose membrane, and PAP7 protein detected by Western blot analysis with the anti-PAP7 antibody as described in the materials and methods. B: effect of iron status on expression of PAP7 in rat tissues. As described in the materials and methods, Belgrade (b/b) (L) rats were used as a model of iron deficiency with +/b litter mates raised on normal chow used as iron replete rats (N) and on a high iron diet used as iron loaded rats (H). Proteins from lysates from various tissues of the N, H, and L rats were separated by SDS-PAGE and PAP7 levels determined by Western blot analysis. Ferritin levels were examined as a control for iron loading and actin was employed as a loading control. Shown is a representative Western blot analysis. C: PAP7 protein levels in iron replete and iron depleted Caco2 cells. Caco2 cells were grown as detailed in materials and methods under conditions of low, normal, or high iron conditions for 2 wk, cell extracts were obtained, the proteins were separated by SDS-PAGE, and PAP7 was detected by Western blot analysis. Shown is a representative Western blot analysis with β-actin used as a loading control.
To determine if PAP7 protein levels as detected by Western blot analysis varied with iron status of animals, we used the Belgrade rat as a model of iron deficiency with +/b litter mates raised on normal chow as iron replete (N) rats and iron loaded (H) +/b rats generated by feeding rodent chow enriched with carbonyl iron for 2 wk (Fig. 1B). In the duodenum, PAP7 levels decreased in the iron overloaded and increased in the low iron rats (Fig. 1B) with ferritin levels, used as a control for iron loading, increasing in the iron-loaded rats as expected. Similar differences were seen in the spleen. We also examined the effect of the cellular iron status on levels of PAP7 protein in Caco2 cells. Under culture conditions that we have shown previously to markedly alter cellular iron levels, the PAP7 protein was decreased in iron loaded Caco2 cells and increased in iron depleted Caco2 cells (Fig. 1C). In neither the rat tissues nor in the Caco2 cells were the PAP7 mRNA levels affected by iron status (data not shown). These observations suggest that PAP7 proteins levels are regulated by a posttranscriptional mechanism.
Subcellular localization of PAP7.
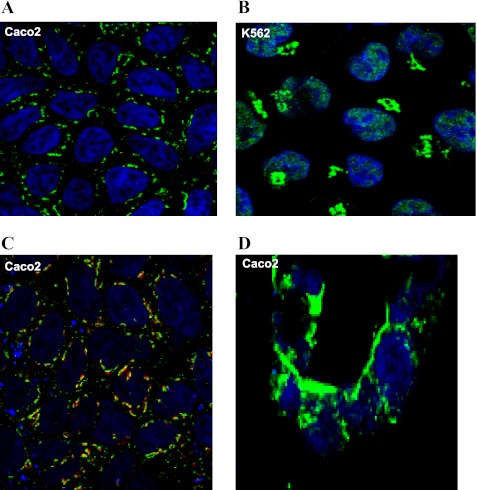
The intracellular localization of PAP7 was examined in Caco2 and K562 cells (Fig. 2) and rat intestine (see Fig. 4) by immunohistochemistry. K562 cells were used as these cells, which can be induced to undergo erythroid differentiation, have a high level of iron uptake. In both cell lines, PAP7 could be readily detected in perinuclear structures consistent with the Golgi complex (Fig. 2, A and B). In addition, in the K562 cells PAP7 was found in the nucleus. The presence of PAP7 in the trans-Golgi was confirmed by staining Caco2 cells both with anti-PAP7 (green) and anti-Golgi-97 (red) antibodies that demonstrated colocalization (yellow) within structures consistent with the Golgi (Fig. 2C). Caco2 cells grown on a semiporous membrane in bicameral chambers are a model used to study intestinal transport including iron transport. To examine the localization of PAP7 in Caco2 cells grown in this model system, the semiporous membranes were removed from bicameral chambers, and frozen sections were obtained and stained with anti-PAP7 antibodies (Fig. 2D). PAP7 was clearly on the BBM of the Caco2 cells as identified by staining adjacent sections with anti-DMT1 antisera. Initial attempts to demonstrate PAP7 in the BBM either of Caco2 cells grown on the semiporous membranes or of tissue slices of rat intestine using paraformaldehyde as a fixative failed (data not shown). However, when Caco2 cells or intestine were prepared with frozen section techniques and were fixed with acetone:methanol (9:1), PAP7 was demonstrated in the BBM (Fig. 2D and see Fig. 4).
Fig. 2.
Intracellular localization of PAP7 in Caco2 and K562 cells. Caco2 cells were grown on glass coverslips (A and C) or on the semiporous membranes of bicameral chambers (D), which were then removed and frozen sections obtained. K562 cells grown in suspension culture were centrifuged onto glass slides (B). Cells were either fixed with 2% paraformaldehyde (A–C) or with acetone:methanol (9:1; D). Cells were stained with anti-PAP7 antiserum as the primary antibody and Alexa 488-labeled anti-rabbit IgG antibody as the secondary antibody (A and B). Golgi were stained with mouse anti-Golgi 97 antibody using Alexa 594-labeled anti-mouse IgG antibody as the second antibody (C), and the nuclei were stained by TOPRO-3. Cells were observed by confocal fluorescence microscopy with a single optical section through the cells at the level of the nuclei shown (A–C) with the merged images being shown for PAP7 (green) and Golgi97 (red; C). A single optical section approximately parallel to the vertical axis through the Caco-2 cells grown on semiporous membrane is shown in D.
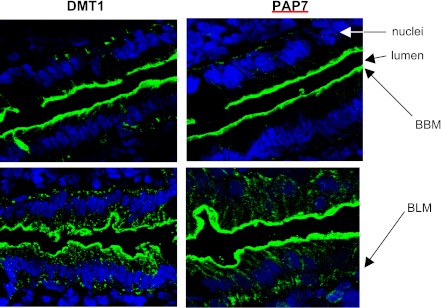
Fig. 4.
Effect of iron feeding on PAP7 localization in the rat duodenum. As described in the materials and methods immunofluorescence staining of PAP7 and divalent metal transporter 1 (DMT1) was conducted on sections of the rat duodenum before (top) and at 1 h (bottom) after iron feeding. Simultaneously, the nuclei were stained with TOPRO-3. Arrows indicate the brush border membrane (BBM) and basolateral membrane (BLM).
Transfections with constructs of various portions of PAP7 showed the expected subcellular localization with EGFP-NLS localizing to the nucleus and cytoplasm, EGFP-GOLD to the Golgi, and EGFP-PAP7 to both the nucleus and Golgi (data not shown). In addition, Western blot analysis of the transfected cells with anti-GFP and anti-PAP7 antibodies confirmed the specificity of the anti-PAP7 antibodies (Fig. 3, A and B).
Effect of feeding iron on the localization of PAP7 in the rat duodenum.
A property of DMT1 is that in the iron-starved state in both the rat duodenum and in Caco2 cells the protein is present in the BBM but not in the cytosol. With iron feeding, DMT1 undergoes internalization and then translocation back to the BBM. If PAP7 interacts with DMT1, it would be expected that PAP7 would colocalize with DMT1 at some point during the internalization-externalization of DMT1. As both the antiserum to PAP7 and DMT1 were raised in rabbits, it was not possible to directly examine rat intestinal epithelium colocalization of PAP7 with DMT1 by immunohistochemistry. When duodenal loops were examined after overnight starvation, PAP7 was found in the BBM similar to the localization of DMT1 (Fig. 4A). With iron feeding, PAP7 translocated to the basal portions of the cells (Fig. 4B) while as previously described DMT1 translocated from the BBM into the apical cytoplasm (Fig. 4B). These results suggest that PAP7 colocalization with DMT1 in the BBM may play a role in preventing DMT1 internalization until the appropriate signal is obtained with iron feeding.
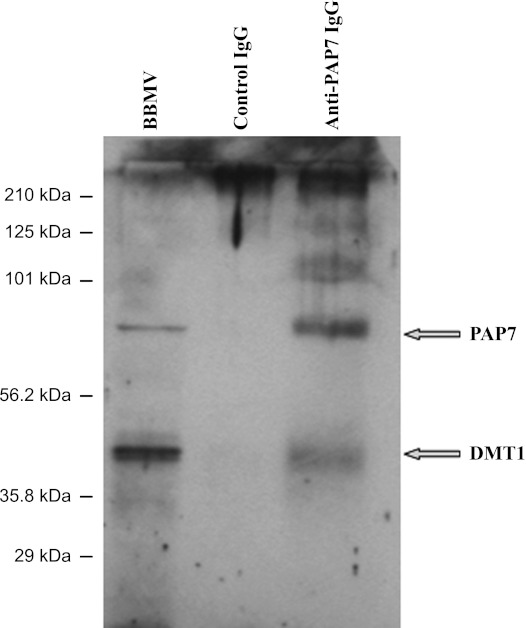
To confirm both the presence of PAP7 in the BBM and the interaction between PAP7 and DMT1, we isolated rat intestine BBMV and demonstrated the presence of PAP7 in the BBMV by Western blot analysis and by the copurification upon antibody-mediated affinity chromatography of the two proteins from detergent-solubilized BBMV (Fig. 5). Additionally, PAP7 could be demonstrated to be a peripheral protein as treatment of the BBMV with 0.1 N NaOH completely dissociated PAP7 from the BBMV into a soluble fraction (data not shown).
Fig. 5.
PAP7 and DMT1 interact in BBM vesicles (BBMV) from rat duodenum. As described in the materials and methods, BBMV were isolated from duodenal epithelium cells of heterozygous Belgrade rats (+/b). Detergent-solubilized BBMV were passed through the columns of anti-PAP7 IgG and control IgG cross-linked to agarose beads as described in materials and methods. After being extensively washed, proteins were eluted from the beads and the eluates and an aliquot of the BBMV were separated by SDS-PAGE followed by Western blot analysis with anti-DMT1 [iron-responsive element (IRE)]. Subsequently, the antibodies were removed from the same membrane as described in the materials and methods and Western blot analysis repeated using anti-PAP7 antiserum.
Effects of PAP7 expression on expression of various genes involved in cellular iron metabolism.
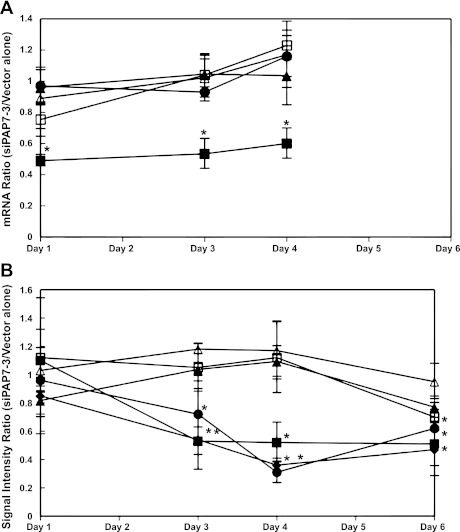
To determine if PAP7 overexpression affected cellular iron metabolism, we examined the effect of overexpression of PAP7 and the constructs expressing the PAP7 NLS and GOLD motifs alone on expression of the transferrin receptor 1 (TfR1), ferritin, DMT1, and endogenous PAP7 as these proteins are affected by altered cellular iron concentration. In these experiments, we transfected K562 cells with EGFP-tagged plasmids, obtaining transfection efficiencies of 70% for EGFP-PAP7(NLS) and 20–30% for EGFP-PAP7(GOLD) and EGFP-PAP7 through 48 h. Following transfection, no effect on expression was seen either at the RNA or protein level for TfR1, ferritin, and DMT1 (IRE) and endogenous PAP7 (data not shown). In the course of these experiments, we did notice that PAP7 protein expression was about twofold greater in cells grown at 2.5 × 105 cells/ml than cells grown at a fourfold higher cell density (data not shown). We then examined the effects of down regulation of PAP7 expression by siRNA on expression of genes involved in cellular iron metabolism. To downregulate PAP7 expression, we used the siRNA constructs siPAP7–3, -8, and -10 that also contained GFP on a separate internal ribosomal entry site to allow us to monitor the efficiency of transfection. All three siRNAs used exhibited ∼80% transfection efficiencies at days 1 and 2, which subsequently decreased to ∼30–50% efficiency at day 6. Gene expression was monitored both by quantitative PCR and Western blot analysis (Fig. 6A). Mock transfection and transfections with vector alone or siPAP7–8 had no effect on PAP7 expression. siPAP7–3 and PAP7–10 both decreased mRNA levels at 24 h after transfection with a decrease of RNA of ∼0.49 ± 0.08-fold (means of fold decrease ± SD of 4 experiments) compared with vector alone from days 1 through 4 after transfection (Fig. 6A). The suppression of PAP7 mRNA was present through at least day 4 after transfection as shown for siPAP7–3 while not affecting mRNA levels of DMT1 (IRE), DMT1 (nonIRE), ferritin H-chains, and transferrin receptor 1 (Fig. 6A). A similar time course of suppression of PAP7 mRNA was seen with siPAP7–10 while neither siPAP7–8 nor siPAP7–10 affected DMT1, ferritin, or transferrin receptor (data not shown). Protein levels of PAP7, DMT1 (IRE), DMT1 (non-IRE), ferritin H-chains, and transferrin receptor 1 were monitored from 24 h up to day 6 after transfection (Fig. 6B). Both siPAP7–3 and PAP7–10 but not mock transfection, vector alone, or PAP7–8 decreased DMT1 (IRE) protein levels by day 3 after transfection. Neither siPAP7–3 nor PAP7–10 affected DMT1 (non-IRE) or TfR1 expression. At 6 days after transfection H-ferritin, TfR1, and DMT1 (non-IRE) protein levels were decreased, but the decreases were neither reproducible nor statistically significant.
Fig. 6.
Effect of downregulation of PAP7 by siRNA on expression of genes involved in cellular iron metabolism. As detailed in the materials and methods, K562 cells at various times after transfection with small interfering (si)PAP7–3 or with vector alone or mock transfection were lysed and protein and mRNA levels were determined. There were no significant differences between mock and vector alone transfections. Hence, we used vector alone as a reference to validate the effects of siPAP7–3 vector on the mRNA and protein expression levels of PAP7 (■), DMT1 total (♦) (Western blot only), DMT1 (IRE) (●), DMT1 (non-IRE) (▲), TfR1 (△), and H-ferritin (□), which were measured either by real-time PCR (A) or by Western blot analysis (B), respectively. Shown are the ratios for each day examined after transfection with siPAP7–3 to vector alone for the mRNA (A) or the signal intensity from scans of the Western blots (mean levels ± SE for 3 experiments). An unpaired t-test was applied to the results: *P < 0.05, significant difference from vector alone transfected cells.
Effects of PAP7 siRNA on cell growth and cell protein levels.
In the course of examining the effects of PAP7 siRNAs, we noticed that the effective siRNAs, i.e., siPAP7–3 and PAP7–10, always decreased cell growth at days 3, 4, and 5 (Table 1). To determine if the inhibition of cell growth by the siRNAs was a consequence of altered cell cycle, cycle status was determined by flow cytometry at day 3 after transfection. A small but significant effect was observed with siRNA PAP7–3 and PAP7–10 increasing the number of cells in G1 and decreasing the number of cells in G2-phase consistent with G1 cycle arrest (Table 2). These small but consistent differences could not be attributed to differences in transfection efficiency. In addition, at day 3 after transfection slight differences were noted in total cellular protein per cell with protein/cell being 70.3 ± 7.5 and 70.0 ± 4.2% for PAP7–3 and PAP7–10, respectively, compared with mock transfection, differences that were statistically significant with P < 0.05.
Table 1.
Effect of downregulation of PAP7 by siRNA on cell growth
| Days After Transfection |
|||||
|---|---|---|---|---|---|
| Transfection | 2 | 3 | 4 | 5 | 6 |
| Mock | 1.3 ± 0.1 | 3.1 ± 0.2 | 6.9 ± 0.5 | 12.3 ± 2.0 | 15.1 ± 2.3 |
| siPAP7-3 | 1.2 ± 0.2 | 1.9 ± 0.3* | 3.8 ± 0.3* | 8.0 ± 0.5* | 14.8 ± 2.2 |
| (P = 0.0006) | (P = 0.003) | (P = 0.02) | |||
| siPAP7-8 | 1.3 ± 0.1 | 2.8 ± 0.4 | 7.1 ± 0.9 | 11.7 ± 1.5 | 17.1 ± 1.9 |
| siPAP7-10 | 1.2 ± 0.2 | 2.2 ± 0.5* | 4.5 ± 0.9* | 9.1 ± 0.8 | 17.3 ± 1.0 |
| (P = 0.001) | (P = 0.0006) | ||||
As detailed in materials and methods, K562 cell numbers were determined at days 2–6 following transfection. Shown are the cell numbers ×105/ml (means ± SD) for 4 experiments for either mock-transfected cells or cells transfected with small interfering (si) peripheral benzodiazepine receptor-associated protein 7 (PAP7)- 3, -8, or -10.
Significant difference from mock-transfected cells on the same day with the P value determined by an unpaired t-test shown in parentheses.
Table 2.
Effect of downregulation of PAP7 by siRNA on cell cycle
| Transfection | G1, % | G2, % |
|---|---|---|
| Mock | 26.7 ± 3.5 | 18.9 ± 2.9 |
| siPAP7-3 | 36.3 ± 6.3* | 7.9 ± 4.4* |
| (P = 0.02) | (P = 0.05) | |
| siPAP7-8 | 26.6 ± 2.8 | 17.9 ± 2.6 |
| siPAP7-10 | 30.9 ± 4.0* | 15.2 ± 2.4* |
| (P = 0.009) | (P = 0.03) |
After transfection of K562 cells with siRNA cells were synchronized to allow for determination of cell cycle as detailed in materials and methods. Shown are the percentages of cells in G1 or G2 (means ± SD for 4 experiments) for either mock-transfected cells or cells transfected with siPAP7-3, -8, or -10.
Significant difference from mock-transfected cells with the P value determined by an unpaired t-test shown in parentheses.
DISCUSSION
DMT1 is a complex protein the function of which is to transport iron across the small intestine epithelial BBM and across endosomes containing the Tf-TfR1 complex. The functions of DMT1 have been well studied. The physiology of DMT1 as a proton-divalent metal cotransporter has been analyzed in studies using transfection of DMT1 (26). The cell biology of DMT1 and the response of DMT1 to a variety of stimuli including iron depletion and iron overload have also been well elucidated (4, 16, 29). In addition, the molecular biology of DMT1 with the definition of multiple isoforms and the differences between tissues in the array of isoforms expressed has been elegantly elucidated (13, 16, 27). However, there are several complexities to DMT1 function that have not been fully explained. First, the functional differences between the multiple isoforms have not been determined; in particular, a function has not been ascribed to the unique COOH-terminal sequence of amino acids in the IRE vs. the non-IRE isoforms except that PAP7 has been demonstrated to interact with DMT1 in a neuronal cell line and may mediate increased iron transport possibly responsible for NMDA toxicity (6). Second, DMT1 is mobile. In the intestine, DMT1 is expressed in the BBM and with iron uptake DMT1 is internalized into the cytosol. In this process, vesicles from the BBM containing DMT1 undergo transcytosis and fuse with vesicles from the basolateral membrane (18). The signal imposed by a bolus of iron to allow this process to occur is unknown. We, and others, (3, 30) have demonstrated that DMT1 (IRE) is the predominant isoform that is expressed on the intestinal BBM. The isoforms appear to dictate tissue-specific expression patterns as well as distinct subcellular localizations (16, 27). The IRE isoform is more often expressed at the plasma membrane of epithelial cells whereas the non-IRE isoform is expressed in erythroid cells and appears essential for the acquisition of iron from transferrin internalized by receptor-mediated endocytosis into acidified endosomes (16). In addition, the isoforms appear to direct DMT1 either into late endosomes and lysosomes for the IRE isoform or into early endosomes in the case of the non-IRE isoform (16, 27). As an initial approach to understanding the two complexities, the function of the unique COOH termini of the DMT1 isoforms and the internalization of DMT1, we undertook the studies reported here to determine those proteins that interacted with DMT1 and that, in turn, might shed light on the COOH-terminal function and DMT1 internalization.
By use of the yeast two-hybrid system, we identified three proteins that interacted with DMT1. One protein, Snapin (accession no. AF086837), is a SNARE-associated protein important for synaptic vesicle docking and fusion and, indeed, has been implicated in vesicle endocytosis and exocytosis and in vesicle-vesicle fusion in many systems (2). A second clone isolated was for fatty acid-binding protein (accession no. M35992), which belongs to a conserved multigene family of intracellular lipid-binding proteins to which both heme and protoporphyrin have been identified as potential ligands (28). Both Snapin and fatty acid-binding protein mRNAs were demonstrable in rat duodenum, but their role in iron uptake has not been further explored.
The third protein, originally designated as DAP (accession no. AY336075), is now referred to as PAP7 in part because PAP7 has been noted in an investigation stimulated by our entry into GenBank to interact with DMT1 in neuronal cells (6). Still, there is only 97.5% identify between DAP and PAP7 and to be determined is whether the lack of complete homology represents strain differences or another phenomenon. PAP7 clearly has an effect on iron uptake as demonstrated in neuronal cells where Dexras1 also interacted with PAP7, and stimulation of NMDA receptors led to activation of Dexras1 through S-nitrosylation and the activated Dexras1 via PAP7 and DMT1 physiologically induced neuronal iron uptake. This signaling pathway appears important in mediating NMDA neurotoxicity as iron chelation prevents NMDA neurotoxicity in cortical cultures (6).
PAP7 was defined as a protein that bound to the PBR, a protein located in the outer membrane of mitochondria. As various functions for PAP7 have been elucidated the protein has been designated as GCP60, GOCAP1, and most recently ACBD3. ACBD3 functions including steriodogenesis, proliferation and apoptosis, immunomodulation, porphyrin transport, and maintenance of Golgi apparatus (8). In the current studies, we show that PAP7 is ubiquitously expressed in the rat and is also expressed in the K562 and Caco2 cell lines.
By immunohistochemistry, PAP7 and DMT1 appear on the BBM of the rat duodenum as well on the BBM of Caco2 cells. In addition, DMT1 copurifies with PAP7 when anti-PAP7 antisera was used for antibody-mediated affinity chromatography of detergent-solubilized intestinal BBMV. Immunohistochemical studies demonstrate that PAP7 is expressed in the duodenum. In the rat-starved overnight, PAP7 is found only on the BBM. Interestingly, with iron feeding PAP7 translocates into the intestinal cell as does DMT1 but the pattern of redistribution of PAP7 differs from that of DMT1 as PAP7 moves primarily to the basolateral membrane. In addition, some PAP7 does translocate to perinuclear structures consistent with the Golgi. These findings suggest that PAP7 may bind to DMT1 until an appropriate signal is generated by the initial iron influx into the cell perhaps then allowing for DMT1 internalization. Similar findings were seen when rats were fed heme (data not shown). Although fixation-related artifacts cannot be excluded in Caco2 cells and K562 cells, PAP7 is demonstrable within the Golgi with colocalization demonstrated using an appropriate Golgispecific marker. Despite the demonstration of PAP7 having been initially identified by binding to the PBR, a mitochondrial membrane protein, we could detect no localization of PAP7 with mitochondria in either Caco2 or K562 cells. While it seems unlikely then that PAP7 is functioning to conduct vesicles carrying iron to the mitochondria, the negative results could also be the result of fixative artifact or an epitope on PAP7 hiddened from our particular antisera in these two cell types.
Considering that PAP7 interacts with an iron transporter, it was unexpected that PAP7 would interact either with the Golgi or be present in the nucleus. The confirmation of the localization was achieved by using constructs that contained either the PAP7 GOLD domain or the NLS domain. The involvement of the Golgi in cellular iron metabolism has been tenuous until two recent reports. In HIV-1, the multifunctional Nef protein has now been demonstrated to downregulate surface myosin heavy chain-I molecules including the myosin heavy chain-like protein HFE, which regulates iron homeostasis and is mutated in the iron-overloading disorder hemochromatosis (7). Nef has been found to direct HFE to the trans-Golgi network, away from the cell surface where it would bind to the transferrin receptor and regulate iron uptake. As a consequence, Nef expression causes cellular iron accumulation. Hepcidin, a liver-expressed protein that plays a major role in iron homeostasis as a negative regulator of iron absorption and release, is localized as prophepcidin in the Golgi of hepatocytes (29). It is unknown if this localization is solely because hepcidin is secreted from the cells or if other interactions occur that are involved in regulation and/or expression of hepcidin. However, hemojuvelin, an essential protein in the regulation of hepcidin synthesis, also undergoes processing in the Golgi apparatus for the formation of soluble hemojuvelin (20).
Using siRNA to downregulate PAP7 levels revealed a surprising function for PAP7. The lower levels of PAP7 produced by the siRNAs PAP7–3 and PAP7–10 also decreased DMT1 (IRE) protein but not RNA levels. The decrease of PAP7 protein seemed to precede the decrease in DMT1 as PAP7 expression was always decreased by day 2 after transfection (data not shown) while the decrease of DMT1 (IRE) was not seen until day 3. There are two possibilities that we are exploring for the effect of PAP7 on DMT1 expression: either PAP7 stabilizes DMT1 or has an effect on translation of DMT1 mRNA. The effect of lowered PAP7 levels on DMT1 expression is specific as decreased levels of PAP7 had no effect on expression of DMT1 (non-IRE), TfR1, and ferritin H-chain. One might have expected that the decreased levels of DMT1 should result in decreased iron uptake with a resulting phenotype of increased TfR1 and decreased ferritin. While we did not measure iron uptake after transfection, the levels of TfR1 and ferritin were not affected following transfection with siPAP7. Presumably, then, the levels of DMT1 (IRE) were adequate for iron uptake or that DMT1 (non-IRE) could compensate and contribute to iron uptake. Interestingly, overexpression of PAP7 and of various portions of PAP7 had no effect on DMT1 (IRE) expression, suggesting that endogenous PAP7 was sufficient to maintain DMT1 expression.
We did observe that the lowered levels of PAP7 had some more protean effects causing decreased cell growth and cell cycle inhibition. Whether the mechanism(s) causing these effects is (are) similar to those effecting on DMT1 (IRE) levels remains to be determined. We also noted that cell density affected PAP7 expression with higher PAP7 levels occurring in cells grown at low cell density. PAP7 as ACBD3 has been demonstrated to bind to a caspase derived fragment of Golgin-160 to prevent nuclear localization of the fragment (24, 25), a phenomenon that suggests that PAP7 (i.e., ACBD3) may be associated with cell growth and apoptosis. The interaction, then of DMT1 (IRE) with PAP7 would integrate the transport of iron, an essential element for growth, with regulation of cell growth.
GRANTS
The study was supported in part by National Institute of Diabetes and Digestive and Kidney Diseases Research Grants DK-065101 (to K. Y. Yeh) and DK-43785 (J. Glass) and by the Carroll W. Feist Professorship and the Feist-Weiller Cancer Center, Louisiana University Health Sciences Center.
DISCLOSURES
No conflicts of interest, financial or otherwise, are declared by the author(s).
AUTHOR CONTRIBUTIONS
Author contributions: Y.O., K.-y.Y., and J.G. conception and design of research; Y.O., Y.M., M.Y., H.Y., and Z.L. performed experiments; Y.O., H.Y., K.-y.Y., and J.G. analyzed data; Y.O. and J.G. interpreted results of experiments; Y.O. drafted manuscript; Y.O. and J.G. approved final version of manuscript; K.-y.Y. and J.G. edited and revised manuscript.
ACKNOWLEDGMENTS
We thank Emiko Okazaki for technical assistance.
Present address of Y. Okazaki: Dept. of Pathology and Biological Responses, Nagoya Univ. Graduate School of Medicine, 65 tsurumai-cho, Showa-ku, Nagoya, 466-8550, Japan.
Present address of Y. Ma: Dept. of Pathology and Laboratory Medicine, Hospital of the Univ. of Pennsylvania, 3400 Spruce Street, Philadelphia, PA 19104.
REFERENCES
- 1. Barr FA, Short B. Golgins in the structure and dynamics of the Golgi apparatus. Curr Opin Cell Biol 15: 405–413, 2003 [DOI] [PubMed] [Google Scholar]
- 2. Cai Q, Lu L, Tian JH, Zhu YB, Qiao H, Sheng ZH. Snapin-regulated late endosomal transport is critical for efficient autophagy-lysosomal function in neurons. Neuron 68: 73–86, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Canonne-Hergaux F, Gruenheid S, Govoni G, Gros P. Cellular and subcellular localization of the Nramp2 iron transporter in the intestinal brush border and regulatory by dietary iron. Blood 93: 4406–4417, 1999 [PubMed] [Google Scholar]
- 4. Canonne-Hergaux F, Levy JE, Fleming MD, Montross LK, Andrews NC, Gros P. Expression of the DMT1 (NRAMP2/DCT1) iron transporter in mice with genetic iron overload disorders. Blood 97: 1138–1140, 2001 [DOI] [PubMed] [Google Scholar]
- 5. Canonne-Hergaux F, Zhang AS, Ponka P, Gros P. Characterization of the iron transporter DMT1 (NRAMP2/DCT1) in red blood cells of normal and anemic mk/mk mice. Blood 98: 3823–3830, 2001 [DOI] [PubMed] [Google Scholar]
- 6. Cheah JH, Kim SF, Hester LD, Clancy KW, Patterson SE, 3rd, Papadopoulos V, Snyder SH. NMDA receptor-nitric oxide transmission mediates neuronal iron homeostasis via the GTPase Dexras1. Neuron 51: 431–440, 2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Drakesmith H, Chen N, Ledermann H, Screaton G, Townsend A, Xu XN. HIV-1 Nef down-regulates the hemochromatosis protein HFE, manipulating cellular iron homeostasis. Proc Natl Acad Sci USA 102: 11017–11022, 2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Fan J, Liu J, Culty M, Papadopoulos V. Acyl-coenzyme A binding domain containing 3 (ACBD3; PAP7; GCP60): an emerging signaling molecule. Prog Lipid Res 49: 218–234, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Fleming MD, Trenor CC, III, Su MA, Foernzler D, Beier DR, Dietrich WF, Andrews NC. Microcytic anemia mice have a mutation in Nramp2, a candidate iron transporter gene. Nat Genet 27: 383–386, 1997 [DOI] [PubMed] [Google Scholar]
- 10. Gunshin H, Mackenzie B, Berger UV, Gunshin Y, Romero MF, Boron WF, Nussberger S, Gollan JL, Hediger MA. Cloning and characterization of a mammalian proton-coupled metal-ion transporter. Nature 388: 482–488, 1997 [DOI] [PubMed] [Google Scholar]
- 11. Hearn PR, Russell RGG. The formation and orientation of brush border vesicles from rat duodenal mucosa. J Cell Sci 47: 227–236, 1981 [DOI] [PubMed] [Google Scholar]
- 12. Hicks SW, Machamer CE. The NH2-terminal domain of Golgin-160 contains both Golgi and nuclear targeting information. J Biol Chem 277: 35833–35939, 2002 [DOI] [PubMed] [Google Scholar]
- 13. Hubert N, Hentze MW. Previously uncharacterized isoforms of divalent metal transporter (DMT)-1: implications for regulation and cellular function. Proc Natl Acad Sci USA 99: 12345–12350, 2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Lacapère JJ, Papadopoulos V. Peripheral-type benzodiazepine receptor: structure and function of a cholesterol-binding protein in steroid and bile acid biosynthesis. Steroids 68: 569–585, 2003 [DOI] [PubMed] [Google Scholar]
- 15. Lam-Yuk-Tseung S, Govoni G, Forbes J, Gros P. Iron transport by Nramp2/DMT1: pH regulation of transport by 2 histidines in transmembrane domain 6. Blood 101: 3699–3707, 2003 [DOI] [PubMed] [Google Scholar]
- 16. Lam-Yuk-Tseung S, Touret N, Grinstein S, Gros P. Carboxyl-terminus determinants of the iron transporter DMT1/SLC11A2 isoform II (-IRE/1B) mediate internalization from the plasma membrane into recycling endosomes. Biochemistry 44: 12149–12159, 2005 [DOI] [PubMed] [Google Scholar]
- 17. Li H, Degenhardt B, Tobin D, Yao ZX, Tasken K, Papadopoulos V. Identification, localization, and function in steroidogenesis of PAP7: a peripheral-type benzodiazepine receptor-and PKA8 RIa-associated protein. Mol Endocrinol 15: 2211–2228, 2001 [DOI] [PubMed] [Google Scholar]
- 18. Ma Y, Specian RD, Yeh KY, Yeh M, Rodriguez-Paris J, Glass J. The transcytosis of divalent metal transporter 1 and apo-transferrin during iron uptake in intestinal epithelium. Am J Physiol Gastrointest Liver Physiol 283: G965–G973, 2002 [DOI] [PubMed] [Google Scholar]
- 19. Mackenzie B, Hediger MA. SLC11 family of H+-coupled metal-ion transporters NRAMP1 and DMT1. Pflügers Arch 447: 571–579, 2004 [DOI] [PubMed] [Google Scholar]
- 20. Maxson JE, Enns CA, Zhang AS. Processing of hemojuvelin requires retrograde trafficking to the Golgi in HepG2 cells. Blood 113: 1786–1793, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Misumi Y, Sohda M, Tashiro A, Sato H, Ikehara Y. An essential cytoplasmic domain for the Golgi localization of coiled-coil proteins with a COOH-terminal membrane anchor. J Biol Chem 276: 6867–6873, 2001 [DOI] [PubMed] [Google Scholar]
- 22. Núñez MT, Tapia V, Rojas A, Aguirre P, Gómez F, Nualart F. Iron supply determines apical/basolateral membrane distribution of intestinal iron transporters DMT1 and ferroportin 1. Am J Physiol Cell Physiol 298: C477–C485, 2010 [DOI] [PubMed] [Google Scholar]
- 23. Papadopoulos V, Baraldi M, Guilarte TR, Knudsen TB, Lacapere Lindemann P JJ, Norenberg DM, Nutt D, Weizman A, Zhang MR, Gavish M. Translocator protein (18kDa): new nomenclature for the peripheral-type benzodiazepine receptor based on its structure and molecular function. Trends Pharmacol Sci 27: 402–409, 2006 [DOI] [PubMed] [Google Scholar]
- 24. Sbodio JI, Hicks SW, Simon D, Machamer CE. GCP60 preferentially interacts with a caspase-generated golgin-160 fragment. J Biol Chem 281: 27924–27931, 2006 [DOI] [PubMed] [Google Scholar]
- 25. Sohda M, Misumi Y, Yamamoto A, Yano A, Nakamura N, Ikehara Y. Identification and characterization of a novel Golgi protein, GCP60, that interacts with the integral membrane protein giantin. J Biol Chem 276: 45298–45306, 2001 [DOI] [PubMed] [Google Scholar]
- 26. Su MA, Trenor CC, Fleming JC, Fleming MD, Andrews NC. The G185R mutation disrupts function of the iron transporter Nramp2. Blood 92: 2157–2163, 1998 [PubMed] [Google Scholar]
- 27. Tabuchi M, Tanaka N, Nishida-Kitayama J, Ohno Kishi F H. Alternative splicing regulates the subcellular localization of divalent metal transporter 1 Isoforms. Mol Biol Cell 13: 4371–4387, 2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Velkov T, Lim ML, Horne J, Simpson JS, Porter CJ, Scanlon MJ. Characterization of lipophilic drug binding to rat intestinal fally acid binding protein. Mol Cell Biochem 326: 87–95, 2009 [DOI] [PubMed] [Google Scholar]
- 29. Wallace DF, Summerville L, Lusby PE, Subramaniam VN. Prohepcidin localizes to the Golgi compartment and secretory pathway in hepatocytes. J Hepatol 43: 720–728, 2005 [DOI] [PubMed] [Google Scholar]
- 30. Yeh KY, Yeh M, Watkins JA, Rodriguez-Paris J, Glass J. Dietary iron induces rapid changes in rat intestinal divalent metal transporter expression. Am J Physiol Gastrointest Liver Physiol 279: G1070–G1079, 2000 [DOI] [PubMed] [Google Scholar]