Abstract
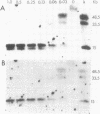
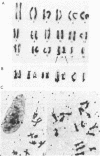
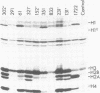
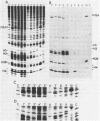
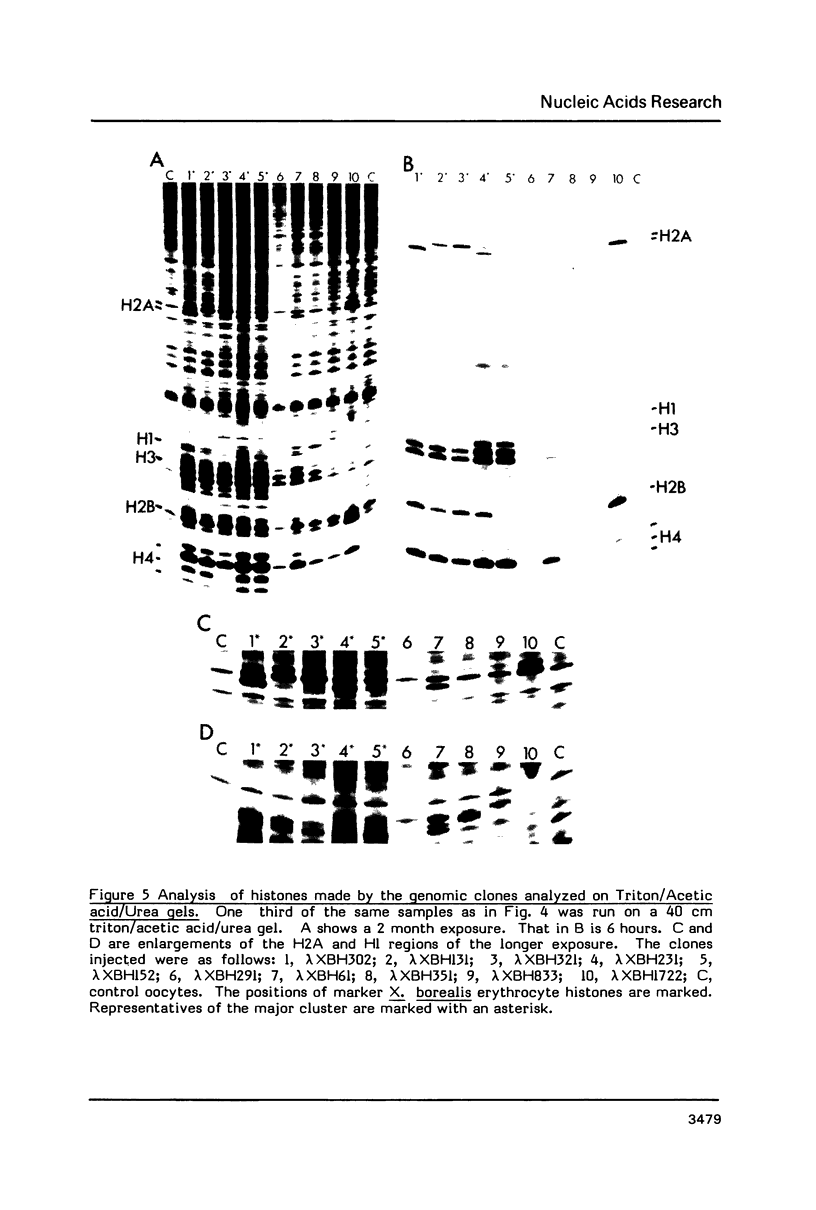
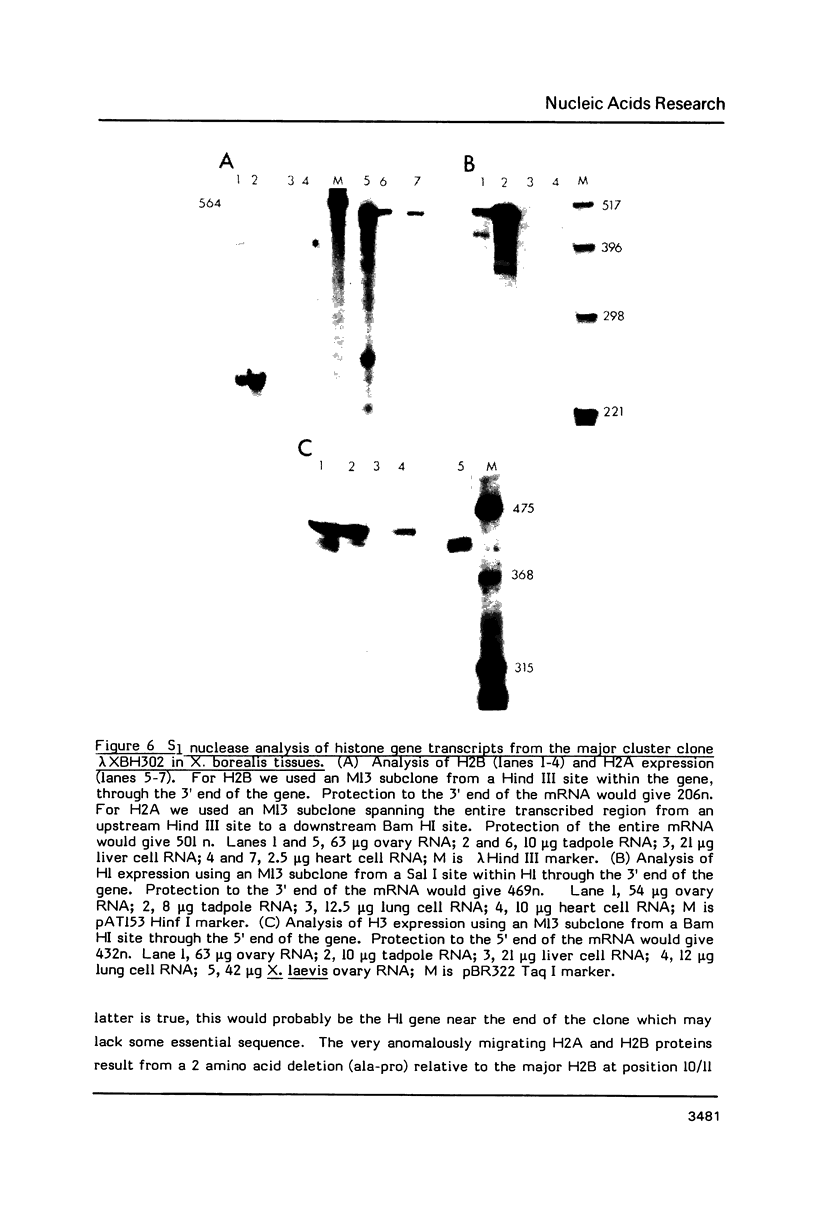
We have isolated genomic clones from Xenopus borealis representing 3 different types of histone gene cluster. We show that the major type (H1, H2B, H2A, H4, H3), present at about 60-70 copies per haploid genome (1), is tandemly reiterated with a repeat length of 15 kb. In situ hybridization to mitotic chromosomes shows that the majority of histone genes in Xenopus borealis are at one locus. This locus is on the long arm of one of the small sub-metacentric chromosomes. A minor cluster type with the gene order H1, H3, H4, H2A is present at about 10-15 copies. The genome also contains rare or unique cluster types present at less than 5 copies having other types of organisation. An isolate of this type had the gene order H1, H4, H2B, H2A, H1 (no H3 cloned). Microinjection of all of the clones into Xenopus laevis oocyte nuclei shows that most of the genes present are functional or potentially functional and a number of variant histone proteins have been observed. S1 mapping experiments confirm that the genes of the major cluster are expressed in all tissues and at all developmental stages examined.
Full text
PDF












Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Benton W. D., Davis R. W. Screening lambdagt recombinant clones by hybridization to single plaques in situ. Science. 1977 Apr 8;196(4286):180–182. doi: 10.1126/science.322279. [DOI] [PubMed] [Google Scholar]
- Berk A. J., Sharp P. A. Sizing and mapping of early adenovirus mRNAs by gel electrophoresis of S1 endonuclease-digested hybrids. Cell. 1977 Nov;12(3):721–732. doi: 10.1016/0092-8674(77)90272-0. [DOI] [PubMed] [Google Scholar]
- Busslinger M., Rusconi S., Birnstiel M. L. An unusual evolutionary behaviour of a sea urchin histone gene cluster. EMBO J. 1982;1(1):27–33. doi: 10.1002/j.1460-2075.1982.tb01119.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Childs G., Maxson R., Cohn R. H., Kedes L. Orphons: dispersed genetic elements derived from tandem repetitive genes of eucaryotes. Cell. 1981 Mar;23(3):651–663. doi: 10.1016/0092-8674(81)90428-1. [DOI] [PubMed] [Google Scholar]
- Destrée O. H., Bendig M. M., De Laaf R. T., Koster J. G. Organization of Xenopus histone gene variants within clusters and their transcriptional expression. Biochim Biophys Acta. 1984 Jun 16;782(2):132–141. doi: 10.1016/0167-4781(84)90016-2. [DOI] [PubMed] [Google Scholar]
- Grandy D. K., Dodgson J. B. Structure and organization of the chicken H2B histone gene family. Nucleic Acids Res. 1987 Feb 11;15(3):1063–1080. doi: 10.1093/nar/15.3.1063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gross K., Schaffner W., Telford J., Birnstiel M. Molecular analysis of the histone gene cluster of Psammechinus miliaris: III. Polarity and asymmetry of the histone-coding sequences. Cell. 1976 Aug;8(4):479–484. doi: 10.1016/0092-8674(76)90215-4. [DOI] [PubMed] [Google Scholar]
- Harvey R. P., Whiting J. A., Coles L. S., Krieg P. A., Wells J. R. H2A.F: an extremely variant histone H2A sequence expressed in the chicken embryo. Proc Natl Acad Sci U S A. 1983 May;80(10):2819–2823. doi: 10.1073/pnas.80.10.2819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hentschel C. C., Birnstiel M. L. The organization and expression of histone gene families. Cell. 1981 Aug;25(2):301–313. doi: 10.1016/0092-8674(81)90048-9. [DOI] [PubMed] [Google Scholar]
- Krieg P. A., Robins A. J., Gait M. J., Titmas R. C., Wells J. R. Chicken histone H5: selection of a cDNA recombinant using an extended synthetic primer. Nucleic Acids Res. 1982 Mar 11;10(5):1495–1502. doi: 10.1093/nar/10.5.1495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lifton R. P., Goldberg M. L., Karp R. W., Hogness D. S. The organization of the histone genes in Drosophila melanogaster: functional and evolutionary implications. Cold Spring Harb Symp Quant Biol. 1978;42(Pt 2):1047–1051. doi: 10.1101/sqb.1978.042.01.105. [DOI] [PubMed] [Google Scholar]
- Loenen W. A., Brammar W. J. A bacteriophage lambda vector for cloning large DNA fragments made with several restriction enzymes. Gene. 1980 Aug;10(3):249–259. doi: 10.1016/0378-1119(80)90054-2. [DOI] [PubMed] [Google Scholar]
- Maxson R., Cohn R., Kedes L., Mohun T. Expression and organization of histone genes. Annu Rev Genet. 1983;17:239–277. doi: 10.1146/annurev.ge.17.120183.001323. [DOI] [PubMed] [Google Scholar]
- Old R. W., Woodland H. R., Ballantine J. E., Aldridge T. C., Newton C. A., Bains W. A., Turner P. C. Organization and expression of cloned histone gene clusters from Xenopus laevis and X. borealis. Nucleic Acids Res. 1982 Dec 11;10(23):7561–7580. doi: 10.1093/nar/10.23.7561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Old R. W., Woodland H. R. Histone genes: not so simple after all. Cell. 1984 Oct;38(3):624–626. doi: 10.1016/0092-8674(84)90256-3. [DOI] [PubMed] [Google Scholar]
- Rigby P. W., Dieckmann M., Rhodes C., Berg P. Labeling deoxyribonucleic acid to high specific activity in vitro by nick translation with DNA polymerase I. J Mol Biol. 1977 Jun 15;113(1):237–251. doi: 10.1016/0022-2836(77)90052-3. [DOI] [PubMed] [Google Scholar]
- Ruberti I., Fragapane P., Pierandrei-Amaldi P., Beccari E., Amaldi F., Bozzoni I. Characterization of histone genes isolated from Xenopus laevis and Xenopus tropicalis genomic libraries. Nucleic Acids Res. 1982 Dec 11;10(23):7543–7559. doi: 10.1093/nar/10.23.7543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephenson E. C., Erba H. P., Gall J. G. Histone gene clusters of the newt notophthalmus are separated by long tracts of satellite DNA. Cell. 1981 Jun;24(3):639–647. doi: 10.1016/0092-8674(81)90090-8. [DOI] [PubMed] [Google Scholar]
- Turner P. C., Aldridge T. C., Woodland H. R., Old R. W. Nucleotide sequences of H1 histone genes from Xenopus laevis. A recently diverged pair of H1 genes and an unusual H1 pseudogene. Nucleic Acids Res. 1983 Jun 25;11(12):4093–4107. doi: 10.1093/nar/11.12.4093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner P. C., Woodland H. R. H3 and H4 histone cDNA sequences from Xenopus: a sequence comparison of H4 genes. Nucleic Acids Res. 1982 Jun 25;10(12):3769–3780. doi: 10.1093/nar/10.12.3769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner P. C., Woodland H. R. Histone gene number and organisation in Xenopus: Xenopus borealis has a homogeneous major cluster. Nucleic Acids Res. 1983 Feb 25;11(4):971–986. doi: 10.1093/nar/11.4.971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodland H. R., Warmington J. R., Ballantine J. E., Turner P. C. Are there major developmentally regulated H4 gene classes in Xenopus? Nucleic Acids Res. 1984 Jun 25;12(12):4939–4958. doi: 10.1093/nar/12.12.4939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu M., Holmes D. S., Davidson N., Cohn R. H., Kedes L. H. The relative positions of sea urchin histone genes on the chimeric plasmids pSp2 and pSp17 as studied by electronmicroscopy. Cell. 1976 Sep;9(1):163–169. doi: 10.1016/0092-8674(76)90061-1. [DOI] [PubMed] [Google Scholar]
- Zernik M., Heintz N., Boime I., Roeder R. G. Xenopus laevis histone genes: variant H1 genes are present in different clusters. Cell. 1980 Dec;22(3):807–815. doi: 10.1016/0092-8674(80)90557-7. [DOI] [PubMed] [Google Scholar]
- van Dongen W., de Laaf L., Zaal R., Moorman A., Destrée O. The organization of the histone genes in the genome of Xenopus laevis. Nucleic Acids Res. 1981 May 25;9(10):2297–2311. doi: 10.1093/nar/9.10.2297. [DOI] [PMC free article] [PubMed] [Google Scholar]