Subsets of microtubules are modified by polyglutamylation, but the precise function of this modification is unknown. A microtubule-binding protein, CSAP, is identified that colocalizes with polyglutamylated tubulin. In zebrafish, CSAP is required for normal brain development and proper left–right asymmetry.
Abstract
The diverse populations of microtubule polymers in cells are functionally distinguished by different posttranslational modifications, including polyglutamylation. Polyglutamylation is enriched on subsets of microtubules including those found in the centrioles, mitotic spindle, and cilia. However, whether this modification alters intrinsic microtubule dynamics or affects extrinsic associations with specific interacting partners remains to be determined. Here we identify the microtubule-binding protein centriole and spindle–associated protein (CSAP), which colocalizes with polyglutamylated tubulin to centrioles, spindle microtubules, and cilia in human tissue culture cells. Reducing tubulin polyglutamylation prevents CSAP localization to both spindle and cilia microtubules. In zebrafish, CSAP is required for normal brain development and proper left–right asymmetry, defects that are qualitatively similar to those reported previously for depletion of polyglutamylation-conjugating enzymes. We also find that CSAP is required for proper cilia beating. Our work supports a model in which polyglutamylation can target selected microtubule-associated proteins, such as CSAP, to microtubule subpopulations, providing specific functional capabilities to these populations.
INTRODUCTION
Much in the same way that a “histone code” of posttranslational modifications specifies different regions of the chromatin to define distinct functional domains, different microtubule populations are functionally distinguished by posttranslational modifications, including glutamylation, glycylation, detyrosination, and acetylation (Verhey and Gaertig, 2007; Wloga and Gaertig, 2010; Janke and Bulinkski, 2011). Tubulin modifications are enriched on the C-terminal tails of tubulin, where most microtubule-binding proteins are known to interact. Polyglutamylation generates glutamate side chains that are linked to the main peptide chain via the γ-carboxy residues of gene-encoded glutamates within these tails. Historically, these microtubule modifications have been studied primarily using modification-specific antibodies. For example, the monoclonal antibody GT335 recognizes either subunit of the tubulin dimer modified by the addition of glutamate side chains of any length (Wolff et al., 1992). The use of the GT335 antibody demonstrated that tubulin polyglutamylation is enriched on the mitotic spindle and centrioles in cycling cells and on the cilia and basal bodies. In addition to localization of specific subpopulations, microtubules may be further distinguished by the length of the glutamate side chain or general levels of glutamylation. For example, the levels of glutamylation increase as cells progress into mitosis (Bobinnec et al., 1998). Even higher levels of glutamate side chains accumulate on stable microtubule structures such as neuronal microtubules, axonemes, and centrioles (Audebert et al., 1994; Fouquet et al., 1994; Plessmann and Weber, 1997; Bobinnec et al., 1998).
Localization-based studies provided insights into potential functions for polyglutamylation, but it was not until the recent identification of glutamate-conjugating and glutamate-deconjugating enzymes that it was possible to test the functional contribution of this modification. Recent work established the tubulin-tyrosine ligase–like (TTLL) family of 13 enzymes, which share a homologous TTL domain (van Dijk et al., 2007), as key players in generating tubulin modifications. Nine of these enzymes were demonstrated to be involved in polyglutamylation (Janke et al., 2005; van Dijk et al., 2007). Individual TTLL enzymes have different preferences for α- or β-tubulin and are involved in either the initiation reaction (addition of the first glutamate residue) or the elongation reaction (further addition of glutamate residues to the growing side chain). The coordination of these enzymes could explain how polyglutamylation is differently regulated on distinct microtubule populations. Recently the roles of individual TTLL enzymes have been explored. For example, TTLL6 is required for ciliogenesis and cilia formation in zebrafish (Pathak et al., 2007), and TTLL1 was shown to be required for the glutamylation of mouse lung cilia necessary for asymmetric beating (Ikegami et al., 2010). Similarly, microtubule deglutamylases of the cytosolic carboxypeptidase (CCP) family, including CCP1, CCP4, CCP6, and CCP5 (which specifically removes the branching-point glutamylase), can reverse this modification (Kimura et al., 2010; Rogowski et al., 2010). However, whether tubulin polyglutamylation alters intrinsic microtubule dynamics or affects extrinsic associations with specific interacting partners remains to be determined.
Here we identify the microtubule-binding protein centriole and spindle–associated protein (CSAP). On the basis of its specific subcellular colocalization with markers for polyglutamylation and the dependence of this localization upon polyglutamylase function, we propose that CSAP is preferentially targeted to polyglutamylated microtubules. In zebrafish, CSAP is required for normal brain development, proper left–right asymmetry, and ciliary beating, defects that are similar to those reported previously for depletion of polyglutamylation-conjugating enzymes. Thus our work suggests that polyglutamylation can target selected microtubule-associated proteins, such as CSAP, to microtubule subpopulations, providing specific functional capabilities to these populations.
RESULTS
Identification of C1orf96 as a centriole, spindle, and cilia–associated protein
In a screen for mitotic components, we identified C1orf96 as a human gene with similar expression in tumors to genes involved in mitosis. Although our analysis found that C1orf96 coclusters with mitotic genes in tumor expression data sets (www.intgen.org/expo), C1orf96 is also up-regulated in cells undergoing ciliogenesis (Tim Stearns, Stanford University, personal communication). C1orf96 is a 30.2-kDa protein with no defined domains or motifs. Homologues of C1orf96 are present in vertebrates and the chordate Ciona intestinalis but are not detectable in other organisms.
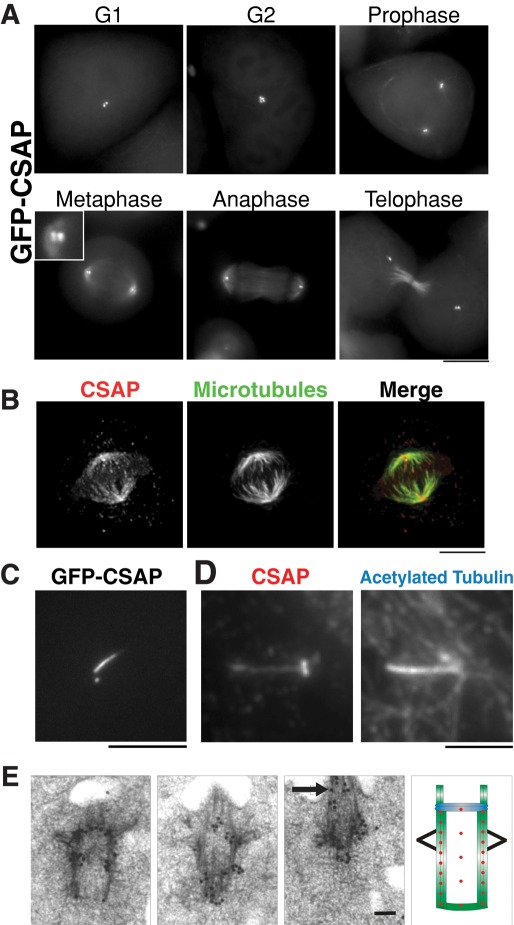
To define the localization of C1orf96, we generated a clonal cell line stably expressing green fluorescent protein (GFP)–C1orf96 in HeLa cells. GFP-C1orf96 localized to two to four foci suggestive of centrioles throughout the cell cycle (Figure 1A). During prometaphase, C1orf96 localized to the mitotic spindle and continued to localize to mitotic spindle microtubules throughout the remainder of mitosis (Figure 1A). C1orf96 was observed at low levels on interphase microtubules but was far more pronounced on mitotic spindle microtubules. To confirm the results from the GFP fusion, we generated an affinity-purified rabbit polyclonal antibody against C1orf96. Immunofluorescence indicated that endogenous C1orf96 displayed similar localization to the GFP fusion (Figure 1B). On the basis of this dual localization of C1orf96 to centrioles and the spindle, we chose to name this protein CSAP for centriole and spindle–associated protein.
FIGURE 1:
CSAP localizes to centrioles, mitotic spindle microtubules, and cilia. (A) Images of HeLa cells stably expressing GFP-CSAP. CSAP localizes to centrioles throughout the cell cycle and to mitotic spindle microtubules. Scale bar, 10 μm. (B) Immunofluorescence images showing the localization of CSAP (anti-CSAP antibodies) to the mitotic spindle (DM1α antibodies). Scale bar, 10 μm. (C) CSAP localizes to basal bodies and cilia. Images show serum-starved hTERT-RPE1 cells stably expressing GFP-CSAP. Scale bar, 10 μm. (D) Immunofluorescence images showing the colocalization of CSAP (anti-CSAP antibodies) with acetylated tubulin, a marker for cilia. Scale bar, 10 μm. (E) Analysis of CSAP centriole and cilia localization by immuno-EM. Totals of 203 and 131 gold particles were quantified for unciliated and ciliated centrioles, respectively. A nearly identical localization pattern was observed with CSAP gold label overlapping with the microtubule cylinder walls for 80% of the gold and 20% in the centriole lumen. A representative example is provided in three serial sections (left). Arrow denotes cilia. The compiled localization pattern is described by the relative distribution of 25 red spots (right). Scale bar, 100 nm.
During interphase, centrioles become the basal bodies from which ciliogenesis occurs. To determine whether CSAP localized to these additional microtubule-based structures, we generated a stable clonal cell line expressing GFP-CSAP in hTERT-RPE1 cells, an established cell line for studying ciliogenesis (Vorobjev and Chentsov Yu, 1982). GFP-CSAP localized to both basal bodies, the transition zone (which connects the basal bodies to ciliary microtubules), and throughout the axoneme structure to the ciliary tip (Figure 1C). Colocalization of anti-CSAP antibodies with antibodies against acetylated tubulin (a marker for ciliary axoneme microtubules) confirmed this localization (Figure 1D). To analyze the ultrastructural localization of CSAP, we performed immuno–electron microscopy (IEM) of CSAP at unciliated (n = 3) and ciliated (n = 3) centrioles in RPE1 cells (Figure 1E). CSAP localized similarly to both classes of centrioles with uniform association along the entire length of centriole triplet microtubules (Figure 1E). In addition to centriole localization, we also found that CSAP localizes to the base and along the length of the microtubules in the ciliary axoneme. In total, these data indicate that CSAP is a cilia, centriole, and spindle–associated protein.
CSAP preferentially localizes to polyglutamylated microtubules
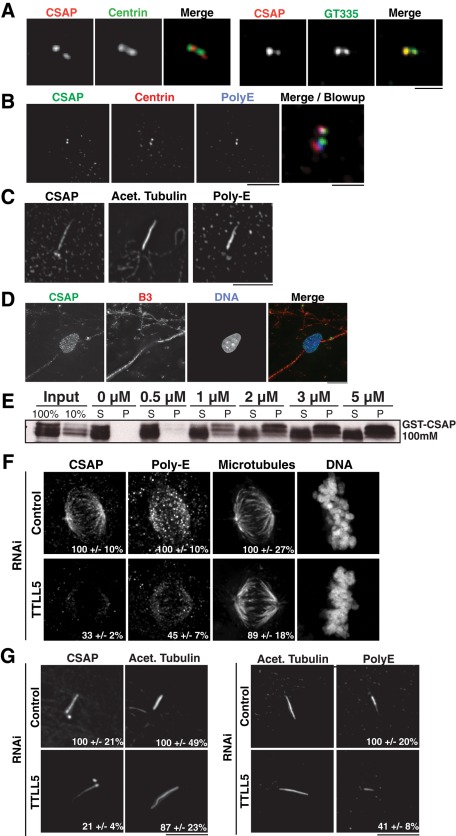
CSAP displays a distinctive localization to centrioles and mitotic spindle microtubules in HeLa cells and to basal bodies and cilia in serum starved hTERT-RPE1 cells. Although we were not able to identify other proteins in the literature that display this specific dual-localization pattern, this localization is similar to that described previously for polyglutamylated microtubules (Wolff et al., 1992; Bre et al., 1994; Fouquet et al., 1994; Weber et al., 1997; Bobinnec et al., 1998; Regnard et al., 1999; Janke et al., 2008). Polyglutamylation is enriched at mitotic spindles, centrioles/basal bodies, and cilia. To evaluate a potential connection between CSAP and polyglutamylation, we used GT335, an antibody that recognizes glutamate chains with at least one glutamate residue (Wolff et al., 1992), and PolyE, an antibody that recognizes glutamate side chains of at least three resides (Shang et al., 2002). To assess the colocalization of CSAP and polyglutamylation, we focused on the centriole. Although CSAP localizes to both the mother and daughter centrioles, it does not precisely overlap with the centriolar marker centrin within the larger structure in HeLa cells by immunofluorescence (Figure 2, A and B). In contrast, CSAP and GT335 or PolyE colocalize within the centriolar structure in HeLa cells (Figure 2, A and B) suggesting a close relationship between CSAP and polyglutamylation. Similarly, in hTERT-RPE1 cells, CSAP and PolyE colocalize along the length of the axoneme (Figure 2C). Finally, we tested CSAP localization in neurons, which are known to be highly polyglutamylated (Audebert et al., 1993). In differentiated neuronal iPS cells (Soldner et al. 2009), CSAP is localized throughout the neuronal tracks and colocalizes with B3, a marker for neuronal tubulin (Figure 2D). CSAP also colocalizes with GT335 along the neuronal axon tracks and to the cilia (unpublished data).
FIGURE 2:
CSAP requires polyglutamylation to target to specific microtubule subpopulations. (A, B) CSAP precisely colocalizes with polyglutamylated tubulin but not centrin. Immunofluorescence images showing CSAP (anti-CSAP antibodies), centrin (a marker for centrioles), and (A) GT335 or (B) PolyE (markers for polyglutamylated microtubules). (C) In hTERT-RPE1 cells, CSAP (anti-CSAP antibodies) colocalizes with acetylated tubulin and PolyE along the length of the cilia. (D) CSAP colocalizes with B3 along neuronal tracks. Immunofluorescence images showing CSAP (anti-CSAP antibodies) and B3 (a marker for neuronal microtubules) in differentiated human iPS cells. (E) Western blot showing the cosedimentation of GST-CSAP with bovine brain microtubules. (F) CSAP localization to the mitotic spindle requires TTLL5. Immunofluorescence images of HeLa cells treated with control siRNAs or siRNAs against TTLL5. Percentages indicate the relative level of CSAP or PolyE compared with microtubule staining in the TTLL5 depletion vs. control. Antibodies to CSAP, PolyE, and DM1α were used to assess localization. n = 10 cells/condition. (G) Immunofluorescence images as in D, showing CSAP and PolyE localization to the cilia in serum-starved RPE-1 cells after control and TTLL5 RNAi. N = 10 cells/condition. Scale bars, 10 μm.
We next sought to test the mechanisms that target CSAP to specific microtubule populations. We found that CSAP does not stably associate with other proteins based on one-step purifications from human cells (unpublished data), suggesting that this localization reflects a direct association of CSAP with microtubules. To test this, we conducted microtubule cosedimentation assays. CSAP bound directly to bovine brain microtubules (Figure 2E), which are highly polyglutamylated (Audebert et al., 1993), with an apparent affinity of 1–2 μM, consistent with established microtubule-binding proteins (Cheeseman et al., 2006). Robust methods for isolating vertebrate microtubules without glutamate side chains are not available, and thus we were unable to test whether CSAP preferentially binds to polyglutamylated microtubules in vitro.
Therefore we next sought to assess the functional relationship between CSAP and polyglutamylation in human cells. Despite repeated attempts with different small interfering RNAs (siRNAs), we were unable to efficiently deplete CSAP in human tissue culture cells. Thus we sought to test the consequences of reducing microtubule polyglutamylation on CSAP localization. Recently specific members of the TTLL family have been identified as the enzymes responsible for microtubule polyglutamylation (van Dijk et al., 2007). To determine whether polyglutamylation can target CSAP to specific microtubule-based structures, we depleted several different TTLL enzymes by RNA interference (RNAi). Of the tested siRNAs, those targeting TTLL5—a polyglutamylation-initiation enzyme specific for α-tubulin (van Dijk et al., 2007)—were most effective in reducing PolyE immunostaining. After TTLL5 depletion in HeLa cells, PolyE staining was reduced by 55% relative to bulk microtubules as assessed by the DM1α antibody (Figure 2E). Depletion of TTLL5 decreased CSAP localization to the mitotic spindle in HeLa cells by 67% relative to the bulk level of tubulin polymer (Figure 2E). Similarly, in serum-starved hTERT-RPE1 cells, depletion of TTLL5 resulted in a decrease of cilia-localized PolyE by 59% and CSAP by 79% but did not change the levels of acetylated tubulin staining in the axoneme (Figure 2F). In these TTLL depletions, CSAP is gradually lost from the distal tip but remains concentrated at the basal bodies (Figure 2F), suggesting that the basal body microtubules are less dynamic. We also found that TTLL11 depletion reduced CSAP localization to microtubule structures (unpublished data), and work from others found that TTLL6 depletion eliminates CSAP ciliary localization (Jagesh Shah, Harvard Medical School, personal communication). In contrast to the effects observed after depletion of TTLL5, we were not able to detect a significant increase in CSAP localization to interphase microtubules upon overexpression of individual TTLL enzymes (Lacroix et al., 2010). This suggests that multiple TTLLs are required to modify tubulin in a way that is sufficient to direct CSAP localization or that there is additional regulation that controls CSAP localization during the cell cycle.
CSAP is predominately expressed in the zebrafish brain and localizes to basal bodies in ciliated cells
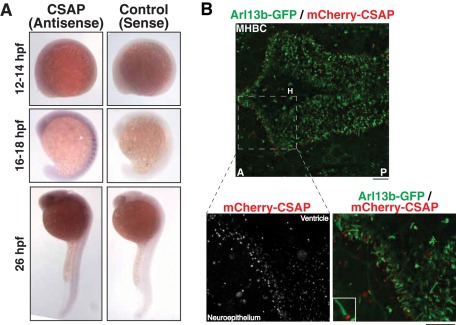
We next sought to assess the functional contribution of CSAP and its relationship to polyglutamylation in an intact organism. Zebrafish (Danio rerio) provide an excellent vertebrate system in which to analyze CSAP function in vivo and test multiple different tissues with high levels of polyglutamylation, including the brain and ciliated tissues (Pathak et al., 2007, 2011). On the basis of reverse transcription (RT)-PCR analysis, CSAP mRNA is maternally expressed and can be detected throughout early development (Supplemental Figure S1B). As assessed by in situ hybridization, CSAP is expressed ubiquitously in the developing embryo and is enriched in the somites and brain in 16- to 26-h postfertilization (hpf) embryos (Figure 3A).
FIGURE 3:
CSAP is expressed throughout the developing zebrafish embryo and localizes to basal bodies. (A) In situ hybridization showing the expression of CSAP in the developing zebrafish embryo. CSAP is expressed throughout the developing embryo but is enriched in brain tissue and somites. The sense probe serves as a negative control. (B) Colocalization of mCherry-CSAP with the ciliary marker Arl13b-GFP in live transgenic zebrafish embryos. CSAP localizes to the basal bodies at the base of cilia along the apical side of the neuroepithelium, shown here on the posterior side of the midbrain–hindbrain boundary. MHBC, Midbrain–hindbrain boundary constriction; H, hindbrain. Embryos are oriented with the anterior to the left. The enlargements are taken from the designated region of the MHBC. Scale bar, 20 μm.
To analyze CSAP localization in zebrafish, we injected mCherry-CSAP mRNA into one-cell-stage, Arl13b-GFP transgenic embryos. Live confocal imaging revealed that mCherry-CSAP localized to specific foci at the apical surface of neuroepithelial cells where cilia form along the brain ventricle lumen (Figure 3B). On the basis of colocalization with the ciliary marker Arl13b-GFP (Borovina et al., 2010), these mCherry-CSAP foci are present at the base of the cilia (Figure 3B), indicating that CSAP localizes to the basal body. In total, these results suggest that CSAP is a basal body component in zebrafish.
CSAP is required for proper brain development
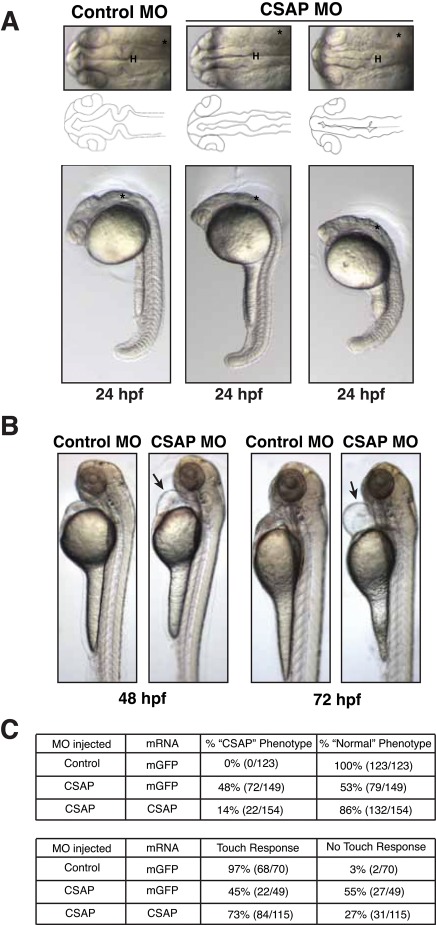
To determine the functional contribution of CSAP to vertebrate development, we depleted CSAP in developing zebrafish embryos using a morpholino antisense oligonucleotide (MO) targeting a splice-site junction. For all the morpholino experiments, the control MO and CSAP MO were coinjected with a MO targeting p53 to avoid established MO-induced side effects (Robu et al., 2007). RT-PCR analysis from the morpholino-injected embryos confirmed that this morpholino induced an incorrect splicing event (Supplemental Figure S1C). CSAP MO-injected embryos (morphants) displayed a range of defects, including forebrain, midbrain, and hindbrain defects, a slight loss of distinct somite boundaries, underdeveloped eyes, dying cells in the ventricle space, movement defects, and a lack of a touch response (Figure 4A and unpublished data). To test for the presence of developmental defects at later time points, we examined 48- and 72-hpf fish. At both time points, there is dramatic heart edema in the morphants as compared with controls (Figure 4B). The morphological brain defect, movement, and touch response defects were all efficiently rescued by coinjection of CSAP mRNA, indicating that these phenotypes are specific to loss of CSAP function (Figure 4C).
FIGURE 4:
CSAP is required for proper zebrafish development. (A) Live brightfield imaging of 24-hpf zebrafish embryos injected with 10 ng of control MO or 10 ng of CSAP splice-site MO in conjunction with p53 MO. Top, a dorsal view of the brain. H, Hindbrain; asterisk, the ear. A tracing of the neuroepithelium and brain ventricles is highlighted below. Bottom, a lateral view of whole embryos. Morphant embryos have abnormal brain development, somite defects, and cell death. Asterisk designates the ear. (B) Lateral view of live brightfield imaging of 48- and 72-hpf zebrafish injected with 10 ng of control MO or 10 ng of CSAP splice-site MO in conjunction with p53 MO. Morphant embryos display a prominent heart edema (arrow) at this time point. These older embryos continue to display brain defects but lack hydrocephaly and kidney cysts and have normal otoliths. (C) Percentages of embryos with the CSAP phenotype (brain defects, heart positioning, tail defects). All phenotypes can be rescued by coinjecting CSAP mRNA with the CSAP MO. The rescue of the touch response is highlighted at the bottom.
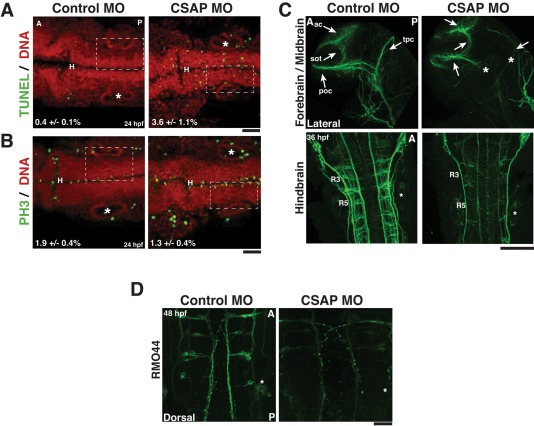
We observed an apparent increase in cell death in the brain of CSAP morphants. By analyzing a section of the neuroepithelium in the hindbrain from the ear to the midline in 24-hpf embryos, we found a 35% reduction in the number of cells (based on the number of nuclei) in CSAP morphants compared with controls (p <0.01; n > 25/condition). To determine whether the reduced cell number was caused by cell death, we performed terminal deoxynucleotidyl transferase dUTP nick-end labeling (TUNEL) staining to score the number of apoptotic cells (Figure 5A). In 24-hpf CSAP morphants, there was an approximately ninefold increase in the percentage of apoptotic cells (p <0.01; n = 15). We also found that a 60% increase in the frequency of mitotic cells (p <0.05; n = 15) based on the localization of mitotic-specific phospho–histone H3 (Ser-10; Figure 5B). This increase in apoptotic cells may contribute to the loss of cell density in the neuroepithelium observed in CSAP morphant embryos.
FIGURE 5:
CSAP is required for neuronal development. (A) Immunofluorescence images of 24-hpf embryos, measuring apoptotic index (indicated by the percentage). Embryos are stained for TUNEL (green) and propidium iodide (red). Cells were counted by placing a 400-nm box around a region from the ear to the midline. N = 15 embryos/condition. Embryos are positioned anterior to posterior. H, Hindbrain; asterisk, the ear. Scale bar, 50 μm. (B) Immunofluorescence images of 24-hpf embryos measuring mitotic index (indicated by the percentage). Embryos are stained for PH3 (green) and propidium iodide (red). Cells were counted as in A. n = 15 embryos/condition. Scale bar, 50 μm. (C) Immunofluorescence images of axon tracts in the brain stained with acetylated tubulin. Top, embryos are oriented laterally with the anterior to the left. Major forebrain/midbrain tracts are labeled as follows: ac (anterior commissure), sot (supraoptic tract), tpc (tract of the posterior commissure), and tpoc (tact of postoptic commissure). CSAP MO-injected embryos have defects in axon tract formation, specifically the tpoc and tpc, as designated by asterisks. The eyes have been removed to easily visualize axon tracts. Bottom, dorsal view of embryos positioned with the anterior at the top. Axon tracts in the hindbrain are labeled with acetylated tubulin. In CSAP morphants, the density of axon tracts is reduced. R3 and R5, rhombomeres 3 and 5, respectively. The ear is noted by an asterisk. Scale bar, 100 μm. (D) The 48-hpf embryos fixed and labeled with RMO44 (a marker for reticulospinal neurons). Dorsal view of the embryos with the anterior at the top. In CSAP morphants, the density of Mauthner interneurons is reduced. The ear is noted by an asterisk. Scale bar, 100 μm.
To further analyze the pronounced brain defects in CSAP-depleted embryos, we visualized neurons with acetylated tubulin immunostaining. CSAP morphants displayed fewer axons and overall axonal fasciculation defects in the forebrain, midbrain, and hindbrain compared with controls (Figure 5C). Similar results were also observed in 48-hpf embryos by immunostaining the reticulospinal neurons (including the Mauthner neurons) with RMO44, an antibody against Neurofilament Medium (Figure 5D). In total, these results suggest that CSAP is required for normal zebrafish development, specifically in brain tissue, where CSAP is prominently expressed.
CSAP is required for left–right lateral asymmetry and ciliary beating
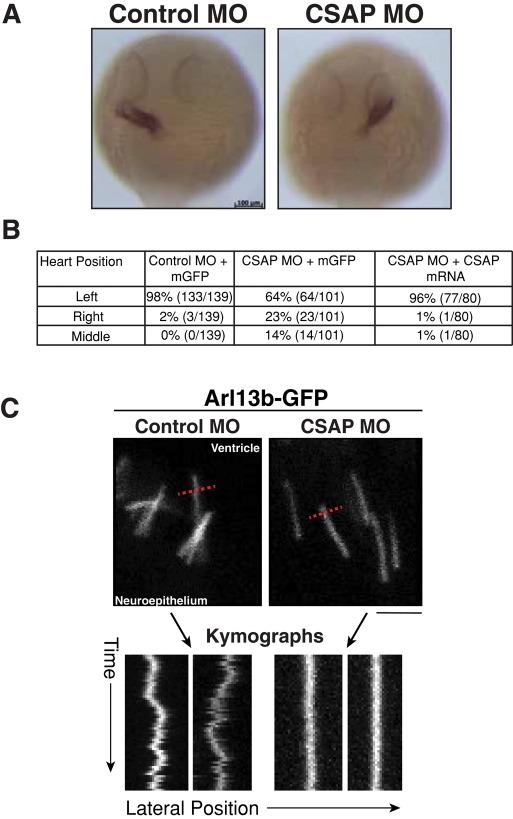
On the basis of the localization of CSAP to basal bodies and the cilia defects previously reported for loss of polyglutamylation (Pathak et al., 2007, 2011), we next tested whether CSAP is necessary for cilia formation and function in vivo. Defects in Kupffer's vesicle cilia disrupt proper development of laterality in zebrafish (Essner et al., 2005; Kramer-Zucker et al., 2005). To determine whether left–right asymmetry is disrupted in CSAP morphants, we performed in situ hybridization using an antisense probe against cardiac myosin light chain 2, a marker for the developing heart (Peterkin et al., 2007). In wild-type embryos, the heart develops on the left. CSAP morphant embryos showed laterality defects, with 23% displaying right heart positioning and 14% medial location (Figure 6A). These defects were rescued by coinjection with CSAP mRNA (Figure 6B). To determine the basis for this altered laterality, we visualized the Kupffer's vesicle cilia directly at the 12-somite stage. The cilia number and length were unchanged (4.08 ± 1.07 μm in controls; 4.18 ± 1.4 μm in CSAP morphants). The combination of the heart laterality defect and the presence of normal length cilia suggests that CSAP contributes to cilia function, but not ciliary assembly or length control, to generate the movement and signaling necessary to establish left–right asymmetry in the developing embryo.
FIGURE 6:
CSAP is required for heart laterality and cilia beating. (A) The 24-hpf embryos fixed and labeled with an antisense probe against cardiac myosin light chain 2 (cmlc2). Control MO-injected embryos display normal heart development on the left side, whereas CSAP MO-injected embryos display heart development on the right side and in the middle of the embryo at a rate higher than controls. This defect is rescued by coinjection of CSAP mRNA. n > 75 embryos/condition. Scale bar, 100 μm. (B) Table of laterality defects with rescue experiments. (C) Still images taken from time-lapse sequences visualizing the beating of cilia along the neuroepithelial lining along the apical surface of the neuroepithelium on the posterior side of the midbrain–hindbrain boundary. Images were acquired at 5 or 6 frames/s. Ciliary movement was visualized by the kymographs shown at the bottom. Kymographs were generated by drawing a line across the tip of cilia (red hashed line) to track movement as a function of time. Also see Supplemental Movies S1–S4.
On the basis of our observation of incorrect laterality in CSAP morphants, we next sought to assess directly whether CSAP depletion affects ciliary function. For technical reasons, we were unable to film cilia movement in the Kupffer's vesicle. Therefore we imaged cilia along the apical surface of the neuroepithelium on the posterior side of the midbrain–hindbrain boundary in the hindbrain ventricle lumen. In CSAP morphant embryos, cilia length and organization in this region appeared normal compared with controls (Figure 6D). The motility of cilia in region was imaged using time-lapse imaging of transgenic zebrafish embryos expressing the cilia marker Arl13b-GFP (Borovina et al., 2010). Images were acquired at 6 frames/s, allowing us to follow ciliary beating. Cilia in CSAP morphant embryos showed a dramatic reduction in beating, as demonstrated by a lack in ciliary tip movement compared with controls (Figure 6D and Supplemental Movies S1–S4). This suggests that even though CSAP is not required for ciliogenesis, it plays a role in brain development, specifically neuron development, and affects cilia beating frequency.
DISCUSSION
CSAP is targeted to polyglutamylated microtubules to contribute to multiple microtubule-based processes
The existence of polyglutamylated microtubules has been known for more than 20 years (Eddé et al., 1990). The use of modification-specific antibodies to detect the presence of polyglutamylated microtubules has facilitated studies on the localization of these modified microtubules. These antibodies, including GT335 and PolyE, provide powerful tools for this cellular analysis but prevent live-cell analysis of this modification. Our work identified the previously uncharacterized protein CSAP as a microtubule-binding protein that preferentially targets to polyglutamylated microtubules in vertebrate cells based on colocalization with polyglutamylated microtubules and a dependence on microtubule polyglutamylating enzymes. The identification of CSAP provides a new potential tool with which to analyze polyglutamylation in live vertebrate cells using a GFP-CSAP fusion. In addition, as CSAP localizes to basal bodies and the complete cilia, it will also provide a useful additional tool to assess cilia assembly and function in live cells. Indeed, other groups have already had great success using GFP-CSAP as a ciliary or polyglutamylation maker (Shah, personal communication; Gislene Pereira, German Cancer Research Centre, personal communication).
Although polyglutamylation was known to display an intriguing range of localization to subpopulations of microtubules, it was not until recently that the discovery of glutamylation-conjugating (Janke et al., 2005; van Dijk et al., 2007) and glutamylation-deconjugating enzymes (Kimura et al., 2010; Rogowski et al., 2010) provided a way to test the functional contribution of these modified microtubules. Reducing polyglutamylation by disrupting specific polyglutamylation-conjugating enzymes reduces cilia beating in Tetrahymena (Suryavanshi et al., 2010), Chlamydomonas reinhardtii (Kubo et al., 2010), and zebrafish (Pathak et al., 2007, 2011). However, in each of these cases, it is important to note that multiple TTLL enzymes contribute to the polyglutamylation of microtubules. Thus the individual TTLL mutants tested thus far do not eliminate all polyglutamylation and may also show partial phenotypes or tissue-specific consequences, depending of the expression of these enzymes.
Despite dramatic consequences of preventing polyglutamylation, the molecular function of polyglutamylation is unclear. The unique localization pattern that we found for CSAP, mirroring that of tubulin polyglutamylation, argues that an important role for polyglutamylation may be to control the association of specific microtubule-binding proteins with subpopulations of microtubules. Consistent with this, the neuronal microtubule-binding protein Tau was shown to have differential affinity for microtubules with different levels of polyglutamylation in vitro (Boucher et al., 1994). Similarly, the microtubule-severing protein spastin has been shown to sever microtubules with higher levels of polyglutamylation more efficiently (Lacroix et al., 2010). Finally, defects in cilia beating after reductions in polyglutamylation have been proposed to act by regulating inner dynein arm activity (Kubo et al., 2010; Suryavanshi et al., 2010), although it was unclear whether this was due to a direct effect on the ability of dynein to contact tubulin. Thus a key role for polyglutamylation may be to target specific microtubule-associated proteins such as CSAP to subsets of microtubules within a cell to achieve specific functions. Other such proteins that preferentially associate with polyglutamylated microtubules may also exist. In this case, our work predicts that these would display a similar subcellular localization to centrioles, spindle microtubules, and cilia when expressed in vertebrate cells.
In addition to its preferential association with polyglutamylated microtubules, the identification of CSAP provides a new player with an important role in vertebrate development. Loss of CSAP in zebrafish embryos causes a variety of developmental defects, including neuronal defects and a loss of proper cilia beating. These defects are qualitatively similar to those previously described for loss of specific polyglutamylating enzymes and are thus consistent with a role in microtubule polyglutamylation. However, there are also some potential differences from previous work on polyglutamylation during zebrafish development. For example, mutants in polyglutamylase modulator Fleer (flr) mutants display pleiotropic cilia defects (Pathak et al. 2007) that are more dramatic than some phenotypes we observed after CSAP depletion. However, we note that in addition to glutamylation, Fleer also regulates tubulin glycylation (Pathak et al., 2011), which may be required to give rise to the phenotypes described for flr mutants. In addition, CSAP is unlikely to be the only downstream target of polyglutamylation, with other microtubule-binding proteins also preferentially recognizing this modification. Finally, CSAP is not expressed uniformly in zebrafish, and is instead enriched in the brain. Thus CSAP may not be required for the function of all cilia. Indeed, although we found that left–right asymmetry was disrupted in CSAP morphants, we did not observe obvious kidney cysts or other defects found when cilia function is completely eliminated (Kramer-Zucker et al., 2005; Sullivan-Brown et al., 2008). Thus the coordinated action of specific levels of polyglutamylation downstream of the multiple TTLL enzymes and the tissue-specific expression of microtubule-binding proteins such as CSAP can provide a combinatorial code to create unique properties to microtubule subpopulations to achieve proper cilia beating, development, and neuronal function.
MATERIALS AND METHODS
Human cell culture experiments
Human cell lines were maintained as described previously (Kline et al., 2006). Stable clonal cells lines expressing GFPLAP fusions were generated in HeLa and hTERT-RPE1 cells as described previously (Cheeseman et al., 2004). A cDNA for the human CSAP (c1orf96; accession number BC071609) was obtained as an IMAGE clone (5264741). siRNAs for TTLL5 were obtained from Dharmacon (Lafayette, CO) as a pool of four sequences; GGUCCUACCUCGAGCAUAA, CGACGGAGUAGCAGAUUGA, GGAUCGUGCUAUCUAAACA, and GAGUAAUUGGAGAACGUUA. Control RNAi was performed with siCONTROL Non-targeting siRNA Pool #1 (Dharmacon). siRNAs were transfected using Lipofectamine RNAi Max (Invitrogen, Carlsbad, CA). For hTERT-RPE1 cells, cilia formation was induced by serum starvation for 48 h. hiPSCs were derived and maintained as described previously (Soldner et al., 2009).
Immunofluorescence and microscopy
Immunofluorescence in human cells was conducted as described previously (Kline et al., 2006). Antibodies against full-length human CSAP and centrin were generated as described previously (Desai et al., 2003). Because we are unable to deplete CSAP effectively from human cells, to validate the CSAP antibody, we conducted Western blots of HeLa and Rpe1 cells with and without expression of GFP-CSAP (Supplemental Figure S1). These blots indicated the presence of a specific band upon expression of GFP-CSAP, demonstrating that this antibody can recognize CSAP. In addition, immunofluorescence using this antibody showed an identical pattern of localization to GFP-CSAP. Other antibodies used were DM1α (Sigma-Aldrich, St. Louis, MO), acetylated tubulin (Sigma-Aldrich), mGT335 (a gift from Carsten Janke, Institut Curie), and rPolyE (a gift from Martin Gorovsky, University of Rochester). Cy2-, Cy3-, and Cy5-conjugated secondary antibodies were obtained from Jackson ImmunoResearch Laboratories (West Grove, PA). DNA was visualized using 10 μg/ml Hoechst. Images were acquired on a DeltaVision Core deconvolution microscope (Applied Precision, Issaquah, WA) equipped with a CoolSnap HQ2 CCD camera. Approximately 40 Z-sections were acquired at 0.2-μm steps using a 100×, 1.3 numerical aperture, Olympus U-PlanApo objective (Olympus, Center Valley, PA). Images were deconvolved using the DeltaVision software. MetaMorph software (Molecular Devices, Sunnyvale, CA) was used to quantify fluorescence intensity.
For zebrafish experiments, embryos were fixed in 2% trichloroacetic acid for 3 h and blocked in phosphate buffer with 0.5% Triton-X100 (PBT), 10% goat serum, and 0.1% bovine serum albumin (BSA). Embryos were incubated in primary antibody (acetylated tubulin, 1:1000) and secondary (Alexa Fluor anti–mouse 488). For phospho–histone H3 (PH3) staining, embryos were fixed in 4% paraformaldehyde overnight, blocked overnight at 4°C in 2% goat serum, 2% BSA, 17% dimethyl sulfoxide (DMSO), and 0.5% triton in PBT, and then incubated in primary antibody (PH3, 1:800) and secondary (Alexa Fluor anti–rabbit 488). Apoptosis was detected via Apoptag Kit (Chemicon, Temecula, CA) according to the given protocol. For the anti-polyglutamylation antibody, embryos were fixed in Dent's fixation (80% methanol, 20% DMSO) at 4°C overnight. Embryos were rehydrated and blocked for 2 h with 5% goat serum and then overnight with primary antibodies (mGT335, 1:400; and rPolyE, 1:800) and then secondary (Alexa Fluor anti–mouse 488, 1:800; and Alexa Fluor anti–rabbit 488, 1:800). All immunochemistry was counterstained with propidium iodide (1:1000) in PBT. Embryos were flat mounted in glycerol and imaged using a Zeiss LSM710 laser-scanning confocal microscope (Carl Zeiss, Jena, Germany). Images were analyzed using LSM software (Zeiss).
Zebrafish maintenance and morpholino experiments
Zebrafish lines were maintained using standard procedures (Kimmel et al., 1995; Westerfield, 2000). Lines included wild-type AB and Arl13b-GFP transgenics (Borovina et al., 2010; a gift from Brian Ciruna, Sick Kids/University of Toronto). A splice-site-blocking morpholino antisense oligonucleotide (MO; 5′-TTGGAAATCACCCCGACTCACCTCT′; Gene Tools, Philomath, OR) that targets the exon 3/intron 3 boundary of zebrafish c1orf96 (LOC402912) was injected into one-cell-stage embryos. A standard control MO (5′-CCTCTTACCTCAGTTACAATTTATA-3′) and zebrafish p53 MO (5′- GCGCCATTGCTTTGCAAGAATTG′) were used (Gene Tools). Primers to identify abnormal splicing in CSAP morphants by RT-PCR were 5′-GGAGCGAGACTGCTTCAAGT′ and 3′-GCAGATCTGGCTCTCTGCTT′. RT-PCR control primers were EF1α forward, 5′-GATGCACCACGAGTCTCTGA-3′, and EF1α reverse, 5′-TGATGACCTGAGCGTTGAAG-3′.
The in situ probe to assess gene expression was generated by cloning zebrafish CSAP cDNA (from IMAGE clone 6997555) into a pCS2+ vector with forward primer 5′-GGAGCGAGACTGCTTCAAGT and reverse primer 3′-GCAGATCTGGCTCTCTGCTT. The fragment was subcloned into pCS2+ and used for in situ hybridization and rescue experiments using standard procedures. The CSAP rescue construct was generated with NotI linearization and Sp6 transcription using mMessage mMachine (Ambion, Austin, TX), as was mCherry-CSAP mRNA for CSAP localization.
Brightfield images of MO-injected embryos were collected using a Zeiss Discovery V8 and analyzed with AxioVision. Arl13b-GFP and mCherry-CSAP live imaging was conducted as described (Gutzman et al., 2008; Graeden and Sive, 2009). Briefly, for cilia time-lapse imaging, embryos were mounted inverted in 0.8% agarose and anesthetized with tricaine. Data were collected at 5–6 frames/s. Movies were analyzed using MetaMorph software. Kymographs of each movie stack were created by drawing a line across cilia tips and recording movement as a function of position over time.
Electron microscopy
RPE1 cells were induced to form primary cilia by transferring cells at ∼50% confluency into 0.25% FBS in DMEM for 48 h. Cells were fixed using high-pressure freezing/freeze substitution, and samples were sectioned, stained, and processed as previously described (Pearson et al., 2009). CSAP primary antibodies were used at a 1:15 dilution. A nonspecific signal of ∼5% of the gold label was observed in all sections. The remaining gold label was observed at centrioles and cilia. Six centrioles were visualized for a total of 334 gold particles.
Protein purification and microtubule cosedimentation assays
A glutathione S-transferase (GST) fusion with full-length CSAP was generated in pGEX-6P-1 for expression in Escherichia coli. GST-CSAP was purified using glutathione agarose (Sigma-Aldrich). Microtubule cosedimentation assays using the purified proteins were conducted as described previously in BRB80 in the presence of 100 mM KCl (Cheeseman et al., 2006).
Supplementary Material
Acknowledgments
We are extremely grateful to Hazel Sive for her generous support to allow us to conduct the zebrafish experiments, Olivier Paugois for help with zebrafish husbandry and care, Frank Soldner for providing the differentiated human neuronal iPS cells, Gary Gorbsky and John Daum for attempting antibody injections for CSAP, and the members of the Sive and Cheeseman labs. We thank Brian Ciruna, Carsten Janke, Iain Drummond, Martin Gorovsky, and Joseph Yost for providing reagents. We thank Mark Winey for financial support (National Institutes of Health Grant GM074746) and Tom Giddings for sectioning and advice on the immuno-EM studies. This work was supported by awards to I.M.C. from the Searle Scholars Program and the Human Frontiers Science Foundation, a grant from the National Institutes of Health/National Institute of General Medical Sciences (GM088313), and a Research Scholar Grant (121776) from the American Cancer Society. I.M.C. is Thomas D. and Virginia W. Cabot Career Development Professor of Biology.
Abbreviations used:
- CCP
cytosolic carboxypeptidase
- CSAP
centriole and spindle–associated protein
- GFP
green fluorescent protein
- GST
glutathione S-transferase
- hpf
hours postfertilization
- MHBC
midbrain hindbrain boundary constriction
- MO
morpholino oligonucleotide
- RT-PCR
reverse transcriptase-PCR
- TTLL
tubulin-tyrosine ligase-like
- TUNEL
terminal deoxynucelotidyl transferase dUTP nick end labeling
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E11-11-0931) on April 4, 2012.
REFERENCES
- Audebert S, Desbruyeres E, Gruszczynski C, Koulakoff A, Gros F, Denoulet P, Edde B. Reversible polyglutamylation of alpha- and beta-tubulin and microtubule dynamics in mouse brain neurons. Mol Biol Cell. 1993;4:615–626. doi: 10.1091/mbc.4.6.615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Audebert S, Koulakoff A, Berwald-Netter Y, Gros F, Denoulet P, Edde B. Developmental regulation of polyglutamylated alpha- and beta-tubulin in mouse brain neurons. J Cell Sci. 1994;107:2313–2322. doi: 10.1242/jcs.107.8.2313. [DOI] [PubMed] [Google Scholar]
- Bobinnec Y, Khodjakov A, Mir LM, Rieder CL, Edde B, Bornens M. Centriole disassembly in vivo and its effect on centrosome structure and function in vertebrate cells. J Cell Biol. 1998;143:1575–1589. doi: 10.1083/jcb.143.6.1575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borovina A, Superina S, Voskas D, Ciruna B. Vangl2 directs the posterior tilting and asymmetric localization of motile primary cilia. Nat Cell Biol. 2010;12:407–412. doi: 10.1038/ncb2042. [DOI] [PubMed] [Google Scholar]
- Boucher D, Larcher JC, Gros F, Denoulet P. Polyglutamylation of tubulin as a progressive regulator of in vitro interactions between the microtubule-associated protein Tau and tubulin. Biochemistry. 1994;33:12471–12477. doi: 10.1021/bi00207a014. [DOI] [PubMed] [Google Scholar]
- Bre MH, de Nechaud B, Wolff A, Fleury A. Glutamylated tubulin probed in ciliates with the monoclonal antibody GT335. Cell Motil Cytoskeleton. 1994;27:337–349. doi: 10.1002/cm.970270406. [DOI] [PubMed] [Google Scholar]
- Cheeseman IM, Chappie JS, Wilson-Kubalek EM, Desai A. The conserved KMN network constitutes the core microtubule-binding site of the kinetochore. Cell. 2006;127:983–997. doi: 10.1016/j.cell.2006.09.039. [DOI] [PubMed] [Google Scholar]
- Cheeseman IM, Niessen S, Anderson S, Hyndman F, Yates JR, III, Oegema K, Desai A. A conserved protein network controls assembly of the outer kinetochore and its ability to sustain tension. Genes Dev. 2004;18:2255–2268. doi: 10.1101/gad.1234104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desai A, Rybina S, Muller-Reichert T, Shevchenko A, Shevchenko A, Hyman A, Oegema K. KNL-1 directs assembly of the microtubule-binding interface of the kinetochore in C. elegans. Genes Dev. 2003;17:2421–2435. doi: 10.1101/gad.1126303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eddé B, Rossier J, Le Caer JP, Desbruyères E, Gros F, Denoulet P. Posttranslational glutamylation of alpha-tubulin. Science. 1990;247:83–85. doi: 10.1126/science.1967194. [DOI] [PubMed] [Google Scholar]
- Essner JJ, Amack JD, Nyholm MK, Harris EB, Yost HJ. Kupffer's vesicle is a ciliated organ of asymmetry in the zebrafish embryo that initiates left-right development of the brain, heart and gut. Development. 2005;132:1247–1260. doi: 10.1242/dev.01663. [DOI] [PubMed] [Google Scholar]
- Fouquet JP, Edde B, Kann ML, Wolff A, Desbruyeres E, Denoulet P. Differential distribution of glutamylated tubulin during spermatogenesis in mammalian testis. Cell Motil Cytoskeleton. 1994;27:49–58. doi: 10.1002/cm.970270106. [DOI] [PubMed] [Google Scholar]
- Graeden E, Sive H. Live imaging of the zebrafish embryonic brain by confocal microscopy. J Vis Exp. 2009;26:e1217. doi: 10.3791/1217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutzman JH, Graeden EG, Lowery LA, Holley HS, Sive H. Formation of the zebrafish midbrain-hindbrain boundary constriction requires laminin-dependent basal constriction. Mech Dev. 2008;125:974–983. doi: 10.1016/j.mod.2008.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikegami K, Sato S, Nakamura K, Ostrowski LE, Setou M. Tubulin polyglutamylation is essential for airway ciliary function through the regulation of beating asymmetry. Proc Natl Acad Sci USA. 2010;107:10490–10495. doi: 10.1073/pnas.1002128107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janke C, Bulinski JC. Post-translational regulation of the microtubule cytoskeleton: mechanisms and functions. Nat Rev Mol Cell Biol. 2011;12:773–786. doi: 10.1038/nrm3227. [DOI] [PubMed] [Google Scholar]
- Janke C, Rogowski K, van Dijk J. Polyglutamylation: a fine-regulator of protein function? “Protein modifications: beyond the usual suspects” review series. EMBO Rep. 2008;9:636–641. doi: 10.1038/embor.2008.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janke C, et al. Tubulin polyglutamylase enzymes are members of the TTL domain protein family. Science. 2005;308:1758–1762. doi: 10.1126/science.1113010. [DOI] [PubMed] [Google Scholar]
- Kimmel CB, Ballard WW, Kimmel SR, Ullmann B, Schilling TF. Stages of embryonic development of the zebrafish. Dev Dyn. 1995;203:253–310. doi: 10.1002/aja.1002030302. [DOI] [PubMed] [Google Scholar]
- Kimura Y, Kurabe N, Ikegami K, Tsutsumi K, Konishi Y, Kaplan OI, Kunitomo H, Iino Y, Blacque OE, Setou M. Identification of tubulin deglutamylase among Caenorhabditis elegans and mammalian cytosolic carboxypeptidases (CCPs) J Biol Chem. 2010;285:22936–22941. doi: 10.1074/jbc.C110.128280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kline SL, Cheeseman IM, Hori T, Fukagawa T, Desai A. The human Mis12 complex is required for kinetochore assembly and proper chromosome segregation. J Cell Biol. 2006;173:9–17. doi: 10.1083/jcb.200509158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kramer-Zucker AG, Olale F, Haycraft CJ, Yoder BK, Schier AF, Drummond IA. Cilia-driven fluid flow in the zebrafish pronephros, brain and Kupffer's vesicle is required for normal organogenesis. Development. 2005;132:1907–1921. doi: 10.1242/dev.01772. [DOI] [PubMed] [Google Scholar]
- Kubo T, Yanagisawa HA, Yagi T, Hirono M, Kamiya R. Tubulin polyglutamylation regulates axonemal motility by modulating activities of inner-arm dyneins. Curr Biol. 2010;20:441–445. doi: 10.1016/j.cub.2009.12.058. [DOI] [PubMed] [Google Scholar]
- Lacroix B, van Dijk J, Gold ND, Guizetti J, Aldrian-Herrada G, Rogowski K, Gerlich DW, Janke C. Tubulin polyglutamylation stimulates spastin-mediated microtubule severing. J Cell Biol. 2010;189:945–954. doi: 10.1083/jcb.201001024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pathak N, Austin CA, Drummond IA. Tubulin tyrosine ligase-like genes ttll3 and ttll6 maintain zebrafish cilia structure and motility. J Biol Chem. 2011;286:11685–11695. doi: 10.1074/jbc.M110.209817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pathak N, Obara T, Mangos S, Liu Y, Drummond IA. The zebrafish fleer gene encodes an essential regulator of cilia tubulin polyglutamylation. Mol Biol Cell. 2007;18:4353–4364. doi: 10.1091/mbc.E07-06-0537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearson CG, Osborn DP, Giddings TH, Jr, Beales PL, Winey M. Basal body stability and ciliogenesis requires the conserved component Poc1. J Cell Biol. 2009;187:905–920. doi: 10.1083/jcb.200908019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterkin T, Gibson A, Patient R. Redundancy and evolution of GATA factor requirements in development of the myocardium. Dev Biol. 2007;311:623–635. doi: 10.1016/j.ydbio.2007.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plessmann U, Weber K. Mammalian sperm tubulin: an exceptionally large number of variants based on several posttranslational modifications. J Protein Chem. 1997;16:385–390. doi: 10.1023/a:1026332621215. [DOI] [PubMed] [Google Scholar]
- Regnard C, Desbruyeres E, Denoulet P, Edde B. Tubulin polyglutamylase: isozymic variants and regulation during the cell cycle in HeLa cells. J Cell Sci. 1999;112:4281–4289. doi: 10.1242/jcs.112.23.4281. [DOI] [PubMed] [Google Scholar]
- Robu ME, Larson JD, Nasevicius A, Beiraghi S, Brenner C, Farber SA, Ekker SC. p53 activation by knockdown technologies. PLoS Genet. 2007;3:e78. doi: 10.1371/journal.pgen.0030078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogowski K, et al. A family of protein-deglutamylating enzymes associated with neurodegeneration. Cell. 2010;143:564–578. doi: 10.1016/j.cell.2010.10.014. [DOI] [PubMed] [Google Scholar]
- Shang Y, Li B, Gorovsky MA. Tetrahymena thermophila contains a conventional gamma-tubulin that is differentially required for the maintenance of different microtubule-organizing centers. J Cell Biol. 2002;158:1195–1206. doi: 10.1083/jcb.200205101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soldner F. Parkinson's disease patient-derived induced pluripotent stem cells free of viral reprogramming factors. Cell. 2009;136:964–977. doi: 10.1016/j.cell.2009.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan-Brown J, Schottenfeld J, Okabe N, Hostetter CL, Serluca FC, Thiberge SY, Burdine RD. Zebrafish mutations affecting cilia motility share similar cystic phenotypes and suggest a mechanism of cyst formation that differs from pkd2 morphants. Dev Biol. 2008;314:261–275. doi: 10.1016/j.ydbio.2007.11.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suryavanshi S, et al. Tubulin glutamylation regulates ciliary motility by altering inner dynein arm activity. Curr Biol. 2010;20:435–440. doi: 10.1016/j.cub.2009.12.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Dijk J, Rogowski K, Miro J, Lacroix B, Edde B, Janke C. A targeted multienzyme mechanism for selective microtubule polyglutamylation. Mol Cell. 2007;26:437–448. doi: 10.1016/j.molcel.2007.04.012. [DOI] [PubMed] [Google Scholar]
- Verhey KJ, Gaertig J. The tubulin code. Cell Cycle. 2007;6:2152–2160. doi: 10.4161/cc.6.17.4633. [DOI] [PubMed] [Google Scholar]
- Vorobjev IA, Chentsov Yu S. Centrioles in the cell cycle. I. Epithelial cells. J Cell Biol. 1982;93:938–949. doi: 10.1083/jcb.93.3.938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber K, Schneider A, Westermann S, Muller N, Plessmann U. Posttranslational modifications of alpha- and beta-tubulin in Giardia lamblia, an ancient eukaryote. FEBS Lett. 1997;419:87–91. doi: 10.1016/s0014-5793(97)01436-1. [DOI] [PubMed] [Google Scholar]
- Westerfield M. The Zebrafish Book. A Guide for the Laboratory Use of Zebrafish (Danio rerio) Eugene: University of Oregon Press; 2000. [Google Scholar]
- Wloga D, Gaertig J. Post-translational modifications of microtubules. J Cell Sci. 2010;123:3447–3455. doi: 10.1242/jcs.063727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolff A, de Nechaud B, Chillet D, Mazarguil H, Desbruyeres E, Audebert S, Edde B, Gros F, Denoulet P. Distribution of glutamylated alpha and beta-tubulin in mouse tissues using a specific monoclonal antibody, GT335. Eur J Cell Biol. 1992;59:425–432. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.