Abstract
The mechanisms by which mutations in the presenilins (PSEN) or the amyloid precursor protein (APP) genes cause familial Alzheimer disease (FAD) are controversial. FAD mutations increase the release of amyloid β (Aβ)42 relative to Aβ40 by an unknown, possibly gain-of-toxic-function, mechanism. However, many PSEN mutations paradoxically impair γ-secretase and ‘loss-of-function’ mechanisms have also been postulated. Here, we use kinetic studies to demonstrate that FAD mutations affect Aβ generation via three different mechanisms, resulting in qualitative changes in the Aβ profiles, which are not limited to Aβ42. Loss of ε-cleavage function is not generally observed among FAD mutants. On the other hand, γ-secretase inhibitors used in the clinic appear to block the initial ε-cleavage step, but unexpectedly affect more selectively Notch than APP processing, while modulators act as activators of the carboxypeptidase-like (γ) activity. Overall, we provide a coherent explanation for the effect of different FAD mutations, demonstrating the importance of qualitative rather than quantitative changes in the Aβ products, and suggest fundamental improvements for current drug development efforts.
Keywords: Alzheimer, amyloid, FAD mutations, γ-secretase, presenilin
Introduction
A central and still unresolved debate with important therapeutic implications in the field of Alzheimer disease (AD) research revolves around the question of how mutations in presenilin (PSEN), the catalytic core of the γ-secretases (De Strooper et al, 1998), cause disease. The γ-secretases are intramembrane cleaving protein complexes (Hebert et al, 2004; Shirotani et al, 2004) responsible for the generation of amyloid β (Aβ) from the amyloid precursor protein (APP). Aβ peptides of different lengths accumulate in amyloid plaques in the AD brain. More than 150 familial Alzheimer disease (FAD) mutations have been mapped to the genes encoding PSEN1 or PSEN2 (http://www.molgen.ua.ac.be/ADMutations), pointing to a crucial role of the γ-secretase complexes in the disease. Apart from PSEN, a mature and active γ-secretase complex consists of three additional subunits: Nicastrin (Nct), PSEN enhancer 2 (Pen-2), and either anterior pharynx 1 (APH-1) A or B (for a review, see Tolia and De Strooper, 2009). The γ-secretase complexes proteolyse type 1 transmembrane proteins, among them the APP, the Notch receptors and ligands, the Erb4 receptor and N-Cadherin (Wakabayashi and De Strooper, 2008).
As a rule, FAD PSEN mutations increase the relative amount of Aβ42 versus Aβ40 in in vivo and in vitro paradigms (Borchelt et al, 1996; Duff et al, 1996; Scheuner et al, 1996; Murayama et al, 1999), which led to propose that PSEN mutations act via a toxic gain-of-function mechanism. However, more refined analyses have made clear that the change in Aβ ratio does not necessarily reflect an increase in Aβ42 production, but can also be the consequence of a decrease in Aβ40 levels. Actually, many mutations reduce one or both products of the γ-secretase in steady-state conditions (Song et al, 1999; Bentahir et al, 2006; Shen and Kelleher, 2007; Shimojo et al, 2007; Heilig et al, 2010). These observations have led to an opposite hypothesis in which FAD mutations in PSEN cause dementia through a loss of function of γ-secretase, resulting in decreased proteolytic processing of different substrates and compromising intracellular signalling pathways (Shen and Kelleher, 2007; Kelleher and Shen, 2010). In fact, the current model for γ-secretase successive proteolysis (Takami et al, 2009) may link a loss of function to misprocessing of APP and abnormal generation of Aβ (De Strooper, 2007; Wolfe, 2007). However, the fact that less efficient proteolytic processing of APP may lead to alterations in the Aβ profile and AD is contraintuitive in the light of the classical amyloid hypothesis, which stresses the importance of quantitative accumulation of either total Aβ or Aβ42 (Hardy and Selkoe, 2002). Moreover, a recent report has shown that reduced γ-secretase activity does not increase the production (accumulation) of longer Aβ peptides (Quintero-Monzon et al, 2011).
Importantly, the biophysical and biochemical properties of Aβ vary strongly with its length. Longer Aβ42 has a much stronger tendency to aggregate than the shorter Aβ40 (Jarrett and Lansbury, 1993; Jarrett et al, 1993). Furthermore, the relative ratio of Aβ40 to Aβ42 influences strongly the biological effects of the Aβ mixture in vitro and in vivo, even when total Aβ amounts are kept equal (Kuperstein et al, 2010). Whereas Aβ40 appears to act protectively in various toxicity assays (Wang et al, 2006; Kim et al, 2007), longer Aβ peptides promote aggregation and neurotoxicity (McGowan et al, 2005). In fact, it has been suggested that the ratio (Aβ42/Aβ40) is more important than the absolute amounts of Aβ42 (Tanzi and Bertram, 2005). Similar to Aβ42, Aβ43 is potently amyloidogenic and neurotoxic (Saito et al, 2011). While it is commonly found in AD brains (Welander et al, 2009), its potential relevance in disease was only recently addressed (Saito et al, 2011). Thus, qualitative changes in Aβ (De Strooper, 2007; Wolfe, 2007) are at least as important as the quantitative alterations proposed by the original amyloid hypothesis (Hardy and Selkoe, 2002).
In contrast, the ‘simple’ loss-of-function hypothesis proposes that Aβ alterations are only an epiphenomenon of the PSEN mutations, and that inefficient cleavage of membrane proteins by γ-secretase complexes is the fundamental upstream cause of the neurodegenerative process (Shen and Kelleher, 2007; Kelleher and Shen, 2010). This hypothesis finds support in (a) experimental results with Psen knockout mice (Saura et al, 2004), where progressive neurodegeneration occurs without Aβ deposition, and (b) in three case reports in which missense mutations in PSEN genes displayed neurodegenerative clinical phenotypes but no Aβ accumulation (discussed in Shen and Kelleher, 2007; Kelleher and Shen, 2010). However, this last argument has been considerably weakened by follow-up studies showing that neurodegeneration was likely caused by a second mutation in the progranulin gene in one case (Boeve et al, 2006), whereas in a second case abundant amyloid deposition in the frontal lobe appeared at autopsy (for further discussion, see Bergmans and De Strooper, 2010).
On the other hand, recent observations in patients suffering from familial acne inversa in China (Wang et al, 2010) and independently in Great Britain (Pink et al, 2011) raise doubts about the validity of the ‘simple’ γ-secretase loss-of-function hypothesis. This condition appears to be associated with the haploinsufficiency of γ-secretase subunit genes (Nicastrin, Pen2) and most likely involves a deficiency in Notch cell signalling. However, none of the acne-affected individuals had AD symptoms. These observations indicate that reduced γ-secretase activity is not sufficient to cause AD, although further follow-up studies in these families are needed. Alternative mechanisms for the loss-of-function hypothesis have been proposed over the years (for an overview, see De Strooper and Annaert, 2010). For instance, several reports indicate alterations in subcellular trafficking or turnover of selected membrane proteins (Wilson et al, 2004; Esselens et al, 2004) or defective acidification of phagolysosomal compartments associated with PSEN loss of function (Lee et al, 2010). In addition, disturbances in cellular Ca2+ homeostasis by direct effects on the Ca2+ leakage function of PSEN (Zhang et al, 2010) or indirect effects on Ca2+ signalling pathways (reviewed in Bezprozvanny and Mattson, 2008) have been associated to PSEN loss of function. However, these hypotheses do not provide an explanation for the mutations in APP and also do not take into account that all tested FAD mutations affect the prime function of PSEN, which is proteolysis.
From this brief overview it is clear that further in-depth investigation of the effects of clinical mutations on the function and structure of γ-secretase is required, especially given the relevance of such analysis for further drug development.
Addressing this important question implies multidisciplinary approaches, in which deep structural and functional studies dissect the mechanisms of FAD mutations. Solving the 3D-structure of the protease complex would allow studying how FAD mutations affect the structure, and possibly the function. However, this is a huge challenge as important technical and experimental barriers need to be overcome. On the other hand, dissecting γ-secretase activity by kinetic analysis can yield important mechanistic insights into how FAD mutants regulate enzyme function.
In vitro reconstitution of γ-secretase activity has provided initial insights into the enzymatic mechanism. Ihara and co-workers have provided compelling evidence for sequential processing of substrates by γ-secretase (Sato et al, 2003; Qi-Takahara et al, 2005; Kakuda et al, 2006; Yagishita et al, 2008). The most direct evidence was the identification of particular tri- and tetra-peptides generated from the APP-CTF stub by the γ-secretase (Takami et al, 2009). Their model proposes that APP can be sequentially cut along two production lines: Aβ49>Aβ46>Aβ43>Aβ40 and Aβ48>Aβ45>Aβ42>Aβ38 (Figure 1A). Accordingly, the endoproteolytic activity (first ε-cleavage) releases the APP intracellular domain (AICD) and Aβ48 or Aβ49. These long Aβs are then shortened by consecutive carboxypeptidase-like γ-cleavages, which progressively decrease Aβ hydrophobicity and increase the probability of its release into the extracellular environment. In agreement, it has been shown that the endoproteolytic cleavage site determines the product line preference of the γ-secretase in vivo (Funamoto et al, 2004), and therefore the series of Aβ products. Also, presenilinase cleavage (the autocatalytic activation of PSEN) results in the generation of tripeptides in accordance with this model (Fukumori et al, 2010). The ε-cleavage in the APP substrate is analogous to the Notch S3 cleavage site (Sastre et al, 2001; Weidemann et al, 2002) and most likely other γ-secretase substrates are processed in similar ways.
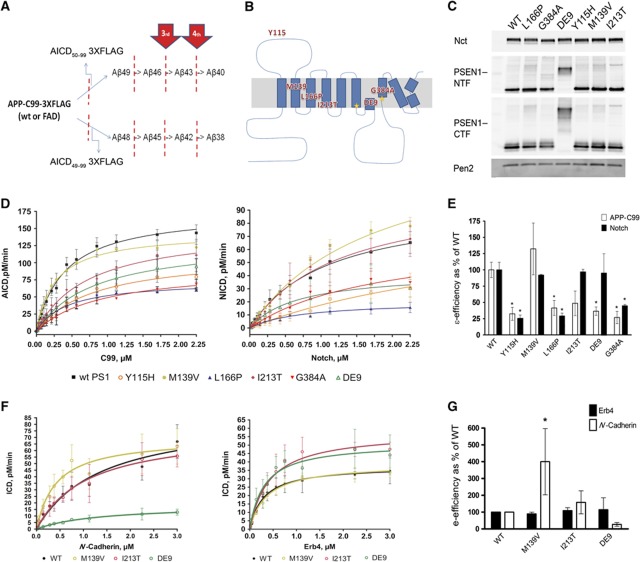
Figure 1.
FAD–PSEN1 mutations do not consistently decrease the enzymatic efficiency of the endopeptidase cleavage. (A, B) Schematic overviews of APP processing and location of FAD–PSEN1 mutations used in the current study. (C) Expression levels of Nct, PSEN1–NTF, PSEN1–CTF and Pen-2 in Psen1/2−/− mEFs transduced with human wt or FAD–PSEN1 mutants using a replication-defective recombinant retroviral expression system (Clontech) and selected with puromycin (5 μg/ml). Western blotting and densitometric analysis of the CHAPSO-solubilized membrane proteins from the different PSEN1 cell lines indicate that wt and mutant PSEN1 rescued γ-secretase complex to similar extents. In order to determine specific activities for the wt or FAD complexes, γ-secretase activities were normalized to PSEN CTF fragment levels or full-length PS1 levels for the DE9 mutant. (D) Kinetic curves for wt and PS1–FAD mutants using purified APP-C99-3XFLAG or Notch-3XFLAG substrates (mean±s.e.) or (F) ErB4-3XFLAG and N-Cadherin-3XFLAG substrates (mean±s.d.). Detergent-extracted membranes were incubated in 0.25% CHAPSO reaction buffer with varying concentrations of purified substrate for 4 h at 37 °C. In vitro generated ICD-3XFLAG were analysed by quantitative western blot analysis (see Materials and methods). (E) FAD–PSEN1 ε-enzymatic efficiencies for APP-C99 and Notch substrates (mean±s.e.). Enzymatic efficiencies unequivocally demonstrate that loss of function at the ε-cleavage is not a constant among PSEN1 mutations. (G) FAD–PSEN1 mutations that did not affect the generation of NICD did not change significantly the processing of ErB4 (mean±s.d.) either. In contrast, N-Cadherin processing was significantly upregulated by the M139V (mean±s.d.). (E, G) Experiments were repeated 3–5 times. Statistical significance of the data was tested with one-way analysis of variance (ANOVA) and Dunnett’s post test, taking the corresponding WT efficiency as control group, *P<0.05.
In the current study, we used a cell-free assay to analyse how clinical mutations in PSEN1, PSEN2 and APP affect the activity of the γ-secretase complex. Dissection of the different activities of the γ-secretase complex allowed us to reach a coherent explanation for the effects of the tested FAD mutations. We coupled kinetic studies of the endopeptidase activity to the analysis of the carboxypeptidase-like cleavage to show that FAD mutations have widely variable effects on the efficiency of the first cleavage, which releases the intracellular signalling domains of substrates. This observation rules out an impairment in the endopeptidase (ε) mechanism as necessary for the pathological effect of FAD mutations. In contrast, all FAD PSEN and APP mutations alter the processing of APP, regulating the generation of Aβ by three different mechanisms.
Results
FAD–PS1 mutations do not consistently impair the endopeptidase activity of the γ-secretase
We analysed the effects of FAD mutations PSEN1-Y115H, -M139V, -L166P, -I213T, -G384A and delta-exon9 (DE9) on the kinetic constants of the ε-cleavage of APP, Notch, ErB4 and N-Cadherin substrates. The selected mutations are spread throughout the PSEN1 primary sequence (Figure 1B). Importantly, blockage by the transition state analogue L-685,458 (TSA, InhX) demonstrated the specificity of the assays (Supplementary Figure 1A). To determine the kinetic constants of wt and FAD γ-secretase complexes, we used CHAPSO-extracted membranes from Psen1/2−/−, rescued with wt or FAD-mutant PSEN1 as source of enzyme (Figure 1C) and purified APP C99–3XFlag, Notch–3XFlag, Erb4–3XFlag or N-Cadherin–3XFlag as substrates.
The kinetic data fit the Michaelis–Menten reaction curves (Figure 1D and F), and Km (affinity constant) as well as Vmax (maximal velocity) were determined (Table I). Since γ-secretase activities are normalized to enzyme levels, Vmax can be taken as kcat and enzymatic efficiencies calculated as kcat/Km. The results reveal diverse effects of the FAD–PSEN1 mutations on this important kinetic parameter. Y115H, L166P and G384A mutants decrease γ-secretase efficiencies by 75% for both APP and Notch, while I213T and DE9 only affect APP, and M139V does not show any effect on the ε-cleavage (Figure 1E). Moreover, FAD–PSEN1 mutations that do not affect Notch endoproteolysis do not impair ErB4 cleavage either, while only the M139V significantly increases the processing of N-Cadherin (Figure 1G). Thus, the tested FAD–PSEN1 mutations have no consistent inhibitory effect on the endoproteolytic cleavage of γ-secretase substrates, indicating that reduced release of intracellular domains and signalling cannot explain their AD-causing effects.
Table 1. Kinetic parameters for human PSEN1 γ-secretase complexes using APP-C99, Notch, ErB4 or N-Cadherin as substrates.
| APP-C99 substrate |
Notch substrate |
Erb4 substrate |
N-Cadherin substrate |
|||||
|---|---|---|---|---|---|---|---|---|
| Km±s.e., μM | Vmax±s.e., pM/min | Km±s.e., μM | Vmax±s.e., pM/min | Km±s.d., μM | Vmax±s.d., pM/min | Km±s.d., μM | Vmax±s.d., pM/min | |
| PS1 wt | 0.40±0.05 | 175.6±8.4 | 1.08±0.17 | 95.7±7.5 | 0.31±0.07 | 37.72±6.18 | 1.46±0.36 | 88.37±10.95 |
| Y115H | 0.81±0.18 | 113.3±11.3a | 3.92±1.97 | 86.49±31.4 | — | — | — | — |
| M139V | 0.27±0.04a | 144.5±6.8a | 1.78±0.21a | 146.9±10.1a | 0.40±0.23 | 39.76±6.36 | 0.42±0.19a | 71.16±20.14 |
| L166P | 0.43±0.07 | 74.03±4.2a | 0.97±0.2 | 23.76±2.4a | — | — | — | — |
| I213T | 0.73±0.18 | 151.1±14.7 | 1.26±0.26 | 106.1±11.2 | 0.45±0.13 | 58.68±5.83a | 1.02±0.11 | 74.95±11.76 |
| DeltaE9 | 0.82±0.18a | 133.5±13.5a | 0.67±0.24 | 42.8±6.5a | 0.33±0.06 | 40.84±8.12 | 1.70±0.43 | 21.97±8.39a |
| G384A | 0.92±0.18 | 93.87±8.7a | 1.85±0.42 | 71.04±9.5 | — | — | — | — |
| Kinetic values are derived from the curves displayed in Figure 1 and were determined by nonlinear curve-fitting using GraphPad Prism 4 software (see Material and methods section). | ||||||||
| aSignificant changes according to the 95% CI (P<0.05). In vitro activity assays were performed using CHAPSO-extracted membranes from Psen1/2−/− mEFs stably transduced with human wt or FAD PSEN1 mutants and purified substrates-3XFlag, n⩾. | ||||||||
FAD–PSEN mutations impair the fourth γ-secretase cleavage in both product lines
Next, we asked whether FAD–PSEN1 mutations lead to APP misprocessing at the γ-cleavage sites. We use Ihara’s model (see Introduction) for further description of our work since it explains very well our observations. Kinetic analysis of the carboxypeptidase-like activity is challenging to perform since controlling substrate concentrations, that is, the intermediary Aβ products, is experimentally not possible yet. Nevertheless, we measured two γ-products in each production line: Aβ43, Aβ42, Aβ40 and Aβ38 (Figure 1A) at saturating APP-substrate concentration. Thus, substrates (Aβ43 and Aβ42) and products (Aβ40 and Aβ38) of the fourth γ-secretase cleavage in both pathways are analysed and provide a relative number for the γ-cleavage efficiency. Importantly, as some of the clinical mutants affect the ε-cleavage, we normalized the Aβ product levels (Aβ38, Aβ40, Aβ42 or Aβ43) towards total AICD (Figure 2A and B). AICD reflects the total initial Aβ substrate (Aβ49+Aβ48) produced and processed in each reaction. Low Aβ40 and Aβ38 levels and high, long Aβ levels (>Aβ42) are found in all the FAD-linked mutations tested, including the M139V, which does not affect the ε-efficiency. Interestingly, the M139V mutation affects the processing of APP only at the level of Aβ, indicating that endo- and carboxypeptidase-like activities of the γ-secretase can be dissociated.
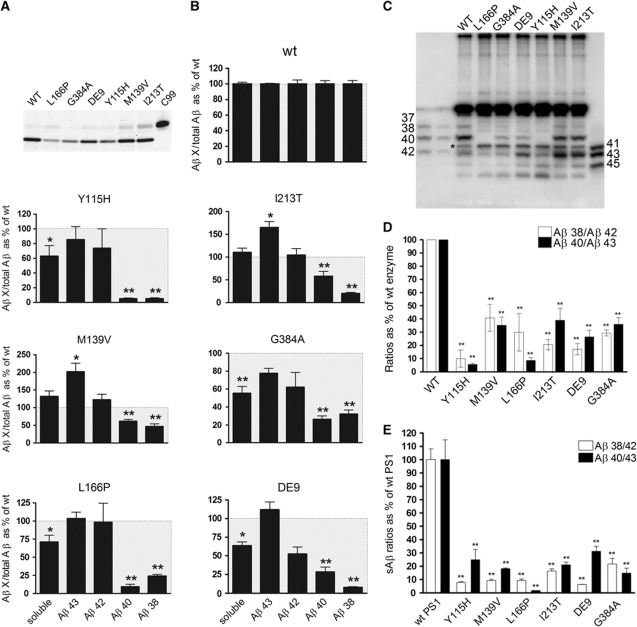
Figure 2.
FAD–PSEN1 mutations impair the fourth enzymatic turnover. AICD levels (moles per min) generated by the wt or FAD mutant complexes (A) were used to normalize Aβ products (moles per min) in order to determine accurately Aβ generation relative to C99 substrate. Aβ profiles (B) thus represent Aβ products corrected for the initial endoprotease activities, plotted as percentage of the wt Aβ products (mean±s.e.). Soluble Aβ (sum of Aβ38, Aβ40, Aβ42 and Aβ43 peptides) gives information about the efficiency of the γ-cleavages: lower levels (<100%, grey box) suggest that longer peptides (>Aβ43) accumulate in the reactions. (C) In agreement with the ELISA quantifications, total Aβ analysed in urea-based gels show increments in Aβ42 and Aβ43, and reductions in Aβ40 and Aβ38 in FAD–PSEN1 mutations, relative to wt. (*) Indicates a non product band that is present in the C99 substrate (see Supplementary Figure 2). (D) Aβ product/substrate ratios determined in vitro for the FAD–PSEN1 mutations show an impairment at the fourth γ-secretase turnover (mean±s.e.). Experiments in (B) and (D) were repeated 4–6 times. Statistical significance of the data tested with one-way ANOVA and Dunnett’s post test taking the corresponding WT as control group; *P<0.05, **P<0.01. (E) Aβ product/substrate ratios determined in vivo confirm impairment at the fourth enzymatic cycle: wt or FAD–PSEN1 mEF cell lines were transiently transduced with APPswe, extracellular media collected at 24 h after infection and sAβ measured by ELISA (mean±s.e.). Statistical significance: n=4, ANOVA and Dunnett’s post test, **P<0.01.
Total ‘secreted’ Aβ, defined as the sum of Aβ38, Aβ40, Aβ42 and Aβ43, decreases significantly in the Y115H, L166P, DE9 and G384A mutations (Figure 2B), implying the concomitant accumulation of longer Aβ precursors generated in cycles 2 and 3. Qualitative analysis of the Aβ profiles in urea-based SDS–PAGE confirmed this observation (Figure 2C and Supplementary Figure 2B). We finally determine product/substrate ratios for the fourth enzymatic turnover (Aβ38/Aβ42 and Aβ40/Aβ43) (Figure 2D), which demonstrates that the FAD–PSEN1 mutations investigated here dramatically impair the fourth γ-secretase cleavage in both product lines. Our data imply that FAD–PSEN1 mutations cause AD by qualitative shifts in Aβ profiles and not by general loss of function of the enzyme complex (Shen and Kelleher, 2007; Wolfe, 2007). The dysfunction at the carboxypeptidase-like activity of the complex not only explains the widely documented increase of the Aβ42/Aβ40 ratio, but also suggests a pathological relevance of an increase in Aβ43, which has been reported recently in vivo with the PSEN–R278I (Saito et al, 2011).
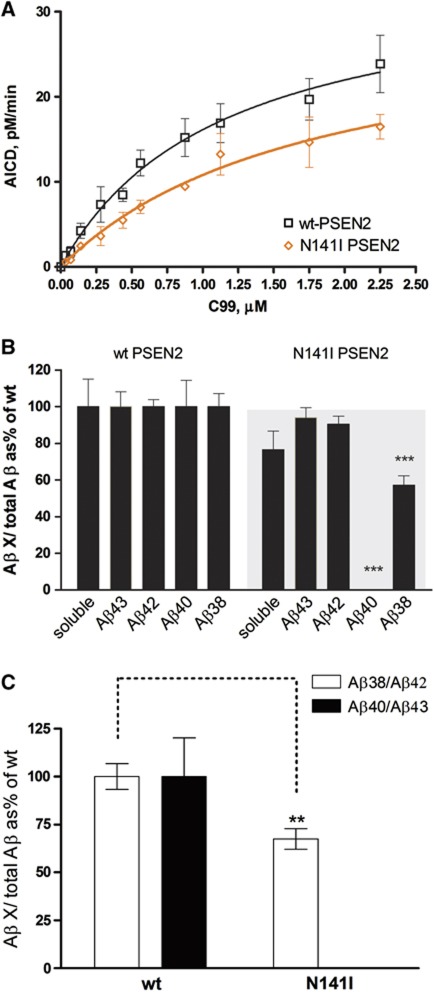
In order to confirm our in vitro data, mouse embryonic fibroblasts (MEFs) derived from Psen1/2−/− mice (Herreman et al, 2000) rescued with human WT– or FAD–PSEN1 mutants were transiently transduced with APPsw. Secreted Aβ levels (sAβ) were quantified by enzyme-linked immunosorbent assay (ELISA). Figure 2E shows drastic reductions in the Aβ38/Aβ42 and Aβ40/Aβ43 ratios for all FAD–PSEN1 mutations, confirming that the fourth enzymatic turnover of the γ-secretase is actually impaired in cells (native γ-secretase conditions). To investigate whether FAD–PSEN2 mutations also affect the fourth enzymatic turnover of the γ-secretase, we performed kinetic analyses with human WT–PSEN2 or FAD N141I–PSEN2 mutant. The effect of the mutation on the endopeptidase efficiency of the γ-secretase complex does not reach statistical significance (Figure 3A) (mean±s.e.: 100±39.9, n=4 or 46.6±3.9, n=3 for WT– or FAD N141I–PSEN2, respectively, two-tailed t-test, P=0.3). Although we cannot discard that this difference is biologically meaningful, the effect on the fourth catalytic cycle is unequivocal. In particular, the Aβ40 product was decreased to undetectable levels (Figure 3B). Similar to the mutations in PSEN1, the N141I–PSEN2 reduces the Aβ38/Aβ42 and Aβ40/Aβ43 ratios (Figure 3C), confirming an impairment in the carboxypeptidase-like activity.
Figure 3.
FAD–PSEN2 N141I impair the fourth enzymatic turnover. (A) Kinetic curves for wt and PSEN2–FAD N141I mutant using purified APP-C99-3XFLAG as substrate (mean±s.e.). (B) Aβ profiles represent Aβ products corrected for the initial endoprotease activities, plotted as % of the wt Aβ products (mean±s.e.). Soluble Aβ (sum of Aβ38, Aβ40, Aβ42 and Aβ43 peptides) suggests accumulation of longer peptides (>Aβ43) in the mutant reactions. (C) Aβ product/substrate ratios determined in vitro for the FAD–PSEN2 mutation show an impairment at the fourth γ-secretase turnover (mean±s.e.). In (B) and (C) statistical significance (two-tailed t-test) is indicated by **P<0.005 and ***P<0.001. Note that N141I abolishes Aβ40 generation.
Since high Aβ43 and Aβ42 (substrates in this cycle) accumulate in vitro or are released in vivo, we speculate that FAD–PSEN mutations promote a premature release of the Aβ43/Aβ42 peptides.
FAD–APP mutations change the product line preference of the γ-secretase
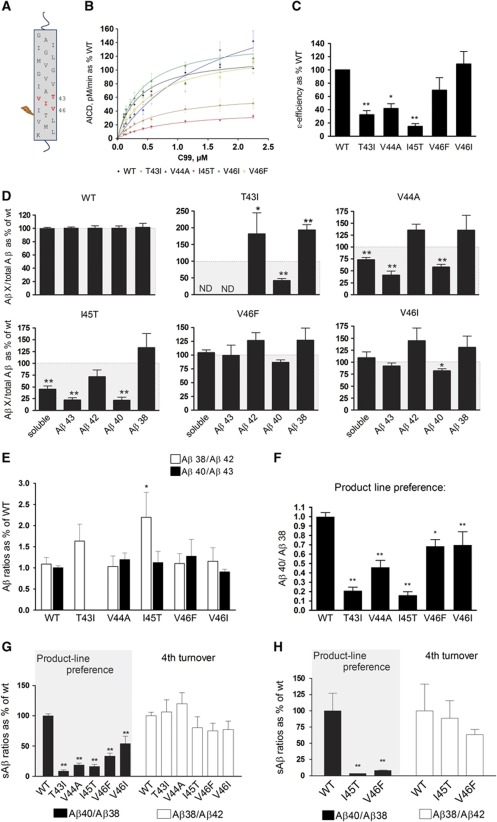
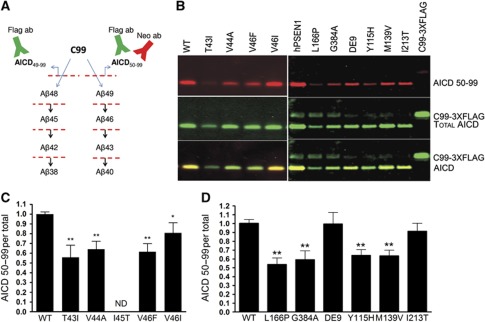
We then asked whether similar mechanisms could explain the effect of mutations in the APP substrate. The tested mutations are located close to the γ-secretase cleavage site, that is, T43I, V44A, I45T, V46F and V46I (Figure 4A) and all produce wild-type Aβ38, Aβ40, Aβ42 and Aβ43 peptides, except for the T43I mutation. Kinetic analyses of the ε-cleavage show that APP-T43I, V44A and -I45T mutants produce less AICD per mol mutant APP compared to wt substrate, while the other mutations do not affect the ε-enzymatic efficiency (Figure 4B and C). In order to analyse accurately the Aβ profiles from wt and FAD substrates, Aβ levels were normalized to the amount of AICD generated in the reaction. FAD Aβ levels, corrected for the initial amounts of substrates, were then plotted as a percentage of the wt enzyme (Figure 4D). Importantly, and in contrast to FAD–PSEN, all the tested APP mutations do not affect Aβ38/Aβ42 or Aβ40/Aβ43 ratios (Figure 4E). The I45T mutant is the exception, showing increased Aβ38/Aβ42 ratio, which would be consistent with an impairment in the processing of Aβ45 (mutant peptide) to Aβ42. However, FAD–APP mutations result in high Aβ40/Aβ38 compared to wt APP (Figure 4F). Thus, all investigated mutations change the product line preference by shifting APP processing towards the Aβ38 production line. The APP-V44A and -I45T substrates in particular show an additional accumulation of longer Aβ precursors (generated in cycles 2 and 3), as deduced from soluble Aβ in Figure 4D. The change in the product line can be explained if these APP mutations shift the position of the ε-cleavage to generate more Aβ48, the initial substrate in the Aβ38 production line. A neo-epitope antibody against AICD50–99 (characterized in Supplementary Figure 3) was generated, and allowed us to confirm the product line preference (Figure 5A and B).
Figure 4.
FAD substrate mutations shift APP processing towards the Aβ38 product line. (A) Schematic overview of FAD–APP mutations used in this study. (B) Kinetic curves for the ε-processing of APP. Detergent-extracted membranes from Psen1/2−/− mEFs rescued with human wt PSEN1 were incubated in 0.25% CHAPSO reaction buffer, with varying concentrations of purified wt or FAD–APP substrates. AICD-3XFLAG levels were analysed by quantitative western blot analysis (see Materials and methods). (C) Enzymatic efficiencies for FAD–APP-C99 substrates (mean±s.e.) prove that AICD generation is affected in three out of five FAD-mutant substrates. (D) FAD Aβ product profiles suggest consistent increments in Aβ42 and Aβ38. Soluble Aβ levels (sum of Aβ38, Aβ40, Aβ42 and Aβ43 peptides) suggest accumulations of longer Aβ peptides in the γ-processing of the V44A and I45T mutants. The T43I mutation disrupts the epitope for the anti-Aβ43-specific antibody, thus neither Aβ43 nor soluble Aβ levels could be determined (ND). (E) Aβ product/substrate ratios reveal that APP mutations do not consistently affect the fourth γ-secretase turnover, but change the product-line preference as indicated by the Aβ40/Aβ38 ratio (F). (G) sAβ in the conditioned media of HEK293 cells transiently transfected with wt or FAD-C99 mutants were quantified by ELISA (see Supplementary Figure 4). sAβ ratios indicate that APP–FAD substrate mutants change the product-line preference towards the Aβ48>Aβ38 (Aβ40/Aβ38) in living cells, but do not affect the fourth catalytic turnover of the γ-secretase (Aβ38/Aβ42) (mean±s.e., n=5). (H) Primary cultured neurons were transduced with SFV expressing WT APP or the indicated mutant substrates (mean±s.d., n=3). (C–H) Statistical significance tested with one-way ANOVA and Dunnett’s post-test, taking the corresponding WT as control group; *P<0.05,**P<0.01.
Figure 5.
Shift in the ε-cleavage position contributes to the FAD-associated phenotype. (A) Detection of AICD50–99 and total AICD using a neo-epitope antibody and the FLAG-M2 antibody, respectively. (B) SDS–PAGE/western blot analysis of AICD products from either wt and FAD substrates (left panel) or wt and FAD–PSEN1mutants (right panel). Signals for the AICD50–99 neo-epitope antibody or the FLAG-M2 antibody are shown in red and green, respectively. Overlapping neo-epitope and FLAG antibody signals are displayed in yellow. (C) AICD50–99/total AICD ratios indicate that FAD–APP mutations promote the Aβ38 product line by shifting the ε-cleavage position. The I45T could not be included in the analysis because of extremely low AICD signals (ND, not determined). (D) This pathogenic mechanism is also observed in some FAD–PSEN1 mutations. Statistical significance of the data (n=5) tested with ANOVA and Dunnett’s post test, taking AICD generated in WT conditions as control group; *P<0.05,**P<0.01.
Figure 5C shows that the FAD–APP mutations consistently shift the position of the ε-cleavage towards AICD49–99, promoting the Aβ38 product line, and therefore causing increments in the Aβ42 and Aβ38 products. Importantly, HEK cells transiently transfected with FAD-mutant C99 substrates increase the Aβ42 and Aβ38 levels in the extracellular medium while decreasing Aβ40, compared to control (wt C99) (Supplementary Figure 4). Figure 4G actually shows that FAD–APP mutations change the product line preference (Aβ40/Aβ38 ratio), but do not alter the fourth enzymatic turnover (Aβ38/Aβ42 ratio). Similar results were obtained from primary neuronal cultures transiently expressing wt-, -I45T- or V46F-APP (Figure 4H). These results indicate that our observations in the cell-free assay can be extrapolated to the in vivo situation. The FAD–APP data imply that promoting the Aβ38 production line is pathogenic.
It has been shown that small changes in the composition of Aβ mixes affect critically their aggregation kinetics and toxic effects (Kuperstein et al, 2010), for example, a minor increase in the Aβ42:Aβ40 ratio stabilizes toxic oligomeric species. Since APP–FAD mutations increase both Aβ42 and Aβ38, we asked whether Aβ38 could have similar biophysical attributes as Aβ40, and therefore could alleviate the potential toxic effects associated to Aβ42. However, and in contrast to Aβ40, Aβ38 has a predicted higher tendency to aggregate (http://www.tango.crg.es) (Fernandez-Escamilla et al, 2004). To validate this prediction, Aβ-aggregation assays were performed using thioflavine T fluorescence as readout. We compared the behaviours of Aβ40 and Aβ38 peptides by analysing the aggregation properties of the Aβ42:Aβ40 (1:9) and Aβ42:Aβ38 (1:9) mixes. In contrast to Aβ40, Aβ38 drives aggregation of Aβ mixes to higher extents (Supplementary Figure 5A). We then compared the effects of different Aβ peptides on spontaneous synaptic transmission in the primary mouse hippocampal neurons. Our results show that Aβ38, similar to Aβ42 and Aβ43, but to a lesser extent, elicits acute synaptotoxicity (Supplementary Figure 5B). Although further work is needed to investigate the effects of Aβ38 in vivo, these data confirm that individual Aβ peptides have widely divergent biophysical and biochemical properties.
Effects of inhibitors and modulators on the γ-secretase activity
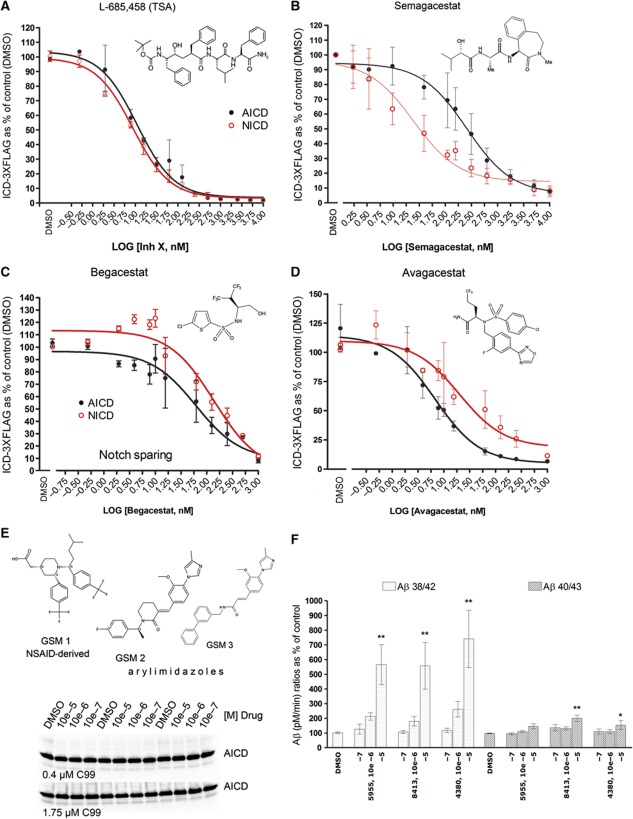
Our data indicate that various mechanisms affecting the Aβ spectrum generated by γ-secretase are responsible for the pathogenic effects of the FAD mutations. Therefore, we asked to what extent γ-secretase inhibitors (GSI) and modulators (GSM) that were tested in clinical trial or are under development (De Strooper et al, 2010) affected the different parameters discussed above. To evaluate γ-secretase inhibition under equal kinetic conditions, we took advantage of the in vitro system and performed activity assays at 1 × Km substrate concentrations for APP C99 or Notch substrates (0.4 and 1.1 μM, respectively). Under these conditions, the GSI semagacestat (LY-450139), begacestat (Notch sparing GSI) and avagacestat (Notch sparing GSI) efficiently inhibit Aβ generation in the two production lines (Supplementary Figure 6). However, semagacestat, which failed in phase III trial (https://www.investor.lilly.com/releasedetail2.cfm?releaseid=592438) because of cognition and skin problems, is more selective for Notch than for APP (AICD IC50=257.8 nM and Notch intracellular domain (NICD) IC50=24.62 nM (95% confidence interval (CI): 190.2–349.5 nM for APP and 15.74–38.51 nM for Notch, n=5), whereas the transition state inhibitor L-685,458 affects both substrates to an equal extent (Figures 6A and B). Surprisingly, and in contrast to previous claims (Martone et al, 2009; Gillman et al, 2011), the selectivity of both of the ‘Notch sparing’ GSI is not significantly different for APP and Notch substrates (Begacestat: AICD IC50=61.71 nM and NICD IC50=138.4 nM (95% CI: 24.77–153.7 nM for AICD and 73.29–261.3 for NICD, n=3) and BMS708163: AICD IC50=6.82 nM and NICD IC50=20.03 nM (95% CI: 4.06–11.46 nM for AICD and 7.76–51.7 nM for NICD, n=3)) (Figure 6C and D).
Figure 6.
Analysis of GSI and GSM. Dose-response inhibitory assays for (A) the transition state analogue (TSA) L-685,458 (InhX), (B) semagacestat, and the Notch-sparing compounds (C) begacestat and (D) avagacestat (see materials in Supplementary data) were performed using CHAPSO-extracted membranes from dKO PSEN1/2 MEFs stably expressing human wt PSEN1 as source of γ-secretase and 1 × Km substrate concentrations (400 nM APP-C99-3XFLAG or 1 μM Notch-3XFLAG). Structures of the different compounds are displayed. In vitro-generated AICD (in black) or NICD (in red) are plotted as percentage of control reaction (DMSO). Error bars indicate s.d. (n=3); except for semagacestat plot (s.e., n=5). (E) Top panel: structures of the GSM tested. Low panel: increasing concentrations of GSM 1–3 did not change in vitro AICD generation, neither at 0.4 μM APP-C99 substrate (1 × Km) nor at saturating conditions (1.75 μM C99-3XFLAG). (F) Effect of increasing concentrations of GSM 1–3 on Aβ production at 1 × Km APP-C99 substrate (0.4 μM): Aβ product/substrate ratios show that GSM 1–3 specifically activate the fourth cycle of the γ-secretase complex. In particular, GSM activate the Aβ38 product line. Panel shows mean±s.e.; statistical significance of the data (n=4) tested with ANOVA and Dunnett’s post test, vehicle (DMSO) as control group; *P<0.05,**P<0.01.
Recently, γ-secretase modulators (GSM) have been evaluated as an alternative to GSI (Weggen et al, 2001). GSM lower Aβ42 and increase Aβ38, but the precise mechanism of action has not been elucidated. One NSAID and two arylimidazole-derived GSM (Oehlrich et al, 2010) (E-2012 and a close analogue) did not affect the endopeptidase activity (Figure 6E and Supplementary Figure 7A) but, as expected, reduced Aβ42 and increased Aβ38. Analysis of the product/substrate ratios for the fourth enzymatic turnover shows that these drugs increase specifically this cycle (Figure 6F and Supplementary Figure 7B) and act, therefore, in the opposite way to the clinical FAD–PSEN mutations. The Aβ ratios indicated that the GSM evaluated in this study act mainly on the fourth cycle of the Aβ38 production line. In fact, our data show them as activators of the γ-secretase (GSA). Taking into account the changes in the FAD–APP Aβ profiles and the possibility that Aβ38 may be part of the pathogenic mechanism, it is crucial to evaluate to what extent the differential effects of the GSA on the Aβ production lines are problematic.
Discussion
This study settles several issues that have been heavily debated in the literature. Dissection of the different activities of the γ-secretase complex allowed us to characterize the mechanisms that are regulated in a consistent fashion by FAD mutations in PSEN and APP. Previous reports have employed steady-state analyses to evaluate the effect of these mutations on the γ-secretase. In general, these studies have employed transfected cells to measure, in culture or in vitro, changes in secreted product levels or follow the intracellular generation of ICD products by coupling it to reporter systems. Although these approaches are informative, they do not truly reflect the kinetic features of the mutated γ-secretase complexes or APP substrates. For instance, substrate concentration and accessibility is not controlled in such assays.
By analysing the catalytic efficiency of the γ-secretase complex (wt or mutated) in an in vitro assay, with both enzyme and substrate in solution, we find here that mutations in PSEN1 and PSEN2 affect γ-secretase at three levels. We see (1) a variable inhibitory effect at the initial endoproteolytic ε-cleavage step, which releases the intracellular domains of substrates such as APP, Notch, Erb4 and N-Cadherin. (2) A consistent effect on the consecutive carboxypeptidase-like γ-cleavage with all PSEN mutations causing a ‘premature’ release of (intermediary) substrates/products, explaining why longer Aβ is generated by these mutants. Interestingly, our data suggest that both Aβ42/Aβ38 and Aβ43/Aβ40 ratio increments are pathologically relevant. (3) Additionally, some of the mutations in PSEN and all mutations investigated here in the APP sequence (selected for their close position to the γ-cleavage site in APP) affect the initial position of the ε-site, that is, whether γ-secretase cleaves preferentially at position 49–50 or 51–50 in the APP sequence. While these three mechanisms explain for the first time the abundantly documented increase in Aβ42/Aβ40 ratio associated with all FAD mutations, they also provide a set of entirely novel insights, as we discuss in the following paragraphs.
Our study gives definite numbers on the catalytic efficiency of the γ-secretase complex at the ε-site and unequivocally shows that ‘loss of function’—lower catalytic efficiency—is not consistent across the FAD mutations tested. In this regard, we wish to draw attention to the fact that point mutations in PSEN might affect protein stability, and therefore solubilization of ‘less stable’ FAD γ-secretase complexes might result in an accentuated ‘loss of function’. Thus, we cannot exclude that the enzymatic efficiencies of particular FAD complexes (less stable) are underestimated in the conditions used in the current work, which would even strengthen our conclusion that ‘loss of function’ is not necessary for the FAD pathogenic mechanism. Taking all into consideration, our results indeed rule out the possibility that loss of intracellular signalling is necessary and sufficient to cause AD, as postulated by the ‘simple’ loss-of-function hypothesis. Interestingly, the effects at the ε-cleavage site are also variable for different substrates tested (Km values for APP, Notch, Erb4 and N-Cadherin, Table I), suggesting that some of the clinical mutations affect the substrate specificity mechanism. This is in particular clear for the M139V and DE9 mutations. DE9 removes part of the hydrophobic domain VII (HDVII) of PSEN1, which is located in the active site of the γ-secretase (Tolia et al, 2008), while the M139 residue is located in the second transmembrane domain of PSEN, which contributes to the formation of the initial substrate-binding site (Watanabe et al, 2010). On the contrary, our results show that the L166P mutation affects the catalytic rate of the enzyme but does not change the substrate specificity of the complex, explaining why steady-state levels of NICD and AICD products in vivo are equally affected by different amino-acid substitutions in the position 166 (Moehlmann et al, 2002).
We analysed in considerable detail the effects of FAD–PSEN1 and FAD–PSEN2 mutations on the γ-secretase (carboxypeptidase-like) mechanism that follows the initial ε-endoproteolytic cleavage of the APP substrate. We find that FAD–PSEN mutations impair dramatically the fourth turnover in both Aβ49>Aβ40 and Aβ48>Aβ38 product lines, resulting in decreased Aβ40/Aβ43 and Aβ38/Aβ42 ratios. Our data therefore give a mechanistic explanation for the decrease in short Aβs (<40) reported in the cerebrospinal fluid of carriers of PSEN-A431E10 (Portelius et al, 2010) or the alterations in the lengths of Aβ peptides produced in vitro by FAD–PSEN-containing complexes (Murphy et al, 2002). Moreover, biophysical observations have shown that FAD–PSEN mutations alter the conformation of the γ-secretase complex (Berezovska et al, 2005). Based on our biochemical data we propose that changes in the active site of FAD–PSEN mutations promote the premature release of the Aβ43 or the Aβ42 peptides from the γ-secretase complexes.
A third mechanism by which FAD–APP mutations act in particular is the shift in the initial ε-cleavage site resulting in an increased Aβ48>Aβ38. Likely, the product preference results from differential docking modes of the APP substrate into the active site. We confirmed the shift towards Aβ48>Aβ38 in living cells expressing wt or mutant APP substrates. This result corroborates our claim that the product line preference is an intrinsic property of the γ-secretase complex that remains unaltered in our cell-free assay. Interestingly, this shift in initial docking and production lines is also observed in four of the six PSEN1 mutants (Figure 5C and D). The fact that some FAD–PSEN1 mutations combine these two mechanisms (impaired fourth cycle and change in the product line preference) explains the direct and indirect correlations between Aβ38 and Aβ42 levels reported in the past (Czirr et al, 2008; Page et al, 2008).
Our study thus demonstrates that FAD mutations cause qualitative changes in the Aβ profiles by various mechanisms (Bentahir et al, 2006; De Strooper, 2007), and that decreased release of intracellular domains (Kelleher and Shen, 2010) is not an essential part of the AD pathogenic mechanism. Nevertheless, as indicated above, the most aggressive PSEN1 mutations, for example, the L166P, negatively impact the endopeptidase activity as well, and therefore it is not unlikely that ‘partial loss’ of γ-secretase function at Notch or other γ-secretase substrates acts as an aggravating factor in FAD.
Moreover, our Aβ product profiles evidence the generation of longer Aβ peptides (>Aβ43) by the most aggressive FAD complexes. However, whether particular changes in the FAD Aβ profiles can be correlated to the age of onset is an interesting but unaddressed question.
In a final series of experiments, we have also assessed to what extent different γ-secretase-directed drugs such as GSI and GSM affect the three mechanisms identified in the current work. When investigating the transition state analogue L-685,458 (InhX), semagacestat, and the Notch-sparing begacestat and avagacestat, we found that the four compounds lowered all γ-cleavages to a similar extent and did not change the Aβ ratios. However, when we assayed the effects of these GSI on Notch processing, which is considered to be the major liability of GSI, we surprisingly found that semagacestat was more effective in inhibiting Notch than APP. This is particularly significant when considering that a phase III clinical trial with semagacestat was interrupted last year because of severe side effects including worsened cognition and increased incidence of skin cancer. Similarly, the Notch-sparing compounds begacestat and avagacestat did not show significant higher selectivity for APP compared to the Notch substrate in our assay. These data raise serious concerns about the interpretation of inhibitory studies that relied on cellular or in vivo data, which in general do not allow direct quantitative comparisons as done with our assay. Importantly, our data do not discard the selective inhibition of APP at the ε-cleavage as a plausible strategy for drug development, but basically indicates that the approaches that have been used to reach this aim need to be revisited.
We also tested GSMs and found that all three candidates keep full functionality at the endopeptidase cleavage and regulate the carboxypeptidase-like activity by activating the fourth cycle of the γ-secretase, resulting in an increased processing of the aggregation-prone Aβs towards shorter Aβ peptides. Our data, however, suggest some caution with this strategy as the tested compounds differentially affect the Aβ48>Aβ38 versus the Aβ49>Aβ40 pathway. The relative increase of Aβ38 observed with all compounds needs further scrutiny, as APP clinical mutations also promote this production line and our initial data provided here (Supplementary Figure 5) suggest that Aβ38 is less benign than Aβ40 with regard to its interaction with Aβ42. However, further research is needed to evaluate whether the weak (but significant) effects we see with Aβ38 translate into an increased toxicity in vivo.
In conclusion, our work provides an important step forward towards the understanding of the mechanisms by which FAD mutations in PSEN and APP cause AD. Our findings support strongly the hypothesis that although these mutations affect γ-secretase in various ways, they all lead to qualitative shifts in the Aβ profiles, which provides a common denominator for the pathogenic effect of all FAD mutations.
Materials and methods
Antibodies
Rabbit polyclonal antibodies human PSEN1–NTF (B19.3), PSEN2–CTF (B24), APH-1a (B80.3), PEN-2 (B126.1) and APP C-terminus (B63.3) and monoclonal 9C3 against Nicastrin have been described (Annaert et al, 2001; Esselens et al, 2004). Rabbit monoclonal neo-epitope AICD was obtained from Lilly Company. Other antibodies purchased were as follows: anti-FLAG M2 from Sigma, goat-anti-mouse IRDye800 from Rockland, goat-anti-rabbit Alexa680 from Invitrogen, 82E1 from Demeditec Diagnostics, MAB5232 and MAB1563 against PSEN1–CTF and human PSEN1–NTF from Chemicon, biotinylated anti-mouse IgG from Vector laboratories, streptavidin-HRP from GE Healthcare and 1E8 from Nanotools, Teningen, Germany. ELISA capturing antibodies purchased were as follows: JRF AB038 for Aβ1-38 from Janssen; JRF/cAb40/28 for Aβ1-40 from Janssen; JRF/cAb42/26 for Aβ1-42 from Janssen; and Aβ1-43 from Signet Labs Inc.. Detection antibody was obtained from Jansen; huAB25-HRPO (Zhou et al, 2011).
GSI and GSM
L-685,458 (Inhibitor X) was purchased from Calbiochem. Begacestat and the GSMs were synthesized according to the procedures reported in either primary publications or patents. GSM 1 was synthesized according to WO2006043064, GSM 2 (E-2012) according to WO2006112552 and WO2006046575, and GSM 3 according to WO2005115990. LY-450139 (semagacestat) and BMS-708163 (avagacestat) were obtained from Haoyuan Chemexpress Co. Limited, Shanghai.
Cell culture
Psen1/Psen2-deficient (−/−) MEF (Psen1/2−/− mEF) (Herreman et al, 2000) were cultured in Dulbecco’s modified Eagle’s medium/F-12 containing 10% fetal bovine serum. Psen1/2−/− mEF rescued with wt (human) PSEN1 or L166P, G384A and DE9 as well as wt (human) PSEN2 or N141I were reported before Bentahir et al (2006). The Y115H, M139V and I213T FAD–PSEN1 cell lines were generated accordingly. mEF–PSEN1 cell lines were transduced with a recombinant adenovirus Ad5/CMV-APP bearing human APP-swe, as previously described (Chavez-Gutierrez et al, 2008). Neuronal cultures derived from E14 embryos and Semiliki Forest virus transfection procedures have been described previously (Annaert et al, 1999). Semliki Forest viruses (SFV) were produced as described (Annaert et al, 1999). Briefly, brains from E14 embryos were trypsinized and plated on 6-cm dishes (Nunc) precoated with poly-L-lysine (Sigma–Aldrich). Cultures were maintained in neurobasal medium (Gibco) supplemented with B27 (GibcoBRL) and 5 mM cytosine arabinoside to prevent glial cell prolification. After 3 days, neurons were transduced with SFV expressing wt or FAD mutant APP. After 1 and 3 h, post-infection media were refreshed. After 24 h, sAβ were analysed by ELISA.
Expression and purification of substrates-3xFLAG
Substrate purification was performed as previously described (Chavez-Gutierrez et al, 2008). Notch-, ErB4- and N-Cadherin-based substrates were designed to be similar in size to the APP substrate (C99–3XFLAG). Purity was assessed by SDS–PAGE and Coomassie staining (GelCode reagent, Pierce).
In vitro activity assays using solubilized γ-secretase
In vitro activity assays were done as previously described (Chavez-Gutierrez et al, 2008), with minor modifications. MEF’s microsomal fractions were prepared in 50 mM citric acid, pH 6.7, 0.25 M sucrose, 1 mM EGTA, complete PI and 1% CHAPSO. In vitro reactions were carried out in 50 mM citric acid, pH 6.7, 0.25 M sucrose, 1 mM EGTA, 1 × EDTA-free complete proteinase inhibitors (Roche), 2.5% DMSO and 0.05% phosphatidylcholine. Reactions were incubated for 4 h at 37 °C unless otherwise mentioned.
Lipids and substrates were extracted by adding 1 volume chloroform/methanol (2:1, v/v). Then, the aqueous fraction (ICD products) was taken and subjected to SDS–PAGE and quantitative western immunoblot. Known amounts of C99-3XFLAG were included as standards for absolute quantifications. ICD-3XFLAG and standards were determined with the anti-FLAG M2 and goat-anti-mouse IR800 antibodies, whereas the AICD50–99 product was determined with a neo-epitope mAb and a goat-anti-rabbit Alexa680 secondary antibody. Infrared signals were detected using the Odyssey Infrared Imaging System.
Calculation of kinetics constants
Kinetic constants were estimated by nonlinear curve-fitting using GraphPad Prism 4 software. The equation V=(Vmax × [S])/(Km+[S]) was used to calculate apparent Km and Vmax values for the different enzymes, where V was experimentally determined using a range of substrate concentrations [S]. γ-Secretase activities were normalized to PSEN–CTF fragment levels or full-length PS1 levels for the DE9 mutant.
Quantification of soluble Aβ using sandwich ELISA
Ninety-six-well plates (NUNC) were coated with 1.5 μg/ml Aβ capture antibody, excepting Aβ43-ab coated at 7.5 μg/ml, in a final volume of 50 μl buffer (10 mMTris HCl, 10 mM NaCl, 10 mM NaN3, pH 8.5). After overnight incubation at 4 °C, the plates were rinsed with PBS+0.05% Tween 20 and blocked with 100 μl per well of casein buffer (1 g casein in 1 l 1 × PBS, pH7.4) for 4 h at room temperature. Samples and standards (synthetic human Aβ1-38, Aβ1-40, Aβ1-42 or Aβ1-43 peptides) were diluted in casein buffer. After overnight incubation at 4 °C, plates were rinsed and developed using 50 μl per well of 100 mM NaAC pH 4.9/TMB (Sigma)/H2O2. Reactions were stopped with 50 μl per well of 2 N H2SO4 and read on a Perkin Elmer Envision 2103 multilabel reader at 450 nm.
Urea gels
Aβ-peptides were analysed by a modified version of the urea-based SDS–PAGE (10% T/5% C instead of 12%T/5% C polyacrylamide and 0.075 M instead of 0.1 M H2SO4 in the separation gel) (Wiltfang et al, 2002). Western immunoblot was done using 1E8, amplifying the signal with biotinylated anti-mouse IgG and streptavidin-HRP. Signals were detected using ECL chemiluminescence with an Intas Imager (Intas, Göttingen, Germany).
Supplementary Material
Acknowledgments
This work was funded by the Fund for Scientific Research, Flanders; the KULeuven; a Methusalem grant from the KULeuven and the Flemisch Government; the Foundation for Alzheimer Research (SAO/FRMA); the Interuniversity Attraction Poles Program of the Belgian Federal Science Policy Office; and the grant PURE (Protein Research Unit Ruhr within Europe) from the State Government North Rhine-Westphalia (JW and HE). BDS is supported by the Arthur Bax and Anna Vanluffelen chair for Alzheimer’s disease. We thank Pat May and Philip Skezeres (Lilly, Indianapolis) for providing us with the AICD neo-epitope antibody, IMEC for access to MEA technology and Michel Vande Kerckhove for helpful discussions.
Author contributions: LC-G and BDS designed the study and wrote the manuscript. LB and LZ contributed to the analysis of FAD–APP mutations. AV, IB, JS, FR and KBcontributed to Aβ aggregation assays and characterized the synaptotoxicity of Aβ peptides on primary neurons. MBo, MBe, SL and LS provided their technical and experimental assistance. SVC contributed to the experimental analysis of FAD–PSEN mutations. HE and JW characterized the Aβ profiles of FAD–PSEN mutants in urea-based gels. HG and EK synthesized and provided the GSI and GSM. All authors have read the manuscript and provided the input.
Footnotes
Harrie Gijsen is an employee of Janssen Pharmaceutica. Bart de Strooper is a consultant for Janssen Pharmaceutica, Envivo Pharmaceutics and Remynd and is supported by research grants from Janssen Pharmaceutics.
References
- Annaert WG, Esselens C, Baert V, Boeve C, Snellings G, Cupers P, Craessaerts K, De Strooper B (2001) Interaction with telencephalin and the amyloid precursor protein predicts a ring structure for presenilins. Neuron 32: 579–589 [DOI] [PubMed] [Google Scholar]
- Annaert WG, Levesque L, Craessaerts K, Dierinck I, Snellings G, Westaway D, George-Hyslop PS, Cordell B, Fraser P, De Strooper B (1999) Presenilin 1 controls gamma-secretase processing of amyloid precursor protein in pre-golgi compartments of hippocampal neurons. J Cell Biol 147: 277–294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bentahir M, Nyabi O, Verhamme J, Tolia A, Horre K, Wiltfang J, Esselmann H, De Strooper B (2006) Presenilin clinical mutations can affect gamma-secretase activity by different mechanisms. J Neurochem 96: 732–742 [DOI] [PubMed] [Google Scholar]
- Berezovska O, Lleo A, Herl LD, Frosch MP, Stern EA, Bacskai BJ, Hyman BT (2005) Familial Alzheimer’s disease presenilin 1 mutations cause alterations in the conformation of presenilin and interactions with amyloid precursor protein. J Neurosci 25: 3009–3017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergmans BA, De Strooper B (2010) Gamma-secretases: from cell biology to therapeutic strategies. Lancet Neurol 9: 2215–226 [DOI] [PubMed] [Google Scholar]
- Bezprozvanny I, Mattson MP (2008) Neuronal calcium mishandling and the pathogenesis of Alzheimer’s disease. Trends Neurosci 31: 454–463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boeve BF, Baker M, Dickson DW, Parisi JE, Giannini C, Josephs KA, Hutton M, Pickering-Brown SM, Rademakers R, Tang-Wai D, Jack CR Jr., Kantarci K, Shiung MM, Golde T, Smith GE, Geda YE, Knopman DS, Petersen RC (2006) Frontotemporal dementia and parkinsonism associated with the IVS1+1G−>a mutation in progranulin: a clinicopathologic study. Brain 129: Pt 113103–3114 [DOI] [PubMed] [Google Scholar]
- Borchelt DR, Thinakaran G, Eckman CB, Lee MK, Davenport F, Ratovitsky T, Prada CM, Kim G, Seekins S, Yager D, Slunt HH, Wang R, Seeger M, Levey AI, Gandy SE, Copeland NG, Jenkins NA, Price DL, Younkin SG, Sisodia SS (1996) Familial Alzheimer’s disease-linked presenilin 1 variants elevate Abeta1-42/1-40 ratio in vitro and in vivo. Neuron 17: 1005–1013 [DOI] [PubMed] [Google Scholar]
- Chavez-Gutierrez L, Tolia A, Maes E, Li T, Wong PC, de Strooper B (2008) Glu(332) in the Nicastrin ectodomain is essential for gamma-secretase complex maturation but not for its activity. J Biol Chem 283: 20096–20105 [DOI] [PubMed] [Google Scholar]
- Czirr E, Cottrell BA, Leuchtenberger S, Kukar T, Ladd TB, Esselmann H, Paul S, Schubenel R, Torpey JW, Pietrzik CU, Golde TE, Wiltfang J, Baumann K, Koo EH, Weggen S (2008) Independent generation of Abeta42 and Abeta38 peptide species by gamma-secretase. J Biol Chem 283: 17049–17054 [DOI] [PubMed] [Google Scholar]
- De Strooper B (2007) Loss-of-function presenilin mutations in Alzheimer disease. Talking Point on the role of presenilin mutations in Alzheimer disease. EMBO Rep 8: 141–146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Strooper B, Annaert W (2010) Novel research horizons for presenilins and gamma-secretases in cell biology and disease. Annu Rev Cell Dev Biol 26: 235–260 [DOI] [PubMed] [Google Scholar]
- De Strooper B, Saftig P, Craessaerts K, Vanderstichele H, Guhde G, Annaert W, Von Figura K, Van Leuven F (1998) Deficiency of presenilin-1 inhibits the normal cleavage of amyloid precursor protein. Nature 391: 387–390 [DOI] [PubMed] [Google Scholar]
- De Strooper B, Vassar R, Golde T (2010) The secretases: enzymes with therapeutic potential in Alzheimer disease. Nat Rev Neurol 6: 99–107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duff K, Eckman C, Zehr C, Yu X, Prada CM, Perez-tur J, Hutton M, Buee L, Harigaya Y, Yager D, Morgan D, Gordon MN, Holcomb L, Refolo L, Zenk B, Hardy J, Younkin S (1996) Increased amyloid-beta42(43) in brains of mice expressing mutant presenilin 1. Nature 383: 710–713 [DOI] [PubMed] [Google Scholar]
- Esselens C, Oorschot V, Baert V, Raemaekers T, Spittaels K, Serneels L, Zheng H, Saftig P, De Strooper B, Klumperman J, Annaert W (2004) Presenilin 1 mediates the turnover of telencephalin in hippocampal neurons via an autophagic degradative pathway. J Cell Biol 166: 1041–1054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez-Escamilla AM, Rousseau F, Schymkowitz J, Serrano L (2004) Prediction of sequence-dependent and mutational effects on the aggregation of peptides and proteins. Nat Biotechnol 22: 1302–1306 [DOI] [PubMed] [Google Scholar]
- Fukumori A, Fluhrer R, Steiner H, Haass C (2010) Three-amino acid spacing of presenilin endoproteolysis suggests a general stepwise cleavage of gamma-secretase-mediated intramembrane proteolysis. J Neurosci 30: 7853–7862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Funamoto S, Morishima-Kawashima M, Tanimura Y, Hirotani N, Saido TC, Ihara Y (2004) Truncated carboxyl-terminal fragments of beta-amyloid precursor protein are processed to amyloid beta-proteins 40 and 42. Biochemistry 43: 13532–13540 [DOI] [PubMed] [Google Scholar]
- Gillman KW, Starrett JE, Parker MF, Xie K, Bronson JJ, Marcin LR, McElhone KE, Bergstrom CP, Mate RA, Williams R, Meredith JE, Burton CR, Barten DM, Toyn JH, Roberts SB, Lentz KA, Houston JG, Zaczek R, Albright CF, Decicco CP et al. (2011) Discovery and evaluation of BMS-708163, a potent, selective and orally bioavailable Î3-secretase inhibitor. ACS Med Chem Lett 1: 120–124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardy J, Selkoe DJ (2002) The amyloid hypothesis of Alzheimer’s disease: progress and problems on the road to therapeutics. Science 297: 353–356 [DOI] [PubMed] [Google Scholar]
- Hebert SS, Serneels L, Dejaegere T, Horre K, Dabrowski M, Baert V, Annaert W, Hartmann D, De Strooper B (2004) Coordinated and widespread expression of gamma-secretase in vivo: evidence for size and molecular heterogeneity. Neurobiol Dis 17: 260–272 [DOI] [PubMed] [Google Scholar]
- Heilig EA, Xia W, Shen J, Kelleher RJ III (2010) A presenilin-1 mutation identified in familial Alzheimer disease with cotton wool plaques causes a nearly complete loss of gamma-secretase activity. J Biol Chem 285: 22350–22359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herreman A, Serneels L, Annaert W, Collen D, Schoonjans L, De Strooper B (2000) Total inactivation of gamma-secretase activity in presenilin-deficient embryonic stem cells. Nat Cell Biol 2: 461–462 [DOI] [PubMed] [Google Scholar]
- Jarrett JT, Berger EP, Lansbury PT Jr. (1993) The carboxy terminus of the beta amyloid protein is critical for the seeding of amyloid formation: implications for the pathogenesis of Alzheimer’s disease. Biochemistry 32: 4693–4697 [DOI] [PubMed] [Google Scholar]
- Jarrett JT, Lansbury PT Jr (1993) Seeding ‘one-dimensional crystallization’ of amyloid: a pathogenic mechanism in Alzheimer’s disease and scrapie? Cell 73: 1055–1058 [DOI] [PubMed] [Google Scholar]
- Kakuda N, Funamoto S, Yagishita S, Takami M, Osawa S, Dohmae N, Ihara Y (2006) Equimolar production of amyloid beta-protein and amyloid precursor protein intracellular domain from beta-carboxyl-terminal fragment by gamma-secretase. J Biol Chem 281: 14776–14786 [DOI] [PubMed] [Google Scholar]
- Kelleher RJ 3rd, Shen J (2010) Genetics. Gamma-secretase and human disease. Science 330: 1055–1056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Onstead L, Randle S, Price R, Smithson L, Zwizinski C, Dickson DW, Golde T, McGowan E (2007) Abeta40 inhibits amyloid deposition in vivo. J Neurosci 27: 627–633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuperstein I, Broersen K, Benilova I, Rozenski J, Jonckheere W, Debulpaep M, Vandersteen A, Segers-Nolten I, Van Der Werf K, Subramaniam V, Braeken D, Callewaert G, Bartic C, D’Hooge R, Martins IC, Rousseau F, Schymkowitz J, De Strooper B (2010) Neurotoxicity of Alzheimer’s disease Abeta peptides is induced by small changes in the Abeta42 to Abeta40 ratio. EMBO J 29: 3408–3420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JH, Yu WH, Kumar A, Lee S, Mohan PS, Peterhoff CM, Wolfe DM, Martinez-Vicente M, Massey AC, Sovak G, Uchiyama Y, Westaway D, Cuervo AM, Nixon RA (2010) Lysosomal proteolysis and autophagy require presenilin 1 and are disrupted by Alzheimer-related PS1 mutations. Cell 141: 1146–1158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martone RL, Zhou H, Atchison K, Comery T, Xu JZ, Huang X, Gong X, Jin M, Kreft A, Harrison B, Mayer SC, Aschmies S, Gonzales C, Zaleska MM, Riddell DR, Wagner E, Lu P, Sun SC, Sonnenberg-Reines J, Oganesian A et al. (2009) Begacestat (GSI-953): a novel, selective thiophene sulfonamide inhibitor of amyloid precursor protein gamma-secretase for the treatment of Alzheimer’s disease. J Pharmacol Exp Ther 331: 598–608 [DOI] [PubMed] [Google Scholar]
- McGowan E, Pickford F, Kim J, Onstead L, Eriksen J, Yu C, Skipper L, Murphy MP, Beard J, Das P, Jansen K, Delucia M, Lin WL, Dolios G, Wang R, Eckman CB, Dickson DW, Hutton M, Hardy J, Golde T (2005) Abeta42 is essential for parenchymal and vascular amyloid deposition in mice. Neuron 47: 191–199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moehlmann T, Winkler E, Xia X, Edbauer D, Murrell J, Capell A, Kaether C, Zheng H, Ghetti B, Haass C, Steiner H (2002) Presenilin-1 mutations of leucine 166 equally affect the generation of the Notch and APP intracellular domains independent of their effect on Abeta 42 production. Proc Natl Acad Sci USA 99: 8025–8030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murayama O, Tomita T, Nihonmatsu N, Murayama M, Sun X, Honda T, Iwatsubo T, Takashima A (1999) Enhancement of amyloid beta 42 secretion by 28 different presenilin 1 mutations of familial Alzheimer’s disease. Neurosci Lett 265: 61–63 [DOI] [PubMed] [Google Scholar]
- Murphy MP, Uljon SN, Golde TE, Wang R (2002) FAD-linked mutations in presenilin 1 alter the length of Abeta peptides derived from betaAPP transmembrane domain mutants. Biochim Biophys Acta 1586: 199–209 [DOI] [PubMed] [Google Scholar]
- Oehlrich D, Berthelot DJ, Gijsen HJ (2011) Gamma-secretase modulators as potential disease modifying anti-alzheimer’s drugs. J Med Chem 54: 669–698 [DOI] [PubMed] [Google Scholar]
- Page RM, Baumann K, Tomioka M, Perez-Revuelta BI, Fukumori A, Jacobsen H, Flohr A, Luebbers T, Ozmen L, Steiner H, Haass C (2008) Generation of Abeta38 and Abeta42 is independently and differentially affected by familial Alzheimer disease-associated presenilin mutations and gamma-secretase modulation. J Biol Chem 283: 677–683 [DOI] [PubMed] [Google Scholar]
- Pink AE, Simpson MA, Brice GW, Smith CH, Desai N, Mortimer PS, Barker JN, Trembath RC (2011) PSENEN and NCSTN mutations in familial hidradenitis suppurativa (acne inversa). J Invest Dermatol 131: 1568–1570 [DOI] [PubMed] [Google Scholar]
- Portelius E, Andreasson U, Ringman JM, Buerger K, Daborg J, Buchhave P, Hansson O, Harmsen A, Gustavsson MK, Hanse E, Galasko D, Hampel H, Blennow K, Zetterberg H (2010) Distinct cerebrospinal fluid amyloid beta peptide signatures in sporadic and PSEN1 A431E-associated familial Alzheimer’s disease. Mol Neurodegener 5: 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi-Takahara Y, Morishima-Kawashima M, Tanimura Y, Dolios G, Hirotani N, Horikoshi Y, Kametani F, Maeda M, Saido TC, Wang R, Ihara Y (2005) Longer forms of amyloid beta protein: implications for the mechanism of intramembrane cleavage by gamma-secretase. J Neurosci 25: 436–445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quintero-Monzon O, Martin MM, Fernandez MA, Cappello CA, Krzysiak AJ, Osenkowski P, Wolfe MS (2011) Dissociation between the processivity and total activity of gamma-secretase: implications for the mechanism of Alzheimer’s disease-causing presenilin mutations. Biochemistry 50: 9023–9035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito T, Suemoto T, Brouwers N, Sleegers K, Funamoto S, Mihira N, Matsuba Y, Yamada K, Nilsson P, Takano J, Nishimura M, Iwata N, Van Broeckhoven C, Ihara Y, Saido TC (2011) Potent amyloidogenicity and pathogenicity of Abeta43. Nat Neurosci 14: 1023–1032 [DOI] [PubMed] [Google Scholar]
- Sastre M, Steiner H, Fuchs K, Capell A, Multhaup G, Condron MM, Teplow DB, Haass C (2001) Presenilin-dependent gamma-secretase processing of beta-amyloid precursor protein at a site corresponding to the S3 cleavage of Notch. EMBO Rep 2: 835–841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato T, Dohmae N, Qi Y, Kakuda N, Misonou H, Mitsumori R, Maruyama H, Koo EH, Haass C, Takio K, Morishima-Kawashima M, Ishiura S, Ihara Y (2003) Potential link between amyloid beta-protein 42 and C-terminal fragment gamma 49-99 of beta-amyloid precursor protein. J Biol Chem 278: 24294–24301 [DOI] [PubMed] [Google Scholar]
- Saura CA, Choi SY, Beglopoulos V, Malkani S, Zhang D, Shankaranarayana Rao BS, Chattarji S, Kelleher RJ III, Kandel ER, Duff K, Kirkwood A, Shen J (2004) Loss of presenilin function causes impairments of memory and synaptic plasticity followed by age-dependent neurodegeneration. Neuron 42: 23–36 [DOI] [PubMed] [Google Scholar]
- Scheuner D, Eckman C, Jensen M, Song X, Citron M, Suzuki N, Bird TD, Hardy J, Hutton M, Kukull W, Larson E, Levy-Lahad E, Viitanen M, Peskind E, Poorkaj P, Schellenberg G, Tanzi R, Wasco W, Lannfelt L, Selkoe D et al. (1996) Secreted amyloid beta-protein similar to that in the senile plaques of Alzheimer’s disease is increased in vivo by the presenilin 1 and 2 and APP mutations linked to familial Alzheimer’s disease. Nat Med 2: 864–870 [DOI] [PubMed] [Google Scholar]
- Shen J, Kelleher RJ III (2007) The presenilin hypothesis of Alzheimer’s disease: evidence for a loss-of-function pathogenic mechanism. Proc Natl Acad Sci USA 104: 403–409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimojo M, Sahara N, Murayama M, Ichinose H, Takashima A (2007) Decreased Abeta secretion by cells expressing familial Alzheimer’s disease-linked mutant presenilin 1. Neurosci Res 57: 446–453 [DOI] [PubMed] [Google Scholar]
- Shirotani K, Edbauer D, Prokop S, Haass C, Steiner H (2004) Identification of distinct gamma-secretase complexes with different APH-1 variants. J Biol Chem 279: 41340–41345 [DOI] [PubMed] [Google Scholar]
- Song W, Nadeau P, Yuan M, Yang X, Shen J, Yankner BA (1999) Proteolytic release and nuclear translocation of Notch-1 are induced by presenilin-1 and impaired by pathogenic presenilin-1 mutations. Proc Natl Acad Sci USA 96: 6959–6963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takami M, Nagashima Y, Sano Y, Ishihara S, Morishima-Kawashima M, Funamoto S, Ihara Y (2009) Gamma-secretase: successive tripeptide and tetrapeptide release from the transmembrane domain of beta-carboxyl terminal fragment. J Neurosci 29: 13042–13052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanzi RE, Bertram L (2005) Twenty years of the Alzheimer’s disease amyloid hypothesis: a genetic perspective. Cell 120: 545–555 [DOI] [PubMed] [Google Scholar]
- Tolia A, De Strooper B (2009) Structure and function of gamma-secretase. Semin Cell Dev Biol 20: 211–218 [DOI] [PubMed] [Google Scholar]
- Tolia A, Horre K, De Strooper B (2008) Transmembrane domain 9 of presenilin determines the dynamic conformation of the catalytic site of gamma-secretase. J Biol Chem 283: 19793–19803 [DOI] [PubMed] [Google Scholar]
- Wakabayashi T, De Strooper B (2008) Presenilins: members of the {gamma}-secretase quartets, but part-time soloists too. Physiology (Bethesda) 23: 194–204 [DOI] [PubMed] [Google Scholar]
- Wang B, Yang W, Wen W, Sun J, Su B, Liu B, Ma D, Lv D, Wen Y, Qu T, Chen M, Sun M, Shen Y, Zhang X (2010) Gamma-secretase gene mutations in familial acne inversa. Science 330: 1065. [DOI] [PubMed] [Google Scholar]
- Wang R, Wang B, He W, Zheng H (2006) Wild-type presenilin 1 protects against Alzheimer disease mutation-induced amyloid pathology. J Biol Chem 281: 15330–15336 [DOI] [PubMed] [Google Scholar]
- Watanabe N, Image I II, Takagi S, Tominaga A, Image Image I, Tomita T, Iwatsubo T (2010) Functional analysis of the transmembrane domains of presenilin 1: participation of transmembrane domains 2 and 6 in the formation of initial substrate-binding site of gamma-secretase. J Biol Chem 285: 19738–19746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weggen S, Eriksen JL, Das P, Sagi SA, Wang R, Pietrzik CU, Findlay KA, Smith TE, Murphy MP, Bulter T, Kang DE, Marquez-Sterling N, Golde TE, Koo EH (2001) A subset of NSAIDs lower amyloidogenic Abeta42 independently of cyclooxygenase activity. Nature 414: 212–216 [DOI] [PubMed] [Google Scholar]
- Weidemann A, Eggert S, Reinhard FB, Vogel M, Paliga K, Baier G, Masters CL, Beyreuther K, Evin G (2002) A novel epsilon-cleavage within the transmembrane domain of the Alzheimer amyloid precursor protein demonstrates homology with Notch processing. Biochemistry 41: 2825–2835 [DOI] [PubMed] [Google Scholar]
- Welander H, Franberg J, Graff C, Sundstrom E, Winblad B, Tjernberg LO (2009) Abeta43 is more frequent than Abeta40 in amyloid plaque cores from Alzheimer disease brains. J Neurochem 110: 697–706 [DOI] [PubMed] [Google Scholar]
- Wilson CA, Murphy DD, Giasson BI, Zhang B, Trojanowski JQ, Lee VM (2004) Degradative organelles containing mislocalized alpha-and beta-synuclein proliferate in presenilin-1 null neurons. J Cell Biol 165: 335–346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiltfang J, Esselmann H, Bibl M, Smirnov A, Otto M, Paul S, Schmidt B, Klafki HW, Maler M, Dyrks T, Bienert M, Beyermann M, Ruther E, Kornhuber J (2002) Highly conserved and disease-specific patterns of carboxyterminally truncated Abeta peptides 1-37/38/39 in addition to 1-40/42 in Alzheimer’s disease and in patients with chronic neuroinflammation. J Neurochem 81: 481–496 [DOI] [PubMed] [Google Scholar]
- Wolfe MS (2007) When loss is gain: reduced presenilin proteolytic function leads to increased Abeta42/Abeta40. Talking Point on the role of presenilin mutations in Alzheimer disease. EMBO Rep 8: 136–140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yagishita S, Morishima-Kawashima M, Ishiura S, Ihara Y (2008) Abeta46 is processed to Abeta40 and Abeta43, but not to Abeta42, in the low density membrane domains. J Biol Chem 283: 733–738 [DOI] [PubMed] [Google Scholar]
- Zhang H, Sun S, Herreman A, De Strooper B, Bezprozvanny I (2010) Role of presenilins in neuronal calcium homeostasis. J Neurosci 30: 8566–8580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou L, Brouwers N, Benilova I, Vandersteen A, Mercken M, Van Laere K, Van Damme P, Demedts D, Van Leuven F, Sleegers K, Broersen K, Van Broeckhoven C, Vandenberghe R, De Strooper B (2011) Amyloid precursor protein mutation E682K at the alternative beta-secretase cleavage beta’-site increases Abeta generation. EMBO Mol Med 3: 291–302 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.