Abstract
Processivity clamps such as proliferating cell nuclear antigen (PCNA) and the checkpoint sliding clamp Rad9/Rad1/Hus1 (9-1-1) act as versatile scaffolds in the coordinated recruitment of proteins involved in DNA replication, cell-cycle control, and DNA repair. Association and handoff of DNA-editing enzymes, such as flap endonuclease 1 (FEN1), with sliding clamps are key processes in biology, which are incompletely understood from a mechanistic point of view. We have used an integrative computational and experimental approach to define the assemblies of FEN1 with double-flap DNA substrates and either proliferating cell nuclear antigen or the checkpoint sliding clamp 9-1-1. Fully atomistic models of these two ternary complexes were developed and refined through extensive molecular dynamics simulations to expose their conformational dynamics. Clustering analysis revealed the most dominant conformations accessible to the complexes. The cluster centroids were subsequently used in conjunction with single-particle electron microscopy data to obtain a 3D EM reconstruction of the human 9-1-1/FEN1/DNA assembly at 18-Å resolution. Comparing the structures of the complexes revealed key differences in the orientation and interactions of FEN1 and double-flap DNA with the two clamps that are consistent with their respective functions in providing inherent flexibility for lagging strand DNA replication or inherent stability for DNA repair.
Flap endonuclease 1 (FEN1) belongs to a class of essential nucleases (the FEN1 5′ nuclease superfamily) present in all domains of life (1). FEN1 catalyzes the endonucleolytic cleavage of bifurcated DNA or RNA structures known as 5′ flaps. These 5′ flaps are generated during lagging strand DNA synthesis or during long-patch base excision repair. The FEN1 substrates are in fact double-flap DNA (dfDNA) with DNA on the opposite side of the 5′ flap, forming a single nucleotide 3′ flap when bound to the enzyme (2, 3). By removing the 5′ ssDNA or RNA flap from such substrates, FEN1 produces a single nicked product that could be sealed by the subsequent action of a DNA ligase (4). Consistent with its crucial role in DNA replication and repair, FEN1 is highly expressed in all proliferative tissues, and its activity is key for the maintenance of genomic integrity (5). FEN1 has been identified as a cancer susceptibility gene, and mutations in it have been linked to a number of genetic diseases, such as EM map myotonic dystrophy, Huntington disease, several ataxias, fragile X syndrome, and cancer (6–10).
The nuclease activity of FEN1 can be stimulated by association with processivity clamps such as proliferating cell nuclear antigen (PCNA), which encircle DNA at sites of replication and repair (11–13). PCNA is a recognized master coordinator of cellular responses to DNA damage and interacts with numerous DNA repair and cell-cycle control proteins. In this capacity, PCNA serves not only as a mobile platform for the attachment of these proteins to DNA but, importantly, plays an active role in the recruitment and release of these crucial participants at the replication fork (14, 15). The dominant mode of interaction for many of these factors is through attachment to the interdomain connector (IDC) loop of PCNA and the PCNA C terminus (2, 13, 16). The trimeric PCNA ring can provide, at most, three binding sites for replication and repair factors. The crystal structure of human FEN1 with PCNA indeed revealed three FEN1 enzymes bound to the sliding clamp in different orientations (16). Additionally, a biochemical study of the Sulfolobus solfataricus proteins supported the idea that distinct protein partners such as DNA polymerase, FEN1, and DNA ligase could simultaneously associate with PCNA (17). The competition among proteins to simultaneously bind to the surface of PCNA as well as to their common DNA substrate has led to the notion of conformational switching and handoffs of repair intermediates (2, 17); these are key processes in PCNA biology, which are incompletely understood from a mechanistic perspective.
In addition to PCNA, FEN1 is known to associate with the alternative checkpoint clamp Rad9-Rad1-Hus1 (9-1-1 complex). Whereas PCNA is comprised of three identical subunits, 9-1-1 is a heterotrimeric sliding clamp (18–20). This fact reflects the different protein partners the two clamps engage and the distinct roles these complexes play in coordinating DNA processing. In contrast to PCNA, the 9-1-1 complex is thought to serve as a recruitment platform to bring checkpoint effector kinases to sites of DNA damage, thus activating checkpoint control, and also functions to stabilize stalled replication forks that have encountered DNA lesions (21–23). It has also been demonstrated that 9-1-1 interacts with and stimulates enzymes involved in base excision repair (BER), such as NEIL1, MYH, TDG, FEN1, and DNA Ligase I, thus potentially linking BER activities to checkpoint coordination (24–27).
In view of their crucial involvement in replication and repair, a detailed structural comparison of the ternary PCNA/FEN1/DNA and 9-1-1/FEN1/DNA complexes would be of great value. Though structural snapshots are available for the individual components of such assemblies (PCNA [Protein Data Bank (PDB) ID code: 1VYM], human 9-1-1 [3GGR]) (Fig. S1) and for two binary complexes [FEN1/DNA (3Q8L) and FEN1/PCNA (1UL1)] (3, 16, 18–20, 28), the larger ternary assemblies present extreme challenges to molecular crystallography (MX). Here we report models for the ternary PCNA/FEN1/DNA and 9-1-1/FEN1/DNA assemblies, which were constructed by combining all available high-resolution MX data for the individual components and subassemblies. The models were refined by multinanosecond atomistic molecular dynamics (MD) simulations. Single-particle electron microscopy (EM) of negatively stained samples indicated that the structure defined by the 9-1-1/FEN1/DNA model exists in solution. Subsequently, the computational model was integrated with the EM data resulting in a 3D reconstruction for the ternary assembly determined at 18-Å resolution. Finally, we present a detailed comparative analysis of the two ternary complexes.
Results and Discussion
Overall Structure of the Ternary FEN1 Complexes.
To shed light on the conformations and structural dynamics of the ternary FEN1 complexes, we carried out multinanosecond MD simulations (Movies S1 and S2) and were able to refine models of PCNA/FEN1/DNA and 9-1-1/FEN1/DNA, which exhibited no structural discrepancies or steric hindrance between the sliding clamps, DNA, and the nuclease (Fig. 1). The initial PCNA ternary complex was based on two structures—the FEN1/DNA complex (PDB ID code 3Q8L) (3) and the FEN1/PCNA structure (PDB ID code 1UL1) (16). The FEN1/PCNA structure showed three bound copies of FEN1 (X, Y, and Z chains) in three orientations relative to the PCNA homotrimer. Initial overlays of FEN1 in these two structures showed that chain Y orientation was the only one which would allow a double-stranded DNA extension on the 3′ flap side (upstream with respect to replication) to pass through the PCNA ring (29). Therefore, only chain Y orientation was used in the model. The range of conformations observed in the crystal structure suggests segmental flexibility whereby a flexibly linked protein (such as FEN1) can adopt several discrete positions (30). To generate the initial model of the 9-1-1 ternary complex we used an overlay of the human 9-1-1 structure with truncated Rad9 subunit (9Δ-1-1; PDB ID code 3GGR) (20) onto the PCNA/FEN1/DNA model (Fig. 1B). FEN1 was modeled bound to the Rad1 subunit based on previous experimental work (20). The final models were selected after pairwise rmsd clustering analysis of the MD trajectories (31, 32), and the structure closest to the centroid of the most populated cluster was chosen as representative for each ternary complex (Fig. 1 A and B).
Fig. 1.
Dominant structures for the PCNA/FEN1/DNA and 9Δ-1-1/FEN1/DNA complexes from MD trajectory clustering analysis. The (A) PCNA and (B) 9Δ-1-1 complex are shown as cartoons. PCNA is shown in green, FEN1 in purple, Rad1 in green, Hus1 in yellow, Rad 9 in blue, and DNA in black. The gray surfaces are the FEN1/DNA from the original starting models. The surfaces for the two clamps in the starting models were omitted for clarity. (C and D) Computed B-factors mapped onto the PCNA and 9Δ-1-1 models. Coloring corresponds to computed B-factor values from high (red) to low (blue). (Insets) Highlight of the B-factor difference for upstream DNA as it passes through the two clamps.
In both PCNA and 9Δ-1-1 ternary models, FEN1 occupied an overall upright position on the polymerase binding face of PCNA or on the corresponding face of the checkpoint clamp. The path of the DNA bends 100° at the position of the double flap bound by FEN1, consistent with the structure of FEN1/DNA (3). Sakurai et al. (16) had postulated that an additional “swing-in” motion of ∼50° would be required to move FEN1 (chain Y) into a productive conformation with DNA passing through the PCNA ring. However, we observe that no such swing-in motion is required due to the 100° bend of the dfDNA by FEN1 and the tilted orientation of the upstream DNA with respect to the plane of PCNA or 9-1-1. In both models the enzyme was facing the central cavity of the PCNA or 9Δ-1-1 ring, a position that would promote DNA exchange with other PCNA-bound proteins such as polymerases or ligases. A major global difference between PCNA and 9Δ-1-1 ternary complexes was a 14-Å shift of the FEN1 core toward Rad1 compared with a more upright position found in the PCNA complex.
EM Analysis and Computational Flexible Fitting of the 9-1-1/FEN1 Binary and Ternary Complexes.
Hybrid methods for structural analysis constitute an emergent area, which could shed light on biological complexes with a high degree of structural plasticity. The FEN1 ternary assembles are examples of systems, wherein the inherent flexibility has so far precluded crystallographic analysis. As a direct visualization technique, single-particle EM could be used to investigate such flexible assemblies (33, 34).
Binary complex formation for FEN1 and 9-1-1 was assessed by incubating both proteins at 30 °C and subsequently running size-exclusion chromatography (SEC). Analysis of the corresponding fractions by SDS/PAGE showed 9-1-1 and FEN1 eluting at different retention times, thus indicating weak interaction under our experimental conditions (Fig. S2). To increase the stability of the complex, the sample was cross-linked before size-exclusion chromatography. Fractions containing the 9-1-1/FEN1 complex were then visualized by EM. In parallel, a 3D EM reconstruction of the 9-1-1 checkpoint clamp was generated that superimposed well with the 9-1-1 crystal structure (Fig. S3). Due to the pseudosymmetry of the 9-1-1 reconstruction at this resolution, it was not possible to definitively discriminate between the 9-1-1 subunits solely based on the EM density.
Reference-free 2D class averages for the binary 9-1-1/FEN1 complex showed shapes that could be classified as either six-lobed rings with an additional density in the periphery of the ring or as compact two-lobed rods with an extra density region to one side (Fig. 2A, Top). Comparing the images to the 3D reconstruction of 9-1-1, the six-lobed rings and two-lobed rods were respectively assigned as top and side views of the checkpoint clamp. The position of the extra density adjacent to the 9-1-1 ring was highly variable and corresponded in all cases to a single FEN1 enzyme bound to the clamp. Our findings indicate that only one of the 9-1-1 subunits is competent to associate with FEN1. Based on previously reported surface plasmon resonance measurements (20) and crystallographic results (19), we assigned Rad1 as the FEN1-interacting subunit. Importantly, the side views clearly showed how FEN1 could swing by at least 90° from a position in-line with the plane of the ring to an upright orientation facing the central hole of 9-1-1. This observation is particularly interesting in light of the multiple FEN1 orientations in the Sakurai et al. (16) binary PCNA structure. In the structure, chain X corresponded to the in-line (“sideways”) position, whereas chains Y and Z corresponded to the “upright” position. Our results show that the binary FEN1/9-1-1 complex displays the same range of conformational variability as the binary FEN1/PCNA complex, suggesting significance to this flexibility. The FEN1 positioning in the Sakurai structure could, in principle, be limited by specific crystal contacts and steric constraints within the crystal. By contrast, our EM results reflect the conformational range for the complexes in solution. Thus, we propose that both the 9-1-1/FEN1 and PCNA/FEN1 binary assemblies are flexibly tethered. The functional role of such flexibility could be to allow initial FEN1 binding in the sideways orientation. In this position FEN1 could stay associated and move along DNA without affecting the function of proteins bound to the front face of the clamp. Upon encountering the correct DNA substrate, FEN1 could swing into the catalytically competent upright orientation and engage in handoffs with proteins simultaneously attached to the other two clamp subunits.
Fig. 2.
Single-particle EM analysis shows how FEN1 interacts flexibly with 9-1-1 and adopts a fixed position on 9Δ-1-1 in the presence of the DNA substrate. (A) Representative reference-free 2D class averages (top and side views) for the 9-1-1/FEN1 binary complex are compared with those corresponding to the 9Δ-1-1/FEN1/DNA ternary complex. Top and side views of the 9-1-1 complex are shown (Bottom) for reference. (B) Side and top views of the 9Δ-1-1/FEN1/DNA 3D reconstruction. (C) MDFF flexible fitting of the 9Δ-1-1/FEN1/DNA complex into the 3D map of the ternary complex.
To obtain a ternary 9-1-1/FEN1/DNA complex, we based the dfDNA substrate on one from the FEN1/DNA crystal (3), except that 10 additional bases were introduced at the 3′ end (details provided in SI Materials and Methods). A band-shift assay was performed in the presence of either full-length 9-1-1 or a truncation construct lacking the extra C-terminal tail of Rad9 (9Δ-1-1). No band shift was observed with the full-length 9-1-1 (Fig. S4A), suggesting that in the absence of the checkpoint clamp loader the Rad9 C-terminal tail may interfere with DNA binding. However, a band shift was observed when FEN1 and DNA were incubated with 9Δ-1-1, indicating formation of a ternary complex (Fig. S4A). After cross-linking and SEC (Fig. S4 B and C), samples were visualized by negative-stain EM (Fig. S5A).
Representative reference-free 2D class averages of the ternary 9Δ-1-1/FEN1/DNA complex showed that in the presence of DNA, the extra density lobe above the 9Δ-1-1 ring adopted a well-defined fixed position (Fig. 2A, Middle). Addition of DNA locked FEN1 into the upright position facing the central cavity of the 9Δ-1-1 clamp, providing direct experimental support for our computational results. Computationally generated projections of our model (filtered at 20-Å resolution) qualitatively resembled the EM reference-free class averages (Fig. S5B). Thus, we used this filtered structure as an initial model to assign relative orientations to the different experimental views of the complex. We did 3D refinement using iterative projection matching in EMAN2 (35, 36), reaching a final 3D reconstruction of the 9Δ-1-1/FEN1/DNA complex at an estimated resolution of 18 Å (Fig. 2B). The 3D map contains two principal features—a hexagonal nonsymmetrical ring with an external diameter of ∼85 Å and a central channel of ∼30 Å corresponding to 9Δ-1-1, and a two-lobed extra density ∼70 Å in length, corresponding to FEN1 and connected to the 9Δ-1-1 ring. Although the presence of DNA was suggested by the clear effect in the relative positioning of FEN1 with respect to the 9Δ-1-1 ring and a strong OD260 peak in SEC after cross-linking, the reconstruction did not show clear density for DNA. This finding was not surprising, given the well-known difficulty in visualizing DNA by negative staining.
As a final stage of our atomistic model generation and refinement, we used flexible fitting into the experimental EM map using the molecular dynamics flexible fitting (MDFF) method (37, 38) (Fig. 2C). MDFF involves a MD simulation wherein external forces proportional to the EM density gradient are applied to bias the atoms of the model into the high-density regions of the EM map. The DNA substrate was excluded from the MDFF fitting procedure. After the refinement and minimization, the 9Δ-1-1/FEN1/DNA atomic model exhibited excellent agreement with the experimental EM map with fewer than 300 atoms found outside the EM density envelope (at a conservative density threshold of 3.6; rigid-body fit for the complex is compared with the flexible fit by MDFF in Fig. S6). All flexible elements, including the surface loops of 9-1-1 and the helical arch of FEN1, were consistent with the EM map. Importantly, the EM map supported the observation in the computational analysis that FEN1 was tilted toward Rad1.
Structural Determinants of Flexibility for the Ternary Assemblies in the Simulations.
To identify regional points of flexibility with functional implications, we plotted the time evolution of rmsd values for each ternary assembly and its constituent parts over 100 ns of dynamics (Fig. S7 A and B). The most adaptable component was the DNA, which underwent the largest displacement in average heavy-atom rmsd [7.14 ± 0.76 Å (SD) for the PCNA and 6.11 ± 0.54 Å (SD) for the 9Δ-1-1 complex, respectively]. By contrast, the core domain of FEN1 and the subunits of PCNA and the 9Δ-1-1 clamp displayed minimal internal displacements [1.90 ± 0.14 Å (SD) for the FEN1 core, 2.31 ± 0.18 Å (SD) for PCNA, and 3.54 ± 0.24 Å (SD) for 9-1-1]. The most significant internal motions in the proteins were confined to surface loops of the clamps (e.g., P-loop of PCNA, IDC loops of Hus1 and Rad9), the extreme C-terminal ends of PCNA, and a few flexible outer helices in FEN1 (α13, α4, and α5, α10, α11, and α12; Fig. S1). Importantly, although motions bringing FEN1 closer to the PCNA central cavity were observed in the simulation (Fig. 1A and Fig. S7 A and B), the overall displacement along this mode was rather subtle. The displacement was reflected in the observed rmsd for the complexes [3.58 ± 0.49 Å (SD) for the PCNA complex and 6.94 ± 0.38 Å (SD) for the 9Δ-1-1 complex excluding DNA]. The rmsd result for the DNA is corroborated by the high computed B-factors mapped onto the structures of the two ternary complexes (Fig. 1 C and D). Not surprisingly, the entire DNA substrate was found to be highly flexible; the one exception was the central portion of the upstream DNA, wherein the DNA backbone was found to form extensive contacts with the positive inner surface of the sliding clamps. Notably, the B-factors for this middle region of the upstream DNA are lower in the case of the 9Δ-1-1 complex, indicating tighter association. Other mobile regions included the H2TH DNA binding motif (helices α10, α11, and α12) and the “helical arch” region (helices α4 and α5), which caps the 5′ flap and the active site of FEN1. The helical arch is disordered in the absence of DNA (16) and is likely less stable even in the presence of DNA compared with the rest of the FEN1 core. The mobility of H2TH may have significance for the dsDNA binding mediated by H2TH/K+ (3).
A pronounced tilt was observed in the two models between the axis of the upstream DNA and the plane of the PCNA or 9Δ-1-1 ring (Fig. S7E): 17.3 ± 2.9° (SD) for PCNA and 27.2 ± 2.0° (SD) for 9Δ-1-1. Our findings carry strong parallels to previous computational and experimental work. Conventionally, DNA is drawn perpendicular to PCNA in cartoon models. However, a tilted orientation was first suggested by our computational studies on PCNA/DNA binary complex (39). Experimentally, the crystal structure of the bacterial β-clamp/DNA complex (40) indicated that DNA passes through the clamp at a sharp angle of 22°. A PCNA crystal structure with a short DNA revealed the DNA at a 40° angle. Because further extension of the DNA would sterically clash with the ring, such a large tilting angle appears unlikely in this case (41). EM analysis of PCNA/Pol-B/DNA and PCNA/ligase/DNA assemblies suggested that DNA was tilted by 13° and 16°, respectively (42, 43). By contrast, in the replication factor C/PCNA/DNA ternary assembly, the DNA axis was found to be nearly perpendicular to the PCNA ring (44). This versatile mode of association between the sliding clamp and DNA appears to be a universal feature observed in bacterial, archaeal, and eukaryotic clamps, and may allow for distinct functionalities with specific protein partners.
DNA Is More Tightly Bound in the 9-1-1 than in the PCNA Ternary Complex.
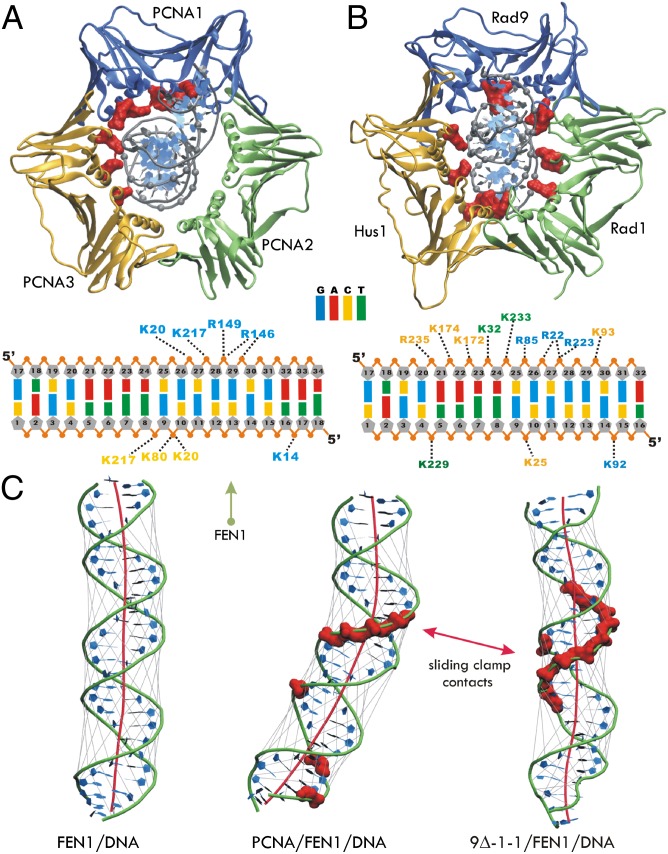
Intriguingly, the mode of association with DNA was strikingly different for PCNA vs. 9Δ-1-1 ternary complex (Fig. 3). The dynamic association of the upstream DNA to the inner cavities of PCNA and 9Δ-1-1 was extensively sampled in the simulations. Despite similarity in overall architecture, PCNA and 9-1-1 present distinct inner surfaces to DNA that are characterized by different shapes, electrostatics, hydrophobic and polar patches, and secondary structure elements. A highly asymmetric mode of association was observed in the PCNA complex, with only one of the PCNA subunits forming the majority of stable contacts with the DNA backbone (K14, K20, K217, R146, and R149). The second subunit was contributing three stable contacts (K20, K80, and K217), and the third subunit made no persistent contacts with DNA at all. Of these, K20, K80, and R149 were found in yeast studies to be functionally important for DNA-mediated replication factor C association, and K14, R146, and R149 were shown to make contact with DNA in the crystal structure (41). On the DNA side, the Arg and Lys residues making salt bridge interactions were evenly distributed between the two DNA strands.
Fig. 3.
In the ternary complexes PCNA and 9-1-1 engage the upstream DNA duplex in two distinct binding modes. (A and B) Cartoon representations of PCNA and 9-1-1 binding to dsDNA, colored in blue for Rad9 and PCNA1, yellow for Hus1 and PCNA3, and green for Rad1 and PCNA2. The dsDNA phosphodiester groups (gray spheres) and basic residues on the inner surface of PCNA and 9-1-1 (red surfaces) are shown. Below we explicitly list all persistent DNA contacts (observed in more than 50% of the frames in the MD trajectories) for the PCNA and the 9-1-1 complex, respectively. (C) Averaged structures for the upstream DNA duplex from FEN1/DNA (Left), PCNA/FEN1/DNA (Center), and 9Δ-1-1/FEN1/DNA (Right) simulations. The DNA is shown in ribbons representation. The DNA axis was computed with the program Curves+ and is shown in red. The gray lines are representative of the widths of the major and minor grooves of DNA.
By contrast, basic residues in the 9Δ-1-1 central hole track along the template strand. The extended interface formed between the template strand and 9Δ-1-1 contains 10 persistent contacts. Three additional contacts (K229 from Rad1, K25 from Hus1, and K92 from Rad9) are made with the opposite strand and are positioned roughly one helical turn apart. Consistent with the previously noted differences in the computed B-factors, 9Δ-1-1 displays significantly tighter association with DNA, forming 13 stable contacts with the DNA backbone (compared with nine contacts in the PCNA complex). Unlike PCNA, the set of contacts with 9-1-1 are more evenly distributed among the three subunits (five contacts with Hus1, three with Rad1, and five with Rad9) and are also more persistent in the MD trajectories compared with the PCNA complex (Fig. S7 C and D). The distinct distribution of basic residue contacts in the two complexes likely determined the overall positioning of the DNA (Fig. S7E).
We also observed differences in the DNA bend between FEN1 and the clamps (Fig. 3C). We averaged the structure of DNA over 80 ns (excluding the first 20 ns as equilibration) from three simulations: (i) FEN1/DNA, (ii) PCNA/FEN1/DNA, and (iii) the 9Δ-1-1/FEN1/DNA. The axis of the upstream duplex was computed with the program Curves+ (45). In the absence of either clamp, the upstream DNA axis is essentially straight and conforms closely to the expected canonical B-form geometry. By contrast, the asymmetric mode of association observed for the PCNA/FEN1/DNA complex results in a moderate bending of the DNA axis by ∼20°. Such structural distortion may introduce strain in the DNA substrate between the 5′ flap and the clamp-binding region, which could assist the dissociation of the substrate after flap removal. Consistent with the more symmetric contact distribution, the 9-1-1 complex exhibits no such tendency for DNA bending. However, the DNA is not entirely free from structural distortion—we observe contraction of the minor groove at the level of the 9-1-1 interface accompanied by major groove widening above and below the interface. The electrostatic properties of 9-1-1 appear to be evolutionarily functional. An analysis of processivity clamps from a variety of organisms showed that the electrostatic properties of Hus1 and, to a lesser degree, Rad9, but not of Rad1, were more similar to nonring viral processivity factors than to the eukaryotic PCNA (46). These viral processivity factors (e.g., UL42, UL44) possess an increased positive charge on the DNA-binding surface compared with the ring-forming clamps (PCNA, PolIII β subunit) and bind to the DNA directly rather than rely on a topological connection to DNA. Our results are consistent with this view that elevated positive potentials on the inner surface of 9-1-1 (specifically for Hus1 and Rad9; Fig. S1) lead to tighter association with DNA.
Specific Contacts with the Clamp Surface Determine the FEN1 Orientation in the Complexes.
The computational models reveal multiple direct interactions between FEN1 and the surface of the two clamps that go beyond the canonical PCNA-interacting protein motif (PIP) motif (Fig. S8). Notably, we have taken care to average along the simulation trajectories and, based on a geometric criterion for each interaction type, to exclude contacts occurring only transiently. Thus, all contacts shown are stable and persistent above a characteristic threshold level. Specifically, we note that the long C-terminal tail of FEN1 makes extensive contacts with Rad1 or PCNA along the IDC loop region. Although the FEN1 C-terminal tail is required for stimulation of FEN1 activity by either PCNA or the checkpoint clamp, intriguingly, the exact C-terminal residues responsible for stimulation by the two clamps appear to be distinct (47). The residues within the PIP box are critical for stimulation by PCNA. By contrast, residues at the extreme C-terminal end (last 21 residues) are important for stimulation by 9-1-1. In our two models, the FEN1/PCNA interface features more hydrophobic contacts, displaying two distinct hydrophobic pockets [(i) P234, V233, A208, I128, L47, and V45, and (ii) M119, L118, A96, M68, A67, and L66]. Residues from these pockets interact directly with the FEN1 PIP box (βA–αA motif). In contrast, the Rad1 surface appears less hydrophobic in this region, lacking both hydrophobic pockets (Fig. S8). Suggestively, the closer proximity of the tilted FEN1 catalytic core to Rad1 might allow the FEN1 C terminus to come back up to interact with the FEN1 catalytic core in the 9-1-1 complex, providing a rationale for the perplexing data that the extreme C terminus is required for 9-1-1 stimulation but not for stimulation by PCNA.
Besides the canonical PIP box/IDC loop interaction, the PCNA C terminus is important for PCNA stimulation of FEN1, although the PCNA C terminus is not required for recruitment (2, 13). In an archaeal system, the FEN1 and PCNA C termini form a structured antiparallel β-sheet (2). Accordingly, we observe that the acidic C termini of both PCNA (A252–E258) and Rad1 (E275–S382) interact with a basic region in the FEN1 catalytic core in our models. Importantly for FEN1 specificity for Rad1, of all three 9-1-1 subunits, only Rad1 has an acidic C terminus similar in charge and length to the one on PCNA. Rad1 also presents a similar acidic surface complementary to the basic surface of the FEN1 catalytic core (Fig. S1).
Concluding Remarks.
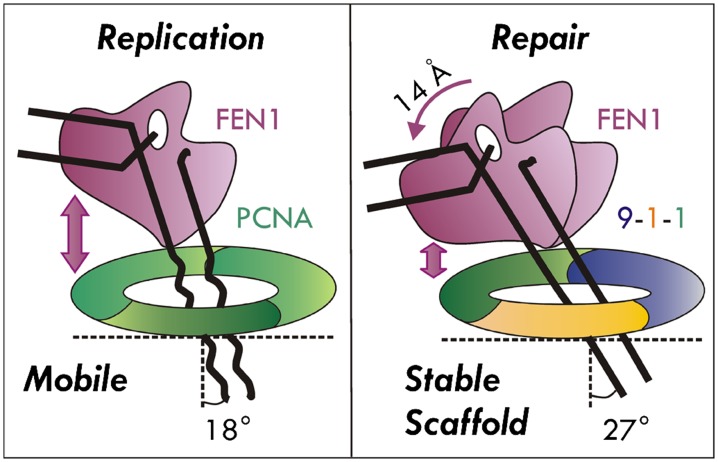
The role of sliding clamps in increasing processivity for DNA-editing enzymes has been long appreciated. Whether these clamps simply act as tethers or play a more active mechanistic role in DNA replication and repair remains controversial. Our combined EM and computational results provide insights into the ternary complexes PCNA/FEN1/DNA and 9-1-1/FEN1/DNA (Fig. 4) and imply an active role for the clamps. The observed FEN1 interactions in the models are consistent with distinct functions for the complexes. Our models show the DNA substrate is functionally bound by FEN1 with the upstream duplex passing through the inner hole of the ringed clamps. Due to segmental flexibility, in the DNA-free complexes FEN1 is able to occupy multiple positions. In the ternary complexes the DNA substrate locks FEN1 into an upright position producing a single incision-capable state. The DNA passes through the rings at a tilt and the tilt angle is larger by 9° in the 9-1-1 complex. The computational analyses also suggest that 9-1-1, which has more positive inner surface than PCNA, has almost 50% more charged interactions with the DNA. It is interesting to note that the more symmetrical PCNA ring shows a biased DNA interaction toward one side of the ring, whereas the 9-1-1 interactions involve all three subunits. The distinct DNA interactions are consistent with different roles for the two clamps in DNA replication and repair. During replication, PCNA must be mobile and slide on DNA along with replicative polymerases. However, the checkpoint clamp 9-1-1 acts as a temporary scaffold during repair of a section of DNA. Such functional differences arguably dictate corresponding differences in the interactions and the mode of association of the clamps in their respective replication/repair complexes. The tethered flexibility observed in the binary complexes of PCNA/FEN1 and 9-1-1/FEN1 would allow FEN1 to swing from an out-of-the-way position on the outside of the ring to the upright functional position. This flexibility is consistent with the rapid interchange between PCNA-bound polymerase and FEN1 during Okazaki fragment maturation.
Fig. 4.
Functionally important differences between the PCNA/FEN1/DNA and 9-1-1/FEN1/DNA complexes. PCNA and 9-1-1 serve different functions and, as a consequence, display different modes of association to FEN1 and DNA. The DNA and FEN1 are more tilted in 9Δ-1-1 complex than in the corresponding PCNA complex. The 9Δ-1-1 ring shows more interactions with the upstream duplex and stabilizes DNA passing through the ring. Such differences at the structural level may translate into functional differences in the ways these complexes engage at the replication fork. PCNA/DNA interactions are linked to processive and mobile sliding, whereas the tighter 9-1-1 interactions would be consistent with a scaffolding role for 9-1-1.
Materials and Methods
Sample Preparation, EM Data Collection, and Processing.
Human 9-1-1/FEN1 binary complex was formed by incubating 9-1-1 with a 3-M excess of FEN1 at room temperature for 15 min in 20 mM Hepes (pH 7.6), 80 mM KCl. The complex was cross-linked and purified by SEC through Superdex 200. 9Δ-1-1/FEN1/DNA ternary complex was cross-linked and further processed as for the binary complex. To visualize the complexes by negative-stain EM, the purified complexes were loaded onto continuous carbon grids and stained either with 2% (wt/vol) phosphotungstic acid (9-1-1/FEN1 binary complex) or 2% (wt/vol) uranyl acetate (9Δ-1-1/FEN1/DNA ternary complex and 9-1-1 alone). Images were collected using the Leginon data collection software (48) on a Fei Tecnai F20 microscope using a 80,000× magnification (1.5 Å/pixel) in low-dose mode (20e−/Å2) on a Gatan 4K × 4K pixel CCD camera (15-μm pixel size). The 2D data were processed using the Appion package (49).
Computational Models and Protocols.
Initial models were constructed based on structures from the Protein Data Bank (accession codes 1UL1 and 3Q8L) (3, 16). The sequences of the DNA substrate (in the EM experiments and the simulation) and the protein sequences for the 9Δ-1-1 subunits are shown in Fig. S9. The FEN1/DNA complex was superimposed onto the PCNA/FEN1 structure by optimally aligning FEN1 to the Y chain in the 1UL1 complex, and the 3′ end of the dfDNA was extended so that it could pass through the PCNA/9-1-1 ring. Hydrogen atoms, counterions (Na+), an additional 100 mM NaCl concentration, and TIP3P solvent (50) were introduced. The systems were then minimized and equilibrated. Production runs were carried out in the isothermal–isobaric ensemble (1 atm and 300 K) for 120 ns for the PCNA/FEN1/DNA and 100 ns for the 9Δ-1-1/ FEN1/DNA complex. The simulations used smooth particle mesh Ewald electrostatics, 10-Å cutoff for short-range nonbonded interactions, and 2-fs integration time step. All of the simulations were performed using the NAMD 2.7 code (51, 52) with the AMBER Parm99SB parameter set (53) with modified nucleic acid parameters (BSC0) (54) on Hopper II, a Cray XE6 system at the National Energy Research Scientific Computing Center. A detailed description of the experimental procedures and the computational modeling protocols is given in SI Materials and Methods.
Supplementary Material
Acknowledgments
The authors thank Dr. Gabriel Lander, Ernesto Arias, and Patricia Grob for advice during EM data collection and processing. Computational resources were provided in part by a National Science Foundation (NSF) Teragrid allocation (CHE110042) and through an allocation at the National Energy Research Scientific Computing Center supported by the Department of Energy (Contract No. DE-AC02-05CH11231). Work on the project is supported by Georgia State University (I.I.), a Cleon C. Arrington research initiation grant (to I.I.), NSF Grant MCB-1149521 (to I.I.), and National Cancer Institute Grants P01 CA092584 and R01 CA081967 (to J.A.T). E.N. is a Howard Hughes Medical Institute Investigator.
Footnotes
The authors declare no conflict of interest.
Data deposition: The data reported in this paper have been deposited in the Electron Microscopy Data Bank, www.ebi.ac.uk/pdbe/emdb/ [accession codes: EMD-2029 (FEN1/DNA/9-1-1 negative-stain EM map) and EMD-2030 (9-1-1 complex) EM map].
*This Direct Submission article had a prearranged editor.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1121116109/-/DCSupplemental.
References
- 1.Grasby JA, Finger LD, Tsutakawa SE, Atack JM, Tainer JA. Unpairing and gating: Sequence-independent substrate recognition by FEN superfamily nucleases. Trends Biochem Sci. 2012;37:74–84. doi: 10.1016/j.tibs.2011.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chapados BR, et al. Structural basis for FEN-1 substrate specificity and PCNA-mediated activation in DNA replication and repair. Cell. 2004;116:39–50. doi: 10.1016/s0092-8674(03)01036-5. [DOI] [PubMed] [Google Scholar]
- 3.Tsutakawa SE, et al. Human flap endonuclease structures, DNA double-base flipping, and a unified understanding of the FEN1 superfamily. Cell. 2011;145:198–211. doi: 10.1016/j.cell.2011.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Finger LD, et al. The 3′-flap pocket of human flap endonuclease 1 is critical for substrate binding and catalysis. J Biol Chem. 2009;284:22184–22194. doi: 10.1074/jbc.M109.015065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zheng L, et al. Functional regulation of FEN1 nuclease and its link to cancer. Nucleic Acids Res. 2011;39:781–794. doi: 10.1093/nar/gkq884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Freudenreich CH, Kantrow SM, Zakian VA. Expansion and length-dependent fragility of CTG repeats in yeast. Science. 1998;279:853–856. doi: 10.1126/science.279.5352.853. [DOI] [PubMed] [Google Scholar]
- 7.Schweitzer JK, Livingston DM. Expansions of CAG repeat tracts are frequent in a yeast mutant defective in Okazaki fragment maturation. Hum Mol Genet. 1998;7:69–74. doi: 10.1093/hmg/7.1.69. [DOI] [PubMed] [Google Scholar]
- 8.Kucherlapati M, et al. Haploinsufficiency of Flap endonuclease (Fen1) leads to rapid tumor progression. Proc Natl Acad Sci USA. 2002;99:9924–9929. doi: 10.1073/pnas.152321699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Henneke G, Friedrich-Heineken E, Hübscher U. Flap endonuclease 1: A novel tumour suppresser protein. Trends Biochem Sci. 2003;28:384–390. doi: 10.1016/S0968-0004(03)00138-5. [DOI] [PubMed] [Google Scholar]
- 10.Stoimenov I, Helleday T. PCNA on the crossroad of cancer. Biochem Soc Trans. 2009;37:605–613. doi: 10.1042/BST0370605. [DOI] [PubMed] [Google Scholar]
- 11.Jónsson ZO, Hindges R, Hübscher U. Regulation of DNA replication and repair proteins through interaction with the front side of proliferating cell nuclear antigen. EMBO J. 1998;17:2412–2425. doi: 10.1093/emboj/17.8.2412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li XY, Li J, Harrington J, Lieber MR, Burgers PMJ. Lagging strand DNA synthesis at the eukaryotic replication fork involves binding and stimulation of FEN-1 by proliferating cell nuclear antigen. J Biol Chem. 1995;270:22109–22112. doi: 10.1074/jbc.270.38.22109. [DOI] [PubMed] [Google Scholar]
- 13.Gomes XV, Burgers PMJ. Two modes of FEN1 binding to PCNA regulated by DNA. EMBO J. 2000;19:3811–3821. doi: 10.1093/emboj/19.14.3811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Moldovan GL, Pfander B, Jentsch S. PCNA, the maestro of the replication fork. Cell. 2007;129:665–679. doi: 10.1016/j.cell.2007.05.003. [DOI] [PubMed] [Google Scholar]
- 15.Maga G, Hubscher U. Proliferating cell nuclear antigen (PCNA): A dancer with many partners. J Cell Sci. 2003;116:3051–3060. doi: 10.1242/jcs.00653. [DOI] [PubMed] [Google Scholar]
- 16.Sakurai S, et al. Structural basis for recruitment of human flap endonuclease 1 to PCNA. EMBO J. 2005;24:683–693. doi: 10.1038/sj.emboj.7600519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dionne I, Brown NJ, Woodgate R, Bell SD. On the mechanism of loading the PCNA sliding clamp by RFC. Mol Microbiol. 2008;68:216–222. doi: 10.1111/j.1365-2958.2008.06150.x. [DOI] [PubMed] [Google Scholar]
- 18.Doré AS, Kilkenny ML, Rzechorzek NJ, Pearl LH. Crystal structure of the rad9-rad1-hus1 DNA damage checkpoint complex—implications for clamp loading and regulation. Mol Cell. 2009;34:735–745. doi: 10.1016/j.molcel.2009.04.027. [DOI] [PubMed] [Google Scholar]
- 19.Sohn SY, Cho Y. Crystal structure of the human rad9-hus1-rad1 clamp. J Mol Biol. 2009;390:490–502. doi: 10.1016/j.jmb.2009.05.028. [DOI] [PubMed] [Google Scholar]
- 20.Xu M, et al. Structure and functional implications of the human rad9-hus1-rad1 cell cycle checkpoint complex. J Biol Chem. 2009;284:20457–20461. doi: 10.1074/jbc.C109.022384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Navadgi-Patil VM, Burgers PM. Cell-cycle-specific activators of the Mec1/ATR checkpoint kinase. Biochem Soc Trans. 2011;39:600–605. doi: 10.1042/BST0390600. [DOI] [PubMed] [Google Scholar]
- 22.Parrilla-Castellar ER, Arlander SJH, Karnitz L. Dial 9-1-1 for DNA damage: the Rad9-Hus1-Rad1 (9-1-1) clamp complex. DNA Repair (Amst) 2004;3:1009–1014. doi: 10.1016/j.dnarep.2004.03.032. [DOI] [PubMed] [Google Scholar]
- 23.Lieberman HB, et al. The role of RAD9 in tumorigenesis. J Mol Cell Biol. 2011;3:39–43. doi: 10.1093/jmcb/mjq039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Balakrishnan L, Brandt PD, Lindsey-Boltz LA, Sancar A, Bambara RA. Long patch base excision repair proceeds via coordinated stimulation of the multienzyme DNA repair complex. J Biol Chem. 2009;284:15158–15172. doi: 10.1074/jbc.M109.000505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gembka A, et al. The checkpoint clamp, Rad9-Rad1-Hus1 complex, preferentially stimulates the activity of apurinic/apyrimidinic endonuclease 1 and DNA polymerase beta in long patch base excision repair. Nucleic Acids Res. 2007;35:2596–2608. doi: 10.1093/nar/gkl1139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Guan X, et al. The human checkpoint sensor Rad9-Rad1-Hus1 interacts with and stimulates DNA repair enzyme TDG glycosylase. Nucleic Acids Res. 2007;35:6207–6218. doi: 10.1093/nar/gkm678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Helt CE, Wang W, Keng PC, Bambara RA. Evidence that DNA damage detection machinery participates in DNA repair. Cell Cycle. 2005;4:529–532. doi: 10.4161/cc.4.4.1598. [DOI] [PubMed] [Google Scholar]
- 28.Krishna TS, Kong XP, Gary S, Burgers PM, Kuriyan J. Crystal structure of the eukaryotic DNA polymerase processivity factor PCNA. Cell. 1994;79:1233–1243. doi: 10.1016/0092-8674(94)90014-0. [DOI] [PubMed] [Google Scholar]
- 29.Tsutakawa SE, Tainer JA. Double strand binding-single strand incision mechanism for human flap endonuclease: Implications for the superfamily. Mech Ageing Dev. 2012 doi: 10.1016/j.mad.2011.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tsutakawa SE, et al. Solution X-ray scattering combined with computational modeling reveals multiple conformations of covalently bound ubiquitin on PCNA. Proc Natl Acad Sci USA. 2011;108:17672–17677. doi: 10.1073/pnas.1110480108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Daura X, Jaun B, Seebach D, van Gunsteren WF, Mark AE. Reversible peptide folding in solution by molecular dynamics simulation. J Mol Biol. 1998;280:925–932. doi: 10.1006/jmbi.1998.1885. [DOI] [PubMed] [Google Scholar]
- 32.Lindahl E, Hess B, van der Spoel D. GROMACS 3.0: A package for molecular simulation and trajectory analysis. J Mol Model. 2001;7:306–317. [Google Scholar]
- 33.Orlova EV, Saibil HR. Methods for three-dimensional reconstruction of heterogeneous assemblies. Methods Enzymol. 2010;482:321–341. doi: 10.1016/S0076-6879(10)82013-0. [DOI] [PubMed] [Google Scholar]
- 34.Zhang W, Kimmel M, Spahn CMT, Penczek PA. Heterogeneity of large macromolecular complexes revealed by 3D cryo-EM variance analysis. Structure. 2008;16:1770–1776. doi: 10.1016/j.str.2008.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ludtke SJ, Baldwin PR, Chiu W. EMAN: Semiautomated software for high-resolution single-particle reconstructions. J Struct Biol. 1999;128:82–97. doi: 10.1006/jsbi.1999.4174. [DOI] [PubMed] [Google Scholar]
- 36.Tang G, et al. EMAN2: An extensible image processing suite for electron microscopy. J Struct Biol. 2007;157:38–46. doi: 10.1016/j.jsb.2006.05.009. [DOI] [PubMed] [Google Scholar]
- 37.Trabuco LG, Villa E, Mitra K, Frank J, Schulten K. Flexible fitting of atomic structures into electron microscopy maps using molecular dynamics. Structure. 2008;16:673–683. doi: 10.1016/j.str.2008.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Trabuco LG, Villa E, Schreiner E, Harrison CB, Schulten K. Molecular dynamics flexible fitting: A practical guide to combine cryo-electron microscopy and X-ray crystallography. Methods. 2009;49:174–180. doi: 10.1016/j.ymeth.2009.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ivanov I, Chapados BR, McCammon JA, Tainer JA. Proliferating cell nuclear antigen loaded onto double-stranded DNA: Dynamics, minor groove interactions and functional implications. Nucleic Acids Res. 2006;34:6023–6033. doi: 10.1093/nar/gkl744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Georgescu RE, et al. Structure of a sliding clamp on DNA. Cell. 2008;132:43–54. doi: 10.1016/j.cell.2007.11.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.McNally R, Bowman GD, Goedken ER, O’Donnell M, Kuriyan J. Analysis of the role of PCNA-DNA contacts during clamp loading. BMC Struct Biol. 2010;10:3. doi: 10.1186/1472-6807-10-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mayanagi K, et al. Architecture of the DNA polymerase B-proliferating cell nuclear antigen (PCNA)-DNA ternary complex. Proc Natl Acad Sci USA. 2011;108:1845–1849. doi: 10.1073/pnas.1010933108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mayanagi K, et al. Mechanism of replication machinery assembly as revealed by the DNA ligase-PCNA-DNA complex architecture. Proc Natl Acad Sci USA. 2009;106:4647–4652. doi: 10.1073/pnas.0811196106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Miyata T, et al. Open clamp structure in the clamp-loading complex visualized by electron microscopic image analysis. Proc Natl Acad Sci USA. 2005;102:13795–13800. doi: 10.1073/pnas.0506447102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lavery R, Moakher M, Maddocks JH, Petkeviciute D, Zakrzewska K. Conformational analysis of nucleic acids revisited: Curves+ Nucleic Acids Res. 2009;37:5917–5929. doi: 10.1093/nar/gkp608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kazlauskas D, Venclovas C. Computational analysis of DNA replicases in double-stranded DNA viruses: Relationship with the genome size. Nucleic Acids Res. 2011;39:8291–8305. doi: 10.1093/nar/gkr564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Friedrich-Heineken E, et al. The two DNA clamps Rad9/Rad1/Hus1 complex and proliferating cell nuclear antigen differentially regulate flap endonuclease 1 activity. J Mol Biol. 2005;353:980–989. doi: 10.1016/j.jmb.2005.09.018. [DOI] [PubMed] [Google Scholar]
- 48.Suloway C, et al. Automated molecular microscopy: The new Leginon system. J Struct Biol. 2005;151:41–60. doi: 10.1016/j.jsb.2005.03.010. [DOI] [PubMed] [Google Scholar]
- 49.Lander GC, et al. Appion: An integrated, database-driven pipeline to facilitate EM image processing. J Struct Biol. 2009;166:95–102. doi: 10.1016/j.jsb.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Jorgensen WL, Chandrasekhar J, Madura JD, Impey RW, Klein ML. Comparison of simple potential functions for simulating liquid water. J Chem Phys. 1983;79:926–935. [Google Scholar]
- 51.Kale L, et al. NAMD2: Greater scalability for parallel molecular dynamics. J Comput Phys. 1999;151:283–312. [Google Scholar]
- 52.Phillips JC, et al. Scalable molecular dynamics with NAMD. J Comput Chem. 2005;26:1781–1802. doi: 10.1002/jcc.20289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hornak V, et al. Comparison of multiple Amber force fields and development of improved protein backbone parameters. Proteins. 2006;65:712–725. doi: 10.1002/prot.21123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Pérez A, et al. Refinement of the AMBER force field for nucleic acids: Improving the description of α/γ conformers. Biophys J. 2007;92:3817–3829. doi: 10.1529/biophysj.106.097782. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.