Abstract
Antibody class defines function in B cell immunity, but how class is propagated into B cell memory remains poorly understood. Here, we demonstrate that memory B cell subsets unexpectedly diverge across antibody class through the differential impact of major transcriptional regulators. Conditional genetic deletion of Tbx21 selectively blocks the formation and antigen-specific response of IgG2a memory B cells in vivo. Cell intrinsic T-bet expression regulates STAT1 expression, steady-state cell survival and IgG2a BCR transcription. In contrast, RORα was differentially expressed in IgA memory B cells with siRNA knockdown and chemical inhibition supporting its selective control in cell survival and IgA BCR transcription. Thus, divergent transcriptional regulators dynamically maintain subset integrity to promote specialized immune function within class-specific memory B cells.
Memory B cells are long-lived antigen-experienced B cells that typically express high-affinity B cell receptor (BCR), rapidly expand and differentiate into plasma cells upon antigen re-challenge1,2. While IgM memory B cells have specialized functions3, many antigen-primed B cells switch to non-IgM isotypes under the antigen-specific regulation of follicular helper T cells4. Furthermore, non-IgM classes of membrane-bound antibody have differing abilities to transduce signals through their BCR based on the expressed constant region5,6. However, little is known about the molecular signals required for class-switched memory B cell survival, activation or differentiation. Several studies have delineated how cytokines differentially affect transcriptional programs in naïve B cells that culminate in class switch recombination7,8. Interleukin 4 (IL-4) and interferon-γ (IFN-γ) reciprocally regulate IgG1 and IgG2a class switch7, while the capacity to express TGFβR2 in naive B cells is required for IgA switch9. However, it remains to be studied whether programmatic differences initiated at the time of class-switch extend into memory B cell compartments to control longevity, cell fate and memory B cell function in an antibody class-specific manner.
Differentiation of immune effector cells relies heavily on transcription factors belonging to three families: T-box, GATA, and Retinoic acid receptor-related Orphan Receptors (RORs)10. Members of all three families share the ability to directly interact with chromatin remodeling machinery to transactivate or repress gene targets in a cell context-dependent manner. Transcriptional regulators from each family induce molecular programs known to specify immune cell lineages into functional subsets11–13. T-bet, a member of the T-box family, plays a critical role inducing IFN-γ production by TH1, natural killer (NK), and CD8+ cells to regulate antiviral immunity12. T-bet expression by naive B cells is sufficient to promote IgG2a class switch and is required for IFN-γ induced production of IgG2a antibody in vivo14,15. In contrast, GATA family members are expressed by TH2 cells, basophils and mast cells, crucial to IL-4 production and immunity against helminthes13. While ROR family members are less characterized, RORγt and RORα are found in TH17 cells16 and known to be involved in mucosal immunity against extracellular bacteria11. While factors belonging to these families have been well characterized in several immune subsets, their roles in memory B cell development and the regulation of memory B cell function remain unknown.
Here, we focus on IgG2a+ and IgA+ memory B cells and provide evidence for the divergent programming of memory B cell function by major transcriptional regulators T-bet and RORα. Temporal deletion of T-bet in IgG2a+ memory B cells established a central and selective role for this regulator in IgG2a+ memory B cell survival and antigen responsiveness in vivo. Differential expression of cytokine receptors, integrins and RORα highlighted the specialized development and unique properties of IgA memory B cells. Importantly, a role for both T-bet and RORα in persistent BCR transcription indicated dynamic and ongoing class-specific requirements for each of these factors in IgG2a+ and IgA+ memory B cells, respectively. Taken together, we propose that expression of non-IgM antibody signifies a transcriptionally regulated commitment event across sub-specialized, class-specific memory B cells.
RESULTS
IgG2a memory formation requires T-bet in B cells
To look for the existence of class-specific transcriptional programs, we first focused on IgG2a+ memory B cells given evidence that B cell intrinsic expression of T-bet is sufficient to induce IgG2a germline transcripts15. Furthermore, IFN-γ provided by transferred OT-1 transgenic CD8+ T cells skewed B cells towards IgG2a class-switch in vivo in ways dependent on non-T cell T-bet expression14. To address the B cell intrinsic requirement for T-bet in IgG2a class switch more directly and in polyclonal animals, we used animals with a germline Tbx21 deletion17. These animals display defects in multiple cell types12 including defects in serum IgG antibody15. Consistent with these antibody results, analysis of the Tbx21−/− animals revealed significant decreases in IgG3, IgG2a and IgG2b class-switched (IgM−IgD−) CD38hi B cells (also CD19+B220+CD138−) compared to intact IgG1+ B cells (Fig. 1a & Supplementary Fig. 1). Thus, development of these putative memory B cell subsets was compromised in the absence of T-bet with no direct or indirect requirement for T-bet in IgG1 memory development.
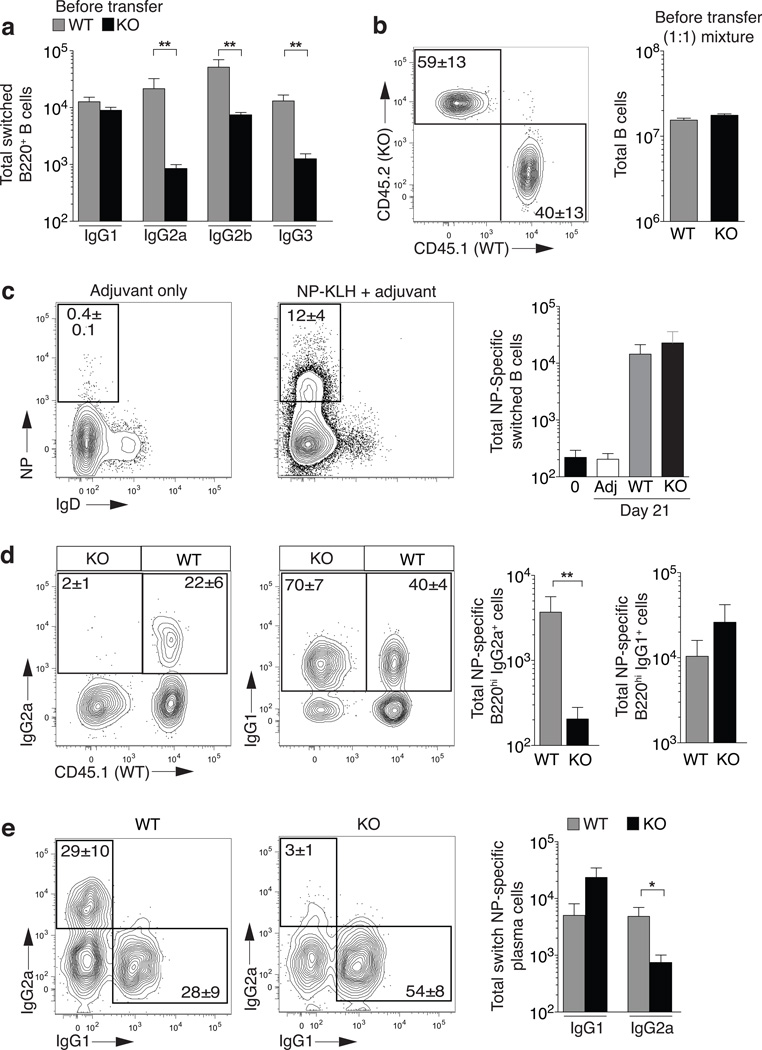
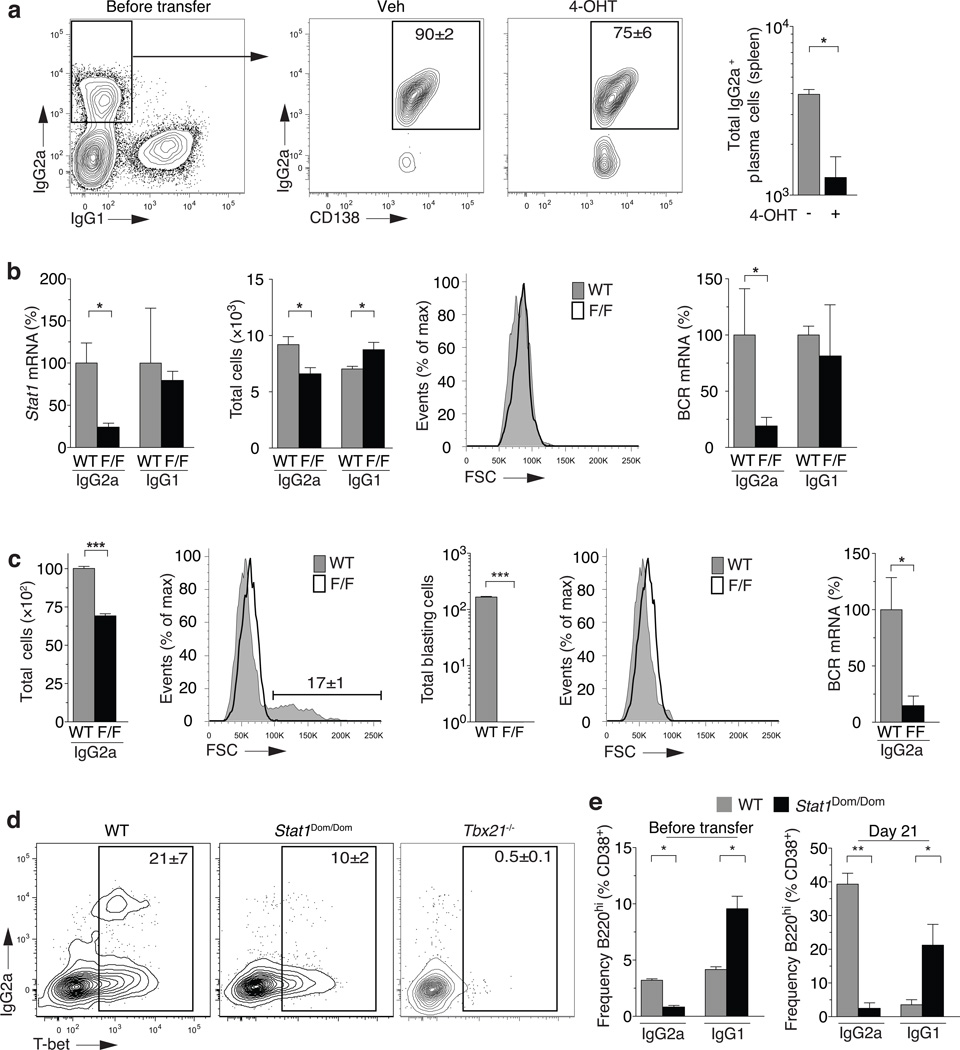
Figure 1. B cell intrinsic T-bet is required for IgG2a formation.
(a) Total numbers of polyclonal splenic B cells (Gr1−CD4−CD8−, CD19+ or CD138+) that are switched (IgM−IgD−) and CD138− B220hi. C57BL/6 (WT, grey bars) or Tbx21−/− mice (KO, black bars). Mean±sem; n=5 **P<0.01. (b) Rag1−/− mice receive 1:1 mixture of splenocytes from CD45.1 (WT) and Tbx21−/− (KO) donors and immediately immunized with NP-KLH in adjuvant. Frequency boxed in FACS profile (left panel) and total cell numbers (right panel) of B cells (Gr1−CD4−CD8−, CD19+ or CD138+) from donors transferred into Rag1−/− recipients. (c) Day 21 splenic NP-specific response of Rag1−/− recipient mice from adoptively transferred and immunized cells in (b). Boxed area indicates frequency (left panels) of switched (IgM−) B cells (Gr1−CD4−CD8− and CD19+ or CD138+). Total number (right panel) of NP-specific cells prior to transfer (0), with adjuvant only (adj), or in WT or KO compartments immunized with NP-KLH in adjuvant. (d) Class-specific frequencies and total cell number of NP-specific switched B220hi B cells. Frequencies represent percentage of same congenic compartment (left panels) and total numbers (right panels) following transfer and immunization in (b). (e) Class-specific frequencies and total cell number of NP-specific plasma cells (Gr1−CD4−CD8−, IgM−IgD−, NP+CD138+), that are switched. Frequencies (boxed in left panels) and total numbers (right panels) following transfer and immunization in (b). (b–e), Mean±sem; n=5 Rag1−/− mice.*P<0.05,**P<0.01.
To provide Tbx21−/− B cells a source of wild-type polyclonal T cell help, we generated Rag1−/− mixed peripheral chimeras containing equal numbers of wild-type and Tbx21−/− spleen cells (Fig. 1b). Upon hapten-protein immunization (nitrophenylacetyl-keyhole limpet hemocyanin; NP-KLH), both wild-type and Tbx21−/− B cell compartments produced robust class-switched NP-specific B cell responses (Fig. 1c). Strikingly, there was >95% reduction in the antigen-specific IgG2a+ B cell response (Fig. 1d) that extended across antigen-specific germinal center (CD38lo), memory B (CD38hi) cell (Supplementary Fig. 2a) and plasma cell (CD138hi) compartments (Fig. 1e) in the B cell-specific absence of T-bet. In contrast, slightly higher IgG1+ NP-specific B cell responses were induced in both donor populations (Fig. 1d) with similar distribution into germinal center, memory B cell (Supplementary Fig. 2a) and plasma cells compartments (Fig. 1e). No defects were found in IgG3 and IgG2b NP-specific B cell compartments in the absence of T-bet (Supplementary Fig. 2b). Furthermore, total class-switched B cells, that would contain the KLH-specific response and polyclonal reactivities in the peripheral chimeras, displayed a similar selective defect in IgG2a with concentrations of IgG1, IgG3 and IgG2b B cell compartments equivalent to wild-type (Supplementary Fig. 3). Therefore, B cell intrinsic expression of T-bet is required selectively for IgG2a class switch and/or survival of IgG2a+ B cells after class switch in vivo.
IgG2a memory B cell survival and function requires T-bet
To enable analysis of T-bet expression in IgG2a+ B cells after class-switch, we crossed Tbx21F/F mice18 to C57BL/6 mice with tamoxifen-inducible Cre in the ubiquitously expressed Rosa26 locus (Rosa26-CreERT2). Treatment of intact CreERT2 Tbx21F/F mice with 4-hydroxy-tamoxifen (4-OHT) induced the loss of T-bet protein in >50% of T-bet expressing splenocytes (Fig. 2a). There were similar reductions in IgG2a+ B cells in treated animals with no change in numbers of IgG1+ B cells. Importantly, all remaining IgG2a+ B cells expressed T-bet protein (Fig. 2b) at levels equivalent to treated Cre-ERT2 Tbx21+/+ animals (not shown). Thus, ablation of Tbx21 in vivo led to significant loss of IgG2a+ memory B cells over 4 days with no exogenous BCR stimulus.
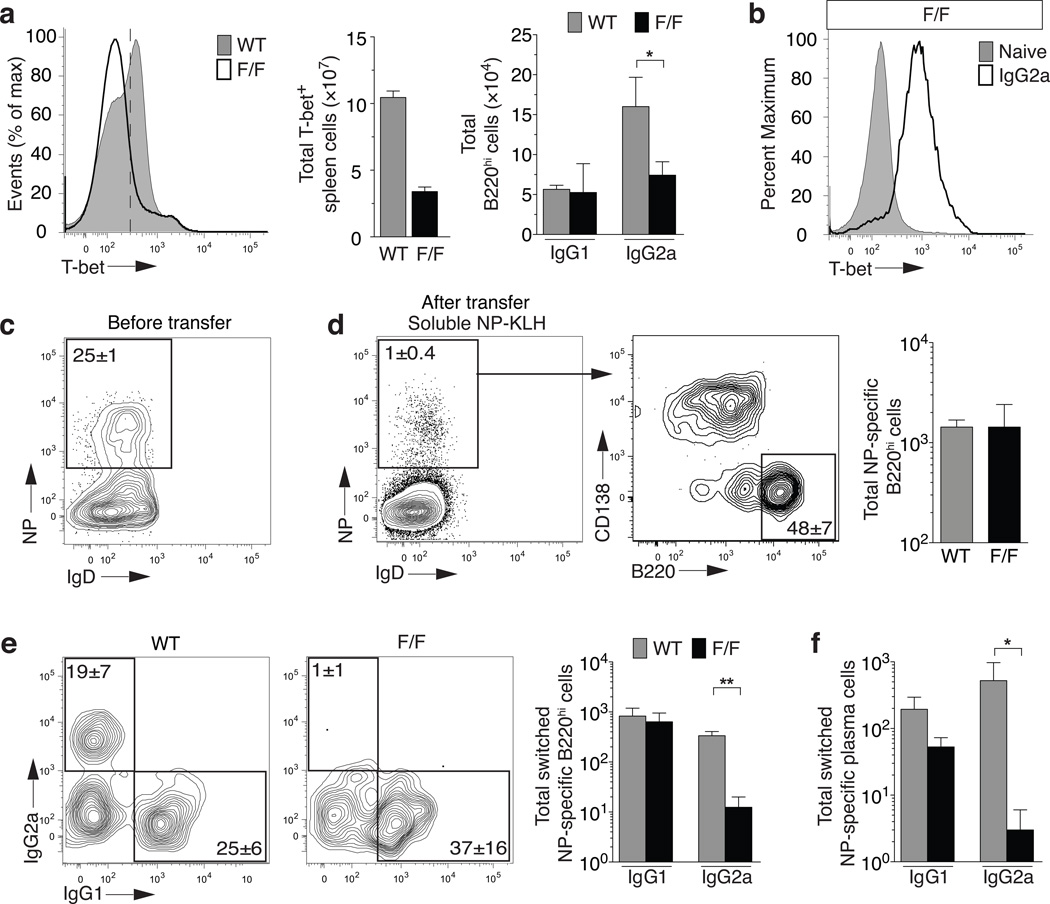
Figure 2. T-bet is required for memory B cell survival and function in vivo.
(a) Mice received daily injections of 4-OHT for 3 days and spleens were harvested on day 4. T-bet expression in total splenocytes (left panel) following a forward and side scatter lymphocyte gate. Total cell numbers of T-bet+ splenocytes (middle panel) or IgG2a+ and IgG1+ B cells following in vivo 4-OHT treatment in Rosa26-CreERT2;Tbx21+/+(WT) or Tbx21F/F(F/F) mice (right panel). (b) T-bet expression in B220hiCD38+IgG2a+ B cells (Gr1−CD4−CD8− IgM−IgD− CD19+CD138−) (white) or naïve B cells (Gr1−CD4−CD8− IgM+IgD+ CD19+B220+) (grey). (a–b), Mean±sem; n=3 WT or F/F mice, *P<0.05, (c) Splenic NP gate from B cells (Gr1−CD4−CD8−, CD19+ or CD138+) that are IgM− and transferred into Rag1−/− recipients. (d) 7 days after transfer in (c) and soluble boost with NP-KLH (right panels). Expression of CD138 and B220 on NP-specific cells following transfer and soluble boost. Total B220hi B cell numbers (right panel). (e) Frequency of IgG2a+ and IgG1+ NP-specific B cells (left panels) and total cell numbers (right panel) after transfer and soluble boost gated on B220hi in (c). (f) Total cell numbers of NP-specific plasma cells (CD138+) from transfer in (c). (c–f) Mean±sem; n=5 Rag1−/− mice. *P<0.05,**P<0.01.
As temporal deletion with 4-OHT in vivo targets all cells, it was important to determine whether B cell-specific loss of T-bet had caused the selective IgG2a deficit. In the next strategy, we induced an NP-specific B cell response in the CreERT2 Tbx21F/F and CreERT2 Tbx21+/+ donor mice that contained antigen-specific B220hi IgG1+ and IgG2a+ compartments (Fig. 2c,d & Supplementary Fig. 4a). Splenocytes from both of these donors were treated in vitro with 4-OHT for 1 h. This treatment excised >90% of the Tbx21F/F allele (Supplementary Fig. 4b) and controls for toxicity of treatment in wild-type cells. Both sets of treated cells were transferred into separate Rag1−/− recipients together with T-bet sufficient splenocytes from NP-KLH immunized wild-type congenic mice. The mixed peripheral chimeras were then challenged 24 h later with soluble antigen without adjuvant (unable to induce a primary NP-specific response; not shown), to promote a secondary NP-specific B cell response in recipients. Both Tbx21F/F and Tbx21+/+ 4-OHT treated sources of NP-specific B cells induced secondary responses to soluble antigen with equivalent NP-binding CD138−B220+ B cells and plasma cells (Fig. 2e). However, the NP-specific IgG2a+ recall response required T-bet with <5% of Tbx21F/F B cells responding after 4-OHT treatment compared to wild-type B cells (Fig. 2f). The NP-specific IgG1 response remained intact and emerged to equivalent extent as seen in the absence of T-bet (Fig. 2f). The same trends were seen across the IgG2a and IgG1 plasma cell compartment (Fig. 2f). Mixed chimeras were constructed 6 months after initial priming of donor animals and displayed the same selective loss of IgG2a+ NP-specific cells upon transfer and immunization (Supplementary Fig. 4c). Thus, IgG2a+ memory B cells selectively require B cell-intrinsic T-bet expression to respond effectively to antigen re-challenge in vivo.
T-bet persists during IgG2a memory B cell development
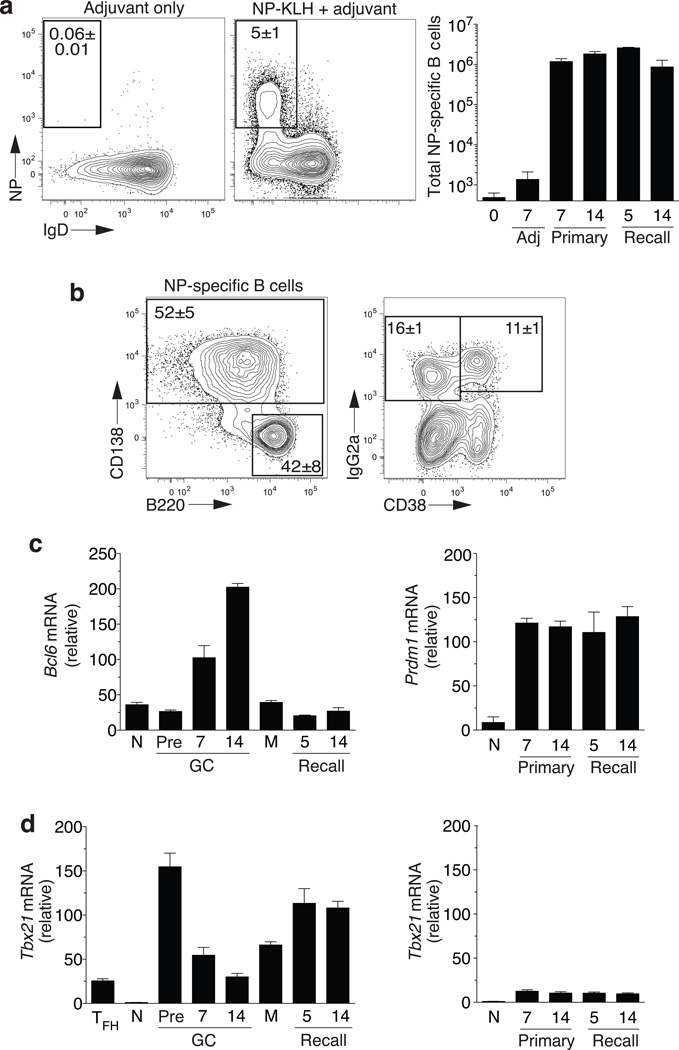
Next, we assessed changes in Tbx21 expression over the course of a primary and memory response in NP-KLH immunized animals19,20 (Fig. 3a). Differential CD138, B220 and CD38 expression on class-switched (IgM−IgD−) antigen-binding (NP+) CD19+ B cells provided direct access to IgG2a+ B cells (Fig. 3b). We used Bcl-6 (ref.21) and Blimp-1 (ref.22) expression together with antibody isotype and phenotypic markers to further distinguish pre-GC (day 7; B220hiCD38hi Bcl6lo), germinal center (day 7, 14 B220hiCD38lo Bcl6hi), memory (day 14 primary, day 5 and 14 secondary, B220hiCD38hi Bcl6lo) and plasma cell (CD138hi, Blimp-1hi) stages of antigen-specific IgG2a+ memory B cell development (Fig. 3c). As anticipated, T-bet was abundantly expressed in primary antigen-responsive IgG2a+ B cells before entry into germinal centers, then decreased significantly in the presence of Bcl-6 within germinal centers at day 7 and 14 after priming (Fig. 3d). Toll-like receptor 4 (TLR4) agonist-based immunization used here promotes negligible IgM+ memory B cells with the majority of NP-specific memory B cells expressing IgG isotypes (not shown). Upon antigen re-challenge, NP-specific IgG2a+ memory B cells expressed elevated amounts of T-bet that remained high for at least 14 days after recall (Fig. 3d, right). In the presence of Blimp-1, IgG2a+ plasma cells from all stages of the primary and memory response expressed low but detectable amounts of T-bet (Fig. 3d, left). Thus, T-bet is expressed early upon initiation of IgG2a class switch and remains expressed at all stages of antigen-specific IgG2a+ memory B cell development and response in vivo.
Figure 3. T-bet expression during antigen-specific IgG2a memory development.
(a) Day 7 NP-specific B cell response in lymph nodes of C57BL/6 mice immunized with adjuvant only or adjuvant with NP-KLH gated Gr1−CD4−CD8−, B cell positive [CD138+ or CD19+] and IgM- (left panels). Timecourse of total NP-specific B cells (right panel). (b) Day 7 representative NP-specific B cell subsets (left panel) and B220hi expression of CD38+ and IgG2a+ (right panel). (c) mRNA for Bcl6 (left panel) or Prdm1 (right panel) after isolation of naïve (N)(day 0, IgM+IgD+CD23+) and NP-specific CD19+B220hiIgG2a+ day 7 pre-GC (CD38+), day 7 GC (CD38−), day 14 GC (CD38−), day 14 memory (M)(CD38+), recall day 5 and 14 memory cells (CD38+). mRNA for Prdm1 (right panel) after isolation of IgG2a+ plasma cells (CD138+) (d) T-bet expression for TH cell samples (antigen-responsive TFH cells) and populations described in (c) (left panel) and for IgG2a+ plasma cells (CD138+, right panel). Mean±sem; n≥5 mice.
T-bet activity in IgG2a memory B cells
As evidence for T-bet activity in antigen-specific IgG2a+ CD38hi memory B cells, transcription for a series of known T-bet target genes23 were elevated at day 5 of the memory response (Fig. 4a, top). Differential expression of these target genes implies that T-bet enables separate functions in IgG2a+ memory B cells compared to naive B cells. Recent studies have shown that Bcl-6 can directly bind to T-bet and repress T-bet target gene expression in T cells24. To test whether something similar occurs in B cells, we assayed the same T-bet target genes in germinal center B cells that contain elevated Bcl-6 and found that expression of these targets was diminished (Fig. 4a, bottom). Furthermore, Blimp-1 has been shown to directly antagonize T-bet expression25. Analysis of T-bet target genes in plasma cells that contain elevated Blimp-1 protein revealed reduced expression for most of the genes tested (Fig. 4b). Although T-bet activity is present in IgG2a+ memory B cells, Bcl-6 and Blimp-1 expression significantly decreases this activity within IgG2a+ GC B cells and IgG2a+ plasma cells respectively.
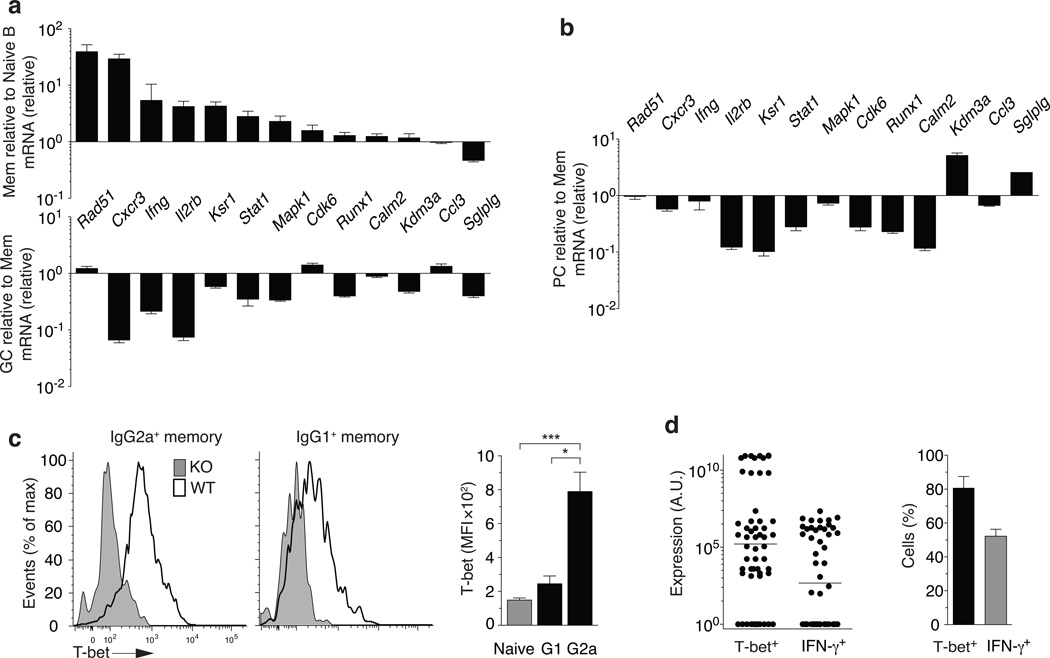
Figure 4. Evidence for T-bet activity in IgG2a+ memory B cells.
(a) mRNA for T-bet targets at recall day 5 IgG2a+ memory versus naive B cells from day 5 memory NP-specific B cells from draining lymph nodes of C57BL/6 mice immunized with adjuvant and NP-KLH are FACS-purified and gated on Gr1−CD4−CD8−, B cell positive [CD138+ or CD19+], switched (IgM−IgD−), and NP+IgG2a+ memory B220hiCD38+CD138−. Naïve B cells are FACS-purified and gated on Gr1−CD4−CD8−, B cell positive (B220+CD19+CD138−) and IgM+IgD+CD23+ (top panel). IgG2a+ day 14 mature-GC (CD38−) versus recall day 5 (bottom panel). Mean±sem, n≥5. (b) mRNA for T-bet targets in recall day 5 IgG2a+ memory versus IgG2a+ plasma cells. NP+IgG2a+ plasma cells FACS-purified by gating on Gr1−CD4−CD8−, B cell positive [CD138+ or CD19+], switched (IgM−IgD−) and CD138+ are compared to recall day 5 IgG2a+ memory FACS-purified as in (a). (c) T-bet MFI in IgG2a+(G2a) and IgG1+(G1) B220hiCD38+ B cells (Gr1−CD4−CD8− IgM−IgD− CD19+CD138−) with control histograms from Tbx21−/− mice in grey. (d) T-bet and IFN-γ mRNA expression from single FACS-sorted cells (left panel) that are Gr1−CD4−CD8−, B cell positive [CD19+or CD138+], switched IgM−IgD−, and B220hiCD138−CD38+IgG2a+. Frequency of total single cells shown in left panel positive for T-bet or IFN-γ signal (right panel).
T-bet protein is expressed broadly across most IgG2a+CD38hi memory B cells with low but detectable amounts in IgG1+CD38hi memory B cells (Fig. 4c). At the single cell level, the majority IgG2a+CD38hi memory B cells expressed intermediate amounts of T-bet mRNA with a minor fraction expressing substantially higher amounts per cell (Fig. 4d). As suspected from the population analysis, many of the T-bet expressing memory cells also expressed detectable IFN-γ mRNA. Thus, it is likely that IgG2a+ memory B cells are a source of IFN-γ in vivo.
BCR-driven IgG2a response requires T-bet expression
As IgM memory B cells were not actively excluded in previous experiments, we purified IgG2a+ B cells from unimmunized CreERT2 Tbx21F/F mice and treated these cells with 4-OHT in vitro. These treated IgG2a+ memory B cells were transferred into Rag1−/− recipients and reactivated through their BCR with anti-IgG2a. This in vivo stimulus induced robust production of IgG2a+ plasma cells with negligible numbers of CD19+ non-plasma cells remaining 4 days after transfer (Fig. 5a). Nevertheless, after treatment with 4-OHT, there was a 75% reduction in IgG2a+ plasma cells with no increase in residual CD19+ B cell compartment. While cell recovery was low, similar trends were obtained in a second series of experiments after sorting NP-specific memory B cells from immunized CreERT2 Tbx21F/F mice, 4-OHT treatment in vitro, transfer into Rag1−/− recipients and antigen re-challenge (Supplementary Fig. 5). Nevertheless, IgG2a-selective defects were seen in total class-switched B cells using this experimental design (data not shown). Thus, B cell intrinsic T-bet was required to permit IgG2a+ BCR-driven plasma cell differentiation in vivo.
Figure 5. B cell intrinsic T-bet is required for survival, IgG2a+ BCR expression and function.
(a) Class-switched memory B cells (GR1−CD4−CD8− IgM−IgD−CD19+B220+CD138−CD38+) were FACS-purified from spleens of unimmunized Tbx21F/F; Rosa26-CreERT2 mice treated ex vivo with 4-OHT and transferred into Rag1−/− recipients. 1 day after transfer, mice were injected with anti-IgG2a antibody and spleens collected 3 days after injection. Frequency and total cell number of IgG2a+ plasma cells (GR1−CD4−CD8−CD138+) from unimmunized Tbx21F/F;Rosa26-CreERT2 mice after transfer into Rag1−/− mice following donor cell treatment with 4-OHT or vehicle (on CD4−CD8−CD138−). Mean±sem; n=3 Rag1−/− mice. *P<0.05 (b) Total cell number and mRNA expression from live PI− FACS-purified Gr1−CD4−CD8−, B cell positive [CD19+orCD138+], switched IgM−IgD−, and class-specific CD138−B220hiCD38+ B cells from unimmunized Rosa26-CreERT2 Tbx21+/+(WT) or Tbx21F/F(F/F) mice treated with 4-OHT (n≥3) after 2 days. Forward scatter of PI− cells sorted after 2 days for mRNA analysis from wells containing Rosa26-CreERT2 Tbx21+/+(WT) or Tbx21F/F(F/F) cells. mRNA expression is displayed as F/F's percentage relative to WT (%) *P<0.05 (c) Total cell number and mRNA expression from IgG2a+ memory B cells sorted as in (b) after 4 days. Cells were re-activated with anti-IgG2a antibody after 48 hours in culture. Forward scatter of total PI− cells and total number of PI− blasting cells from wells containing Rosa26-CreERT2 Tbx21+/+(WT) or Tbx21F/F (F/F) cells (left panels). Forward scatter of actual PI− cells sorted for mRNA analysis from wells containing Rosa26-CreERT2 Tbx21+/+(WT) or Tbx21F/F(F/F) cells (right panels). *P<0.05 (d) T-bet expression in splenic switched B220hiCD38+IgG2a+ B cells from wild-type mice (C57BL/6), STAT1Dom/Dom and Tbx21−/− mice on the C57BL/6 background. (e) Frequency of IgG2a+ and IgG1+ B220hiCD38+ B cells before (left panel) and 21 days following generation of peripheral chimeras (right panel). Mean±sem; n≥5 Rag1−/− mice. *P<0.05, **P<0.01.
STAT-1, survival and IgG2a BCR expression requires T-bet
To examine how T-bet exerted its impact on IgG2a B cell fate, we used IgG1+ and IgG2a+ memory B cells from wild-type or CreERT2 Tbx21F/F mice for in vitro studies. Deletion of Tbx21 using 4-OHT in vitro resulted in a significant loss of STAT1 transcription, the signal transducer of IFN-γ, selectively in IgG2a+ B cells and not IgG1+ B cells, on a per cell basis (Fig. 5b, left). In the absence of T-bet, there was also a small but significant impact on cell survival in short-term cultures in the presence of BAFF with loss of IgG2a+ and increase of IgG1+ B cells (Fig. 5b, middle). However, over the same period, there was >75% loss of transcription for mature IgG2a+ BCR transcripts on a per cell basis (Fig. 5b, right). Secondary cultures using anti-BCR coated plates induced significant numbers of B cell blasts over 48 h in vitro while IgG2a+ B cells remained small by forward scatter in the absence of T-bet (Fig. 5c). Small live cells from both culture conditions were selected for quantitative PCR analysis of mature IgG2a+ BCR transcripts with exaggerated losses detected in the absence of T-bet (Fig. 5c, right). Thus, in the absence of T-bet in vitro IgG2a+ BCR down-regulation and loss of STAT1 transcription were more pronounced than overall loss in IgG2a+ memory B cells.
Next, we investigated the IgG2a+ B cell compartment in an N-ethyl-N-nitrosourea (ENU)-generated animal strain with a phosphorylation defect that prevents nuclear translocation in STAT1 named Domino (Stat1Dom/Dom)26. While T-bet expression was decreased by 50%, there was almost complete loss of IgG2a+ B cells (Fig. 5d) with a compensatory increase in IgG1+ B cells within these mice (Fig. 5e, left). Both trends become exaggerated upon adoptive transfer into Rag1−/− mice in a mixed peripheral chimera without immunization (Fig. 5e, right). Thus, both T-bet expression and STAT-1 activation appear necessary for continued IgG2a BCR expression and/or IgG2a+ memory B cell survival in vivo.
Separable programs for IgA and IgG2a memory B cells
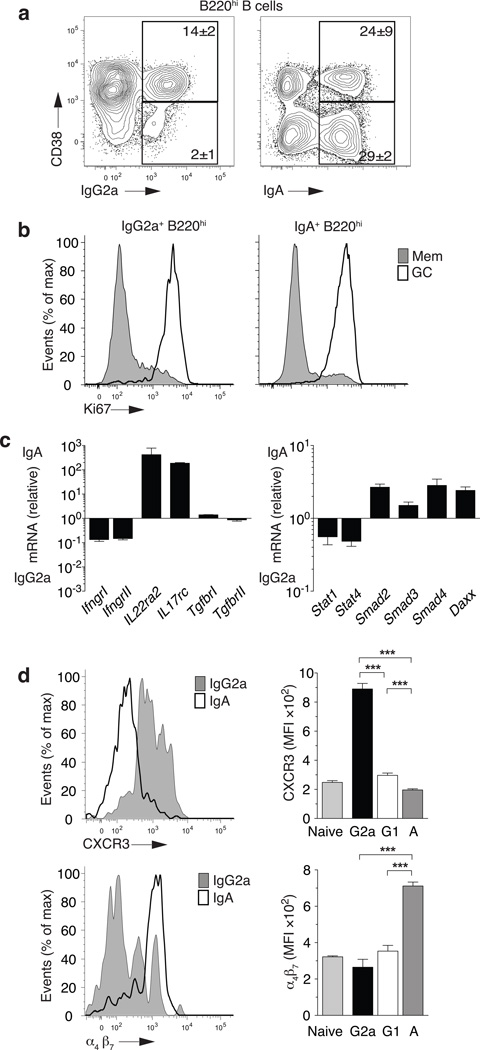
IgA antibody plays a dominant role at mucosal surfaces, binding to a variety of innate immune cells, enhancing phagocytosis and triggering the local release of cytokines and inflammatory mediators27,28. TGFβRII expression by B cells is required for IgA class switch9 with some evidence for TH17 cell involvement co-coordinating mucosal IgA responses29. Transcription factors in the Runx and Smad family have been shown to promote germline IgA transcripts towards IgA class switch8,30. To contrast the transcriptional program of IgG2a+ and IgA+ memory B cells, we isolated isotype-switched (IgM−IgD−) CD38+ B cells (also CD19+CD138−B220+) that were largely Ki67− (non-cycling) as quiescent memory B cell compartments (Fig. 6a,b from spleen or Peyer’s patches). IgG2a+ memory B cells differentially expressed IFN-γRI and IFN-γRII while IgA+ memory B cells differentially expressed IL17Rc andIL22Rα2 suggesting differential responsiveness to IL-17 and IL-22. Furthermore, while the TGFβR was expressed similarly in both memory subsets, increased TGFβ signaling intermediates of the Smad family members Smad2/3/4 and TGFβ associated adapter Daxx was increased in IgA+ memory B cells (Fig. 6c). CXCR3 protein was differentially expressed by IgG2a+ memory B cells while most IgA+ memory B cells expressed high amounts of integrin α4β7 (Fig. 6d). These data suggest that IgG2a and IgA memory B cells have separable growth factor requirements, signaling propensity and tissue homing potential (Supplementary Fig. 6).
Figure 6. Separable programs for IgG2a+ and IgA+ memory B cells.
(a) Frequency of Gr1−CD4−CD8−, B cell positive [CD19+orCD138+], switched IgM−IgD−, class-specific CD138−B220hi B cells in unimmunized C57BL/6 mice. Representative staining from spleen (left panel) and Peyers Patches (right panel) (b) Ki67 staining on cells from (a). Shaded histograms are from CD38+ (memory) cells gated as in (a), and white histograms are CD38− (GC) cells gated as in (a). (c) mRNA expression of IgG2a+ or IgA+ splenic B cells (a) for cytokine receptors (left panel) or signal transduction molecules (right panel). (d) Surface staining (MFI) of IgG2a+, IgG1+, IgA+ or naive splenic B cells for CXCR3 (left panels) and α4β7 (right panel). (a–d) Mean±sem; n≥4 mice. ***P<0.001.
Survival and BCR expression in IgA memory requires RORα
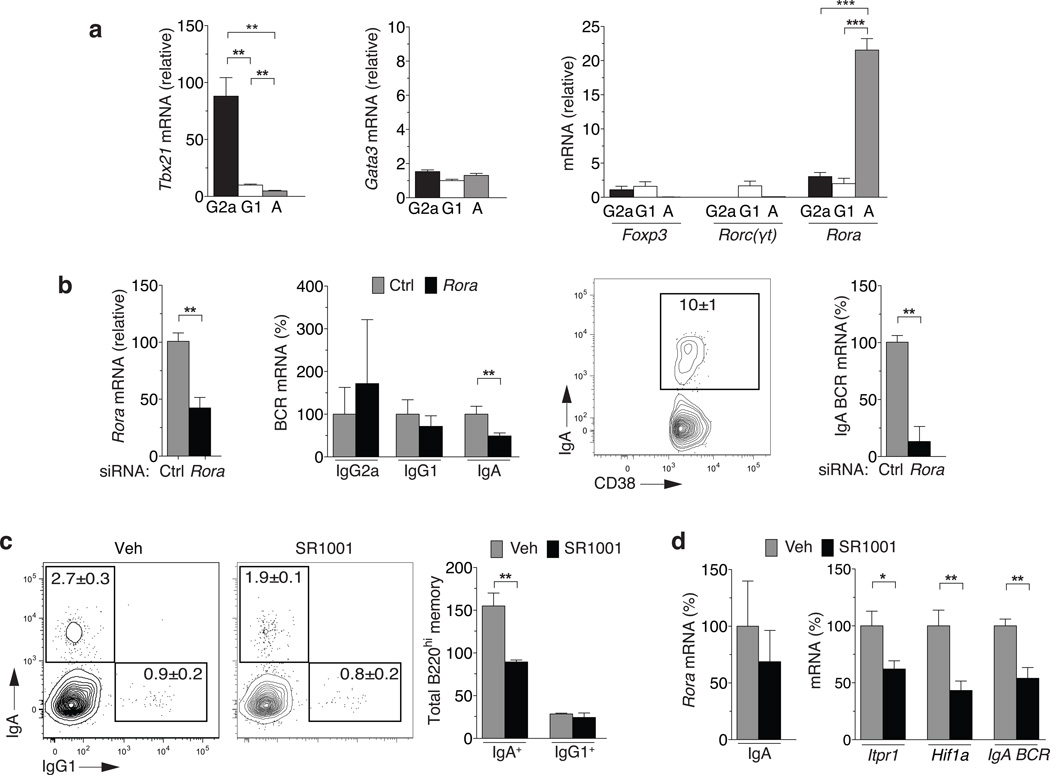
Of the major transcription factors downstream from TGF-β and IL-17 signaling, RORα was highly expressed in IgA memory cells as compared to other memory and naïve B cells (Fig. 7a). GATA-3, Foxp3 and RORγt were detectable at only low amounts in naive B cells, IgG1+, IgG2a+ and IgA+ memory B cells. Germline deletion of RORα in mice produces a complex phenotype with neurological defects and early fatality31. Hence, to probe the function of RORα in IgA memory B cells, we silenced expression in normal IgA+ B cells using targeted siRNA. Whole mesenteric lymph node and Peyer’s patch cells were nucleofected with RORα siRNA, resulting in ~50% decrease in RORα mRNA after 4 days culture in vitro (Fig. 7b, left). This level of RORα knockdown resulted in ~50% decrease in mature IgA mRNA in the total cell population (Fig. 7c, middle left). Isolating IgA+ memory B cells from these cultures revealed a >80% reduction of mature IgA BCR transcripts on a per cell basis (Fig. 7b, right). Thus, RORα is differentially expressed in IgA+ B cells with evidence for control of dynamic BCR expression.
Figure 7. RORα regulates survival and BCR expression in IgA+ Memory B cells.
(a) mRNA expression for indicated gene products relative to naive B cells from IgG2a+, IgG1+, or IgA+ cells gated on Gr1−CD4−CD8−, B cell positive [CD19+orCD138+], switched IgM−IgD−, class-specific CD38+B220hiCD138− B cells in unimmunized C57BL/6 mice. (b) mRNA from pooled Peyer’s Patches and mesenteric lymph nodes 4 days after nucleofection with a Rorα-specific (Rorα) (shown in black) or control (Ctrl) (shown in grey) siRNA in samples from total cells (left panels) or from sorted IgA+CD38+B220hi switched B cells (right panels). mRNA expression is displayed as F/F's percentage relative to WT (%) (c) Frequency and total number of class-specific CD38+B220hi splenocytes after treatment with 5µM SR1001 or vehicle for 6 days. (d) mRNA for indicated gene products from FACS-purified CD38+IgA+ cells from (a) treated with 5µM SR1001 (black bars) or vehicle (grey bars) for 24 hours. Mean±sem; n≥4; *P<0.05, **P<0.01, ***P<0.001.
To interfere with ROR protein function, we used the compound SR1001 recently described to inhibit RORα and RORγt transcriptional activity32. The presence of this functional inhibitor selectively decreased the number of IgA+ memory B cells that survived in culture for 6 days without impacting IgG1+ B cells (Fig. 7c). To establish the influence of the inhibitor on IgA+ memory B cells, we treated purified IgA+ memory B cells with SR1001 over short-term cultures. As the drug only inhibits protein function, there was no significant impact on RORα mRNA expression (Fig. 7d, right). However, there was a significant decrease in transcription of some known RORα targets, Itpr1 and Hif1a. Interestingly, there were significant decreases in the abundance of mature IgA BCR transcripts per cell (Fig. 7d, left). Thus, RORα can impact IgA+ memory B cell function with evidence that it is required for IgA+ B cell survival and control of IgA BCR mRNA transcription.
DISCUSSION
Antigen-specific B cell memory is central to long-term immune protection and develops across many different antibody classes. Here, we focus on unique properties of IgG2a+ and IgA+ memory B cells to demonstrate that divergent transcriptional regulators maintain class-specific memory B cell subset integrity. Both T-bet and RORα regulate mature BCR transcription, which appears necessary for memory B cell survival in IgG2a+ and IgA+ memory B cells respectively. In addition, our study utilizes conditional genetic models that temporally delete T-bet to demonstrate severe defects in the capacity of IgG2a+ memory B cells to respond to antigen. Both IgG2a+ and IgA+ memory B cells expressed different transcriptional programs that reflect separate potentials in cytokine secretion, trafficking and survival that permit flexibility in long-term antigen-specific immune protection. We propose that the events that lead to class-switch after antigen-experience lead to the imprinting of molecular programs whose persistence is essential for maintaining memory B cell subset identity.
The dynamic regulation of BCR transcription was a common component of both RORα and T-bet programs. We found consensus binding sites for both transcription factors within the immunoglobulin 3' regulatory region known to control class switch and antibody secretion33. Thus, separable T-bet and RORα dependent transcriptional programs within IgG2a+ and IgA+ memory B cells also control the central recognition properties of antigen-specific B cell memory.
Cognate TFH cells regulate multiple facets of antigen-specific memory B cell development4. Bcl-6 programs TFH development and movement to B cell areas for cognate control of B cell immunity34. Cytokine production occurs in these follicular regions upon contact with antigen-primed B cells in ways that can initiate antibody class-switch35. Within germinal centers, TFH cell contact36 regulates the clonal composition of germinal center B cells and export into B cell memory compartments37. In germinal center B cells, Bcl-6 can antagonize T-bet function and this dynamic molecular interplay may underpin germinal center B cell fate and IgG2a+ memory B cell development. There is evidence for cytokine-producing germinal center TFH cells that engage germinal center B cells in an antibody class-specific manner35. In this scenario, antibody class-specific regulation by cognate TFH cells may streamline affinity maturation and efficiently promote memory B cell development.
T-bet regulates critical cellular functions within IgG2a+ memory B cells. In naïve T cells, signaling through the antigen receptor synergizes with IFN-γ–induced STAT1 activation to promote initial T-bet expression12. We demonstrate a similar IFN-γ, STAT1 and T-bet dependent regulatory axis in IgG2a+ memory B cells. Ablating T-bet expression interferes with this program, resulting in down-regulation of BCR transcription and loss of IgG2a+ memory B cells. While antigen binding by memory BCR is not required for survival38, we propose that memory B cells rely on tonic signals through BCR for survival as found in naïve B cells39. Similar to antigen-specific plasma cells20, memory B cells may also retain antigen-presenting capacity for extended periods in vivo. In this manner, continued BCR expression may also indirectly permit ongoing local contact with antigen-specific TFH cells40.
T-bet activity establishes a molecular framework that governs IgG2a B cell functions. While T-bet binds to target promoters regardless of cell type, its ability to transactivate and remodel loci is cell context-dependent23. The regulation of genes by T-bet is also coupled to the dynamic expression of its molecular antagonists Bcl-6 and Blimp-1 (refs.24, 25). Interestingly, T-bet can assort asymmetrically in T cells at cell division41 and more recently Bcl-6 was shown to assorted unevenly across GC B cells42. Our studies show that the co-expression of Bcl-6 with T-bet in GC B cells results in repression of transcription of T-bet targets. However, rather than abolishing a T-bet-defined subset, Bcl-6 transiently and reversibly alters IgG2a+ programming to allow productive GC activity and IgG2a+ memory B cell development.
RORαlike T-bet, is able to recruit chromatin remodeling machinery and directly transactivate loci11. However, unlike T-bet, the interactions of RORα with Bcl-6 and Blimp-1 have not been characterized. Although some studies point to Bcl-6 as repressing RORγt and TH17 differentiation43,44, the cell-intrinsic effects on RORα is unknown. Mice deficient in RORα have severe defects and die early due to the early expression of RORα during development11. Studies on RORα targets in immune cells provide connections to activation of IL-17, IL-17F, IL-22, and IL-23R45, and in repression of IL-6 and tumor necrosis factor46. It is unknown whether activation of these targets is direct or indirect, and we found no differential expression of these targets in IgA+ memory B cells (not shown). However, RORα is involved in several other pathways including calcium signaling, circadian rhythm and cellular metabolism11 with evidence for an impact of the SR1001 on Iptr1 and HIF-1α that might indicate other pathways in which IgA+ memory B cells may be unique from other classes.
Antigen-specific memory B cell responses may also require sub-specialized regulatory programs organized by antibody class. Upon antigen re-challenge, memory B cells require cognate T cell help to expand and differentiate into plasma cells47. IgM memory B cells express separable functions and capacity to respond compared to their IgG counterparts3,48. Expression of downstream antibody classes engage different signaling pathways based on the constant region of the BCR5,6. Other class-specific memory B cell properties control migration, such as T-bet driving IgG2a+ memory B cells to inflammatory sites via CXCR3 expression and α4β7 integrin guiding IgA+ memory cells to mucosal tissues27. Enhanced STAT1 and IFN-γR transcription may uniquely sensitize IgG2a+ memory B cells to IFN-γ signals. Similarly, RORα can enhance calcium sensitivity within cells49 and CamKIV can enhance RORα expression50. These types of changes in IgA+ memory B cells may lower BCR activation thresholds in a class-specific manner. Therefore, multiple attributes of class-specific B cell memory that are introduced upon development may be further reinforced upon antigen recall under the cognate guidance of memory TFH cells40.
Memory B cells are generally considered functionally equivalent, differing only by the antibody isotype they express. In the current study, we reveal unexpectedly that B cell memory is organized in class-specific subsets each with separate central transcriptional regulators. Specifically, transcriptional regulators T-bet and RORα control divergent IgG2a and IgA memory B cell subsets respectively to control separate functions within these unique class-specific memory B cell compartments. T-bet is used by many cell types in response to inflammatory stimuli with focus on the clearance of intracellular pathogen12. We now reveal that IgG2a+ B cell memory also relies selectively on a T-bet dependent program to establish and maintain subset integrity. Similarly, IgA+ B cell memory is specialized to protect the mucosal surfaces27 and the selective use of transcriptional regulator RORα enhances this unique memory B cell function. Importantly, these unique developmental programs can be exploited for directed immunotherapeutic applications, future class-skewing vaccine formations and the treatment of cancer and autoimmunity.
METHODS
Animals
Tbx21−/− (purchased from Jackson), C57BL/6 (B6), Rag1−/−, B6.CD45.1, Tbx21F/F, STAT1Dom/Dom and Tbx21F/F;Rosa26-CreERT2 were housed in pathogen-free conditions. All experiments were performed in compliance with federal laws and institutional guidelines as approved by the Scripps Research Institutional Animal Care and Use Committee. We generated Rosa26-CreERT2;Tbx21F/F by crossing Tbx21F/F mice with Rosa26-CreERT2 mice (purchased from Jackson). For genotyping of mutants, the following primers were used at 0.4µM final concentration in a standard FastStart Reaction Mix (Roche) using manufacturer’s guidelines: oIMR1719 (Common) (5'-TGGGCATACAGGAGGCAGCAACAAATA-3 '), oIMR1718 (Wild-Type) (5'-GACTGAAGCCCCGACCCCCACTCCTAAG-3'), oIMR1717 (Mutant) (5'-GCGCGAAGGGGCCACCAAAGAACGGAG-3') for genotyping of B6.129S6-Tbx21tm1Glm/J (Tbx21−/−). oIMR8545 (common) (5'-AAAGTCGCTCTGAGTTGTTAT-3'), oIMR8546 (wild-type reverse) (5'-GGAGCGGGAGAAATGGATATG-3'), oIMR8547 (mutant reverse) (5'-CCTGATCCTGGCAATTTC G-3') were used for genotyping of B6.129-Gt(ROSA)26Sortm1(cre/ERT2)Tyj/J (Rosa26-CreERT2) mice. For the detection of non-excised locus in Tbx21F/F animals and cells primers A (sense) (5' – TATGATTACACTGCAGCTGTCTTC AG-3') and B (anti-sense) (5'-CAGGAATGGGAACATTCGCCTGTG-3') were used and for detection deletion within locus deltaF (sense) (5'-AGCCATCTCTCCAGCCTA CA-3') and C2 (anti-sense) (5'-CTCTGCCTCCCATCTCTTAGGAGC-3') were used for amplification.
Flow Cytometry
Draining lymph nodes and spleen were removed from unimmunized or immunized animals and single cell suspensions in PBS with 5% (vol/vol) FBS prepared. 4 × 108 cells per mL were incubated for 15 min with anti-CD16/32 (Fc block, 2.4G2) followed for 45 min at 4°C with fluorophore-labeled or biotin-labeled monoclonal antibodies: allophycocyanin-conjugated anti-CD138 (281-2), allophycocyanin-Cy7-conjugated anti-CD19 (1D3), phycoerythrin-conjugated anti-CD138 (281-2), phycoerythrin texas red-conjugated anti-B220 (RA3-6B2), fluorescein isothiocyanate-conjugated anti-IgG1 (A85-1), anti-IgG3 (R40-8L), Horizon V500-conjugated anti-CD8 (53-6.7) and anti-CD4 (Rm4-5), biotin-conjugated anti IgG2ab (5.7) [all from BD Biosciences]. Phycoerythrin-conjugated anti-T-bet (eBio4B10), Phycoerythrin-cy7-conjugated anti-CD45.1 (A20), Phycoerythrin-cy7-conjugated anti-CXCR3 (CXCR3-173), fluorescein isothiocyanateconjugated anti-CD45.2 (104), Biotin-conjugated anti-IgG2b (RMG2b-1), Biotin-conjugated anti-CD45.2 (104), and Biotin-conjugated anti-IgA (RMA-1) [all from Biolegend, Inc]. Alexa Fluor 700-conjugated anti-CD38 (90), phycoerythrin-cy5-conjugated anti-Gr-1 (Ly6G/C, RB68C5) [all from eBiosciences, Inc]. Allophycocyanin-conjugated anti-NP, phycoerythrin-conjugated NP, peridinin-chlorophyll protein-cy5.5-conjugated anti-IgM (331.12), Pacific Blue-conjugated IgD (11.26) [all from MMW Lab]. Immunoglobulin-specific antibodies were added in a separate step for 45 min at 4°C with normal mouse serum (1:50) before other reagents were added. Streptavidin-conjugated Qdot 655 (Invitrogen) were used as a second step visualization reagent. For intracellular staining, cells were fixed, permeabilized, and stained using the protocol in the Foxp3 Staining Buffer Set (eBioscience). Cells were washed and re-suspended in PBS/FBS and analyzed with a FACSAria III with FACSDiva software (BD Biosciences). Data were analyzed with FlowJo software (Tree Star, Inc).
Quantitative PCR
cDNA was prepared as described previously (51). Briefly, 5000 cells were sorted directly into lysis buffer (Qiagen) and mRNA was purified using RNeasy Kit (Qiagen). cDNA was generated using the First-Strand Synthesis System for RT-PCR (Invitrogen) using random hexamers. SYBR Green qPCR was conducted using the Platinum SYBR Green Supermix UDG reaction mix (Invitrogen) on the StepOnePlus Real-time PCR system and analyzed using the Step One Software (Applied Biosystems). Primers were used at a final concentration of 0.25µM and can be found in Supplementary Table 1. GAPDH was used as an endogenous control. For measurement of T-bet expression levels, naïve B cells were assigned a value of 1, and the relative expression assigned accordingly. For measurement of STAT1 and IgG2a post-switch mRNA levels, cells receiving 4-OHT treatment were assigned a value of 1, and the relative expression assigned accordingly. For cells treated with RORa siRNA or SR1001, cells receiving the drug or siRNA were assigned a value of 1, and the relative expression assigned accordingly. For single cell qPCR on mRNA, single cells were directly sorted into 2× Reaction buffer containing SSIII RT Platinum Taq found in the SuperScript III One-Step RT-PCR System with Platinum Taq DNA Polymerase Kit (Invitrogen) and containing outside primers for IFNg, T-bet, and GAPDH at a final concentration of 0.25µM. 96-well plates were placed in PCR cycler for 15 minutes at 50C, followed by 2 minutes at 95C. 22 cycles of 15 seconds at 95C followed by 4 minutes at 60C were then performed. 1µL of product was used in a standard qPCR reaction described above.
Adoptive transfers
Draining lymph nodes and spleen were removed from unimmunized or immunized animals and single cell suspensions in PBS. For germline knockout peripheral chimeras, 3.5×107 cells from Tbx21−/− or STAT1Dom/Dom and B6.CD45.1 mice where mixed 1:1 (7×107 cells total) and injected intraperitoneally (i.p.) into Rag1−/− hosts. Mice were then immunized subcutaneously at the base of tail with 400µg NP-KLH in MPL-based adjuvant. For conditional knockout peripheral chimeras, splenocytes from Rosa26-CreERT2;Tbx21F/F, Rosa26-CreERT2;Tbx21+/+, and B6.CD45.1 were treated with 4-hydroxytamoxifen (4-OHT) (Sigma). In brief, 1×107 from Rosa26-CreERT2;Tbx21F/F or Rosa26-CreERT2;Tbx21+/+ unfractionated splenocytes were treated 1 hour in 4-OHT-containing media (RPMI, 10% FBS, 2mM L-gluatmine, and 1µM 4-OHT), filtered, and mixed 1:1 with 1×107 cells from B6.CD45.1 mice (2×107 total) and transferred i.p. into Rag1−/− hosts. Mice were immunized 1 day later with 400µg NP-KLH in PBS. For experiments with depletion of plasma cells and naïve B cells, cells were FACS-purified and 1×106 cells from Rosa26-CreERT2;Tbx21F/F mice were treated with 4-OHT or vehicle for 1 hour and transferred i.p. into Rag1−/− hosts. For IgG2a+ B cell transfers, cells were FACS-purified and 1×104 cells from Rosa26-CreERT2;Tbx21F/F mice were treated with 4-OHT or vehicle for 1 hour and transferred i.p. into Rag1−/− hosts. Mice were given 75µg anti-IgG2a antibody (R19-15, BD Biosciences) i.p. 1 day after transfer into empty hosts.
Immunizations
Mice were immunized subcutaneously at the base of the tail with 400µg 4-hydroxy-3-nitrophenylacetyl (Biosearch) conjugated to keyhole limpet hemocyanin (Pierce, NP-KLH) in monophosphoryl lipid A-based adjuvant supplemented with trehalose dimycolate (1mg per 1mg MPL, Sigma). Antigen re-challenge was done 120 days or more after priming with 400 µg NP-KLH in MPL-based adjuvant. Soluble boost was done by mixing 400 µg NP-KLH in PBS.
4-Hydroxytamoxifen treatment of mice
4-hydroxytamoxifen (4-OHT) was dissolved in 100% ETOH at a final concentration of 20mg/mL, and was injected intraperitoneally (i.p) into unimmunized Rosa26-CreERT2 or Tbx21F/F;Rosa26-CreERT2 mice at a dose of 0.5mg/mouse for the first injection, and 0.25mg/mL for the 2 subsequent injections.
Cell culture
IgG2a+ or IgG1+ cells were FACS-purified with a FACSAria III. For survival studies, 8×103 cells were placed in media (DMEM, 10% FBS, 2mM L-gluatmine, and 1µM 4-OHT) and given 200ng/mL BAFF (R&D systems) for 48 hours and total numbers were analyzed with a FACSAria III. For gene expression analysis, 2×104 cells were placed in media (DMEM, 10% FBS, 2mM L-gluatmine, 50µM beta-mercaptoethanol and 1µM 4-OHT) and given 200ng/mL BAFF (R&D systems) for 48 hours and live (PI−) cells were sorted directly into lysis buffer and processed as stated above. For BCR-stimulation studies, 2×104 cells were placed in media (DMEM, 10% FBS, 2mM L-gluatmine, and 1µM 4-OHT) and given 200ng/mL BAFF (R&D systems) for 48 hours, and then transferred into plates coated overnight at 4°C with biotin-conjugated anti-IgG2ab (5.7) [BD Biosciences] or biotin-conjugated anti-IgG1 (RMG1-1) [Biolegend, Inc], and supplemented with a further 200ng/mL of BAFF. Live (PI−) cells were sorted directly into lysis buffer 48 hours after transfer to coated plates and processed as stated above for gene expression analysis
SiRNA
For siRNA experiments, Peyer’s Patches and mesenteric lymph nodes were homogenized in media (DMEM, 10% FBS, 2mM L-glutamine, 50µM beta-mercaptoethanol) from unimmunized C57BL/6 animals. 3×106 cells were nucleofected using the Amaxa Mouse B cell Nucleofector Kit (Lonza) according to manufacterer’s protocol. Each well was nucleofected with 300pmol of either Silencer Select Pre-designed siRNA against RORα (Ambion) or a scrambled control. Cells were collected whole or sorted to purity 4 days after nucleofection, and mRNA was isolated as above.
SR1001 Treatment
SR1001 was dissolved in 10% DMSO, 10% Tween-80, and 80% water. Whole splenocytes were homogenized and 2×107 cells were treated with 5µM SR1001 for 6 days in media (DMEM, 10% FBS, 2mM L-glutamine, 50µM beta-mercaptoethanol). For IgA memory cultures, Peyer’s Patches and mesenteric lymph nodes were homogenized and IgA memory cells were FACS-purified and placed into culture with media and 5µM SR1001 for 24 hours. Live cells were collected and mRNA was obtained and analyzed as above.
Statistics
Mean values, standard error of the mean, Student’s T-test, and Mann-Whitney test were calculated with GraphPad Prism (GraphPad Software). A P value <0.05 was considered to be statistically significant (*<0.05, **<0.01, ***<0.001).
Supplementary Material
ACKNOWLEDGEMENTS
We thank B. Beutler (The Scripps Research Institute) for providing the Stat1Dom/Dom (Domino) mice, N. Cereb and S. Y. Yang (Histogenetics) and L. Boring (Xenogen Biosciences) for assistance with construction of the conditional Tbx21 allele. N.S.W. is supported by NIH TL1 RR025772-03. S.L.O. received fellowships from the Swiss National Science Foundation, Novartis Jubliaeumsstiftung and Roche Research Foundation. The work was supported by National Institutes of Health (AI042370 and AI076458 to S.L.R; DK080201 and MH092769 to T.P.B. and AI047231, AI040215 and AI071182 to M.G.M-W.). This is TSRI manuscript #21154.
Footnotes
AUTHOR CONTRIBUTIONS
N.S.W., L.J.M-W and M.G.M-W conceived and designed the project. N.S.W. executed and analyzed the data for all experiments. L.J.M-W identified T-bet in memory B cells and contributed to the manuscript preparation. S.L.O. contributed to experimental design, S.L.R. provided the Tbx21F/F mice and T.P.B. provided the SR1001. N.S.W. and M.G.M-W wrote the manuscript.
COMPETING FINANCIAL INTEREST
The authors declare no competing financial interests.
References
- 1.Kurosaki T, Aiba Y, Kometani K, Moriyama S, Takahashi Y. Unique properties of memory B cells of different isotypes. Immunol Rev. 2010;237:104–116. doi: 10.1111/j.1600-065X.2010.00939.x. [DOI] [PubMed] [Google Scholar]
- 2.McHeyzer-Williams MG, Okitsu SL, Wang NS, McHeyzer-Williams LJ. Molecular programming of B cell memory. Nat Rev Immunol. 2012;12:24–34. doi: 10.1038/nri3128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dogan I, et al. Multiple layers of B cell memory with different effector functions. Nat Immunol. 2009;10:1292–1299. doi: 10.1038/ni.1814. [DOI] [PubMed] [Google Scholar]
- 4.Fazilleau N, Mark L, McHeyzer-Williams LJ, McHeyzer-Williams MG. Follicular helper T cells: lineage and location. Immunity. 2009;30:324–335. doi: 10.1016/j.immuni.2009.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Engels N, et al. Recruitment of the cytoplasmic adaptor Grb2 to surface IgG and IgE provides antigen receptor-intrinsic costimulation to class-switched B cells. Nat Immunol. 2009;10:1018–1025. doi: 10.1038/ni.1764. [DOI] [PubMed] [Google Scholar]
- 6.Martin SW, Goodnow CC. Burst-enhancing role of the IgG membrane tail as a molecular determinant of memory. Nat Immunol. 2002;3:182–188. doi: 10.1038/ni752. [DOI] [PubMed] [Google Scholar]
- 7.Snapper CM, Paul WE. Interferon-gamma and B cell stimulatory factor-1 reciprocally regulate Ig isotype production. Science. 1987;236:944–947. doi: 10.1126/science.3107127. [DOI] [PubMed] [Google Scholar]
- 8.Stavnezer J, Guikema JE, Schrader CE. Mechanism and regulation of class switch recombination. Annu Rev Immunol. 2008;26:261–292. doi: 10.1146/annurev.immunol.26.021607.090248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cazac BB, Roes J. TGF-beta receptor controls B cell responsiveness and induction of IgA in vivo. Immunity. 2000;13:443–451. doi: 10.1016/s1074-7613(00)00044-3. [DOI] [PubMed] [Google Scholar]
- 10.Miller SA, Weinmann AS. Common themes emerge in the transcriptional control of T helper and developmental cell fate decisions regulated by the T-box, GATA and ROR families. Immunology. 2009;126:306–315. doi: 10.1111/j.1365-2567.2008.03040.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jetten AM. Retinoid-related orphan receptors (RORs): critical roles in development, immunity, circadian rhythm, and cellular metabolism. Nucl Recept Signal. 2009;7:e003. doi: 10.1621/nrs.07003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lazarevic V, Glimcher LH. T-bet in disease. Nat Immunol. 2011;12:597–606. doi: 10.1038/ni.2059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhu J, Yamane H, Cote-Sierra J, Guo L, Paul WE. GATA-3 promotes Th2 responses through three different mechanisms: induction of Th2 cytokine production, selective growth of Th2 cells and inhibition of Th1 cell-specific factors. Cell Res. 2006;16:3–10. doi: 10.1038/sj.cr.7310002. [DOI] [PubMed] [Google Scholar]
- 14.Mohr E, et al. IFN-{gamma} produced by CD8 T cells induces T-bet-dependent and -independent class switching in B cells in responses to alum-precipitated protein vaccine. Proc Natl Acad Sci U S A. 2010;107:17292–17297. doi: 10.1073/pnas.1004879107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Peng SL, Szabo SJ, Glimcher LH. T-bet regulates IgG class switching and pathogenic autoantibody production. Proc Natl Acad Sci U S A. 2002;99:5545–5550. doi: 10.1073/pnas.082114899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yang XO, et al. T helper 17 lineage differentiation is programmed by orphan nuclear receptors ROR alpha and ROR gamma. Immunity. 2008;28:29–39. doi: 10.1016/j.immuni.2007.11.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Szabo SJ, et al. Distinct effects of T-bet in TH1 lineage commitment and IFN-gamma production in CD4 and CD8 T cells. Science. 2002;295:338–342. doi: 10.1126/science.1065543. [DOI] [PubMed] [Google Scholar]
- 18.Intlekofer AM, et al. Anomalous type 17 response to viral infection by CD8+ T cells lacking T-bet and eomesodermin. Science. 2008;321:408–411. doi: 10.1126/science.1159806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.McHeyzer-Williams LJ, Cool M, McHeyzer-Williams MG. Antigen-specific B cell memory: expression and replenishment of a novel B220-memory b cell compartment. J Exp Med. 2000;191:1149–1166. doi: 10.1084/jem.191.7.1149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pelletier N, et al. Plasma cells negatively regulate the follicular helper T cell program. Nat Immunol. 2010;11:1110–1118. doi: 10.1038/ni.1954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dent AL, Shaffer AL, Yu X, Allman D, Staudt LM. Control of inflammation, cytokine expression, and germinal center formation by BCL-6. Science. 1997;276:589–592. doi: 10.1126/science.276.5312.589. [DOI] [PubMed] [Google Scholar]
- 22.Martins G, Calame K. Regulation and functions of Blimp-1 in T and B lymphocytes. Annu Rev Immunol. 2008;26:133–169. doi: 10.1146/annurev.immunol.26.021607.090241. [DOI] [PubMed] [Google Scholar]
- 23.Beima KM, et al. T-bet binding to newly identified target gene promoters is cell type-independent but results in variable context-dependent functional effects. J Biol Chem. 2006;281:11992–12000. doi: 10.1074/jbc.M513613200. [DOI] [PubMed] [Google Scholar]
- 24.Oestreich KJ, Mohn SE, Weinmann AS. Molecular mechanisms that control the expression and activity of Bcl-6 in TH1 cells to regulate flexibility with a TFH-like gene profile. Nat Immunol. 2012;13:405–411. doi: 10.1038/ni.2242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cimmino L, et al. Blimp-1 attenuates Th1 differentiation by repression of ifng, tbx21, and bcl6 gene expression. J Immunol. 2008;181:2338–2347. doi: 10.4049/jimmunol.181.4.2338. [DOI] [PubMed] [Google Scholar]
- 26.Crozat K, et al. Analysis of the MCMV resistome by ENU mutagenesis. Mamm Genome. 2006;17:398–406. doi: 10.1007/s00335-005-0164-2. [DOI] [PubMed] [Google Scholar]
- 27.Cerutti A, Rescigno M. The biology of intestinal immunoglobulin A responses. Immunity. 2008;28:740–750. doi: 10.1016/j.immuni.2008.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fagarasan S, Kawamoto S, Kanagawa O, Suzuki K. Adaptive immune regulation in the gut: T cell-dependent and T cell-independent IgA synthesis. Annu Rev Immunol. 2010;28:243–273. doi: 10.1146/annurev-immunol-030409-101314. [DOI] [PubMed] [Google Scholar]
- 29.Jaffar Z, Ferrini ME, Herritt LA, Roberts K. Cutting edge: lung mucosal Th17-mediated responses induce polymeric Ig receptor expression by the airway epithelium and elevate secretory IgA levels. J Immunol. 2009;182:4507–4511. doi: 10.4049/jimmunol.0900237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lebman DA, Nomura DY, Coffman RL, Lee FD. Molecular characterization of germ-line immunoglobulin A transcripts produced during transforming growth factor type beta-induced isotype switching. Proc Natl Acad Sci U S A. 1990;87:3962–3966. doi: 10.1073/pnas.87.10.3962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hamilton BA, et al. Disruption of the nuclear hormone receptor RORalpha in staggerer mice. Nature. 1996;379:736–739. doi: 10.1038/379736a0. [DOI] [PubMed] [Google Scholar]
- 32.Solt LA, et al. Suppression of TH17 differentiation and autoimmunity by a synthetic ROR ligand. Nature. 2011;472:491–494. doi: 10.1038/nature10075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cogne M, et al. A class switch control region at the 3' end of the immunoglobulin heavy chain locus. Cell. 1994;77:737–747. doi: 10.1016/0092-8674(94)90057-4. [DOI] [PubMed] [Google Scholar]
- 34.Crotty S. Follicular helper CD4 T cells (TFH) Annu Rev Immunol. 2011;29:621–663. doi: 10.1146/annurev-immunol-031210-101400. [DOI] [PubMed] [Google Scholar]
- 35.Reinhardt RL, Liang HE, Locksley RM. Cytokine-secreting follicular T cells shape the antibody repertoire. Nat Immunol. 2009;10:385–393. doi: 10.1038/ni.1715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Allen CD, Okada T, Tang HL, Cyster JG. Imaging of germinal center selection events during affinity maturation. Science. 2007;315:528–531. doi: 10.1126/science.1136736. [DOI] [PubMed] [Google Scholar]
- 37.Victora GD, et al. Germinal center dynamics revealed by multiphoton microscopy with a photoactivatable fluorescent reporter. Cell. 2010;143:592–605. doi: 10.1016/j.cell.2010.10.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Maruyama M, Lam KP, Rajewsky K. Memory B-cell persistence is independent of persisting immunizing antigen. Nature. 2000;407:636–642. doi: 10.1038/35036600. [DOI] [PubMed] [Google Scholar]
- 39.Lam KP, Kuhn R, Rajewsky K. In vivo ablation of surface immunoglobulin on mature B cells by inducible gene targeting results in rapid cell death. Cell. 1997;90:1073–1083. doi: 10.1016/s0092-8674(00)80373-6. [DOI] [PubMed] [Google Scholar]
- 40.Fazilleau N, et al. Lymphoid reservoirs of antigen-specific memory T helper cells. Nat Immunol. 2007;8:753–761. doi: 10.1038/ni1472. [DOI] [PubMed] [Google Scholar]
- 41.Chang JT, et al. Asymmetric T lymphocyte division in the initiation of adaptive immune responses. Science. 2007;315:1687–1691. doi: 10.1126/science.1139393. [DOI] [PubMed] [Google Scholar]
- 42.Barnett BE, et al. Asymmetric B Cell Division in the Germinal Center Reaction. Science. 2011;335:342–344. doi: 10.1126/science.1213495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nurieva RI, et al. Bcl6 mediates the development of T follicular helper cells. Science. 2009;325:1001–1005. doi: 10.1126/science.1176676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mondal A, Sawant D, Dent AL. Transcriptional repressor BCL6 controls Th17 responses by controlling gene expression in both T cells and macrophages. J Immunol. 2010;184:4123–4132. doi: 10.4049/jimmunol.0901242. [DOI] [PubMed] [Google Scholar]
- 45.Yang XO, et al. Molecular antagonism and plasticity of regulatory and inflammatory T cell programs. Immunity. 2008;29:44–56. doi: 10.1016/j.immuni.2008.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Dzhagalov I, Giguere V, He YW. Lymphocyte development and function in the absence of retinoic acid-related orphan receptor alpha. J Immunol. 2004;173:2952–2959. doi: 10.4049/jimmunol.173.5.2952. [DOI] [PubMed] [Google Scholar]
- 47.Aiba Y, et al. Preferential localization of IgG memory B cells adjacent to contracted germinal centers. Proc Natl Acad Sci U S A. 2010;107:12192–12197. doi: 10.1073/pnas.1005443107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pape KA, Taylor JJ, Maul RW, Gearhart PJ, Jenkins MK. Different B cell populations mediate early and late memory during an endogenous immune response. Science. 2011;331:1203–1207. doi: 10.1126/science.1201730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gold DA, et al. RORalpha coordinates reciprocal signaling in cerebellar development through sonic hedgehog and calcium-dependent pathways. Neuron. 2003;40:1119–1131. doi: 10.1016/s0896-6273(03)00769-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kane CD, Means AR. Activation of orphan receptor-mediated transcription by Ca(2+)/calmodulin-dependent protein kinase IV. EMBO J. 2000;19:691–701. doi: 10.1093/emboj/19.4.691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Fazilleau N, et al. Nat Immunol. 2009;10:375–384. doi: 10.1038/ni.1704. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.