Abstract
Epigenetic modifications, such as DNA and histone methylation, are responsible for regulatory pathways that affect disease. Current epigenetic analyses use bisulfite conversion to identify DNA methylation and chromatin immunoprecipitation to collect molecules bearing a specific histone modification. In this work, we present a proof-of-principle demonstration for a new method using a nanofluidic device that combines real-time detection and automated sorting of individual molecules based on their epigenetic state. This device evaluates the fluorescence from labeled epigenetic modifications to actuate sorting. This technology has demonstrated up to 98% accuracy in molecule sorting and has achieved postsorting sample recovery on femtogram quantities of genetic material. We have applied it to sort methylated DNA molecules using simultaneous, multicolor fluorescence to identify methyl binding domain protein-1 (MBD1) bound to full-duplex DNA. The functionality enabled by this nanofluidic platform now provides a workflow for color-multiplexed detection, sorting, and recovery of single molecules toward subsequent DNA sequencing.
Keywords: epigenomic, DNA–protein interaction, single molecule detection, fluorescence microscopy, lab on a chip
In chromatin, chemical modifications to histone proteins and DNA alter the status of the epigenome and influence gene regulation and normal development. Their aberrant placement has been linked to the onset of cancer (1, 2) and other diseases. Bisulfite conversion and immunoprecipitation (IP) have been used extensively to examine these modifications on locus-specific and genome-wide scales. These approaches have limitations in terms of material handling or multiplexed detection. Conventional chromatin immunoprecipitation (ChIP) requires an abundance of input material, often 103–106 cells for genome-wide studies, to compensate for > 99% material loss during processing (3, 4). This problem compounds for sequential re-ChIP reactions, limiting the study of multivalent modifications (4), which could provide a clear view of epigenetic coordination. Whereas DNA methylation analysis using bisulfite conversion can operate on picogram quantities of DNA (5–7), the conversion causes degradation of > 90% of the input DNA. Methods that combine ChIP and bisulfite sequencing in a sequential process (8) have demonstrated progress in multiplexed epigenetic analysis. There continues to be active research in reducing the input material requirements and in automation of the processes (9–11) for epigenetic analysis. Furthermore, there is interest in additional capability for simultaneous detection of multiple epigenetic modifications in the same material.
Miniaturized fluidic devices offer a compelling toolset for multiplexed detection and efficient sample handling in analytical and preparatory systems. Microfluidics have performed complicated workflows that include nanoliter sample handling (12) and incorporate electrodes (13–16) or valves (12, 17) for sophisticated processing. Nanofluidics have achieved attoliter-scale fluid volume confinement to isolate and quantify the attributes of individual molecules that can be obscured during ensemble measurements. These devices have recently been demonstrated for single molecule analysis on native chromatin (18); however, adaptation of these devices to include fluorescence-activated sorting would enable color-multiplexed detection in real-time and collection of molecules with specific epigenetic modifications. Existing fluorescence-activated techniques for cell (19, 20) and droplet (21) sorting lack the volume confinement necessary for sorting individual, single fluorophore-labeled molecules at picomolar concentrations and above that are favorable for protein or antibody binding and chromatin stability (22, 23).
Here we present a bifurcated nanofluidic device for real-time detection and automated sorting of individual methylated DNA molecules. These molecules were selected from a mixture that included unmethylated DNA and were recognized using a fluorescently labeled methyl binding domain protein-1 (MBD1) that binds specifically to double-stranded, methylated DNA (24). We discuss the technologies that enabled us to identify methylated DNA bound with MBD1 using its fluorescence signature. We demonstrate accurate evaluation of this fluorescence signature to actuate sorting of these molecules from the mixture. We show recovery of sorted material and independently verify molecule enrichment by quantitative polymerase chain reaction (qPCR) to elucidate opportunities for postsorting analysis. We report the proof-of-principle demonstration of our single molecule sorter for color-multiplexed epigenetic analysis and sample recovery of limited genetic material.
Results and Discussion
Constructing a Fluorescence-Activated, Single Molecule Sorter.
We designed our nanofluidic sorter to operate using materials prepared by established methods to ensure its integration within an epigenetic analysis workflow. For this design, we implemented a process (Fig. 1) wherein fluorescently labeled probes such as antibodies or proteins are bound to epigenetic modifications located on histone proteins or DNA. This mixture is then loaded into our nanofluidic device to identify these molecules and their corresponding modifications by fluorescence color signature. Each molecule with a color signature that matches the criteria for collection is then sorted during a brief actuation, or pulse, that redirects flow to the sorted output. The flow direction returns to the default output following a sort event. Sorted molecules are then collected for subsequent qPCR analysis.
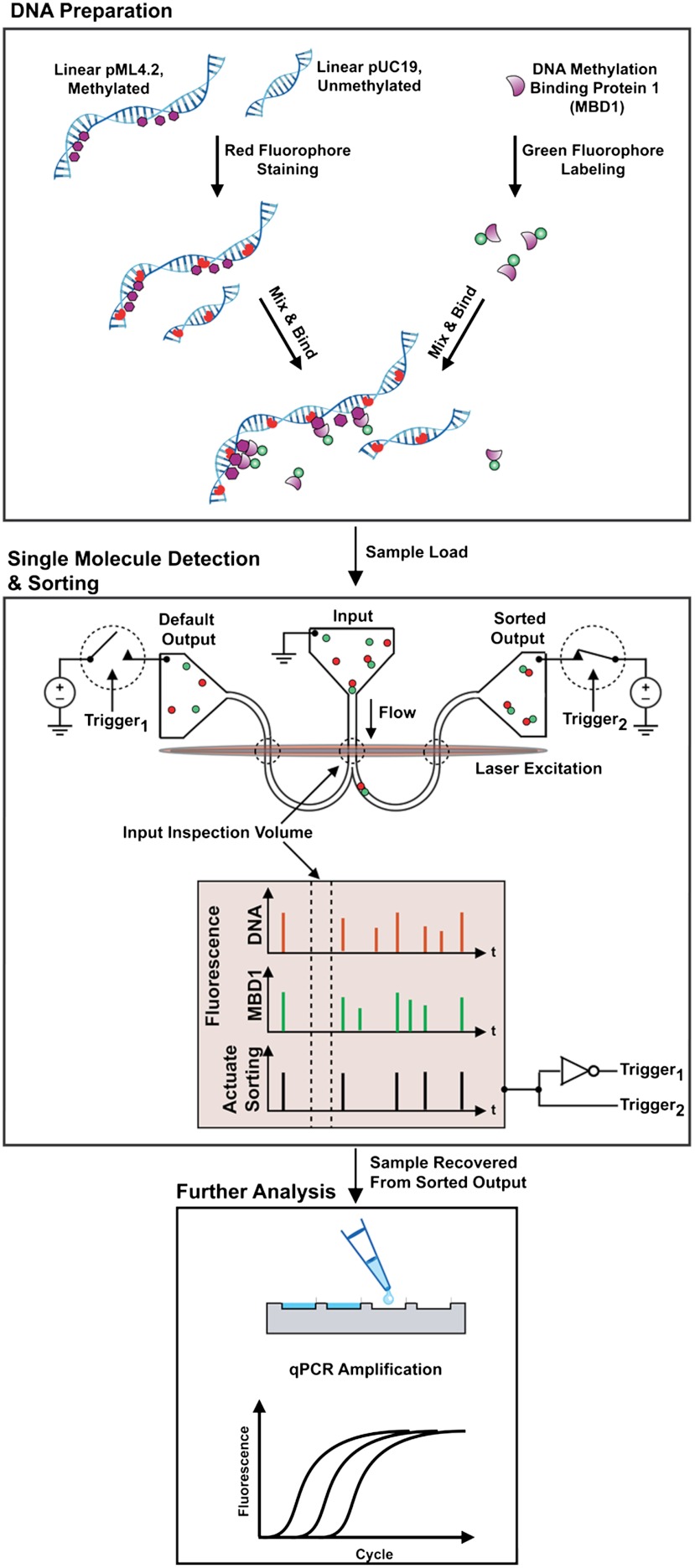
Fig. 1.
An epigenetic analysis workflow using fluorescence-activated, single molecule sorting. (Top) DNA preparation. Linearized DNA plasmids, unmethylated pUC19, and methylated pML4.2 were fluorescently labeled with a red stain and then mixed with green-labeled methyl binding domain protein-1 (MBD1) during a bulk reaction. (Middle) Single molecule detection and sorting. Samples were loaded into the input of a bifurcated nanofluidic device. An applied voltage flowed molecules through the device. As each fluorescently labeled molecule passed through the input inspection volume its fluorescence signature was detected and then evaluated in real time. In this panel, an MBD1 bound to methylated DNA was identified by its two-color fluorescence signature. This signature actuated a sorting trigger and a pair of opposing switches to direct the molecule toward the sorted output. After a molecule was delivered to the sorted output, the flow was redirected to the default output. (Bottom) Further analysis. At the conclusion of a sorting experiment, material in the sorted output was recovered by pipette and the amounts of pUC19 and pML4.2 were measured by qPCR analysis.
In order to develop a sorting device, we modified the nanofluidic device previously described (18) in several ways. First, we added a bifurcation to create two outflow paths for molecules and then adapted the optics to detect input molecules flowing towards the bifurcation and output molecules flowing away from the bifurcation in both paths; second, we developed computational hardware to evaluate the fluorescence signal from each input molecule in real-time and then actuate sorting; third, we developed a circuit to rapidly direct the flow of molecules to either outflow path during sorting and optimized sorting accuracy by tuning the circuit parameters. These modifications are discussed in the following paragraphs.
To perform in situ optimization of sorting performance, we developed a method to monitor multiple nanofluidic paths simultaneously for tracking each molecule within the device. This detection method uses a linear array of confocal apertures whose spacing is geometrically matched to the branches of the m-shaped nanofluidic channel (Figs. 1 and 2A). As fluorescence is emitted from each molecule, it is imaged within the inspection volume formed by the overlap of one confocal aperture to a region of nanofluidic channel. The confocal apertures are formed by a linear array of optical fibers (Fig. 2B and Fig. S1), which are aligned simultaneously to all branches of the nanofluidic channel. To ensure the fluorescence from a given inspection volume does not affect those adjacent, we measured a low optical cross talk, -13 dB, for apertures spaced 500 μm apart (Fig. S1). We evaluated the parallel fluorescence detection and tracking accuracy of this method by monitoring the input and outputs of a bifurcated nanofluidic device during flow of a fluorescently labeled plasmid, pML4.2. To track uniquely one molecule at a time within the path between input and output, detection was performed at a 50 pM sample concentration; we calculate a < 2.6 × 10-3 probability of multiple molecules cooccupying this path. In this experiment, 99.7% of all molecules detected at the input were tracked to the output. The molecules not tracked had fluctuated above or below the detection threshold, but were distinct from noise, suggesting slight photobleaching during transit and differences in beam illumination uniformity. Therefore, our design demonstrated facile alignment and simultaneous detection of single molecule fluorescence from multiple fluidic channels to enable in situ optimization of sorting performance.
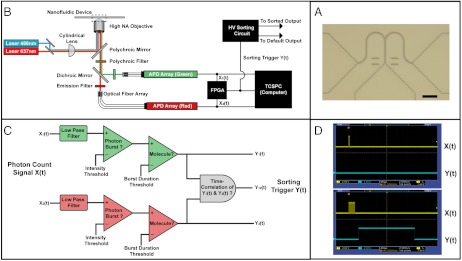
Fig. 2.
Constructing a fluorescence-activated, single molecule sorter. (A) A brightfield photomicrograph of a bifurcated nanofluidic channel with cross-section 500 nm wide by 250 nm deep was designed to isolate individual molecules for sorting. Eighteen of these devices were formed on a 100-mm diameter fused silica wafer. The scale bar is 10 μm. (B) The nanofluidic device was mounted onto a confocal fluorescence microscope and illuminated by two overlapped lasers to excite molecule fluorescence simultaneously. Single molecule fluorescence was imaged using an optical fiber array connected to single photon counting avalanche photodiodes (APDs). A field programmable gate array (FPGA) performed real-time fluorescence analysis at the input inspection volume and triggered a high voltage (HV) sorting circuit to direct the electrokinetic flow of a molecule to the sorted or default output. All detection events were recorded using time-correlated single photon counting (TCSPC) hardware operated in synchrony with the FPGA. (C) An FPGA-based electrical circuit was used to evaluate photon count signals, X1(t) and X2(t), each reporting a different fluorescence color. Detection noise was reduced using a digital low-pass filter to improve molecule identification and switching accuracy. Photon detection events arriving within a short interval, a photon burst, were compared against an intensity threshold to identify signal from noise. Above-threshold bursts that also satisfied a minimum time duration were identified as single molecules that actuated the trigger signal Y1(t) or Y2(t). Molecules bound with multiple fluorescence colors caused time-correlation of Y1(t) and Y2(t) evaluated by a logical AND gate that output Y12(t). (D) An oscilloscope photomicrograph illustrates FPGA operation using a 5-pulse train and a 25-pulse train, shown as X(t), to simulate a photon burst. Only the simulated burst in the bottom panel exceeded the 5-pulse/50 μs threshold for more than 100 μs and, thereby, actuated the trigger for a programmed, 2 ms duration.
Real-time analysis is required for automated, fluorescence-activated sorting to be practical over a wide-range of molecule flow speeds. To perform real-time analysis, we selected a field programmable gate array (FPGA) for its capabilities in digital signal processing (DSP) and facile integration with existing avalanche photodiode (APD) and time-correlated single photon counting (TCSPC) hardware (Fig. 2B). The process of real-time detection within the FPGA began with digital electrical signals representative of individual photons measured by the APDs. These signals were counted, low-pass filtered, and then compared against a burst intensity and duration threshold to identify a single molecule (Fig. 2C). We found low-pass filtering was necessary to smooth and reduce noise fluctuations around the intensity threshold that otherwise caused some molecules to be multiple-counted and actuated for sorting. The low-pass filter designs we implemented, a boxcar and a Gaussian-shaped filter, were used during subsequent DNA-size and methylation sorting experiments. Whereas DSP with these filters introduced detection delay, the time necessary to analyze single molecule fluorescence scaled proportionally with flow rate and often occurred in < 1 ms. (Fig. 2D). To verify the accuracy of our real-time detection method, we flowed fluorescently labeled pML4.2 molecules because of their high (∼200) signal to noise ratio (SNR) during detection and then compared a time-resolved record of their fluorescence against real-time, FPGA-detected events. We measured > 98% agreement between real-time and postexperiment detection methods using the same analysis conditions. Because these detection methods operated in clock-synchrony and using similar algorithms, we attribute differences in detection primarily to the dead-time specification of TCSPC hardware as compared to the FPGA. In summary, our FPGA-based analysis has shown highly accurate, real-time single molecule detection.
For sorting individual molecules into the nanofluidic output paths, we used electrokinetic flow because it can be actuated by rapidly switched, externally applied voltages. To exert precise flow control, we began with an electric circuit model for the nanofluidic device that used channel geometry and buffer ion concentration to describe the equivalent channel resistance and double layer capacitance, respectively, and then characterized the electrical operation of the nanofluidic device. The electrical resistance of the nanofluidic device filled with 1x Tris-EDTA (1x TE) buffer measured 3.5 ± 0.1 GΩ (Fig. S2), which indicated that relays with low off-state leakage current were necessary to disconnect the device electrically during flow switching. We also found that external voltage balancing minimized fabrication-induced variations in channel geometry and further sculpted the electric field at the bifurcation. These adjustments lead to symmetric sorter operation and a misdirection rate of approximately 10-3 molecules when operated in a fixed voltage configuration. We then assessed electrokinetic flow response to a voltage impulse to mimic sorting actuation. The impulse response of the nanofluidic demonstrated an upper limit of 10 kHz on switching speed based upon a transient settling time of approximately 50 μs (Fig. S3). Switching at this frequency could allow sorting at rates up to 100,000 molecules/ min using the present device geometry.
Single Molecule Sorting for Enrichment and Recovery.
A 50 pM mixture of DNA molecules was loaded into the nanofluidic device to evaluate molecule counting and tracking during a single-color sorting experiment to size-separate the molecules. The mixture was composed of linearized pUC19 (2.7 kbp) and pML4.2 (15.2 kbp) DNA plasmids intercalated with TOTO-3. The approximate fivefold difference in molecule size was observed proportionally in fluorescence intensity (25) and allowed molecule identification for sorting. A threshold value of 50 photons/50 μs distinguished the DNA molecule sizes (Fig. S4) and was programmed into the FPGA to identify and actuate sorting of the pML4.2 molecules. A time-resolved record of this molecule sorting experiment (Fig. 3A) illustrated the bursts of fluorescence from pML4.2 matched to the sorting actuation trigger. This record confirmed successful sorting by in situ tracking of each above-threshold molecule from input to sorted output. A transit time of 4.3 ± 0.3 ms through the bifurcated region was calculated by fitting a time-correlated measurement of the delay between molecule observations (Fig. S4). These measurements successfully demonstrated real-time detection and in situ tracking of individual DNA molecules during fluorescence-activated sorting at > 500 molecules/ min.
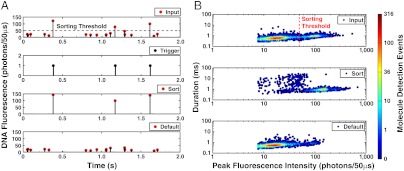
Fig. 3.
Optimization of molecule sorting efficiency. A 1∶4 molar ratio mixture of red-intercalated pML4.2 (15.2 Kb) and pUC19 (2.7 Kb) DNA was loaded into the nanofluidic device and sorted on differences in fluorescence intensity to collect pML4.2. (A) A time-resolved record of single molecule events, black pulses marked with a red dot, illustrates 2 s from a 30 min sorting experiment. At the nanofluidic channel input, a molecule with fluorescence > 50 photons/50 μs actuated the sorting trigger to direct pML4.2 molecules to the sorted output; whereas, pUC19 molecules were directed to the default output. Sorting performance was optimized by tracking each molecule’s trajectory in situ to either of the output inspection volumes and then adjusting electrical actuation parameters to improve sorting accuracy. (B) The nanofluidic sorter evaluated a mixture of 15,848 molecules at the input and sorted 3,394 pML4.2 molecules to the sort output. At the device output inspection volumes, in situ measurements revealed a false-positive rate of 1.9 ± 0.1% (± 1 SD) and 49 ± 3 (± 1 SD) fold enrichment of pML4.2 in single step.
We proceeded to investigate sorting accuracy by measuring the rate of false-positive and negative sorting events. These figures of merit determine the limit of molecule enrichment that can be attained by sorting. Our sorting device was configured to collect the pML4.2 molecules from mixtures with pUC19 at molar ratios of 1∶4 and 1∶12, respectively. To allow for sufficient sampling of false-positive and negative events, over 15,000 molecules were observed in both experiments. False negative events, defined as input pML4.2 molecules directed to the default output, were present as 8.2 ± 0.5% and 3.4 ± 0.4% of all pML4.2 molecules for the 1∶4 and 1∶12 mixtures, respectively. This false negative rate was attributed to molecules not identified or sorted by the FPGA. False-positive events, pUC19 molecules directed to the sorted output, are significant for the purpose of maximizing enrichment purity. Using bivariate analysis with a scatter plot (Fig. 3B), we categorized false-positive sorting events as “cooccupant” or “uncontrolled” molecules. Cooccupant molecules are a consequence of the Poisson-distributed arrival of two or more molecules in the inspection volume at the same time, which can cause a single trigger actuation to direct multiple molecules to the sorted output. By contrast, uncontrolled molecules migrate to the sorted output without actuation and exhibit widely varying 2–20 ms burst durations. Because symmetric operation of the device is calibrated at the start of an experiment, uncontrolled molecules occurred as the result of ion concentration effects and fouling of the channel surface that accumulated over time and led to slightly asymmetric operation. Together, these sources of false-positive events occurred as 1.9 ± 0.1% and 1.0 ± 0.1% of all pUC19 molecules for the 1∶4 and 1∶12 mixtures, respectively. This translates into a pML4.2 enrichment of 49 ± 3 and 88 ± 5 fold in a single sorting step. Sorted material was then recovered and analyzed by qPCR, which confirmed the enrichment of pML4.2 observed in situ and demonstrated successful recovery of a sample enriched by automated sorting (Tables S1, S2, and S3). The levels of enrichment measured in situ are commensurate with other fluorescence-activated sorting methods (20, 26, 27) presently limited to the study of cells and large particles.
Application to Single Molecule Sorting of Methylated DNA.
To demonstrate our single molecule sorter for epigenetic analysis, we proceeded to develop its capability for identifying DNA methylation using two-color fluorescence detection. Similar to the experiments described above, we generated linearized pUC19 and pML4.2 plasmids from methylation deficient hosts, but then in vitro methylated pML4.2 to form a mixture of methylated and unmethylated DNA. Both DNA species were red labeled with TOTO-3 and then mixed with green-labeled MBD1. MBD1 was used for its ability to bind methylcytosines on full-duplex DNA, the native configuration of DNA in chromatin (24). We then analyzed the fluorescence of individual molecules within a mixture of unmethylated pUC19: methylated pML4.2: MBD1, prepared at a 2.8∶1∶200 molar ratio. The FPGA identified input molecules with a two-color fluorescence signature as MBD1 bound to methylated DNA (MBD1-DNA) and directed these molecules to the sorted output. Two-color fluorescence detection was achieved by reconfiguring the FPGA design with Gaussian filter coefficients and evaluating the outputs from multiple copies of the design using a logical AND gate (Fig. 2C); and copropagating a second laser emitting at 488 nm with the existing 640 nm laser (Fig. 2B). Proper operation of the FPGA for multicolor detection was verified during a separate experiment (Fig. S5). Using independent thresholds of 3 and 6 photons/50 μs to identify MBD1 and DNA molecules, our device identified and sorted MBD1-DNA molecules (Fig. 4A).
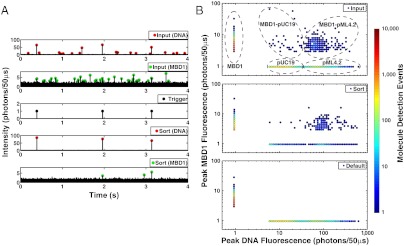
Fig. 4.
Single molecule detection and sorting of methylated DNA. DNA methylation state was identified using a green-labeled methyl binding domain-1 (MBD1) protein incubated in a mixture of red-labeled DNAs including unmethylated-pUC19 and methylated-pML4.2. Methylation-specific binding was confirmed by two-color fluorescence identification, which actuated sorting collection of bound MBD1-DNA complexes from the original mixture during a 20 minute sorting experiment. (A) This 4 s, time-resolved record illustrates three two-color molecules identified by the FPGA trigger at the device input. After sorting, the fluorescence from two of the three MBD1 molecules remained paired with DNA, while the third had fluctuated below the intensity threshold. (B) A total of 5,723 DNA molecules were analyzed at the device input during this experiment and 297 MBD1-DNA events were identified in this analysis. These MBD1-DNA events were composed of 27 MBD1-pUC19 and 270 MBD1-pML4.2, which demonstrated specific binding of MBD1 to methylated pML4.2 molecules. Unbound molecules were plotted along fluorescence = 1 photon/50 μs to accommodate logarithmic plotting. In situ molecule tracking demonstrated a false-positive rate of 5.6 ± 0.4% (± 1 SD) and 3.5 ± 0.3 (± 1 SD) fold enrichment of methylated pML4.2 at the sorted output.
To investigate MBD1 binding specificity, we characterized the molecules from a 20-minute sorting experiment (Fig. 4B) according to their two-color status and then DNA fluorescence intensity. This postexperiment analysis measured 297 MBD1-DNA events at the device input composed of 27 unmethylated pUC19 and 270 methylated pML4.2 molecules. We then verified genuine detection of MBD1-DNA events using a time-coincidence histogram analysis (Fig. S6) and measured 294 ± 21 molecules above a background of randomly correlated, two-color detection events. These results confirm that MBD1 was specifically bound to methylated DNA.
In situ fluorescence detection was used to evaluate sorting accuracy by tracking molecules from device input to output. During the 20-minute sorting experiment (Fig. 4B), all 5723 DNA molecules entering the device input were analyzed, and 420 MBD1-DNA molecules were identified and directed toward the sorted output by the FPGA. In our postexperiment analysis, only 297 of the 420 events were identified as MBD1-DNA due to the low SNR (< 20) of MBD1 that accentuated photon counting hardware differences. The fluctuations and low-intensity of green fluorescence from MBD1 indicative of unbound or singly bound MBD1 to pML4.2 limited our ability to track MBD1-DNA complexes (Fig. S7). Therefore, we used DNA fluorescence to successfully track over 90% of all DNA molecules from input to output. From DNA tracking measurements, we evaluated the false negative and false-positive sorting rates (81 ± 3% and 5.6 ± 0.4%, respectively) for this experiment. The high false negative sorting rate was the result of the low concentration of MBD1 and pML4.2 in the analyte and the high KD value for their binding, which resulted in a low concentration of the complex at equilibrium. Based upon experimental observations of MBD1-DNA binding at the device input, we calculate MBD1 binding to an average of 0.15 of 737 methylcytosine sites on pML4.2; this result corroborates with our theoretical estimate for MBD1 binding to an average of 0.95 of 737 methylcytosine sites on pML4.2, given experimental conditions and a 3 μM KD (24). These observations emphasize the need for improved detection through enhanced binding conditions. By contrast, the low false-positive rate indicates control over the molecule sorting process, yielding 3.5 ± 0.3 fold enrichment of methylated pML4.2 with a 39% background of pUC19 at the sorted output. Our results for sorting DNA plasmids compare well with existing literature for immunoprecipitation reactions that report typical 2–8 fold enrichment levels (3, 28), binding-limited methylation detection (28), and collection of 60–99% background DNA (29).
Conclusion
We have described the development of a nanofluidic device that can be used as part of an epigenetic analysis workflow. This device provides nanofluidic confinement to isolate individual molecules for color-multiplexed detection and to sort these molecules by voltage-actuated flow. These attributes have enabled us to perform real-time fluorescence detection and automated sorting on individual molecules based upon their fluorescence signature. We have optimized the sorting operation using time-correlated, in situ detection to achieve up to 98% accuracy in sorting and confirmed sample recovery of sorted material by qPCR to suggest opportunities for postsorting sample analysis by microarray or DNA sequencing methods. We have applied this technology to detect and sort methylated DNA molecules bound with MBD1 protein from within 11 fg of total genetic material.
We envision several goals for continued development of this device. The first goal is to improve detection efficiency by using different methylcytosine binding probes or modified conditions that enhance MBD1 binding to DNA. This could be achieved using high affinity antibody probes with nanomolar KD values to enhance binding to methylated ssDNA or chemical cross-linking at high sample concentrations to enhance MBD1 binding to dsDNA (30, 31). Yet another goal is to increase sample throughput toward studying DNA or chromatin sources at a whole genome scale. Throughput may be increased in several ways including: (i) disabling the in situ single molecule tracking used in this work to increase input sample concentration and throughput by 20-fold; (ii) removing unbound fluorescent probes to preconcentrate the collection of informative molecules; and, (iii) scaling the system to include a collection of devices operated in parallel and monitored using alternative fluorescence readout methods (32). A third goal could explore workflow integration by joining the single molecule sorter with recent advances in cell processing (12, 27, 33, 34). The sorter operates using small quantities of input material, a feature that pairs well with targeted extraction of genetic material from specific and limited (1–100) cell populations. Such integration could provide an alternative to the off-chip material preparation steps used in this work and lead to more efficient sample workflows. The fourth goal would further develop methods for postsorting material extraction to allow library preparation and downstream microarray or deep sequencing by adapting single cell sequencing methods (35, 36). These goals for development establish abundant opportunity to explore the application of single molecule sorting for epigenomic analysis and a variety of molecule separation workflows.
Materials and Methods
Nanofluidic Device Fabrication.
Nanofluidic devices were constructed in a 100 mm diameter fused silica substrate (Mark Optics). The device pattern was defined by photolithography and reactive ion etched 250 nm into the substrate. Fluid reservoirs of the device were formed using a focused jet of alumina abrasive. The device was cleaned and then touch bonded with a 170 μm-thin fused silica substrate to enclose the fluidic channels. The wafer stack was annealed at 1080 °C. Additional details are described elsewhere (18).
Optical Measurement Setup.
Optical measurements were performed using an inverted microscope (Olympus IX-71). Fluorescence was induced using lasers emitting at 488 nm and 637 nm (Coherent). Their collinear beams were elongated using a cylindrical lens (Thorlabs) and focused onto the nanofluidic device using a 40x water immersion objective (Olympus). Fluorescence was collected through a polychoric mirror and emission filter pair (Chroma) then chromatically split and passband filtered at 525/40 nm and 685/40 nm (Semrock). A linear array of confocal apertures, formed by multimode optical fibers, collected and delivered the fluorescence to single photon counting APDs (Perkin Elmer). Photon counts were assessed using a custom-programmed Altera FPGA (Terasic Technologies DE0) and recorded by two TCSPC cards (FastComTec P7888).
Real-Time and Postexperiment Single Molecule Analysis.
Real-time photon counting and signal analysis was performed using an FPGA operating with an adjustable, 50 μs time base or bin. Photon events were counted during each bin, processed by a digital low-pass filter, and compared against an intensity threshold to identify fluorescence bursts. To eliminate spurious bursts, a two-bin minimum duration was enforced. Single molecule events satisfied both thresholds and their detection actuated the sorting trigger. Postexperiment single molecule analysis was performed on the photon record file from the TCSPC hardware using a custom Matlab routine. For multicolor detection, an AND gate evaluated the time-coincidence of each color event and actuated the sorting trigger. This FPGA trigger was time-correlated with detection events during postexperiment analysis using Matlab.
Electrical Characterization of Sorter Hardware.
Operation of the FPGA and high voltage switch were verified using an arbitrary waveform generator (Tektronix AFG 3252) and oscilloscope (Tektronix DPO 3052). Nanofluidic channel resistance and solid-state relay (Vishay) leakage current were measured using a picoammeter with built-in voltage source (Keithley 6487). Impulse response of the nanofluidic channel was measured within a Faraday cage using a low-noise current preamplifier (Ithaco 1201) and visualized using an oscilloscope.
DNA and MBD1 Sample Preparation.
pUC19 and pML4.2 were grown in dam-/dcm- Escherichia coli (New England Biolabs—C2925) and purified using a QIAGEN Plasmid Midi Kit. Purified DNAs from pML4.2 and pUC19 were linearized with AscI and EcoRI to generate molecules measuring 15,156 bp and 2,687 bp, respectively. pML4.2 was in vitro methylated using M.SssI, and the degree of methylation was confirmed using HhaI and DdeI (Fig. S8). DNA was stained with TOTO-3 (Invitrogen) at a dye to base pair ratio of 1∶5. The 1xMBD1 probe was expressed in E. coli, purified and then labeled with an Alexa-488 dye (Invitrogen A20000) as described (18). Labeled MBD1 was purified by size exclusion chromatography using a Superdex 75 resin (GE healthcare) in phosphate-buffered saline (PBS) buffer and then confirmed for probe activity (Fig. S9). The MBD1-DNA binding reaction was performed in 1x Tris-buffered saline (TBS) at pH 8.0 in 0.5% bovine-serum-albumin and 0.1% Triton X-100. DNA components in this TBS buffer were then mixed with MBD1 in a PBS buffer with 50% glycerol. The mixture was covered and slow rotated overnight at 4 °C. All biological samples were diluted into a “flow buffer,” 10 mM Tris, and 1 mM EDTA buffered to pH 8.0 (1x TE buffer) that also contained 0.1% Triton X-100 and 0.3% polyvinylpyrrolidone (Sigma Alrich) to flow them through the nanofluidic device.
Sample Recovery and qPCR Analysis.
Samples were extracted from the fluid reservoir by pipette, transferred into low-adhesion microcentrifuge tubes, and stored at -20 °C. qPCR was performed on an ABI 7500 system using SybrGreen mastermix (Applied Biosystems) with primers specific for pUC19 (F-,R-) or pML4.2 (F-,R-). Standard curves were established using purified plasmid DNA. qPCR measurements were performed in triplicate using 1 μL of material collected from the sorter. qPCR results were compared with in situ measurements of the total molecules sorted to calculate molecule enrichment.
Supplementary Material
Acknowledgments.
The authors acknowledge Adrian Bird for the MBD1 expression vector and Robert Barton and Tobias Furhman for helpful discussions. This work was supported by the National Institute of Health (Grant: DA025722), the Cornell Center for Vertebrate Genomics, and the National Cancer Institute (award number: U54CA143876). Nanofabrication was performed at the Cornell NanoScale Facility, a member of the National Nanotechnology Infrastructure Network, which is supported by the National Science Foundation (Grant: ECS-0335765).
Footnotes
Conflict of interest statement: The authors H.G.C., P.D.S., and S.L.L. declare a financial interest in the company (Odyssey Molecular) attempting to commercialize the technology described in this manuscript.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1117549109/-/DCSupplemental.
References
- 1.Jones PA, Baylin SB. The fundamental role of epigenetic events in cancer. Nat Rev Genet. 2002;3:415–428. doi: 10.1038/nrg816. [DOI] [PubMed] [Google Scholar]
- 2.Robertson KD. DNA methylation and human disease. Nat Rev Genet. 2005;6:597–610. doi: 10.1038/nrg1655. [DOI] [PubMed] [Google Scholar]
- 3.Lee TI, Johnstone SE, Young RA. Chromatin immunoprecipitation and microarray-based analysis of protein location. Nat Protoc. 2006;1:729–748. doi: 10.1038/nprot.2006.98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bernstein BE, et al. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell. 2006;125:315–326. doi: 10.1016/j.cell.2006.02.041. [DOI] [PubMed] [Google Scholar]
- 5.Clark SJ, Harrison J, Paul CL, Frommer M. High-Sensitivity mapping of methylated cytosines. Nucleic Acids Res. 1994;22:2990–2997. doi: 10.1093/nar/22.15.2990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Clark SJ, Statham A, Stirzaker C, Molloy PL, Frommer M. DNA methylation: bisulphite modification and analysis. Nat Protoc. 2006;1:2353–2364. doi: 10.1038/nprot.2006.324. [DOI] [PubMed] [Google Scholar]
- 7.Xiong ZG, Laird PW. COBRA: A sensitive and quantitative DNA methylation assay. Nucleic Acids Res. 1997;25:2532–2534. doi: 10.1093/nar/25.12.2532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Li Y, Tollefsbol TO. Combined Chromatin Immunoprecipitation and Bisulfite Methylation Sequencing Analysis Epigenetics Protocols Methods in Molecular Biology. Vol 791. Clifton, NJ: Humana Press; 2011. pp. 239–251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wu AR, et al. Automated microfluidic chromatin immunoprecipitation from 2,000 cells. Lab Chip. 2009;9:1365–1370. doi: 10.1039/b819648f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Flusberg BA, et al. Direct detection of DNA methylation during single-molecule, real-time sequencing. Nat Methods. 2010;7:461–U472. doi: 10.1038/nmeth.1459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Laird PW. Principles and challenges of genome-wide DNA methylation analysis. Nat Rev Genet. 2010;11:191–203. doi: 10.1038/nrg2732. [DOI] [PubMed] [Google Scholar]
- 12.White AK, et al. High-throughput microfluidic single-cell RT-qPCR. Proc Natl Acad Sci USA. 2011;108:13999–14004. doi: 10.1073/pnas.1019446108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.McKnight TE, Culbertson CT, Jacobson SC, Ramsey JM. Electroosmotically induced hydraulic pumping with integrated electrodes on microfluidic devices. Anal Chem. 2001;73:4045–4049. doi: 10.1021/ac010048a. [DOI] [PubMed] [Google Scholar]
- 14.Kralj JG, Lis MTW, Schmidt MA, Jensen KF. Continuous dielectrophoretic size-based particle sorting. Anal Chem. 2006;78:5019–5025. doi: 10.1021/ac0601314. [DOI] [PubMed] [Google Scholar]
- 15.Wang L, Flanagan LA, Jeon NL, Monuki E, Lee AP. Dielectrophoresis switching with vertical sidewall electrodes for microfluidic flow cytometry. Lab Chip. 2007;7:1114–1120. doi: 10.1039/b705386j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Studer V, Pepin A, Chen Y, Ajdari A. An integrated AC electrokinetic pump in a microfluidic loop for fast and tunable flow control. Analyst. 2004;129:944–949. doi: 10.1039/b408382m. [DOI] [PubMed] [Google Scholar]
- 17.Thorsen T, Maerkl SJ, Quake SR. Microfluidic large-scale integration. Science. 2002;298:580–584. doi: 10.1126/science.1076996. [DOI] [PubMed] [Google Scholar]
- 18.Cipriany BR, et al. Single Molecule Epigenetic Analysis in a Nanofluidic Channel. Anal Chem. 2010;82:2480–2487. doi: 10.1021/ac9028642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dittrich PS, Schwille P. An integrated microfluidic system for reaction, high-sensitivity detection, and sorting of fluorescent cells and particles. Anal Chem. 2003;75:5767–5774. doi: 10.1021/ac034568c. [DOI] [PubMed] [Google Scholar]
- 20.Fu AY, Spence C, Scherer A, Arnold FH, Quake SR. A microfabricated fluorescence-activated cell sorter. Nat Biotechnol. 1999;17:1109–1111. doi: 10.1038/15095. [DOI] [PubMed] [Google Scholar]
- 21.Baret JC, et al. Fluorescence-activated droplet sorting (FADS): efficient microfluidic cell sorting based on enzymatic activity. Lab Chip. 2009;9:1850–1858. doi: 10.1039/b902504a. [DOI] [PubMed] [Google Scholar]
- 22.Claudet C, Angelov D, Bouvet P, Dimitrov S, Bednar J. Histone octamer instability under single molecule experiment conditions. J Biol Chem. 2005;280:19958–19965. doi: 10.1074/jbc.M500121200. [DOI] [PubMed] [Google Scholar]
- 23.Hagerman TA, et al. Chromatin Stability at Low Concentration Depends on Histone Octamer Saturation Levels. Biophys J. 2009;96:1944–1951. doi: 10.1016/j.bpj.2008.10.070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jorgensen HF, Adie K, Chaubert P, Bird AP. Engineering a high-affinity methyl-CpG-binding protein. Nucleic Acids Res. 2006;34:e96. doi: 10.1093/nar/gkl527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Foquet M, Korlach J, Zipfel W, Webb WW, Craighead HG. DNA fragment sizing by single molecule detection in submicrometer-sized closed fluidic channels. Anal Chem. 2002;74:1415–1422. doi: 10.1021/ac011076w. [DOI] [PubMed] [Google Scholar]
- 26.Cho SH, et al. Review Article: Recent advancements in optofluidic flow cytometer. Biomicrofluidics. 2010;4:043001. doi: 10.1063/1.3511706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Brouzes E, et al. Droplet microfluidic technology for single-cell high-throughput screening. Proc Natl Acad Sci USA. 2009;106:14195–14200. doi: 10.1073/pnas.0903542106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Keshet I, et al. Evidence for an instructive mechanism of de novo methylation in cancer cells. Nat Genet. 2006;38:149–153. doi: 10.1038/ng1719. [DOI] [PubMed] [Google Scholar]
- 29.Pepke S, Wold B, Mortazavi A. Computation for ChIP-seq and RNA-seq studies. Nat Methods. 2009;6:S22–S32. doi: 10.1038/nmeth.1371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schmiedeberg L, Skene P, Deaton A, Bird A. A Temporal Threshold for Formaldehyde Crosslinking and Fixation. PLoS One. 2009;4:e4636. doi: 10.1371/journal.pone.0004636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Solomon MJ, Varshavsky A. Formaldehyde-Mediated Dna Protein Crosslinking—A Probe For Invivo Chromatin Structures. Proc Natl Acad Sci USA. 1985;82:6470–6474. doi: 10.1073/pnas.82.19.6470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lundquist PM, et al. Parallel confocal detection of single molecules in real time. Opt Lett. 2008;33:1026–1028. doi: 10.1364/ol.33.001026. [DOI] [PubMed] [Google Scholar]
- 33.Di Carlo D, Wu LY, Lee LP. Dynamic single cell culture array. Lab Chip. 2006;6:1445–1449. doi: 10.1039/b605937f. [DOI] [PubMed] [Google Scholar]
- 34.Geng T, et al. Histone modification analysis by chromatin immunoprecipitation from a low number of cells on a microfluidic platform. Lab Chip. 2011;11:2842–2848. doi: 10.1039/c1lc20253g. [DOI] [PubMed] [Google Scholar]
- 35.Dean FB, Nelson JR, Giesler TL, Lasken RS. Rapid amplification of plasmid and phage DNA using phi29 DNA polymerase and multiply-primed rolling circle amplification. Genome Res. 2001;11:1095–1099. doi: 10.1101/gr.180501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhang K, et al. Sequencing genomes from single cells by polymerase cloning. Nat Biotechnol. 2006;24:680–686. doi: 10.1038/nbt1214. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.