Abstract
Oxaliplatin, satraplatin, and picoplatin are cisplatin analogs that interact with DNA forming intrastrand and interstrand DNA cross-links (ICLs). Replicative bypass of cisplatin DNA adducts requires the cooperative actions of at least three translesion DNA synthesis (TLS) polymerases: Polη, REV1, and Polζ. Because oxaliplatin, satraplatin, and picoplatin contain bulkier chemical groups attached to the platinum core compared with cisplatin, we hypothesized that these chemical additions may impede replicative bypass by TLS polymerases and reduce tolerance to platinum-containing adducts. We examined multiple responses of cancer cells to oxaliplatin, satraplatin, or picoplatin treatment under conditions where expression of a TLS polymerase was limited. Our studies revealed that, although Polη contributes to the tolerance of cisplatin adducts, it plays a lesser role in promoting replication through oxaliplatin, satraplatin, and picoplatin adducts. REV1 and Polζ were necessary for tolerance to all four platinum analogs and prevention of hyperactivation of the DNA damage response after treatment. In addition, REV1 and Polζ were important for the resolution of DNA double-stranded breaks created during replication-associated repair of platinum-containing ICLs. Consistent with ICLs being the predominant cytotoxic lesion, depletion of REV1 or Polζ rendered two different model cell systems extremely sensitive to all four drugs, whereas Polη depletion had little effect. Together, our data suggest that REV1 and Polζ are critical for promoting resistance to all four clinically relevant platinum-based drugs by promoting both translesion DNA synthesis and DNA repair.
Introduction
Cisplatin is widely used for the treatment of a broad range of malignant diseases, including testicular, ovarian, lung, and bladder cancers (Kelland, 2007). The antitumor effect of cisplatin is through its ability to covalently interact with guanine residues in DNA resulting in the formation of both intra- and interstrand DNA cross-links (ICLs). Although cisplatin is an effective anticancer drug in several tumor types, its usefulness can become limited due to severe dose-limiting side effects and acquired resistance (Rabik and Dolan, 2007; Köberle et al., 2010). Since the introduction of cisplatin, additional analogs have been developed with the goal of reducing toxicity, broadening the spectrum of activity, and circumventing acquired resistance (Kelland, 2007). Modifications to the leaving groups or the two amine ligands of cisplatin resulted in the development of carboplatin and oxaliplatin, respectively, both currently approved for use in the United States. Carboplatin was developed to lower the toxicity profile of cisplatin by replacing the dichloride-leaving groups with 1,1-cyclobutanedicarboxylate. This resulted in a cisplatin-like compound that generates the same DNA adducts but is more stable and undergoes aquation at a slower rate. Oxaliplatin was designed with a 1,2-diaminocyclohexane group in place of the two amine ligands based on the prediction that a bulkier platinum-DNA adduct would interfere with DNA repair and overcome cisplatin resistance.
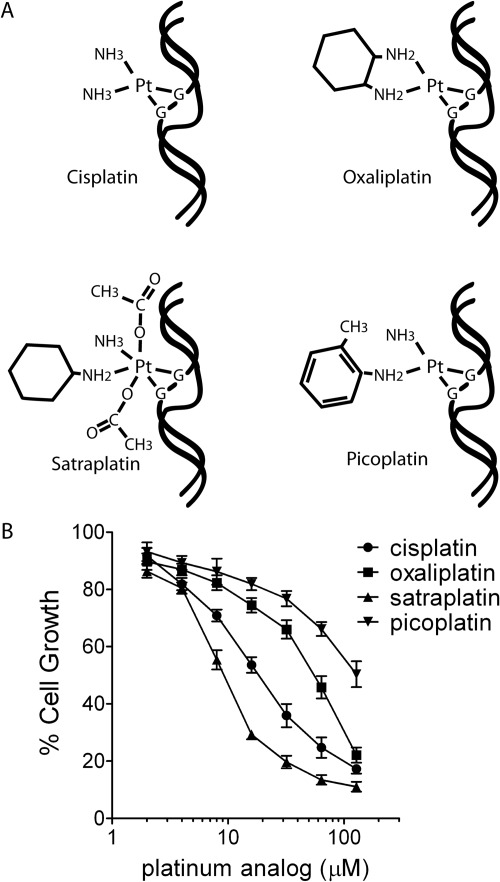
Continuous efforts to improve efficacy and bioavailability of platinum-based chemotherapeutic agents resulted in the development of two newer analogs that are currently being evaluated in clinical trials. Satraplatin (JM216) was developed to circumvent acquired resistance by replacing one of the amine ligands of cisplatin with a bulkier cyclohexylamine group. Picoplatin (AMD473) contains a pyrimidine ring substituted for one of the amine ligands of cisplatin to prevent the platinum center from being inactivated by glutathione. Even though all of these analogs interact with DNA in a similar manner (Fig. 1A), it is becoming increasingly clear that the different adducts produced by these compounds are associated with distinct spectrums of activity and, in some cases, an inability of specific proteins to recognize the lesion, thus leading to altered cellular responses (Nehmé et al., 1999; Raymond et al., 2002; Chaney et al., 2005; Lewis et al., 2009).
Fig. 1.
A, the different adducts created by cisplatin, oxaliplatin, satraplatin, and picoplatin are illustrated. B, dose-response relationships between platinum analogs and growth inhibition. HeLa cells were treated with different doses of platinating agent for 1 h, washed, and then cultured for 4 days. The number of cells present was counted and normalized to the number of cells present when grown in the absence of drug. Results shown are the mean ± S.E.M. of at least four independent experiments.
The overall cellular response to platinum-DNA intrastrand cross-links and ICLs involves multiple processes that ultimately determine cell fate. We and others have identified TLS as an important pathway influencing cisplatin-induced cytotoxicity (Simpson and Sale, 2003; Sonoda et al., 2003; Bassett et al., 2004; Niedzwiedz et al., 2004; Wu et al., 2004; Albertella et al., 2005; Nojima et al., 2005; Okuda et al., 2005; Chen et al., 2006; Doles et al., 2010; Hicks et al., 2010). The TLS pathway promotes tolerance to various DNA lesions that block replicative polymerases and is triggered by the monoubiquitination of proliferating cell nuclear antigen (PCNA) by the RAD6 (E2)·RAD18 (E3) complex (Waters et al., 2009). Monoubiquitination of PCNA on Lys164 recruits TLS polymerases through a combination of protein-protein interacting motifs that include PCNA-interacting peptide and ubiquitin-binding domains. TLS polymerases possess accommodating active sites and are capable of replicating DNA containing bulky DNA lesions, even when the DNA template is distorted by adducts such as those created by cisplatin (Alt et al., 2007; Waters et al., 2009; Washington et al., 2010; Bhattacharyya et al., 2011).
Multiple TLS polymerases are implicated in the lesion bypass of DNA intrastrand cross-links, including those generated by cisplatin. Both DNA polymerase η (Polη) and DNA polymerase ζ (Polζ, composed of catalytic subunit REV3 and accessory subunit REV7) are believed to cooperate together when synthesizing DNA opposite cisplatin adducts, and this activity requires the TLS polymerase REV1 (Shachar et al., 2009; Hicks et al., 2010). Current models suggest that REV1 facilitates polymerase switching during TLS through its ability to bind multiple TLS polymerases (Guo et al., 2003; Ohashi et al., 2004; Tissier et al., 2004). It is currently unclear whether Polη and/or REV1/Polζ-dependent TLS mediate resistance to the newer platinum-based drugs, which based on their structure could create larger obstructions to DNA replication, including TLS. Here we demonstrate that cancer cells lacking REV1 or REV3 are highly sensitive to these agents, show markedly reduced survival, and fail to resolve replication-associated DNA double-stranded breaks (DSBs) after treatment with cisplatin, oxaliplatin, satraplatin, or picoplatin. In contrast, we found that Polη seemed to play a relatively small role in promoting resistance to these drugs. Together our findings are consistent with REV1 and Polζ being key factors in promoting resistance to platinum-based chemotherapy and, hence, support the rationale of TLS inhibition as an adjuvant therapy for treating malignancies that develop chemoresistance.
Materials and Methods
Reagents.
Cisplatin [cis-diamminedichloroplatinum(II)], oxaliplatin [1R,2R-diaminocyclohexane oxalatoplatinum(II)], and picoplatin [cis-amminedichloro, 2-methylpyridine, platinum(II)] were purchased from LC Laboratories (Woburn, MA). Satraplatin [bis-acetato-ammine-dichloro-cyclohexylamine platinum(IV)] was purchased from Sequoia Research Products (Pangbourne, UK). Rabbit polyclonal anti-Polη (H-300), anti-53BP1 (H-300), anti-GAPDH (FL-335), and anti-Rad51 (H92) were purchased from Santa Cruz Biotechnology (Santa Cruz, CA). Rabbit polyclonal anti-phospho-replication factor A (RPA32; S4/S8), anti-phospho-Ser345 CHK1, anti-phospho-Ser139 histone H2AX, and anti-RAD18 were purchased from Bethyl Laboratories (Montgomery, TX), Cell Signaling Technology (Danvers, MA), Active Motif, Inc. (Carlsbad, CA), and Proteintech Group (Chicago, IL), respectively. The following mouse monoclonal antibodies were used: anti-phospho-Ser1981 ATM (Rockland Immunochemicals, Gilbertsville, PA), anti-GFP (Roche Applied Science, Indianapolis, IN), anti-Chk1 (G-4; Santa Cruz Biotechnology), anti-RPA32 (Ab-3; Calbiochem/EMD Chemicals, San Diego, CA), and anti-topoisomerase 1 (BD Biosciences, Franklin Lakes, NJ).
Cell Lines, siRNA, and Culture Conditions.
HeLa cells were obtained from the American Tissue Culture Collection (Manassas, VA) and cultured in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum. The BL2 human Burkitt's lymphoma cell lines were cultured in RPMI 1640 medium supplemented with 10% fetal bovine serum (Gueranger et al., 2008). All siRNA duplexes were purchased from QIAGEN (Valencia, CA) and transfected into HeLa cells using X-tremeGENE Transfection Reagent (Roche Applied Science) as described previously (Hicks et al., 2010). The gene sequences used for designing siRNA used in this study were as follows: siControl (5′-AATTCTCCGAACGTGTCACGT-3′), siREV1 (5′-ATCGGTGGAATCGGTTTGGAA-3′), siREV3 (5′-CCCACTGGAATTAATGCACAA-3′), Polη (5′-CAGCCAAATGCCCATTCGCAA-3′), siRAD18 (5′-ATGGTTGTTGCCCGAGGTTAA-3′), and siRAD51 (5′-AAGCTGAAGCTATGTTCGCCA-3′).
Measurement of Cell Growth and Loss in Viability.
To measure the extent of growth inhibition caused by treatment with platinating agents, HeLa cells were seeded 10,000 cells/well in 12-well plates and exposed to drug for 1 h. Four days later, cells were harvested and counted using an Accuri C6 flow cytometer (BD Accuri Cytometers, Ann Arbor, MI). For survival assays, HeLa cells were transfected overnight with each siRNA as described previously (Hicks et al., 2010). The next day, cells were seeded at known densities (500–2000 cells/well) in 12-well plates, allowed to attach to culture plates overnight, and then cultured with two different doses of cisplatin, oxaliplatin, satraplatin, or picoplatin until the untreated well approached confluence (7–10 days). Cellular survival was determined using a crystal violet assay, as described previously (Taniguchi et al., 2002). BL2 lymphoma cells were treated with the different platinum analogs continuously for 48 h. The cells were then washed with phosphate-buffered saline, and viability was determined by measuring the percentage of BL2 cells excluding propidium iodide using an Accuri C6 flow cytometer.
Immunofluorescence.
For γ-H2AX immunofluorescence, HeLa cells grown on coverslips in 12-well plates were transfected overnight with the different siRNAs and allowed to recover another 24 h. The transfected cells were treated for 1 h with the drug concentration that inhibited cell growth by 35 to 40% (as measured using the growth inhibition assay; 10 μM cisplatin, 32 μM oxaliplatin, 6 μM satraplatin, and 64 μM picoplatin). Cells were fixed with 100% methanol 24 h later and stained for γ-H2AX as described previously (Hicks et al., 2010). To analyze the presence of DSBs by immunofluorescence, HeLa cells were transfected with siRNA as above and then treated with 5 μM cisplatin, 15 μM oxaliplatin, 2 μM satraplatin, or 25 μM picoplatin for 1 h. Cells were fixed with 100% methanol 24 and 48 h after drug treatment and stained for S1981P-ATM and 53BP1 as described previously (Hicks et al., 2010). The doses chosen were based on experiments included in Supplemental data and Figs. 2 through 5.
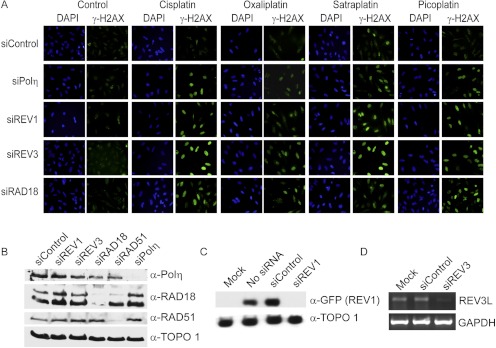
Fig. 2.
The TLS pathway limits the DNA damage response after treatment with platinating agents. A, HeLa cells were transfected overnight with the indicated siRNAs and allowed to recover for 24 h. Cells were treated with platinum-containing drug for 1 h using concentrations expected to inhibit cell proliferation by 35 to 40% (10 μM cisplatin, 32 μM oxaliplatin, 6 μM satraplatin, and 64 μM picoplatin). Cells were fixed 24 h after drug treatment and stained for γ-H2AX to measure activation of the DNA damage response. B, HeLa cells were transfected with the indicated siRNAs and harvested 2 days later to assess knockdown efficiencies. Whole-cell lysates were subjected to SDS-PAGE and immunoblotted for endogenous RAD18, RAD51, Polη, or topoisomerase 1 (loading control). Rad18 levels were reduced to 14.1 ± 10.7%; RAD51 levels were reduced to 36.8 ± 7.5%; and Polη levels were reduced to 18.0 ± 3.0% of control levels as determined by densitometry analysis (n = 3, mean ± S.D.). C, 293T cells were cotransfected with GFP-tagged REV1 and the indicated siRNAs. Two days later, whole-cell lysates were subjected to SDS-PAGE and immunoblotted for GFP or topoisomerase 1 (α-TOPO 1). REV1-specific siRNA reduced GFP-REV1 protein levels to 22 ± 7% of control (n = 3, mean ± S.D.). D, HeLa cells were transfected with control or REV3-specific siRNA. Two days later, REV3L and GAPDH mRNA levels were determined by reverse transcriptase-polymerase chain reaction using gene-specific primers. Shown is a representative ethidium bromide-stained agarose gel. REV3L mRNA levels were reduced by 35 ± 1.3% compared with control as determined by densitometry analysis (n = 3, mean ± S.D.). DAPI, 4,6-diamidino-2-phenylindole.
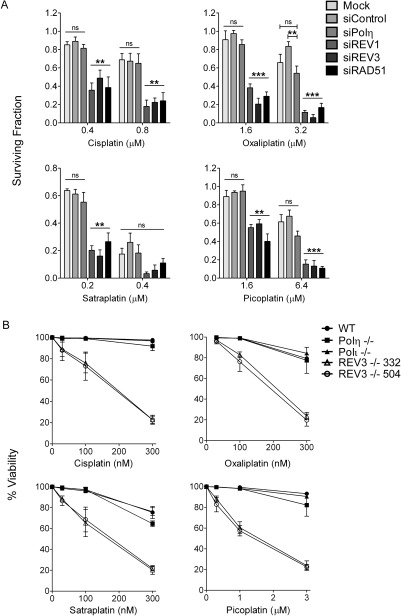
Fig. 5.
Cancer cells lacking REV1 or REV3 are hypersensitive to cisplatin, oxaliplatin, satraplatin, and picoplatin. A, HeLa cells were mock transfected or transfected with control siRNA or siRNA targeting REV1, REV3, or Polη. Cells were then seeded at known densities cultured with or without two different doses of cisplatin, oxaliplatin, satraplatin, or picoplatin until the untreated wells approached confluence (7–10 days). The relative surviving fraction was determined by measuring absorbance of solubilized crystal violet staining in each well normalized to the corresponding untreated well. Cells transfected with siRNA specific for RAD51 and treated with individual drugs are shown for comparison. Data represent the mean ± S.E.M. from at least three independent experiments. Open lines indicate comparisons of the means among all columns. Bracketed lines indicate comparisons between two means: ns, not significant; **, P < 0.01; ***, P < 0.001; one-way analysis of variance. B, wild type, Polη−/−, Polι−/−, and REV3−/− (clone 332 or 504) were cultured in the presence of three different doses of cisplatin, oxaliplatin, satraplatin, or picoplatin for 48 h. Viability was determined by measuring the percentage of cells excluding propidium iodide by flow cytometry. Data represent the mean ± S.E.M. of at least three independent experiments.
Flow Cytometry.
HeLa cells were transfected overnight with individual siRNAs and allowed 24 h to recover. Cells were then treated with 10 μM cisplatin, 32 μM oxaliplatin, 6 μM satraplatin, and 64 μM picoplatin for 1 h and washed, harvested 24 h later, and fixed in 70% methanol. For two parameter flow cytometry, cells were blocked with 5% bovine serum albumin plus 1% goat serum and 0.05% Tween 20, and then stained with γ-H2AX monoclonal antibody. Cells were washed, incubated with the goat anti-mouse fluorescein isothiocyanate-conjugated secondary antibody (Sigma-Aldrich, St. Louis, MO), and then counterstained with propidium iodide in phosphate-buffered saline containing RNase A. Cells were acquired on an Accuri C6 flow cytometer.
Immunoblotting.
Cells were lysed in SDS sample buffer: 10 mM Tris, pH 8.0, 2% SDS, 1× protease inhibitor cocktail (Roche Applied Science), and 1× phosphatase inhibitor cocktails 1 and 2 (Sigma-Aldrich). Lysates were sonicated and then denatured by heating to 95°C for 5 min. Equal amounts of protein were separated on SDS-PAGE gels, transferred onto nitrocellulose membranes, and then probed with the appropriate primary antibodies followed by secondary horseradish peroxidase-conjugated goat anti-rabbit or mouse antibodies (Thermo Fisher Scientific, Waltham, MA). Proteins were visualized using SuperSignal West Pico Chemiluminescent Substrate (Thermo Fisher Scientific).
Statistics.
Statistically significant differences were determined using one-way analysis of variance with the Tukey post-comparison test using GraphPad Prism version 5 (GraphPad Software, Inc., San Diego, CA).
Results
RAD18, Polη, REV1, and REV3 Promote Different Degrees of Tolerance to Cisplatin Analogs.
We have previously determined that exposing HeLa cells to 10 μM cisplatin for 1 h inhibits cell proliferation by approximately 35 to 40%. After this treatment, we were able to measure significant differences in survival and activation of the DNA damage response pathway in cells depleted of various components of the TLS pathway (RAD18, Polη, REV1, or REV3) (Hicks et al., 2010). Because platinum analogs can form DNA adducts with different efficiencies after entering a cell and could pose different degrees of replication blockade, we first compared the ability of HeLa cells to proliferate after treating cells with oxaliplatin, satraplatin, and picoplatin for 1 h to identify comparable doses that would lead to 35 to 40% inhibition of cell growth (Fig. 1B). On the basis of this analysis, we chose to treat siRNA-transfected HeLa cells for 1 h with 32 μM oxaliplatin, 6 μM satraplatin, or 64 μM picoplatin and compared responses of cells to 10 μM cisplatin using various endpoints that indirectly measure the stalling of replication forks. We presumed that, under these conditions, we were comparing responses of cells to equivalent levels of replication blockade generated by each analog.
Activation of the ATR kinase is a well characterized response to replication blockade (Cimprich and Cortez, 2008). Therefore, we measured the phosphorylation of ATR substrates (e.g., H2AX and Chk1) as surrogate markers for replication stress as a result of deficient TLS (Cruet-Hennequart et al., 2008; Hicks et al., 2010). We previously determined that HeLa cells treated with cisplatin exhibit intense hyperphosphorylation of H2AX on serine 139 (γ-H2AX) when cells are depleted of REV1, REV3, RAD18, or Polη (Hicks et al., 2010). This exaggerated response indicates that each protein is necessary for efficient bypass of replication-blocking cisplatin adducts and the avoidance of ATR activation in response to replication fork stalling. We tested whether RAD18, REV1, REV3, or Polη deficiency (validated in Fig. 2, B–D) leads to replication stalling and activation of ATR after treatment with the different platinum analogs. As expected, HeLa cells deficient in REV1, REV3, RAD18, or Polη expression exhibited robust γ-H2AX staining in comparison with control siRNA-transfected cells after exposure to cisplatin (Fig. 2A). In contrast, oxaliplatin did not seem to induce the same extent of H2AX phosphorylation in TLS-deficient cells, the exception being siREV3-transfected cells. This experiment also revealed that depletion of Polη was not associated with an exaggerated γ-H2AX response after treatment with oxaliplatin, picoplatin, or satraplatin. We extended these observations by comparing both time-dependent and dose-dependent induction of H2AX phosphorylation in control- and REV3-specific siRNA-transfected cells (Supplementary Fig. S1). The results show that Polζ deficiency resulted in prolonged and extensive H2AX phosphorylation after short-term treatment with each cisplatin analog.
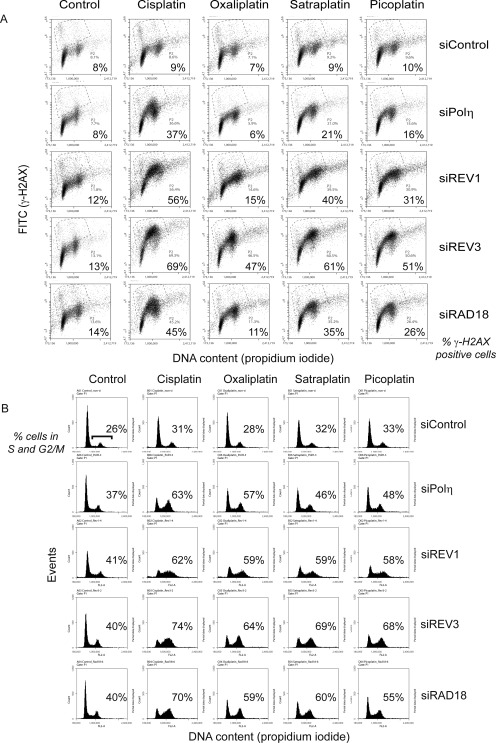
To better differentiate intensities of γ-H2AX staining observed by immunofluorescence among the different treatment groups in Fig. 2, we analyzed cells treated under identical conditions and stained for γ-H2AX and DNA content by flow cytometry (Fig. 3, A and B). Differences in both the level of γ-H2AX staining and changes in cell cycle distribution after treatment with the different platinum analogs were revealed more clearly. It is noteworthy that only cisplatin seems to induce an exaggerated γ-H2AX response in HeLa cells when Polη is depleted (Fig. 3A). When these cells are treated with oxaliplatin, satraplatin, or picoplatin, there are relatively small increases in γ-H2AX positive cells (Fig. 3, A and B). In contrast, the greatest altered cell-cycle profiles and intensities of γ-H2AX staining were observed in cells depleted of REV3, the catalytic subunit of Polζ, regardless of the platinum analog used for treatment (Fig. 3, A and B). Consistent with REV1 and REV3 cooperating to perform lesion bypass, cells depleted of REV1 exhibited similar cell-cycle profiles and cell-cycle patterns of γ-H2AX staining (mainly cells residing in S and G2/M) as REV3-depleted cells, the exception being oxaliplatin. REV3-depleted cells showed the greatest shift in γ-H2AX staining after treatment with each of the four platinum analogs.
Fig. 3.
REV1 and Polζ are essential for tolerance of platinum adducts. HeLa cells were transfected with siRNA and treated with the indicated platinum-containing drugs for 1 h as in Fig. 2A. Cells were fixed 24 h later, fixed, stained for γ-H2AX and DNA content (propidium iodide), and then analyzed by flow cytometry (A and B). Dot plots depicting the level of γ-H2AX staining versus DNA content are shown in A, and the corresponding histograms depicting the DNA content per event are shown in B. Each dot plot is labeled with the percentage of cells displaying enhanced γ-H2AX staining. Each histogram is labeled with the percentage of cells residing in S and G2/M at the time of fixation. Cells can progress through the cell cycle and avoid replication stalling more efficiently in the absence of Polη compared with REV1 and Polζ after treatment with oxaliplatin, satraplatin, or picoplatin. Shown are representative dot plots and histograms from three independent experiments. FITC, fluorescein isothiocyanate.
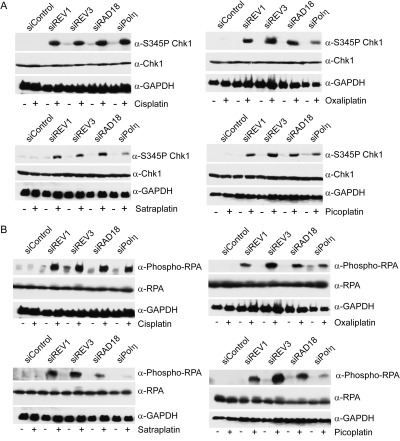
We and others have previously found that depleting cells of TLS polymerases (Polη or Polζ), as well as abrogating the monoubiquitination of PCNA by RAD18, leads to enhanced ATR-dependent phosphorylation of the Chk1 protein kinase after UV irradiation or cisplatin treatment (Bomgarden et al., 2006; Cruet-Hennequart et al., 2008; Hicks et al., 2010). Therefore, we compared the response of TLS-deficient HeLa cells after treatment with oxaliplatin, picoplatin, and satraplatin as shown in Figs. 2 and 3. Consistent with the degree of γ-H2AX formation and cell-cycle arrests in S and G2/M after cisplatin treatment, depletion of REV1, REV3, RAD18, or Polη resulted in enhanced phosphorylation of Chk1 on Ser345 by the ATR kinase (Fig. 4A and Supplemental Fig. S2). However, the extent of Chk1 phosphorylation-observed Polη-depleted cells was notably less after treatment with oxaliplatin, picoplatin, or satraplatin compared with REV3-depleted cells. Similar results were observed when phosphorylation of RPA32 on serines 4 and 8, another marker for replication stress, was analyzed. (Fig. 4B and Supplemental Fig. S2). Again, cells depleted of Polη displayed significantly less phosphorylated RPA compared with REV3-depleted cells after drug treatment, especially in response to satraplatin. It is also apparent from these analyses that RAD18 is important for most lesion bypass events induced by oxaliplatin, picoplatin, or satraplatin since cells depleted of RAD18 exhibited enhanced phosphorylation of Chk1 and RPA.
Fig. 4.
Platinating agents induce enhanced Chk1 and RPA phosphorylation in the absence of REV1 and Polζ. HeLa cells transfected with the indicated siRNAs were treated with 10 μM cisplatin, 32 μM oxaliplatin, 6 μM satraplatin, and 64 μM picoplatin for 1 h and harvested 24 h later. Whole-cell lysates were immunoblotted for phospho-Ser345-Chk1 or total Chk1 protein (A) or phospho-RPA32 (S4/S8) or total RPA protein (B). Immunoblots showing GAPDH immunostaining demonstrate equal loading. Immunoblots are representative of two independent experiments.
REV1 and Polζ Promote Resistance to Multiple Platinating Agents.
We next examined the roles of REV1, Polζ, and Polη in preventing drug-induced cytotoxicity using two different model systems. REV1, REV3, or Polη-depleted HeLa cells were cultured in the presence of two different concentrations of cisplatin, oxaliplatin, satraplatin, or picoplatin for approximately 8 days, and the relative surviving fraction was calculated based on crystal violet staining (Taniguchi et al., 2002). We observed that REV1 and REV3 siRNA-transfected HeLa cells exhibited the greatest sensitivities to cisplatin-, satraplatin-, oxaliplatin-, and picoplatin-induced loss in cell survival, particularly at the lower drug concentrations (Fig. 5A). Polη depletion did not seem to sensitize HeLa cells to either platinating agent, at least under the conditions used here. Consistent with the homologous recombination pathway playing an essential role in repairing interstrand DNA cross-links (ICLs), depletion of RAD51 caused HeLa cells to be hypersensitive to all four platinating agents, thus validating our experimental approach (Deans and West, 2011). To confirm the predominant role of REV3 in protecting cells from platinum adduct-induced cytotoxicity, we examined the sensitivity of human BL2 lymphoma cells and several different BL2 lines devoid of the REV3, Polη, or Polι genes to each platinum analog (Gueranger et al., 2008). Consistent with the results observed in REV3-depleted HeLa cells, two different BL2 clones lacking REV3 were significantly more sensitive to each drug (Fig. 5B). These results confirm that Polη and Polι (a member of the Y-family TLS polymerases) play relatively minor roles in promoting resistance to platinum adducts compared with Polζ.
REV1 and REV3 Depletion Leads to Defective Interstrand Cross-Link Repair.
The extreme sensitivity of REV3-deficient cells to cisplatin has been linked to defective repair of ICLs (Niedzwiedz et al., 2004; Räschle et al., 2008; Hicks et al., 2010). DSBs can be detected after cells are exposed to ICL-inducing agents such as mitomycin C or cisplatin and represent an intermediate step during ICL repair after a replication fork encounters the lesion (Rothfuss and Grompe, 2004; Hanada et al., 2006; Hicks et al., 2010; Kratz et al., 2010; Liu et al., 2010; MacKay et al., 2010). Cleavage on either side of the ICL by the coordinated actions of 5′- and the 3′-flap endonucleases is thought to generate replication-associated DSBs that are subsequently resolved by homologous recombination repair coordinated by the Fanconi anemia complex of proteins (Deans and West, 2011). REV1 and REV3 are believed to promote ICL repair by inserting nucleotides opposite the unhooked ICL, a step that is essential for preparing the damaged DNA template for homologous recombination repair. The appearance and resolution of ICL-induced DSBs can be followed using immunofluorescence microscopy after staining cells with antibodies that specifically detects proteins that associate with chromatin surrounding DSBs forming visible foci. Localization of activated ATM protein kinase (specifically detected as the S1981P-modified form) and 53BP1 to DSBs are both well characterized surrogate markers of DSBs (Schultz et al., 2000; Bakkenist and Kastan, 2003). Therefore, we examined cisplatin analog-induced colocalization of these two proteins as a sensitive means to follow the appearance and resolution of DSBs created during ICL repair.
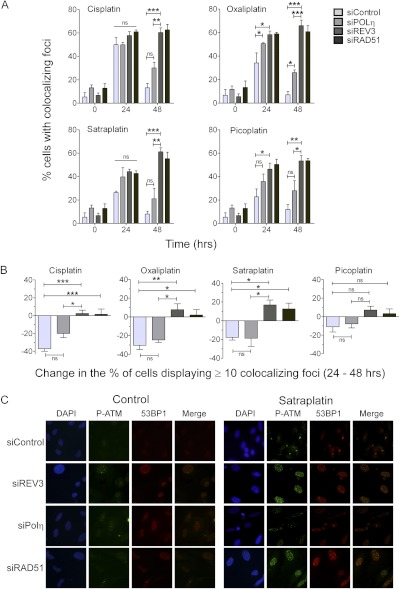
HeLa cells were transfected with individual siRNAs, allowed to recover, and then treated with relatively low doses of a drug: 5 μM cisplatin, 15 μM oxaliplatin, 2 μM satraplatin, or 25 μM picoplatin for 1 h. Cells were fixed 24 or 48 h after drug exposure and stained for activated ATM and 53BP1 to follow the induction and resolution of replication-associated DSBs. Lower doses were chosen for this study based on a preliminary experiment assessing the appearance and resolution of DSBs after treatment of control or REV3 knockdown cells exposed to three different doses of platinating agent (Supplemental Figs. S3–S5). After drug treatment, we typically observed the accumulation of DSBs by 24 h, such that approximately 30 to 50% of cells displayed 10 or more colocalized foci marked by S1981P-ATM and 53BP1 (Fig. 6A). The percentage of cells displaying 10 or more foci was notably higher in Polη-, REV3-, or RAD51-deficient cells. By 48 h, the majority of siControl-transfected cells displayed less than 10 S1981-ATM- and 53BP1-colocalized foci, indicating that most DSBs have been resolved. In contrast, Rad51-depleted HeLa cells, which are defective in homologous recombination repair (Sharma et al., 2012), failed to resolve DSBs within the 48-h time period as expected, thus validating our experimental approach. Similar to control cells, Polη-depleted HeLa cells resolved the majority of DSBs within a 48-h period after treatment with cisplatin, satraplatin, or picoplatin. However, we observed significant differences in the percentage of cells displaying DSBs induced by oxaliplatin in Polη-depleted cells (Fig. 6A).
Fig. 6.
RAD51 and Polζ are necessary for resolving platinating agent-induced DSBs. REV3- and Polη-depleted HeLa cells were treated with cisplatin (5 μM), oxaliplatin (15 μM), satraplatin (2 μM), and picoplatin (25 μM) for 1 h; washed; and then fixed 24 and 48 h later. Cells were stained for S1981P-ATM and53BP1 as surrogate markers of DNA DSBs. Nuclear DNA was stained with 4,6-diamidino-2-phenylindole (DAPI). A, the percentage of cells exhibiting 10 or more colocalized foci containing both S1981P-ATM and 53BP1 was determined. Data represent the mean ± S.E.M. from four independent experiments where >100 cells were counted in each experiment. B, the difference in percentage of cells displaying ≥10 colocalized foci containing both S1981P-ATM and 53BP1 between 24 and 48 h is shown. C, representative images of control-, REV3- and Polη-depleted cells treated with satraplatin are shown. The presence or absence of Polη had little impact on the efficiency of ICL-induced DSB resolution.
The percentage of drug treated REV3-depleted HeLa cells displaying S1981P-ATM- and 53BP1-colocalized foci did not decrease at the 48-h time point, similar to the RAD51-depleted cells (Figs. 6A). Greater than 50% of REV3-depleted cells failed to resolve foci marking DSBs induced by all four drugs within the 48-h period, indicating that DSB repair in these cells was affected to a large degree (see also Supplemental Figs. S3–S5). Given that the percentage of drug-treated cells exhibiting DSBs varied at the 24-h time point, we analyzed the differences in percentage of cells displaying ≥10 colocalized foci between 24 and 48 h to examine the impact of DNA polymerase deficiency on DSB resolution during this time period (Fig. 6B). This analysis clearly shows that Polη-deficient cells were capable of resolving DSBs, whereas the percentage of Polζ-depleted cells displaying ≥10 foci increased rather than decreased.
Discussion
In the present study, we examined the roles of REV1, Polζ, and Polη in protecting cells from the antiproliferative effects of cisplatin in comparison with three cisplatin analogs that produce bulkier adducts on DNA. Previous studies have focused on Polη and its ability to replicate cisplatin and oxaliplatin adducts in template DNA and promote resistance to these agents (Vaisman et al., 2000; Bassett et al., 2004; Alt et al., 2007; Cruet-Hennequart et al., 2008). Our data agree with the concept that Polη is necessary for replicative bypass through cisplatin adducts. However, our data also indicate that Polη does not seem to play a large role in the bypass of oxaliplatin, satraplatin, and picoplatin adducts, at least in comparison with REV1 and Polζ in the context of two different cancer cell lines. It is important to note here that DNA polymerase κ performs error-free bypass of benzo[a]pyrene adducts and error-prone bypass of cisplatin adducts in the absence of Polη (Bi et al., 2006; Shachar et al., 2009; Ziv et al., 2009). It will be important to test whether Polκ can fulfill the role of Polη in bypassing bulkier platinum-containing adducts during DNA replication with respect to drug-induced mutagenesis and the promotion of cell survival. Regardless, both lesion bypass events initiated by Polη or Polκ require Polζ as the universal extender beyond nucleotides inserted opposite DNA adducts during TLS, probably in collaboration with REV1. Overall, our data point to REV1 and Polζ as being essential for lesion bypass and tolerance in many of the platinating agents that are used clinically today.
The importance of REV1 and Polζ in facilitating replicative bypass of platinum DNA adducts has important clinical implications. It is well accepted that most mutations induced by DNA-damaging agents result from error-prone TLS that is attributed to the activities of REV1 and Polζ (Waters et al., 2009). The emergence of drug resistance to cisplatin and cyclophosphamide has recently been linked to the activities of REV3 and REV1 in murine models of B-cell lymphoma and lung adenocarcinoma (Doles et al., 2010; Xie et al., 2010). Rendering tumor cells REV1 or REV3 deficient using short hairpin RNA significantly sensitized these tumors to treatment and limited the emergence of drug resistance. Taken together, these data suggest that inhibition of REV1 or Polζ may have dual anti-cancer effects—sensitizing tumors to therapy and preventing the emergence of chemoresistance by limiting drug-induced mutagenesis. Our findings suggest that this targeted approach would be applicable to oxaliplatin, satraplatin, and picoplatin, in addition to cisplatin.
The chemosensitizing effects of targeting REV1 or Polζ are probably attributed to interference with TLS across intrastrand cross-links and TLS associated with ICL repair. Disruption of REV1 or Polζ is associated with far greater sensitivities to cisplatin and other ICL-inducing agents compared with disrupting other TLS polymerases, and these observations are consistent with both genetic and biochemical evidence, implicating REV1 and Polζ in promoting ICL repair (Niedzwiedz et al., 2004; Nojima et al., 2005; Räschle et al., 2008; Hicks et al., 2010). Repair of ICLs requires a complex interplay between the TLS, Fanconi anemia, and the homologous recombination pathways (Deans and West, 2011). The data presented here support the concept that targeting REV1 or Polζ would also interfere with repair of the structurally diverse adducts created by all four platinating agents when present as an ICL as demonstrated by inefficient resolution of ICL-induced DSBs, in addition to limiting DNA replication through these distorting lesions on a single strand of DNA. It is important to note that following the appearance and resolution of ICL-induced DSBs is an indirect measure of actual ICL repair. The phenotypes we observed in Polζ-depleted cells are very similar to cells defective in homologous recombination repair (e.g., RAD51 knockdown cells) and cells deficient in ICL repair because of defects in the Fanconi anemia pathway, suggesting that ICL repair is affected to a significant degree (Rothfuss and Grompe, 2004; Kratz et al., 2010; Liu et al., 2010; MacKay et al., 2010).
It is noteworthy that recent biochemical studies have characterized Polζ as being inefficient at mediating TLS across artificial “unhooked” cross-links (Ho et al., 2011). This is unexpected based on the model for ICL repair and because the majority of genetic evidence suggests a prominent role for REV3 in protecting from ICL-induced cytotoxicity and genomic instability. Although REV1 and Polζ are believed to be essential for replicating across unhooked ICL in preparation for homologous recombination, we propose an alternative model where REV1 and Polζ participate downstream during homologous recombination repair (Sharma et al., 2012). Recent evidence implicate both REV1 and Polζ in promoting DSB repair and genomic stability (Wittschieben et al., 2006; Schenten et al., 2009; Sharma et al., 2012). It is also becoming increasingly clear that Polζ contributes to tumor suppression (Wittschieben et al., 2010). Before REV1 or Polζ can be considered as targets for adjuvant therapy with platinating agents, these additional roles will need to be better understood.
Supplementary Material
 The online version of this article (available at http://molpharm.aspetjournals.org) contains supplemental material.
The online version of this article (available at http://molpharm.aspetjournals.org) contains supplemental material.
This work was supported by the National Institutes of Health National Cancer Institute through the University of Michigan's Cancer Center [Support Grant 5P30-CA46592]; and the National Institutes of Health National Cancer Institute [Grant CA133046].
Article, publication date, and citation information can be found at http://molpharm.aspetjournals.org.
- ICL
- interstrand DNA cross-link
- TLS
- translesion DNA synthesis
- AMD473
- picoplatin
- Polη
- DNA polymerase η
- Polζ
- DNA polymerase ζ
- Polι
- DNA polymerase ι
- DSBs
- DNA double-stranded breaks
- RPA
- replication protein A
- RPA32
- replication factor A-32
- JM216
- satraplatin
- ATM
- ataxia telangiectasia mutated
- H2AX
- histone H2A.x
- ATR
- ataxia telangiectasia and Rad3 related
- siRNA
- small interfering RNA
- PAGE
- polyacrylamide gel electrophoresis
- PCNA
- proliferating cell nuclear antigen
- GAPDH
- glyceraldehyde-3-phosphate dehydrogenase
- GFP
- green fluorescent protein.
Authorship Contributions
Participated in research design: Sharma and Canman.
Conducted experiments: Sharma, Shah, Joiner, Roberts, and Canman.
Performed data analysis: Sharma and Canman.
Wrote or contributed to the writing of the manuscript: Sharma and Canman.
References
- Albertella MR, Green CM, Lehmann AR, O'Connor MJ. (2005) A role for polymerase eta in the cellular tolerance to cisplatin-induced damage. Cancer Res 65:9799–9806 [DOI] [PubMed] [Google Scholar]
- Alt A, Lammens K, Chiocchini C, Lammens A, Pieck JC, Kuch D, Hopfner KP, Carell T. (2007) Bypass of DNA lesions generated during anticancer treatment with cisplatin by DNA polymerase eta. Science 318: 967–970 [DOI] [PubMed] [Google Scholar]
- Bakkenist CJ, Kastan MB. (2003) DNA damage activates ATM through intermolecular autophosphorylation and dimer dissociation. Nature 421:499–506 [DOI] [PubMed] [Google Scholar]
- Bassett E, King NM, Bryant MF, Hector S, Pendyala L, Chaney SG, Cordeiro-Stone M. (2004) The role of DNA polymerase eta in translesion synthesis past platinum-DNA adducts in human fibroblasts. Cancer Res 64:6469–6475 [DOI] [PubMed] [Google Scholar]
- Bhattacharyya D, Ramachandran S, Sharma S, Pathmasiri W, King CL, Baskerville-Abraham I, Boysen G, Swenberg JA, Campbell SL, Dokholyan NV, et al. (2011) Flanking bases influence the nature of DNA distortion by platinum 1,2-intrastrand (GG) cross-links. PLoS ONE 6:e23582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bi X, Barkley LR, Slater DM, Tateishi S, Yamaizumi M, Ohmori H, Vaziri C. (2006) Rad18 regulates DNA polymerase kappa and is required for recovery from S-phase checkpoint-mediated arrest. Mol Cell Biol 26:3527–3540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bomgarden RD, Lupardus PJ, Soni DV, Yee MC, Ford JM, Cimprich KA. (2006) Opposing effects of the UV lesion repair protein XPA and UV bypass polymerase eta on ATR checkpoint signaling. EMBO J 25:2605–2614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaney SG, Campbell SL, Bassett E, Wu Y. (2005) Recognition and processing of cisplatin- and oxaliplatin-DNA adducts. Crit Rev Oncol Hematol 53:3–11 [DOI] [PubMed] [Google Scholar]
- Chen YW, Cleaver JE, Hanaoka F, Chang CF, Chou KM. (2006) A novel role of DNA polymerase eta in modulating cellular sensitivity to chemotherapeutic agents. Mol Cancer Res 4:257–265 [DOI] [PubMed] [Google Scholar]
- Cimprich KA, Cortez D. (2008) ATR: an essential regulator of genome integrity. Nat Rev Mol Cell Biol 9:616–627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cruet-Hennequart S, Glynn MT, Murillo LS, Coyne S, Carty MP. (2008) Enhanced DNA-PK-mediated RPA2 hyperphosphorylation in DNA polymerase eta-deficient human cells treated with cisplatin and oxaliplatin. DNA Repair 7:582–596 [DOI] [PubMed] [Google Scholar]
- Deans AJ, West SC. (2011) DNA interstrand crosslink repair and cancer. Nat Rev Cancer 11:467–480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doles J, Oliver TG, Cameron ER, Hsu G, Jacks T, Walker GC, Hemann MT. (2010) Suppression of Rev3, the catalytic subunit of Pol{zeta}, sensitizes drug-resistant lung tumors to chemotherapy. Proc Natl Acad Sci U S A 107:20786–20791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gueranger Q, Stary A, Aoufouchi S, Faili A, Sarasin A, Reynaud CA, Weill JC. (2008) Role of DNA polymerases eta, iota and zeta in UV resistance and UV-induced mutagenesis in a human cell line. DNA Repair 7:1551–1562 [DOI] [PubMed] [Google Scholar]
- Guo C, Fischhaber PL, Luk-Paszyc MJ, Masuda Y, Zhou J, Kamiya K, Kisker C, Friedberg EC. (2003) Mouse Rev1 protein interacts with multiple DNA polymerases involved in translesion DNA synthesis. EMBO J 22:6621–6630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanada K, Budzowska M, Modesti M, Maas A, Wyman C, Essers J, Kanaar R. (2006) The structure-specific endonuclease Mus81-Eme1 promotes conversion of interstrand DNA crosslinks into double-strand breaks. EMBO J 25:4921–4932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hicks JK, Chute CL, Paulsen MT, Ragland RL, Howlett NG, Guéranger Q, Glover TW, Canman CE. (2010) Differential roles for DNA polymerases eta, zeta, and REV1 in lesion bypass of intrastrand versus interstrand DNA cross-links. Mol Cell Biol 30:1217–1230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho TV, Guainazzi A, Derkunt SB, Enoiu M, Schärer OD. (2011) Structure-dependent bypass of DNA interstrand crosslinks by translesion synthesis polymerases. Nucleic Acids Res 39:7455–7464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelland L. (2007) The resurgence of platinum-based cancer chemotherapy. Nat Rev Cancer 7:573–584 [DOI] [PubMed] [Google Scholar]
- Köberle B, Tomicic MT, Usanova S, Kaina B. (2010) Cisplatin resistance: preclinical findings and clinical implications. Biochim Biophys Acta 1806:172–182 [DOI] [PubMed] [Google Scholar]
- Kratz K, Schöpf B, Kaden S, Sendoel A, Eberhard R, Lademann C, Cannavó E, Sartori AA, Hengartner MO, Jiricny J. (2010) Deficiency of FANCD2-associated nuclease KIAA1018/FAN1 sensitizes cells to interstrand crosslinking agents. Cell 142:77–88 [DOI] [PubMed] [Google Scholar]
- Lewis KA, Lilly KK, Reynolds EA, Sullivan WP, Kaufmann SH, Cliby WA. (2009) Ataxia telangiectasia and rad3-related kinase contributes to cell cycle arrest and survival after cisplatin but not oxaliplatin. Mol Cancer Ther 8:855–863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu T, Ghosal G, Yuan J, Chen J, Huang J. (2010) FAN1 acts with FANCI-FANCD2 to promote DNA interstrand cross-link repair. Science 329:693–696 [DOI] [PubMed] [Google Scholar]
- MacKay C, Déclais AC, Lundin C, Agostinho A, Deans AJ, MacArtney TJ, Hofmann K, Gartner A, West SC, Helleday T, et al. (2010) Identification of KIAA1018/FAN1, a DNA repair nuclease recruited to DNA damage by monoubiquitinated FANCD2. Cell 142:65–76 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nehmé A, Baskaran R, Nebel S, Fink D, Howell SB, Wang JY, Christen RD. (1999) Induction of JNK and c-Abl signalling by cisplatin and oxaliplatin in mismatch repair-proficient and -deficient cells. Br J Cancer 79:1104–1110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niedzwiedz W, Mosedale G, Johnson M, Ong CY, Pace P, Patel KJ. (2004) The Fanconi anaemia gene FANCC promotes homologous recombination and error-prone DNA repair. Mol Cell 15:607–620 [DOI] [PubMed] [Google Scholar]
- Nojima K, Hochegger H, Saberi A, Fukushima T, Kikuchi K, Yoshimura M, Orelli BJ, Bishop DK, Hirano S, Ohzeki M, et al. (2005) Multiple repair pathways mediate tolerance to chemotherapeutic cross-linking agents in vertebrate cells. Cancer Res 65:11704–11711 [DOI] [PubMed] [Google Scholar]
- Ohashi E, Murakumo Y, Kanjo N, Akagi J, Masutani C, Hanaoka F, Ohmori H. (2004) Interaction of hREV1 with three human Y-family DNA polymerases. Genes Cells 9:523–531 [DOI] [PubMed] [Google Scholar]
- Okuda T, Lin X, Trang J, Howell SB. (2005) Suppression of hREV1 expression reduces the rate at which human ovarian carcinoma cells acquire resistance to cisplatin. Mol Pharmacol 67:1852–1860 [DOI] [PubMed] [Google Scholar]
- Rabik CA, Dolan ME. (2007) Molecular mechanisms of resistance and toxicity associated with platinating agents. Cancer Treat Rev 33:9–23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Räschle M, Knipscheer P, Knipsheer P, Enoiu M, Angelov T, Sun J, Griffith JD, Ellenberger TE, Schärer OD, Walter JC. (2008) Mechanism of replication-coupled DNA interstrand crosslink repair. Cell 134:969–980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raymond E, Faivre S, Chaney S, Woynarowski J, Cvitkovic E. (2002) Cellular and molecular pharmacology of oxaliplatin. Mol Cancer Ther 1:227–235 [PubMed] [Google Scholar]
- Rothfuss A, Grompe M. (2004) Repair kinetics of genomic interstrand DNA cross-links: evidence for DNA double-strand break-dependent activation of the Fanconi anemia/BRCA pathway. Mol Cell Biol 24:123–134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schenten D, Kracker S, Esposito G, Franco S, Klein U, Murphy M, Alt FW, Rajewsky K. (2009) Pol zeta ablation in B cells impairs the germinal center reaction, class switch recombination, DNA break repair, and genome stability. J Exp Med 206:477–490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schultz LB, Chehab NH, Malikzay A, Halazonetis TD. (2000) p53 binding protein 1 (53BP1) is an early participant in the cellular response to DNA double-strand breaks. J Cell Biol 151:1381–1390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shachar S, Ziv O, Avkin S, Adar S, Wittschieben J, Reissner T, Chaney S, Friedberg EC, Wang Z, Carell T, et al. (2009) Two-polymerase mechanisms dictate error-free and error-prone translesion DNA synthesis in mammals. EMBO J 28:383–393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma S, Hicks JK, Chute CL, Brennan JR, Ahn JY, Glover TW, Canman CE. (2012) REV1 and polymerase æ facilitate homologous recombination repair. Nucleic Acids Res 40:682–691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simpson LJ, Sale JE. (2003) Rev1 is essential for DNA damage tolerance and non-templated immunoglobulin gene mutation in a vertebrate cell line. EMBO J 22:1654–1664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonoda E, Okada T, Zhao GY, Tateishi S, Araki K, Yamaizumi M, Yagi T, Verkaik NS, van Gent DC, Takata M, et al. (2003) Multiple roles of Rev3, the catalytic subunit of polzeta in maintaining genome stability in vertebrates. EMBO J 22:3188–3197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taniguchi T, Garcia-Higuera I, Xu B, Andreassen PR, Gregory RC, Kim ST, Lane WS, Kastan MB, D'Andrea AD. (2002) Convergence of the fanconi anemia and ataxia telangiectasia signaling pathways. Cell 109:459–472 [DOI] [PubMed] [Google Scholar]
- Tissier A, Kannouche P, Reck MP, Lehmann AR, Fuchs RP, Cordonnier A. (2004) Co-localization in replication foci and interaction of human Y-family members, DNA polymerase pol eta and REVl protein. DNA Repair (Amst) 3:1503–1514 [DOI] [PubMed] [Google Scholar]
- Vaisman A, Masutani C, Hanaoka F, Chaney SG. (2000) Efficient translesion replication past oxaliplatin and cisplatin GpG adducts by human DNA polymerase eta. Biochemistry 39:4575–4580 [DOI] [PubMed] [Google Scholar]
- Washington MT, Carlson KD, Freudenthal BD, Pryor JM. (2010) Variations on a theme: eukaryotic Y-family DNA polymerases. Biochim Biophys Acta 1804:1113–1123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waters LS, Minesinger BK, Wiltrout ME, D'Souza S, Woodruff RV, Walker GC. (2009) Eukaryotic translesion polymerases and their roles and regulation in DNA damage tolerance. Microbiol Mol Biol Rev 73:134–154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wittschieben JP, Patil V, Glushets V, Robinson LJ, Kusewitt DF, Wood RD. (2010) Loss of DNA polymerase zeta enhances spontaneous tumorigenesis. Cancer Res 70:2770–2778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wittschieben JP, Reshmi SC, Gollin SM, Wood RD. (2006) Loss of DNA polymerase zeta causes chromosomal instability in mammalian cells. Cancer Res 66:134–142 [DOI] [PubMed] [Google Scholar]
- Wu F, Lin X, Okuda T, Howell SB. (2004) DNA polymerase zeta regulates cisplatin cytotoxicity, mutagenicity, and the rate of development of cisplatin resistance. Cancer Res 64:8029–8035 [DOI] [PubMed] [Google Scholar]
- Xie K, Doles J, Hemann MT, Walker GC. (2010) Error-prone translesion synthesis mediates acquired chemoresistance. Proc Natl Acad Sci U S A 107:20792–20797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziv O, Geacintov N, Nakajima S, Yasui A, Livneh Z. (2009) DNA polymerase ζ cooperates with polymerases κ and ι in translesion DNA synthesis across pyrimidine photodimers in cells from XPV patients. Proc Natl Acad of Sci U S A 106:11552–11557 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.