Background: Fungal metabolites, zaragozic acids, are potent inhibitors against squalene synthase and S. aureus dehydro-squalene synthase.
Results: Here we reported the zaragozic acid A binding mode for the head-to-head prenyltransferase drug targets.
Conclusion: Zaragozic acid A presents an unexpected binding mode from the cyclopropyl-containing intermediate in both enzymes.
Significance: Our structures provide a structural basis for development of potential drugs for hyperlipidemias and bacterial infection treatments.
Keywords: Drug Design, Staphylococcus aureus, Steroid, Structural Biology, X-ray Crystallography, Fungal Metabolite, Hyperlipidemias, Staphyloxanthin
Abstract
Zaragozic acids (ZAs) belong to a family of fungal metabolites with nanomolar inhibitory activity toward squalene synthase (SQS). The enzyme catalyzes the committed step of sterol synthesis and has attracted attention as a potential target for antilipogenic and antiinfective therapies. Here, we have determined the structure of ZA-A complexed with human SQS. ZA-A binding induces a local conformational change in the substrate binding site, and its C-6 acyl group also extends over to the cofactor binding cavity. In addition, ZA-A effectively inhibits a homologous bacterial enzyme, dehydrosqualene synthase (CrtM), which synthesizes the precursor of staphyloxanthin in Staphylococcus aureus to cope with oxidative stress. Size reduction at Tyr248 in CrtM further increases the ZA-A binding affinity, and it reveals a similar overall inhibitor binding mode to that of human SQS/ZA-A except for the C-6 acyl group. These structures pave the way for further improving selectivity and development of a new generation of anticholesterolemic and antimicrobial inhibitors.
Introduction
Natural compounds isolated from plants, animals, and microbes are a valuable source of lead compounds for disease treatment and medical uses. The quest for such secondary metabolites has been widely practiced in the pharmaceutical industry (1, 2). The fungal metabolite zaragozic acids (ZAs)2 or squalestatins have attracted high profile attention by their potent inhibitory activities on squalene synthase (SQS) in human and other species at nano- to picomolar concentration and possess in vivo activity. Because SQS is highly conserved across various species and represents the first dedicated step succeeding HMG-CoA reductase in sterol biosynthetic pathway, pharmacologists regard SQS inhibitors as promising lead compounds in the development of potential therapeutic agents to treat hyperlipoproteinemia (3–5), and fungal and Trypanosomatid infections (6, 7). The biology, chemistry, and synthetic studies of the structurally complex unprecedentedly ZA family have been reviewed and discussed (7–10). Surprisingly, ZA members have also been identified as ras farnesyl transferase and geranylgeranyl transferases inhibitors, and also reduce dengue viral replication and the PrP-induced neuronal injury (11–14).
Human SQS (EC 2.5.1.21) is an endoplasmic reticulum-bound enzyme that catalyzes the NADPH-dependent condensation of two farnesyl diphosphate (FPP) molecules into squalene, via presqualene diphosphate (PSPP) (15). A similar chemical change is catalyzed by Staphylococcus aureus dehydrosqualene synthase (CrtM) to form dehydrosqualene, the precursor of staphyloxanthin, by a nonreductive rearrangement reaction (Fig. 1A). Blocking reactive oxygen species detoxification by inhibiting staphyloxanthin biosynthesis presents a novel antimicrobial strategy (16, 17). Our previous work has shown that several inhibitors of SQS as cholesterol-lowering drugs decrease S. aureus survival in animal models (18). In a recent study, López and Kolter also found that a ZA member curtails S. aureus staphyloxanthin and biofilm formations (19).
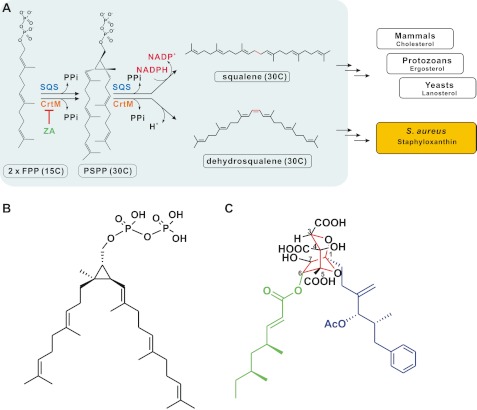
FIGURE 1.
A, reaction mechanism catalyzed by SQS and CrtM. The first reaction step for the two enzymes is similar, producing a cyclopropyl-containing intermediate, PSPP. The second reaction step differs from that of squalene synthesis, but not dehydrosqualene synthesis, proceeding through an NADPH-utilizing reductive rearrangement reaction. SQS and CrtM catalyze the first committed step of sterols and carotenoid biosynthetic pathways. ZA blocks sterols and staphyloxanthin formations. B and C, PSPP (B) and ZA-A (C) share partial structural similarities. The central 2,8-dioxabicyclo[3.2.1]octane core of ZA-A with six contiguous stereocenters is depicted in red. Its two lipophilic groups are highlighted in blue (the C-1 alkyl side chain) and green (the C-6 acyl side chain).
ZA family has a complex fused “bicyclic core,” a highly oxygenated 2,8-dioxabicyclo[3,2,1]octane-4,6,7-trihydroxy-3,4,5-tricarboxylic acid ring, with two variable hydrophobic tails, termed the C-1 alkyl and the C-6 acyl side chains, and displays diverse effects on target enzymes. For example, the C-6 short chain derivatives retain only 2–15% SQS inhibitory activity of ZA-A. However, substitution of the C-1 alkyl group of the ω-phenyl group by a ω-phenoxy group improves the activity further (7, 20, 21). So far, clear three-dimensional quantitative structure activity relationships have not been established.
In this report we provide x-ray crystal structures of the ligand-free human SQS with two flexible regions for ligand binding, ZA-A in complex with human SQS and S. aureus CrtM, and we analyze the binding properties in vitro. These structures provide the detailed, high resolution view of the binding interactions and conformational changes of ZA-A with these two important enzymes. An alternative region in human SQS active site might be specifically responsible for cofactor/inhibitor occupancy. The refined structures not only help to resolve the longstanding ambiguity on the binding mode of ZA-A to SQS, but also increase scientific understanding of the distinct binding features of ZA-A in two similar enzymes. Finally, our work provides the structural basis for further inhibitor optimization and drug development. Possible therapeutic applications include cholesterol-lowering agents, antiprotozoan and antimicrobial therapy.
EXPERIMENTAL PROCEDURES
Protein Expression, Purification, and Crystallization
The human SQS was cloned from a human liver cDNA library. The DNA fragment corresponding to human SQS(31–370) was inserted into a pET28a expression vector (Novagen), yielding pET28a-hSQS(31–370). The final construct encoded a truncated human SQS with an N-terminal His6 tag and a thrombin cleavage site. The protein was expressed and purified by slightly modified procedures of Thompson et al. (22) and Pandit et al. (31). The His6 tag was then removed by using thrombin. Human SQS (31–370) was crystallized by mixing an equal volume of protein solution (15 mg/ml) with precipitating solution (20% PEG2K-MME, 0.01 m NiCl2, 0.1 m Tris, pH 8.5; 1.4 m sodium citrate tribasic dehydrate, 0.1 m Na-HEPES, pH 7.5; 2 m K2HPO4/NaH2HPO4, pH 6.5) at room temperature. A crystal-seeding process improved the crystal size and quality. The wild-type S. aureus CrtM and the mutant Y248A were expressed, purified, and crystallized as described previously (23). The ZA-A complexes were prepared by incubation of the enzyme with ZA-A for 30 min on ice to yield a final enzyme:ZA-A molar ratio of 1:1. Both complex crystals were grown at 25 °C by vapor diffusion in sitting and hanging drops.
Data Collection, Structure Determination, and Refinement
The diffraction data of native human SQS(31–370) and its complex crystals were collected at the National Synchrotron Radiation Center (NSRRC) of Taiwan and beamline BL44XU of the SPring-8 in Japan. All diffraction data were processed and scaled using the HKL2000 package (24). The structures of the ligand-free human SQS(31–370) and its complex with ZA-A were solved by molecular replacement using MolRep (25) in which the human SQS(31–370)-inhibitor complex served as a search model (Protein Data Bank ID code 1EZF). Iterative model building and computational refinement were performed using COOT (26) and REFMAC (27). Manual rebuilding of the models also used the COOT based on the 2Fo− Fc and Fo− Fc electron density maps. RAMPAGE (28) was used to calculate a Ramachandran plot, identify and correct rotamer outliers, and identify potential steric clashes in the models. The figures illustrating the crystal structures and superpositions were prepared by using PyMOL. Data collection and refinement statistics can be found in supplemental Table S1.
CrtM Enzyme inhibition Assay
The activity of CrtM was determined by measuring the release of pyrophosphate (PPi) from the conversion of FPP to dehydrosqualene in the presence of Mg2+ (29, 30). The PiPer Pyrophosphate assay (Invitrogen) was employed as described by the manufacturer in the “Assaying for Enzyme Activity” setting with some modifications. The phosphate release reaction was measured in a 96-well plate using the continuous spectrophotometric method at 565 nm.
Isothermal Titration Calorimetry
Thermodynamic parameters of ZA-A (Sigma) binding were determined using a MicroCal iTC200 (GE Healthcare). All samples were filtered with 0.22-μm cutoff filters (Millipore). The buffer for all enzymes and inhibitor solutions consisted of 20 mm Tris-HCl, pH 7.5, 100 mm sodium chloride, and 2 mm DTT. A 10 μm solution of ZA-A was directly titrated into a solution of 100 μm human SQS(31–370), CrtM, and CrtM mutant in 200-μl aliquots. The experiments were performed at 25 °C. Each experiment was repeated at least three times. Data were then analyzed using the software Origin.
RESULTS
Overall Structure of Apo-human SQS
The active, doubly truncated human SQS(31–370) used in this study has been characterized previously (22). We obtained the structures of the ligand-free human SQS and its complex with ZA-A in four different crystal forms (forms I–IV), and the refinement statistics are shown in supplemental Table S1. Human SQS shows a typical “isoprenoid synthase fold” with a large, hydrophobic active site. The four conserved regions shared among SQS are assumed to be involved in substrate/cofactor recognition and catalysis (Fig. 2A, and supplemental Fig. S1). SQS contains two noncanonical aspartate-rich motifs of DXXED (80DXXED84 and 219DXXED223), which reside on opposite sides of the active site cleft where the donor and acceptor FPP molecules undergo 1′-1 condensation reaction. The central cavity bears two flexible regions, 51SRSF54 and 315KIRKGQAVTLMMD327, which constitute part of the substrate and cofactor binding sites (black box and yellow circle in Fig. 2A). The ligand-free structures in different crystal forms are essentially identical, with root mean square difference of 0.31–0.71 Å. Notably, the crystal form IV structure shows an outward extended flap (αK) for trimeric packing, and the asymmetric unit indeed contains two similar trimers (supplemental Fig. S2, A and B). Analysis of the B-factors for each structure of apo-human SQS in different crystals reveals that the most flexible region is located in the αJαK loop and αK (supplemental Fig. S2C).
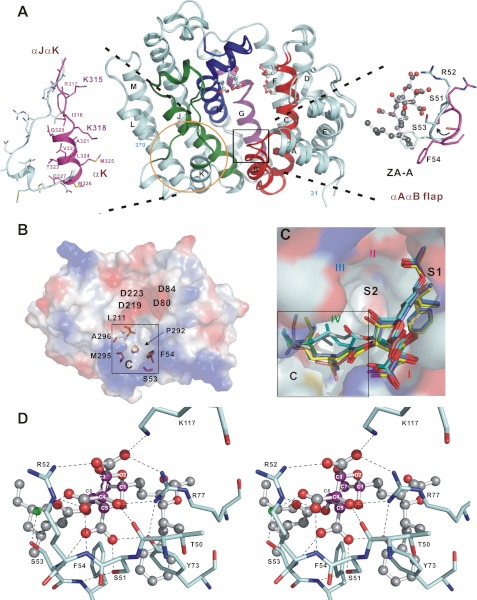
FIGURE 2.
Active site structures of human SQS. A, overall structure of human SQS(31–370). Regions I (red), II (magenta), III (blue), and IV (green), common to all SQS around the active site, are shown. The truncated N and C termini are labeled. The ball-and-stick models show the Asp and Glu residues in the two aspartate-rich motifs located in the active site. The two flexible regions are indicated by a black box and a yellow circle. B, top view of an electrostatic surface model showing a broad active site pocket in ligand-free human SQS. The conserved residues Ser53, Phe54, Leu211, Met295, and Ala296 constitute a hydrophobic pocket, denoted the C site. C, ZA-A binding modes. The ZA-A molecules in different chains of the human SQS–ZA-A complex are shown as stick models. The C-1 alkyl group stays in the S1 site, and the extensive C site is occupied by the C-6 acyl group with a dramatically different binding pattern. The S2 site is empty. D, close-up view of the detailed interactions of ZA-A central core with human SQS. Carbon atoms in the bicyclic core are colored in magenta in ball-and-stick representation. Water molecule is depicted as a small green ball, and some important residues are labeled.
Of the four regions that are highly conserved from yeast to human SQS, regions I, II, and III share sequence homology with S. aureus CrtM. Based on the similarity of structure, catalytic reaction, and bisphosphonate binding mode between human SQS and CrtM (23), we assume that both enzymes adopt comparable FPP and PSPP binding patterns (two FPP molecules in S1 and/or S2 sites, and PSPP in both sites). The regions I, II, and III mainly form the two nonidentical FPP binding sites. However, human SQS further reduces PSPP by NADPH in the second step to form squalene. Although there is no sequence resemblance to the known pyridine dinucleotide binding α/β-motifs of a classic “Rossmann fold,” a series of modifications in region IV prevented SQS from undergoing the second half-reaction, indicating that this region may reasonably constitute a functional NADPH binding site.
From our observation, the residues Ser53, Phe54, Leu211, Pro292, Met295, and Ala296 located on regions I, III, and IV of human SQS, which are completely conserved among all species, constitute an alternative pocket in human SQS active site (denoted cofactor site or C site, Fig. 2B). Indeed, this cavity is not found in S. aureus CrtM, underscoring its structural and catalytic importance. We also suspect that the highly flexible αJαK loop may act as a lid that regulates the access of NADPH to its binding site during catalysis.
Crystal Structure of Human SQS Complexed with ZA-A
Thompson et al. previously demonstrated that ZA-A is a competitive inhibitor of FPP with human SQS and exhibits an IC50 value of 0.7–1.2 nm (22). Based on the gross structural similarities, it has been speculated that ZAs inhibit SQS activity by mimicking the binding of PSPP to the enzyme (Fig. 1, B and C). In human SQS–ZA-A complex crystal, each of the six protein molecules is bound to a single molecule of ZA-A in the active site. Fig. 2C displays the ZA-A binding mode of human (31–370)/ZA-A monomers (chains A–F). The Fourier maps show well defined electron or difference density that corresponds to the ZA position (supplemental Fig. S3A). Superposition of this complex with the previously determined human SQS structure with a substrate-mimetic inhibitor (BPH-652) shows that the C-1 alkyl side chain of ZA-A binds onto the S1 site, and the binding pattern of the C-4′ terminal groups on the C-1 tail fits well to the biphenyl side chain of the phosphonosulfonate. Moreover, the terminal phenyl group is located at the bottom of the S1 site (supplemental Fig. S3B). This binding induces a local conformational change in the active site compared with the apo structure. We observe that the hydrophobic moiety of the C-1 bulky side chain induces an inward shift of the αAαB flap (51SRSF54) and positions Tyr73 toward the surface. This situation creates a highly polar environment for stabilizing the fully oxidized core. The large core is located in the upper part of cavity, and the level of low nanomolar affinity is reflected by sufficient binding interactions for stabilizing the flexible 51SRSF54 flap (detailed in supplemental Table S2. Fig. 2D presents the key ZA-A bicyclic core interactions with human SQS monomer (chain A). The polyacidic charge of the bicyclic core elicits strong binding and inhibitory activity against human SQS, accompanied by more extensive hydrogen bonds than those found from other inhibitors bearing carboxylic or phosphonic acid groups (23, 31). Various degrees of substitutions along the C-3, C-4, and C-5 in rat liver microsomal SQS and in vivo models have been discussed (9, 33–35).
A comparison of the active sites of human SQS and CrtM indicates that the binding site of the PSPP pyrophosphate group would overlap with the ZA-A bicyclic core region. Surprisingly, unlike the binding pattern of PSPP hydrophobic tails, the other 6-O-acyl side chain of ZA-A does not reside in the S2 site. Instead, it binds to human SQS in the other hydrophobic pocket, the C site (Fig. 2C). Hydrophobic interactions between the two lipophilic side chains and human SQS are given in supplemental Table S3. The residues Phe54, Leu211, Phe292, and Met295 in the αAαB loop, αH, and αJ constitute the second hydrophobic region for the C-6 group binding (supplemental Fig. S3C). Further, this site presents a spacious cavity for accommodating the α,β-unsaturated ester chain of the C-6 acyl unit in various binding patterns. This result may explain how the increase of the C-6 chain length in previous work improved or retained potency at the subnano- to nanomolar level in rat SQS and in modulating regiono- and stereoselective behavior of these compounds (20, 21, 36). Furthermore, unlike most phosphonosulfonate or bisphosphonate inhibitors that bind to the allylic substrate site and interact with the DDXXD motif through a metal-mediated interaction, ZA-A binds directly to human SQS in a metal-independent manner.
Binding Analysis of ZA-A to Human SQS and S. aureus CrtM
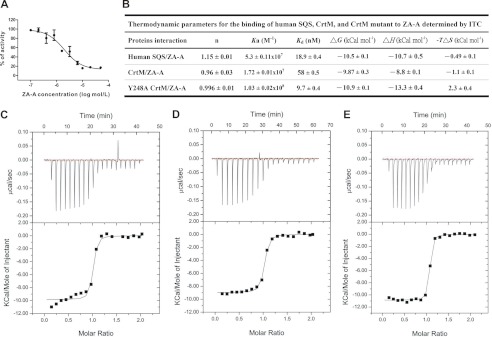
Many SQS inhibitors that mimic either FPP or PSPP can also greatly diminish CrtM activity and staphyloxanthin biosynthesis (18, 23, 29, 30). Recently, López and Kolter also reported that ZA-A impaired golden pigment formation and was a potent biofilm inhibitor in S. aureus and Bacillus subtilis without causing cell death (19). These results imply that ZA-A and its analogues can serve as selective antibacterial drugs. Indeed, ZA-A exhibits potent inhibition of CrtM activity (IC50 = 10.3 ± 0.4 μm) (Fig. 3A). Using isothermal titration calorimetry, we further analyzed the thermodynamics of ZA-A binding to CrtM. ZA-A binds to CrtM with a very low Kd value (high binding constant) of 58 nm, and it binds to human SQS even more efficiently (Kd = 18.9 nm) (Fig. 3, B–D). As with CrtM, the binding stoichiometry of ZA-A to human SQS is 1:1, consistent with the structural data. However, crystals of the wild-type CrtM–ZA-A complex were difficult to obtain; the main reason was revealed by further molecular docking studies.
FIGURE 3.
Effects of ZA-A on human SQS and S. aureus CrtM. A, inhibitory effect of ZA-A on S. aureus CrtM activity. The IC50 of ZA-A for CrtM is 10 μm. B, thermodynamic parameters from isothermal titration calorimetry data for binding of ZA-A to human SQS(31–370), S. aureus CrtM, and CrtM mutant. C, binding between ZA-A and human SQS. D, binding between ZA-A and S. aureus CrtM. E, binding between ZA-A and Y248A CrtM. The enzymes bind to ZA-A with a 1:1 stoichiometry, indicating that the proteins are properly folded and that the concentration is determined accurately.
A modeling experiment showed that the active site of CrtM is well adapted to bind both the hydrophobic C-1 side chain and the polynegative-charged bicyclic core. However, the bulky C-6 group would cause a steric crash with the side chain of Tyr248 in CrtM, which has a location similar to that of Ala296 in region IV of human SQS. Of note, the interactions with Tyr248, Phe22 (Phe54 in human SQS), and Leu164 (Leu211 in human SQS) crucially drive CrtM to fold into a compact shape with narrow gated substrate binding channels. When Tyr248 is replaced with alanine, the CrtM retains 20–30% of its original enzyme activity. We observed that the B-values for the 19SKSF22 and J-K loop in the Y248A mutant structure were elevated, suggesting that the reduced activity may be a result of disturbing the Tyr248-Phe22-Leu164 interaction that leads to a destabilized enzyme conformation.
Compared with the wild-type CrtM, the Y248A mutant interestingly showed an enhanced affinity to ZA-A (Fig. 3, B and E). The dissociation constant (Kd) of the ZA-A binding to the CrtM mutant was significantly decreased compared with that of the wild type. Moreover, the mutation altered the binding enthalpy (ΔH), −8.8 ± 0.1 kCal/mol for the wild-type and −13.3 ± 0.4 kCal/mol for Y248A. This indicates that more specific interactions are involved in the CrtM mutant binding to ZA-A than in the wild-type protein. Our results are also consistent with the above modeling analysis, which suggests that Tyr248 located on nonhomologous region IV may spatially obstruct the C-6 group binding.
Crystal Structure of S. aureus Dehydrosqualene Synthase (CrtM) Complexed with ZA-A
The Y248A CrtM–ZA-A complex crystal grew in the hexagonal space group P3121 and contains two molecules per asymmetric unit (designated as chains A and B in supplemental Fig. S4A). The active site of each protein was occupied by a ZA-A molecule, and this complex was refined at 2.12 Å to an R-factor of 20% (supplemental Table S1). For each structure, the fit of the inhibitors to the electron density maps was unambiguously defined for the final structure (supplemental Fig. S4B). The orientation and interactions of ZA-A bound to the Y248A CrtM indeed clearly resemble those in the human SQS complex (Fig. 4A). The highly oxygenated core structure contacts residues 19SKSF22 (equivalent to the 51SRSF54 flap in human SQS). In addition, the C-1 lipophilic tail extends into the narrow pocket which is lined with hydrophobic residues that help to stabilize the interaction with the isoprenoid moiety of the donor FPP (S1 site). The crystal structure reveals that the side chains of Phe22 and Phe26 are moved toward the bottom of the active site, and the orientation of the Tyr41 side chain provides sufficient space for stabilization of the ZA-A C-1 unit in the S1 site (Fig. 4B).
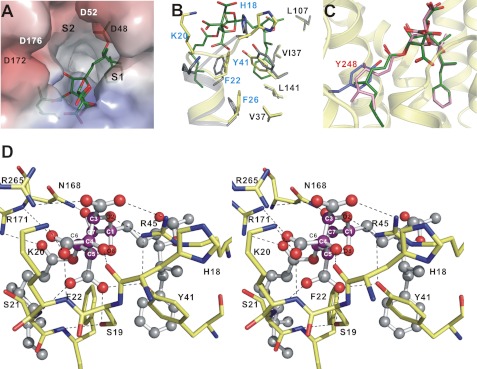
FIGURE 4.
Overall structure of the Y248A CrtM–ZA-A complex. A, electrostatic surface representation of Y248A CrtM binding site with ZA-A (green). Here, the S2 site is empty. B, superimposition of apo-CrtM (gray) and Y248A CrtM–ZA-A (yellow) showing the local conformational changes of the S1 site upon binding to ZA-A. C, ZA-A binds to the Y248A CrtM active site. If present, the C-6 acyl group would crash into Tyr248 (in slate). D, close-up view of the detailed interactions of ZA-A central core (magenta carbons) with Y248A CrtM shown in stereo. The ZA-A molecule is shown as a ball-and-stick representation. The important residues are labeled. Potential hydrogen bonds between the side chains of Y248A CrtM and ZA-A are shown as black dashes.
Structural comparison further illustrates that PSPP and ZA-A adopt distinct binding modes in CrtM, although the bicyclic core and the PSPP pyrophosphate share an overlapping binding site (supplemental Fig. S4C). The highly acidic core is fastened by a large network of hydrogen-bonding and ionic interactions with the functional groups of residues including Ser19, Lys20, Tyr41, Arg45 (corresponding to residues Ser51, Arg52, Tyr73, Arg77 in human SQS), and the backbone of His18, Lys20, Ser21. Moreover, the hydrogen-bonding forces between the amide group of Asn168 and the C-7 hydroxyl are found in Y248A CrtM–ZA-A complex (Fig. 4D). In the nonhomologous region IV of CrtM, the guanidinium group of Arg265 also provides a hydrogen bond to the C-3 carboxylic acid. Notably, the binding orientations of the C-6 group on CrtM and human SQS differ radically. In addition, the carbonyl oxygen of the C-6 side chain closes onto the functional group of Arg171, inducing hydrogen bonding. Supplemental Tables S2 and S3 summarize the intricate interactions between the inhibitor and the mutant protein. Finally, as expected, the presence of the Tyr248 phenol ring of the wild-type CrtM would block the C6-dimethyloctenoate group binding (Fig. 4C).
DISCUSSION
Natural products with huge structural diversity are considered a valuable source of drug leads for improvement of their pharmacological activity by structural modification. More than a half of commercialized drugs are natural product-based compounds. As ZAs exhibit fascinating biological activity for SQS, much work has focused on total synthesis by Merck, Glaxo, and other groups in an attempt to identify the key structural features of SQS-inhibitory activity (9, 20, 21, 33–37). However, variations of C-3, C-4, and C-5 tricarboxylic acids, C-4, C-7 dihydroxyl groups, and the C-1 and C-6 mimetics are often at odds with regard to which structural changes make the compounds more active or less. Because of the therapeutic potential of those active metabolites on sterol and staphyloxanthin biosynthesis, the determination of their binding modes is of critical importance for further drug development.
The structures of the bound ZA-A in human SQS and CrtM elucidate how the 2,8-dioxabicyclo[3.2.1]octane-3,4,5-tricarboxylic acid core fine-tunes the selective effects against the two enzymes. These structures also support a model where the lipophilic substrate/inhibitor binds to the S1 site and triggers local conformational change in the active site. Indeed, substrate-induced conformational changes are often observed in cis- and trans-prenyltransferases. Further, the anionic groups of SQS inhibitors are either predominantly stabilized by the aspartate-rich motifs via divalent metals, or they are directly bound by the positively charged residues in the vicinity of the active site entryway. Because human SQS and CrtM structures share a similar scaffold but CrtM lacks the ZA-A C-6 group binding cavity, the above information hinted at several important possibilities to develop effective inhibitors to these targets. One is that ZA derivatives coupled with shortened C-6 length (e.g. C-6 deoxy H1) may enhance the inhibitory specificity against CrtM. In a previous report, Ponpipom et al. found that the C-6 short chain derivatives show enhanced activity in vivo than the C-6 long chain analogues. Earlier work on these natural products also reported their effects on microbial population. Fairlamb et al. showed that ZA-A exerts no distinct effects on bacterial growth (38). In a later study, López and Kolter demonstrated ZA-A impaired bacterial biofilm formation but not cell viability, indicating no additional selective pressure on infection treatment. Our data, together with these previous findings, encourage the development of ZA derivatives into some antivirulence agents based on the complex structures.
By observing the active site interactions of a series of inhibitors to both enzymes, it seems that the ZA central core is not critically essential for inhibition and may only serve as a polynegative determinant. For example, a group of Trichoderma viride Pers metabolites (the viridiofungins or VFs isolated by Merck and derived from amino alkyl citrates with acyclic tricarboxylic acids) (32) and the ZA analogues (those bearing acyclic or monocyclic ring with polycarboxylic acids found by Glaxo group) (9) possess reduced but still considerable enzyme-inhibitory activity for rat SQS. These results, taken together, suggest that the identical orientation of the acidic moiety of these compounds is a critical feature for the inhibitory activity. In other words, different ring size can be considered as a means of modification. Various structural modifications of C-1 (such as peptidomimetic, a fatty acyl moiety, or unnatural amino acids) could be tested. Moreover, it should also be feasible to design new ZA derivates bearing C-7 lipophilic group as dual SQS/CrtM inhibitors that are selective for both S1+S2 sites.
Until now, no human SQS inhibitor has been marketed as an antihyperlipidemic drug, and so an effort continues in the development of SQS inhibitors with improved pharmacological properties. In conclusion, our findings shed light on the possibility of designing and developing new generation of sterol-lowering agents and antimicrobial drugs that would especially target pathogenic S. aureus, without affecting the de novo cholesterol biosynthesis pathway.
Supplementary Material
Acknowledgments
We thank the National Synchrotron Radiation Research Center in Taiwan and SPring-8 in Japan for beamtime allocations and data collection assistance.
This work was supported by Academia Sinica and National Science Council, Taiwan Grants NSC97-3112-B-001-017, NSC98-3112-B-001-024, and NSC99-3112-B-001-026 (to A. H.-J. W.).

This article contains supplemental Tables S1–S3 and Figs. S1–S4.
The atomic coordinatesandstructure factors (codes 3VJ8, 3VJ9, 3VJA, 3VJB, 3VJC, 3VJD, and 3VJE) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).
- ZA
- zaragozic acid
- CrtM
- dehydrosqualene synthase
- FPP
- farnesyl diphosphate
- PSPP
- presqualene diphosphate
- SQS
- squalene synthase.
REFERENCES
- 1. Newman D. J., Cragg G. M. (2007) Natural products as sources of new drugs over the last 25 years. J. Nat. Prod. 70, 461–477 [DOI] [PubMed] [Google Scholar]
- 2. Ji H. F., Li X. J., Zhang H. Y. (2009) Natural products and drug discovery: can thousands of years of ancient medical knowledge lead us to new and powerful drug combinations in the fight against cancer and dementia? EMBO Rep. 10, 194–200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Do R., Kiss R. S., Gaudet D., Engert J. C. (2009) Squalene synthase: a critical enzyme in the cholesterol biosynthesis pathway. Clin. Genet. 75, 19–29 [DOI] [PubMed] [Google Scholar]
- 4. Charlton-Menys V., Durrington P. N. (2007) Squalene synthase inhibitors: clinical pharmacology and cholesterol-lowering potential. Drugs 67, 11–16 [DOI] [PubMed] [Google Scholar]
- 5. Toutouzas K., Drakopoulou M., Skoumas I., Stefanadis C. (2010) Advancing therapy for hypercholesterolemia. Expert Opin. Pharmacother. 11, 1659–1672 [DOI] [PubMed] [Google Scholar]
- 6. de Souza W., Rodrigues J. C. (2009) Sterol biosynthesis pathway as target for anti-trypanosomatid drugs. Interdiscip. Perspect. Infect. Dis. 2009, 642502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Bergstrom J. D., Dufresne C., Bills G. F., Nallin-Omstead M., Byrne K. (1995) Discovery, biosynthesis, and mechanism of action of the zaragozic acids: potent inhibitors of squalene synthase. Annu. Rev. Microbiol. 49, 607–639 [DOI] [PubMed] [Google Scholar]
- 8. Koert U. (1995) Total syntheses of zaragozic acid. Angew. Chem. 34, 773–778 [Google Scholar]
- 9. Watson N. S., Procopiou P. A. (1996) Squalene synthase inhibitors: their potential as hypocholesterolaemic agents. Prog. Med. Chem. 33, 331–378 [DOI] [PubMed] [Google Scholar]
- 10. Wang Y., Metz P. (2011) A general access to zaragozic acids: total synthesis and structure elucidation of zaragozic acid D and formal syntheses of zaragozic acids A and C. Chemistry 17, 3335–3337 [DOI] [PubMed] [Google Scholar]
- 11. Dufresne C., Wilson K. E., Singh S. B., Zink D. L., Bergstrom J. D., Rew D., Polishook J. D., Meinz M., Huang L., Silverman K. C. (1993) Zaragozic acids D and D2: potent inhibitors of squalene synthase and of Ras farnesyl-protein transferase. J. Nat. Prod. 56, 1923–1929 [DOI] [PubMed] [Google Scholar]
- 12. Bate C., Salmona M., Diomede L., Williams A. (2004) Squalestatin cures prion-infected neurons and protects against prion neurotoxicity. J. Biol. Chem. 279, 14983–14990 [DOI] [PubMed] [Google Scholar]
- 13. Bate C., Williams A. (2007) Squalestatin protects neurons and reduces the activation of cytoplasmic phospholipase A2 by Aβ1–42. Neuropharmacology 53, 222–231 [DOI] [PubMed] [Google Scholar]
- 14. Rothwell C., Lebreton A., Young Ng C., Lim J. Y., Liu W., Vasudevan S., Labow M., Gu F., Gaither L. A. (2009) Cholesterol biosynthesis modulation regulates dengue viral replication. Virology 389, 8–19 [DOI] [PubMed] [Google Scholar]
- 15. Tansey T. R., Shechter I. (2000) Structure and regulation of mammalian squalene synthase. Biochim. Biophys. Acta 1529, 49–62 [DOI] [PubMed] [Google Scholar]
- 16. Liu G. Y., Essex A., Buchanan J. T., Datta V., Hoffman H. M., Bastian J. F., Fierer J., Nizet V. (2005) Staphylococcus aureus golden pigment impairs neutrophil killing and promotes virulence through its antioxidant activity. J. Exp. Med. 202, 209–215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Clauditz A., Resch A., Wieland K. P., Peschel A., Götz F. (2006) Staphyloxanthin plays a role in the fitness of Staphylococcus aureus and its ability to cope with oxidative stress. Infect. Immun. 74, 4950–4953 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Liu C. I., Liu G. Y., Song Y., Yin F., Hensler M. E., Jeng W. Y., Nizet V., Wang A. H., Oldfield E. (2008) A cholesterol biosynthesis inhibitor blocks Staphylococcus aureus virulence. Science 319, 1391–1394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. López D., Kolter R. (2010) Functional microdomains in bacterial membranes. Genes Dev. 24, 1893–1902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Ponpipom M. M., Girotra N. N., Bugianesi R. L., Roberts C. D., Berger G. D., Burk R. M., Marquis R. W., Parsons W. H., Bartizal K. F., Bergstom J. D. (1994) Structure-activity relationships of C1 and C6 side chains of zaragozic acid A derivatives. J. Med. Chem. 37, 4031–4051 [DOI] [PubMed] [Google Scholar]
- 21. Procopiou P. A., Bailey E. J., Bamford M. J., Craven A. P., Dymock B. W., Houston J. G., Hutson J. L., Kirk B. E., McCarthy A. D., Sareen M. (1994) The squalestatins: novel inhibitors of squalene synthase. Enzyme inhibitory activities and in vivo evaluation of C1-modified analogues. J. Med. Chem. 37, 3274–3281 [DOI] [PubMed] [Google Scholar]
- 22. Thompson J. F., Danley D. E., Mazzalupo S., Milos P. M., Lira M. E., Harwood H. J., Jr. (1998) Truncation of human squalene synthase yields active, crystallizable protein. Arch. Biochem. Biophys. 350, 283–290 [DOI] [PubMed] [Google Scholar]
- 23. Lin F. Y., Liu C. I., Liu Y. L., Zhang Y., Wang K., Jeng W. Y., Ko T. P., Cao R., Wang A. H., Oldfield E. (2010) Mechanism of action and inhibition of dehydrosqualene synthase. Proc. Natl. Acad. Sci. U.S.A. 107, 21337–21342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Otwinowski Z., Minor W. (1997) Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 276, 307–326 [DOI] [PubMed] [Google Scholar]
- 25. Collaborative Computational Project, Number 4 (1994) The CCP4 suite: programs for protein crystallography. Acta Crystallogr. D Biol. Crystallogr. 50, 760–763 [DOI] [PubMed] [Google Scholar]
- 26. Emsley P., Cowtan K. (2004) COOT: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 [DOI] [PubMed] [Google Scholar]
- 27. Winn M. D., Murshudov G. N., Papiz M. Z. (2003) Macromolecular TLS refinement in REFMAC at moderate resolutions. Methods Enzymol. 374, 300–321 [DOI] [PubMed] [Google Scholar]
- 28. Lovell S. C., Davis I. W., Arendall W. B., 3rd, de Bakker P. I., Word J. M., Prisant M. G., Richardson J. S., Richardson D. C. (2003) Structure validation by Cα geometry: φ, ψ, and Cβ deviation. Proteins 50, 437–450 [DOI] [PubMed] [Google Scholar]
- 29. Song Y., Lin F. Y., Yin F., Hensler M., Rodrígues Poveda C. A., Mukkamala D., Cao R., Wang H., Morita C. T., González Pacanowska D., Nizet V., Oldfield E. (2009) Phosphonosulfonates are potent, selective inhibitors of dehydrosqualene synthase and staphyloxanthin biosynthesis in Staphylococcus aureus. J. Med. Chem. 52, 976–988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Song Y., Liu C. I., Lin F. Y., No J. H., Hensler M., Liu Y. L., Jeng W. Y., Low J., Liu G. Y., Nizet V., Wang A. H., Oldfield E. (2009) Inhibition of staphyloxanthin virulence factor biosynthesis in Staphylococcus aureus: in vitro, in vivo, and crystallographic results. J. Med. Chem. 52, 3869–3880 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Pandit J., Danley D. E., Schulte G. K., Mazzalupo S., Pauly T. A., Hayward C. M., Hamanaka E. S., Thompson J. F., Harwood H. J., Jr. (2000) Crystal structure of human squalene synthase: a key enzyme in cholesterol biosynthesis. J. Biol. Chem. 275, 30610–30617 [DOI] [PubMed] [Google Scholar]
- 32. Mandala S. M., Thornton R. A., Frommer B. R., Dreikorn S., Kurtz M. B. (1997) Viridiofungins, novel inhibitors of sphingolipid synthesis. J. Antibiot. 50, 339–343 [DOI] [PubMed] [Google Scholar]
- 33. Procopiou P. A., Cox B., Kirk B. E., Lester M. G., McCarthy A. D., Sareen M., Sharratt P. J., Snowden M. A., Spooner S. J., Watson N. S., Widdowson J. (1996) The squalestatins: inhibitors of squalene synthase. Enzyme inhibitory activities and in vivo evaluation of C3-modified analogues. J. Med. Chem. 39, 1413–1422 [DOI] [PubMed] [Google Scholar]
- 34. Chan C., Andreotti D., Cox B., Dymock B. W., Hutson J. L., Keeling S. E., McCarthy A. D., Procopiou P. A., Ross B. C., Sareen M., Scicinski J. J., Sharratt P. J., Snowden M. A., Watson N. S. (1996) The squalestatins: decarboxy and 4-deoxy analogues as potent squalene synthase inhibitors. J. Med. Chem. 39, 207–216 [DOI] [PubMed] [Google Scholar]
- 35. Biftu T., Acton J. J., Berger G. D., Bergstrom J. D., Dufresne C., Kurtz M. M., Marquis R. W., Parsons W. H., Rew D. R., Wilson K. E. (1994) Selective protection and relative importance of the carboxylic acid groups of zaragozic acid A for squalene synthase inhibition. J. Med. Chem. 37, 421–424 [DOI] [PubMed] [Google Scholar]
- 36. Giblin G. M. P., Bell R., Hancock A. P., Hartley C. D., Inglis G. G. A., Payne J. J., Procopiou P. A., Shingler A. H., Smith C., Spooner S. J. (1993) Semisynthetic squalestatins: squalene synthase inhibition and antifungal activity, the SAR of C6 and C7 modifications. Bioorg. Med. Chem. Lett. 3, 2605–2610 [Google Scholar]
- 37. Naito S., Escobar M., Kym P. R., Liras S., Martin S. F. (2002) Novel approach to the zaragozic acids. Enantioselective total synthesis of 6,7-dideoxysqualestatin H5. J. Org. Chem. 67, 4200–4208 [DOI] [PubMed] [Google Scholar]
- 38. Fairlamb I. J., Dickinson J. M., O'Connor R., Higson S., Grieveson L., Marin V. (2002) Identification of novel mammalian squalene synthase inhibitors using a three-dimensional pharmacophore. Bioorg. Med. Chem. 10, 2641–2656 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.