A Drosophila model of myosin-based inclusion body myopathy type 3 is presented. Muscle function, ATPase activity, and actin sliding velocity were dramatically reduced. The mutant myosin is prone to aggregate, likely accounting for the observed cytoplasmic inclusions and disorganized muscle filaments reminiscent of the human disease.
Abstract
Hereditary myosin myopathies are characterized by variable clinical features. Inclusion body myopathy 3 (IBM-3) is an autosomal dominant disease associated with a missense mutation (E706K) in the myosin heavy chain IIa gene. Adult patients experience progressive muscle weakness. Biopsies reveal dystrophic changes, rimmed vacuoles with cytoplasmic inclusions, and focal disorganization of myofilaments. We constructed a transgene encoding E706K myosin and expressed it in Drosophila (E701K) indirect flight and jump muscles to establish a novel homozygous organism with homogeneous populations of fast IBM-3 myosin and muscle fibers. Flight and jump abilities were severely reduced in homozygotes. ATPase and actin sliding velocity of the mutant myosin were depressed >80% compared with wild-type myosin. Light scattering experiments and electron microscopy revealed that mutant myosin heads bear a dramatic propensity to collapse and aggregate. Thus E706K (E701K) myosin appears far more labile than wild-type myosin. Furthermore, mutant fly fibers exhibit ultrastructural hallmarks seen in patients, including cytoplasmic inclusions containing aberrant proteinaceous structures and disorganized muscle filaments. Our Drosophila model reveals the unambiguous consequences of the IBM-3 lesion on fast muscle myosin and fibers. The abnormalities observed in myosin function and muscle ultrastructure likely contribute to muscle weakness observed in our flies and patients.
INTRODUCTION
Myosin is a major structural component of muscle. It is the chemomechanical motor responsible for generating force. Myosin-containing thick filaments interact with F-actin containing thin filaments throughout repetitive ATP-dependent actomyosin contractile cycles. As a result, thin filaments slide relative to thick filaments, resulting in muscle shortening. During filament sliding the subfragment 1 (S-1) head domain of the myosin motor undergoes a series of requisite conformational changes. Defects in myosin can disrupt the structural changes necessary for effective work production. They are known to perturb the motor's ability to interact with F-actin and affect filament movement and muscle contraction, and they can cause human muscle disease (Oldfors et al., 2004; Oldfors, 2007). Deciphering unambiguous molecular consequences of myosin mutations is critical for elucidating pathogenic mechanisms and developing effective therapies to improve muscle performance and disease prognosis.
Autosomal dominant inclusion-body myopathy type 3 (IBM-3) is an inherited muscle disorder caused by an E706K amino acid substitution in the MYH2 myosin gene (Darin et al., 1998; Martinsson et al., 2000). MYH2 is largely expressed in human fast skeletal muscle type IIA. The clinical course of IBM-3 appears nonprogressive in childhood, but most adults experience deterioration of muscle function (Darin et al., 1998; Martinsson et al., 2000). The major histopathological change in skeletal muscle is focal disorganization of myofilaments. In adults with progressive muscle weakness, muscle biopsies reveal dystrophic changes and rimmed vacuoles with cytoplasmic inclusions and accumulation of aberrant proteins (Darin et al., 1998; Martinsson et al., 2000).
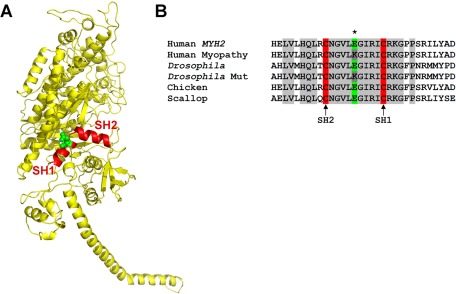
The E706K IBM-3 mutation is localized to the SH1-SH2 α-helix of the myosin head (Figure 1A). The SH1 (Cys-707) and SH2 (Cys-697) residues are the two most reactive cysteines of S-1 and are located at opposite ends of the short, kinked α-helix (Rayment et al., 1993; Bobkova et al., 1999). They are separated by ∼19 Å. The SH1-SH2 helix is believed to play a key role in the conformational changes of myosin during force generation. It is highly flexible at different actomyosin states (Huston et al., 1988), and when the cysteines are cross-linked, S-1 shows weakened ATPase activity and lower actin-binding affinity and exhibits rotational disorder (Thompson et al., 2008). Chemical modification of the helix results in decreased myosin ATPase activity and a loss of the motor function in in vitro motility assays (Bobkova et al., 1999). Finally, mutations in this area affect myosin motor performance (Hu et al., 2002; Kad et al., 2007; Preller et al., 2011) and reduce myosin thermal stability (Iwai et al., 2006). Thus the SH1-SH2 reactive thiol region plays key roles in determining force generation by myosin motors (Preller et al., 2011).
FIGURE 1:
The E706K IBM-3 mutation is localized to the conserved SH1-SH2 α-helix of the myosin head. (A) Ribbon model of MHC subfragment 1 (S-1) of chicken skeletal muscle myosin (PDB: 2MYS). The site of the mutation (E706K) in the SH1 helical region (red) is indicated in green (Martinsson et al., 2000). (B) Sequence alignment reveals conservation of the Glu residue among human, chicken, Drosophila, and scallop myosin isoforms. Transgenic Drosophila were created that express the mutant Lys residue at the corresponding amino acid position, position 701 (dark gray). Accession numbers for aligned sequences are as follows: human,MYH2, Q9UKX2; Drosophila, myosin heavy chain, muscle, P05661; chicken, myosin heavy chain, fast skeletal muscle type IIA, adult, P13538; scallop, myosin heavy chain striated muscle, P24733.
Efforts at understanding the mechanism of autosomal dominant IBM-3 have had limited success. In studies of human biopsies, in vitro motility assays suggested decreased myosin-based motility (Li et al., 2006). However, conclusions were hampered due to poor quality of the tissue, a paucity of material, and the presence of wild-type, nonmutant myosin in the biopsied muscles. Use of a Caenorhabditis elegans body wall muscle model to express an analogous mutant myosin resulted in thick filament assembly along with paralysis, indicating that the mutation does not severely affect myosin assembly properties but did impair motor function (Tajsharghi et al., 2005). However, organisms transformed with extrachromosomal arrays of the wild-type myosin gene sometimes were paralyzed, suggesting that the observed effects may have been due to other factors, such as expression levels. At the molecular level, it was reported that a Dictyostelium nonmuscle myosin containing a mutation analogous to E706K shows decreased actin-activated ATPase and an increased duty ratio, suggesting that the myosin lesion might slow contractile velocity if expressed in muscle (Zeng et al., 2004). Overall, however, the forced expression of the mutant isoform in relatively slow muscle fibers or in nonmuscle myosin backbones can substantially confound data interpretation.
Here we developed the first Drosophila melanogaster model system to study IBM-3. Drosophila is a useful system for investigating myosin myopathies. Its use benefits from well-developed genetic tools, including germline transformation, and, importantly, a single Mhc gene that encodes all muscle myosin heavy chain (MHC) isoforms (Bernstein et al., 1983; Rozek and Davidson, 1983). MHC-null organisms serve as excellent genetic recipients for transgenic expression of myopathic Mhc genes (Swank et al., 2000). Thus we can control for presence of a nonmutated, wild-type allele, other myosin isoforms, and modifier genes. This permits biochemical and ultrastructural analysis of homogeneous myosin isoforms from fast muscle samples. We find that the SH1-SH2 point mutation depresses the chemomechanical properties of purified E706K myosin and increases the structural lability of the motor. In addition, we show that homozygosity results in severe disruption of muscle fiber ultrastructure and production of inclusion bodies, as found in human biopsies. We propose that the chemomechanical and structural perturbations of the mutant motors and tissue are primary contributors to muscle weakness observed in adult IBM-3 patients.
RESULTS
Amino acid alignment shows that the SH1-SH2 helix is 100% conserved in both conventional and unconventional myosins among different species (Figure 1B; Martinsson et al., 2000). The D. melanogaster Mhc gene has 19 exons (George et al., 1989; Swank et al., 2000). The E701 residue that corresponds to E706 in humans is located in constitutive exon 10. A DNA construct containing the entire Drosophila myosin heavy chain gene with the analogous E701K mutation was cloned into a P element vector and microinjected into embryos. We obtained four lines that express the E701K mutation in the Mhc10 MHC-null background. Because Mhc10 flies lack the ability to accumulate MHC isoforms in the indirect flight muscle (IFM) and jump muscle (Collier et al., 1990; Cripps et al., 1994), the introduction of E701K allows for exclusive expression of mutated myosin in these two muscle types. The E701K mutations in lines E701K-3, E701K-5, and E701K-11 were verified via reverse transcriptase-PCR (RT-PCR; see Materials and Methods). A fly line transformed with a wild-type genomic Mhc gene, known as PwMhc2 (Swank et al., 2000), was used as a transgenic control.
The MHC expression levels relative to actin accumulation in the upper thoraces of 2-h-old flies were determined for the transgenic lines via SDS–PAGE. Levels of MHC expression were nearly equivalent with that of the PwMhc2 control in all lines tested except for the E701K-2 line (Table 1). Thus we successfully generated novel fly surrogates that permit integrative analysis of homozygous IBM-3 E701K myosin from the single-molecule through the tissue level.
TABLE 1:
Ratio of MHC to actin expression in 2-h-old transgenic flies relative to control.
| Line | Percentage protein expression ± SEM (n) |
|---|---|
| PwMhc2 (control) | 100 ± 2.4 (6) |
| Mhc10 | 13 ± 3.2 (5) |
| E701K-2 | 82 ± 5.2 (5) |
| E701K-3 | 92 ± 6.3 (5) |
| E701K-5 | 95 ± 4.8 (5) |
| E701K-11 | 89 ± 4.6 (5) |
n, sample number.
To determine muscle function of homozygous E701K flies in the Mhc10 background, we performed flight and jump assays with 2-d-old adult female flies at 22°C. Test results show that homozygous E701K flies completely lack flight ability when compared with the PwMhc2 flies (unpublished data). The E701K flies were unable to beat their wings, and a wings-up phenotype was present in 2-d-old flies, indicating that the IFM might be compromised. The jump ability of young flies was also severely reduced compared with controls (<0.6 vs. 5.4 cm control value), suggesting that the jump muscle might also be seriously impaired by the E701K MHC mutation (Table 2). Therefore homozygous expression of IBM-3 myosin severely compromises the function of two distinct fast muscle types in Drosophila.
TABLE 2:
Jump ability of control and homozygous E701K transgenic flies.
| Line | Jump distance ± SEM (cm) (n) |
|---|---|
| PwMhc2 (control) | 5.4 ± 0.2 (100) |
| E701K-2 | 0.4 ± 0.2 (100)a |
| E701K-3 | 0.3 ± 0.2 (103)a |
| E701K-5 | 0.6 ± 0.1 (112)a |
| E701K-11 | 0.3 ± 0.1 (105)a |
Measurements were done at room temperature (22°C). n, sample number.
aStudent's t tests showed significant differences between jump abilities of PwMhc2 and E701K-2, 3, 5, and 11 (p < 0.001).
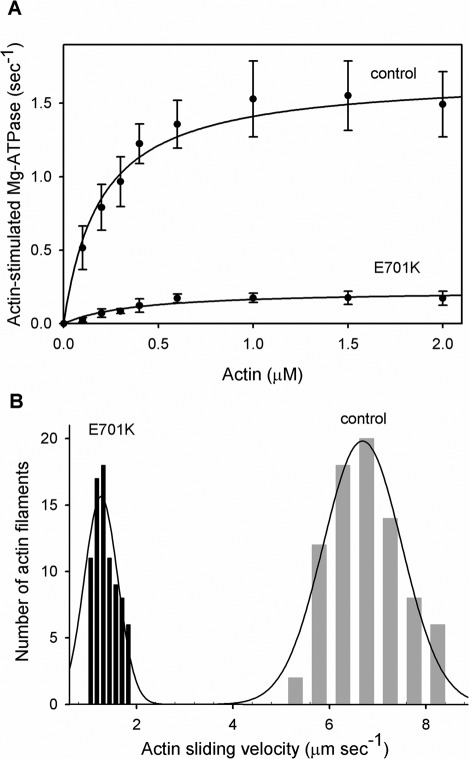
To determine the effects of the IBM-3 mutation on fast muscle myosin molecular performance, we isolated myosin molecules from E701K homozygous IFM and from PwMhc2 control IFM. Myosin CaATPase, basal MgATPase, and actin-stimulated MgATPase activities (Vmax), actin affinity relative to ATPase (Km), and catalytic efficiency (Vmax/Km ratio) of E701K and control myosin are shown in Figure 2A and Table 3. E701K catalytic activity was significantly reduced compared with control myosin. CaATPase activity of E701K myosin was reduced roughly 12-fold compared with control (0.84 ± 0.36 vs. 10.35 ± 1.14 s−1, p < 0.001; Table 3). Similarly, basal MgATPase activity of the mutant showed a 10-fold reduction relative to control myosin (0.03 ± 0.01 vs. 0.25 ± 0.02 s−1, p < 0.001). Actin-stimulated Vmax of E701K was lowered more than eightfold compared with control (0.23 ± 0.06 vs. 1.84 ± 0.32 s−1, p < 0.001). The higher Km value for the mutant myosin represents poorer actin affinity compared with control myosin (0.39 ± 0.11 vs. 0.25 ± 0.09 μM, p < 0.05; Figure 2A and Table 3). The ratio Vmax/Km, a measure of catalytic efficiency, shows a ∼13-fold reduction in the mutant compared with control myosin (0.61 ± 0.16 vs. 7.86 ± 1.84, p < 0.001; Table 3). These results confirm that the IBM-3 mutation induces poor ATPase activity, weak actin affinity, and significant reduction in catalytic efficiency in a fast skeletal myosin isoform.
FIGURE 2:
Actin-stimulated MgATPase activity and actin sliding velocity of control (PwMhc2) and IBM-3 mutant (E701K) myosin. (A) ATPase activity of control and mutant myosin was determined in the absence and in the presence of F-actin. For the determination of Vmax and Km (Table 3), basal Mg-ATPase activities obtained in the absence of actin were subtracted from all data points. The data were fitted with the Michaelis–Menten equation to determine Vmax and Km (actin). The ratio of Vmax/Km from all assays was used to determine catalytic efficiency. CaATPase, MgATPase, actin-stimulated MgATPase activity, and the Vmax/Km ratio were significantly reduced (p < 0.001), whereas Km was significantly enhanced (p < 0.05) in mutant relative to control myosin (Table 3). (B) Actin sliding velocity stimulated by IBM-3 mutant (E701K) myosin was significantly reduced (p < 0.001) compared with that of control myosin. Our data suggest that the IBM-3 mutation depresses the enzymatic activity of the molecular motor.
TABLE 3:
Chemomechanical properties of PwMhc2 (control) and mutant (E701K) myosin.
| Myosin type | CaATPase ± SD (s−1) | Basal MgATPase ± SD (s−1) | Actin-stimulated Vmax ± SD (s−1) | Km (actin) ± SD (μM) | Catalytic efficiency Vmax/Km ± SD | Actin sliding velocity ± SD (μm s−1) |
|---|---|---|---|---|---|---|
| PwMhc2 (control) | 10.35 ± 1.14 (n = 8) | 0.25 ± 0.02 (n = 8) | 1.84 ± 0.32 (n = 8) | 0.25 ± 0.09 (n = 8) | 7.86 ± 1.84 (n = 8) | 6.67 ± 0.97 (n = 6) |
| E701K-5 | 0.84 ± 0.36a (n = 5) | 0.03 ± 0.01a (n = 5) | 0.23 ± 0.06a (n = 5) | 0.39 ± 0.11b (n = 5) | 0.61 ± 0.16a (n = 5) | 1.34 ± 0.05a (n = 4) |
aSignificantly different from control (p < 0.001).
bSignificantly different from control (p < 0.05).
In vitro motility assays were used to examine the unloaded sliding velocity of fluorescently labeled actin filaments generated by E701K and by PwMhc2 wild-type control myosin. On the basis of four independent preparations, we found that actin sliding velocity stimulated by E701K myosin was significantly decreased by 80% when compared with the control. The PwMhc2 myosin moves actin filaments at a velocity of 6.67 ± 0.97 μm s−1 at 22°C (Figure 2B and Table 3). In contrast, E701K myosin drives actin filament movement at a rate of 1.34 ± 0.05 μm s−1 (Figure 2B and Table 3). These results illustrate that the IBM-3 myosin mutation depresses fast skeletal muscle myosin motility, possibly as a result of poor ATPase activity and weak actin affinity.
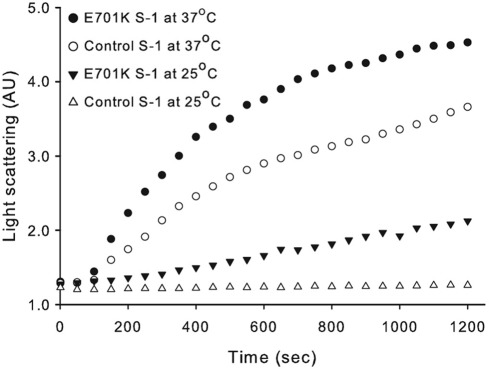
Because inclusion bodies with aggregated proteinaceous structures are hallmarks of the human IBM-3 phenotype, we wanted to assess E701K myosin's aggregation-prone nature. We examined the aggregation propensity of IBM-3 myosin S-1 by evaluating increased light scattering at 25 and at 37°C over time (Figure 3); 37°C is the Drosophila heat shock temperature, as compared with the human heat shock temperature of 43°C (Lindquist, 1986). The results reveal an increased propensity for mutant myosin aggregation. The elevated aggregation kinetics and increased aggregation detected for IBM-3 myosin at 25 and at 37°C suggest enhanced lability of S-1 as a result of the IBM-3 SH1-SH2 helix lesion.
FIGURE 3:
Aggregation kinetics of control (PwMhc2) and IBM-3 (E701K) myosin S-1 fragment as detected by light scattering. The top two curves represent light scattering of S-1 prepared from the IBM-3 mutant (E701K) and PwMhc2 control myosin at 37°C, respectively. Second from bottom and bottom curves represent light scattering of S-1 prepared from the IBM-3 mutant (E701K) and control myosin at 25°C, respectively. These data suggest that the mutant S-1 has a higher propensity to aggregate at both room and elevated temperatures relative to control S-1.
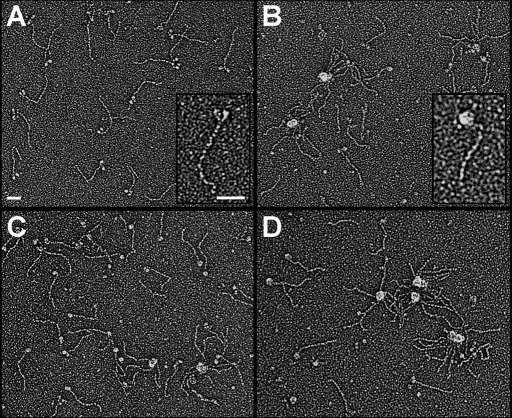
To directly determine whether the IBM-3 mutation alters the normal two-headed myosin structure exhibited by wild-type molecules, we generated electron micrographs of rotary shadowed control and E701K myosin maintained at 23°C and after a 4-min incubation at 37°C (Figure 4). PwMhc2 myosin showed typical two-headed structures connected by a long, ∼150-nm tail segment at 23°C. However, at the same temperature, E701K molecules often showed collapsed heads that frequently packed into aggregated clumps reminiscent of wild-type motors exposed to elevated temperatures. Discrete S-1 heads of a given mutant molecule were seldom resolved. Two-headed structures were rarely observed when either PwMhc2 or E701K myosin was briefly incubated at 37°C. We quantitatively assessed the lability of myosin in micrographs by counting molecules at each temperature that appeared to have two well-resolved heads and compared the number of molecules that formed intramolecular or intermolecular aggregates (Table 4). At either temperature, PwMhc2 myosin exhibited a substantially higher percentage of molecules with normal structure. The finding that at 23°C only 22.5% of IBM-3 myosin appeared normal suggests that these molecules are inherently less stable than control molecules. To our knowledge, this is the first direct structural evidence of altered myosin stability resulting from an SH1-SH2 mutation.
FIGURE 4:
The IBM-3 myosin mutation increases the molecular motor's structural lability. To directly determine whether the IBM-3 mutation alters the normal two-headed myosin structure exhibited by rotary shadowed wild-type molecules, we generated electron micrographs of PwMhc2 control and E701K myosin maintained at 23°C and after a 4-min incubation at 37°C. PwMhc2 myosin showed typical two-headed structures connected by a long, ∼150-nm tail segment (A). However, at the same temperature, E701K molecules often showed collapsed heads, and they frequently packed into aggregated clumps (B) reminiscent of wild-type motors exposed to elevated temperatures (C), where discrete S-1 heads of a given molecule were seldom resolved. Two-headed structures were rarely observed when either PwMhc2 or E701K myosin was briefly incubated at 37°C. (C, D). Insets show enlarged, individual PwMhc2 (A) or E701K (B) myosin at 23°C and clearly demonstrate intramolecular head aggregation of the mutant molecule. Bars, 50 nm.
TABLE 4:
Effect of temperature on myosin head morphology.
| Myosin type | Temperature (ºC) | Percentage of molecules exhibiting well-resolved heads (n) | Percentage of molecules aggregated |
|---|---|---|---|
| PwMhc2 | 23 | 77.5 (1146) | 22.5 |
| E701K-5 | 23 | 22.5 (1041) | 77.5 |
| PwMhc2 | 37 | 19.1 (1181) | 80.9 |
| E701K-5 | 37 | 4.7 (1192) | 95.3 |
n, number of molecules counted for each condition.
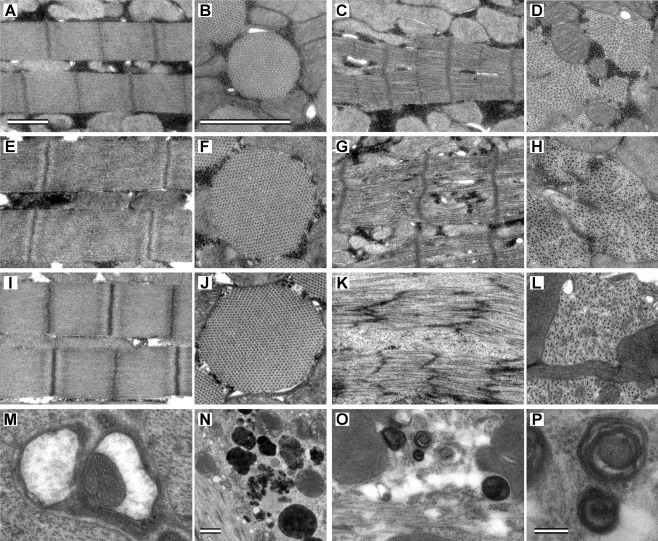
To address whether expression of E701K myosin produced ultrastructural defects, the IFMs of late-stage pupae, young (∼2 h old), and 2-d-old adults were examined by transmission electron microscopy and compared with those of the PwMhc2 transgenic control flies (Figure 5). The myofibril ultrastructure of PwMhc2 shows wild-type hexagonal packing of thick and thin filaments containing well-organized Z-bands at each age (Figure 5, A, B, E, F, I, and J). Although M-lines and Z-disks are distinguishable, E701K pupal myofibrils show disruption of integrity and lack round shape (Figure 5, C and D). Mutant myofibrillar structure becomes progressively worse with age. Young adult E701K myofibrils show a loss of M-lines, with further decreases in myofibril integrity and hexagonal packing (Figure 5, G and H). Myofibril structure of 2-d-old E701K shows severe ultrastructural deterioration, with broken and streaming Z-disks and a loss of thick filaments and M-lines (Figure 5, K and L). E701K fibers display sarcolemmal membrane invaginations and protrusions. Thus the E701K mutation in the myosin SH1-SH2 helix domain severely affects IFM myofibril assembly and stability, which likely contribute to the flightless phenotype.
FIGURE 5:
Ultrastructure of PwMhc2 control and E701K mutant IFM over time. Longitudinal sections through pupal PwMhc2 (A) and E701K IFM (C). Control sarcomeres are highly organized, showing a characteristic pattern of straight myofilaments running parallel to each other and perpendicular to straight M-lines surrounded by denser-staining Z-disks. Mutant myofibrils show assembly defects. E701K sarcomeres are less organized than control, display cracking between myofilaments, and have jagged M-lines and Z-disks. Transverse sections through pupal PwMhc2 (B) and E701K (D) IFM myofibrils. Control myofibrils are round, with a consistent pattern of hexagonally packed hollow thick filaments each surrounded by six thin filaments. Mutant myofibrils lack round shape and show disrupted hexagonal packing between thick and thin filaments. Bars, 1 μm in A and B. Longitudinal sections through 2-h-old adult PwMhc2 (E) and E701K (G) IFM. Mutant sarcomeres lack visible M-lines and are disorganized. Myofilaments are losing their parallel relationship to one another and appear broken. Transverse sections through 2-h-old adult PwMhc2 (F) and E701K (H) IFM myofibrils. Thin and thick filaments are dissociated in approximately half of the myofibril surface area. Longitudinal sections through 2-d-old adult PwMhc2 (I) and E701K (K, M–P) IFM. Sarcomeric structure in mutants is nearly absent. E701K myofibrils show Z-disk streaming and loss of Z-disk spacing and thick filaments. Thin filaments were observed in the sarcoplasm in the absence of any sarcomeric structures. Transverse sections through 2-d-old adult PwMhc2 (J) and E701K (L) IFM myofibrils. Hexagonal packing is absent in mutants; sarcoplasmic reticulum is broken down into vesicles surrounded by a soup of thick and thin filaments. Mutant fibers also display rimmed vacuoles fused with additional membranes (M, N), suggesting the formation of type 2 autophagic vacuoles (Fernandez et al., 2005). Bar, 1 μm in N. Cylindrical spiral membranes (O, P) found in E701K fibers appear similar to sarcolemmal inclusions in human skeletal myopathy (Carpenter and Karpati, 1979). Bar, 0.1 μm in P. Magnification is the same in A, C, E, G, I, K, and O. Magnification is the same in B, D, F, H, J, L, and M.
Most interestingly, we observed abnormal vacuoles in various sizes and shapes in young and older E701K flies (Figure 5). Mitochondrial structures were observed inside some of the vacuoles of older E701K-5 flies (Figure 5, M–P). Cylindrical spiral membranes found in E701K fibers appear similar to sarcolemmal inclusions in human skeletal myopathy (Carpenter et al., 1979). Mutant fibers also display rimmed vacuoles fused with additional membranes, suggesting the formation of type 2 autophagic vacuoles (Fernandez et al., 2005). Such vesicles were not reported in the C. elegans IBM-3 model (Tajsharghi et al., 2005). These findings imply an evolutionarily well-conserved pathogenic reaction to defective fast skeletal muscle myosin.
DISCUSSION
Autosomal dominant IBM-3 is an inherited muscle disorder caused by a point mutation in the SH1-SH2 helical region of the MYH2 myosin head (Darin et al., 1998; Martinsson et al., 2000). Adult patients experience deterioration of muscle function with focal disorganization of myofilaments, dystrophic changes, and rimmed vacuoles with cytoplasmic inclusions and accumulation of aberrant proteins. Here we developed a novel viable Drosophila model system to investigate unambiguous homozygous effects of the myopathic myosin on fast skeletal muscles. We demonstrated that the mutant myosin severely compromises the structure and function of the IFM and jump muscles, two distinct fast muscle types in Drosophila. Pure populations of IBM-3 myosin display poor enzymatic behavior, with a dramatic reduction in catalytic efficiency and decreased in vitro F-actin sliding speeds. Our studies corroborate and extend previous efforts, which used material from IBM-3 biopsies (Li et al., 2006) and from Dictyostelium (Zeng et al., 2004) and C. elegans (Tajsharghi et al., 2005) models to define the effects of the mutation on motor performance. Overall, studies of IBM-3 myosin highlight the critical role the mutated residue of the SH1-SH2 helix plays during the myosin ATPase cycle. The importance of this domain is further documented by studies showing that perturbations in regions near the IBM-3 locus also depress the chemomechanical activity of the myosin motor (Bobkova et al., 1999; Hu et al., 2002; Iwai et al., 2006; Thompson et al., 2008).
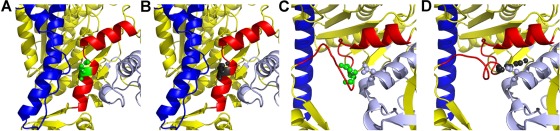
The SH1-SH2 helix that contains the E701K mutation is a critical element of the chemomechanical cycle of myosin–actin interaction (Bobkova et al., 1999). Recent crystallographic evidence reveals extensive, intimate interactions that couple the reactive thiol regions of the SH1-SH2 helix to surrounding structural elements of the myosin head (Preller et al., 2011). Earlier findings suggest that melting of the helix after nucleotide binding uncouples the linkage of the myosin motor domain from the myosin converter domain and its adjacent lever arm, which allows the lever arm to swing while actin is bound, thereby generating power for muscle contraction (Gourinath et al., 2003). Molecular modeling of the IBM-3 mutation in pre–power stroke and post–power stroke states (Figure 6) indicates that the mutation alters the orientation of the melted SH1-SH2 helix in the post–power stroke state, allowing interaction with the N-terminal portion of the protein. This abnormal interaction may prevent reformation of the SH1-SH2 helix and cause the defects in ATPase function and in vitro motility described. Furthermore, consistent with our single-molecule electron microscopy and protein aggregation results, increased propensity for helix melting might reduce overall structural integrity and stability of the mutant myosin.
FIGURE 6:
Modeling of wild-type (Glu-706) and IBM-3 mutant (Lys-706) myosin (A) In the pre–power stroke state (PDB: 1QVI), Glu-706 (green) is part of the SH1-SH2 helix of myosin (red). For orientation the relay helix is shown in blue (B) Substitution of the IBM-3 mutation (Lys-706, gray spheres) does not have a major effect on the pre–power stroke structure (C) The wild-type SH1 helix containing Glu-706 (green spheres) melts during the power stroke, as shown here (red) in the post–power stroke state (PDB: 1KK8). (D) The IBM-3 mutation (Lys-706; gray spheres) is predicted to alter the orientation of the melted helix (red) in the post–power stroke state, bringing it in contact (2.8 Å) with Asp-98 (light blue spheres) in the N-terminal region. This new charge-based interaction may prevent reformation of the SH1 helical region of the SH1-SH2 helix after ATP binding.
Our imaging results of single molecules, myofibrils, and muscle fibers provide unparalleled direct structural evidence of altered myosin and myofibrillar stability and the presence of vacuolar structures similar to those seen in the human disease resulting from the IBM-3 SH1-SH2 mutation. The major histopathological change in IBM-3 skeletal muscle is disorganization of myofilaments (Darin et al., 1998; Martinsson et al., 2000). Similarly, Drosophila E701K sarcomeres are less organized than control, display cracks between myofilaments, and have disrupted M- and Z-lines. The mutant myofibrils lack round shape, are disorganized, and exhibit perturbed hexagonal packing between myofilaments. Thus the depressed muscle performance in the IBM-3 Drosophila model likely results from impaired motor function, increased myosin lability, and ultrastructural abnormalities similar to those found in human skeletal myopathy. It is important to note that the aggregation-prone nature of the mutant myosin correlates with the formation of the membrane-bound vesicles. Such vesicles are commonly seen during autophagic, proteosomal, and/or ubiquitination processes, and their presence suggests a possible link between mutant motor misfolding and degradation responses (Kumamoto et al., 1998; Eble et al., 1999; Kimura et al., 2009).
In most instances IBM-3 has a mild phenotypic expression in children but progressive course in some adults. There is a relationship between the level of expression of mutated MYH2 and muscle pathology (Tajsharghi et al., 2002; Oldfors et al., 2004). Thus, as mutant MYH2 expression increases with age, there is an expected increase in mutant myosin content with depressed motor properties and stability, as well as elevated myosin aggregates and membrane-bound inclusions. Together these molecular consequences are presumed to promote muscle debility in a dose-dependent manner. It should be noted, however, that humans express other fast myosin isoforms that may partially compensate for the IBM-3 mutation, which is unlike the situation in Drosophila.
Our Drosophila model is the first to show the unambiguous consequences of the E706K mutation on fast muscle myosin performance and myosin structure. In addition, it is the first to faithfully reproduce membrane-bound inclusions. These potentially conserved responses and disease traits suggest the Drosophila model should serve as an effective tool to better understand the molecular composition and function of these vacuolar structures. Purification and characterization of the molecular constituents of the inclusions will permit identification of possible therapeutic targets to alleviate the development and accumulation of these myopathic hallmarks. Furthermore, our ongoing characterization of the effects of heterozygous IBM-3 myosin expression on muscle function in flies will provide insights into the confounding effects of mixed populations of wild-type and mutant molecular motors and into potential dose-dependent effects of mutant myosin expression. Overall, our model reveals insights into mechanisms of muscle weakness that may be relevant to human IBM-3, which include depressed myosin performance and stability, myofilamentous disorganization, and sarcoplasmic inclusions.
MATERIALS AND METHODS
Creation of the E701K construct
The E701K mutant Mhc gene was generated via site-directed mutagenesis. D. melanogaster has a single muscle myosin heavy chain gene. The construct containing the entire D. melanogaster Mhc gene with the E701K mutation was cloned into a P element vector with the miniwhite gene, w+, as a selectable eye-color marker (Swank et al., 2000). Construction of the mutant myosin gene was initiated by subcloning a 0.6-kb BamHI–EagI fragment that includes constitutive exon 10 (which encodes residue 701). Site-directed mutagenesis was performed on the subclone using Stratagene's QuikChange II Kit (Stratagene, La Jolla, CA). On sequence confirmation of the E701K site-directed mutagenesis product, the mutated subclone fragment was inserted to replace the wild-type version in a 2.5-kb PstI–EagI subclone containing the exon 9–11 region. The resulting subclone was cut with EagI and replaced back into the wild-type construct PwMhc2 at its EagI site. The resulting clone was digested with EagI, and the 12-kb fragment of the 3′ end of the Mhc gene was removed from its vector by EagI digestion and inserted into the linearized 5′ fragment using DNA ligase. Ligation sites were confirmed by DNA sequencing, as were all splice junctions and coding regions of the final E701K plasmid. The E701K construct was purified using QIAfilter Plasmid Maxi Kit (Qiagen, Valencia, CA) according to the manufacturer's instructions.
Drosophila transformation
BestGene (Chino Hills, CA) produced transgenic lines by P element–mediated germline transformation (Rubin and Spradling, 1982). The E701K plasmid, along with the helper plasmid Δ2-3, was injected into embryos (generation G0) collected from wild-type yw Drosophila. G0 larvae were raised in vials and crossed with yw flies. The offspring (G1 generation) were screened for orange eye color, indicating the presence of the miniwhite (w+) marker and E701K transgene. A total of 2200 embryos were injected, resulting in 15 viable lines.
Drosophila genetic crosses
Transformant lines were crossed with w1118; CyO/Bl1; TM2/TM6B balancer flies to map the chromosomal location of the transgene of each line using standard genetic techniques. Among the 15 lines, three lines mapped to the X chromosome and three lines mapped to the third chromosome. Lines containing insertions on the X and third chromosome were subsequently crossed into the Mhc10 background, a null allele for the indirect flight and jump muscle myosin isoforms (Collier et al., 1990). Two of the X-linked lines were homozygous lethal in the Mhc10 background. Therefore four viable homozygous E701K lines in the Mhc10 background—named E701K-2, E701K-3, E701K-5, and E701K-11—and the PwMhc2 control fly line (Swank et al., 2000) were used in all further studies. The presence of the mutant transcript in the transgenic lines was verified by RT-PCR.
Quantification of transgene expression levels
Protein expression levels of transgenes were determined by SDS–PAGE (Laemmli, 1970). Six upper thoraces from 2-h-old virgin female flies from each transgenic line were individually homogenized in 30 μl of Laemmli sample buffer. PwMhc2 flies were used as a control. Samples were run on 10% polyacrylamide Tris-glycine gels (Bio-Rad, Hercules, CA). Gels were stained with Coomassie blue, destained, scanned, and analyzed using ImageJ (National Institutes of Health, Bethesda, MD). The myosin-to-actin ratio for each sample was determined and expressed in relative proportions to the average myosin-to-actin ratio for control PwMhc2 samples.
Molecular modeling
Modeling of the MHC subfragment 1 (S-1) of chicken skeletal muscle myosin (Protein Data Bank [PDB]: 2MYS) and of scallop S-1 in the pre–power stroke state (PDB: 1QVI) or in the post–power stroke state (PDB: 1KK8) was performed using the Swiss-Model homology modeling server (http://swissmodel.expasy.org). PyMOL (www.pymol.org) was used to visualize output.
Jump and flight tests
Flight tests (Drummond et al., 1991) were performed at room temperature. Individual adult flies were released into the center of a Plexiglas box (43 cm high × 27.5 cm wide × 43 cm long), with a light source positioned at the top. Each fly was assigned a flight index (FI) value based on its ability to fly upward (6.0), horizontally (4.0), downward (2.0), or not at all (0.0). The average flight index was calculated by dividing the sum of the individual FI values by the number of individuals tested for each line. Jump testing was performed as previously described (Swank et al., 2002). Newly eclosed flies had their wings removed and were placed into glass vials for 2 d at 25°C. Individual flies were persuaded with a paint brush to jump from the edge of a 9.5-cm-tall inverted vial onto a piece of paper with concentric rings printed 0.5 cm apart. The three farthest jump distances out of 10 trials per fly were averaged for 50 flies per genotype.
Myosin isolation and purification
Myosin was purified from dorsolongitudinal IFMs (Swank et al., 2000). These fibers were isolated from >150 young female transgenic flies and incubated in a glycerol and 2% Triton X-100 solution. Myosin was extracted and purified by a series of high-salt suspensions and low-salt precipitations (Swank et al., 2001). The protein concentration was determined by the absorbance of 280 nm (1 OD280-310/0.53 = 1 mg/ml; Margossian and Lowey, 1982). Single-molecule imaging, ATPase, and in vitro motility assays were performed immediately after myosin preparation.
Actin preparation
G-actin was isolated from acetone powder of chicken skeletal muscle as recently reported (Swank et al., 2001). Briefly, after multiple cycles of polymerization–depolymerization, soluble G-actin was obtained after dialysis against 2 mM Tris-Cl (pH 8), 0.2 mM ATP, 2 mM CaCl2, and 1 mM dithiothreitol (DTT) and quantified spectrophotometrically using an extinction coefficient of 0.62 cm−1 (A310nm − A290nm) for 1 mg of actin ml−1. F-Actin was prepared by adding 1 volume of 10× polymerization buffer (50 mM Tris-Cl, pH 8, 0.5 mM KCl, 20 mM MgCl2, and 10 mM ATP) to 9 volumes of G-actin. Actin was stored on ice at 4°C and used for actin-activated Mg2+-ATPase assays. For in vitro motility assays, G-actin was polymerized and labeled with rhodamine-phalloidin before analysis.
Determination of catalytic activity of control and mutant (E701K) myosin
ATPase activities of control and E701K mutant myosin were determined using [γ-32P]ATP as previously described in detail (Swank et al., 2001; Kronert et al., 2010). Briefly, 1–2 μg of freshly prepared myosin was added to CaATPase buffer (10 mM imidazole pH 6.0, 0.1 M KCl, 10 mM CaCl2). CaATPase activity of myosin was initiated at 25°C by addition of [γ-32P]ATP. The reaction was quenched after incubation for 15 min by addition of 1.8 M HClO4. ATPase measurements were performed as reported (Swank et al., 2001). Basal MgATPase was carried out using MgATPase buffer (10 mM imidazole, pH 6.0, 20 mM KCl, 2 mM MgCl2, 0.1 mM CaCl2, and 1mM [γ-32P]ATP). Actin-stimulated MgATPase was performed in MgATPase buffer in the presence of increasing concentrations of F-actin (0.1–2 μM). After 25 min the reaction was quenched using 1.8 M HClO4. Basal MgATPase activities obtained in the absence of actin were subtracted from all data points. Mean ± SD of CaATPase or MgATPase from at least five experiments was used to determine average ATPase activities. Vmax and actin affinity (Km) were obtained by fitting all data points from several preparations of control and mutant myosin with the Michaelis–Menten equation using SigmaPlot software (Systat, San Jose, CA). Catalytic efficiency of control and E701K myosin was determined from the Vmax/Km ratio as recently described for myosin S-1 (Bloemink et al., 2011). Values were averaged to give mean ± SD. Statistical comparisons of ATPase data were carried out using Student's t tests.
Determination of in vitro motility
In vitro actin sliding velocity assays were carried out as previously described (Swank et al., 2001; Kronert et al., 2010). At least four sets of assays were performed for each isoform studied. Analysis of captured video sequences was performed as described (Swank et al., 2001; Kronert et al., 2010). Distribution curves for control and mutant myosin sliding velocity were generated using SigmaPlot. Statistical differences of actin sliding velocity between IBM-3 mutant and IFI myosin were determined using Student's t tests.
Light scattering of IBM-3 mutant (E701K) and IFI myosin subfragment-1
S-1 fragments of control and IBM-3 mutant (E701K) myosin were produced by proteolysis with chymotrypsin as previously reported (Miller et al., 2003). Aggregation kinetics of IBM-3 mutant (E701K) and IFI myosin S-1 at 37°C were measured with light scattering as previously described (Melkani et al., 2010). Briefly, light scattering of S-1 (0.01 mg/ml) in low-salt buffer (30 mM KCl, 5 mM MgCl2, 20 mM 3-(N-morpholino)propanesulfonic acid, pH 7.0, and 4 mM DTT) was measured at 25 or 37°C using a PTI spectrofluorometer equipped with a temperature-controlled cell holder (Photon Technology International, Birmingham, NJ). The excitation and emission wavelengths were 360 nm.
Myosin molecular and IFM ultrastructure
Electron microscopy of myosin was carried out by modification of the methods previously described (Suggs et al., 2007). The samples were examined at a magnification of 15,000× with an FEI Tecnai 12 transmission electron microscope (FEI, Hillsboro, OR) operating at 120 kV. Digital images were taken with a TVIPS 214 high-resolution digital camera (Tietz Video and Image Processing Systems, Gauting, Germany) and imaged in reverse contrast. Structural analysis consisted of counting particles that resembled normal two-headed myosin molecules at room temperature or at elevated temperatures. The number of molecules that showed intermolecular or intramolecular associations at each temperature was distinguished from the molecules that did not show such associations.
Thoraces from different age female E701K flies (late-stage pupae, <1 d old, and 2 d old) were isolated and prepared for transmission electron microscopy by adapting previously published protocols (Reedy and Beall, 1993a, b). The upper thorax tissue was fixed in 3% glutaraldehyde and 3% paraformaldehyde in 100 mM sodium cacodylate buffer, pH 7.4, on ice for 2 d. Thoraces were washed several times with 100 mM sodium phosphate buffer, pH 6.1, and fixed secondarily in 1% osmium tetroxide and 10 mM MgCl2 in 100 mM sodium phosphate buffer, pH 6.1, for 2 h on ice in a dark room. Samples were washed several times with water and dehydrated with a series of washes containing increasing concentrations of acetone, from 50, 75, and 90 to 100% acetone. Samples were then infiltrated with Embed812 resin (Electron Microscopy Science, Hatfield, PA) by incubation in resin–acetone mixtures with increasing concentrations of resin from 50 to 100%. Samples were placed into molds with fresh resin, degassed overnight under vacuum at room temperature, oriented, and then heated in a 60°C oven under vacuum for 24–48 h to polymerize the resin. Thin sections from samples were collected and stained with uranyl acetate and lead citrate. Images were collected on a Tecnai 12 transmission electron microscope operating at 120 kV using a TVIPS 214 high-resolution digital camera.
Acknowledgments
We appreciate the assistance of Adam Bialobrodski in the analysis of myosin molecules viewed by electron microscopy. This work was supported by Muscular Dystrophy Association Research Grant 68954 and National Institutes of Health Grant R01GM32243 to S.I.B. A.C. was funded by an American Heart Association Western States Affiliate Postdoctoral Fellowship and American Heart Association 10SDG4180089.
Abbreviations used:
- FI
flight index
- IBM-3
inclusion body myopathy type 3
- IFM
indirect flight muscle
- MHC
myosin heavy chain
- RT-PCR
reverse transcriptase-PCR
- SH
sulfhydryl
- S-1
subfragment 1
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E12-02-0120) on April 11, 2012.
REFERENCES
- Bernstein SI, Mogami K, Donady JJ, Emerson CP., Jr Drosophila muscle myosin heavy chain encoded by a single gene in a cluster of muscle mutations. Nature. 1983;302:393–397. doi: 10.1038/302393a0. [DOI] [PubMed] [Google Scholar]
- Bloemink MJ, Melkani GC, Dambacher CM, Bernstein SI, Geeves MA. Two Drosophila myosin transducer mutants with distinct cardiomyopathies have divergent ADP and actin affinities. J Biol Chem. 2011;286:28435–28443. doi: 10.1074/jbc.M111.258228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bobkova EA, Bobkov AA, Levitsky DI, Reisler E. Effects of SH1 and SH2 modifications on myosin: similarities and differences. Biophys J. 1999;76:1001–1007. doi: 10.1016/S0006-3495(99)77264-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carpenter S, Karpati G. Duchenne muscular dystrophy: plasma membrane loss initiates muscle cell necrosis unless it is repaired. Brain. 1979;102:147–161. doi: 10.1093/brain/102.1.147. [DOI] [PubMed] [Google Scholar]
- Carpenter S, Karpati G, Robitaille Y, Melmed C. Cylindrical spirals in human skeletal muscle. Muscle Nerve. 1979;2:282–287. doi: 10.1002/mus.880020407. [DOI] [PubMed] [Google Scholar]
- Collier VL, Kronert WA, O'Donnell PT, Edwards KA, Bernstein SI. Alternative myosin hinge regions are utilized in a tissue-specific fashion that correlates with muscle contraction speed. Genes Dev. 1990;4:885–895. doi: 10.1101/gad.4.6.885. [DOI] [PubMed] [Google Scholar]
- Cripps RM, Becker KD, Mardahl M, Kronert WA, Hodges D, Bernstein SI. Transformation of Drosophila melanogaster with the wild-type myosin heavy-chain gene: rescue of mutant phenotypes and analysis of defects caused by overexpression. J Cell Biol. 1994;126:689–699. doi: 10.1083/jcb.126.3.689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darin N, Kyllerman M, Wahlstrom J, Martinsson T, Oldfors A. Autosomal dominant myopathy with congenital joint contractures, ophthalmoplegia, and rimmed vacuoles. Ann Neurol. 1998;44:242–248. doi: 10.1002/ana.410440215. [DOI] [PubMed] [Google Scholar]
- Drummond DR, Hennessey ES, Sparrow JC. Characterisation of missense mutations in the Act88F gene of Drosophila melanogaster. Mol Gen Genet. 1991;226:70–80. doi: 10.1007/BF00273589. [DOI] [PubMed] [Google Scholar]
- Eble DM, Spragia ML, Ferguson AG, Samarel AM. Sarcomeric myosin heavy chain is degraded by the proteasome. Cell Tissue Res. 1999;296:541–548. doi: 10.1007/s004410051315. [DOI] [PubMed] [Google Scholar]
- Fernandez C, Figarella-Branger D, Meyronet D, Cassote E, Tong S, Pellissier JF. Electron microscopy in neuromuscular disorders. Ultrastruct Pathol. 2005;29:437–450. doi: 10.1080/01913120500323175. [DOI] [PubMed] [Google Scholar]
- George EL, Ober MB, Emerson CP., Jr Functional domains of the Drosophila melanogaster muscle myosin heavy-chain gene are encoded by alternatively spliced exons. Mol Cell Biol. 1989;9:2957–2974. doi: 10.1128/mcb.9.7.2957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gourinath S, Himmel DM, Brown JH, Reshetnikova L, Szent-Gyorgyi AG, Cohen C. Crystal structure of scallop myosin S1 in the pre-power stroke state to 2.6 Å resolution: flexibility and function in the head. Structure. 2003;11:1621–1627. doi: 10.1016/j.str.2003.10.013. [DOI] [PubMed] [Google Scholar]
- Hu A, Wang F, Sellers JR. Mutations in human nonmuscle myosin IIA found in patients with May-Hegglin anomaly and Fechtner syndrome result in impaired enzymatic function. J Biol Chem. 2002;277:46512–46517. doi: 10.1074/jbc.M208506200. [DOI] [PubMed] [Google Scholar]
- Huston EE, Grammer JC, Yount RG. Flexibility of the myosin heavy chain: direct evidence that the region containing SH1 and SH2 can move 10 Å under the influence of nucleotide binding. Biochemistry. 1988;27:8945–8952. doi: 10.1021/bi00425a011. [DOI] [PubMed] [Google Scholar]
- Iwai S, Hanamoto D, Chaen S. A point mutation in the SH1 helix alters elasticity and thermal stability of myosin II. J Biol Chem. 2006;281:30736–30744. doi: 10.1074/jbc.M605365200. [DOI] [PubMed] [Google Scholar]
- Kad NM, Patlak JB, Fagnant PM, Trybus KM, Warshaw DM. Mutation of a conserved glycine in the SH1-SH2 helix affects the load-dependent kinetics of myosin. Biophys J. 2007;92:1623–1631. doi: 10.1529/biophysj.106.097618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura N, Kumamoto T, Oniki T, Nomura M, Nakamura K, Abe Y, Hazama Y, Ueyama H. Role of ubiquitin-proteasome proteolysis in muscle fiber destruction in experimental chloroquine-induced myopathy. Muscle Nerve. 2009;39:521–528. doi: 10.1002/mus.21223. [DOI] [PubMed] [Google Scholar]
- Kronert WA, Melkani GC, Melkani A, Bernstein SI. Mutating the converter-relay interface of Drosophila myosin perturbs ATPase activity, actin motility, myofibril stability and flight ability. J Mol Biol. 2010;398:625–632. doi: 10.1016/j.jmb.2010.03.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumamoto T, Fujimoto S, Nagao S, Masuda T, Sugihara R, Ueyama H, Tsuda T. Proteasomes in distal myopathy with rimmed vacuoles. Intern Med. 1998;37:746–752. doi: 10.2169/internalmedicine.37.746. [DOI] [PubMed] [Google Scholar]
- Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Li M, Lionikas A, Yu F, Tajsharghi H, Oldfors A, Larsson L. Muscle cell and motor protein function in patients with a IIa myosin missense mutation (Glu-706 to Lys) Neuromuscul Disord. 2006;16:782–791. doi: 10.1016/j.nmd.2006.07.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindquist S. The heat-shock response. Annu Rev Biochem. 1986;55:1151–1191. doi: 10.1146/annurev.bi.55.070186.005443. [DOI] [PubMed] [Google Scholar]
- Margossian SS, Lowey S. Structural and contractile proteins. In: Cunningham LW, Frederiksen DW, editors. Methods in Enzymology. New York: Academic Press; 1982. pp. 55–70. 85. [Google Scholar]
- Martinsson T, Oldfors A, Darin N, Berg K, Tajsharghi H, Kyllerman M, Wahlstrom J. Autosomal dominant myopathy: missense mutation (Glu-706 → Lys) in the myosin heavy chain IIa gene. Proc Natl Acad Sci USA. 2000;97:14614–14619. doi: 10.1073/pnas.250289597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melkani GC, Lee CF, Cammarato A, Bernstein SI. Drosophila UNC-45 prevents heat-induced aggregation of skeletal muscle myosin and facilitates refolding of citrate synthase. Biochem Biophys Res Commun. 2010;396:317–322. doi: 10.1016/j.bbrc.2010.04.090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller BM, Nyitrai M, Bernstein SI, Geeves MA. Kinetic analysis of Drosophila muscle myosin isoforms suggests a novel mode of mechanochemical coupling. J Biol Chem. 2003;278:50293–50300. doi: 10.1074/jbc.M308318200. [DOI] [PubMed] [Google Scholar]
- Oldfors A. Hereditary myosin myopathies. Neuromuscul Disord. 2007;17:355–367. doi: 10.1016/j.nmd.2007.02.008. [DOI] [PubMed] [Google Scholar]
- Oldfors A, Tajsharghi H, Darin N, Lindberg C. Myopathies associated with myosin heavy chain mutations. Acta Myol. 2004;23:90–96. [PubMed] [Google Scholar]
- Preller M, Bauer S, Adamek N, Fujita-Becker S, Fedorov R, Geeves MA, Manstein DJ. Structural basis for the allosteric interference of myosin function by reactive thiol region mutations G680A and G680V. J Biol Chem. 2011;286:35051–35060. doi: 10.1074/jbc.M111.265298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rayment I, Rypniewski WR, Schmidt-Base K, Smith R, Tomchick DR, Benning MM, Winkelmann DA, Wesenberg G, Holden HM. Three-dimensional structure of myosin subfragment-1: a molecular motor. Science. 1993;261:50–58. doi: 10.1126/science.8316857. [DOI] [PubMed] [Google Scholar]
- Reedy MC, Beall C. Ultrastructure of developing flight muscle in Drosophila. I. Assembly of myofibrils. Dev Biol. 1993a;160:443–465. doi: 10.1006/dbio.1993.1320. [DOI] [PubMed] [Google Scholar]
- Reedy MC, Beall C. Ultrastructure of developing flight muscle in Drosophila. II. Formation of the myotendon junction. Dev Biol. 1993b;160:466–479. doi: 10.1006/dbio.1993.1321. [DOI] [PubMed] [Google Scholar]
- Rozek CE, Davidson N. Drosophila has one myosin heavy-chain gene with three developmentally regulated transcripts. Cell. 1983;32:23–34. doi: 10.1016/0092-8674(83)90493-2. [DOI] [PubMed] [Google Scholar]
- Rubin GM, Spradling AC. Genetic transformation of Drosophila with transposable element vectors. Science. 1982;218:348–353. doi: 10.1126/science.6289436. [DOI] [PubMed] [Google Scholar]
- Suggs JA, Cammarato A, Kronert WA, Nikkhoy M, Dambacher CM, Megighian A, Bernstein SI. Alternative S2 hinge regions of the myosin rod differentially affect muscle function, myofibril dimensions and myosin tail length. J Mol Biol. 2007;367:1312–1329. doi: 10.1016/j.jmb.2007.01.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swank DM, Bartoo ML, Knowles AF, Iliffe C, Bernstein SI, Molloy JE, Sparrow JC. Alternative exon-encoded regions of Drosophila myosin heavy chain modulate ATPase rates and actin sliding velocity. J Biol Chem. 2001;276:15117–15124. doi: 10.1074/jbc.M008379200. [DOI] [PubMed] [Google Scholar]
- Swank DM, Knowles AF, Suggs JA, Sarsoza F, Lee A, Maughan DW, Bernstein SI. The myosin converter domain modulates muscle performance. Nat Cell Biol. 2002;4:312–316. doi: 10.1038/ncb776. [DOI] [PubMed] [Google Scholar]
- Swank DM, Wells L, Kronert WA, Morrill GE, Bernstein SI. Determining structure/function relationships for sarcomeric myosin heavy chain by genetic and transgenic manipulation of Drosophila. Microsc Res Tech. 2000;50:430–442. doi: 10.1002/1097-0029(20000915)50:6<430::AID-JEMT2>3.0.CO;2-E. [DOI] [PubMed] [Google Scholar]
- Tajsharghi H, Pilon M, Oldfors A. A Caenorhabditis elegans model of the myosin heavy chain IIa E706K mutation. Ann Neurol. 2005;58:442–448. doi: 10.1002/ana.20594. [DOI] [PubMed] [Google Scholar]
- Tajsharghi H, Thornell LE, Darin N, Martinsson T, Kyllerman M, Wahlstrom J, Oldfors A. Myosin heavy chain IIa gene mutation E706K is pathogenic and its expression increases with age. Neurology. 2002;58:780–786. doi: 10.1212/wnl.58.5.780. [DOI] [PubMed] [Google Scholar]
- Thompson AR, Naber N, Wilson C, Cooke R, Thomas DD. Structural dynamics of the actomyosin complex probed by a bifunctional spin label that cross-links SH1 and SH2. Biophys J. 2008;95:5238–5246. doi: 10.1529/biophysj.108.138982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng W, Conibear PB, Dickens JL, Cowie RA, Wakelin S, Malnasi-Csizmadia A, Bagshaw CR. Dynamics of actomyosin interactions in relation to the cross-bridge cycle. Philos Trans R Soc Lond B Biol Sci. 2004;359:1843–1855. doi: 10.1098/rstb.2004.1527. [DOI] [PMC free article] [PubMed] [Google Scholar]