Abstract
Cyclin D1 is a cell cycle machine, a sensor of extracellular signals and plays an important role in G1-S phase progression. The human cyclin D1 promoter contains multiple transcription factor binding sites such as AP-1, NF-қB, E2F, Oct-1, and so on. The extracellular signals functions through the signal transduction pathways converging at the binding sites to active or inhibit the promoter activity and regulate the cell cycle progression. Different signal transduction pathways regulate the promoter at different time to get the correct cell cycle switch. Disorder regulation or special extracellular stimuli can result in cell cycle out of control through the promoter activity regulation. Epigenetic modifications such as DNA methylation and histone acetylation may involved in cyclin D1 transcriptional regulation.
Keywords: Promoter, Transcription factor, Signal transduction pathway, Epigenetic regulation
Introduction
During the G1 phase, cells will response to the extracellular signals that influence cell division, growth, and differentiation. Cyclin D1 is thought to play pivotal roles in G1-S phase transition. Mistakes in G1 phase may lead to cell cycle out of control and cause tumorigenesis. Cyclin D1 is a sensor to integrate extracellular signals with the cell cycle machinery, with functions through CDK4/6 to trigger cell cycle progression. In recent years, accumulating evidence suggests cyclin D1 also convey cell cycle or CDK-independent functions, and cell can do without cyclin D1 (Coqueret 2002; Fu et al. 2004; Lamb et al. 2003; Pestell et al. 1999).
The cyclin D1 promoter sequence was studied and subcloned in several different laboratories (Albanese et al. 1995; Herber et al. 1994b; Motokura and Arnold 1993; Nagata et al. 2001). The promoter sequence, GenBank number Z29078 (Herber et al. 1994b), contains no obvious TATA box with TF (transcription factor) binding sites such as AP-1, SP-1, E2F, OCT-1, and so on. In this review, the structure of cyclin D1 promoter is discussed with such binding sites, and regulation from signal transduction pathway converging at the binding site.
The elements of the cyclin D1 promoter
Cyclin D1 promoter popularly studied is 1,810 bp about with many cis-elements that can mediate signals activate or inactivate the promoter activity. From −1,309 (NFAT binding site) to −10 (Ets binding site), there are many regulatory elements reported. And if searching by computer program, there are some more elements that have not yet been studied. To compare the cyclin D1 promoter with rat and mouse, homologues region were found (Eto 2000), which can lead us to find new elements in human cyclin D1 promoter. The elements reviewed here only include the elements that have been studied (Fig. 1; Table 1).
Fig. 1.
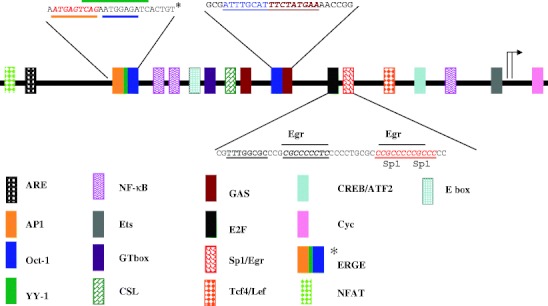
Schematic representation of the elements of human cyclin D1 promoter. Elements of human cyclin D1 promoter are represented by different colors. Detail sequences of complex motifs such as ERGE (including AP-1, Oct-1, and YY-1) and Oct1-GAS are showed. Sequence of Egr-1 and Sp1 are also showed. Starting site of transcription is by two arrows due to the data of the referenced papers, and the sequences are showed in the manuscript
Table 1.
Positions of TF elements in human cyclin D1 promoter
| elements | location | reference |
|---|---|---|
| CREB/ATF2 | −58 | J Biol Chem, 1999 .274(11):7341 |
| Lef/Tcf 4 | −82 | J Boil Chem, 2002 .277(48):45847 |
| Sp-1 | −113, −119 | J Biol Chem, 1997 .272(52):33181 |
| Egr | −137, −118 | J Biol Chem, 1997 .272(52):33181 |
| E2F | −148 | Mol Cell Biol, 2000. 20(2): 672 |
| GAS | −478, −144 | Mol Cell Biol, 2003. 23(24): 8934–45 |
| Oct_1 | −252 | Mol Cell Biol, 2003. 23(24): 8934–45 |
| GT-boxA | −494 | Mol Cell, 2003.11(6):1503 |
| CSL | −525 | Mol Cell Biol, 2001. 21(17): 5925 |
| E-box | −588 | Epigenetics, 2009.4(7):487–99 |
| Ets | −779 | Mol Biol Cell, 2001.12(12):4066 |
| Oct_1 | −941 | Mol Cell Biol, 2004.24(16):7260 |
| YY-1 | −945 | Mol Cell Biol, 2004.24(16):7260 |
| AP-1 | −952 | Mol Cell Biol, 2004.24(16):7260 |
| ERGE | −952 | Mol Cell Biol, 2004.24(16):7260 |
| NFAT | −1,309 | J Biol Chem, 2009.284:36302-36311 |
Elements locations in human cyclin D1 promoter here are normalized to the paper (Motokura and Arnold 1993) and may be some different to the reference showed in the table
AP1
AP1 site was identified in the promoter, which locates in −954 (Albanese et al. 1999). The site may be assigned TRE (12-O-tetradecanoylphorbol-13-acetate, TPA) response elements. AP1 family protein, such as c-Fos, c-Jun, JunB, JunD, ATF, Fra-1, Fra-2, and so on, can bind at this site by forming homo- or heter-dimer (Fig. 1).
c-Jun (Albanese et al. 1995; Cicatiello et al. 2004; Mechta et al. 1997; Soh and Weinstein 2003) Fra-1 (Burch et al. 2004; Mechta et al. 1997) Fra-2 (Balmanno and Cook 1999) can activate cyclin D1 transcription. ATF3 (Allan et al. 2001) activate cyclin D1 promoter activity requiring cAMP response element-binding (CREB) site involved although it directly bind to AP-1 site. c-Fos may sometime depress (Albanese et al. 1995) or activate (Brown et al. 1998; Cicatiello et al. 2004; Watanabe et al. 1996b) cyclin D1 promoter. JunB usually inhibits cyclin D1 promoter and can antagonized the c-Jun activation of cyclin D1 promoter (Shaulian and Karin 2001). A change of AP-1 composition toward an increase of JunB results in downregulation of cyclin D1 (Grosch et al. 2003). So generally c-Jun is an activator and JunB a repressor of cyclin D1 promoter. c-Fos is expressed rapidly and transiently (Balmanno and Cook 1999), so the inhibition effect by c-Fos overexpression (Albanese et al. 1995) probably cannot function in real cell cycle, except for c-Fos prolonged binding by some stimulation, e.g., oxidative stress (Burch et al. 2004).
Not only protein level but also the phosphorylated modification status is important to AP-1 proteins. c-Jun activation of cyclin D1 promoter requires phosphorylated on Ser63/73-Pro motifs (Wulf et al. 2001). Phosphorylation of JunB results in decreased JunB protein levels in mitotic and early G1 cells. In contrast, c-Jun levels remain constant with N-terminal phosphorylation. And the modifications of AP-1 proteins may regulate cyclin D1 transcription temporally to control cell cycle progression (Bakiri et al. 2000).
Some TFs in addition to Ap-1 family may regulate cyclin D1 promoter activity through AP-1 site directly (Roche et al. 2004) or indirectly, e.g., by protein interaction (Albanese et al. 1999), cooperation with other TF binding sites such as CREB (Watanabe et al. 1996a).
GAS
Among the STATs, only STAT3 and STAT5 can bring about the activation of cyclin D1 (Bromberg et al. 1999; Calo et al. 2003; Leslie et al. 2006).
Literatures showed that activated form of STAT3 was accompanied by increased expression levels of cyclin D1 (Bromberg et al. 1999; Kijima et al. 2002; Leslie et al. 2006; Masuda et al. 2001, 2002). And some paper showed that STAT3 can inhibit cyclin D1expression (Zhang et al. 2003). And during the liver regeneration after partial hepatectomy, the cyclin D1 induction was repressed, but STAT3 was unchanged in mice (Chen et al. 2004), which may suggest that modification of STAT3 is important to its activity. Data also showed cyclin D1 overexpression and STAT3 activation were, mutually exclusive events in MM (Quintanilla-Martinez et al. 2003).
But there was no evidence showing STAT3 can directly function through the cyclin D1 promoter, lacking data such as EMSA, ChIP and so on (Masuda et al. 2001, 2002). Moreover, cyclin D1 repression may due to CDKN1A or CDKN1B promoter induction. There are some evidences shows that STAT3 can active CDKN1B or CDKN1A promoter through PI3K pathway. Clearly, PI3K pathway can induce cyclin D1 promoter, and new evidences (Bienvenu et al. 2005) show that cyclin D1 is recruited to the CDKN1A promoter by a STAT3-NcoA complex leading to an inhibition of the p21waf1 gene (Bienvenu et al. 2005). In conclusion, in some context STAT3 and cyclin D1 balanced in cell cycle regulation but generally the relation between cyclin D1 and STAT3 may due to cell type and now is unclear.
Unlike STAT3, STAT5 can directly bind cyclin D1 promoter in which there are two STAT binding sites, one called GAS1 the other is GAS2 (Magne et al. 2003). The GAS1 site (distal) can bind stat5a/b which can activate cyclin D1 promoter (Brockman et al. 2002; Magne et al. 2003; Matsumura et al. 1999).The phosphorylated modification of STAT5b at Tyr679 induces STAT5b activation and then activate cyclin D1 promoter through interaction with other transcription factors, such as LEF1 and CREB/ATF2 (Kabotyanski and Rosen 2003). STAT5a lacks the Tyr679 site which can explain why only STAT5a/5b heterdimer or STAT5b/5b homodimer but not STAT5a/5a homodimer bind to the cyclin D1 promoter (Magne et al. 2003). Unlike GAS1, the GAS2 site, accurately composite Oct-GAS element, may be masked by Oct-1 protein which binding site overlap with GAS2. The binding of STAT5 to this site is required both GAS2 and OCT-1 element, with the interaction between STAT and PAU domain of Oct-1 (Magne et al. 2003).
E2F
The E2F binding sites in cyclin D1 promoter illustrate Fig. 1. Among five members of the E2F family, including E2F1, 2, 3, 4, and 5, only E2F1and E2F4 can bind this promoter (Watanabe et al. 1998). E2F transcription factors are bound to RB protein, and when RB is phosphalated by cyclin D1/CDK4, 6, E2Fs are released free. The free E2Fs then regulate their target genes promoting cell cycle progression.
Cyclin D1/CDK4, 6, RB and E2F cooperate together to enter cell cycle and progression in normal cell or to be transformed in tumor cell lines. Although cyclin D1 is upstream upon E2F protein during cell cycle, there are three feedbacks loop between cyclin D1 and E2F to facilitate the progression. E2F4 expresses at early G1 phase (Muller et al. 1997) which can activate cyclin D1 and results in more E2F4 protein level. This is a positive feedback loop, which occurs at early phase of cell cycle and let cell enter cell cycle quickly. There are also two other feedback loops, which respectively result in cell cycle arrest or progression depending cell types. E2F4 and E2F1 are functionally different which also express at different time in cell cycle (Muller et al. 1997). Contrast to E2F4, E2F1 expresses at late G1 phase (Muller et al. 1997). E2F1 regulates a set of genes that can let cell cycle progression. Depending different cell context, E2F1 can activate (Inoshita et al. 1999) or depress (Watanabe et al. 1998) cyclin D1 expression. High level of free E2F1 protein can induce proliferation then apoptosis (Knezevic and Brash 2004). Transgenetic mice expressing high level E2F1 also induce apoptosis (Pierce et al. 1998a). In this context, free E2F1 can depress cyclin D1, which formed a negative feedback loop to avoid apoptosis (Watanabe et al. 1998). The last feedback loop is that free E2F1 proteins can active cyclin D1 (Fan and Bertino 1997). The high level free E2F1 protein can activate another set of genes, e.g., FGFR which let cell cycle progression or transformed cells (Tashiro et al. 2003). The affinity of E2F1 to cyclin D1 promoter is higher than E2F4 (Lee et al. 2000). E2F1 has more potent activator activity than E2F4 (Pierce et al. 1998b). E2F-4 is located in nucleus from G0 until mid-G1 phase and mainly cytoplasmic in late G1, S, and G2 phases. In contrast, endogenous E2F-1 is absent from resting cells and is predominantly nuclear in late G1 and S (Muller et al. 1997). Due to the different affinity, at early stage E2F4 bounding that induce cell cycle entrance, and at late stage E2F1 take place of E2F4 results in cell cycle progression or transformation.
When depression of cyclin D1 promoter by E2F1, the SP1/2/3 is needed (Watanabe et al. 1998). Sp1 is inducible in early–mid-G1 phase (Nagata et al. 2001). In conclusion, E2F4 activate cyclin D1 promoter whereas E2F1 can activate or depress cyclin D1 promoter due to cell context.
NF-қB
NF-қB contributes to cell cycle progression, and one of its targets might be cyclin D1in T47D cell (Hinz et al. 1999). Dbl and Dbs regulated transcription from the cyclin D1 promoter in a NF-қB-dependent manner (Whitehead et al. 1999). Examination of the sequence from the human cyclin D1 promoter identified potential NF-қB-binding sites at positions −858, −749, and −39 that matched the NF-қB consensus binding sequence, GGG(G/A)NNYYCC (Guttridge et al. 1999). Different NF-қB complex members, p65 p50 and p52, can bind these sites (Guttridge et al. 1999; Westerheide et al. 2001). Although there are three NF-қB-binding sites, only the proximal site (−39) may be functional (Guo et al. 2009).
Generally NF-қB binding can induce cyclin D1 promoter(Joyce et al. 1999), whereas PKC delta depress cyclin D1 through NF-қB binding to −39 site (Page et al. 2002). Bcl-3, a co-activator with NF-қB p52 homodimers, was demonstrated to directly activate the cyclin D1 promoter through an NF-қB binding site (Westerheide et al. 2001), whereas p53 represses cyclin D1 transcription through this site under UV treatment downregulating of Bcl-3 (Rocha et al. 2003). Data also showed p53 can inhibit cyclin D1 promoter under heat shock (Guo et al. 2009). So stress stimulations may depress cyclin D1 through the proximal NF-қB binding site.
CREB/ATF2
The cAMP can inhibit or induce cell cycle progression and cyclin D1 expression. The CRE/ATF2 binding consensus site in cyclin D1 promoter locates at −57 (D’Amico et al. 2000; Lee et al. 1999; Musa et al. 1999; Watanabe et al. 1996a), which can bind CRE/ATF2 (D’Amico et al. 2000; Lee et al. 1999; McMahon et al. 1999; Musa et al. 1999), c-Jun (ATF-2/c-Jun heterodimers; Sabbah et al. 1999), CREM1 (Page et al. 2002), ATF1 (Schneider et al. 2002), and c-Fos (Brown et al. 1998). The effecter of Wnt signal transduction pathway, β-catenin/Tcf4, can also bind this site to induce cyclin D1 promoter (Pradeep et al. 2004). Galectin-3 (Lin et al. 2002) and G-17 (Pradeep et al. 2004) induces the cyclin D1 promoter also via the CREB site. Cyclosporine effective element overlaps the element and confers cyclosporine sensitivity to the cyclin D1 promoter (Schneider et al. 2002). PPAR gamma2 (Sharma et al. 2004), PKCdelta (Page et al. 2002) and p16INK4a (D’Amico et al. 2004) can inhibit cyclin D1 promoter through this site.
CREB Ser 133 phosphorylation is necessary for induction of cyclin D1 promoter through this site (D’Amico et al. 2000; Lee et al. 1999; Sharma et al. 2004), but the POU domain of oct-1 can potent its activation without Ser 133 phosphorylation by protein interaction (Boulon et al. 2002). But CREB Ser 133 phosphorylation may result in repression of cyclin D1 due to cell type (Musa et al. 1999).
TCF4
Nuclear β-catenin expression was correlated with cyclin D1 overexpression (Saito et al. 2001; Shtutman et al. 1999; Utsunomiya et al. 2001) with promoting malignant transformation by triggering cyclin D1 expression (Behrens 2000; Brabletz et al. 1999, 2000, Graham and Asthagiri 2004; Jung et al. 2004; Lepourcelet et al. 2004; Morin 1999; Muller-Tidow et al. 2004). TCF4 (Graham and Asthagiri 2004) and β-catenin (Shtutman et al. 1999) activate the cyclin D1 promote via the consensus TCF/LEF-binding sites (Grueneberg et al. 2003; Holnthoner et al. 2002; Tetsu and McCormick 1999). In addition to TCF4 mainly (Gotoh et al. 2003), LEF-1 (Grueneberg et al. 2003), HBP1 (Sampson et al. 2001), and UBF2 (Grueneberg et al. 2003) can also affect the cyclin D1 promoter activity by interaction with β-catenin or LEF-1.
β-catenin is a key component in the canonical Wnt pathway (D’Amico et al. 2000). Some molecules in addition to wnt, such as IKKα (Albanese et al. 2003), PTEN (Persad et al. 2001b), and SOX17 (Lange et al. 2009) can also regulate it. In addition to Wnt pathway, PI3k signal transduction pathway (Albanese et al. 2003; D’Amico et al. 2000), ILK (D’Amico et al. 2000; Persad et al. 2001b), RA (Shah et al. 2002), caveolin-1 (Hulit et al. 2000) can also regulate cyclin D1 promoter activity via consensus this site in cyclin D1 promoter, and CREB site (Pradeep et al. 2004) may be needed.
E box
There is an E box element at −558 in human cyclin D1 promoter (Eto 2000; Magne et al. 2003; Zhang et al. 2002). The E box can bind Myc or other transcription factor, so some paper may assigned it c-myc element. Myc proteins bind to cyclin D1 promoter to inhibit its activity (Chien et al. 2008; Gonzalez-Mariscal et al. 2009; Philipp et al. 1994), probably inducing DNA methylation (Hervouet et al. 2009). The element may activate cyclin D1 promoter by different protein interaction with myc, e.g., Max (Yang et al. 2009).
Ets
In the proximal region of cyclin D1 promoter, an Ets (c-Ets2) site was first identified in 1995 (Albanese et al. 1995). There several putative Ets binding site in cyclin D1 promoter. Tetsu and McCormick (1999) demonstrated four other Ets sites which they named Ets A B C D, but only the B box is mediated by P21RAS. Zhao et al. (2001) demonstrated that the EtsB binding site mediated cyclin D1 promoter regulation by FAK. The proximal box can mediated PKC delta activity (Page et al. 2002), and RAS induced MAPK signal transduction (Albanese et al. 1995).
CSL
Notch, an evolution-conserved membrane crossed-signal molecular (for review, see Artavanis-Tsakonas et al. 1999) encoding a family of transmembrane proteins that are involved in many cellular processes such as differentiation, proliferation, and apoptosis, can activated cyclin D1 promoter transcription through a CSL site (Jeffries et al. 2002; Ronchini and Capobianco 2001; Stahl et al. 2006).
GT box
There are four GT box in cyclin D1 promoter but only the GT box A was active which was responsible for the inhibition effect of KLF8 to cyclin D1 promoter (Zhao et al. 2003).
Egr-1
In cyclin D1 promoter,Egr-1 site which overlaps two sp1 sites, can mediate TGFβ (Yan et al. 1997) and Ang II (Guillemot et al. 2001)-induced cyclin D1 upregulation. But unexpectedly the Egr-1 binding activity to the cyclin D1 promoter is not influenced by SP1 binding (Yan et al. 1997).
Sp1
The transcription factor SP1 is a DNA-binding protein which interacts with a variety of gene promoters containing GC-box elements. Among many possible SP1 sites, the site studied in the promoter overlaps with Egr-1. Induction of the cyclin D1 promoter activity in the early to mid G 1 phase is via the SP1 sites by the Ras-dependent pathway (Nagata et al. 2001). NeuT can induce cyclin D1 promoter by Sp1/3 binding in cooperation with E2F site (Lee et al. 2000). In PC12 cells NGF can induce neurite outgrowth and cyclin D1 transcription via Sp1 and NF-қB binding site in the proximal region of the cyclin D1 promoter (Marampon et al. 2008).
CycY
The motif is SP1-like but bind basic transcription element binding factor (BTEB) whose molecular weight is smaller than SP1 (Hsiang and Straus 2002).But binding BTEB on CycY site is not responsible for cyclopentenon (Hsiang and Straus 2002).
ARE
p19ARF repressed cyclin D1 through a novel distal cis-element ARE at −1137, which bound p53 revealed by chromatin-immunoprecipitation assays (D’Amico et al. 2004). P53 can also repress cyclin D1 promoter in −39 NF-қB site and HADC1 may involved (Guo et al. 2009; Rocha et al. 2003).
Complex motif
Complex motif here means that two elements in a promoter are very close, sometimes joined together. In this promoter, e.g., E2F and sp1, stat and oct-1 are close to form complex motifs. More often, the proteins that bound to complex motif could interact with each other. So we can deduce that proteins which can interact with each other may result in DNA sequence rearrangement. The protein and DNA sequence can co-evolve.
Starting site
Different groups studied the transcription star site with different methods (Herber et al. 1994a; Hsiang and Straus 2002; Motokura and Arnold 1993; Philipp et al. 1994). Among these, CCTCCAGAGGGCTGT (Motokura and Arnold 1993) and CCTCCAGAGGGCTGT (Hsiang and Straus 2002; transcription star site is underlined) were prevalently accepted. In this review, elements positions were normalized to CCTCCAGAGGGCTGT (Motokura and Arnold 1993).
Signal transduction pathway
There are mainly three signal transduction pathways involved in cyclin D1 promoter regulation, which are MAPK, PI3K/Akt, and Wnt. Others such as ER, NF-κB, JAK/STAT, Rac1/NADPH oxidase are also involved. Here, we discuss the main three pathways: MAPK, PI3K/Akt and Wnt including its molecules, response elements and cross-talk points (Fig. 2).
Fig. 2.

The main three signal transduction pathways on human cyclin D1 promoter. Three pathways are MAPK, PI3k/Akt, and Wnt. MAPK pathways regulate cyclin D1 promoter via AP-1 and Ets elements. Elements such as LEF/TCF and CRE are responsible for PI3k/Akt and Wnt pathways and GSK3β is their cross-talk point. PETN can cross-talk between MAPK and PI3k/Akt pathway
ERK1/2 cascade can activted cyclin D1 promoter activity. Raf/Mek/Erk pathway usually activates cyclin D1 promoter (Chu et al. 2005; Greulich and Erikson 1998; Page et al. 1999a, b; Ramakrishnan et al. 1998; Watanabe et al. 1996a, b; Weber et al. 1997a, b). There are two phases of ERK activation, of which the second sustained phase is required for activation of cyclin D1 promoter (Fassett et al. 2003; Talarmin et al. 1999; Treinies et al. 1999; Weber et al. 1997b). But the prolonged ERK activation results in downregulation cyclin D1 due to inhibition of CREB activity, including its DNA binding ability and Ser-133 phosphorylation (Wang et al. 2003), or due to ERK nuclear location (Burch et al. 2004; Clark et al. 2004). P38 usually inhibits (Catalano et al. 2004; Ellinger-Ziegelbauer et al. 1999; Kintscher et al. 2003; Lavoie et al. 1996; Lee et al. 1999; Page et al. 2001; Pruitt et al. 2002; Todd et al. 2004; Westwick et al. 1998) and sometimes activated (Klein et al. 2003; Lee et al. 2000, 1999; Recio and Merlino 2002) cyclin D1.SV 40 small antigen can induce cyclin D1 promoter by ERK and SAPK pathway (Watanabe et al. 1996a). JNK can activate cyclin D1 promoter via activating c-jun (Oktay et al. 1999; Wulf et al. 2001) and ATF2, which can by binding CREB/ATF2 site (Lee et al. 1999). But some literatures reported JNK also inhibited cyclin D1 promoter (Grosch et al. 2003) or had no function on cyclin D1 expression in bovine tracheal myocytes (Page et al. 1999a).
ERK pathway induces cyclin D1 promoter by Ets or AP-1 (Chu et al. 2005) elements in cyclin D1 promoter (Albanese et al. 1995; Chu et al. 2005; Guillemot et al. 2001). V-src activation of cyclin D1 involved the ERK, p38, and JNK via CREB site (Lee et al. 1999), but in CCL39 cells, ERK5 but not the ERK1/2 cascade regulate cyclin D1 promoter via this site (Mulloy et al. 2003).
The Wnt signaling pathway is conserved in various organisms from worms to mammals, and plays important roles in development, cellular proliferation, and differentiation. Wnt stabilizes cytoplasmic β-catenin and then β-catenin is translocated into the nucleus where it stimulates the expression of genes including cyclin D1 (Kikuchi 2000; Shtutman et al. 1999; Tetsu and McCormick 1999). PI3k/Akt signal transduction pathway can inhibit GSK3β and then promote β-catenin to activate cyclin D1 promoter via the TCF site (Albanese et al. 2003). PI3k/Akt signal transduction pathway can also activate cyclin D1 promoter by modulating CREB via its binding site. But this may be weaker than that by inhibition of GSK3β (Xie et al. 2003). ILK and PDK1 can activate Akt by phasphation at different amino acid site, ser-473 and ser-308, respectively (Persad et al. 2001a), which all take part in Akt activation which consequently then inhibits GSK3β at ser-9 (Troussard et al. 2003). In some cell type, PKC but not Akt can inhibit GS3Kβ (Xie et al. 2003). Rac1 which can form positive regulation loop with PI3k (Welch et al. 2003), can activate cyclin D1 by NF-қB (Joyce et al. 1999) and CREB site (Bauerfeld et al. 2001; Joyce et al. 1999; Page et al. 2000) independent of ERK (Page et al. 1999b, 2000). Wnt pathway regulation whereby activation of Rac1 amplifies the signaling activity of stabilized/mutated β-catenin by promoting its accumulation in the nucleus, and synergizing with β-catenin to augment TCF/LEF-dependent gene transcription (Esufali and Bapat 2004). PI3k/Akt signal transduction pathway plays important role in regulation of cyclin D1 promoter. The pathway may induce cyclin D1 by CREB site in the promoter and can modulate GSK3β to activate β-catenin, which can induce cyclin D1. There are many interlinks between PI3k and wnt pathway in regulation of cyclin D1 promoter.
It is usually thought that MAPK, unlike wnt, distinct from PI3k signal transduction pathway (Page et al. 2000), but there are still many cross-talks between them. The ERK pathway modulated AKT phosphorylation by acting on the PTEN levels (Marino et al. 2003). Persad et al. (2001b) define a pathway that ILK and GSK-3 can regulate β-catenin stability, nuclear β-catenin expression, and its transcriptional activity. Wnt-transactivated ErbB1 was responsible for MAPK activation and the increased levels of cyclin D1 present in the Wnt-expressing HC11 cells (Civenni et al. 2003). TGF-β1 also first decreases and later potentiates the levels of EGF-activated MEK1/MAPK and PKB, which results in initially suppresses EGF-induced cyclin D1 expression then later releases the inhibition (Yan et al. 2000) implying there are other cross-talks between MAPK and PI3k/Akt.
Taken together, there are cross-talks between Wnt and PI3k usually converging at GSK3β. MAPK pathway is generally distinct from PI3k, but they can cross-talk, e.g., by PTEN (Marino et al. 2003; Weng et al. 2001), TGF-β1 (Yan et al. 2000), PAK (Nheu et al. 2004), or others. PTEN is also involved in the regulation of nuclear β-catenin accumulation and TCF transcriptional activation in an APC-independent manner (Persad et al. 2001b). Sometimes in the mammary gland Wnt pathway can activate cyclin D1 by MAPK activation (Civenni et al. 2003).
The temporal expression of cyclin D1
Cell cycle progression requires different signal molecules function at the right time. The stimulation from growth factor is temporal, biphasic (Jones and Kazlauskas 2001). So what pathway function at what time is critical for cell cycle progression. Rac/Cdc42 signaling induces cyclin D1 expression in an early G1 phase. In the mid-G1 phase, cyclin D1 is induced by sustained ERK, which can be promoted by Rho kinase. At the same time, Rho kinase suppresses Rac/Cdc42 activity (Roovers and Assoian 2003; Roovers et al. 2003; Welsh et al. 2001). MKP, as an inhibitor of ERK, can form a feedback loop to a flexibly balanced ERK activity (Bennett and Tonks 1997; Bhalla et al. 2002; Ryser et al. 2004). MKP overexpression can result in downregulation of cyclin D1 (Kawanaka et al. 2001; Lavoie et al. 1996; Qin et al. 2005). In the later stages of G1, PI3k pathways instead of ERK to sustain cyclin D1 expression to perform S phase entry (Gille and Downward 1999; Marino et al. 2003). Akt/PKB, an important downstream of PI3k, is expressed in late G1phase (Gille and Downward 1999; Paramio et al. 1999), but it only influences partly cyclin D1 expression (Gille and Downward 1999). So there may be multiple signal molecules involved.
Epigenetic regulation of the cyclin D1 transcription
Epigenetic regulation means a heritable alteration in gene expression without the primary DNA sequence changing. The major mechanisms involved in epigenetic changes are modification of DNA and histone protein such as DNA methylation at cytosine bases and histone acetylation.
Epigenetic modification sites involved in cyclin D1 transcriptional regulation include (1) GC-rich Sp1/CRE binding site, (2) remote upstream region mainly in chromosome translocation, a common cause of blood tumor, (3) 1 kb upstream including E-box element, and (4) other DNA methylation sites which have not been studied. Actually, function of DNA methylation and histone modification are commonly studied together.
DNA methylation at Sp1/CRE binding sites of rat cyclin D1 promoter may be essential for keeping a number of the stromal cells in the basal layer live (Kitazawa et al. 1999). In hamster cell, using human cyclin D1 promoter, data showed that DNA methylation was found at Sp1/CRE binding sites (Hilton et al. 2005). However, the epigenetic modification including DNA methylation at cytosine bases and H3/H4 acetylation at Sp1/CRE binding sites may not be essential for transcriptional regulation of cyclin D1 (Krieger et al. 2005). Chromosome translocation, a common cause of blood tumor, is thought to transcriptional regulation of cyclin D1. Data showed that such epigenetic modifications mainly were found in the translocation region, distal upstream region of cyclin D1 promoter (120 kb from the transcriptional start site; Liu et al. 2004) and demethylation may due to CTCF and NPM (Liu et al. 2008a). Different group found the DNA methylation or histone acetylation in this region from different blood tumor including MCL, MM, and NHL and so on. Although the epigenetic modification may be essential in gene transcriptional regulation, it was thought that the epigenetic modification have no effect on cyclin D1 transcription. No DNA methylation was found in cyclin D1 promoter by genomewide methylation analysis in MCL patients (Leshchenko et al. 2010). The endogenous cyclin D1 promoter may be inaccessible to the transcription factor and cyclin D1 transcription may be control through other different manner. Actually the MYEOV gene which located approach to cyclin D1 was transregulated by this epigenetic modification (Janssen et al. 2002), which showed that epigenetic regulation may need a proper transcriptional status. Interestingly, in some MM and MCL samples that did not express cyclin D1, the cyclin D1 promoter was hypomethylated and hyperacetylated, which suggested that DNA methylation in the promoter may be related to malignant phase rather than to cyclin D1 regulation (Liu et al. 2004). And this agreed with the data in NHL research, in which the DNA methylation was identified as a tumor maker, although it is not involved in cyclin D1 transcription (Shi et al. 2007), which showed that the region was proven to be methylated. Genes other than cyclin D1 may be regulated by DNA methylation which can then regulated cylcin D1 including CDKN2A (Vonlanthen et al. 1998; Kawauchi et al. 2004; Takahira et al. 2004; Hutter et al. 2006; Liu et al. 2008b; Matsuda 2008; Takahira et al. 2004; Kawauchi et al. 2004; Hashiguchi et al. 2001; Hutter et al. 2006; Dominguez et al. 2002), wnt (Fox et al. 2008; Martin et al. 2009), and miRNA (Ilnytskyy et al. 2008). Data from blood tumor, epigenetic modification in 1 kb region upstream from transcription start site may not affect cyclin D1 transcription (Liu et al. 2004). Although data from blood tumor cell mainly showed that epigenetic modification may not involved in cyclin D1 transcription, in glioma cells Hhervouet et al. (2009) showed a DNA methylation mechanism in depression of cyclin D1 transcription via E-box, a site-specific DNA methylation site in the 1 kb upstream region of cyclin D1.And different from blood tumor research which showed treatment of TSA or 5-Aza had no effect on cyclin D1 transcription (Krieger et al. 2005), data showed that the epigenetic regent can regulate its transcription or translation in glioma cell,H1299 cell, follicular lymphoma (also blood tumor) cell and MCF-7 cell (Alao 2007; Alao et al. 2006a, b; Bennett et al. 2009; Hervouet et al. 2009; Rocha et al. 2003). Data from HCC (primary liver cancer) showed DNA methylation in cyclin D1 promoter (Matsuda 2008) and in lung cancer, DNA methylation of CDKN2A promoter in regulation of cyclin D1 may be different (Zhou et al. 2001). So epigenetic regulation may be different due to cell types. Other than histone acetylation, histone methylation of H3k9 may inhibit human (Krieger et al. 2005) and mouse (Shirato et al. 2009) cyclin D1 transcription, and this may function in development (Ait-Si-Ali et al. 2004). Considering CpG islands identifying, other sites may be studied to reveal the epigenetic regulation mechanism involved in cyclin D1 for example there are many other CpG inlands (Krieger et al. 2005) except for the region mentioned above.
In conclusion, epigenetic modification (DNA methylation and histone modification) involved in cyclin D1 transcriptional regulation may be cell type-specific. In most blood tumor, cyclin D1 transcription is not due to DNA or histone modification, but this was not the barrier for the DNA methylation to be used as a putative tumor marker. Other gene (especially CDKN2A) may be regulated by epigenetic modification. There may be other epigenetic modification which can be studied to provide insight into a new mechanism of epigenetic transcriptional regulation of cyclin D1, for there are other CpG islands not studied yet.
Conclusion
Cell cycle control is complex, in which cyclin D1 transcription regulation may be important. But firstly, cell cycle control is not only in transcription level but also in post-transcriptionally regulated manner, e.g., protein degradation, modification, which all play an important role in cell cycle control. For example, GSK3β can also increases cyclin D1 protein degradation (Hamelers et al. 2002; Jirmanova et al. 2002; Kim et al. 2002; Zou et al. 2004), and cyclin D1 mRNA half-life becomes shorter when serum is removed (Guo et al. 2005). And secondly, much study got from synchronized cell by serum deprivation, which cannot reflect the real cycle. In actively cycling cells, cyclin D1 may be induced to high levels in G2 phase, and the expression levels of cyclin D1 in G2 phase determine the fate of the next cell cycle (Guo et al. 2005; Stacey 2003). Thirdly, some cell can proliferfy and organ developed without cyclin D1 (Kozar et al. 2004; Malumbres et al. 2004). Taken together, the regulation of cyclin D1 promoter is important in cell cycle control, but it is not all.
Acknowledgment
The research was supported by foundation from National Nature Science Foundation of China (31050010), Hebei Education Department (2010154) and North China Medical College (BS06002).
References
- Ait-Si-Ali S, Guasconi V, Fritsch L, Yahi H, Sekhri R, Naguibneva I, Robin P, Cabon F, Polesskaya A, Harel-Bellan A. A Suv39h-dependent mechanism for silencing S-phase genes in differentiating but not in cycling cells. EMBO J. 2004;23:605–615. doi: 10.1038/sj.emboj.7600074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alao JP. The regulation of cyclin D1 degradation: roles in cancer development and the potential for therapeutic invention. Mol Cancer. 2007;6:24. doi: 10.1186/1476-4598-6-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alao JP, Gamble SC, Stavropoulou AV, Pomeranz KM, Lam EW, Coombes RC, Vigushin DM. The cyclin D1 proto-oncogene is sequestered in the cytoplasm of mammalian cancer cell lines. Mol Cancer. 2006;5:7. doi: 10.1186/1476-4598-5-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alao JP, Stavropoulou AV, Lam EW, Coombes RC, Vigushin DM. Histone deacetylase inhibitor, trichostatin A induces ubiquitin-dependent cyclin D1 degradation in MCF-7 breast cancer cells. Mol Cancer. 2006;5:8. doi: 10.1186/1476-4598-5-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albanese C, Johnson J, Watanabe G, Eklund N, Vu D, Arnold A, Pestell RG. Transforming p21ras mutants and c-Ets-2 activate the cyclin D1 promoter through distinguishable regions. J Biol Chem. 1995;270:23589–23597. doi: 10.1074/jbc.270.31.18301. [DOI] [PubMed] [Google Scholar]
- Albanese C, D’Amico M, Reutens AT, Fu M, Watanabe G, Lee RJ, Kitsis RN, Henglein B, Avantaggiati M, Somasundaram K, et al. Activation of the cyclin D1 gene by the E1A-associated protein p300 through AP-1 inhibits cellular apoptosis. J Biol Chem. 1999;274:34186–34195. doi: 10.1074/jbc.274.48.34186. [DOI] [PubMed] [Google Scholar]
- Albanese C, Wu K, D’Amico M, Jarrett C, Joyce D, Hughes J, Hulit J, Sakamaki T, Fu M, Ben-Ze’ev A, et al. IKKalpha regulates mitogenic signaling through transcriptional induction of cyclin D1 via Tcf. Mol Biol Cell. 2003;14:585–599. doi: 10.1091/mbc.02-06-0101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allan AL, Albanese C, Pestell RG, LaMarre J. Activating transcription factor 3 induces DNA synthesis and expression of cyclin D1 in hepatocytes. J Biol Chem. 2001;276:27272–27280. doi: 10.1074/jbc.M103196200. [DOI] [PubMed] [Google Scholar]
- Artavanis-Tsakonas S, Rand MD, Lake RJ. Notch signaling: cell fate control and signal integration in development. Science. 1999;284:770–776. doi: 10.1126/science.284.5415.770. [DOI] [PubMed] [Google Scholar]
- Bakiri L, Lallemand D, Bossy-Wetzel E, Yaniv M. Cell cycle-dependent variations in c-Jun and JunB phosphorylation: a role in the control of cyclin D1 expression. EMBO J. 2000;19:2056–2068. doi: 10.1093/emboj/19.9.2056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balmanno K, Cook SJ. Sustained MAP kinase activation is required for the expression of cyclin D1, p21Cip1 and a subset of AP-1 proteins in CCL39 cells. Oncogene. 1999;18:3085–3097. doi: 10.1038/sj.onc.1202647. [DOI] [PubMed] [Google Scholar]
- Bauerfeld CP, Hershenson MB, Page K. Cdc42, but not RhoA, regulates cyclin D1 expression in bovine tracheal myocytes. Am J Physiol Lung Cell Mol Physiol. 2001;280:L974–L982. doi: 10.1152/ajplung.2001.280.5.L974. [DOI] [PubMed] [Google Scholar]
- Behrens J. Control of beta-catenin signaling in tumor development. Ann NY Acad Sci. 2000;910:21–33. doi: 10.1111/j.1749-6632.2000.tb06698.x. [DOI] [PubMed] [Google Scholar]
- Bennett AM, Tonks NK. Regulation of distinct stages of skeletal muscle differentiation by mitogen-activated protein kinases. Science. 1997;278:1288–1291. doi: 10.1126/science.278.5341.1288. [DOI] [PubMed] [Google Scholar]
- Bennett LB, Schnabel JL, Kelchen JM, Taylor KH, Guo J, Arthur GL, Papageorgio CN, Shi H, Caldwell CW. DNA hypermethylation accompanied by transcriptional repression in follicular lymphoma. Genes Chromosom Cancer. 2009;48:828–841. doi: 10.1002/gcc.20687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhalla US, Ram PT, Iyengar R. MAP kinase phosphatase as a locus of flexibility in a mitogen-activated protein kinase signaling network. Science. 2002;297:1018–1023. doi: 10.1126/science.1068873. [DOI] [PubMed] [Google Scholar]
- Bienvenu F, Barre B, Giraud S, Avril S, Coqueret O. Transcriptional regulation by a DNA-associated form of cyclin D1. Mol Biol Cell. 2005;16(4):1850–1858. doi: 10.1091/mbc.E04-08-0654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boulon S, Dantonel JC, Binet V, Vie A, Blanchard JM, Hipskind RA, Philips A. Oct-1 potentiates CREB-driven cyclin D1 promoter activation via a phospho-CREB- and CREB binding protein-independent mechanism. Mol Cell Biol. 2002;22:7769–7779. doi: 10.1128/MCB.22.22.7769-7779.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brabletz T, Jung A, Dag S, Hlubek F, Kirchner T. Beta-catenin regulates the expression of the matrix metalloproteinase-7 in human colorectal cancer. Am J Pathol. 1999;155:1033–1038. doi: 10.1016/S0002-9440(10)65204-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brabletz T, Herrmann K, Jung A, Faller G, Kirchner T. Expression of nuclear beta-catenin and c-myc is correlated with tumor size but not with proliferative activity of colorectal adenomas. Am J Pathol. 2000;156:865–870. doi: 10.1016/S0002-9440(10)64955-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brockman JL, Schroeder MD, Schuler LA. PRL activates the cyclin D1 promoter via the Jak2/Stat pathway. Mol Endocrinol. 2002;16:774–784. doi: 10.1210/me.16.4.774. [DOI] [PubMed] [Google Scholar]
- Bromberg JF, Wrzeszczynska MH, Devgan G, Zhao Y, Pestell RG, Albanese C, Darnell JE., Jr Stat3 as an oncogene. Cell. 1999;98:295–303. doi: 10.1016/S0092-8674(00)81959-5. [DOI] [PubMed] [Google Scholar]
- Brown JR, Nigh E, Lee RJ, Ye H, Thompson MA, Saudou F, Pestell RG, Greenberg ME. Fos family members induce cell cycle entry by activating cyclin D1. Mol Cell Biol. 1998;18:5609–5619. doi: 10.1128/mcb.18.9.5609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burch PM, Yuan Z, Loonen A, Heintz NH. An extracellular signal-regulated kinase 1- and 2-dependent program of chromatin trafficking of c-Fos and Fra-1 is required for cyclin D1 expression during cell cycle reentry. Mol Cell Biol. 2004;24:4696–4709. doi: 10.1128/MCB.24.11.4696-4709.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calo V, Migliavacca M, Bazan V, Macaluso M, Buscemi M, Gebbia N, Russo A. STAT proteins: from normal control of cellular events to tumorigenesis. J Cell Physiol. 2003;197:157–168. doi: 10.1002/jcp.10364. [DOI] [PubMed] [Google Scholar]
- Catalano A, Caprari P, Rodilossi S, Betta P, Castellucci M, Casazza A, Tamagnone L, Procopio A. Cross-talk between vascular endothelial growth factor and semaphorin-3A pathway in the regulation of normal and malignant mesothelial cell proliferation. FASEB J. 2004;18:358–360. doi: 10.1096/fj.03-0513fje. [DOI] [PubMed] [Google Scholar]
- Chen L, Zeng Y, Yang H, Lee TD, French SW, Corrales FJ, Garcia-Trevijano ER, Avila MA, Mato JM, Lu SC. Impaired liver regeneration in mice lacking methionine adenosyltransferase 1A. FASEB J. 2004;18:914–916. doi: 10.1096/fj.03-0899com. [DOI] [PubMed] [Google Scholar]
- Chien J, Narita K, Rattan R, Giri S, Shridhar R, Staub J, Beleford D, Lai J, Roberts LR, Molina J, et al. A role for candidate tumor-suppressor gene TCEAL7 in the regulation of c-Myc activity, cyclin D1 levels and cellular transformation. Oncogene. 2008;27:7223–7234. doi: 10.1038/onc.2008.360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu M, Guo J, Chen CY. Long-term exposure to nicotine, via ras pathway, induces cyclin D1 to stimulate G1 cell cycle transition. J Biol Chem. 2005;280:6369–6379. doi: 10.1074/jbc.M408947200. [DOI] [PubMed] [Google Scholar]
- Cicatiello L, Addeo R, Sasso A, Altucci L, Petrizzi VB, Borgo R, Cancemi M, Caporali S, Caristi S, Scafoglio C, et al. Estrogens and progesterone promote persistent CCND1 gene activation during G1 by inducing transcriptional derepression via c-Jun/c-Fos/estrogen receptor (progesterone receptor) complex assembly to a distal regulatory element and recruitment of cyclin D1 to its own gene promoter. Mol Cell Biol. 2004;24:7260–7274. doi: 10.1128/MCB.24.16.7260-7274.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Civenni G, Holbro T, Hynes NE. Wnt1 and Wnt5a induce cyclin D1 expression through ErbB1 transactivation in HC11 mammary epithelial cells. EMBO Rep. 2003;4:166–171. doi: 10.1038/sj.embor.embor735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark JA, Black AR, Leontieva OV, Frey MR, Pysz MA, Kunneva L, Woloszynska-Read A, Roy D, Black JD. Involvement of the ERK signaling cascade in protein kinase C-mediated cell cycle arrest in intestinal epithelial cells. J Biol Chem. 2004;279:9233–9247. doi: 10.1074/jbc.M312268200. [DOI] [PubMed] [Google Scholar]
- Coqueret O. Linking cyclins to transcriptional control. Gene. 2002;299:35–55. doi: 10.1016/S0378-1119(02)01055-7. [DOI] [PubMed] [Google Scholar]
- D’Amico M, Hulit J, Amanatullah DF, Zafonte BT, Albanese C, Bouzahzah B, Fu M, Augenlicht LH, Donehower LA, Takemaru K, et al. The integrin-linked kinase regulates the cyclin D1 gene through glycogen synthase kinase 3beta and cAMP-responsive element-binding protein-dependent pathways. J Biol Chem. 2000;275:32649–32657. doi: 10.1074/jbc.M000643200. [DOI] [PubMed] [Google Scholar]
- D’Amico M, Wu K, Fu M, Rao M, Albanese C, Russell RG, Lian H, Bregman D, White MA, Pestell RG. The inhibitor of cyclin-dependent kinase 4a/alternative reading frame (INK4a/ARF) locus encoded proteins p16INK4a and p19ARF repress cyclin D1 transcription through distinct cis elements. Cancer Res. 2004;64:4122–4130. doi: 10.1158/0008-5472.CAN-03-2519. [DOI] [PubMed] [Google Scholar]
- Dominguez G, Carballido J, Silva J, Silva JM, Garcia JM, Menendez J, Provencio M, Espana P, Bonilla F. p14ARF promoter hypermethylation in plasma DNA as an indicator of disease recurrence in bladder cancer patients. Clin Cancer Res. 2002;8:980–985. [PubMed] [Google Scholar]
- Ellinger-Ziegelbauer H, Kelly K, Siebenlist U. Cell cycle arrest and reversion of Ras-induced transformation by a conditionally activated form of mitogen-activated protein kinase kinase kinase 3. Mol Cell Biol. 1999;19:3857–3868. doi: 10.1128/mcb.19.5.3857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esufali S, Bapat B. Cross-talk between Rac1 GTPase and dysregulated Wnt signaling pathway leads to cellular redistribution of beta-catenin and TCF/LEF-mediated transcriptional activation. Oncogene. 2004;23:8260–8271. doi: 10.1038/sj.onc.1208007. [DOI] [PubMed] [Google Scholar]
- Eto I. Molecular cloning and sequence analysis of the promoter region of mouse cyclin D1 gene: implication in phorbol ester-induced tumour promotion. Cell Prolif. 2000;33:167–187. doi: 10.1046/j.1365-2184.2000.00176.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan J, Bertino JR. Functional roles of E2F in cell cycle regulation. Oncogene. 1997;14:1191–1200. doi: 10.1038/sj.onc.1200940. [DOI] [PubMed] [Google Scholar]
- Fassett JT, Tobolt D, Nelsen CJ, Albrecht JH, Hansen LK. The role of collagen structure in mitogen stimulation of ERK, cyclin D1 expression, and G1-S progression in rat hepatocytes. J Biol Chem. 2003;278:31691–31700. doi: 10.1074/jbc.M300899200. [DOI] [PubMed] [Google Scholar]
- Fox KE, Colton LA, Erickson PF, Friedman JE, Cha HC, Keller P, MacDougald OA, Klemm DJ. Regulation of cyclin D1 and Wnt10b gene expression by cAMP-responsive element-binding protein during early adipogenesis involves differential promoter methylation. J Biol Chem. 2008;283:35096–35105. doi: 10.1074/jbc.M806423200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu M, Wang C, Li Z, Sakamaki T, Pestell RG. Minireview: Cyclin D1: normal and abnormal functions. Endocrinology. 2004;145:5439–5447. doi: 10.1210/en.2004-0959. [DOI] [PubMed] [Google Scholar]
- Gille H, Downward J. Multiple ras effector pathways contribute to G(1) cell cycle progression. J Biol Chem. 1999;274:22033–22040. doi: 10.1074/jbc.274.31.22033. [DOI] [PubMed] [Google Scholar]
- Gonzalez-Mariscal L, Tapia R, Huerta M, Lopez-Bayghen E. The tight junction protein ZO-2 blocks cell cycle progression and inhibits cyclin D1 expression. Ann NY Acad Sci. 2009;1165:121–125. doi: 10.1111/j.1749-6632.2009.04024.x. [DOI] [PubMed] [Google Scholar]
- Gotoh J, Obata M, Yoshie M, Kasai S, Ogawa K. Cyclin D1 over-expression correlates with beta-catenin activation, but not with H-ras mutations, and phosphorylation of Akt, GSK3 beta and ERK1/2 in mouse hepatic carcinogenesis. Carcinogenesis. 2003;24:435–442. doi: 10.1093/carcin/24.3.435. [DOI] [PubMed] [Google Scholar]
- Graham NA, Asthagiri AR. Epidermal growth factor-mediated T-cell factor/lymphoid enhancer factor transcriptional activity is essential but not sufficient for cell cycle progression in nontransformed mammary epithelial cells. J Biol Chem. 2004;279:23517–23524. doi: 10.1074/jbc.M314055200. [DOI] [PubMed] [Google Scholar]
- Greulich H, Erikson RL. An analysis of Mek1 signaling in cell proliferation and transformation. J Biol Chem. 1998;273:13280–13288. doi: 10.1074/jbc.273.21.13280. [DOI] [PubMed] [Google Scholar]
- Grosch S, Tegeder I, Schilling K, Maier TJ, Niederberger E, Geisslinger G. Activation of c-Jun-N-terminal-kinase is crucial for the induction of a cell cycle arrest in human colon carcinoma cells caused by flurbiprofen enantiomers. FASEB J. 2003;17:1316–1318. doi: 10.1096/fj.02-0919fje. [DOI] [PubMed] [Google Scholar]
- Grueneberg DA, Pablo L, Hu KQ, August P, Weng Z, Papkoff J. A functional screen in human cells identifies UBF2 as an RNA polymerase II transcription factor that enhances the beta-catenin signaling pathway. Mol Cell Biol. 2003;23:3936–3950. doi: 10.1128/MCB.23.11.3936-3950.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guillemot L, Levy A, Raymondjean M, Rothhut B. Angiotensin II-induced transcriptional activation of the cyclin D1 gene is mediated by Egr-1 in CHO-AT(1A) cells. J Biol Chem. 2001;276:39394–39403. doi: 10.1074/jbc.M103862200. [DOI] [PubMed] [Google Scholar]
- Guo Y, Harwalkar J, Stacey DW, Hitomi M. Destabilization of cyclin D1 message plays a critical role in cell cycle exit upon mitogen withdrawal. Oncogene. 2005;24:1032–1042. doi: 10.1038/sj.onc.1208299. [DOI] [PubMed] [Google Scholar]
- Guo Z, Wang J, Yang J, Wu NH, Zhang Y, Shen YF. An inhibitory role of p53 via NF-kappaB element on the cyclin D1 gene under heat shock. Biochim Biophys Acta. 2009;1789:758–762. doi: 10.1016/j.bbagrm.2009.09.011. [DOI] [PubMed] [Google Scholar]
- Guttridge DC, Albanese C, Reuther JY, Pestell RG, Baldwin AS., Jr NF-kappaB controls cell growth and differentiation through transcriptional regulation of cyclin D1. Mol Cell Biol. 1999;19:5785–5799. doi: 10.1128/mcb.19.8.5785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamelers IH, Schaik RF, Sipkema J, Sussenbach JS, Steenbergh PH. Insulin-like growth factor I triggers nuclear accumulation of cyclin D1 in MCF-7 S breast cancer cells. J Biol Chem. 2002;277:47645–47652. doi: 10.1074/jbc.M208727200. [DOI] [PubMed] [Google Scholar]
- Hashiguchi Y, Tsuda H, Yamamoto K, Inoue T, Ishiko O, Ogita S. Combined analysis of p53 and RB pathways in epithelial ovarian cancer. Hum Pathol. 2001;32:988–996. doi: 10.1053/hupa.2001.27115. [DOI] [PubMed] [Google Scholar]
- Herber B, Truss M, Beato M, Muller R. Inducible regulatory elements in the human cyclin D1 promoter. Oncogene. 1994;9:1295–1304. [PubMed] [Google Scholar]
- Herber B, Truss M, Beato M, Muller R. Inducible regulatory elements in the human cyclin D1 promoter. Oncogene. 1994;9:2105–2107. [PubMed] [Google Scholar]
- Hervouet E, Vallette FM, Cartron PF. Dnmt3/transcription factor interactions as crucial players in targeted DNA methylation. Epigenetics. 2009;4:487–499. doi: 10.4161/epi.4.7.9883. [DOI] [PubMed] [Google Scholar]
- Hilton TL, Li Y, Dunphy EL, Wang EH. TAF1 histone acetyltransferase activity in Sp1 activation of the cyclin D1 promoter. Mol Cell Biol. 2005;25:4321–4332. doi: 10.1128/MCB.25.10.4321-4332.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinz M, Krappmann D, Eichten A, Heder A, Scheidereit C, Strauss M. NF-kappaB function in growth control: regulation of cyclin D1 expression and G0/G1-to-S-phase transition. Mol Cell Biol. 1999;19:2690–2698. doi: 10.1128/mcb.19.4.2690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holnthoner W, Pillinger M, Groger M, Wolff K, Ashton AW, Albanese C, Neumeister P, Pestell RG, Petzelbauer P. Fibroblast growth factor-2 induces Lef/Tcf-dependent transcription in human endothelial cells. J Biol Chem. 2002;277:45847–45853. doi: 10.1074/jbc.M209354200. [DOI] [PubMed] [Google Scholar]
- Hsiang CH, Straus DS. Cyclopentenone causes cell cycle arrest and represses cyclin D1 promoter activity in MCF-7 breast cancer cells. Oncogene. 2002;21:2212–2226. doi: 10.1038/sj.onc.1205293. [DOI] [PubMed] [Google Scholar]
- Hulit J, Bash T, Fu M, Galbiati F, Albanese C, Sage DR, Schlegel A, Zhurinsky J, Shtutman M, Ben-Ze’ev A, et al. The cyclin D1 gene is transcriptionally repressed by caveolin-1. J Biol Chem. 2000;275:21203–21209. doi: 10.1074/jbc.M000321200. [DOI] [PubMed] [Google Scholar]
- Hutter G, Scheubner M, Zimmermann Y, Kalla J, Katzenberger T, Hubler K, Roth S, Hiddemann W, Ott G, Dreyling M. Differential effect of epigenetic alterations and genomic deletions of CDK inhibitors [p16(INK4a), p15(INK4b), p14(ARF)] in mantle cell lymphoma. Genes Chromosom Cancer. 2006;45:203–210. doi: 10.1002/gcc.20277. [DOI] [PubMed] [Google Scholar]
- Ilnytskyy Y, Zemp FJ, Koturbash I, Kovalchuk O. Altered microRNA expression patterns in irradiated hematopoietic tissues suggest a sex-specific protective mechanism. Biochem Biophys Res Commun. 2008;377:41–45. doi: 10.1016/j.bbrc.2008.09.080. [DOI] [PubMed] [Google Scholar]
- Inoshita S, Terada Y, Nakashima O, Kuwahara M, Sasaki S, Marumo F. Roles of E2F1 in mesangial cell proliferation in vitro. Kidney Int. 1999;56:2085–2095. doi: 10.1046/j.1523-1755.1999.00799.x. [DOI] [PubMed] [Google Scholar]
- Janssen JW, Imoto I, Inoue J, Shimada Y, Ueda M, Imamura M, Bartram CR, Inazawa J. MYEOV, a gene at 11q13, is coamplified with CCND1, but epigenetically inactivated in a subset of esophageal squamous cell carcinomas. J Hum Genet. 2002;47:460–464. doi: 10.1007/s100380200065. [DOI] [PubMed] [Google Scholar]
- Jeffries S, Robbins DJ, Capobianco AJ. Characterization of a high-molecular-weight Notch complex in the nucleus of Notch(ic)-transformed RKE cells and in a human T-cell leukemia cell line. Mol Cell Biol. 2002;22:3927–3941. doi: 10.1128/MCB.22.11.3927-3941.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jirmanova L, Afanassieff M, Gobert-Gosse S, Markossian S, Savatier P. Differential contributions of ERK and PI3-kinase to the regulation of cyclin D1 expression and to the control of the G1/S transition in mouse embryonic stem cells. Oncogene. 2002;21:5515–5528. doi: 10.1038/sj.onc.1205728. [DOI] [PubMed] [Google Scholar]
- Jones SM, Kazlauskas A. Growth factor-dependent signaling and cell cycle progression. FEBS Lett. 2001;490:110–116. doi: 10.1016/S0014-5793(01)02113-5. [DOI] [PubMed] [Google Scholar]
- Joyce D, Bouzahzah B, Fu M, Albanese C, D’Amico M, Steer J, Klein JU, Lee RJ, Segall JE, Westwick JK, et al. Integration of Rac-dependent regulation of cyclin D1 transcription through a nuclear factor-kappaB-dependent pathway. J Biol Chem. 1999;274:25245–25249. doi: 10.1074/jbc.274.36.25245. [DOI] [PubMed] [Google Scholar]
- Jung C, Kim RS, Lee SJ, Wang C, Jeng MH. HOXB13 homeodomain protein suppresses the growth of prostate cancer cells by the negative regulation of T-cell factor 4. Cancer Res. 2004;64:3046–3051. doi: 10.1158/0008-5472.CAN-03-2614. [DOI] [PubMed] [Google Scholar]
- Kabotyanski EB, Rosen JM. Signal transduction pathways regulated by prolactin and Src result in different conformations of activated Stat5b. J Biol Chem. 2003;278:17218–17227. doi: 10.1074/jbc.M301578200. [DOI] [PubMed] [Google Scholar]
- Kawanaka H, Tomikawa M, Baatar D, Jones MK, Pai R, Szabo IL, Sugimachi K, Sarfeh IJ, Tarnawski AS. Despite activation of EGF-receptor-ERK signaling pathway, epithelial proliferation is impaired in portal hypertensive gastric mucosa: relevance of MKP-1, c-fos, c-myc, and cyclin D1 expression. Life Sci. 2001;69:3019–3033. doi: 10.1016/S0024-3205(01)01409-6. [DOI] [PubMed] [Google Scholar]
- Kawauchi S, Liu XP, Kawasaki K, Hirakawa T, Amada S, Furuya T, Oga A, Sasaki K. Significance of beta-catenin and pRB pathway components in malignant ovarian germ cell tumours: INK4A promoter CpG island methylation is associated with cell proliferation. J Pathol. 2004;204:268–276. doi: 10.1002/path.1629. [DOI] [PubMed] [Google Scholar]
- Kijima T, Niwa H, Steinman RA, Drenning SD, Gooding WE, Wentzel AL, Xi S, Grandis JR. STAT3 activation abrogates growth factor dependence and contributes to head and neck squamous cell carcinoma tumor growth in vivo. Cell Growth Differ. 2002;13:355–362. [PubMed] [Google Scholar]
- Kikuchi A. Regulation of beta-catenin signaling in the Wnt pathway. Biochem Biophys Res Commun. 2000;268:243–248. doi: 10.1006/bbrc.1999.1860. [DOI] [PubMed] [Google Scholar]
- Kim AL, Gautier J, Bickers DR, Athar M. Reduced cyclin D1 ubiquitination in UVB-induced murine squamous cell carcinomas. Biochem Biophys Res Commun. 2002;298:377–382. doi: 10.1016/S0006-291X(02)02435-X. [DOI] [PubMed] [Google Scholar]
- Kintscher U, Bruemmer D, Blaschke F, Unger T, Law RE. p38 MAP kinase negatively regulates angiotensin II-mediated effects on cell cycle molecules in human coronary smooth muscle cells. Biochem Biophys Res Commun. 2003;305:552–556. doi: 10.1016/S0006-291X(03)00802-7. [DOI] [PubMed] [Google Scholar]
- Kitazawa S, Kitazawa R, Maeda S. Transcriptional regulation of rat cyclin D1 gene by CpG methylation status in promoter region. J Biol Chem. 1999;274:28787–28793. doi: 10.1074/jbc.274.40.28787. [DOI] [PubMed] [Google Scholar]
- Klein A, Guhl E, Tzeng YJ, Fuhrhop J, Levrero M, Graessmann M, Graessmann A. HBX causes cyclin D1 overexpression and development of breast cancer in transgenic animals that are heterozygous for p53. Oncogene. 2003;22:2910–2919. doi: 10.1038/sj.onc.1206539. [DOI] [PubMed] [Google Scholar]
- Knezevic D, Brash DE. Role of E2F1 in apoptosis: a case study in feedback loops. Cell Cycle. 2004;3:729–732. doi: 10.4161/cc.3.6.907. [DOI] [PubMed] [Google Scholar]
- Kozar K, Ciemerych MA, Rebel VI, Shigematsu H, Zagozdzon A, Sicinska E, Geng Y, Yu Q, Bhattacharya S, Bronson RT, et al. Mouse development and cell proliferation in the absence of D-cyclins. Cell. 2004;118:477–491. doi: 10.1016/j.cell.2004.07.025. [DOI] [PubMed] [Google Scholar]
- Krieger S, Grunau C, Sabbah M, Sola B. Cyclin D1 gene activation in human myeloma cells is independent of DNA hypomethylation or histone hyperacetylation. Exp Hematol. 2005;33:652–659. doi: 10.1016/j.exphem.2005.03.011. [DOI] [PubMed] [Google Scholar]
- Lamb J, Ramaswamy S, Ford HL, Contreras B, Martinez RV, Kittrell FS, Zahnow CA, Patterson N, Golub TR, Ewen ME. A mechanism of cyclin D1 action encoded in the patterns of gene expression in human cancer. Cell. 2003;114:323–334. doi: 10.1016/S0092-8674(03)00570-1. [DOI] [PubMed] [Google Scholar]
- Lange AW, Keiser AR, Wells JM, Zorn AM, Whitsett JA. Sox17 promotes cell cycle progression and inhibits TGF-beta/Smad3 signaling to initiate progenitor cell behavior in the respiratory epithelium. PLoS ONE. 2009;4:e5711. doi: 10.1371/journal.pone.0005711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavoie JN, L’Allemain G, Brunet A, Muller R, Pouyssegur J. Cyclin D1 expression is regulated positively by the p42/p44MAPK and negatively by the p38/HOGMAPK pathway. J Biol Chem. 1996;271:20608–20616. doi: 10.1074/jbc.271.34.20608. [DOI] [PubMed] [Google Scholar]
- Lee RJ, Albanese C, Stenger RJ, Watanabe G, Inghirami G, Haines GK, 3rd, Webster M, Muller WJ, Brugge JS, Davis RJ, Pestell RG. pp 60(v-src) induction of cyclin D1 requires collaborative interactions between the extracellular signal-regulated kinase, p38, and Jun kinase pathways. A role for cAMP response element-binding protein and activating transcription factor-2 in pp60(v-src) signaling in breast cancer cells. J Biol Chem. 1999;274:7341–7350. doi: 10.1074/jbc.274.11.7341. [DOI] [PubMed] [Google Scholar]
- Lee RJ, Albanese C, Fu M, D’Amico M, Lin B, Watanabe G, Haines GK, 3rd, Siegel PM, Hung MC, Yarden Y, et al. Cyclin D1 is required for transformation by activated Neu and is induced through an E2F-dependent signaling pathway. Mol Cell Biol. 2000;20:672–683. doi: 10.1128/MCB.20.2.672-683.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lepourcelet M, Chen YN, France DS, Wang H, Crews P, Petersen F, Bruseo C, Wood AW, Shivdasani RA. Small-molecule antagonists of the oncogenic Tcf/beta-catenin protein complex. Cancer Cell. 2004;5:91–102. doi: 10.1016/S1535-6108(03)00334-9. [DOI] [PubMed] [Google Scholar]
- Leshchenko VV, Kuo PY, Shaknovich R, Yang DT, Gellen T, Petrich A, Yu Y, Remache Y, Weniger MA, Rafiq S, et al. Genomewide DNA methylation analysis reveals novel targets for drug development in mantle cell lymphoma. Blood. 2010;116(7):1025–1034. doi: 10.1182/blood-2009-12-257485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leslie K, Lang C, Devgan G, Azare J, Berishaj M, Gerald W, Kim YB, Paz K, Darnell JE, Albanese C, et al. Cyclin D1 is transcriptionally regulated by and required for transformation by activated signal transducer and activator of transcription 3. Cancer Res. 2006;66:2544–2552. doi: 10.1158/0008-5472.CAN-05-2203. [DOI] [PubMed] [Google Scholar]
- Lin HM, Pestell RG, Raz A, Kim HR. Galectin-3 enhances cyclin D(1) promoter activity through SP1 and a cAMP-responsive element in human breast epithelial cells. Oncogene. 2002;21:8001–8010. doi: 10.1038/sj.onc.1205820. [DOI] [PubMed] [Google Scholar]
- Liu H, Wang J, Epner EM. Cyclin D1 activation in B-cell malignancy: association with changes in histone acetylation, DNA methylation, and RNA polymerase II binding to both promoter and distal sequences. Blood. 2004;104:2505–2513. doi: 10.1182/blood-2004-02-0483. [DOI] [PubMed] [Google Scholar]
- Liu H, Huang J, Wang J, Jiang S, Bailey AS, Goldman DC, Welcker M, Bedell V, Slovak ML, Clurman B, et al. Transvection mediated by the translocated cyclin D1 locus in mantle cell lymphoma. J Exp Med. 2008;205:1843–1858. doi: 10.1084/jem.20072102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu T, Niu Y, Feng Y, Niu R, Yu Y, Lv A, Yang Y. Methylation of CpG islands of p16(INK4a) and cyclinD1 overexpression associated with progression of intraductal proliferative lesions of the breast. Hum Pathol. 2008;39:1637–1646. doi: 10.1016/j.humpath.2008.04.001. [DOI] [PubMed] [Google Scholar]
- Magne S, Caron S, Charon M, Rouyez MC, Dusanter-Fourt I. STAT5 and Oct-1 form a stable complex that modulates cyclin D1 expression. Mol Cell Biol. 2003;23:8934–8945. doi: 10.1128/MCB.23.24.8934-8945.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malumbres M, Sotillo R, Santamaria D, Galan J, Cerezo A, Ortega S, Dubus P, Barbacid M. Mammalian cells cycle without the D-type cyclin-dependent kinases Cdk4 and Cdk6. Cell. 2004;118:493–504. doi: 10.1016/j.cell.2004.08.002. [DOI] [PubMed] [Google Scholar]
- Marampon F, Casimiro MC, Fu M, Powell MJ, Popov VM, Lindsay J, Zani BM, Ciccarelli C, Watanabe G, Lee RJ, Pestell RG. Nerve Growth factor regulation of cyclin D1 in PC12 cells through a p21RAS extracellular signal-regulated kinase pathway requires cooperative interactions between Sp1 and nuclear factor-kappaB. Mol Biol Cell. 2008;19:2566–2578. doi: 10.1091/mbc.E06-12-1110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marino M, Acconcia F, Trentalance A. Biphasic estradiol-induced AKT phosphorylation is modulated by PTEN via MAP kinase in HepG2 cells. Mol Biol Cell. 2003;14:2583–2591. doi: 10.1091/mbc.E02-09-0621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin V, Valencia A, Agirre X, Cervera J, San Jose-Eneriz E, Vilas-Zornoza A, Rodriguez-Otero P, Sanz MA, Herrera C, Torres A, et al. Epigenetic regulation of the non-canonical Wnt pathway in acute myeloid leukemia. Cancer Sci. 2009;101:425–432. doi: 10.1111/j.1349-7006.2009.01413.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masuda M, Suzui M, Weinstein IB. Effects of epigallocatechin-3-gallate on growth, epidermal growth factor receptor signaling pathways, gene expression, and chemosensitivity in human head and neck squamous cell carcinoma cell lines. Clin Cancer Res. 2001;7:4220–4229. [PubMed] [Google Scholar]
- Masuda M, Suzui M, Yasumatu R, Nakashima T, Kuratomi Y, Azuma K, Tomita K, Komiyama S, Weinstein IB. Constitutive activation of signal transducers and activators of transcription 3 correlates with cyclin D1 overexpression and may provide a novel prognostic marker in head and neck squamous cell carcinoma. Cancer Res. 2002;62:3351–3355. [PubMed] [Google Scholar]
- Matsuda Y. Molecular mechanism underlying the functional loss of cyclindependent kinase inhibitors p16 and p27 in hepatocellular carcinoma. World J Gastroenterol. 2008;14:1734–1740. doi: 10.3748/wjg.14.1734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsumura I, Kitamura T, Wakao H, Tanaka H, Hashimoto K, Albanese C, Downward J, Pestell RG, Kanakura Y. Transcriptional regulation of the cyclin D1 promoter by STAT5: its involvement in cytokine-dependent growth of hematopoietic cells. EMBO J. 1999;18:1367–1377. doi: 10.1093/emboj/18.5.1367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMahon C, Suthiphongchai T, DiRenzo J, Ewen ME. P/CAF associates with cyclin D1 and potentiates its activation of the estrogen receptor. Proc Natl Acad Sci USA. 1999;96:5382–5387. doi: 10.1073/pnas.96.10.5382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mechta F, Lallemand D, Pfarr CM, Yaniv M. Transformation by ras modifies AP1 composition and activity. Oncogene. 1997;14:837–847. doi: 10.1038/sj.onc.1200900. [DOI] [PubMed] [Google Scholar]
- Morin PJ. beta-catenin signaling and cancer. Bioessays. 1999;21:1021–1030. doi: 10.1002/(SICI)1521-1878(199912)22:1<1021::AID-BIES6>3.0.CO;2-P. [DOI] [PubMed] [Google Scholar]
- Motokura T, Arnold A. PRAD1/cyclin D1 proto-oncogene: genomic organization, 5′ DNA sequence, and sequence of a tumor-specific rearrangement breakpoint. Genes Chromosom Cancer. 1993;7:89–95. doi: 10.1002/gcc.2870070205. [DOI] [PubMed] [Google Scholar]
- Muller H, Moroni MC, Vigo E, Petersen BO, Bartek J, Helin K. Induction of S-phase entry by E2F transcription factors depends on their nuclear localization. Mol Cell Biol. 1997;17:5508–5520. doi: 10.1128/mcb.17.9.5508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller-Tidow C, Steffen B, Cauvet T, Tickenbrock L, Ji P, Diederichs S, Sargin B, Kohler G, Stelljes M, Puccetti E, et al. Translocation products in acute myeloid leukemia activate the Wnt signaling pathway in hematopoietic cells. Mol Cell Biol. 2004;24:2890–2904. doi: 10.1128/MCB.24.7.2890-2904.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulloy R, Salinas S, Philips A, Hipskind RA. Activation of cyclin D1 expression by the ERK5 cascade. Oncogene. 2003;22:5387–5398. doi: 10.1038/sj.onc.1206839. [DOI] [PubMed] [Google Scholar]
- Musa NL, Ramakrishnan M, Li J, Kartha S, Liu P, Pestell RG, Hershenson MB. Forskolin inhibits cyclin D1 expression in cultured airway smooth-muscle cells. Am J Respir Cell Mol Biol. 1999;20:352–358. doi: 10.1165/ajrcmb.20.2.3160. [DOI] [PubMed] [Google Scholar]
- Nagata D, Suzuki E, Nishimatsu H, Satonaka H, Goto A, Omata M, Hirata Y. Transcriptional activation of the cyclin D1 gene is mediated by multiple cis-elements, including SP1 sites and a cAMP-responsive element in vascular endothelial cells. J Biol Chem. 2001;276:662–669. doi: 10.1074/jbc.M005522200. [DOI] [PubMed] [Google Scholar]
- Nheu T, He H, Hirokawa Y, Walker F, Wood J, Maruta H. PAK is essential for RAS-induced upregulation of cyclin D1 during the G1 to S transition. Cell Cycle. 2004;3:71–74. doi: 10.4161/cc.3.1.593. [DOI] [PubMed] [Google Scholar]
- Oktay M, Wary KK, Dans M, Birge RB, Giancotti FG. Integrin-mediated activation of focal adhesion kinase is required for signaling to Jun NH2-terminal kinase and progression through the G1 phase of the cell cycle. J Cell Biol. 1999;145:1461–1469. doi: 10.1083/jcb.145.7.1461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Page K, Li J, Hershenson MB. Platelet-derived growth factor stimulation of mitogen-activated protein kinases and cyclin D1 promoter activity in cultured airway smooth-muscle cells. Role of Ras. Am J Respir Cell Mol Biol. 1999;20:1294–1302. doi: 10.1165/ajrcmb.20.6.3597. [DOI] [PubMed] [Google Scholar]
- Page K, Li J, Hodge JA, Liu PT, Vanden Hoek TL, Becker LB, Pestell RG, Rosner MR, Hershenson MB. Characterization of a Rac1 signaling pathway to cyclin D(1) expression in airway smooth muscle cells. J Biol Chem. 1999;274:22065–22071. doi: 10.1074/jbc.274.31.22065. [DOI] [PubMed] [Google Scholar]
- Page K, Li J, Wang Y, Kartha S, Pestell RG, Hershenson MB. Regulation of cyclin D(1) expression and DNA synthesis by phosphatidylinositol 3-kinase in airway smooth muscle cells. Am J Respir Cell Mol Biol. 2000;23:436–443. doi: 10.1165/ajrcmb.23.4.3953. [DOI] [PubMed] [Google Scholar]
- Page K, Li J, Hershenson MB. p38 MAP kinase negatively regulates cyclin D1 expression in airway smooth muscle cells. Am J Physiol Lung Cell Mol Physiol. 2001;280:L955–L964. doi: 10.1152/ajplung.2001.280.5.L955. [DOI] [PubMed] [Google Scholar]
- Page K, Li J, Corbit KC, Rumilla KM, Soh JW, Weinstein IB, Albanese C, Pestell RG, Rosner MR, Hershenson MB. Regulation of airway smooth muscle cyclin D1 transcription by protein kinase C-delta. Am J Respir Cell Mol Biol. 2002;27:204–213. doi: 10.1165/ajrcmb.27.2.20010016oc. [DOI] [PubMed] [Google Scholar]
- Paramio JM, Navarro M, Segrelles C, Gomez-Casero E, Jorcano JL. PTEN tumour suppressor is linked to the cell cycle control through the retinoblastoma protein. Oncogene. 1999;18:7462–7468. doi: 10.1038/sj.onc.1203151. [DOI] [PubMed] [Google Scholar]
- Persad S, Attwell S, Gray V, Mawji N, Deng JT, Leung D, Yan J, Sanghera J, Walsh MP, Dedhar S. Regulation of protein kinase B/Akt-serine 473 phosphorylation by integrin-linked kinase: critical roles for kinase activity and amino acids arginine 211 and serine 343. J Biol Chem. 2001;276:27462–27469. doi: 10.1074/jbc.M102940200. [DOI] [PubMed] [Google Scholar]
- Persad S, Troussard AA, McPhee TR, Mulholland DJ, Dedhar S. Tumor suppressor PTEN inhibits nuclear accumulation of beta-catenin and T cell/lymphoid enhancer factor 1-mediated transcriptional activation. J Cell Biol. 2001;153:1161–1174. doi: 10.1083/jcb.153.6.1161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pestell RG, Albanese C, Reutens AT, Segall JE, Lee RJ, Arnold A. The cyclins and cyclin-dependent kinase inhibitors in hormonal regulation of proliferation and differentiation. Endocr Rev. 1999;20:501–534. doi: 10.1210/er.20.4.501. [DOI] [PubMed] [Google Scholar]
- Philipp A, Schneider A, Vasrik I, Finke K, Xiong Y, Beach D, Alitalo K, Eilers M. Repression of cyclin D1: a novel function of MYC. Mol Cell Biol. 1994;14:4032–4043. doi: 10.1128/mcb.14.6.4032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pierce AM, Fisher SM, Conti CJ, Johnson DG. Deregulated expression of E2F1 induces hyperplasia and cooperates with ras in skin tumor development. Oncogene. 1998;16:1267–1276. doi: 10.1038/sj.onc.1201666. [DOI] [PubMed] [Google Scholar]
- Pierce AM, Schneider-Broussard R, Philhower JL, Johnson DG. Differential activities of E2F family members: unique functions in regulating transcription. Mol Carcinog. 1998;22:190–198. doi: 10.1002/(SICI)1098-2744(199807)22:3<190::AID-MC7>3.0.CO;2-P. [DOI] [PubMed] [Google Scholar]
- Pradeep A, Sharma C, Sathyanarayana P, Albanese C, Fleming JV, Wang TC, Wolfe MM, Baker KM, Pestell RG, Rana B. Gastrin-mediated activation of cyclin D1 transcription involves beta-catenin and CREB pathways in gastric cancer cells. Oncogene. 2004;23:3689–3699. doi: 10.1038/sj.onc.1207454. [DOI] [PubMed] [Google Scholar]
- Pruitt K, Pruitt WM, Bilter GK, Westwick JK, Der CJ. Raf-independent deregulation of p38 and JNK mitogen-activated protein kinases are critical for Ras transformation. J Biol Chem. 2002;277:31808–31817. doi: 10.1074/jbc.M203964200. [DOI] [PubMed] [Google Scholar]
- Qin L, Li X, Ko JK, Partridge NC. Parathyroid hormone uses multiple mechanisms to arrest the cell cycle progression of osteoblastic cells from G1 to S phase. J Biol Chem. 2005;280:3104–3111. doi: 10.1074/jbc.M409846200. [DOI] [PubMed] [Google Scholar]
- Quintanilla-Martinez L, Kremer M, Specht K, Calzada-Wack J, Nathrath M, Schaich R, Hofler H, Fend F. Analysis of signal transducer and activator of transcription 3 (Stat 3) pathway in multiple myeloma: Stat 3 activation and cyclin D1 dysregulation are mutually exclusive events. Am J Pathol. 2003;162:1449–1461. doi: 10.1016/S0002-9440(10)64278-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramakrishnan M, Musa NL, Li J, Liu PT, Pestell RG, Hershenson MB. Catalytic activation of extracellular signal-regulated kinases induces cyclin D1 expression in primary tracheal myocytes. Am J Respir Cell Mol Biol. 1998;18:736–740. doi: 10.1165/ajrcmb.18.6.3152. [DOI] [PubMed] [Google Scholar]
- Recio JA, Merlino G. Hepatocyte growth factor/scatter factor activates proliferation in melanoma cells through p38 MAPK, ATF-2 and cyclin D1. Oncogene. 2002;21:1000–1008. doi: 10.1038/sj.onc.1205150. [DOI] [PubMed] [Google Scholar]
- Rocha S, Martin AM, Meek DW, Perkins ND. p53 represses cyclin D1 transcription through down regulation of Bcl-3 and inducing increased association of the p52 NF-kappaB subunit with histone deacetylase 1. Mol Cell Biol. 2003;23:4713–4727. doi: 10.1128/MCB.23.13.4713-4727.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roche KC, Wiechens N, Owen-Hughes T, Perkins ND. The FHA domain protein SNIP1 is a regulator of the cell cycle and cyclin D1 expression. Oncogene. 2004;23:8185–8195. doi: 10.1038/sj.onc.1208025. [DOI] [PubMed] [Google Scholar]
- Ronchini C, Capobianco AJ. Induction of cyclin D1 transcription and CDK2 activity by Notch(ic): implication for cell cycle disruption in transformation by Notch(ic) Mol Cell Biol. 2001;21:5925–5934. doi: 10.1128/MCB.21.17.5925-5934.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roovers K, Assoian RK. Effects of rho kinase and actin stress fibers on sustained extracellular signal-regulated kinase activity and activation of G(1) phase cyclin-dependent kinases. Mol Cell Biol. 2003;23:4283–4294. doi: 10.1128/MCB.23.12.4283-4294.2003. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Roovers K, Klein EA, Castagnino P, Assoian RK. Nuclear translocation of LIM kinase mediates Rho-Rho kinase regulation of cyclin D1 expression. Dev Cell. 2003;5:273–284. doi: 10.1016/S1534-5807(03)00206-5. [DOI] [PubMed] [Google Scholar]
- Ryser S, Massiha A, Piuz I, Schlegel W. Stimulated initiation of mitogen-activated protein kinase phosphatase-1 (MKP-1) gene transcription involves the synergistic action of multiple cis-acting elements in the proximal promoter. Biochem J. 2004;378:473–484. doi: 10.1042/BJ20031022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabbah M, Courilleau D, Mester J, Redeuilh G. Estrogen induction of the cyclin D1 promoter: involvement of a cAMP response-like element. Proc Natl Acad Sci USA. 1999;96:11217–11222. doi: 10.1073/pnas.96.20.11217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito T, Oda Y, Tanaka K, Matsuda S, Tamiya S, Iwamoto Y, Tsuneyoshi M. beta-catenin nuclear expression correlates with cyclin D1 overexpression in sporadic desmoid tumours. J Pathol. 2001;195:222–228. doi: 10.1002/path.942. [DOI] [PubMed] [Google Scholar]
- Sampson EM, Haque ZK, Ku MC, Tevosian SG, Albanese C, Pestell RG, Paulson KE, Yee AS. Negative regulation of the Wnt-beta-catenin pathway by the transcriptional repressor HBP1. EMBO J. 2001;20:4500–4511. doi: 10.1093/emboj/20.16.4500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider G, Oswald F, Wahl C, Greten FR, Adler G, Schmid RM. Cyclosporine inhibits growth through the activating transcription factor/cAMP-responsive element-binding protein binding site in the cyclin D1 promoter. J Biol Chem. 2002;277:43599–43607. doi: 10.1074/jbc.M204787200. [DOI] [PubMed] [Google Scholar]
- Shah S, Pishvaian MJ, Easwaran V, Brown PH, Byers SW. The role of cadherin, beta-catenin, and AP-1 in retinoid-regulated carcinoma cell differentiation and proliferation. J Biol Chem. 2002;277:25313–25322. doi: 10.1074/jbc.M203158200. [DOI] [PubMed] [Google Scholar]
- Sharma C, Pradeep A, Pestell RG, Rana B. Peroxisome proliferator-activated receptor gamma activation modulates cyclin D1 transcription via beta-catenin-independent and cAMP-response element-binding protein-dependent pathways in mouse hepatocytes. J Biol Chem. 2004;279:16927–16938. doi: 10.1074/jbc.M309045200. [DOI] [PubMed] [Google Scholar]
- Shaulian E, Karin M. AP-1 in cell proliferation and survival. Oncogene. 2001;20:2390–2400. doi: 10.1038/sj.onc.1204383. [DOI] [PubMed] [Google Scholar]
- Shi H, Guo J, Duff DJ, Rahmatpanah F, Chitima-Matsiga R, Al-Kuhlani M, Taylor KH, Sjahputera O, Andreski M, Wooldridge JE, Caldwell CW. Discovery of novel epigenetic markers in non-Hodgkin’s lymphoma. Carcinogenesis. 2007;28:60–70. doi: 10.1093/carcin/bgl092. [DOI] [PubMed] [Google Scholar]
- Shirato H, Ogawa S, Nakajima K, Inagawa M, Kojima M, Tachibana M, Shinkai Y, Takeuchi T. A jumonji (Jarid2) protein complex represses cyclin D1 expression by methylation of histone H3-K9. J Biol Chem. 2009;284:733–739. doi: 10.1074/jbc.M804994200. [DOI] [PubMed] [Google Scholar]
- Shtutman M, Zhurinsky J, Simcha I, Albanese C, D’Amico M, Pestell R, Ben-Ze’ev A. The cyclin D1 gene is a target of the beta-catenin/LEF-1 pathway. Proc Natl Acad Sci USA. 1999;96:5522–5527. doi: 10.1073/pnas.96.10.5522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soh JW, Weinstein IB. Roles of specific isoforms of protein kinase C in the transcriptional control of cyclin D1 and related genes. J Biol Chem. 2003;278:34709–34716. doi: 10.1074/jbc.M302016200. [DOI] [PubMed] [Google Scholar]
- Stacey DW. Cyclin D1 serves as a cell cycle regulatory switch in actively proliferating cells. Curr Opin Cell Biol. 2003;15:158–163. doi: 10.1016/S0955-0674(03)00008-5. [DOI] [PubMed] [Google Scholar]
- Stahl M, Ge C, Shi S, Pestell RG, Stanley P. Notch1-induced transformation of RKE-1 cells requires up-regulation of cyclin D1. Cancer Res. 2006;66:7562–7570. doi: 10.1158/0008-5472.CAN-06-0974. [DOI] [PubMed] [Google Scholar]
- Takahira T, Oda Y, Tamiya S, Yamamoto H, Kawaguchi K, Kobayashi C, Iwamoto Y, Tsuneyoshi M. Alterations of the p16INK4a/p14ARF pathway in clear cell sarcoma. Cancer Sci. 2004;95:651–655. doi: 10.1111/j.1349-7006.2004.tb03324.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talarmin H, Rescan C, Cariou S, Glaise D, Zanninelli G, Bilodeau M, Loyer P, Guguen-Guillouzo C, Baffet G. The mitogen-activated protein kinase kinase/extracellular signal-regulated kinase cascade activation is a key signalling pathway involved in the regulation of G(1) phase progression in proliferating hepatocytes. Mol Cell Biol. 1999;19:6003–6011. doi: 10.1128/mcb.19.9.6003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tashiro E, Maruki H, Minato Y, Doki Y, Weinstein IB, Imoto M. Overexpression of cyclin D1 contributes to malignancy by up-regulation of fibroblast growth factor receptor 1 via the pRB/E2F pathway. Cancer Res. 2003;63:424–431. [PubMed] [Google Scholar]
- Tetsu O, McCormick F. Beta-catenin regulates expression of cyclin D1 in colon carcinoma cells. Nature. 1999;398:422–426. doi: 10.1038/18884. [DOI] [PubMed] [Google Scholar]
- Todd DE, Densham RM, Molton SA, Balmanno K, Newson C, Weston CR, Garner AP, Scott L, Cook SJ. ERK1/2 and p38 cooperate to induce a p21(CIP1)-dependent G(1) cell cycle arrest. Oncogene. 2004;23:3284–3295. doi: 10.1038/sj.onc.1207467. [DOI] [PubMed] [Google Scholar]
- Treinies I, Paterson HF, Hooper S, Wilson R, Marshall CJ. Activated MEK stimulates expression of AP-1 components independently of phosphatidylinositol 3-kinase (PI3-kinase) but requires a PI3-kinase signal To stimulate DNA synthesis. Mol Cell Biol. 1999;19:321–329. doi: 10.1128/mcb.19.1.321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Troussard AA, Mawji NM, Ong C, Mui A, St-Arnaud R, Dedhar S. Conditional knock-out of integrin-linked kinase demonstrates an essential role in protein kinase B/Akt activation. J Biol Chem. 2003;278:22374–22378. doi: 10.1074/jbc.M303083200. [DOI] [PubMed] [Google Scholar]
- Utsunomiya T, Doki Y, Takemoto H, Shiozaki H, Yano M, Sekimoto M, Tamura S, Yasuda T, Fujiwara Y, Monden M. Correlation of beta-catenin and cyclin D1 expression in colon cancers. Oncology. 2001;61:226–233. doi: 10.1159/000055379. [DOI] [PubMed] [Google Scholar]
- Vonlanthen S, Heighway J, Tschan MP, Borner MM, Altermatt HJ, Kappeler A, Tobler A, Fey MF, Thatcher N, Yarbrough WG, Betticher DC. Expression of p16INK4a/p16alpha and p19ARF/p16beta is frequently altered in non-small cell lung cancer and correlates with p53 overexpression. Oncogene. 1998;17:2779–2785. doi: 10.1038/sj.onc.1202501. [DOI] [PubMed] [Google Scholar]
- Wang Z, Zhang B, Wang M, Carr BI. Persistent ERK phosphorylation negatively regulates cAMP response element-binding protein (CREB) activity via recruitment of CREB-binding protein to pp 90RSK. J Biol Chem. 2003;278:11138–11144. doi: 10.1074/jbc.M209108200. [DOI] [PubMed] [Google Scholar]
- Watanabe G, Howe A, Lee RJ, Albanese C, Shu IW, Karnezis AN, Zon L, Kyriakis J, Rundell K, Pestell RG. Induction of cyclin D1 by simian virus 40 small tumor antigen. Proc Natl Acad Sci USA. 1996;93:12861–12866. doi: 10.1073/pnas.93.23.12861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe G, Lee RJ, Albanese C, Rainey WE, Batlle D, Pestell RG. Angiotensin II activation of cyclin D1-dependent kinase activity. J Biol Chem. 1996;271:22570–22577. doi: 10.1074/jbc.271.37.22570. [DOI] [PubMed] [Google Scholar]
- Watanabe G, Albanese C, Lee RJ, Reutens A, Vairo G, Henglein B, Pestell RG. Inhibition of cyclin D1 kinase activity is associated with E2F-mediated inhibition of cyclin D1 promoter activity through E2F and Sp1. Mol Cell Biol. 1998;18:3212–3222. doi: 10.1128/mcb.18.6.3212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber JD, Hu W, Jefcoat SC, Jr, Raben DM, Baldassare JJ. Ras-stimulated extracellular signal-related kinase 1 and RhoA activities coordinate platelet-derived growth factor-induced G1 progression through the independent regulation of cyclin D1 and p27. J Biol Chem. 1997;272:32966–32971. doi: 10.1074/jbc.272.52.32966. [DOI] [PubMed] [Google Scholar]
- Weber JD, Raben DM, Phillips PJ, Baldassare JJ. Sustained activation of extracellular-signal-regulated kinase 1 (ERK1) is required for the continued expression of cyclin D1 in G1 phase. Biochem J. 1997;326(Pt 1):61–68. doi: 10.1042/bj3260061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welch HC, Coadwell WJ, Stephens LR, Hawkins PT. Phosphoinositide 3-kinase-dependent activation of Rac. FEBS Lett. 2003;546:93–97. doi: 10.1016/S0014-5793(03)00454-X. [DOI] [PubMed] [Google Scholar]
- Welsh CF, Roovers K, Villanueva J, Liu Y, Schwartz MA, Assoian RK. Timing of cyclin D1 expression within G1 phase is controlled by Rho. Nat Cell Biol. 2001;3:950–957. doi: 10.1038/ncb1101-950. [DOI] [PubMed] [Google Scholar]
- Weng LP, Smith WM, Brown JL, Eng C. PTEN inhibits insulin-stimulated MEK/MAPK activation and cell growth by blocking IRS-1 phosphorylation and IRS-1/Grb-2/Sos complex formation in a breast cancer model. Hum Mol Genet. 2001;10:605–616. doi: 10.1093/hmg/10.6.605. [DOI] [PubMed] [Google Scholar]
- Westerheide SD, Mayo MW, Anest V, Hanson JL, Baldwin AS., Jr The putative oncoprotein Bcl-3 induces cyclin D1 to stimulate G(1) transition. Mol Cell Biol. 2001;21:8428–8436. doi: 10.1128/MCB.21.24.8428-8436.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westwick JK, Lee RJ, Lambert QT, Symons M, Pestell RG, Der CJ, Whitehead IP. Transforming potential of Dbl family proteins correlates with transcription from the cyclin D1 promoter but not with activation of Jun NH2-terminal kinase, p38/Mpk2, serum response factor, or c-Jun. J Biol Chem. 1998;273:16739–16747. doi: 10.1074/jbc.273.27.16739. [DOI] [PubMed] [Google Scholar]
- Whitehead IP, Lambert QT, Glaven JA, Abe K, Rossman KL, Mahon GM, Trzaskos JM, Kay R, Campbell SL, Der CJ. Dependence of Dbl and Dbs transformation on MEK and NF-kappaB activation. Mol Cell Biol. 1999;19:7759–7770. doi: 10.1128/mcb.19.11.7759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wulf GM, Ryo A, Wulf GG, Lee SW, Niu T, Petkova V, Lu KP. Pin1 is overexpressed in breast cancer and cooperates with Ras signaling in increasing the transcriptional activity of c-Jun towards cyclin D1. EMBO J. 2001;20:3459–3472. doi: 10.1093/emboj/20.13.3459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie Z, Zeng X, Waldman T, Glazer RI. Transformation of mammary epithelial cells by 3-phosphoinositide- dependent protein kinase-1 activates beta-catenin and c-Myc, and down-regulates caveolin-1. Cancer Res. 2003;63:5370–5375. [PubMed] [Google Scholar]
- Yan YX, Nakagawa H, Lee MH, Rustgi AK. Transforming growth factor-alpha enhances cyclin D1 transcription through the binding of early growth response protein to a cis-regulatory element in the cyclin D1 promoter. J Biol Chem. 1997;272:33181–33190. doi: 10.1074/jbc.272.52.33181. [DOI] [PubMed] [Google Scholar]
- Yan S, Krebs S, Leister KJ, Wenner CE. Perturbation of EGF-activated MEK1 and PKB signal pathways by TGF-beta1 correlates with perturbation of EGF-induced cyclin D1 and DNA synthesis by TGF-beta1 in C3H 10 T1/2 cells. J Cell Physiol. 2000;185:107–116. doi: 10.1002/1097-4652(200010)185:1<107::AID-JCP10>3.0.CO;2-D. [DOI] [PubMed] [Google Scholar]
- Yang H, Li TW, Ko KS, Xia M, Lu SC. Switch from Mnt-Max to Myc-Max induces p53 and cyclin D1 expression and apoptosis during cholestasis in mouse and human hepatocytes. Hepatology. 2009;49:860–870. doi: 10.1002/hep.22720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang ZK, Davies KP, Allen J, Zhu L, Pestell RG, Zagzag D, Kalpana GV. Cell cycle arrest and repression of cyclin D1 transcription by INI1/hSNF5. Mol Cell Biol. 2002;22:5975–5988. doi: 10.1128/MCB.22.16.5975-5988.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang F, Li C, Halfter H, Liu J. Delineating an oncostatin M-activated STAT3 signaling pathway that coordinates the expression of genes involved in cell cycle regulation and extracellular matrix deposition of MCF-7 cells. Oncogene. 2003;22:894–905. doi: 10.1038/sj.onc.1206158. [DOI] [PubMed] [Google Scholar]
- Zhao J, Pestell R, Guan JL. Transcriptional activation of cyclin D1 promoter by FAK contributes to cell cycle progression. Mol Biol Cell. 2001;12:4066–4077. doi: 10.1091/mbc.12.12.4066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao J, Bian ZC, Yee K, Chen BP, Chien S, Guan JL. Identification of transcription factor KLF8 as a downstream target of focal adhesion kinase in its regulation of cyclin D1 and cell cycle progression. Mol Cell. 2003;11:1503–1515. doi: 10.1016/S1097-2765(03)00179-5. [DOI] [PubMed] [Google Scholar]
- Zhou JX, Niehans GA, Shar A, Rubins JB, Frizelle SP, Kratzke RA. Mechanisms of G1 checkpoint loss in resected early stage non-small cell lung cancer. Lung Cancer. 2001;32:27–38. doi: 10.1016/S0169-5002(00)00210-5. [DOI] [PubMed] [Google Scholar]
- Zou Y, Ewton DZ, Deng X, Mercer SE, Friedman E. Mirk/dyrk1B kinase destabilizes cyclin D1 by phosphorylation at threonine 288. J Biol Chem. 2004;279:27790–27798. doi: 10.1074/jbc.M403042200. [DOI] [PubMed] [Google Scholar]


