Abstract
An unresolved issue about many neurodegenerative diseases is why neurons are particularly sensitive to defects in ubiquitous cellular processes. One example is Niemann Pick type C1, caused by defects in cholesterol trafficking in all cells, but where neurons are preferentially damaged. Understanding this selective failure is limited by the difficulty in obtaining live human neurons from affected patients. To solve this problem, we generated neurons with decreased function of NPC1 from human embryonic stem cells and used them to test the hypothesis that defective cholesterol handling leads to enhanced pathological phenotypes in neurons. We found that human NPC1 neurons have strong spontaneous activation of autophagy, and, contrary to previous reports in patient fibroblasts, a block of autophagic progression leading to defective mitochondrial clearance. Mitochondrial fragmentation is an exceptionally severe phenotype in NPC1 neurons compared with fibroblasts, causing abnormal accumulation of mitochondrial proteins. Contrary to expectation, these abnormal phenotypes were rescued by treatment with the autophagy inhibitor 3-methyladenine and by treatment with the potential therapeutic cyclodextrin, which mobilizes cholesterol from the lysosomal compartment. Our findings suggest that neurons are especially sensitive to lysosomal cholesterol accumulation because of autophagy disruption and accumulation of fragmented mitochondria, thus defining a new route to effective drug development for NPC1 disease.
INTRODUCTION
Niemann Pick type C1 (NPC1) is a fatal progressive childhood neurodegenerative disease caused by loss-of-function mutations of NPC1 and characterized by the accumulation of cholesterol and other lipids in the lysosomal compartment (1,2). Most of our knowledge of NPC1 has been derived from studies of animal models and human fibroblasts; however, the cellular mechanisms underlying neurodegeneration in NPC1 remain unclear. In particular, it is unknown why neurons are more sensitive to the effects of mutations of NPC1, despite the ubiquitous nature of the cholesterol-trafficking pathway these mutations affect. Accumulation of cholesterol has been suggested to cause neuronal failure; however, studies in NPC1−/− mice have not consistently shown an increase in total neuronal cholesterol content (3–5). Therefore, a more relevant feature of NPC1-deficient neurons may be abnormal distribution of cholesterol due to sequestration in the lysosomal compartment. Many other potential pathogenic phenotypes have been suggested based on work in NPC1 mice and human fibroblasts, including increased spontaneous activation of the autophagic pathway (6–8). Although lysosomal dysfunction has been shown to impair autophagic flow in several lysosomal storage diseases (LSDs) (9,10), progression of autophagy has been argued to be normal in NPC1 (6,11). This atypical behavior of NPC1 relative to other LSDs has important implications not only on the resulting pathological phenotypes, but also on the therapeutic strategies that can be used to ameliorate these phenotypes.
NPC1 function is highly conserved in evolution (12,13), and no differences have been found between its fundamental role in lipid trafficking in mice and humans. Despite these similarities, the pathological consequences of NPC1 dysfunction are unlikely to be the same for mouse and human neurons. In these regards, an important motivation for the development of a human neuronal model of NPC1 is that the widely used mouse model of NPC1 does not reproduce human pathology accurately. Despite continued efforts, strategies using NPC1 mice have not yet revealed how mutations of NPC1 cause neuronal failure in humans and have not yielded a clear therapeutic avenue. Mouse and human neurons have obvious biochemical and physiological differences. Specifically, in NPC1, mouse tau protein does not readily form neurofibrillary tangles, which do form in human mutant NPC1 neurons. In addition, mice lack the apoE genotypes found in human subjects, which also impact disease course, and neuronal loss in NPC1 mice is mostly limited to the cerebellum, whereas cortical and thalamic involvement are important in humans (2,3,14).
Analysis of disease-specific human neurons could significantly advance our understanding of early pathogenic events in NPC1 and resolve the question of how abnormal handling of cholesterol preferentially causes neuronal failure and neuronal death. Recently, human embryonic stem cells (hESCs) have been used as a powerful alternative to model and test treatments for neurologic disorders (15,16). Here we report the generation of human neurons with decreased function of NPC1 from hESCs to probe mechanisms of neuronal dysfunction in NPC1.
RESULTS
Generation of NPC1 knockdown hESC lines
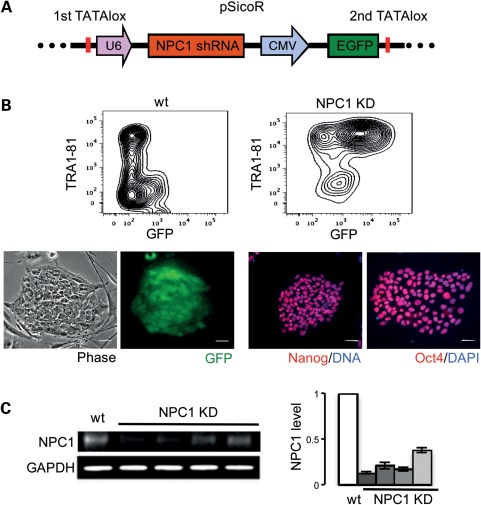
Because the most severe forms of NPC1 are caused by loss-of-function or null mutations of NPC1, we modeled NPC1 disease by generating NPC1 knockdown (KD) hESCs from the HUES9 hESC line (17) by shRNA-mediated silencing of NPC1. Three shRNA sequences were designed to specifically and exclusively silence human NPC1. A pSicoR cre-repressible lentiviral vector for stable RNA interference (18) carrying a green fluorescent protein (GFP) reporter was used for efficient delivery to hESCs and generation of stable cell lines (Fig. 1A). Ten independent NPC1 KD hESC lines were generated and four were chosen for further analysis. Analyzed lines were karyotyped to confirm euploidy. To ensure that NPC1 KD hESCs maintain stem-cell properties, we documented typical morphology and expression of pluripotency markers by immunofluorescence (IF) and flow cytometry (Fig. 1B). We measured NPC1 KD by quantitative real-time-polymerase chain reaction (qRT-PCR) and quantitative western blot (WB) (Fig. 1C, Supplementary Material, Fig. S1A). In addition, we found that reduction of NPC1 in hESCs does not interfere with pluripotency or differentiation capacity. When plated in suspension culture, both wild-type (wt) and NPC1 KD hESCs can form embryoid bodies that stain for markers representative of the three embryonic germ layers (Supplementary Material, Fig. S1B). Moreover, reduction of NPC1 in hESCs does not affect formation of neurospheres that can be differentiated further to generate mixed neuronal cultures essential for the study of disease phenotypes in NPC1 (Supplementary Material, Fig. S1C).
Figure 1.
Generation of NPC1 KD hESC lines. Clonal GFP-positive HUES9 hESC lines generated after transduction with PSicoR NPC1 shRNA lentiviral vector (A) have normal stem-cell morphology, maintain expression of the pluripotency markers TRA1-81, Oct4 and Nanog (B) and show reduction of NPC1 levels (C). Scale bar is 50 μm. Levels of NPC1 from four clones are quantified by WB and are normalized to wt. Glyceraldehyde 3-phosphate dehydrogenase (GAPDH) was used as a loading control.
NPC1 KD hESCs and derived neurons replicate known phenotypes of NPC1
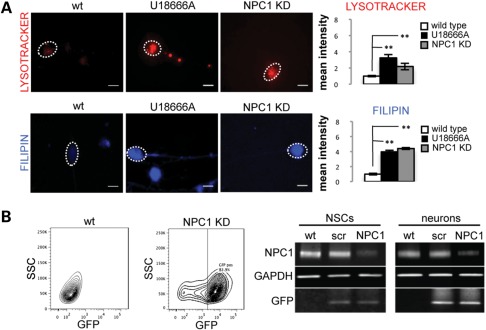
To determine whether typical pathogenic phenotypes associated with NPC1 can be recapitulated in vitro, we analyzed wt and NPC1 KD hESC and neuronal cultures. Treatment with the amphiphilic amino-steroid U18666A, an inhibitor of intracellular cholesterol trafficking used widely to mimic NPC1 disease phenotypes, was used as a positive control (19,20) and consistently recapitulated phenotypes of NPC1 fibroblasts and NPC1 KD neurons reported in this study. We found that hESCs and neurons with reduced levels of NPC1 accumulate large LysoTracker-positive organelles (Fig. 2A, Supplementary Material, Fig. S2A). We reconstituted human high-density lipoproteins (HDLs) with the fluorescent cholesterol analog Bodipy-cholesteryl and found NPC1 KD hESCs have increased accumulation of internalized HDL particles when these are added to the culture media (Supplementary Material, Fig. S2A). We then stained human NPC1 KD neurons with the macrolide antibiotic filipin, which specifically binds cholesterol, and found NPC1 KD causes increased filipin staining of neuronal cell bodies (Fig. 2A).
Figure 2.
Characterization of NPC1 KD hESC-derived neural cells. (A) Human NPC1 KD neurons accumulate large LysoTracker-positive organelles and have increased filipin staining. U18666A-treated neurons were used as positive control for all experiments and replicated phenotypes of NPC1 reported in this study. Quantification shown is normalized to wt (**P < 0.001, n = 15 from three independent experiments). Scale bar is 10 μm. (B) wt NSCs were induced to express NPC1 shRNA by stable transduction with pSicoR-GFP lentiviral vector. Expression of GFP is evident 48 h after infection and is maintained after several passages in populations enriched for GFP-positive cells. WB showing reduced levels of NPC1 in KD NSCs and derived neurons, and unaffected scramble (scr) controls.
Generation of NPC1 KD neural stem-cell lines
Although NPC1 KD hESCs can generate mixed neuronal cultures through formation of neurospheres, this method is inefficient and time-consuming. To induce more efficient neuronal differentiation of hESCs, we co-cultured wt and NPC1 KD HUES9 hESCs with the PA6 stromal cell line (21) and then attempted to isolate a population enriched for neural stem cells (NSCs) using cell surface markers and flow cytometry as shown in Supplementary Material, Figure S2B (22). We were able to efficiently generate homogeneous populations of NSCs from wt but not from NPC1 KD hESCs (data not shown). The significance of this finding remains to be determined. Yang et al. (23) found NSCs derived from NPC1−/− mice had significantly impaired self-renewal and differentiation capacity when compared with wt mice, but neural developmental abnormalities are not typically described in NPC1 patients. In contrast, variation in the response of different hESC lines to directed differentiation has been documented and thought to be secondary to inherited variation in the sex, stage and genetic background of embryos used for hESC line generation (24,25). Therefore, difficulty of generating NSCs in our study may be related to genetic background of the HUES9 hESC line.
To overcome this limitation, wt NSCs were transduced with the lentiviral vector pSicoR carrying an shRNA sequence directed against NPC1. GFP-positive cells were selected by flow cytometry to generate two NPC1 KD NSC lines and one scramble control (Fig. 2B). NSC lines generated by this method have excellent proliferative capacity, express lineage specific markers and can efficiently differentiate into neuronal cultures (Supplementary Material, Fig. S2C). NPC1 KD NSCs maintain stable GFP expression and reduced levels of NPC1 over several passages and upon neuronal differentiation as shown by WB (Fig. 2B).
Accumulation of mitochondrial fragments in NPC1 KD human neurons
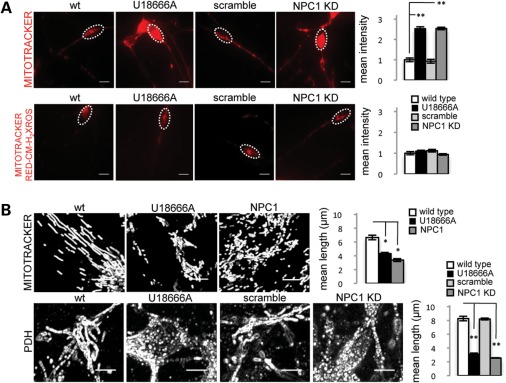
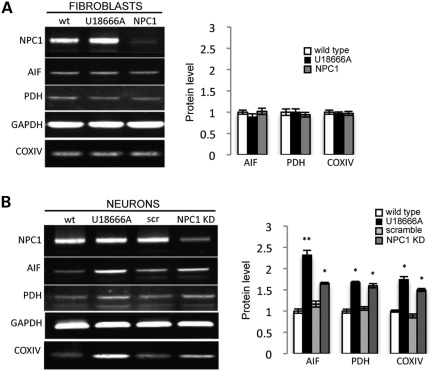
Mitochondrial abnormalities have been reported in several neurodegenerative diseases including NPC1, in which abnormal morphology, deficient ATP production and depolarization of mitochondrial membranes have been described (26–28). An untested hypothesis is that diseases such as NPC1 impair autophagy, leading to downstream defects in mitochondria that are more severe in neurons. To test this hypothesis, we evaluated mitochondrial abnormalities in NPC1 KD neurons. We found that human neurons with reduced levels of NPC1 have increased MitoTracker signal suggestive of mitochondrial accumulation (Fig. 3A). However, no significant difference was observed between wt and NPC1 KD neurons when incubated with the membrane potential-dependent MitoTracker Red-CM-H2XRos (Fig. 3A). We hypothesized that the increase in MitoTracker staining, which is not dependent on mitochondrial membrane potential, may indicate accumulation of depolarized mitochondria that fail to be cleared, which is consistent with Yu et al.'s findings of decreased mitochondrial membrane potential in cultured mouse Npc1−/− neurons (28). Interestingly, these mitochondrial phenotypes were exceptionally severe in human NPC1 neurons and only inconsistently observed in NPC1 patient fibroblasts used as controls. Since damaged mitochondria have been shown to be removed by autophagy, we performed MitoTracker and MitoTracker Red-CM-H2XRos signal measurements in NPC1 fibroblasts grown under serum deprivation, which normally induces autophagy (29,30). Under these conditions, we observed a greater increase of MitoTracker signal in NPC1 fibroblasts compared with wt controls, but no significant differences in MitoTracker Red-CM-H2XRos signal, replicating our findings in human neurons grown under baseline conditions (Supplementary Material, Fig. S3A). To test the possibility that the increase in MitoTracker staining may signal an increase in the metabolically inactive pool of mitochondria, we measured mitochondrial length in MitoTracker-stained human NPC1 KD neurons and NPC1 patient fibroblasts and found accumulation of short mitochondrial fragments compared with wt cells (Fig. 3B). Accumulation of mitochondrial fragments in NPC1 fibroblasts and neurons was aggravated by induction of autophagy with serum deprivation supporting a role for autophagy in mitochondrial fragmentation. No significant difference in length was found in the metabolically active mitochondrial pools stained with MitoTracker Red-CM-H2XRos (Supplementary Material, Fig. S3B). Pyruvate dehydrogenase (PDH) staining confirmed the mitochondrial identity of the short fragments seen with MitoTracker but not with MitoTracker Red-CM-H2XRos. As mitochondrial clearance normally depends on autophagy, we tested whether the observed mitochondrial fragments are associated with the autophagic vesicle marker LC3-GFP. As transfection of human neurons is particularly challenging, and because mitochondrial fragmentation is observed in NPC1 patient fibroblasts upon induction of autophagy, we performed co-localization analysis in NPC1 patient fibroblasts under conditions of serum deprivation. We found that MitoTracker-positive fragments co-localize with the autophagosome marker LC3-GFP in NPC1 but not in wt fibroblasts (Supplementary Material, Fig. S4A). Interestingly, total levels of the mitochondrial markers PDH, AIF and COXIV measured by WB were unchanged in NPC1 patient fibroblasts but were increased in NPC1 KD human neurons (Fig. 4A and B). Mitochondrial fragmentation was also present in NSCs without obvious accumulation of mitochondrial protein (data not shown), and therefore it is possible that neurons may be more likely to accumulate metabolically inactive mitochondria because they cannot rely on cell division to dilute the load of damaged cellular components (31,32). Thus, we found that accumulation of mitochondrial fragments in NPC1 is a previously unrecognized phenotype that is much more severe in human neurons compared with patient fibroblasts.
Figure 3.
NPC1 KD is associated with accumulation of mitochondrial fragments in human neurons. (A) Live imaging of wt and NPC1 KD neurons grown under standard conditions, and stained with membrane potential independent MitoTracker and membrane potential dependent MitoTracker CM-XH2-Ros. Quantification shown is normalized to wt. MitoTracker signal is increased in NPC1 KD neurons at baseline, no change is observed with MitoTracker CM-XH2-Ros (**P < 0.001, n = 15 from three independent experiments). Scale bar is 10 μm. (B) MitoTracker (fibroblast) and PDH (neuron) staining shows mitochondrial fragmentation in NPC1 fibroblasts and neurons grown under standard conditions. Images have been magnified and processed to outline mitochondrial borders (**P < 0.001, n = 10 from two independent experiments). Scale bar is 10 μm.
Figure 4.
NPC1 KD human neurons accumulate mitochondrial protein. Mitochondrial protein levels measured by quantitative WB are unchanged in NPC1 fibroblasts compared with wt control (A), but are increased in NPC1 KD neurons (B). GAPDH was used as a loading control. Quantification is normalized to wt (**P < 0.001, *P < 0.05, n = 6 from three independent experiments).
Autophagy deficits in NPC1 KD human neurons
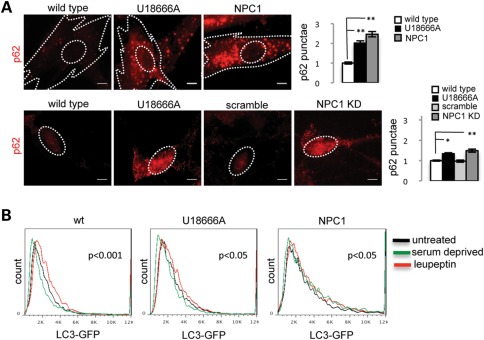
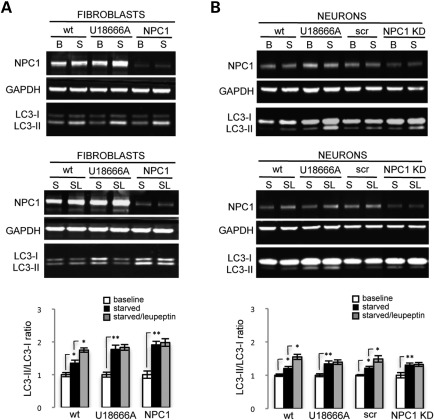
We tested the hypothesis that accumulation of mitochondrial fragments in NPC1 neurons and fibroblasts may be caused by abnormal autophagy. We measured levels of the autophagic markers LC3 and p62 in wt and NPC1 fibroblasts and neurons. We found that under baseline conditions, decreased function of NPC1 leads to increased LC3-GFP signal in NPC1 fibroblasts and increased numbers of p62 punctae in NPC1 neurons and fibroblasts (Fig. 5A, Supplementary Material, Fig. S4B), which suggests that autophagy is spontaneously activated in NPC1 fibroblasts and neurons. This is consistent with prior work from Pacheco et al. (6,11), who found increased autophagy in mouse and human NPC1-deficient primary fibroblasts. To test whether autophagy progression is abnormal, we transfected NPC1 fibroblasts with LC3-GFP and measured LC3-GFP turnover by flow cytometry. Induction of autophagy by serum deprivation normally increases LC3-GFP turnover, which is measured by a decrease in GFP signal intensity. We observed that compared with wt fibroblasts (P < 0.001), serum deprivation causes a less pronounced but still significant decrease of LC3-GFP in U18666A-treated fibroblasts (P < 0.05) and a paradoxical increase of LC3-GFP in NPC1 patient fibroblasts (P < 0.05). In contrast, treatment with the lysosomal poison leupeptin, which blocks autophagic progression and clearance of LC3, induces a normal response on wt fibroblasts by increasing LC3-GFP signal (P < 0.001), and a blunted response in U18666A-treated or NPC1 fibroblasts (P < 0.05; Fig. 5B). These findings suggest that lysosomal turnover of LC3-GFP is less efficient in NPC1 fibroblasts, causing abnormal progression of autophagic flow. We confirmed this possibility by measuring levels of LC3-II, the active form of LC3, in NPC1 fibroblasts and neurons by quantitative WB after serum deprivation and leupeptin treatment. Induction of autophagy by serum deprivation produces an increase of LC3-II/LC3-I ratio in both wt and NPC1 fibroblasts and neurons (Fig. 6A and B). However, LC3-II/LC3-I ratios are more significantly increased in NPC1 fibroblasts and neurons compared with wt, consistent with less efficient lysosomal turnover of LC3-II. In contrast, leupeptin treatment combined with serum deprivation induces a significant increase of LC3-II/LC3-I ratio in wt but not in NPC1 fibroblasts and neurons (Fig. 6A and B). Our data thus support a mixed scenario of autophagy induction and impaired autophagic flow in NPC1, which cause a relative blockage of lysosomal turnover of LC3-II. These findings are in contrast with previous reports in NPC1 patient fibroblasts that argued against deficits of progression of the autophagic pathway (6,11) and establish a potential mechanism of accumulation of mitochondrial fragments in NPC1 neurons by defective autophagic clearance. Impaired autophagic flow may have a preferential damaging effect on neurons as these cells rely on an ordinarily exceptionally efficient autophagic pathway and thus autophagic cargo can accumulate rapidly when late stages in autophagy are impaired (33,34).
Figure 5.
NPC1 fibroblasts and NPC1 KD neurons have increased spontaneous induction and abnormal progression of autophagy. (A) Levels of p62 are increased in NPC1 fibroblasts and neurons grown under standard conditions as shown by IF. Quantification of p62 punctae is shown for fibroblasts and neurons and normalized to wt (**P < 0.001, *P < 0.05, n = 10 from two independent experiments). Scale bar is 10 μm. (B) Flow cytometry analysis of LC3-GFP turnover in human fibroblasts. wt and NPC1 fibroblasts were transfected with LC3-GFP by electroporation, after 72 h transfected fibroblasts were either untreated, treated with serum deprivation or leupeptin for 12 h, dissociated and analyzed by flow cytometry. U18666A-treated fibroblasts were used as controls. Analysis of wt fibroblasts show normal decrease of LC3-GFP signal upon induction of autophagy by serum deprivation relative to the untreated sample, and increase of LC3-GFP signal after lysosomal poisoning with leupeptin, indicative of normal turnover of LC3-GFP. In contrast, NPC1 fibroblasts have a paradoxical increase of LC3-GFP signal induced by serum deprivation and minimal change of LC3-GFP signal after leupeptin treatment, suggestive of impaired clearance of LC3-GFP (n = 6 from three independent experiments).
Figure 6.
NPC1 fibroblasts and NPC1 KD neurons have increased spontaneous induction and abnormal progression of autophagy. Levels of LC3-I and LC3-II measured by quantitative WB in human fibroblasts and neurons are consistent with increased activation and abnormal progression of autophagy. wt and NPC1 fibroblasts and neurons were grown under baseline conditions (B), serum deprivation (S) or a combination of serum deprivation and leupeptin (SL) for 12 h, dissociated, processed and analyzed by WB. U18666A-treated fibroblasts and neurons were used as controls. Quantification is normalized to corresponding baseline sample. LC3-II/LC3-I ratio is increased by serum deprivation in wt and NPC1 fibroblasts (A) and neurons (B); however, this increase is greater in NPC1 fibroblasts and neurons compared with wt, suggesting increased activation of autophagy. In contrast, leupeptin treatment combined with serum deprivation (SL) causes an increase of LC3-II/LC3-I ratio in wt but not in NPC1 fibroblasts (A) and neurons (B), suggesting abnormal autophagy progression in NPC1. GAPDH was used as a loading control (**P < 0.001, *P < 0.05, n = 3 from three independent experiments).
Therapeutic rescue of autophagy defects and mitochondrial fragmentation
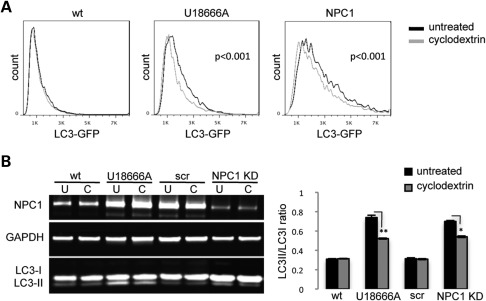
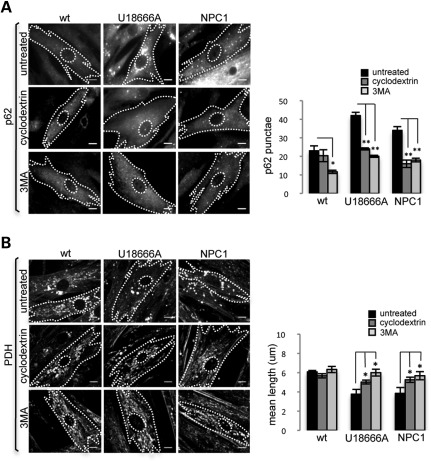
The potential therapeutic agent cyclodextrin (CD) ameliorates lysosomal accumulation of cholesterol in NPC1 mice and human fibroblasts (35,36). CDs have been used for this effect in NPC1-cultured cells and in NPC1−/− mice for more than a decade (37,38). To explore a potential link between lysosomal cholesterol accumulation and disrupted autophagy, we tested whether CDs can correct impaired autophagy in NPC1 fibroblasts and neurons. We measured autophagy levels in NPC1 KD neurons and patient fibroblasts after mobilizing lysosomal cholesterol with CD treatment. Both LC3-GFP signal measured by flow cytometry in NPC1 fibroblasts (Fig. 7A) and LC3-II/LC3-I ratio measured by WB in NPC1 KD neurons (Fig. 7B) are reduced after treatment with CD, suggesting that mobilization of cholesterol from the lysosomal compartment may increase turnover of LC3. To determine whether impaired autophagy contributes to mitochondrial fragmentation and whether CDs can restore mitochondrial length by rescuing normal autophagy, we measured p62 punctae and mitochondrial length in NPC1 fibroblasts after inhibition of autophagy with 3-methyladenine (3MA) or mobilization of cholesterol with CDs. Interestingly, treatment with either 3MA or CDs independently reduces p62 punctae (Fig. 8A) and mitochondrial fragmentation (Fig. 8B) in NPC1 fibroblasts. Thus, our findings suggest that lysosomal cholesterol accumulation leads to autophagy disruption, which in turn leads to mitochondrial fragmentation in NPC1. Rescue of autophagy disruption and mitochondrial fragmentation by CD treatment could be mediated by affecting early stages of autophagosome formation and regulation of autophagy, by causing enhanced turnover at the lysosomal level, or a combination of both factors.
Figure 7.
CD partially restores normal autophagy in NPC1 fibroblasts and NPC1 KD neurons. (A) LC3-GFP-transfected fibroblasts were grown under standard conditions and treated with CDs for 48 h and analyzed by flow cytometry. LC3-GFP was specifically reduced in NPC1 and U18666A-treated fibroblasts by CD treatment. No change was observed in wt fibroblasts (P < 0.001, n = 6 from three independent experiments). (B) Human neurons were treated with CDs for 48 h, harvested and analyzed by quantitative WB. LC3-II/LC3-I ratio was specifically reduced by CDs in NPC1 KD and U18666A-treated neurons (**P < 0.001, *P < 0.05, n = 6 from three independent experiments).
Figure 8.
Mitochondrial fragmentation is ameliorated by inhibition of autophagy and CD treatment in NPC1 fibroblasts. CD treatment reduces the number of p62 punctae (A) and increases mean mitochondrial length (B) in NPC1 and U18666A-treated NPC1 fibroblasts. A similar effect is induced by treatment with the autophagy inhibitor 3MA. Note that CDs and 3MA have no significant effect on wt fibroblasts (**P < 0.001, *P < 0.05, n = 9 from three independent experiments). Scale bar is 10 μm.
DISCUSSION
The inherent capacity of hESCs for unlimited self-renewal and for the production of large numbers of differentiated cells has significant potential for the development of cell replacement therapies and to provide insights into the early events involved in disease pathogenesis. We have used this approach to generate a human neuronal model of NPC1, to explore early cellular phenotypes that are likely to affect neuronal health and function and to uncover a mechanism that causes preferential neuronal failure in NPC1.
Subtle mitochondrial defects have been described in NPC1, including abnormal mitochondrial morphology, depolarization of mitochondrial membranes and impaired ATP production (27,28). However, our study is the first to report accumulation of partially degraded, metabolically inactive mitochondrial fragments in NPC1 human neurons. Despite the fact that low levels of mitochondrial fragmentation can also be observed in NPC1 patient fibroblasts, the preferential sensitivity of neurons to the effects of a block in autophagy progression provides the first definitive test that diseases such as NPC1 cause selective neuronal misbehavior by interfering with normal autophagy.
Attention has recently been given to a potential association between disrupted lipid trafficking and increased autophagy in NPC1 (6–8,11). Indeed, Pacheco et al. (11) showed that NPC1 deficiency leads to increased basal autophagy in human and mouse NPC1-deficient fibroblasts, and Ko et al. (8) found that loss of NPC1 function within mouse Purkinje cells leads to increased autophagy and cell death. Our data are consistent with increased spontaneous activation of the autophagic pathway, but contrary to prior observations suggesting that autophagy progression is normal, our findings support a mixed scenario of autophagy induction and impaired autophagic flow in human NPC1 fibroblasts and neurons. Therefore, our work goes beyond extrapolating pre-existing data to human neurons, to provide quantitative measurements of autophagic level and progression, and reveals a fundamental difference in the mechanism of autophagic disruption in human NPC1 KD neurons. This has important implications not only on the resulting pathological phenotypes, but also on the therapeutic strategies that can be used to ameliorate these phenotypes.
Our data support a model in which the combination of strong induction and impaired progression of autophagy conspires to cause mitochondrial fragmentation in NPC1 neurons and fibroblasts. Deficient turnover of LC3-II and other autophagic cargoes such as mitochondria in NPC1 neurons and fibroblasts may be caused by accumulation of cholesterol in the lysosomal compartment and can be partially rescued by mobilizing lysosomal cholesterol with CDs or by treatment with the autophagy inhibitor 3MA.
Our work provides evidence that mitochondrial fragmentation is downstream of autophagy as it can be rescued by pharmacological inhibition of autophagy. This conclusion from our work is important because it delineates the mechanistic pathway underlying mitochondrial fragmentation and it defines a new route to effective drug development for NPC1 disease. Pharmacological induction of autophagy is possible and has been advocated for treatment of various neurodegenerative diseases, including NPC1 (39,40). However, our findings of impaired turnover of mitochondria adds a potential complication to this therapeutic avenue as NPC1 KD-related accumulation of partially degraded mitochondria is a more severe phenotype in human neurons than in fibroblasts and may account for the preferential susceptibility of neurons to NPC1 defects. In our experiments, inhibition of autophagy rescues NPC1 fibroblasts from accumulation of mitochondrial fragments; however, turnover of other cellular components can potentially be affected by this intervention and needs to be examined further. Our observation that CDs and 3MA, two compounds with very distinct effects, are able to independently rescue the autophagy defects we observe in NPC1 fibroblasts and neurons may provide some insights into the mechanism underlying these defects. 3MA is a class III phosphatidylinositol 3-kinase inhibitor that acts at the level of autophagosome formation (41). It is possible that loss of function of NPC1 directly or indirectly causes abnormal formation of autophagosome membranes and that treatment with 3MA ameliorates the autophagy defects observed in NPC1 by inhibiting this process. Alternatively, CD may increase the pool of cholesterol available for autophagosome assembly or it may act at a later step in the autophagic pathway by decreasing the burden of lysosomal cholesterol and restoring normal autophagic progression in NPC1.
Other potentially neuroprotective effects of 3MA may contribute to ameliorate mitochondrial fragmentation, specifically by inhibition of the apoptotic pathway. However, the effect of 3MA on apoptosis may be closely related to its effect as an inhibitor of autophagy. Canu et al. (42) provide evidence that autophagy mediates a process of programmed cell death by activation of caspase-3 in cultured rat cerebellar neurons deprived of depolarizing concentrations of potassium. Although it has been reported previously that this manipulation causes classical apoptotic cell death, the data presented by Cantu et al. suggest that there is a transition within each cell from an autophagic to an apoptotic process. This is consistent with the view in the NPC1 field that both processes are likely to contribute to neuronal death caused by NPC1 dysfunction. 3MA-mediated rescue of mitochondrial fragmentation in our system could be caused by a direct effect on inhibition of autophagy or by an indirect effect on inhibition of apoptosis. However, independent rescue of autophagy activation and mitochondrial fragmentation by mobilizing cholesterol from the lysosomal compartment with CDs points to autophagy as the primary underlying factor.
The role of glia in the pathogenesis of NPC1 has been elegantly addressed by Ko et al. (8) and more recently by Zhang et al. (43). However, to our knowledge, autophagy and mitochondrial fragmentation have not been studied in glial cells. These pathological phenotypes may also contribute to the effects of NPC1 mutations observed in other tissues, such as the lung and liver (44,45). In this regard, it is interesting to note that NPC1−/− mouse hepatocytes have increased susceptibility to tumor necrosis factor-mediated damage due to mitochondrial dysfunction (46). Whether these mitochondrial abnormalities are linked to disrupted autophagy in the NPC1 liver remains to be investigated.
Further analysis of human NPC1-deficient neurons will reveal whether disrupted autophagy and mitochondrial fragmentation ultimately have an effect on neuronal viability. Indeed, the mechanism of cell death remains unclear in NPC1. Both apoptosis and autophagy have been postulated as potential mechanisms of neuronal death in NPC1 mice, and extensive work in the field suggests that both processes are likely to be at work in NPC1 neurons (47–50). Although the mechanism of neuronal death is of fundamental interest in the study of NPC1 and related neurodegenerative disorders, neuronal dysfunction must precede neuronal death. In this respect, it is interesting that Ko et al. (8) find increased autophagic markers only in degenerating Purkinje neurons of the NPC1−/− mouse. In contrast, our observations suggest that autophagy disruption is an early and universal feature of NPC1 KD human neurons in vitro despite no obvious decrease in short-term viability. Identifying early events leading to neuronal dysfunction is critical for the development of new therapies that may prevent or slow the progression of neurodegeneration in NPC1 before significant cell death has occurred.
Our data highlight the central role that autophagy failure caused by lysosomal cholesterol accumulation plays in the selective neuronal failure observed in NPC1 and provides the first example of a process that causes preferential neuronal defects in this devastating childhood neurodegenerative disease. Accumulation of mitochondria and other substrates that are ordinarily rapidly cleared by the autophagic pathway as well as mitochondrial fragmentation may cause defective energy production and generation of toxic oxygen species that can ultimately affect neuronal health and function. Additionally, our approach establishes a cell-based platform for the high-throughput screening of potential therapeutic compounds that can revert accumulation of cholesterol, lysosomes, mitochondrial fragments and autophagic intermediates in NPC1 and related neurodegenerative diseases.
MATERIALS AND METHODS
Cell culture
HUES9 hESCs were seeded on a mouse embryonic fibroblast (MEF) layer and grown under standard ES cell culture conditions (15). Human fibroblasts from NPC1 patients and control individuals were obtained from Coriell Cell Repository.
Generation of NPC1 KD hESC lines
HUES9 hESCs were transduced with a pSicoR cre-repressible lentiviral vector for stable RNA interference (16) carrying an shRNA-targeted specifically against NPC1 and a GFP marker. Transduced hESCs were selected by flow cytometry and single-GFP-positive cells were plated onto 96-well plates containing irradiated MEFs and expanded under standard culturing conditions (12) to generate clonal hESC lines. Karyotype analysis of NPC1 KD hESC lines was performed by Cell Line Genetics (Madison, WI, USA).
RNA expression and immunoblotting
To compare NPC1 RNA levels between samples, RNA was purified (PARIS Kit; Ambion, Grand Island, NY, USA), DNase-treated (Ambion) and reverse-transcribed (Superscript II; Invitrogen, Grand Island, NY, USA). Quantitative PCR (qPCR) was performed using FastStart Universal SYBR Green Master Mix (Roche, Indianapolis, IN, USA) and primers that amplify various regions of NPC1, HPRT and NONO (primer sequences available on request). Reactions were performed and analyzed on an Applied Biosystems 7300 Real Time PCR System using the ΔΔCt method. NPC1 levels were normalized to mean HPRT/NONO. For immunoblotting, cells were lysed using NP40 buffer with protease inhibitors. After electrophoresis on NuPAGE 4% or 4–12% Bis–Tris gels (Invitrogen), proteins were transferred onto polyvinylidene fluoride membranes (Millipore, Billerica, MA, USA) and then incubated overnight in primary antibody (see Antibodies) followed by IRDye 800CW Goat Anti-Rabbit or Anti-Mouse IgG secondary antibody (LI-COR Biosciences, Lincoln, NE, USA). Blots were developed using the Odyssey Infrared Imaging System (LI-COR Biosciences) for quantitative WB.
IF and FACS
Cells were fixed in 4% paraformaldehyde with 4% sucrose, permeabilized with buffer containing Triton X-100 and stained with primary and secondary antibodies (see Antibodies). Samples were imaged on a Nikon TE2000-U inverted microscope and acquired using Metamorph software (Molecular Devices, Sunnyvale, CA, USA). ImageJ software (NIH) was used to pseudo-color images, adjust contrast, add scale bars and to perform punctae counts and mitochondrial length measurements. FACS experiments were carried out on a FACSAria II cytometer (BD Biosciences, San Jose, CA, USA) and analyzed using FlowJo software (Tree Star, Ashland, OR, USA).
Antibodies
The antibodies used for FACS purification of cells were CD184-APC, CD15-FITC, CD24-PECy7, CD44-PE and CD271-PE (all from BD Biosciences, except CD24-PeCy7, which was purchased from Biolegend, San Diego, CA, USA) and were used at a concentration of one test per 1 × 106 cells. The following antibodies were used for IF and/or WB: NPC1 (Abcam, 1:5000 WB), SOX2 (Millipore, 1:2000 IF), nanog (Santa-Cruz, 1:200 IF), Oct3/4 (Santa-Cruz, 1:1000 IF), SMA (Chemicon, 1:50 IF), AFP (Dako, 1:1000 IF), MAP2a/b (Millipore, 1:500 IF, 1:1000 WB), α-tubulin (Sigma, 1:250k WB), glyceraldehyde 3-phosphate dehydrogenase (GAPDH) (Ambion, 1:1000 WB), nestin (Millipore, 1:200 IF), TUJ1 (Covance, 1:500 IF, 1:5000 WB), PDH (MitoSciences, 1:1000 IF, 1:500 WB), AIF (Santa-Cruz, 1:500 WB), COXIV (Cell Signaling, 1:1000), LC3B (Novus, 1:1000), p62 (Abnova, 1:350 IF, American Research Products, 1:1000). Secondary antibodies for IF were respectively conjugated to anti-mouse or anti-rabbit Alexa Fluor 488, Alexa Fluor 568 or Alexa Fluor 594 (Invitrogen, 1:200).
Live imaging
Cells were grown on coverslips and incubated with LysoTracker Red DND-99 (50 nm), MitoTracker 594 (25 nm), MitoTracker CM-XH2-ROS (50 nm) or filipin (25 μg/ml) for 15–30 min. Cells were then washed with phosphate-buffered saline, imaged on a Nikon TE2000-U inverted microscope and acquired using Metamorph software (Molecular Devices). ImageJ software (NIH) was used to pseudo-color images, adjust contrast, add scale bars and to perform mitochondrial length measurements. Filipin was purchased from Sigma, and all other probes were purchased from Invitrogen.
Neuronal differentiation and culture
Differentiation from hESCs to NSCs and neurons followed Yuan et al. (20). Differentiation began with a confluent 10 cm dish of undifferentiated HUES9 hESCs. For each plate, ∼1 × 105 hESCs were seeded onto 3 × 10 cm2 plates that were seeded the previous day with 5 × 105 PA6 cells (16). On day 12, cells were dissociated with Accutase (Innovative Cell Technologies, San Diego, CA, USA) and CD184 + CD15 + CD44-CD271- NSCs were FACS-purified and plated onto poly-ornithine/laminin-coated plates and cultured with basic fibroblast growth factor. For neuronal differentiation, NSCs were cultured with BDNF, GDNF and dbcAMP (Sigma, St Louis, MO, USA). After 3 weeks of differentiation, mixed neuronal cultures were obtained and harvested for live imaging, immunoblotting or immunocytochemical analysis.
LC3-GFP transfection
Control and patient fibroblasts were grown under standard conditions. Prior to transfection, cells were dissociated with trypsin and counted with a hemocytometer. For each sample, ∼2 × 105 cells and 3 μg of LC3-GFP DNA were mixed in an electroporation cuvette. Electroporation was performed using program X01 following Amaxa electroporation system nucleofection protocol. Cells were replated, grown for 72 h and then harvested for flow cytometry or imaging analysis.
Drug treatments and serum deprivation
Length of drug treatment and final concentrations are as follows: U18666A (3 μg/ml for 12 h), methyl-β-cyclodextrin (30 μm for 48 h), 3MA (5 mm for 24 h), leupeptin (20 μm for 12 h). All compounds were from Sigma. For serum deprivation, cells were grown in supplement-free high-glucose Dulbecco's modified Eagle medium (Gibco, Grand Island, NY, USA) for 12 h.
Statistics
Mean fluorescence intensity and co-localization analyses were performed with ImageJ software. Analysis of flow data was performed using FlowJo software. Data were analyzed using JMP software (SAS Institute, Cary, NC, USA). Statistical analysis was calculated by two-tailed Student's t-test or Tukey's HSD test. Comparisons between hESC lines were made by performing analysis of variance followed by Tukey's honestly significant difference (HSD) post-hoc test. Response to serum deprivation or drug treatments was compared with control by Dunnett method. Bar graphs display mean ± SEM. P < 0.05 was considered statistically significant.
SUPPLEMENTARY MATERIAL
Supplementary Material is available at HMG online.
ACKNOWLEDGEMENTS
We thank J. Young at UCSD and J. Lavine at Columbia University for scientific advice, T. Jacks at MIT for pSicoR vector, A. La Spada at UCSD for LC3-GFP plasmid, B. Balderas at BD Biosciences for antibodies, UCSD Neuroscience Microscopy Shared Facility (grant P30NS047101). M.P.O. and L.S.B.G. conceived the project and designed the experiments; M.P.O., E.A.R., C.U.K., S.H.Y. and W.C.P. performed the experiments; M.P.O., E.A.R. and S.Y. developed differentiation methods; M.P.O. and L.S.B.G. wrote the manuscript.
Conflict of Interest statement: None of the authors has declared any conflict of interest.
FUNDING
This work was funded by CIRM Training Grant TG2-01154 (L.S.B.G., M.P.O.), CIRM Comprehensive Grant RC1-00116-1 (L.S.B.G.), Ara Parseghian Medical Research Foundation Grant (M.P.O., L.S.B.G.) and NIH Challenge Grant 1RC1-NS068705 (M.P.O., L.S.B.G.). L.S.B.G. is an investigator at the Howard Hughes Medical Institute.
REFERENCES
- 1.Niemann A. Ein unbekanntes Krankheitsbild. Jahrbuch Für Kinderheilkunde. 1914;79:1–10. [Google Scholar]
- 2.Karten B., Peake K.B., Vance J.E. Mechanisms and consequences of impaired lipid trafficking in Niemann Pick type C1-deficient mammalian cells. Biochim. Biophys. Acta. 2009;7:659–670. doi: 10.1016/j.bbalip.2009.01.025. [DOI] [PubMed] [Google Scholar]
- 3.Vance J.E., Karten B., Hayashi H. Lipid dynamics in neurons. Biochem. Soc. Trans. 2006;34:399–403. doi: 10.1042/BST0340399. doi:10.1042/BST0340399. [DOI] [PubMed] [Google Scholar]
- 4.Karten B., Vance D., Campenot R., Vance J. Trafficking of cholesterol from cell bodies to distal axons in Niemann Pick type C1-deficient neurons. J. Biol. Chem. 2003;278:4168–4175. doi: 10.1074/jbc.M205406200. doi:10.1074/jbc.M205406200. [DOI] [PubMed] [Google Scholar]
- 5.Xie C., Lund E., Turley S., Russell D., Dietschy J. Quantitation of two pathways for cholesterol excretion from the brain in normal mice and mice with neurodegeneration. J. Lipid Res. 2003;44:1780–1789. doi: 10.1194/jlr.M300164-JLR200. doi:10.1194/jlr.M300164-JLR200. [DOI] [PubMed] [Google Scholar]
- 6.Pacheco C.D., Lieberman A.P. Lipid trafficking defects increase Beclin-1 and activate autophagy in Niemann Pick type C disease. Autophagy. 2007;5:487–489. doi: 10.4161/auto.4586. [DOI] [PubMed] [Google Scholar]
- 7.Ishibashi S., Yamazaki T., Okamoto K. Association of autophagy with cholesterol-accumulated compartments in Niemann-Pick disease type C cells. J. Clin. Neurosci. 2009;16:954–959. doi: 10.1016/j.jocn.2008.09.020. doi:10.1016/j.jocn.2008.09.020. [DOI] [PubMed] [Google Scholar]
- 8.Ko D.C., Milenkovic L., Beier S.M., Manuel H., Buchanan J., Scott M.P. Cell-autonomous death of cerebellar Purkinje neurons with autophagy in Niemann-Pick type C disease. PLoS Genet. 2005;1:81–95. doi: 10.1371/journal.pgen.0010007. doi:10.1371/journal.pgen.0010081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Settembre C., Fraldi A., Rubinztein D.C., Ballabio A. Lysosomal storage diseases as disorders of autophagy. Autophagy. 2008;4:113–114. doi: 10.4161/auto.5227. [DOI] [PubMed] [Google Scholar]
- 10.Fukuda T., Roberts A., Ahearn M., Zaal K., Ralston E., Plotz P.H., Raben N. Autophagy and lysosomes in Pompe disease. Autophagy. 2006;2:318–320. doi: 10.4161/auto.2984. [DOI] [PubMed] [Google Scholar]
- 11.Pacheco C.D., Kunkel R., Lieberman A.P. Autophagy in Niemann Pick C disease is dependent upon Beclin-1 and responsive to lipid trafficking defects. Hum. Mol. Genet. 2007;16:1495–1503. doi: 10.1093/hmg/ddm100. doi:10.1093/hmg/ddm100. [DOI] [PubMed] [Google Scholar]
- 12.Malathi K., Higaki K., Tinkelenberg A.H., Balderes D.A., Almanzar-Paramio D., Wilcox L.J., Erdeniz N., Redican F., Padamsee M., Liu Y., et al. Mutagenesis of the putative sterol-sensing domain of yeast Niemann Pick C-related protein reveals a primordial role in subcellular sphingolipid distribution. J. Cell. Biol. 2004;164:547–556. doi: 10.1083/jcb.200310046. doi:10.1083/jcb.200310046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schwend T., Loucks E.J., Snyder D., Ahigren S.C. Requirement of NPC1 and availability of cholesterol for early embryonic cell movements in zebrafish. J. Lipid. Res. 2011;52:1328–1344. doi: 10.1194/jlr.M012377. doi:10.1194/jlr.M012377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Love S., Bridges L.R., Case C.P. Neurofibrillary tangles in Niemann Pick type C. Brain. 1995;118:119–129. doi: 10.1093/brain/118.1.119. doi:10.1093/brain/118.1.119. [DOI] [PubMed] [Google Scholar]
- 15.Di Giorgio P., Carrasco M., Siao M., Maniatis T., Eggan K. Non-cell autonomous effect of glia on motor neurons in an embryonic stem cell-based ALS model. Nat. Neurosci. 2007;10:608–614. doi: 10.1038/nn1885. doi:10.1038/nn1885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lindvall O., Kokaia Z. Stem cells for the treatment of neurological disorders. Nature. 2006;441:1094–1096. doi: 10.1038/nature04960. doi:10.1038/nature04960. [DOI] [PubMed] [Google Scholar]
- 17.Cowan C.A., Klimanskaya I., McMahon J., Atienza J., Witmyer J., Zucker J.P., Wang S., Morton C.C., McMahon A.P., Powers D., Melton D.A. Derivation of embryonic stem-cell lines from human blastocysts. N. Engl. J. Med. 2004;350:1353–1356. doi: 10.1056/NEJMsr040330. doi:10.1056/NEJMsr040330. [DOI] [PubMed] [Google Scholar]
- 18.Ventura A., Meissner A., Dillon C., McManus M., Sharp P., Parijs L., Jaenisch R., Jacks T. Cre-lox-regulated conditional RNA interference from transgenes. Proc. Natl Acad. Sci. 2004;101:10380–10385. doi: 10.1073/pnas.0403954101. doi:10.1073/pnas.0403954101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cenedella R.J. Cholesterol synthesis inhibitor U18666A and the role of sterol metabolism and trafficking in numerous pathophysiological processes. Lipids. 2009;44:477–487. doi: 10.1007/s11745-009-3305-7. doi:10.1007/s11745-009-3305-7. [DOI] [PubMed] [Google Scholar]
- 20.Koh C.H., Cheung N.S. Cellular mechanism of U198666A mediated apoptosis in cultured murine cortical neurons: bridging Niemann Pick disease type C and Alzheimer's disease. Cell Signal. 2006;18:1844–1853. doi: 10.1016/j.cellsig.2006.04.006. doi:10.1016/j.cellsig.2006.04.006. [DOI] [PubMed] [Google Scholar]
- 21.Morizane A., Takahashi J., Shinoyama M., Ideguchi M., Takagi Y., Fukuda H., Koyanagi M., Sasai Y., Hashimoto N. Generation of graftable dopaminergic neuron progenitors from mouse ES cells by a combination of co-culture and neurosphere methods. J. Neurosci. Res. 2006;83:1015–1027. doi: 10.1002/jnr.20799. doi:10.1002/jnr.20799. [DOI] [PubMed] [Google Scholar]
- 22.Yuan S., Martin J., Elia J., Flippin J., Paramban R.I., Hefferan M.P., Vidal J.G., Mu Y., Killian R.L., Israel M.A., et al. Cell-surface marker signatures for the isolation of neural stem cells, glia and neurons derived from human pluripotent stem cells. PLoS One. 2011;6:E17540. doi: 10.1371/journal.pone.0017540. doi:10.1371/journal.pone.0017540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yang S., Kim S., Byun K., Hutchinson B., Lee B., Michikawa M., Lee Y., Kang K. NPC1 gene deficiency leads to lack of neural stem cell self-renewal and abnormal differentiation through activation of p38 mitogen-activated protein kinase signaling. Stem Cells. 2006;24:292–298. doi: 10.1634/stemcells.2005-0221. doi:10.1634/stemcells.2005-0221. [DOI] [PubMed] [Google Scholar]
- 24.Adewumi O., Aflatoonian B., Ahrlund-Richter L., Amit M., Andrews P.W., Beighton G., Bello P.A., Benvenisty N., Berry L.S., Bevan S., et al. Characterization of human embryonic stem cell lines by the International Stem Cell Initiative. Nat. Biotechnol. 2007;25:803–816. doi: 10.1038/nbt1318. doi:10.1038/nbt1318. [DOI] [PubMed] [Google Scholar]
- 25.Tavakoli T., Xu X., Derby E., Serebryakova Y., Reid Y., Rao M.S., Mattson M.P., Ma W. Self-renewal and differentiation capabilities are variable between human embryonic stem cell lines I3, I6 and BG01V. BMC Cell Biol. 2009;10:44–59. doi: 10.1186/1471-2121-10-44. doi:10.1186/1471-2121-10-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kiselyov K., Jennigs J.J., Rbaibi Y., Chu C.T. Autophagy, mitochondria and cell death in lysosomal storage diseases. Autophagy. 2007;3:259–262. doi: 10.4161/auto.3906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yu W., Gong J.S., Ko M., Garver W.S., Yanagisawa K., Michikawa M. Altered cholesterol metabolism in Niemann Pick type mouse brains affects mitochondrial function. J. Biol. Chem. 2005;280:11731–11739. doi: 10.1074/jbc.M412898200. doi:10.1074/jbc.M412898200. [DOI] [PubMed] [Google Scholar]
- 28.Fernandez A., Llacuna L., Fernandez-Checa J.C., Colell A. Mitochondrial cholesterol loading exacerbates amyloid beta peptide-induced inflammation and neurotoxicity. J. Neurosci. 2009;29:6394–6405. doi: 10.1523/JNEUROSCI.4909-08.2009. doi:10.1523/JNEUROSCI.4909-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rabinowitz J.D., White E. Autophagy and metabolism. Science. 2010;330:1344–1348. doi: 10.1126/science.1193497. doi:10.1126/science.1193497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shang L., Chen S., Du F., Li S., Zhao L., Wang X. Nutrient starvation elicits an acute autophagic response mediated by Ulk1 dephosphorylation and its subsequent dissociation from AMPK. Proc. Natl Acad. Sci. USA. 2011;108:4788–4793. doi: 10.1073/pnas.1100844108. doi:10.1073/pnas.1100844108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Terman A., Gustafsson B., Brunk U.T. Mitochondrial damage and intralysosomal degradation in cellular aging. Mol. Aspects Med. 2006;27:471–482. doi: 10.1016/j.mam.2006.08.006. doi:10.1016/j.mam.2006.08.006. [DOI] [PubMed] [Google Scholar]
- 32.Brunk U.T., Terman A. The mitochondrial-lysosomal axis theory of aging: accumulation of damaged mitochondria as a result of imperfect autophagocytosis. Eur. J. Biochem. 2002;269:1996–2002. doi: 10.1046/j.1432-1033.2002.02869.x. doi:10.1046/j.1432-1033.2002.02869.x. [DOI] [PubMed] [Google Scholar]
- 33.Boland B., Kumar A., Lee S., Platt F., Wegiel J., Haung Y., Nixon R. Autophagy induction and autophagosome clearance in neurons: relationship to autophagic pathology in Alzheimer's disease. J. Neurosci. 2008;28:6926–6937. doi: 10.1523/JNEUROSCI.0800-08.2008. doi:10.1523/JNEUROSCI.0800-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lee J., Yu W.H., Kumar A., Lee S., Mohan P.S., Peterhoff C.M., Wolfe D.M., Martinez-Vicente M., Massey A.C., Sovak G., et al. Lysosomal proteolysis and autophagy require presenilin 1 and are disrupted by Alzheimer-related PS1 mutations. Cell. 2010;141:1146–1158. doi: 10.1016/j.cell.2010.05.008. doi:10.1016/j.cell.2010.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Abi-Mosleh L., Infante R.E., Radhakrishnan A., Goldstein J.L., Brown M.S. Cyclodextrin overcomes deficient lysosome to endoplasmic reticulum transport of cholesterol in Niemann Pick type C cells. Proc. Natl Acad. Sci. 2009;106:19316–19321. doi: 10.1073/pnas.0910916106. doi:10.1073/pnas.0910916106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rosenbaum A., Zhang G., Warren J.D., Maxfield F.R. Endocytosis of beta-cyclodextrins is responsible for cholesterol reduction in Niemann-Pick type C mutant cells. Proc. Natl Acad. Sci. 2010;107:5477–5482. doi: 10.1073/pnas.0914309107. doi:10.1073/pnas.0914309107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Neufeld E.B., Cooney A.M., Pitha J., Dawidowicz E.A., Dwyer N.K., Pentchev P.G., Blanchette-Mackie E.J. Intracellular trafficking of cholesterol monitored with a cyclodextrin. J. Biol. Chem. 1996;271:21604–21613. doi: 10.1074/jbc.271.35.21604. doi:10.1074/jbc.271.35.21604. [DOI] [PubMed] [Google Scholar]
- 38.Camargo F., Erickson R.P., Garver W.S., Hossain G.S., Carbone P.N., Heidenreich R.A., Blanchard J. Cyclodextrin in the treatment of a mouse model of Niemann Pick C disease. Life Sci. 2001;70:131–142. doi: 10.1016/s0024-3205(01)01384-4. doi:10.1016/S0024-3205(01)01384-4. [DOI] [PubMed] [Google Scholar]
- 39.Pacheco C.D., Lieberman A.P. The pathogenesis of Niemann Pick type C disease: a role for autophagy? Expert Rev. Mol. Med. 2008;10:e26. doi: 10.1017/S146239940800080X. doi:10.1017/S146239940800080X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Harrison C. Neurodegenerative disease: a new pathway to autophagy. Nat. Rev. Drug Discov. 2008;7:476–477. doi:10.1038/nrd2598. [Google Scholar]
- 41.He C., Klionsky D.J. Regulation mechanisms and signaling pathways of autophagy. Annu. Rev. Genet. 2009;43:67–93. doi: 10.1146/annurev-genet-102808-114910. doi:10.1146/annurev-genet-102808-114910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Canu M., Tufi R., Serafino A.L., Amadoro G., Ciotti M.T., Calissano P. Role of the autophagic lysosomal system on low potassium induced apoptosis in cultured cerebellar granule cells. J. Neurochem. 2005;92:1228–1242. doi: 10.1111/j.1471-4159.2004.02956.x. doi:10.1002/jnr.21730. [DOI] [PubMed] [Google Scholar]
- 43.Zhang M., Strnatka D., Donohue C., Hallows J., Vincent I., Erickson R.P. Astrocyte-only NPC1 reduces neuronal cholesterol and triples life span of NPC1 -/- mice. J. Neurosci. Res. 2008;86:2848–2856. doi: 10.1002/jnr.21730. doi:10.1111/j.1471-4159.2004.02956.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yerushlami B., Sokol R.J., Narkewicz M.R., Smith D., Ashmead J.W., Wenger D.A. Niemann Pick disease type C in neonatal cholestasis at a North American center. J. Pediatr. Gastroenterol. Nutr. 2002;35:44–50. doi: 10.1097/00005176-200207000-00011. doi:10.1097/00005176-200207000-00011. [DOI] [PubMed] [Google Scholar]
- 45.Erickson R.P., Bhattacharyya A., Hunter R.J., Heidenreich R.A., Cherrington N.J. Am. J. Physiol. Gastrointest. Liver Physiol. 2005;289:G300–G307. doi: 10.1152/ajpgi.00568.2004. doi:10.1152/ajpgi.00568.2004. [DOI] [PubMed] [Google Scholar]
- 46.Mari M., Caballero F., Colell A., Morales A., Caballeria J., Fernandez A., Enrich C., Fernandez-Checa J.C., Garcia-Ruiz C. Mitochondrial free cholesterol loading sensitizes to TNF and Fas mediated steatohepatitis. Cell Metab. 2006;4:185–198. doi: 10.1016/j.cmet.2006.07.006. doi:10.1016/j.cmet.2006.07.006. [DOI] [PubMed] [Google Scholar]
- 47.Wu Y.P., Mizukami H., Matsuda J., Saito Y., Proia R.L., Suzuki K. Apoptosis accompanied by up-regulation of TNF-alpha death pathway genes in the brain of Niemann Pick type C disease. Mol. Genet. Metab. 2005;84:9–17. doi: 10.1016/j.ymgme.2004.08.017. doi:10.1016/j.ymgme.2004.08.017. [DOI] [PubMed] [Google Scholar]
- 48.Erickson R.P., Bernanrd O. Studies on neuronal death in the mouse model of Niemann Pick type C. J. Neurosci. Res. 2002;68:738–744. doi: 10.1002/jnr.10257. doi:10.1002/jnr.10257. [DOI] [PubMed] [Google Scholar]
- 49.Hoyer-Hansen M., Bastholm L., Szyniarowski P., Campanella M., Szabadkai G., Farkas T., Bianchi K., Fehrenbacher N., Elling F., Rizzuto R., et al. Control of macroautophagy by calcium, calmodulin-dependent kinase-beta, and Bcl-2. Mol. Cell. 2007;25:193–205. doi: 10.1016/j.molcel.2006.12.009. doi:10.1016/j.molcel.2006.12.009. [DOI] [PubMed] [Google Scholar]
- 50.Pipalia N.H., Cosner C.C., Huang A., Chatterjee A., Bourbon P., Farley N., Helquist P., Wiest O., Maxfield F.R. Histone deacetylase inhibitor treatment dramatically reduces cholesterol accumulation in Niemann-Pick type C1 mutant human fibroblasts. Proc. Natl Acad. Sci. USA. 2011;108:5620–5625. doi: 10.1073/pnas.1014890108. doi:10.1073/pnas.1014890108. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.